Abstract
Proliferative vitreo retinopathy (PVR) is associated with extracellular matrix membrane (ECM) formation on the neural retina and disruption of the multilayered retinal architecture leading to distorted vision and blindness. During disease progression in PVR, retinal pigmented epithelial cells (RPE) lose cell-cell adhesion, undergo epithelial-to-mesenchymal transition (EMT), and deposit ECM leading to tissue fibrosis. The EMT process is mediated via exposure to vitreous cytokines and growth factors such as TGF-β2. Previous studies have shown that Na,K-ATPase is required for maintaining a normal polarized epithelial phenotype and that decreased Na,K-ATPase function and subunit levels are associated with TGF-β1-mediated EMT in kidney cells. In contrast to the basolateral localization of Na,K-ATPase in most epithelia, including kidney, Na,K-ATPase is found on the apical membrane in RPE cells. We now show that EMT is also associated with altered Na,K-ATPase expression in RPE cells. TGF-β2 treatment of ARPE-19 cells resulted in a time-dependent decrease in Na,K-ATPase β1 mRNA and protein levels while Na,K-ATPase α1 levels, Na,K-ATPase activity, and intracellular sodium levels remained largely unchanged. In TGF-β2–treated cells reduced Na,K-ATPase β1 mRNA inversely correlated with HIF-1α levels and analysis of the Na,K-ATPase β1 promoter revealed a putative hypoxia response element (HRE). HIF-1α bound to the Na,K-ATPase β1 promoter and inhibiting the activity of HIF-1α blocked the TGF-β2 mediated Na,K-ATPase β1 decrease suggesting that HIF-1α plays a potential role in Na,K-ATPase β1 regulation during EMT in RPE cells. Furthermore, knockdown of Na,K-ATPase β1 in ARPE-19 cells was associated with a change in cell morphology from epithelial to mesenchymal and induction of EMT markers such as α-smooth muscle actin and fibronectin, suggesting that loss of Na,K-ATPase β1 is a potential contributor to TGF-β2-mediated EMT in RPE cells.
Keywords: Na, K-ATPase beta subunit, Retinal pigmented Epithelium (RPE), Proliferative Vitreoretinopathy (PVR), Epithelial Mesenchymal Transition (EMT), TGF-β2, HIF-1α
1. Introduction
Fibrotic diseases in the eye disrupt the normal tissue architecture required to maintain a clear visual axis and lead to compromised vision and blindness. Fibrotic EMT follows rhegmatogenous retinal detachment (RRD) where retinal pigmented epithelial cells (RPE) undergo a change to a fibroblastic morphology via a process known as epithelial-to-mesenchymal transition (EMT) and start to produce extracellular matrix (ECM) membranes leading to aberrant wound healing and scar formation around the site of injury [1]. These contractile fibrocellular membranes lead to tractional retinal detachments characteristic of proliferative vitreoretinopathy (PVR) and complicate the prognosis for primary retinal detachment surgery [2]. During the progression of PVR and fibrosis, an increase in the total and active levels of TGF-β2, the predominant isoform in the eye, has been reported in the vitreous, the subretinal fluid and retinal membranes correlating with the severity of the disease [3-7]. TGF-β is known to mediate fibrotic EMT in a number of tissues via Smad-dependent and Smad-independent pathways. Receptor regulated Smads (R-Smads2 and 3) are recruited to the transphosphorylated type II and type I receptor complex after ligand activation. R-Smads form a complex with Smad4 to translocate to the nucleus and bind to transcription factors that act as coactivators or repressors to directly or indirectly regulate gene transcription [8, 9]. Smad independent signaling of TGF-β recruits members of the mitogen activated protein kinase (MAPK) signaling pathways such as Erk1/2, p38 MAP kinase and JNK kinases [8, 9]. TGF-β2 is also associated with RPE cell dedifferentiation, ECM accumulation and contraction of retinal membranes during fibrosis and wound healing [3, 10]. The exact mechanisms by which TGF-β2 induces RPE cell EMT are still under investigation.
The Na,K-ATPase, also known as sodium pump, is vital for several RPE cell functions such as, vectorial transport of ions and solutes from the choroid to the photoreceptors and maintaining a high concentration of Na+ in the subretinal space for normal function of the light-dark cycle [11-13]. Recent studies from our lab have implicated a role for Na,K-ATPase in kidney fibrosis [14]. But while in kidney and most other epithelia the sodium pump is expressed basolaterally, RPE cells are unique in that they contain apical Na,K-ATPase [15, 16] and it remains to be determined whether Na,K-ATPase has similar roles in retinal fibrosis. The pump itself is composed of two non-covalently linked α and β subunits [17] and like most epithelia RPE cells express the α1 and β1 isoforms [16-18]. While the α-subunit is the catalytic subunit of the enzyme, the β subunit functions in translation, folding and membrane insertion, membrane trafficking and stability of the α-subunit and enzymatic function of the protein [19-22]. More recently the β1 subunit has been implicated as a potential cell adhesion molecule [23-28]. Studies from our group have shown that Na,K-ATPase subunits co-localize at epithelial adherens junctions with other junctional proteins and that Na,K-ATPase activity is required for tight junction formation and epithelial polarity [29, 30]. Inhibition of the pump activity increases tight junction permeability of RPE cells [31] and causes epithelial cell monolayers to detach from their substratum [32, 33]. Reduced β1 levels prevent tight junction formation and restoring β1 levels is sufficient to establish cell-cell adhesion in non-polarized cells [23-25, 29]. Interestingly, the β1 subunit levels decrease in renal fibrosis, in renal epithelial cells that undergo TGF-β1-mediated fibrotic EMT and in several cancers that undergo EMT [14, 34-36]. Whether loss of Na,K-ATPase expression and function is associated with TGF-β2 mediated injury and fibrotic EMT of RPE cells as observed during PVR is not known.
We now show that EMT in RPE cells is associated with a significant reduction in Na,K-ATPase β1-subunit levels and is specific to TGF-β signaling. Na,K-ATPase α1-subunit expression, its activity and intracellular Na+ levels largely remained unaltered during TGF-β2-mediated EMT in ARPE-19 cells. shRNA-mediated knockdown of the β1 subunit was sufficient to induce an EMT-like response by altering cell morphology and inducing fibronectin accumulation in ARPE-19 cells. Promoter analysis of the β1-subunit revealed putative binding sites for HIF and Smad transcription factors in close proximity and both, HIF-1α and Smad3 bound to the β1 gene promoter during TGF-β2 mediated EMT suggesting that they could function as potential transcriptional repressors for the Na,K-ATPase β1-subunit. Together, we show that Na,K-ATPase β1-subunit is regulated by TGF-β2 signaling and might be an important contributor to the TGF-β2-mediated EMT response in RPE cells.
2. Materials and methods
2.1. Cell lines and reagents
ARPE-19 cells were obtained from American Type Culture Collection (ATCC, Manassas, VA) and cultured in DMEM-F12 media with 10% FBS, 1mM glutamine and 100U/ml of penicillin and streptomycin. ARPE-19 cells from passage number 7 to 20 were used for this study. Sub-confluent ARTPE-19 cells were serum starved for 4-12 hours prior to treatment and treated daily after media replacement with 4ng/ml of TGF-β2 for short term (2, 4, 8, 12, 24 hours) and long term (24, 48, 72 and 96 hours) time points. Control cells were serum starved and treated with the vehicle for the duration of the entire treatment. All treatment sets were lysed simultaneously. Recombinant human TGF-β2 was obtained from R&D Systems, Inc. (Minneapolis, MN), epidermal growth factor (EGF) and basic fibroblast growth factor (bFGF) were obtained from Invitrogen (Carlsbad, CA) and stock solutions were prepared as recommended by the manufacturer. Cells were treated for 48 and 96 hours with 10ng/ml of EGF and 10ng/ml of bFGF. Ouabain was obtained from Fluka (Steinheim, Germany). Cells were treated for 48 and 96 hours with 10μM of ouabain in ethanol. Control cells were treated with the vehicle (HCl-BSA for TGF-β2 and ethanol for EGF, bFGF and ouabain). Echinomycin (NSC-13502) was obtained from A.G. Scientific, Inc. (San Diego, CA) and stock solutions were prepared in DMSO. Cells were treated once for 24, 48 and 72 hours with 0.3μM echinomycin. Control cells were treated with the vehicle (DMSO). The cell titer blue cell viability assay was obtained from Promega Corporation (Madison, WI) and performed as per manufacturer’s recommendation.
For knockdown studies the shRNA sequence (5′-GTGATGCTGCTCACCATCA-3′) that targets human Na,K-ATPase β1 and cloned into the pSilencer 5.1 (Ambion, Austin, TX) was described earlier [14]. Na,K-ATPase β1-shRNA transfection of ARPE-19 cells was performed using the Amaxa cell line Nucleofector Kit V (Lonza, Walkersville, MD) as per manufacturer’s instructions. Single clones were selected after two to three weeks of treatment with 10μg/ml of puromycin (Sigma-Aldrich Co. LLC, St. Louis, MO) and knockdown of Na,K-ATPase β1 was confirmed by immunoblotting. Representative clones transfected with the control vector in parallel were selected as vector control. All vector control cells selected displayed similar morphology and Na,K-ATPase β1 expression (data not shown).
Primary antibodies used for immunoblotting or immunofluorescence were fibronectin and HIF-1α (BD Biosciences, San Jose, CA), α-smooth muscle actin (α-SMA) and β-actin (Sigma-Aldrich Co. LLC, St. Louis, MO), ZO-1 (Zymed, San Francisco, CA), Na,K-ATPase α1 (M7-PB-E9) and β1 (M17-P5-F11, kind gifts of Dr. J. Ball), Na,K-ATPase α2, α3 and β2 (all Upstate Biotechnology, Lake Placid, NY), glyceraldehyde 3-phosphate dehydrogenase (GAPDH) and EGFR (Cell Signaling Technology, Beverly, MA). HRP-conjugated secondary antibody was obtained from Cell Signaling Technology, Beverly, MA, FITC-and Cy5-conjugated secondary antibodies from Jackson ImmunoResearch Laboratories (West Grove, PA) and Texas red-conjugated phalloidin from Sigma Chemical Co.
2.2. Immunoblotting and cell surface biotinylation
Immunoblotting was performed as described previously [25]. Briefly, after treatment, the cells were washed with ice cold phosphate-buffered saline (PBS) and lysed in 1X SDS lysis buffer (95mM NaCl, 25mM Tris pH 7.4, 0.5mM EDTA, 2% SDS). The lysates were sonicated and clarified by centrifugation at 13,000 rpm for 10 min. Equal amount of protein was loaded and separated on 10% SDS-PAGE gels and transferred onto nitrocellulose membranes. Nitrocellulose membranes were blocked using 5% blocking solution (5% nonfat dry milk in Tris buffered saline with 0.1% Tween-20 (TBS-T)). The blots were incubated overnight at 4°C with primary antibodies diluted in 5% blocking solution, washed with TBS-T and incubated for one hour with HRP-conjugated secondary antibody in 5% blocking solution. After TBS-T washes, bound antibody was detected by enhanced chemiluminescence western lighting system (Perkin-Elmer, Waltham, MA). Densitometric analysis was performed using the Tina image analysis software.
For cell surface biotinylation TGF-β2 treated cells were washed in ice cold PBS-CM. Cell impermeable biotin (EZ-Link Sulfo-NHS-LC-Biotin, ThermoFisher, Hudson, NH) was freshly prepared in anhydrous DMSO. Cell surface biotinylation was performed as described earlier [19] and Na,K-ATPase α1 and β1 subunit and EGFR levels were determined by immunoblotting as described above.
2.3. Immunocytochemistry and laser scanning confocal microscopy
Cells plated on 12 well dishes inlaid with glass coverslips were treated with TGF-β2 as indicated. The coverslips were washed with PBS containing 1mM of CaCl2 and 1mM of MgCl2 (PBS-CM) and fixed in 4% paraformaldehyde for 30 minutes in the dark at room temperature. After quenching with 50mM of NH4Cl, cells were permeabilized with 0.075% Saponin in PBS-CM. Coverslips were incubated for 1 hour with fibronectin antibody followed by incubation for 30 minutes with FITC-conjugated secondary antibody. To visualize F-actin, cells were processed as described above and stained with Texas red-conjugated phalloidin. For staining with ZO-1, cells were fixed in ice-cold methanol at −20°C for 30 minutes. After washing with PBS-CM containing 0.5% bovine serum albumin (PBS-CM-BSA), cells were incubated for one hour with primary antibody followed by 30 minute incubation with Cy5-conjugated secondary antibody. After washing coverslips were mounted on glass slides with ProLong Gold antifade reagent (Invitrogen, Carlsbad, CA). Confocal microscopy was performed using a Leica TCS SP5 confocal microscope. Fibronectin samples labeled with FITC were excited at 488 nm with an Argon laser, Texas red-labeled F-actin samples and Cy5 labeled ZO-1 samples were excited at 594 nm and 633 nm, respectively, with a HeNe laser. The emitted light was captured and images were processed using the LAS AF imaging software (version 2.5.1.6757).
2.4. RNA isolation, cDNA synthesis and quantitative PCR (qPCR)
RNA was isolated from treated cells using the Ambion RNAqueous RNA isolation Kit (Ambion, Inc., Austin, TX) an cDNA was synthesized using the iScript cDNA synthesis kit (Bio-Rad, Hercules, CA) according to manufacturers’ instructions. cDNA was amplified via real-time PCR using the SYBR Green PCR Master Mix (Applied Biosystems, Warrington, UK). The following primers were used: Na,K-ATPase β1 subunit (forward: 5′-ACCAATCTTACCATGGACACTGAA-3′ and reverse: 5′-CGGTCTTTCTCACTGTACCCAAT-3′), HIF-1α (forward: 5′-ATCCATGTGACCATGAGGAAATG-3′ and reverse: 5′-CTCGGCTAGTTAGGGTACACTT-3′), GAPDH (forward: 5′-GCTGTCCAACCACA TCTCCTC-3′ and reverse: 5′-TGGGGCCGAAGATCCTGTT-3′). qPCR was performed in a 384 well format on a 7900HT Fast Real-Time PCR system (Applied Biosystems, Foster City, CA). Samples were assayed in triplicate or quadruplicate. RNA was quantified using the ΔΔCt method of relative quantification (RQ) and the transcript levels were normalized to the endogenous control GAPDH.
2.5. Ouabain sensitive rubidium uptake assay and intracellular Na+ ion measurement
The ouabain sensitive ion transport determined by 86Rb+ uptake was described earlier [37]. Briefly, to measure the ouabain-sensitive 86Rb+ transport, one set of samples was pretreated with 50μM ouabain for 30 minutes at 37°C uptake measurements. Cells were washed with ice cold wash buffer (144mM NaCl, 10mM HEPES pH 7.4 and 0.5mM CaCl2) and uptake buffer (144mM NaCl, 10mM HEPES pH 7.4, 0.5mM MgCl2, 0.5mM CaCl2, 1mM RbCl2 and 1mg/ml glucose) containing 1μCi/ml of 86Rb+ (PerkinElmer, Waltham, MA) was added to the plates and incubated for 10 minutes at 37°C. After three washes, cells were lysed with 0.5N NaOH and 86Rb+ levels were determined using a Beckman LS 6500 Scintillation Counter (Beckman Coulter, Fullerton, CA). The ouabain sensitive 86Rb+ flux was calculated as the difference between 86Rb+ counts for ouabain insensitive and total cellular uptake and the uptake was normalized to the total protein concentration.
Intracellular sodium ion concentration was determined by atomic emission spectrometry as described previously [30] and the intracellular concentrations for Na+ and Mg2+ were measured at 589 nm and 279 nm, respectively. Na+ concentrations were normalized to the total internal Mg2+ content (internal control).
2.6. Promoter analysis and chromatin immunoprecipitation assay (ChIP assay)
The MatInspector software tool (Genomatix Software GmbH, Munich, Germany) was used to examine the 1141bp Na,K-ATPase β1 promoter region for potential transcription factor binding sites. Binding sites with a quality based rating of 0.80 and higher were examined for determining potential transcription factor binding sites in the Na,K-ATPase β1 promoter region.
ChIP assay was performed as described previously [38] with the following changes. Cells were lysed in 1X SDS lysis buffer followed by sonication to shear the DNA into 200 to 2000 bp fragments. Diluted chromatin solution was precleared using a salmon sperm-protein A agarose slurry (Salmon sperm: Upstate Biotechnology, Lake Placid, NY, Protein A: GE Healthcare Life Sciences, Piscataway, NJ). 2μg of HIF-1α ChIP grade antibody (Abcam, Cambridge, MA), total Smad3 or phosphorylated Smad3 (pSmad3) (Cell Signaling Technology, Beverly, MA) was added to the supernatant followed by overnight rotation at 4°C. Antibody against Histone H3 was used as positive control IP and rabbit anti-mouse IgG was used as the negative control immunoprecipitation. Salmon sperm-protein A agarose slurry was added to the lysate-antibody mixture and incubated at 4°C for one hour. The beads were pelleted and washed once for five minutes each with low salt wash buffer, high salt wash buffer, LiCl wash buffer and TE wash buffer. DNA was recovered via phenol-chloroform extraction and ethanol precipitation. 2μl of DNA was used as template for PCR reaction. The following primers, forward: 5′-CCAAGAATTTTACAGCAGAACAAAGG -3′ and reverse: 5′-GCAGTGAGTTGTCCCGTAGAAACC -3′, designed to amplify the HRE and Smad containing regions of the Na,K-ATPase β1 promoter were used for PCR amplification of the DNA. The 186 bp amplified product was run on a 1.5% agarose gel and the images were taken using the Perkin Elmer Geliance 600 imaging system and the GeneSnap™ Image Acquisition Software.
2.7. Statistics
Data are represented as mean ± SE. Statistical significance was determined via one way ANOVA followed by Dunnett’s multiple comparison test. P-value < 0.05 was considered significant.
3. Results
3.1. Regulation of Na,K-ATPase β1 subunit in ARPE-19 cells undergoing TGF-β2-mediated EMT
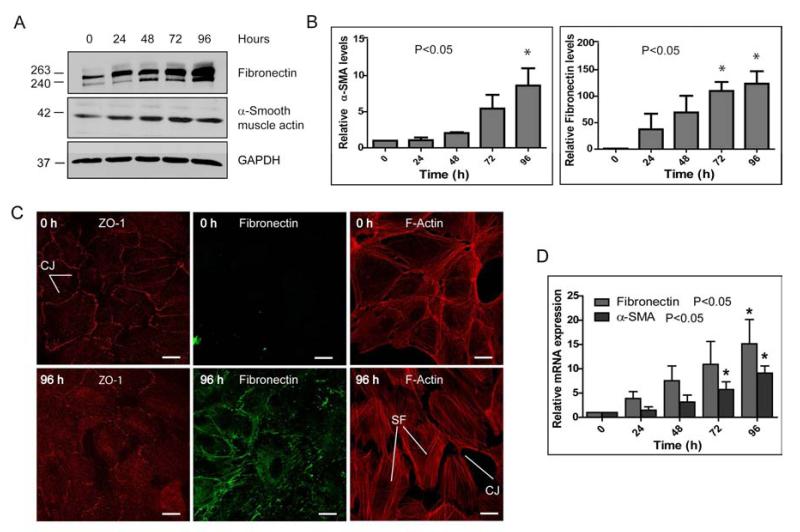
In addition to its traditional role in transepithelial ion transport, it has been found more recently that Na,K-ATPase is required for maintaining the epithelial phenotype, including junctional complexes and polarity [39]. Other studies from our lab have shown that Na,K-ATPase subunits may serve as markers for EMT in cancer and fibrosis and that in TGF-β1-mediated fibrotic EMT in the kidney reduced Na,K-ATPase expression contributes to the mesenchymal phenotype [14]. Whether TGF-β2 affects Na,K-ATPase α1 and β1 subunits during fibrotic EMT of RPE cells as observed in PVR is currently unknown. To address this question, we first confirmed that human RPE cells (ARPE-19) in culture undergo EMT upon TGF-β2 treatment. To model the disrupted RPE monolayer and RPE cell dispersal following retinal detachment we used subconfluent ARPE-19 cells for our studies. In addition, we confirmed that under our experimental conditions (serum starvation and/or TGF-β2 treatment up to 96 hours) RPE cell viability was not affected significantly (Supplementary Fig. 1). In our initial experiments we found that upon TGF-β2 treatment both protein and mRNA levels of the EMT markers α-SMA and fibronectin significant increased over time in TGF-β2 treated cells when compared to control cells (Fig. 1A-D). While the increase in protein and mRNA levels occurred already after 24 hours and was more obvious for fibronectin (Figs. 1B and D), the increase in α-SMA, although delayed, was statistically significant (Figs. 1B and D). Furthermore, the junctional marker protein ZO-1 that usually localizes to sites of cell-cell junctions was relocalized predominantly to the cytoplasm (Fig. 1C). This was accompanied by a reorganization of the actin cytoskeleton, leading to stress fiber formation and decreased cell-cell contact points between adjacent cells (Fig. 1C). Together these data confirmed that ARPE-19 cells were undergoing TGF-β2-mediated EMT in culture.
Figure 1. ARPE-19 cells undergo TGF-β2-mediated EMT.
(A) Immunoblot for EMT markers fibronectin and α-SMA. Blots are representative of three different experiments. GAPDH was used as loading control. (B) Quantitative analysis of fibronectin and α-SMA levels normalized to the loading control. Error bars represent mean ± SE of three independent experiments. (C) Immunofluorescence images of ARPE-19 cells treated with TGF-β2 for 96 hours. Decrease in staining for the epithelial marker ZO-1 at the cell junctions (CJ) and increase in cytoplasmic staining by 96 hours. An increase in fibronectin staining and stress fiber (SF) formation was observed. Scale bars: 30 μm. (D) qPCR analysis of the mRNA levels of EMT markers α-SMA and fibronectin normalized to untreated cells (0 h). Error bars represent mean RQ ± SE of five independent experiments.
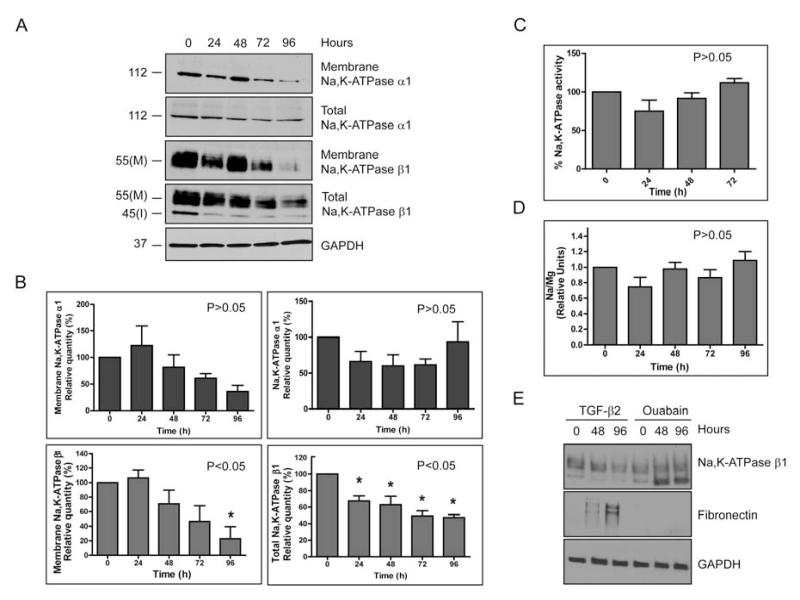
Next we tested whether TGF-β2-mediated EMT in ARPE-19 cells affects Na,K-ATPase α1 and β1 subunits. We consistently found a significant decrease of 47.6 ± 3.4% (P<0.05) in the total Na,K-ATPase β1 levels by 96 hours when compared to untreated cells. However, the levels of total Na,K-ATPase α1 varied and quantitative analysis obtained from five independent experiments revealed no significant change (P>0.05) (Fig. 2A, B). Similarly, the membrane levels of the β1-subunit were significantly decreased by 77.2 ± 16% (P<0.05) after 96 hours of treatment as determined by cell surface biotinylation. While we seemed to observe a trend in decreasing membrane α1-subunit levels the changes were not statistically significant (P>0.05) (Fig, 2A, B). No apparent change in other Na,K-ATPase isoforms (α2, α3 and β2) that might have compensated for the altered expression in β1 was detected (Supplementary Fig. 2). In addition, the membrane levels of EGFR did not decrease over time (Supplementary Fig. 3), suggesting that the changes are specific to the β1-subunit. Consistent with the unaltered levels of α-subunit, we did not find any significant change in the ouabain-sensitive uptake of 86Rb+ as a measure of the enzymatic activity (Fig. 2C) or in intracellular Na+ levels as determined by atomic emission spectrometry (Fig. 2D). In addition, inhibition of the Na,K-ATPase enzymatic activity with the cardiac glycoside ouabain did not induce fibronectin accumulation in ARPE-19 cells (Fig. 2E). Together these data suggest that inhibition of the pump activity does not mediate the EMT response in ARPE-19 cells. It is interesting to note that the decrease in Na,K-ATPase β1 and extensive fibronectin accumulation was observed only with TGF-β2 treatment and not with other EMT inducing growth factors such as EGF and bFGF (Supplementary Fig. 4) suggesting that factors specific to the TGF-β2 signaling pathway are involved in regulating Na,K-ATPase β1 levels during EMT in RPE cells.
Figure 2. Reduced Na,K-ATPase subunit levels in TGF-β2-mediated EMT.
ARPE-19 cells were treated with TGF-β2 for 0, 24, 48, 72 and 96 hours. (A) Total Na,K-ATPase α1- and β1-subunit levels were determined by immunoblotting. I indicates the immature high mannose ER form (45kDa) and M the mature fully glycosylated form with complex N-glycans (55kDa) of the β1-subunit. Plasma membrane levels of α1- and β1-subunit were determined after cell surface biotinylation. GAPDH was used as loading control. (B) Quantitative analysis of α1- and β1-subunit levels normalized to the loading control. Error bars represent mean ± SE of five independent experiments for total and membrane α1-subunits and membrane β1-subunit and seven independent experiments for total β1-subunit. Statistically significant changes are indicated, P<0.05. (C) Ouabain-sensitive 86Rb+ uptake assay. Data represent the average percentage Na,K-ATPase activity of three independent experiments done in triplicates ± SE. Assay did not reveal significant differences in pump activity (P>0.05, one way ANOVA). (D) Intracellular Na+ levels as measured by atomic emission spectrometry. Na+ concentrations were normalized to the total internal Mg2+ content (internal control). The data represent the mean of three independent experiments performed in triplicates ± SE. Data analysis did not show any significant change in Na+ levels upon TGF-β2 treatment (P>0.05, one way ANOVA). (E) ARPE-19 cells were treated for 48 and 96 hours with 10 μM of ouabain and β1-subunit and fibronectin levels were determined by immunoblotting. GAPDH was used as loading control. Image is representative of three different experiments.
3.2. shRNA-mediated knockdown of Na,K-ATPase β1 induces EMT markers
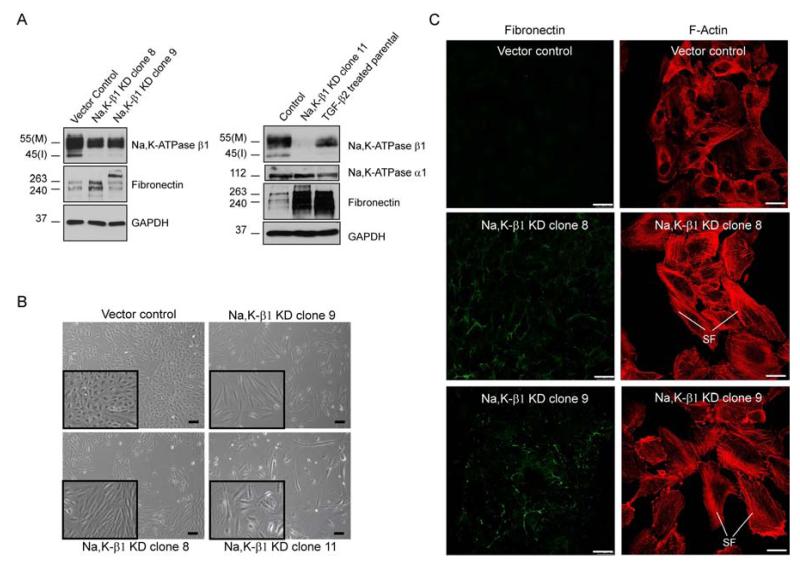
Since Na,K-ATPase β1 can function as a cell-cell adhesion molecule and loss of adhesion molecules may lead to EMT we next tested whether knockdown of the β1 subunit can induce EMT in ARPE-19 cells. Using a shRNA approach we selected three stable Na,K-ATPase β1 knockdown clones, Na,K-β1 KD clone 8, 9 and 11, which expressed 77%, 64% and 4% of the β1-subunit compared to cells transfected with control vector, respectively, for further characterization (Fig. 3A). While the α1-subunit protein levels remained unchanged in knockdown cells, loss of the β1-subunit induced a mesenchymal, spindle shaped morphology compared to vector transfected control cells (Fig. 3B). Furthermore, we observed an accumulation of actin stress fibers and a decrease in cell-cell contact points in β1-subunit knockdown cells (Fig. 3C) accompanied by increased expression of fibronectin (Fig. 3A, C). Together these data suggest that a reduced expression of β1-subunit of Na,K-ATPase could induce an EMT-like phenotype independent of the α1-subunit, and thus might contribute to the pathogenesis of PVR.
Figure 3. Knockdown of Na,K-ATPase β1-subunit induces EMT markers.
(A) Immunoblot of Na,K-ATPase subunits and fibronectin in shRNA-mediated β1-subunit knockdown ARPE-19 clones (Na, K-β1 KD clones 8, 9 and 11) and control cells transfected with vector alone. TGF-β2-treated cells (96 hours) are included for comparison. GAPDH was used as a loading control. (B) Phase contrast images showing an elongated, spindle-shaped morphology of Na,K-ATPase β1-subunit knockdown cells compared to vector transfected control cells (vector control). Inset images represent magnified regions showing differences in morphology. Scale bars: 200 μm. (C) Immunofluorescence images showing fibronectin accumulation and stress fiber (SF) formation in Na, K-β1 KD clones 8 and 9 compared to vector control. Scale bars: 30 μm.
3.3. Transcriptional regulation of Na,K-ATPase β1 during TGF-β2-mediated EMT
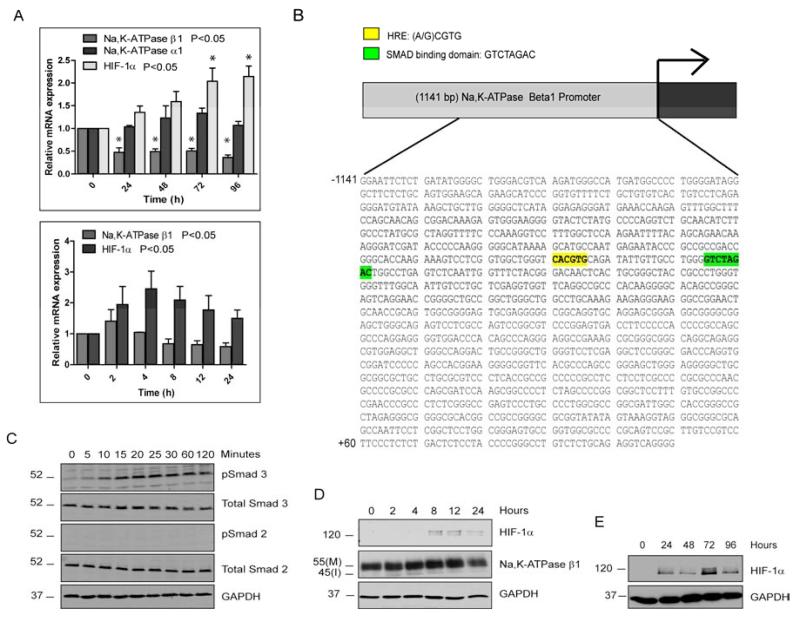
TGF-β induces EMT through both transcriptional and post-transcriptional mechanisms. Thus, we determined the mRNA levels of Na,K-ATPase β1 in TGF-β2 treated ARPE-19 cells by quantitative real time PCR (qPCR) analysis. β1 mRNA levels were reduced as early as eight hours after TGF-β2 treatment when compared to untreated control cells and remained low for the duration of treatment (Fig. 4A) suggesting that activation of the TGF-β2 signaling pathway leads to transcriptional repression of Na,K-ATPase β1. Consistent with our observations on Na,K-ATPase α1 protein levels, TGF-β2 treatment did not result in any significant decrease in α1 subunit mRNA levels (P>0.05) (Fig. 4A). Thus, TGF-β2 signaling may activate transcription factors that specifically regulate the β1-subunit. We have previously shown that the transcription factor Snail that is also associated with TGF-β-induced EMT, can transcriptionally repress Na,K-ATPase β1 [36]. However, we were not able to detect Snail protein in control or TGF-β2 treated ARPE-19 cells (Supplementary Fig. 5) indicating that other transcription factors activated by TGF-β2 signaling may play a role in Na,K-ATPase β1 regulation. Analysis of the 1141 bp Na,K-ATPase β1 gene promoter described by Derfoul et al., [40] for potential transcription factor binding sites revealed among others, a putative hypoxia response element (HRE) (−750 and −746 bp) in close proximity to a Smad binding domain (SBD) (−728 and −721 bp) upstream to the Na,K-ATPase β1 gene transcriptional start site (Fig. 4B), suggesting that hypoxia inducible factors (HIFs) that bind to HREs or Smads that bind to the palindromic SBD [9, 41] could be transcriptional regulators of Na,K-ATPase β1.
Figure 4. Analysis of potential transcriptional regulators of Na,K-ATPase β1.
(A) Relative mRNA levels of Na,K-ATPase β1, Na,K-ATPase α1 and HIF-1α after TGF-β2 treatment. The graph represents mean RQ ± SE of five independent experiments for Na,K-ATPase β1 and three independent experiments for Na,K-ATPase α1 and HIF-1α, all performed in triplicates. P<0.05 was considered statistically significant. (B) Promoter analysis of the 1141 bp Na,K-ATPase β1 gene promoter using the MatInspector software tool revealed a putative hypoxia response element (HRE, yellow) and Smad binding domain (SBD, green). (C) Immunoblots for phospho-Smad2 and phospho-Smad3 in ARPE-19 cells treated for 0, 5, 10, 15, 20, 25, 30, 60 and 120 minutes with TGF-β2. Total Smad2, total Smad3, and GAPDH were used as loading controls. (D) Immunoblot of HIF-1α and Na,K-ATPase β1 in ARPE-19 cells treated with TGF-β2-treated for 0, 2, 4, 8, 12 and 24 hours. GAPDH was used as loading control. Images are representative of three different experiments. (E) Immunoblot of HIF-1α in ARPE-19 cells treated with TGF-β2-treated for 0, 24, 48, 72 and 96 hours. GAPDH was used as loading control. Images are representative of three different experiments.
HIF-1α and Smads, major transcription factors that cooperate and activate multiple genes during TGF-β signaling [42], have been implicated in mediating EMT and renal fibrosis [43-47]. In ARPE-19 cells TGF-β2 treatment resulted in activation of Smad3 but not Smad2 (Fig. 4C) suggesting that Smad3 is the predominant R-Smad activated in response to TGF-β2 signaling in these cells. HIF-1α, the HIF isoform predominantly expressed in RPE cells [48] is usually stabilized under hypoxia (1% O2) but can also be upregulated under normoxia (15% O2) by TGF-β [49, 50]. Indeed, TGF-β2 treatment of ARPE-19 cells resulted in an increase in HIF-1α mRNA (Fig. 4A) and protein (Fig. 4D, E) levels. The increase in activated Smad3 and HIF-1α protein levels prior to (Smad 3) or around the same time as (HIF-1α) the decrease in Na,K-ATPase β1 mRNA levels further suggests that these factors may play a role in the transcriptional repression of Na,K-ATPase β1.
3.4. HIF-1α and Smad3 bind to the Na,K-ATPase β1 promoter
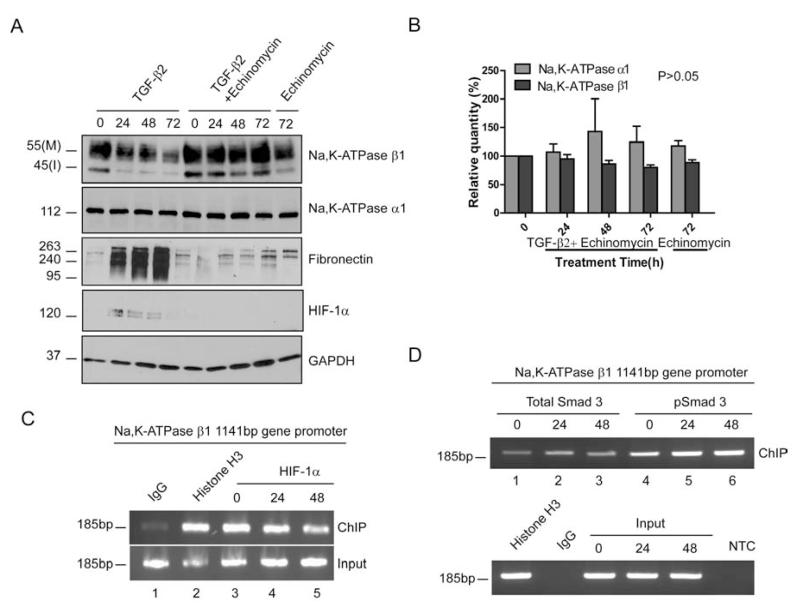
To further test HIF-1α’s potential role in repressing Na,K-ATPase β1 transcription we treated ARPE-19 cells with TGF-β2 in the presence of the drug echinomycin (NSC-13502) and determined the β1-subunit levels. Echinomycin is a peptide antibiotic that intercalates into the DNA specifically at the HRE, blocking HIF-1α binding to its response element [51, 52]. We found that in the presence of 0.3μM echinomycin, TGF-β2 treatment failed to decrease Na,K-ATPase β1-subunit levels and fibronectin accumulation was blocked (Fig. 5A and B), suggesting that HIF-1α DNA binding is important for down regulating Na,K-ATPase β1 upon activation of the TGF-β2 signaling cascade.
Figure 5. HIF-1α and Smad3 bind to the Na,K-ATPase β1 promoter.
(A) Na,K-ATPase subunit, fibronection and HIF-1α expression in ARPE-19 cells treated with 4 ng/ml of TGF-β2 and 0.3μM of echinomycin for 0, 24, 48 and 72 hours. GAPDH was used as loading control. (B) Na,K-ATPase α1 and β1 levels after 0, 24, 48 and 72 hours of TGF-β2 and Echinomycin treatment and 72 hours of Echinomycin treatment. Graph represents mean ± SE of three independent experiments. (C and D) ChIP assay in ARPE-19 cells treated for 0, 24 and 48 hours with TGF-β2. IgG, histone H3 and HIF-1α, Smad3 and pSmad3 represent antibodies used to pull down chromatin fragments (ChIP). Input represents the total fragmented chromatin before IP. Anti-mouse IgG was used as negative control IP. IP using Histone H3 was used as positive control. PCR primers were designed to amplify the specific region in the Na,K-ATPase β1 promoter containing the HRE and SBD after ChIP and the 185bp PCR product was run on a 1.5% agarose gel. NTC represents no template PCR control. Images are representative of three independent experiments.
Next, to determine if HIF-1α and Smad3 directly bind to the Na,K-ATPase β1 promoter, we performed a ChIP assay. Antibodies against HIF-1α, total Smad3 and phosphorylated Smad3 (pSmad3) were used to pull down fragmented chromatin. Histone H3 served as a positive control. Primers that encompass the region containing the HRE and SBD in the 1141bp Na,K-ATPase β1 promoter were used for amplification of the precipitated DNA fragments. We found that HIF-1α (Fig. 5C, lanes 3-5) and Smad3 (Fig. 5D, lanes 1-3) bound to the Na,K-ATPase β1 promoter in stimulated and in control cells. Interestingly, the activated, phosphorylated form of Smad3 appeared to bind more efficiently to the β1 promoter in TGF-β2-treated cells (Fig. 5D, compare lanes 5 and 6 with lane 4). While these data support the potential for HIF-1α and Smad3 to act as transcriptional regulators of Na,K-ATPase β1, it remains to be determined whether HIF-1α and Smad3 cooperate to transcriptionally repress Na,K-ATPase β1 in TGF-β2-treated RPE cells.
4. DISCUSSION
4.1. TGF-β2-induced EMT in RPE cells is associated with reduced Na,K-ATPase β1 subunit expression
Na,K-ATPase was historically known as an ion pump that maintains intracellular ion homeostasis and drives transepithelial transport. In recent years it has become evident that both Na,K-ATPase expression and function are required for maintaining a normal polarized epithelial phenotype. Decreased subunit levels, especially of the β1-subunit have been associated with EMT in cancer and fibrosis [14, 25, 30, 35]. We have previously shown that despite its apical localization in RPE cells and opposed to the basolateral localization in most other epithelia, Na,K-ATPase activity is crucial for proper tight junction structure and function in primary cultures of human RPE cells [31]. However, whether Na,K-ATPase plays a role in RPE EMT and fibrosis as observed in pathogenic processes such as PVR is not known. In this study, using cultured human ARPE-19 cells, we show that loss of Na,K-ATPase β1 expression may contribute to the disease process in TGF-β2 mediated EMT and fibrosis in the retinal pigment epithelium. Upon activation of TGF-β2 signaling, a major factor involved in fibrotic eye diseases, EMT markers such as fibronectin, α-SMA and actin stress fibers were induced while β1-subunit expression decreased. This effect was likely mediated by suppression of β1 transcription by the transcription factors HIF-1α and/or Smad3 that are known to be induced by TGF-β. The effect of TGF-β2 on Na,K-ATPase seemed to be specific to the β1-subunit since α1-subunit levels or the enzymatic activity of the pump were not affected. In support of this, knockdown of Na,K-ATPase β1 induced an EMT-like process with change of the epithelial to a mesenchymal morphology and the induction of fibronection and actin stress fibers. There are no specific pharmacological treatments available to date that can be used to prevent the development of RPE cell EMT. Identification of potential Na,K-ATPase β1 regulators and determining the mechanisms by which TGF-β2 controls Na,K-ATPase β1 levels in ARPE-19 cells might help in the development of novel treatments for fibrotic eye diseases.
4.2. Na,K-ATPase β1 subunit as a possible mediator of TGF-β2-induced EMT in RPE cells
In addition to mesenchymal EMT (type 1) that occurs during normal development in gastrulation and neuronal crest formation, pathological EMT is divided into two major subtypes, EMT type 2 in inflammation and fibrosis and type 3 in cancer, invasion and metastasis. In cancer, changes in Na,K-ATPase subunit levels and activity, either as an increase or inhibition, have been reported in numerous studies [39] suggesting a tissue and cell type-specific mechanism of regulation. In fibrotic EMT in the kidney we recently showed that both Na,K-ATPase α1 and β1 were reduced [14]. However, in the present study TGF-β2-mediated EMT in ARPE-19 cells affected only the β1 but not α1-subunit (Fig. 2), suggesting that not only in cancerous but also in fibrotic EMT tissue-specific factors might be involved in Na,K-ATPase regulation. In addition, in EMT of several cancers such as bladder, colon, and pancreatic cancer that are poorly differentiated, the intracellular Na+ levels were elevated when compared to well-differentiated cells [14, 36, 53]. Similarly, we found in TGF-β1-mediated EMT in the kidney increased intracellular Na+ [14] but in ARPE-19 cells the pump activity was unaltered and there was no associated increase in Na+ (Fig. 2). At this point we don’t know whether these differences are due to intrinsic variations in the TGF-β1 and TGF-β2-associated signaling pathways or other innate differences in kidney and RPE cells.
It is well established that the α-subunit is the catalytic subunit of the Na,K-ATPase and the β1-subunit acts as a chaperone protein for the α1 subunit for its transport to and stabilization at the cell membrane [21, 22, 29]. In addition, the β1 subunit functions as a cell-adhesion molecule [23-28] and our earlier studies showed that knockdown of Na,K-ATPase β1 in kidney epithelial cells can induce EMT [14]. Similarly, knockdown of β1 in ARPE-19 cells gave rise to a mesenchymal cell morphology, actin stress fibers and the accumulation of fibronection that are usually observed during EMT. Interestingly, while knockdown of β1 in kidney was accompanied by reduced α1 levels, we did not observe a significant change of α1 in ARPE-19 cells with β1 knockdown even though the cells expressed only 4% of the β1 subunit found in vector control cells (Fig. 3B). While this seems contradictory to the notion that the β-subunit is required for the α-subunit to be transported to the plasma membrane, we cannot exclude the possibility that due to the unique location of the Na,K-ATPase in the apical plasma membrane in RPE cells other apical proteins may take over the chaperone function of the β1 in these cells.
Our previous studies in kidney cells have shown that both high β1-subunit levels and the pump activity are required for functional tight junctions and epithelial polarization [14, 25, 30]. However, and in contrast to kidney cells, in ARPE-19 cells the Na,K-ATPase activity was not affected during TGF-β2-induced EMT, which is consistent with our observation that the α1 subunit levels were not significantly reduced. Interestingly, in our previous studies in primary cultures of human RPE cells, inhibition of the pump activity decreased the tight junction contact points and increased permeability [31]. Nevertheless, the cells did not lose tight junctions or polarization per se, both of which are hallmarks of EMT. In addition, while in kidney and RPE cells inhibition of Na,K-ATPase activity reduced actin stress fibers [30, 31], TGF-β2 treatment of ARPE-19 cells induced the formation of stress fibers further supporting our finding that the pump activity was not affected in these cells. Thus, while in certain tissues such as kidney, reduced Na,K-ATPase may be contributing to EMT, loss of the Na,K-ATPase enzymatic function appears to be neither sufficient nor required to induce EMT in RPE cells. The apical Na,K-ATPase α1 in RPE cells functions normally to maintain a high concentration of Na+ in the subretinal fluid required for photoreceptor excitability and their ability to convert light to electrical signals [12]. Thus, this function might be preserved despite RPE cell EMT in an attempt to maintain normal photoreceptor function and vision even during wound healing.
4.3. Transcriptional regulation of the Na,K-ATPase β1 subunit during TGF-β2-mediated EMT
TGF-β induces EMT through mechanisms that can occur both at the transcriptional and the post-transcriptional levels. Surprisingly, while TGF-β treatment resulted in reduced β1 expression in both kidney and RPE cells, the β1 mRNA levels were only reduced in RPE cells (Fig. 4) but not in kidney cells [14]. Thus, the regulation of Na,K-ATPase β1 in kidney epithelial cells treated with TGF-β1 seemed to occur at the post-translational level and in RPE cells treated with TGF-β2 by transcriptional repression. While at this point we cannot exclude additional post-translational regulation of β1 in RPE cells it appears that depending on the cell type, TGF-β signaling can regulate the same molecule by different mechanisms.
We previously showed in various cell lines that the transcription factor Snail suppresses Na,K-ATPase β1 [36]. But although TGF-β2 treatment reduced Na,K-ATPase β1 mRNA levels we did not find Snail to be induced in ARPE-19 cells indicating that other transcription factors activated by TGF-β2 signaling may play a role in Na,K-ATPase β1 downregulation. Na,K-ATPase β1 promoter analysis revealed among others, a putative hypoxia response element and a Smad binding domain upstream to the transcription start site. Smad transcription factors mediate TGF-β signaling by recruiting co-activators or co-repressor proteins to the nucleus to modulate target gene transcription [8, 46, 54, 55]. HIF-1α is a transcription factor well-known to be induced by hypoxia but the HIF signaling cascade can also be activated by growth factors such as TGF-β. Interestingly, HIF-1α expression displayed a biphasic pattern of upregulation (Fig. 4) a pattern that was also reported in earlier studies examining HIF-1α levels in RPE cells. In these studies it was shown that HIF-1α can modulate its own levels during hypoxia via transcriptional repression and by upregulating PHD proteins [49]. Since the mRNA and protein levels of HIF-1α seemed to decrease by 24 hours and by 96 hours of TGF-β2 treatment in ARPE-19 cells, it is possible that HIF-1α regulates its own levels via transcriptional repression during TGF-β2 treatment. However, it remains to be verified if TGF-β2 increases HIF-1α stability by affecting PHD levels and if their levels are upregulated during the increase in HIF-1α expression as well in ARPE-19 cells.
Treatment of ARPE-19 cells with echinomycin suppressed the TGF-β2 induced increase in HIF-1α but also prevented the TGF-β2 induced reduction in Na,K-ATPase β1 and increase in fibronectin levels suggesting that a common signaling pathway may link HIF-1α to Na,K-ATPase β1 and fibronectin regulation. However, our ChIP analysis revealed that HIF-1α bound to the Na,K-ATPase β1 promoter even in the absence of TGF-β2 suggesting that other factors might be involved in TGF-β2 induced Na,K-ATPase β1 suppression. Smad3 and HIF-1α are known to interact and act as transcriptional co-activators of gene transcription; however, their ability to interact as co-repressors has not been described. Nevertheless, we found that the active phosphorylated form of Smad3 binds to the β1-subunit promoter and binding appeared to increase in TGF-β2-treated cells. Studies are in progress to confirm whether HIF-1α and pSmad3 bind to the putative HRE and SBD identified in the Na,K-ATPase β1 promoter and transcriptionally repress Na,K-ATPase β1.
5. ConclusionS
EMT is a multi-step process that occurs in many fibrotic diseases and is characterized by epithelial cells losing their well-differentiated polarized phenotype, changing into mesenchymal cells and thereby losing their normal function. We found that in TGF-β mediated EMT that is often observed in fibrotic diseases, repression of Na,K-ATPase β1 is not only associated with EMT but seems to directly contribute to the disease process. To date, in primary retinal detachment no specific pharmacological treatments are available that could be used to prevent the development of PVR [56]. Most of the drugs used in conjunction with primary and secondary retinal detachment surgeries are steroidal or chemotherapeutic drugs to block inflammation and cell proliferation [57],[56]. These drugs lack specificity and as a result, can affect the normal surrounding ocular tissue and also have systemic side effects on the patient [2].
With what appears to be a quite specific process in RPE cells, determining the mechanisms by which TGF-β2 controls Na,K-ATPase β1 levels in cells undergoing EMT could thus provide the basis to develop suitable drugs to improve the prognosis and visual outcome of primary RRD surgery and subsequently prevent PVR development.
Supplementary Material
TGF-β2 treatment of RPE cells results in reduced Na,K-ATPase β1-subunit expression
Loss of Na,K-ATPase β1-subunit may contribute to EMT and fibrosis in RPE cells.
HIF-1α and Smad3 bind to the Na,K-ATPase β1-subunit promoter.
Transforming growth factor (TGF)-β2-mediated epithelial-to-mesenchymal transition in retinal pigmented epithelial (RPE) cells results in reduced Na,K-ATPase β1-subunit expression.
Loss of Na,K-ATPase β1-subunit is implicated in contributing to EMT and fibrosis in RPE cells.
Hypoxia-inducible factor (Hif)-1α and Smad3 bind to the Na,K-ATPase β1-subunit promoter.
ACKNOWLEDGEMENTS
We thank Dr. William Ball, Jr., University of Cincinnati, for providing the Na,K-ATPase α1 and β1 primary antibodies. This work was supported by the American Cancer Society Research Grant RSG-09-021-01-CNE, NIH grants NCRR-5P20RR016472-12 and NIGMS-8P20GM103446-12 and funds from the Nemours Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Friedlander M. Fibrosis and diseases of the eye. J Clin Invest. 2007;117(3):576–86. doi: 10.1172/JCI31030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pastor JC. Proliferative vitreoretinopathy: an overview. Surv Ophthalmol. 1998;43(1):3–18. doi: 10.1016/s0039-6257(98)00023-x. [DOI] [PubMed] [Google Scholar]
- 3.Connor TB, Jr., et al. Correlation of fibrosis and transforming growth factor-beta type 2 levels in the eye. J Clin Invest. 1989;83(5):1661–6. doi: 10.1172/JCI114065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guerin CJ, et al. Transforming growth factor beta in experimentally detached retina and periretinal membranes. Exp Eye Res. 2001;73(6):753–64. doi: 10.1006/exer.2001.1095. [DOI] [PubMed] [Google Scholar]
- 5.Hirase K, et al. Transforming growth factor beta(2) increases in subretinal fluid in rhegmatogenous retinal detachment with subretinal strands. Ophthalmologica. 2005;219(4):222–5. doi: 10.1159/000085731. [DOI] [PubMed] [Google Scholar]
- 6.Kita T, et al. Role of TGF-beta in proliferative vitreoretinal diseases and ROCK as a therapeutic target. Proc Natl Acad Sci U S A. 2008;105(45):17504–9. doi: 10.1073/pnas.0804054105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kon CH, et al. Expression of vitreous cytokines in proliferative vitreoretinopathy: a prospective study. Invest Ophthalmol Vis Sci. 1999;40(3):705–12. [PubMed] [Google Scholar]
- 8.Derynck R, Zhang YE. Smad-dependent and Smad-independent pathways in TGF-beta family signalling. Nature. 2003;425(6958):577–84. doi: 10.1038/nature02006. [DOI] [PubMed] [Google Scholar]
- 9.Massague J, Seoane J, Wotton D. Smad transcription factors. Genes Dev. 2005;19(23):2783–810. doi: 10.1101/gad.1350705. [DOI] [PubMed] [Google Scholar]
- 10.Saika S, et al. Fibrotic disorders in the eye: targets of gene therapy. Prog Retin Eye Res. 2008;27(2):177–96. doi: 10.1016/j.preteyeres.2007.12.002. [DOI] [PubMed] [Google Scholar]
- 11.Tombran-Tink J, Barnstable CJ. Ophthalmology research. xi. Humana Press; Totowa, N.J.: 2008. Visual transduction and non-visual light perception; p. 509. [Google Scholar]
- 12.Hodson S, Armstrong I, Wigham C. Regulation of the retinal interphotoreceptor matrix Na by the retinal pigment epithelium during the light response. Experientia. 1994;50(5):438–41. doi: 10.1007/BF01920742. [DOI] [PubMed] [Google Scholar]
- 13.Kaplan JH. Biochemistry of Na,K-ATPase. Annu Rev Biochem. 2002;71:511–35. doi: 10.1146/annurev.biochem.71.102201.141218. [DOI] [PubMed] [Google Scholar]
- 14.Rajasekaran SA, et al. Na,K-ATPase subunits as markers for epithelial-mesenchymal transition in cancer and fibrosis. Mol Cancer Ther. 2010;9(6):1515–24. doi: 10.1158/1535-7163.MCT-09-0832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marmorstein AD. The polarity of the retinal pigment epithelium. Traffic. 2001;2(12):867–72. doi: 10.1034/j.1600-0854.2001.21202.x. [DOI] [PubMed] [Google Scholar]
- 16.Gundersen D, Orlowski J, Rodriguez-Boulan E. Apical polarity of Na,K-ATPase in retinal pigment epithelium is linked to a reversal of the ankyrin-fodrin submembrane cytoskeleton. J Cell Biol. 1991;112(5):863–72. doi: 10.1083/jcb.112.5.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Blanco G, Mercer RW. Isozymes of the Na-K-ATPase: heterogeneity in structure, diversity in function. Am J Physiol. 1998;275(5 Pt 2):F633–50. doi: 10.1152/ajprenal.1998.275.5.F633. [DOI] [PubMed] [Google Scholar]
- 18.Ruiz A, Bhat SP, Bok D. Characterization and quantification of full-length and truncated Na,K-ATPase alpha 1 and beta 1 RNA transcripts expressed in human retinal pigment epithelium. Gene. 1995;155(2):179–84. doi: 10.1016/0378-1119(94)00812-7. [DOI] [PubMed] [Google Scholar]
- 19.Rajasekaran SA, et al. Na,K-ATPase beta1-subunit increases the translation efficiency of the alpha1-subunit in MSV-MDCK cells. Mol Biol Cell. 2004;15(7):3224–32. doi: 10.1091/mbc.E04-03-0222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chow DC, Forte JG. Functional significance of the beta-subunit for heterodimeric P-type ATPases. J Exp Biol. 1995;198(Pt 1):1–17. doi: 10.1242/jeb.198.1.1. [DOI] [PubMed] [Google Scholar]
- 21.Ackermann U, Geering K. Mutual dependence of Na,K-ATPase alpha- and beta-subunits for correct posttranslational processing and intracellular transport. FEBS Lett. 1990;269(1):105–8. doi: 10.1016/0014-5793(90)81130-g. [DOI] [PubMed] [Google Scholar]
- 22.McDonough AA, Geering K, Farley RA. The sodium pump needs its beta subunit. FASEB J. 1990;4(6):1598–605. doi: 10.1096/fasebj.4.6.2156741. [DOI] [PubMed] [Google Scholar]
- 23.Shoshani L, et al. The polarized expression of Na+,K+-ATPase in epithelia depends on the association between beta-subunits located in neighboring cells. Mol Biol Cell. 2005;16(3):1071–81. doi: 10.1091/mbc.E04-03-0267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vagin O, Tokhtaeva E, Sachs G. The role of the beta1 subunit of the Na,K-ATPase and its glycosylation in cell-cell adhesion. J Biol Chem. 2006;281(51):39573–87. doi: 10.1074/jbc.M606507200. [DOI] [PubMed] [Google Scholar]
- 25.Rajasekaran SA, et al. Na,K-ATPase beta-subunit is required for epithelial polarization, suppression of invasion, and cell motility. Mol Biol Cell. 2001;12(2):279–95. doi: 10.1091/mbc.12.2.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kitamura N, et al. Mouse Na+/K+-ATPase beta1-subunit has a K+-dependent cell adhesion activity for beta-GlcNAc-terminating glycans. Proc Natl Acad Sci U S A. 2005;102(8):2796–801. doi: 10.1073/pnas.0409344102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Barwe SP, et al. Janus model of the Na,K-ATPase beta-subunit transmembrane domain: distinct faces mediate alpha/beta assembly and beta-beta homo-oligomerization. J Mol Biol. 2007;365(3):706–14. doi: 10.1016/j.jmb.2006.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vagin O, et al. The Na,K-ATPase alpha1beta1 heterodimer as a cell adhesion molecule in epithelia. Am J Physiol Cell Physiol. doi: 10.1152/ajpcell.00456.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vagin O, Sachs G, Tokhtaeva E. The roles of the Na,K-ATPase beta 1 subunit in pump sorting and epithelial integrity. J Bioenerg Biomembr. 2007;39(5-6):367–72. doi: 10.1007/s10863-007-9103-0. [DOI] [PubMed] [Google Scholar]
- 30.Rajasekaran SA, et al. Na,K-ATPase activity is required for formation of tight junctions, desmosomes, and induction of polarity in epithelial cells. Mol Biol Cell. 2001;12(12):3717–32. doi: 10.1091/mbc.12.12.3717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rajasekaran SA, et al. Na,K-ATPase inhibition alters tight junction structure and permeability in human retinal pigment epithelial cells. Am J Physiol Cell Physiol. 2003;284(6):C1497–507. doi: 10.1152/ajpcell.00355.2002. [DOI] [PubMed] [Google Scholar]
- 32.Contreras RG, et al. Relationship between Na(+),K(+)-ATPase and cell attachment. J Cell Sci. 1999;112(Pt 23):4223–32. doi: 10.1242/jcs.112.23.4223. [DOI] [PubMed] [Google Scholar]
- 33.Larre I, et al. Contacts and cooperation between cells depend on the hormone ouabain. Proc Natl Acad Sci U S A. 2006;103:10911–6. doi: 10.1073/pnas.0604496103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rajasekaran SA, et al. Reduced expression of beta-subunit of Na,K-ATPase in human clear-cell renal cell carcinoma. J Urol. 1999;162(2):574–80. [PubMed] [Google Scholar]
- 35.Espineda C, et al. Analysis of the Na,K-ATPase alpha- and beta-subunit expression profiles of bladder cancer using tissue microarrays. Cancer. 2003;97(8):1859–68. doi: 10.1002/cncr.11267. [DOI] [PubMed] [Google Scholar]
- 36.Espineda CE, et al. Repression of Na,K-ATPase beta1-subunit by the transcription factor snail in carcinoma. Mol Biol Cell. 2004;15(3):1364–73. doi: 10.1091/mbc.E03-09-0646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lambrecht N, et al. Identification of the site of inhibition by omeprazole of a alpha-beta fusion protein of the H,K-ATPase using site-directed mutagenesis. J Biol Chem. 1998;273(22):13719–28. doi: 10.1074/jbc.273.22.13719. [DOI] [PubMed] [Google Scholar]
- 38.Fujimura N, et al. Spatial and temporal regulation of Wnt/beta-catenin signaling is essential for development of the retinal pigment epithelium. Dev Biol. 2009;334(1):31–45. doi: 10.1016/j.ydbio.2009.07.002. [DOI] [PubMed] [Google Scholar]
- 39.Rajasekaran SA, Rajasekaran AK. Na,K-ATPase and epithelial tight junctions. Front Biosci. 2009;14:2130–48. doi: 10.2741/3367. [DOI] [PubMed] [Google Scholar]
- 40.Derfoul A, et al. Regulation of the human Na/K-ATPase beta1 gene promoter by mineralocorticoid and glucocorticoid receptors. J Biol Chem. 1998;273(33):20702–11. doi: 10.1074/jbc.273.33.20702. [DOI] [PubMed] [Google Scholar]
- 41.Shi Y, et al. Crystal structure of a Smad MH1 domain bound to DNA: insights on DNA binding in TGF-beta signaling. Cell. 1998;94(5):585–94. doi: 10.1016/s0092-8674(00)81600-1. [DOI] [PubMed] [Google Scholar]
- 42.Sanchez-Elsner T, et al. Synergistic cooperation between hypoxia and transforming growth factor-beta pathways on human vascular endothelial growth factor gene expression. J Biol Chem. 2001;276(42):38527–35. doi: 10.1074/jbc.M104536200. [DOI] [PubMed] [Google Scholar]
- 43.Basu RK, et al. Interdependence of HIF-1alpha and TGF-beta/Smad3 signaling in normoxic and hypoxic renal epithelial cell collagen expression. Am J Physiol Renal Physiol. 300(4):F898–905. doi: 10.1152/ajprenal.00335.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Higgins DF, et al. Hypoxia promotes fibrogenesis in vivo via HIF-1 stimulation of epithelial-to-mesenchymal transition. J Clin Invest. 2007;117(12):3810–20. doi: 10.1172/JCI30487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Higgins DF, et al. Hypoxia-inducible factor signaling in the development of tissue fibrosis. Cell Cycle. 2008;7(9):1128–32. doi: 10.4161/cc.7.9.5804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lan HY. Diverse roles of TGF-beta/Smads in renal fibrosis and inflammation. Int J Biol Sci. 7(7):1056–67. doi: 10.7150/ijbs.7.1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sato M, et al. Targeted disruption of TGF-beta1/Smad3 signaling protects against renal tubulointerstitial fibrosis induced by unilateral ureteral obstruction. J Clin Invest. 2003;112(10):1486–94. doi: 10.1172/JCI19270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Forooghian F, Razavi R, Timms L. Hypoxia-inducible factor expression in human RPE cells. Br J Ophthalmol. 2007;91(10):1406–10. doi: 10.1136/bjo.2007.123125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Haddad JJ, Harb HL. Cytokines and the regulation of hypoxia-inducible factor (HIF)-1alpha. Int Immunopharmacol. 2005;5(3):461–83. doi: 10.1016/j.intimp.2004.11.009. [DOI] [PubMed] [Google Scholar]
- 50.Dery MA, Michaud MD, Richard DE. Hypoxia-inducible factor 1: regulation by hypoxic and non-hypoxic activators. Int J Biochem Cell Biol. 2005;37(3):535–40. doi: 10.1016/j.biocel.2004.08.012. [DOI] [PubMed] [Google Scholar]
- 51.Kong D, et al. Echinomycin, a small-molecule inhibitor of hypoxia-inducible factor-1 DNA-binding activity. Cancer Res. 2005;65(19):9047–55. doi: 10.1158/0008-5472.CAN-05-1235. [DOI] [PubMed] [Google Scholar]
- 52.Vlaminck B, et al. Dual effect of echinomycin on hypoxia-inducible factor-1 activity under normoxic and hypoxic conditions. FEBS J. 2007;274(21):5533–42. doi: 10.1111/j.1742-4658.2007.06072.x. [DOI] [PubMed] [Google Scholar]
- 53.Rajasekaran SA, et al. Na-K-ATPase regulates tight junction permeability through occludin phosphorylation in pancreatic epithelial cells. Am J Physiol Gastrointest Liver Physiol. 2007;292(1):G124–33. doi: 10.1152/ajpgi.00297.2006. [DOI] [PubMed] [Google Scholar]
- 54.Reiner JE, Datta PK. TGF-beta-dependent and -independent roles of STRAP in cancer. Front Biosci. 16:105–15. doi: 10.2741/3678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Branton MH, Kopp JB. TGF-beta and fibrosis. Microbes Infect. 1999;1(15):1349–65. doi: 10.1016/s1286-4579(99)00250-6. [DOI] [PubMed] [Google Scholar]
- 56.Asaria RH, Gregor ZJ. Simple retinal detachments: identifying the at-risk case. Eye (Lond) 2002;16(4):404–10. doi: 10.1038/sj.eye.6700189. [DOI] [PubMed] [Google Scholar]
- 57.Nagasaki H, Shinagawa K, Mochizuki M. Risk factors for proliferative vitreoretinopathy. Prog Retin Eye Res. 1998;17(1):77–98. doi: 10.1016/s1350-9462(97)00007-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.