Abstract
Signaling pathways are re-used for multiple purposes in plant and animal development. The Hippo pathway in mammals and Drosophila coordinates proliferation and apoptosis via the co-activator and oncogene, YAP/Yorkie (Yki), which is homeostatically regulated through negative feedback. In the Drosophila eye, cross-repression between the Hippo pathway kinase, LATS/Warts (Wts), and growth regulator, Melted, generates mutually exclusive photoreceptor subtypes. Here, we show that this all-or-nothing neuronal differentiation results from Hippo pathway positive feedback: Yki both represses its negative regulator, warts, and promotes its positive regulator, melted. This post-mitotic Hippo network behavior relies on a tissue-restricted transcription factor network—including a conserved Otx/Orthodenticle-Nrl/Traffic Jam feedforward module—that allows Warts-Yki-Melted to operate as a bistable switch. Altering feedback architecture provides an efficient mechanism to co-opt conserved signaling networks for diverse purposes in development and evolution.
Core signaling pathways are re-used for different purposes during development, allowing extraordinary cell-type diversity (1). For example, the TGF-β, Notch, RTK/MAPK, and Wnt signaling pathways each act repeatedly, from embryogenesis to adulthood, to coordinate tissue patterning, growth and specification throughout the animal. The Hippo pathway is best known for its role in growth control in both flies and mammals, where it regulates the balance between division and death in mitotic cells (2). But the Hippo pathway also regulates post-mitotic events such as photoreceptor subtype specification in the Drosophila eye (3, 4). How the same signaling network can be regulated for context-appropriate outcomes as diverse as proliferation and differentiation is not well understood.
The Drosophila eye comprises about 800 unit eyes (ommatidia), each containing eight photoreceptors (R1-R8) (5). Two main ommatidial subtypes are defined by light-sensing Rhodopsin (Rh) proteins expressed in the color vision photoreceptors, R7 and R8: ‘p’ ommatidia, with UV-sensitive Rh3 in R7 and blue-Rh5 in R8, and ‘y’ ommatidia with longer UV-Rh4 in R7 and green-Rh6 in R8 (Fig. 1A and reviewed in (6)). p and y subtypes are distributed randomly in the retina in a p:y ratio of ~30:70, following stochastic expression of the transcription factor Spineless in the R7 of subtype y (yR7s). pR7s, which lack Spineless, signal to underlying R8s to induce pR8/Rh5 fate, whereas the remaining R8s become yR8/Rh6 by default (6). p vs. y fate in R8s is established by a bistable transcriptional feedback loop between Melted (Melt), a pleckstrin homology-domain protein that specifies pR8/Rh5 fate, and Wts, a kinase in the Hippo pathway that specifies yR8/Rh6 fate (Fig. 1B-C) (3).
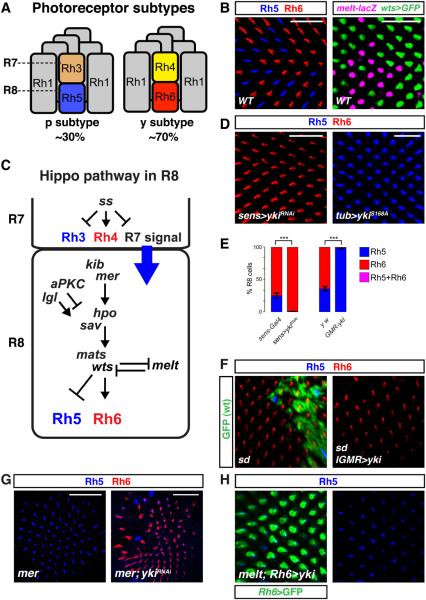
Fig. 1. Yki and Sd instruct mutually exclusive R8 neuron subtypes.
(A) Main photoreceptor subtypes and Rhodopsin coupling in the Drosophila eye.
(B) Confocal images of whole-mounted, wild-type adult retinas. Left: R8 subtypes visualized with antibodies to Rh5 (blue) and Rh6 (red); right: R8 subtypes labeled with transcriptional reporters for melt (β−gal antibody, magenta) and wts (GFP antibody, green). Scale bar, 25μm.
(C) Hippo pathway regulation of R8 subtypes. R7 signals to R8 to induce pR8s (melt and Rh5). yR8 cells express wts and Rh6. wts and melt act in a double negative transcriptional feedback loop. Additional Hippo pathway members required to specify yR8 fate include the entire “core complex” (Wts, the Hpo kinase, the adapter protein Salvador (Sav), and Wts co-factor, Mats) and the upstream regulators Lethal Giant Larvae (Lgl), the FERM-domain protein and NF-2 ortholog, Merlin (Mer), and the WW-domain protein, Kibra (Kib) (4). atypical Protein Kinase C (aPKC) antagonizes yR8 fate. Black arrows and lines represent genetic regulatory interactions.
(D) yki is necessary and sufficient to specify pR8 fate. yki knock-down (ykiRNAi) (left). Pan-photoreceptor expression of activated Yki (right), induced by GMR-flp/FRT mediated excision of a transcriptional STOP between tubulin promoter and ykiS168A (GMR-flp, tub-FRT-STOP-FRT- ykiS168A). Scale bar, 25μm.
(E) Effect of yki manipulations on percentage of R8 cells expressing Rh5 or Rh6 (y-axis). Yki is necessary (left) and sufficient (right) to induce Rh5. Wild-type range is ~20-40% Rh5. From left to right in graph: sens-Gal4 (n=10, N=2998 R8 neurons), sens>ykiRNAi (n=8, N=2112), y,w (n=14 retinas, N=2790), GMR-yki (n=4, N=510). Error bars are ± standard deviation (SD); two-tailed, unpaired Student’s t-test; ** denotes p < 0.01, *** denotes p <0.001.
(F) sd is required for pR8 fate. Left: sd47M mutant clone (GFP absence). The total number of ommatidia was not reduced, indicating R8 cells were mis-specified into yR8 rather than pR8 cells being lost. Right: Yki mis-expression in sd mutant background.
(G) Yki acts downstream of the Hippo pathway and melt to control Rh5 and Rh6. Left: mer4 (left); right: lGMR>ykiRNAi suppress the mer4 phenotype. Scale bar, 50μm.
(H) Ectopic expression of yki in the opposite subtype with Rh6-Gal4 induces Rh5 in melt?1 mutants. Rh6-Gal4 expressing cells co-labeled by expression of GFP (green). Right: Rh5 channel only. Note: Except where noted, in all manuscript figures Rh5 and Rh6 are labeled in blue (Rh5) and red (Rh6).
In its canonical role as a tumor suppressor, Wts is activated by the Hippo kinase (Hpo) and phosphorylates Yki, the Drosophila ortholog of the human oncogene YAP, to sequester Yki in the cytoplasm (2). In the absence of Hippo signaling, non-phosphorylated Yki enters the nucleus and binds as a co-activator to transcription factors like Scalloped (Sd), Homothorax (Hth) and Mothers against Dpp (Mad) (7-11) to activate target proliferation and anti-apoptotic genes. Yki can also induce its negative regulators expanded, merlin, kibra, or dmyc to provide negative feedback onto itself during growth control (12-14).
Here we show that in post-mitotic R8s, as in growth, Yki and its DNA-binding partner Sd mediate transcriptional output of the Hippo pathway. However, the R8 regulatory architecture is fundamentally different, as Yki promotes positive feedback onto itself. This regulation requires a tissue-specific transcription factor network that includes Orthodenticle (Otd) and Traffic Jam (Tj), orthologs of the mammalian photoreceptor determination genes Crx and Nrl (15), respectively, as well as Pph13 and Gfi1/Senseless (Sens). This network generates the post-mitotic context for the Hippo pathway to regulate an all-or-nothing fate decision and ensure robust terminal differentiation of sensory neuron subtypes.
Yki and Sd regulate R8 subtype specification
To test whether Yki functions in R8 neurons, we manipulated yki and assayed Rh5 and Rh6 expression. yki null mutant eye progenitor cells do not divide and are eliminated by apoptosis (16). We therefore used Gal4 drivers to express yki-targeted RNA interference (RNAi) in post-mitotic photoreceptors. Knockdown of yki in all photoreceptors early (lGMR>ykiRNAi), only in adults (using Gal80ts), or in all R8s (and some R1-R6) (sens>ykiRNAi) caused almost all R8s to express Rh6, whereas Rh5 was nearly absent (Fig. 1D and fig. S1A, C). Conversely, over-expressing wild-type or activated yki/YAP (ykiS168A or human YAPS127A) (17, 18) in all photoreceptors (lGMR>yki) transformed almost all R8s into Rh5-expressing pR8s (Fig. 1D and fig. S5C). Ectopic Yki did not require the pR7 signal to induce pR8 fate since Yki induced Rh5 even in the absence of R7s (sev; GMR>yki) (fig. S2A). Furthermore, mis-expressing yki only in yR8s after the fate decision (Rh6>yki) also induced Rh5 (fig. S2C). yki manipulations did not affect general neuronal fate, specific photoreceptor fate, or expression of other Rhodopsins (fig. S2B, D-F). Thus, yki is necessary and sufficient in R8s to specify pR8/Rh5 and prevent yR8/Rh6 subtypes.
Yki is a co-factor of DNA-binding transcription factors such as Sd, Hth, or Mad (7-11) to activate Hippo target genes. hth or mad loss-of-function (lGMR>hthDN (19) and lGMR+ey>madRNAi) (fig. S3D) did not noticeably affect Rh5 or Rh6. In contrast, retinas with sd loss-of-function (sd△B, sd△C, sd47M, or lGMR>sdRNAi), like yki loss-of-function eyes, expressed Rh6 in almost all R8s with a corresponding loss of Rh5 (Fig. 1F and fig. S3E) and did not affect other photoreceptor subtypes (fig. S3A-C). sd mutants or sdRNAi also suppressed the yki gain-of-function phenotype (sd; lGMR>yki) (Fig. 1F and fig. S3E-F), indicating that sd is required for yki function and likely encodes the Yki partner required for Rh5 and Rh6 regulation.
We next confirmed that yki acts canonically downstream of Wts to regulate Rhodopsin expression. In yR8s, Merlin (Mer) constitutively activates Hippo signaling to promote Wts activity (4). Their loss (mer4 or wtsRNAi) led to Rh5 expansion, but ykiRNAi suppressed these phenotypes (Fig. 1G and fig. S4A); furthermore, ectopic yki (GMR-yki) strongly suppressed Hippo pathway-induced Rh6 (GMR-wts, GMR-hpo, or GMR-wts+sav) (fig. S4B-C). We also tested whether yki and melt require each other to activate Rh5. Strong (ey+lGMR>ykiRNAi) or even mild (ykiB5/+) yki loss-of-function suppressed the ability of ectopic melt to induce Rh5 (fig. S4D). However, ectopic yki still induced Rh5 when melt was absent, even when expressed late in yR8s (Rh6>yki; melt) (Fig. 1H), consistent with yki functioning downstream of Melt and the Hippo pathway to regulate Rhodopsins. This regulation occurred through Wts-dependent Yki phosphorylation and inactivation because mis-expression of dominant negative kinase-dead (KD) forms of Wts or Hpo in the retina (lGMR>hpoKD or lGMR>wtsKD) (3) or constitutive gain-of-function alleles that reduce phosphorylation by Wts (ykiDbo1, ykiDbo2) (20) resulted in Rh5 expression in >70% of R8s (fig. S5 and ref. 3). Thus, the molecular relationship among Hippo pathway members in growth also exists in R8 fate.
Yki creates network-level Hippo pathway positive feedback by regulating wts and melt
We next assessed whether yki-dependent feedback exists with its upstream regulators in R8s by removing retinal yki and sd function and assaying expression of wts (yR8s/Rh6) and melt (pR8s/Rh5). ykiRNAi or sd47M mutant clones caused wts-lacZ expansion and melt-lacZ lossin most R8s (Fig. 2A-B, D), mirroring the gain of Rh6 and loss of Rh5. Conversely, activated yki (ykiDbo1/+, lGMR>yki, or lGMR>ykiS168A:GFP ) expanded melt-lacZ into >85% of R8s, with correspondingly decreased wts-lacZ (Fig. 2C, fig. S6A-C), and this function required sd (fig. S6A-C). Therefore, in post-mitotic photoreceptors Yki promotes its own activity with positive network-level feedback by activating melt and repressing its direct negative regulator, wts—a regulation opposite from that in growth control.
Fig. 2. Yki and Sd regulate wts and melt expression in Hippo pathway positive feedback.
(A) ykiRNAi retinas (top-right) contain wts-lacZ (green) in almost all R8s, compared to about two-thirds of R8s in wild-type controls (top-left); melt-lacZ (blue) is absent from most R8s in ykiRNAi retinas (bottom-right) compared with controls (bottom-left). Top panels: antibodies to β−gal (wts-lacZ) and Rh6 (red). Bottom panels: β−gal (melt-lacZ), Rh5 (green) and Rh6 (red). Images at R8 nuclei focal plane. Scale bar, 10μm.
(B) sd mutant clones (absence of GFP) labeled with antibodies to β−gal to mark wts-lacZ (top, green) and melt-lacZ (bottom, magenta) transcriptional reporters. Dashed circles in top panels show wild-type R8s without wts-lacZ, whereas sd mutant R8s contain wts-lacZ. Dashed lines indicate clone boundary. Bottom panel also stained for Rh6. Scale bar, 10μm.
(C) A heterozygous yki gain-of-function allele is sufficient to induce melt expression in most R8s. Images in focal plane of R8 nuclei. Antibodies to β−gal (blue), Rh5 and Rh6. Scale bar, 10μm.
(D) Sagittal sections of adult eyes with nuclei stained for the R8 marker, Sens (blue) and wts (β−gal; green) or melt (β−gal; magenta) expression. Left: melt requires yki and sd to repress wts. wts-lacZ is absent from R8s in lGMR>melt retinas, but de-repressed when yki or sd are simultaneously removed (bottom two panels). Right: melt-lacZ is expressed in most R8s in lGMR>wtsRNAi retinas, but is lost when yki or sd are removed. White bracket denotes R8 layer. Scale bar, 50μm.
(E) Top: melt; lGMR>yki:GFP adult retina labeled for Rh5 and Rh6 (left). Yki can repress wts (β−gal, green in right) in most R8s in the absence of melt. Right panels shows R8 nuclear layer. A minority of R8s still express wts, but most do not. Compare to melt mutant retina, where wts is expressed in almost all R8s (bottom right). Scale bar, 25μm.
(F) Model of Wts-Yki-Melt feedback circuit in R8 subtypes. Regulatory arrows are genetic and show transcriptional control, except for Wts inhibition of Yki, which is biochemical and post-transcriptional. Dashed lines indicate non-mutually exclusive interactions.
(G) Lineage-tracing experiment for wts-Gal4 expression in R8 subtypes using G-TRACE (see Methods). Adult retinas stained for: RFP (red), GFP (green), and phalloidin (blue). RFP alone labels contemporary, adult wts-Gal4 expression. GFP labels wts-Gal4 expression lineage. Right two panels are grayscale of RFP and GFP, respectively. Dashed circles show that pR8 cells did not express wts-Gal4 in their history. Scale bar, 10μm.
Yki/Sd could regulate the wts-melt cross-repression by activating melt, repressing wts, or both. To determine the feedback mechanism, we first performed epistasis analysis between yki and wts while monitoring melt expression. In wtsRNAi retinas, melt expanded into all R8s, yet when yki was simultaneously removed (lGMR>wtsRNAi+ykiRNAi), melt expression was lost (Fig. 2D). Conversely, ectopic wts failed to repress melt in the presence of ectopic yki (fig. S6C). Thus, yki does not activate melt by repressing wts; rather, it acts downstream of wts to promote melt.
If Yki repressed wts exclusively by inducing melt, wts should be de-repressed in melt mutants even if upstream Hippo signaling is inactive. However, wts-lacZ was absent in mer;melt double mutant R8s (fig. S6D), suggesting that a melt independent factor(s) represses wts in mer mutants. We hypothesized that the factor(s) includes Yki. Indeed, melt failed to repress wts in the absence of yki or sd (Fig. 2D), and ectopic yki largely retained the ability to repress wts expression in the absence of melt (melt; GMR>yki) (Fig. 2E). Thus, Yki not only functions downstream of wts to activate melt, but also functions downstream of melt to repress wts.
If ectopic Yki can repress wts independently of melt, what is the role of melt? Although all R8s expressed Rh6 in melt mutants, up to 20% of R8s still co-expressed Rh5 in 1-day old adults, which decreased to <8% after three weeks (fig. S7). Because no such delay was observed when yki was removed, and because yki activation consistently induces Rh5 in 100% of R8s, we infer that the transient Rh5 expression in young melt mutant flies reflects transient Yki activity. The above data suggest that Melt and Yki are temporally separated: Melt is not required to initiate pR8 fate in response to the pR7 signal, but instead likely consolidates Wts inactivity and Yki activity immediately after the fate decision to ensure robust Yki function in pR8s.
Altogether, these data indicate that Yki regulates both wts and melt to promote pR8 specification, and reveal that Yki network-level positive feedback occurs through two mechanisms (Fig. 2F): (i) a Wts-Yki double negative feedback loop, wherein Wts inactivates Yki biochemically whereas Yki represses wts transcription; and (ii) a double positive feedback loop between Melt and Yki, wherein Yki activates melt expression, and Melt promotes Yki (or inhibits the Hippo pathway to activate Yki). This combination ensures a complete switch from the default (yR8) to the induced (pR8) fate.
The R8 Hippo network is distinct from the Hippo growth regulation network
To evaluate the context-specificity of the R8 Hippo network, we asked whether wts transcriptional regulation is specific to post-mitotic R8 cells. We used the G-TRACE lineage reporter (21) to simultaneously label historical (GFP) and contemporary (RFP) wts-Gal4 expression and found that GFP was only co-expressed with RFP in yR8s (Fig. 2G). This indicates that wts-Gal4 was not expressed earlier in mitotic pR8 progenitor cells, and instead is activated post-mitotically to control R8 subtype specification.
We also asked whether the novel Hippo regulatory relationships in R8—melt repressing wts, yki/sd repressing wts and activating melt—also exist in growth contexts. First, we examined wts-lacZ expression following Hippo pathway manipulation in the posterior compartment of the larval wing disc using an engrailed (en)-Gal4 driver. Unlike in R8s, neither ykiRNAi nor wts mis-expressionincreased wts-lacZ expression. Furthermore, in third instar larval eye discs, we did not detect up-regulation of wts-lacZ in sd mutant clones (fig. S8A) or of melt-lacZ in wts mutant clones (fig. S8B). This suggests that the Hippo pathway does not regulate the expression of wts or melt in dividing epithelial cells of the wing or eye disc.
We next tested whether the Wts-Yki-Melt regulatory circuit exists in growth control by manipulating melt and assaying the Yki growth target Ex. Whereas ectopic yki or wtsRNAi (which phenocopy melt gain-of-function in R8) autonomously induced Ex levels when expressed in the wing, ectopic melt did not increase Ex protein or ex transcription (ex-lacZ) (fig. S8D). Although en>melt adult wings were slightly larger than en-Gal4 control wings, strong over-expression of melt in the developing and adult eye did not noticeably affect eye size or morphology (fig. S8C). This difference is likely due to melt’s known role in the wing as a growth promoter in the Insulin/TOR pathway (22), which is dispensable for melt-dependent R8 subtype determination (3). Collectively, these experiments indicate that the regulatory architecture and transcriptional feedback mechanism of the Hippo network differ between R8 subtype specification and growth regulation in at least two mitotically active tissues.
A conserved feedforward Otd-Tj (Crx–Nrl) module regulates photoreceptor subtypes
Given the differences in Yki-mediated feedback in growth and R8 fate, we investigated how the Wts-Yki-Melt regulatory circuit is established specifically in R8s but not in growth. Because melt is a context-specific inhibitor of the Hippo pathway, we focused on mechanisms underlying melt regulation in the eye. We generated serial deletions of the melt first intron (4kb), previously shown to confer expression in pR8s (3), and identified a 450bp (melt450-lacZ) element that drives reporter expression in pR8s (and pR7s) and responds appropriately to wts in R8 (Fig. 3A-B and fig. S9A-B). melt450 contains two conserved K50 homeodomain (HD) binding sites whose mutations abolished reporter expression (Fig. 3A-B and fig. S9A), suggesting that a K50 factor directly promotes melt expression. A good candidate was the pan-photoreceptor K50 HD transcription factor, Otd (23), which directly activates Rh5 in pR8s and controls proper pR8:yR8 ratios (24-26) (fig. S9C). Consistent with otd being required for melt transcription, melt-lacZ was lost from all R8s when otd was removed (sens>otdRNAi) (Fig. 3B), and melt450-lacZ was lost in eye-specific otduvi mutants (fig. S9B). Moreover, Otd was sufficient to activate a melt450-luciferase (luc) reporter 3-fold in cultured Drosophila S2 cells, requiring intact K50 sites (Fig. 3C). Thus, Otd directly activates expression of both the fate determinant, melt, and its downstream output, Rh5, generating feedforward regulation to promote pR8 fate (Fig. 3C).
Fig. 3. The photoreceptor regulator Otd promotes melt expression in pR8s.
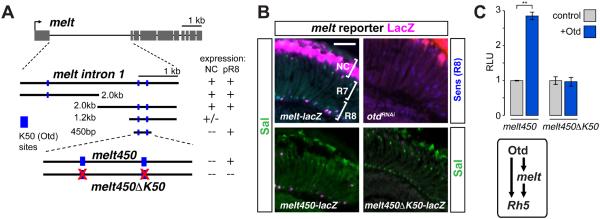
(A) Schematic diagram of melt locus and genomic DNA fragments tested for pR8 expression. Blue boxes: K50/Otd binding sites (TAATCC). Red Xs: Otd site mutations.
(B) Top-left: A 4 kb intronic melt-lacZ reporter expresses in subset of Sal-positive R8 cells and distal non-neuronal cells (‘NC’). Bottom-left: a 450 bp melt enhancer is expressed in pR8s (and some pR7s). Top right: Adult otd loss-of-function retinas immunostained for β−gal (magenta) and the R7/R8 marker Sal (green). Bottom-right: Mutation of two K50/Otd binding sites in the melt450 enhancer (melt450⊿K50-lacZ)abolishes reporter expression. Scale bar, 50μm.
(C) Luciferase reporter assays in S2 cells. y-axis: relative luciferase units (RLU) normalized to a control that represents cells transfected with empty expression vector (=1 RLU). Otd activates the melt450 enhancer ~3-fold, but does not activate melt450⊿K50. Error bars are ±SD; n=3; ** p≤ 0.01. Bottom box: Otd and Melt form a feedforward loop to promote Rh5 expression (pR8 fate).
Because Otd is expressed in all photoreceptors, we posited that other factor(s) act with Otd to regulate the Hippo pathway in R8. We performed a photoreceptor-specific RNAi screen and identified traffic jam (tj), a basic leucine zipper (bZIP) transcription factor (27). tj knockdown in all photoreceptors, or a null tj allele we generated (tj⊿1), each led to loss of Tj protein and a significantly reduced Rh5:Rh6 ratio (Fig. 4A-C and fig. S10B-C) without affecting R7 opsins (fig. S10D), suggesting that Tj is required for pR8 fate.
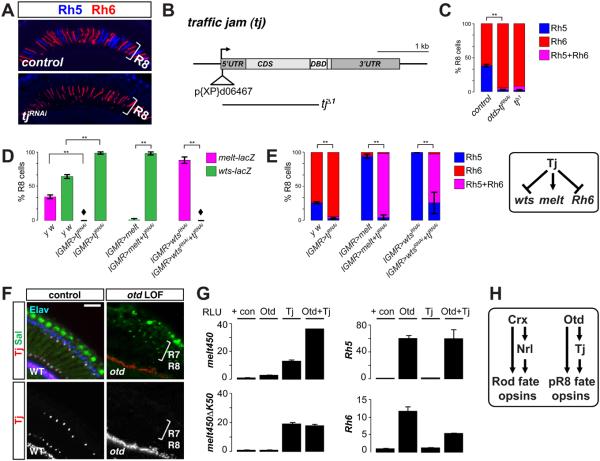
Fig 4. Tissue-restricted transcription factors control melt and Rhodopsins with regulatory logic conserved in mammalian eye development.
(A) Retinas expressing tjRNAi lose Rh5 and gain Rh6. Scale bar, 50μm.
(B) Diagram of the tj locus. tj⊿1 was generated by imprecise excision of a P-element (triangle) in the 5’ UTR. The tj⊿1 null mutant deletion is indicated by the black line.
(C) Quantification of Rh5 and Rh6-expressing R8s. From left to right in graph: control (otd-GAL4): (n=5, N=997), otd>tjRNAi (n=4, N=898), tj?1 (n=6 retinas, N=1230 R8 neurons). Error bars are SD; ** p < 0.01.
(D) Tj regulates wts and melt: Quantification of wts and melt-expressing R8s. From left to right: control (yw): (n=4, N=920), lGMR>tjRNAi (n=7, N=1411), lGMR>melt (n=4, N=980), lGMR>melt+tjRNAi (n=4, N=892), lGMR>wtsRNAi (n=5, N=912), lGMR>tjRNAi+ wtsRNAi (n=4, N=760); error bars are SD; **p<0.01. Diamonds denote complete loss of melt-lacZ.
(E) Tj regulates Rh6 downstream of wts and melt. From left to right: y w control (n=6, N=1185), lGMR>tjRNAi (n=8, N=1675), lGMR>melt (n=5, N=1081), lGMR>melt+tjRNAi (n=5, N=1021), lGMR>wtsRNAi (n=4, N=782), lGMR>tjRNAi+ wtsRNAi (n=4, N=774). %Rh5 compared; error bars are SD. ** p < 0.01. Right diagram: Tj regulates wts, melt, and Rh6.
(F) Tj is expressed in R7 and R8, but not in otd mutants. Adult sections, top-left: wild-type retinas labeled for the neuronal marker Elav (blue), the R7/R8 marker Sal (green), and Tj (red). Bottom-left: Tj alone in grayscale in wild-type retinas. Right: otd mutants (otduvi) lose TJ (red); Sal (green). Scale bar, 50μm.
(G) Luciferase reporter assays in S2 cells. From top-left: Tj activates melt450 expression ~12-fold, while Otd and Tj synergistically activate melt450 ~37-fold; bottom left: mutating K50/Otd sites in melt450 abolishes Otd-dependent, but not Tj-dependent activation. Top-right: Otd, but not Tj, activates Rh5 promoter expression. Bottom right: Tj can repress Rh6. Error bars are ±SD; n=3.
(H) A OTX/CRX/Otd—MAF/NRL/Tj coherent feedforward motif instructs photoreceptor fate in flies and mammals. Crx(Otd) promotes expression of Nrl(Tj), and both promote a specific photoreceptor fate and Rhodopsin expression. Left, mammalian motif; right, fly motif.
Tj retinal expression preceded Rhodopsin expression and was restricted to all R7s and R8s from ~60 hours after pupal formation (APF) into adulthood (Fig. 4F and fig. S10A). tj knockdown in R8 (sens>tjRNAi) phenocopied tj⊿1 mutants, whereas R7 knockdowns (sev>tjRNAi) had wild-type Rh5:Rh6 ratios (fig. S10B-C, E), indicating that tj autonomously affects R8 fate.
Consistent with Tj’s role in promoting pR8 fate, melt-lacZ was lost and wts-lacZ wasexpandedinto most tj mutant R8s (Fig. 4D and fig. S11A). Epistasis experiments revealed that tj bothpromotes melt, independently from wts (wtsp1; lGMR>tjRNAi; melt-lacZ) (Fig. 4D and fig. S11A), and is necessary for melt to fully repress wts (tj⊿1; lGMR>melt; warts-lacZ) (Fig. 4D and fig. S11A). Furthermore, although Rh5 was uniquely expressed in all R8s in melt gain-of-function or wts loss-of-function retinas, simultaneous removal of tj in either situation resulted in Rh5:Rh6 co-expression in most R8s (Fig. 4E and fig. S11B). This indicates that tj functions downstream of wts and melt torepress Rh6, but does not activate Rh5. Combined, these experiments reveal that Tj controls pR8 fate through three distinct mechanisms: Tj promotes melt, represses wts, and represses Rh6 (Fig. 4E, right).
Tj is the single Drosophila ortholog of the four mammalian MAF-bZIP transcription factors (28). One MAF factor—Nrl (Neural Retina Leucine zipper)—is a target of the Otd orthologs, Otx2 and Crx, and functions synergistically with Crx in the mouse retina to promote rod photoreceptor formation at the expense of cones (29-31). Given that otd and tj also promote one photoreceptor fate (pR8s) at the expense of another (yR8s), we examined the genetic relationship between otd and tj in the fly retina. Otd was unaffected in tjRNAi retinas(fig. S11C), but Tj was absent in otduvi mutants (Fig. 4F). Thus, Otd promotes tj in fly photoreceptors, analogous to how Otx2/Crx promotes Nrl in mammalian rods. Otd-dependent pR8 gene expression, however, is not just a consequence of activating tj, as re-supplying tj in otduvi mutants did not restore melt (fig. S11D-E). Hence, otd acts upstream of tj, but both are required to promote melt expression. Thus, similar to Crx and Nrl regulation of mammalian photoreceptor fate, otd and tj form a coherent feedforward loop that promotes pR8 fate (Fig. 4H).
This model predicts that Otd and Tj cooperate to promote melt. Indeed, although no MAF consensus DNA binding sites were detected in the melt450 enhancer, Tj was sufficient to induce melt450-luc 12-fold and synergistically increased Otd-dependent activation from ~3-fold to 35-fold in S2 cells (Fig. 4G). Similar to our in vivo results, Tj did not induce Rh5 expression in S2 cells, but did repress Rh6 promoter activity (Fig. 4G). Combined, our in vivo and in vitro results uncover a second and conserved feedforward system in pR8s, wherein Otd induces tj, and Otd and Tj together activate melt expression in pR8s.
Yki requires the conserved Otd-Tj module to induce pR8 fate
Since Otd and Tj are expressed in all R8s, whereas Yki/Sd function is biochemically restricted to pR8s by Wts, we asked whether these transcriptional regulators integrate to promote pR8 fate. Consistent with otd and tj being essential for melt activation, yki mis-expression failed to induce melt in otduvi or tjRNAi eyes (Fig. 5A). Moreover, yki failed to activate Rh5 in otduvi flies, or repress Rh6 in tjRNAi eyes (fig. S12A). Thus, yki requires otd and tj activity to exert its pR8 specification functions: (i) yki requires otd and tj to induce melt, (ii) yki requires otd to activate Rh5, and iii) yki requires tj to repress Rh6 (Fig. 5B, right).
Fig. 5. The Hippo pathway requires photoreceptor specification factors to regulate R8 subtypes.
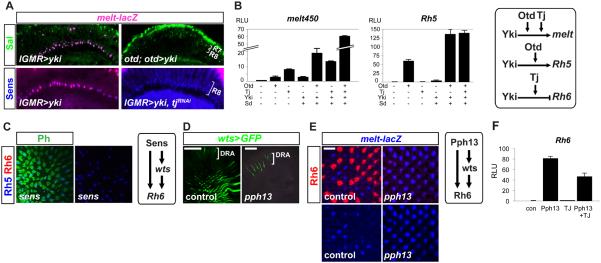
(A) Yki requires Otd and Tj to induce Melt. Adult sections of (top): lGMR>yki (left) and otduvi; lGMR-GAL4>UAS-yki (right) retinas labeled for melt-lacZ (β−gal, magenta) and Sal (green), or (bottom): lGMR>UAS-yki (left), and lGMR>UAS-tjRNAi+UAS-yki labeled for melt-lacZ (magenta) and Sens (blue). Scale bar, 50μm.
(B) Yki, Sd, Otd, and Tj synergistically activate melt and Rh5. Luciferase reporter assays for melt and Rh5 enhancer activity in S2 cells. Cells were transfected with indicated combinations of Otd, Tj, Yki and Sd. Right: Yki requires Otd and Tj to activate melt, and Yki requires Otd to activate Rh5. Error bars are ±SD; n=3.
(C) sens late-mutant retinas (see Methods) lose Rh6 and wts expression; some Rh5 remains.
(D) pph13 mutant retinas lose wts expresssion. wts>GFP remains expressed in R8s of the Dorsal Rim Area (DRA) R8s (bracket) at the margin of the retina, but is absent in yR8s. Scale bars, 50μm.
(E) Left: pph13 mutants gain melt-lacZ (blue) in most R8s. Right: Pph13 is required for wts and Rh6 expression, and to specify yR8 fate. Scale bar, 10μm
(F) Tj reduces Pph13-mediated activation of Rh6 promoter. Error bars are ±SD; n=3.
To test this integration molecularly, we analyzed the ability of Yki+Sd to influence Otd and Tj-dependent activation of melt and Rh5 in S2 cells. Yki+Sd weakly activated melt (~3-fold), additively increased Tj-dependent activation (from 8-fold to 12-fold), and synergistically increased Otd-dependent activation of melt (from ~3-fold to 20-fold) (Fig. 5B). However, the highest melt activation was observed with Otd, Tj, Sd, and Yki together (60-fold), consistent with the requirement of all four factors for pR8 fate in vivo. Similarly, Yki and Sd minimally activated the Rh5 promoter (~3-fold), but largely increased Otd-dependent activation (from 60- to 125-fold) (Fig. 5B). Although the K50/Otd sites were necessary for expression of melt and Rh5, mutating potential Sd sites in the Rh5 promoter did not decrease reporter expression in vivo, suggesting that Yki/Sd-dependent activation of Rh5 occurs indirectly. These studies support the model that Otd, Tj, and Yki/Sd cooperate to promote pR8-specific gene expression.
Altogether, our results indicate that the pR8 state depends on two overlapping feedforward regulatory networks: (i) Otd directly promotes melt and Rh5 expression (Fig. 3A-C) (24, 25). Melt then further promotes Rh5 by antagonizing the Hippo pathway and promoting Yki activity; (ii) Otd promotes tj expression, and Tj and Otd then synergistically induce melt, while Tj also represses wts and Rh6. Because Yki requires Otd and Tj to induce melt (which ultimately promotes Yki), Otd/Tj provide a critical transcriptional context for Yki positive feedback in R8.
Photoreceptor- and R8-restricted transcription factors promote yR8 fate.
We next investigated the mechanisms controlling the yR8 ‘default’ state (active Hippo pathway and Rh6). Sens, an R8-restricted zinc finger transcription factor necessary early for R8 specification (32), and later for terminal R8 differentiation (25, 32, 33), and Pph13, a pan-photoreceptor Q50 homeodomain transcription factor, are both essential for Rh6 expression (34). Thus, we tested whether these factors also promote yR8 fate. Removing sens late (sens>sensRNAi) reduced wts expression (fig. S13A), indicating that Sens functions in yR8s to promote wts and Rh6 expression (Fig. 5C). Sens did not, however, strongly affect pR8 fate as Rh5 was only mildly affected (fig. 5C). In pph13hazy null mutant retinas, not only were wts and Rh6 expression lost, but melt was also expanded into most R8s (Fig. 5D-E). In addition, the Rh6 promoter required the Pph13/Q50 HD binding site forits in vivo activity (fig. S13C), supporting the notion that Pph13 binds to and activates the Rh6 promoter (34). Rh5 protein was difficult to assess due to pph13’s role in forming rhabdomeres, where Rh5 protein localizes. Nevertheless, Rh5-GFP remained expressed (fig. S13B), confirming that Otd, but not Pph13, is required for Rh5 promoter activity (34). Therefore, pph13 regulates yR8 fate determinants (it promotes wts and represses melt) and directly activates Rh6 (Fig. 5E), in another feedforward loop resembling Otd/Tj/Melt and Otd/Melt/Rh5 regulation in the alternate subtype.
Since both Sens and Pph13 are expressed in all R8s, what prevents these factors from activating yR8 gene expression in pR8s? Tj plays this role for Rh6, as Pph13 strongly activates Rh6 promoter activity (~80-fold) and Tj represses this activation (Fig. 5F). Since Rh5 persists in sens and pph13 mutants, these factors are likely to be permissive to promote wts and Rh6 inall R8s.
Together, these results indicate that the R8 Hippo network topology requires Otd, Tj, Pph13, and Sens activity to permissively promote both subtypes in all R8s. Such regulation endows any R8 with competence to respond to the stochastically expressed signal from pR7. Ultimately, the Yki-Wts-Melt feedback module provides the instructive switch that decides between default (yR8) and acquired (pR8) states (Fig. 6A).
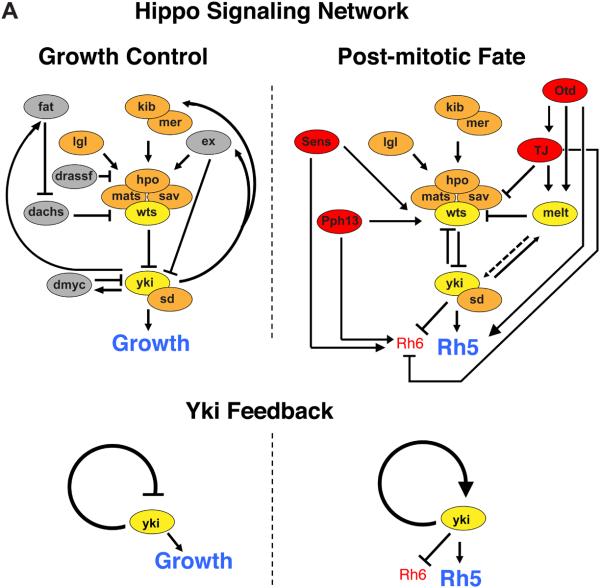
Fig 6. Tissue-restricted transcription factors promote re-use of Hippo pathway with positive feedback for post-mitotic neuronal fate.
(A) Model; Top: A functionally conserved cassette of genes from Kib/Mer to Yki/Sd is re-wired for a context-specific purpose through changes in network-level feedback. Comparison of network-level feedback switch in Hippo pathway for growth vs. post-mitotic R8 fate. In growth control (left), at least four negative feedbacks onto Yki generate homeostatic regulation for Yki’s growth promoting function. In post-mitotic R8 fate specification, positive feedback onto Yki induces an all-or-nothing decision to become pR8 and express Rh5. Four tissue-specific transcription factors (red ovals), including the conserved Otd-Tj (OTX-NRL) module, are co-expressed only in R8 in the eye and generate the permissive context for a Wts-Yki-Melt regulatory circuit and Yki positive feedback. Gray ovals are Hippo growth pathway genes not involved in R8 subtype specification. Orange ovals are genes involved in both contexts. Yellow ovals are genes that create the R8 feedback mechanism, and are yellow in the growth pathway for comparison. Bottom: Effect of Hippo pathway regulatory interactions on network-level feedback onto Yki, in growth control (left; negative feedback) or post-mitotic neural differentiation (right, positive feedback).
Discussion
A fundamental strategy in animal development is to re-purpose the same signaling pathways for a diversity of functions. We identified a tissue-specific transcription factor network that enables the otherwise homeostatic Hippo growth control pathway to act as a bistable switch for terminal cell fate. This alteration in network level properties—such as positive vs. negative feedback—within biochemically conserved pathways is an efficient means to re-use a signaling network in contexts as distinct as proliferation and terminal differentiation.
How is the R8-specific Hippo regulatory circuit achieved? The two interlinked positive feedback loops (one with wts, one with melt) provide the R8 Hippo pathway with multiple points of potential regulation. Context-specific expression of wts and melt is defined by overlapping expression of four transcription factors: Otd, Tj, Pph13, and Sens (Fig. 6A). Otd and Pph13 are expressed in all photoreceptors and generate a permissive context that endows the initially equipotent R8s with the competence to become either subtype: Otd promotes melt/Rh5 whereas Pph13 promotes wts/Rh6 expression. This competence is further restricted by expression of Tj in R7 and R8, and Sens in R8s, which ensures that melt and wts cross-regulation is restricted to R8s. Importantly, it is the status of Yki activity and resulting feedback that assures the outcome of p vs. y fate: in pR8s, Yki functions with Otd and Tj to promote melt and Rh5; in yR8s, wts inhibits Yki, preventing melt and Rh5 expressionand allowing ‘default’ wts and Rh6 expression by Pph13 and Sens. Each of these four transcription factors regulates a partially overlapping subset of R8 subtype fate genes, and together, the network cooperates at multiple regulatory nodes to provide the specific context for repurposing the Hippo pathway.
While other instances of pathways with both positive and negative feedback exist, these are conceptually different from R8 Hippo regulation. For example, in Sprouty (hSpry) regulation of Ras/MAPK-mediated EGFR signaling, EGFR induces hSpry2 expression but hSpry2 inhibits EGFR function (negative feedback); however, hSpry2 also promotes EGFR activity by preventing Cbl-dependent EGFR inhibition (positive feedback) (35, 36). hSpry2 positive feedback is likely coupled to its negative feedback to fine-tune the length and amplitude of receptor activation (36). In contrast, the opposite Hippo pathway feedbacks occur in vastly different cell types (mitotic epithelial cells vs. post-mitotic neurons), and both forms of feedback cannot co-exist in R8 since Yki’s repression of wts expression (positive feedback) would make Yki up-regulation of Hippo regulators (negative feedback) inconsequential.
Gaining positive feedback or losing negative feedback within Hippo signaling could permit oncogenesis. Indeed, the Yki ortholog, YAP, is an oncogene (37, 38) and is amplified in multiple tumors, and LATS1/2 (Wts) down-regulation is associated with non-small cell lung carcinomas, soft tissue sarcoma, metastatic prostate cancers, retinoblastoma, and acute lymphoblastic leukemia (39). Otx and MAF factors are also oncogenic in a number of tissues (40, 41). Thus, understanding the regulatory networks identified here in other contexts will be crucial for deciphering how normal signaling pathways can go awry.
Our findings also reveal that a Crx/Otd-Nrl/Tj feedforward module plays a conserved role in post-mitotic photoreceptor fate specification in both flies and mammals. Both induce one photoreceptor fate at the expense of another, and both regulate opsins with a feedforward loop wherein Crx/Otd activates Nrl/Tj expression and Crx-Nrl or Otd-Tj synergistically activate downstream targets (31). Given such deep evolutionary conservation, this module may be critical for generating photoreceptor diversity in other complex visual systems.
This work has two main implications. First, although positive feedback is well documented in other switch-like, irreversible cell fate decisions such as in Xenopus oocyte maturation or cell cycle entry (42-44), our work suggests that positive feedback could have a broad role in terminal neuronal differentiation, which often requires permanent fate decisions to maintain a neuron’s functional identity. Second, the changes in network topology in R8 photoreceptors allows a finely tuned growth control pathway to be used as a switch in a permanent binary cell fate decision. Context-specific regulation allows the feedback architecture to change in an otherwise conserved signaling module. This may be a general mechanism to endow signaling networks with new systems properties and diversify cell fates in development and evolution.
Supplementary Material
Footnotes
References and Notes
- 1.Pires-daSilva A, Sommer RJ. The evolution of signaling pathways in animal development. Nat Rev Genet. 2003;4:39–49. doi: 10.1038/nrg977. [DOI] [PubMed] [Google Scholar]
- 2.Halder G, Johnson RL. Hippo signaling: growth control and beyond. Development. 2011;138:9–22. doi: 10.1242/dev.045500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mikeladze-Dvali T, et al. The growth regulators warts/lats and melted interact in a bistable loop to specify opposite fates in Drosophila R8 photoreceptors. Cell. 2005;122:775–787. doi: 10.1016/j.cell.2005.07.026. [DOI] [PubMed] [Google Scholar]
- 4.Jukam D, Desplan C. Binary regulation of Hippo pathway by Merlin/NF2, Kibra, Lgl, and Melted specifies and maintains postmitotic neuronal fate. Dev Cell. 2011;21:874–887. doi: 10.1016/j.devcel.2011.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hardie RC. In: Progress in Sens Physiol. Ottoson D, editor. Vol. 5. Springer, Berlin, Heidelberg; New York, Toronto: 1985. pp. 1–79. [Google Scholar]
- 6.Rister J, Desplan C, Vasiliauskas D. Establishing and maintaining gene expression patterns: insights from sensory receptor patterning. Development. 2013;140:493–503. doi: 10.1242/dev.079095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goulev Y, et al. SCALLOPED interacts with YORKIE, the nuclear effector of the hippo tumor-suppressor pathway in Drosophila. Current Biology. 2008;18:435–441. doi: 10.1016/j.cub.2008.02.034. [DOI] [PubMed] [Google Scholar]
- 8.Zhang L, et al. The TEAD/TEF family of transcription factor Scalloped mediates Hippo signaling in organ size control. Dev Cell. 2008;14:377–387. doi: 10.1016/j.devcel.2008.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu S, Liu Y, Zheng Y, Dong J, Pan D. The TEAD/TEF family protein Scalloped mediates transcriptional output of the Hippo growth-regulatory pathway. Dev Cell. 2008;14:388–398. doi: 10.1016/j.devcel.2008.01.007. [DOI] [PubMed] [Google Scholar]
- 10.Peng HW, Slattery M, Mann RS. Transcription factor choice in the Hippo signaling pathway: homothorax and yorkie regulation of the microRNA bantam in the progenitor domain of the Drosophila eye imaginal disc. Genes Dev. 2009;23:2307–2319. doi: 10.1101/gad.1820009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Oh H, Irvine KD. Cooperative regulation of growth by Yorkie and Mad through bantam. Dev Cell. 2011;20:109–122. doi: 10.1016/j.devcel.2010.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hamaratoglu F, et al. The tumour-suppressor genes NF2/Merlin and Expanded act through Hippo signalling to regulate cell proliferation and apoptosis. Nat Cell Biol. 2005;8:27–36. doi: 10.1038/ncb1339. [DOI] [PubMed] [Google Scholar]
- 13.Genevet A, Wehr MC, Brain R, Thompson BJ, Tapon N. Kibra is a regulator of the Salvador/Warts/Hippo signaling network. Dev Cell. 2010;18:300–308. doi: 10.1016/j.devcel.2009.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Neto-Silva RM, de Beco S, Johnston LA. Evidence for a growth-stabilizing regulatory feedback mechanism between Myc and Yorkie, the Drosophila homolog of Yap. Dev Cell. 2010;19:507–520. doi: 10.1016/j.devcel.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Swaroop A, Kim D, Forrest D. Transcriptional regulation of photoreceptor development and homeostasis in the mammalian retina. Nat Rev Neurosci. 2010;11:563–576. doi: 10.1038/nrn2880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang J, Wu S, Barrera J, Matthews K, Pan D. The Hippo signaling pathway coordinately regulates cell proliferation and apoptosis by inactivating Yorkie, the Drosophila Homolog of YAP. Cell. 2005;122:421–434. doi: 10.1016/j.cell.2005.06.007. [DOI] [PubMed] [Google Scholar]
- 17.Dong J, et al. Elucidation of a universal size-control mechanism in Drosophila and mammals. Cell. 2007;130:1120–1133. doi: 10.1016/j.cell.2007.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Oh H, Irvine KD. In vivo regulation of Yorkie phosphorylation and localization. Development. 2008;135:1081–1088. doi: 10.1242/dev.015255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wernet MF, et al. Homothorax switches function of Drosophila photoreceptors from color to polarized light sensors. Cell. 2003;115:267–279. doi: 10.1016/s0092-8674(03)00848-1. [DOI] [PubMed] [Google Scholar]
- 20.Zhao B, et al. Inactivation of YAP oncoprotein by the Hippo pathway is involved in cell contact inhibition and tissue growth control. Genes Dev. 2007;21:2747–2761. doi: 10.1101/gad.1602907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Evans CJ, et al. G-TRACE: rapid Gal4-based cell lineage analysis in Drosophila. Nat Meth. 2009;6:603–605. doi: 10.1038/nmeth.1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Teleman AA, Chen Y-W, Cohen SM. Drosophila Melted modulates FOXO and TOR activity. Dev Cell. 2005;9:271–281. doi: 10.1016/j.devcel.2005.07.004. [DOI] [PubMed] [Google Scholar]
- 23.Vandendries ER, Johnson D, Reinke R. orthodenticle is required for photoreceptor cell development in the Drosophila eye. Dev Biol. 1996;173:243–255. doi: 10.1006/dbio.1996.0020. [DOI] [PubMed] [Google Scholar]
- 24.Tahayato A, et al. Otd/Crx, a dual regulator for the specification of ommatidia subtypes in the Drosophila retina. Dev Cell. 2003;5:391–402. doi: 10.1016/s1534-5807(03)00239-9. [DOI] [PubMed] [Google Scholar]
- 25.Xie B, Charlton-Perkins M, McDonald E, Gebelein B, Cook T. Senseless functions as a molecular switch for color photoreceptor differentiation in Drosophila. Development. 2007;134:4243–4253. doi: 10.1242/dev.012781. [DOI] [PubMed] [Google Scholar]
- 26.McDonald EC, et al. Separable transcriptional regulatory domains within Otd control photoreceptor terminal differentiation events. Dev Biol. 2010;347:122–132. doi: 10.1016/j.ydbio.2010.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li MA, Alls JD, Avancini RM, Koo K, Godt D. The large Maf factor Traffic Jam controls gonad morphogenesis in Drosophila. Nat Cell Biol. 2003;5:994–1000. doi: 10.1038/ncb1058. [DOI] [PubMed] [Google Scholar]
- 28.Kataoka K. Multiple mechanisms and functions of maf transcription factors in the regulation of tissue-specific genes. J. Biochem. 2007;141:775–781. doi: 10.1093/jb/mvm105. [DOI] [PubMed] [Google Scholar]
- 29.Swaroop A, et al. A conserved retina-specific gene encodes a basic motif/leucine zipper domain. Proc Natl Acad Sci USA. 1992;89:266–270. doi: 10.1073/pnas.89.1.266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mears AJ, et al. Nrl is required for rod photoreceptor development. Nature Genetics. 2001;29:447–452. doi: 10.1038/ng774. [DOI] [PubMed] [Google Scholar]
- 31.Hao H, et al. Transcriptional regulation of rod photoreceptor homeostasis revealed by in vivo NRL targetome analysis. PLoS Genet. 2012;8:e1002649. doi: 10.1371/journal.pgen.1002649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Frankfort BJ, Nolo R, Zhang Z, Bellen H, Mardon G. senseless repression of rough is required for R8 photoreceptor differentiation in the developing Drosophila eye. Neuron. 2001;32:403–414. doi: 10.1016/s0896-6273(01)00480-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Morey M, et al. Coordinate control of synaptic-layer specificity and rhodopsins in photoreceptor neurons. Nature. 2008;456:795–799. doi: 10.1038/nature07419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mishra M, et al. Pph13 and orthodenticle define a dual regulatory pathway for photoreceptor cell morphogenesis and function. Development. 2010;137:2895–2904. doi: 10.1242/dev.051722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hall AB, et al. hSpry2 is targeted to the ubiquitin-dependent proteasome pathway by c-Cbl. Current Biology. 2003;13:308–314. doi: 10.1016/s0960-9822(03)00086-1. [DOI] [PubMed] [Google Scholar]
- 36.Rubin C, et al. Sprouty fine-tunes EGF signaling through interlinked positive and negative feedback loops. Current Biology. 2003;13:297–307. doi: 10.1016/s0960-9822(03)00053-8. [DOI] [PubMed] [Google Scholar]
- 37.Overholtzer M, et al. Transforming properties of YAP, a candidate oncogene on the chromosome 11q22 amplicon. Proc Natl Acad Sci USA. 2006;103:12405–12410. doi: 10.1073/pnas.0605579103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zender L, et al. Identification and validation of oncogenes in liver cancer using an integrative oncogenomic approach. Cell. 2006;125:1253–1267. doi: 10.1016/j.cell.2006.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhao B, Li L, Lei Q, Guan K-L. The Hippo-YAP pathway in organ size control and tumorigenesis: an updated version. Genes Dev. 2010;24:862–874. doi: 10.1101/gad.1909210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Eychène A, Rocques N, Pouponnot C. A new MAFia in cancer. Nat Rev Cancer. 2008;8:683–693. doi: 10.1038/nrc2460. [DOI] [PubMed] [Google Scholar]
- 41.Bunt J, et al. OTX2 directly activates cell cycle genes and inhibits differentiation in medulloblastoma cells. Int. J. Cancer. 2012;131:E21–32. doi: 10.1002/ijc.26474. [DOI] [PubMed] [Google Scholar]
- 42.Xiong W, Ferrell JE. A positive-feedback-based bistable “memory module” that governs a cell fate decision. Nature. 2003;426:460–465. doi: 10.1038/nature02089. [DOI] [PubMed] [Google Scholar]
- 43.Pomerening JR, Sontag ED, Ferrell JE. Building a cell cycle oscillator: hysteresis and bistability in the activation of Cdc2. Nat Cell Biol. 2003;5:346–351. doi: 10.1038/ncb954. [DOI] [PubMed] [Google Scholar]
- 44.Skotheim JM, Di Talia S, Siggia ED, Cross FR. Positive feedback of G1 cyclins ensures coherent cell cycle entry. Nature. 2008;454:291–296. doi: 10.1038/nature07118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.We thank J. Bell, S. Britt, R. Carthew, S. Cohen, I. Davis, B. Dickson, R. Fehon, D. Godt, G. Halder, I. Hariharan, K. Irvine, J. Jiang, DJ. Pan, N. Tapon, J. Treisman, T. Xu, C. Zuker, the Bloomington Stock Center, the Kyoto Stock Center, the Vienna Drosophila RNAi Center, and the Exelixis Collection at Harvard Medical School for providing fly stocks and antibodies. G. Mardon generously provided sens-Gal4 flies and S. Sprecher kindly shared Pph13 results prior to publication. We thank T. Blackman and C. Tsanis for transgenic injections, and members of the Desplan and Cook labs for discussions and comments. Supported by an NYU Dean’s Dissertation Award (D.J.), a Univ. of Cincinnati Post-doctoral Research Fellowship (B.X.), EMBO long-term fellowships (ALTF 506-2002 &ALTF 462-2008) (D.P. and J.R.), NIH grants RO1 EY13012 (C.D.) and RO1-EY017907 (T.C.), and Research to Prevent Blindness (T.C.).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.