Abstract
Plants monitor changes in photoperiod and temperature to synchronize their flowering with seasonal changes to maximize fitness. In the Arabidopsis photoperiodic flowering pathway, the circadian clock-regulated components, such as FLAVIN-BINDING, KELCH REPEAT, F-BOX 1 and CONSTANS, both of which have light-controlled functions, are crucial to induce the day-length specific expression of the FLOWERING LOCUS T (FT) gene in leaves. Recent advances indicate that FT transcriptional regulation is central for integrating the information derived from other important internal and external factors, such as developmental age, amount of gibberellic acid, and the ambient temperature. In this review, we describe how these factors interactively regulate the expression of FT, the main component of florigen, in leaves.
Keywords: photoperiod, ambient temperature, gibberellic acid, flowering, FLOWERING LOCUS T, florigen
Photoperiodic flowering mechanism in Arabidopsis
Seasonal changes in day length (photoperiod) are consistent from year to year. Therefore, many plants use photoperiod information to predict upcoming environmental changes and precisely align the timing of flowering with favorable conditions [1]. Another important environmental factor that influences flowering is surrounding temperature. Ambient temperature changes arising from global climate change have already altered the phenology of plants, including the timing of flowering [2]. Therefore, understanding the mechanisms by which plants integrate both photoperiod and temperature cues to control seasonal flowering is necessary.
In Arabidopsis (Arabidopsis thaliana), long-day (LD) conditions accelerate flowering through the function of FLOWERING LOCUS T (FT) protein [3–5]. FT protein is a major component of florigen, a long-sought systemic floral inducing substrate [6]. Once it is synthesized in the leaf vasculature, it moves through the phloem to the shoot apical meristem [6]. Recent studies have reported that various factors, including photoperiod, temperature, plant age, and gibberellic acid (GA), converge to regulate FT expression for flowering (Figure 1). The amount of FT transcript, which is directly induced by the transcriptional activator CONSTANS (CO) protein, strongly influences the timing of flowering [3,5,7]. The circadian clock and light signaling tightly control CO protein activity throughout the day in the companion cells of the leaf phloem [5,8]. There are several recent reviews describing the function of FT proteins in various plants and the molecular events of floral induction initiated by FT at the shoot apical meristem [3,4,9]. Therefore, in this review, we focus on examining how photoperiod and ambient temperature, two influential environmental parameters, integrate to regulate the expression of FT in leaves in Arabidopsis. We first discuss how photoperiodic information is processed through the spatiotemporal regulation of CO transcription and its protein function, which controls FT expression under LD conditions
Figure 1.
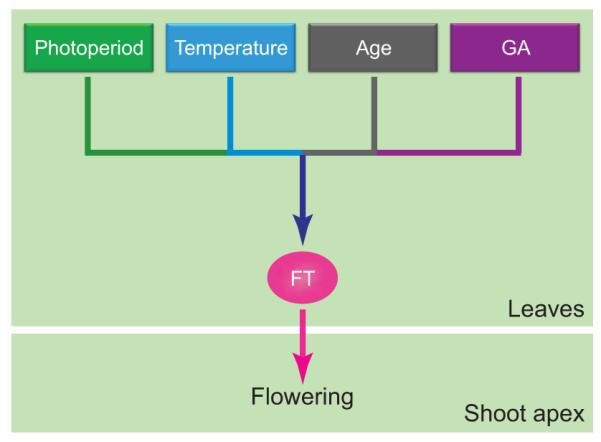
Integration of external and internal signals for flowering. External stimuli (photoperiod and temperature) and internal conditions (plant age and amount of GA) converge in the regulation of FT gene expression and they all affect FT protein output from the leaves. FT protein moves to the shoot apex and induces flowering.
CO transcriptional regulation
The circadian clock-regulated FLAVIN-BINDING, KELCH REPEAT, F-BOX 1 (FKF1), GIGANTEA (GI), and CYCLING DOF FACTOR (CDF) proteins play major roles in regulating daily CO expression profiles (Figure 2) [10–12]. CDF proteins (CDF1, CDF2, CDF3, and CDF5) are transcription factors that repress CO transcription during the morning [10–12]. FKF1 is a photoreceptor E3 ubiquitin ligase (Figure 3) [10,13]. GI is a large nuclear protein [14]. The expression patterns of FKF1 and GI proteins synchronize in the afternoon under LD conditions, but not under short day (SD) conditions. They form a complex in a blue light-dependent manner [11], and the complex degrades CDF proteins in the afternoon, facilitating the expression of CO at that time of day [11,12] (Figure 2). The distribution pattern of GI protein in the nucleus also affects CO expression. EARLY FLOWERING 4 (ELF4) protein directly binds to GI and targets GI to subnuclear compartments in the nucleus [15]. The distribution pattern of GI in the nucleus changes throughout the day. Especially during the night, more GI proteins are localized in the subnuclear compartments, and time-dependent change is regulated by ELF4. The ELF4-dependent targeting of GI to the compartments sequesters GI from the CO promoter in the nucleus, affecting CO expression level [15]. It is not known whether FKF1 is also found in the same subnuclear compartments.
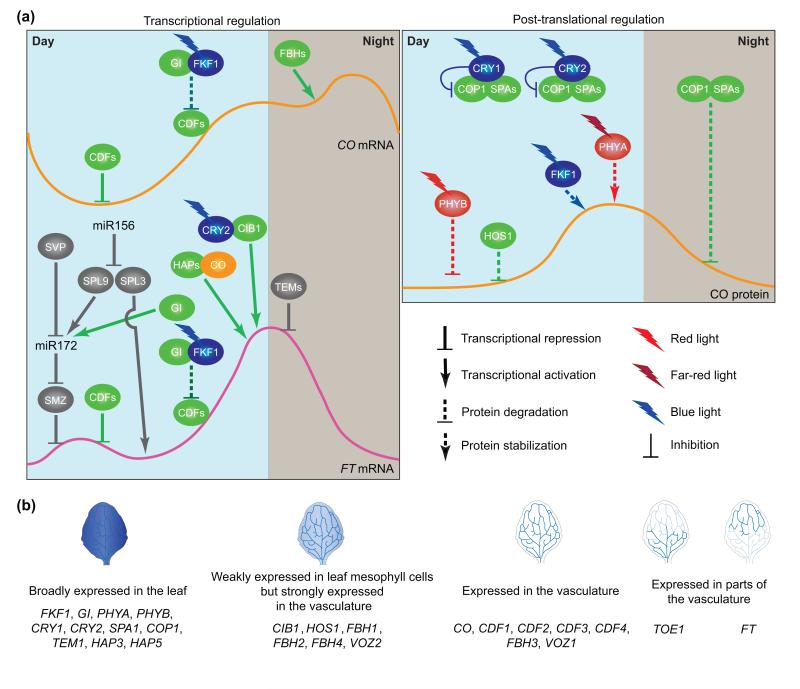
Figure 2.
Photoperiodic regulation of FT expression under LD conditions. (a) Transcriptional regulation of CO and FT genes (left panel) and post-translational regulation of CO protein (right panel). High levels of CDF proteins accumulate on CO and FT promoters in the morning, resulting in repression of CO and FT expression simultaneously. FKF1 and GI form a protein complex in the afternoon when FKF1 protein is expressed and absorbs blue light. The protein complex promotes degradation of CDF proteins on CO and FT promoters. Removal of CDF repression allows other DNA-binding proteins that act as activators to access these promoters. The FBHs (bHLH transcription factors) bind to the CO promoter and activate CO transcription throughout the day. CO protein is post-translationally regulated by the COP1–SPAs complex, photoreceptors, and HOS1 (right panel). The COP1–SPAs complex actively degrades CO protein in the dark. In addition, COP1 degrades PHYA and PHYB under far-red and red light conditions, respectively. Blue light-absorbed CRY1 and CRY2 interact with COP1 and SPAs and inhibit COP1–SPA activity, which increases CO stability. In the morning, HOS1 and red light-absorbed PHYB mediate degradation of CO. In the afternoon, blue light-absorbed FKF1 and far-red light-absorbed PHYA stabilize CO, which, in turn, activates FT transcription. In the regulation of FT expression (left panel), CO protein directly binds to the FT promoter and/or is recruited to the promoter by interactions with HAP and/or other DNA-binding proteins. CIB1 interacts with blue-light-activated CRY2 and directly binds to the FT promoter. CO and CIB1 activate FT transcription in the late afternoon. In addition to photoperiod, FT expression is regulated by plant developmental age. miR156 reduces the amount of SPL9 and SPL3 transcripts in younger plants. miR156 expression is decreased in older plants, resulting in up-regulation of SPL3 and SPL9 expression. SPL3 directly activates FT expression by binding to the FT promoter. SPL9 directly promotes MIR172 expression, which subsequently reduces the amount of AP2-related transcripts, including the SMZ transcript. miR172 expression is also regulated positively by GI and negatively by SVP. Expression of TEM genes encoding direct repressors of FT is also decreased in older plants. TEMs and SMZ repress FT expression throughout the day. FT expression is regulated by both photoperiodic and developmental pathways. The red/far-red photoreceptors are depicted in red, blue-light photoreceptors in blue, CO in orange, other photoperiodic pathway components in green, and developmental age-related components in gray. (b) Spatial expression patterns of the genes that play roles in the photoperiodic pathways under LD conditions. These data are based on promoter:GUS analyses. GI, COP1, SPA1, TEM1, HAP3, HAP5, and photoreceptors, including FKF1, PHYA, PHYB, CRY1, and CRY2, are broadly expressed in the leaf. CIB1 and FBH1, FBH2, FBH4, HOS1, and VOZ2 are strongly expressed in the vasculature of the leaf but weakly expressed in the mesophyll cells. Expression of CO, FBH3, VOS1, and the transcriptional repressors CDF1, CDF2, CDF3, and CDF4 is mainly observed in the leaf vascular tissues. FT is expressed in the distal part of the leaf vasculature whereas TOE1 is inversely expressed in the proximal part of the leaf vasculature.
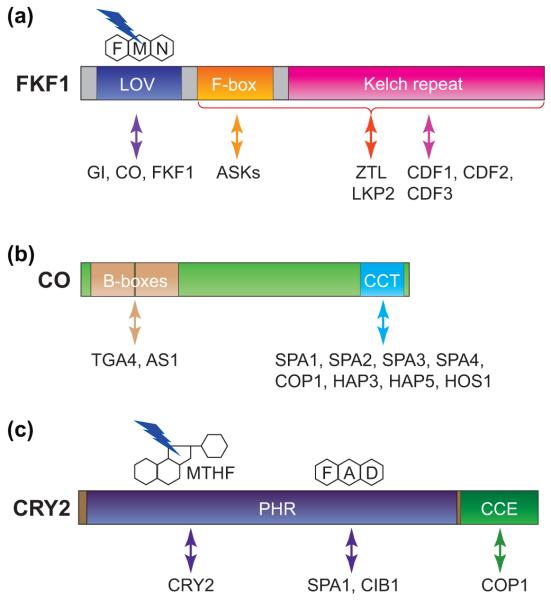
Figure 3.
Functional domains and their interactors of FKF1, CO, and CRY2 proteins. (a) FKF1 functions as a blue-light photoreceptor and possesses E3 ubiquitin ligase activity. The LOV absorbs blue light through the chromophore, flavin mononucleotide (FMN), and is responsible for light-induced protein–protein interaction with GI and CO. FKF1 homodimerizes through its LOV domain in vitro. FKF1 also binds to proteolytic targets, CDFs, through the Kelch repeat domain. FKF1 forms an SCF complex by binding to Arabidopsis SKP1-like (ASK) proteins through the F-box domain. Both F-box and Kelch repeat domains are important for interactions with ZTL and LKP2. (b) CO contains two conserved domains, a tandem repeat of two B-box zinc-finger domains and a CCT domain. CO forms protein complexes with TGA4 and AS1 through the B-box domain and with COP1, SPAs, HAPs, and HOS1 through the CCT domain. (c) CRY2 possesses a blue light-sensing domain, called the Photolyase Homology Region (PHR) that binds two chromophores, methenyltetrahydrofolate (MTHF) and flavin adenine dinucleotide (FAD). MTHF and FAD in the PHR domain are important for CRY2 homodimerization and heterodimerization with SPA1 and CIB1 proteins, respectively. The CRY C-terminal Extension (CCE) domain is responsible for COP1 interaction.
In addition to FKF1, a couple of the E3 ubiquitin ligases are involved in CO transcriptional regulation, although their substrates for ubiquitination have not yet been identified. A RING-finger-type E3 ubiquitin ligase, DAY NEUTRAL FLOWERING (DNF), functions as a negative regulator of flowering by repressing daytime CO expression under SD conditions [16]. EID1-LIKE PROTEIN 3 (EDL3) is an F-box protein, a putative E3 ubiquitin ligase, involved in abscisic acid signaling [17]. It is a potential positive regulator of CO transcription under stress conditions.
Reverse genetics approaches have revealed the mechanism for controlling the levels of CO messenger RNA (mRNA) (Figure 2). A small family of basic helix–loop–helix (bHLH) transcription factors, named FLOWERING BHLH (FBH1, FBH2, FBH3, and FBH4), activates CO transcription throughout the day [18]. FBH proteins bind to the E-box elements near the CO transcriptional start site, and overexpression of FBH genes drastically increases CO mRNA levels regardless of photoperiod. Interestingly, even though the peak CO expression levels in the FBH overexpressors were almost 20 times higher those in wild-type plants, the daily expression patterns of CO (i.e., lower CO expression in the morning and higher expression in the afternoon and night) in the FBH overexpressors remained similar [18]. Given that the FBH1 protein was constitutively expressed throughout the day in these transgenic plants, these findings suggest that FBH is regulated through a post-translational mechanism(s). FBH genes may also be functionally conserved in other plants. Overexpression of rice (Oryza sativa) or poplar (Populus trichocarpa) FBH homologs in Arabidopsis also resulted in greatly increased CO levels [18]. The analysis of the fbh quadruple mutants indicated the presence of other unknown regulator(s) for CO transcription [18]. To further understand how the CO expression pattern is regulated in the phloem companion cells, it is necessary to investigate the spatiotemporal relationships among transcriptional activators and repressors (FBHs, CDFs, and as yet unidentified factors), and modifiers (FKF1, GI, and others) (Figure 2).
CO protein and its post-translational regulation
CO protein possesses several protein–protein interaction domains: two tandem B-box domains at the N-terminus and the CCT (CO, CO-like, and TOC1) motif at the C-terminus [19] (Figure 3). The CCT motif, which contains a nuclear localization signal [19], is also a DNA-binding domain that interacts, in vitro, with the cis-element called CO responsive element (CORE) [20]. CO protein, in vivo, binds near the FT transcription start site, where the CORE sequences are located [21]. Given that the CO protein has a weak binding affinity to the CORE sequences in vitro [20], the CO protein may also bind to the FT promoter by forming complexes with other transcription factors. CO binds to two subunits of the HEME ACTIVATOR PROTEIN (HAP) complex [also known as CCAAT box factor (CBF) or NUCLEAR FACTOR-Y (NF-Y)], HAP3 (NF-YB, or CBF-A) and HAP5 (NF-YC, or CBF-C), through the CCT motif [22,23] (Figure 3). Overexpression of HAP3 and HAP5 promotes flowering, whereas their multiple mutations strongly delay flowering under LD conditions [24–27]. CO also interacts with TGA4 and ASYMMETRIC LEAVES 1 (AS1) through the B-box domains [28,29] (Figure 3). AS1 is highly expressed in the phloem, directly binds to the FT promoter, and is involved in CO-dependent FT induction [29]. Whether CO always interacts with these transcription factors for FT regulation is unknown.
Post-translational regulation of the CO protein is also important for restricting FT expression under LD conditions. CO protein stability is controlled by various light signals during the day [30,31]. At night, CO is actively degraded by CONSTITUTIVE PHOTOMORPHOGENESIS 1 (COP1), a RING-finger E3 ubiquitin ligase, and SUPRESSOR OF PHYA-105 (SPA1, SPA3 and SPA4) complex [32–34] (Figure 2). Two phytochromes antagonistically regulate CO protein stability: phytochrome B (PHYB) facilitates CO degradation in the morning, whereas PHYA mediates CO stabilization in the afternoon under LD conditions [30] (Figure 2). Photoactivated blue-light photoreceptor cryptochromes (CRY1 and CRY2) preferentially bind to SPA1 [31,35,36]. The light-dependent interactions between cryptochromes and SPA1 initiate the repression of COP1–SPA1 function (Figure 2), but CRY1 and CRY2 use different mechanisms for the repression [31,35,36]. CRY1 binds to the C-terminal domain of SPA1 through CRY1 C-terminal domain under blue light, and this interaction prevents the COP1–SPA1 complex formation [35,36]. The N-terminal domain of CRY2 interacts with the SPA1 N-terminus and this interaction facilitates the formation of the CRY2–COP1–SPA1 tripartite complex [31] (Figure 3). In the complex, CRY2 directly represses COP1–SPA1 activity, resulting in the stabilization of CO [31] (Figures 2 and 3). Another RING-finger E3 ubiquitin ligase, HIGH EXPRESSION OF OSMOTICALLY RESPONSIVE GENE 1 (HOS1), also regulates CO protein stability [37]. HOS1 mediates degradation of INDUCER OF C-REPEAT BINDING FACTOR (CBF) EXPRESSION1 (ICE1), a bHLH transcription factor that positively regulates CBF expression during cold stress [38]. Like COP1 and SPA1, HOS1 binds to the CO CCT motif; however, HOS1 degrades CO during the morning, instead of at night [37] (Figure 2). Although several CO regulators have been identified, the mechanism by which the CO protein was stabilized only in the afternoon under LD conditions remained elusive until recently. PHYA and CRY2 proteins are involved in CO stabilization [30]. However, both proteins are expressed constitutively throughout the day [39]. FKF1 protein expression occurs under light in the LD afternoon. FKF1 interacts with the CO protein through its LOV domain to stabilize CO. Blue light absorbed by the LOV domain enhances this interaction (Figures 2 and 3) [21]. Constitutive expression of FKF1 stabilizes CO during the entire part of day [21]. Therefore, both FKF1 expression and light induction of the FKF1–CO interaction determine the timing of CO stabilization. In the external coincidence model proposed by Colin Pittendriph [40], organisms induce photoperiodic responses when light is present in the photoinducible phase, which is regulated by the circadian clock. In Arabidopsis, the timing of FKF1 expression and light-dependent FKF1 function can be the main factor that determines the photoinducible phase for flowering time regulation.
Several interesting questions regarding the CO stabilization mechanism remain. (i) How does FKF1 increase CO stability by direct binding? (ii) What is the relationship between phyB and HOS1, both of which mediate CO degradation during the morning? (iii) FKF1 and its homologs, ZEITLUPE (ZTL) and LOV KELCH PROTEIN2 (LKP2), interact with GI and share their target proteins, including CDFs [11,12,41,42]. However, unlike FKF1, ZTL and LKP2 overexpressors show late flowering phenotypes under LD conditions, probably as a result of capturing FKF1 in the cytosol by direct interaction [43]. How, then, do FKF1, ZTL, and LKP2 proteins synergistically increase destabilization of CDFs? (iv) What are the roles of ZTL and LKP2 in photoperiodic flowering regulation? Answering these questions should help us to further understand the mechanisms of CO post-transcriptional regulation.
FT transcriptional regulation
Light signaling pathways and the circadian clock coordinately control CO protein activity to induce FT under favorable conditions (Figure 2). Because FT is a floral integrator, various factors also regulate FT expression. Several transcriptional repressors, such as FLOWERING LOCUS C (FLC) [44], SHORT VEGETATIVE PHASE (SVP) [45,46], TEMPRANILLO 1 (TEM1) [47], and SCHLAFMÜTZE (SMZ) [48], bind to specific cis-elements in the FT locus. These repressors, as well as their related transcription factors, prevent precocious flowering by repressing FT either under unfavorable conditions for flowering or during the juvenile developmental phases. FLC and SVP are MADS-box transcription factors that form a heterodimeric complex [44] (Figure 4). The amount of the SVP–FLC complex formation is larger in younger leaves (i.e. 3 to 7 days old) than in older leaves (11 days old) [49]. Both FLC and SVP are involved in FT repression under a wide range of cold conditions [46]. The expression levels of TEM1, SMZ, SCHNARCHZAPFEN (SNZ), TARGET OF EAT1 (TOE1), TOE2, and TOE3 are all regulated by developmental stages. The TEM1 expression level decreases after the 8-day-old seedling stage [47]. MicroRNA172 (miR172) decreases the abundance of the miR172 target transcripts, including APETALA2 (AP2)-related transcription factor transcripts (SMZ, SNZ, TOE1, TOE2, and TOE3) [48,50]. The amount of miR172 increases as the plants develop [50]. The miR172 level is also higher under LD conditions than under SD conditions, and GI is involved in this photoperiodic miR172 induction [50]. In addition, SVP protein also directly binds to the CArG motifs in the MIR172a promoter [51], and the level of miR172 in svp mutants is about five times higher than that in wild-type plants [52], indicating that SVP reduces miR172 expression under LD conditions. Furthermore, the miR172 level is negatively regulated by another microRNA, miR156 [53]. The miR172 expression levels are inversely correlated with the miR156 levels during development because miR156 levels are high in early developing seedlings and are reduced as plants grow [53–55]. The miR156 targets are SQUAMOSA PROMOTER BINDING PROTEIN-LIKE (SPL) transcripts [56]. The miR156 target genes, SPL9 and likely SPL10, directly activate the transcription of MIR172 [53]. In addition, SPL3, which has its transcript cleaved by a miR156-dependent mechanism, directly binds to the FT promoter to induce FT expression (see details in a later section) [57] (Figure 2). These regulators are likely to influence the overall expression level of FT over the developmental stages.
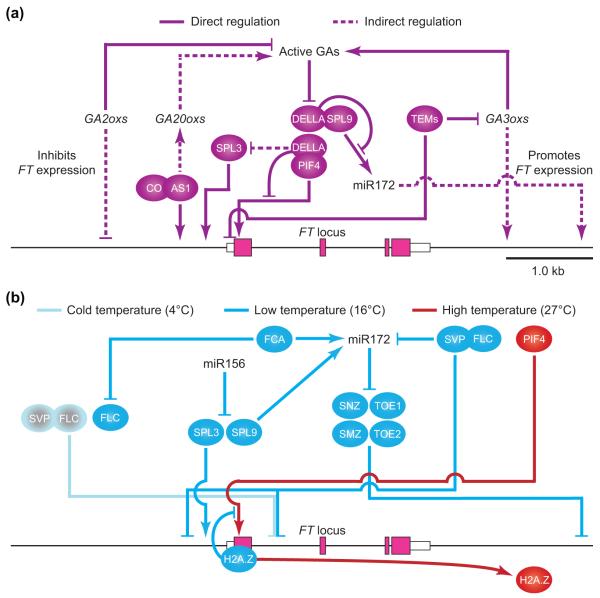
Figure 4.
GA and ambient temperature-dependent FT regulation for flowering. (a) Regulation of FT expression by GA signaling. AS1 positively regulates expression of GA biosynthesis genes, GA20oxs, which encode oxidase enzymes that oxidize the precursors of bioactive GAs. AS1 also directly binds to the FT promoter and may recruit CO to the promoter by a physical protein interaction. Active GAs are synthesized by GA3ox from the GA products catalyzed by GA20ox. TEM1 proteins directly repress GA3ox expression by association with the GA3ox1 and GA3ox2 loci, and reduce FT expression. In addition, TEM1 directly binds to 5′-UTR of the FT gene and represses FT expression. GA promotes degradation of DELLA proteins that inhibit PIF4 and SPL9 activities by directly binding to them, which negatively regulates SPL3 expression indirectly. This allows PIF4 to activate FT expression and SPL9 to indirectly induce FT expression through up-regulation of MIR172 expression. GA2ox genes encode oxidases that deactivate active GAs, resulting in inhibition of FT expression. The bars from DELLA indicate inhibition of transcriptional activities of PIF4 and SPL9. Arrows and bars represent positive and negative regulation, respectively. White and pink boxes represent untranslated regions (UTRs) and exons of the FT gene, respectively. (b) Regulation of FT expression by temperature responsive regulators. Expression of FLC, miR156, and SPL3 is attenuated by a rise in temperature, whereas expression of FCA, miR172, PIF4, and FT is increased. At cold temperatures (4°C), FLC binds to the first intron of the FT gene and represses FT expression. SVP interacts with FLC and binds to the FT locus through the same cis-elements that bind FLC. SVP also negatively regulates MIR172 transcription by directly binding to the MIR172 promoter at 16°C. FCA negatively regulates FLC accumulation and positively regulates miR172 accumulation. miR172 targets and post-transcriptionally represses AP2-like genes SNZ, TOE1, TOE2, and SMZ, which encode FT repressors. Among them, only SMZ is known to associate with the 3′-region of the FT gene. miR156 reduces SPL3 and SPL9 transcripts. SPL3 protein binds to the FT promoter and induces FT expression. SPL9 expression increases when miR156 expression is reduced in the later stages of plant development. The SPL9 protein directly activates MIR172 expression. The occupancy of H2A.Z-nucleosomes on the FT promoter reduces the accessibility of the PIF4-binding site at the promoter. However, the occupancy of H2A.Z is decreased at high temperatures where PIF4 expression is elevated. Therefore, more PIF4 can access G-box elements, resulting in increased FT expression. Arrows and bars indicate transcriptional (including post-transcriptional) activation and repression, respectively.
Others regulate the daily expression profiles of FT. The circadian-regulated CO repressor, CDF1, also represses FT by binding to the FT promoter near the transcription start site. Other CDFs (CDF2, CDF3, and CDF5) are also likely to be involved in FT repression during the morning [21] (Figure 2). FKF1 and GI also associate with the FT promoter, and the presence of CDF1 on the FT promoter in the afternoon is FKF1-dependent, indicating that CDF1 is removed by the FKF1–GI complex on the FT promoter [21]. In addition, GI protein binds to the FT repressors, TEM1, TEM2, and SVP in tobacco (Nicotiana benthamiana) [58]. Presumably, this regulation may change the activities of these FT repressors during specific parts of the day. The activity of another FT activator, cryptochrome-interacting basic-helix–loop–helix 1 (CIB1), is also restricted at a specific time of day. CIB1, which binds to blue-light-absorbing CRY2, directly associates with regions in the FT locus and induces FT expression [59] (Figure 2). The effect of constitutive CIB1 overexpression on FT transcription is restricted from the afternoon to early night, when FT peaks.
PHYB signaling components may also regulate FT expression. Two classes of PHYB-binding transcription factors, PHYTOCHROME INTERACTING FACTOR4 (PIF4) and VASCULAR PLANT ONE-ZINC FINGER1 (VOZ1) and VOZ2, induce FT expression [60,61]. The vos1 vos2 double mutations completely suppress the early flowering phenotype of the phyB mutant under both LD and SD conditions [61]. In the vos1 vos2 mutant, FT level was severely repressed throughout the day without changing the CO expression profile; however, the FLC level was also largely increased [61]. As FLC is a direct repressor of FT [44], it remains elusive whether VOS1 and VOS2 directly activate FT transcription. The component of the mediator complex, PHYTOCHROME AND FLOWERING TIME1 (PFT1), promotes flowering through positively regulating FT expression in CO-dependent and CO-independent pathways [62].
Here we described how the components of the photoperiodic pathway interact to regulate the diurnal patterns of FT, and how the developmental stage-dependent regulation, in which microRNAs (miRNAs) play important roles, modulates the output of photoperiodic flowering by changing the expression levels of FT. In addition to the FT transcriptional regulators discussed here, chromatin modifications on the FT locus also play important roles in FT transcription (see the recent review, [63]). In the next section, we will introduce the interactions that occur between the photoperiodic pathway and the phytohormone gibberellic acid (GA) pathway, both of which regulate flowering through the regulation of FT expression in leaves.
Interaction between photoperiodic and gibberellic acid pathways
GA affects diverse biological processes, including flowering time. Recent studies have reported the interactions occur between photoperiodic and GA pathways to regulate FT expression under both LD and SD conditions [29,64–66] (Figure 4). The bioactive GA4 is synthesized through multiple oxidation steps catalyzed by GA 20-oxidase (GA20ox) and GA 3-oxidase (GA3ox) [64]. The amount of active GA4 is tightly regulated through synthesis as well as through deactivation catalyzed by GA 2-oxidase (GA2ox) [64]. In leaves, the MYB-type transcription factor ASYMMETRIC LEAVES 1 (AS1), which is an important factor for leaf pattern formation [65], positively regulates expression of the GA biosynthesis gene GA20ox1 [29]. The as1 alleles and ga20ox1 mutant show delayed flowering phenotypes regardless of photoperiod. AS1 concomitantly forms a complex with CO protein and regulates FT expression by directly binding to the FT promoter [29] (Figure 4). Therefore, AS1 has dual roles to accelerate flowering by increasing the amount of GA4 and facilitating CO to induce FT. [29]. Another example of flowering time regulators that have roles in both photoperiodic and GA pathways is the FT repressors TEM1 and TEM2, which repress the expression of the GA4 biosynthesis genes GA20ox1, GA3ox1 and GA3ox2 under SD conditions [66]. TEM1 protein directly binds to the promoters of both GA3ox genes. A strain over-expressing TEM1 shows lower levels of expression of GA20ox and GA3ox genes and late flowering under SD conditions. Exogenous application of GA3 to this strain induces earlier flowering under SD conditions. Even under LD conditions, TEM-dependent repression of GA3ox expression contributes to flowering time determination, as the ga3ox1 mutation delays the early flowering phenotype of the tem1 tem2 mutant [66]. Moreover, recent studies indicate that changes in bioactive GA levels in leaf phloem may contribute to FT expression. When the GA2ox7 gene is overexpressed in the leaf phloem companion cells using the SUC2:GA2ox7 construct under LD conditions, the expression levels of photoperiodic-induced FT and TWIN SISTER OF FT (TSF) genes are reduced and the SUC2:GA2ox7 plants show delayed flowering, indicating that GA is involved in FT induction under LD conditions [67]. These results suggest that AS1 and TEM1 may control FT expression partly through changing the amount of bioactive GA in the leaf phloem.
How does GA regulate FT transcription in the leaf? Recent studies have furthered our understanding of the roles played by DELLA proteins (negative regulators in GA signaling) and may partially answer this question. DELLA proteins expressed in companion cells of leaf phloem delay flowering with the reduction of expression of FT and TSF under LD conditions [68,69]. The degradation of DELLA proteins is induced when bioactive GA is perceived by the GA receptors, GIBBERELLIC ACID-INSENSITIVE DWARF 1 (GID1a, GID1b, and GID1c) [70]. The gid1a gid1b gid1c triple mutant shows low levels of expression of FT and TSF without changing CO and GI expression, and never flowers under LD conditions [68]. DELLA represses the expression of FT activator, SPL3, in leaves and SPL3, SPL4, and SPL5 at the shoot apex [68]. DELLA also delays flowering partly by reducing miR172 levels in leaves under LD conditions [68,69]. One of the DELLA proteins, REPRESSOR OF GA1-3 (RGA), binds to the C-terminus of the SPL9 protein and this interaction is likely to attenuate SPL9 transcriptional activity [69]. In addition to regulating SPL activities, DELLA proteins may regulate FT through PIF4 because DELLAs regulate PIF4 binding activity [71] and PIF4 activates FT expression under high temperatures [60]. Through these DELLA-dependent mechanisms, bioactive GA levels directly affect the expression levels of FT in leaves under LD conditions.
Effects of temperature changes on flowering regulation
In addition to day-length changes, leaves sense information about temperature fluctuation. Studies on the effects of temperature changes on flowering time have mostly focused on vernalization responses [72]. The key regulator of the vernalization response in Arabidopsis is the FLC gene, which encodes a transcription repressor of FT. Vernalization represses the expression of FLC by regulating the chromatin status of the FLC locus; therefore, FLC repression is removed in the spring. In contrast to vernalization mechanisms, the molecular mechanisms by which ambient temperature governs the timing of floral transition (i.e. the thermosensory flowering pathway) have just begun to be elucidated (see details below). The thermo-regulation of flowering also converges on the regulation of FT gene expression in leaves recruiting the components and mechanisms used for other flowering regulations.
Responses to lower temperature
Arabidopsis plants flower later when grown under LD conditions kept at 16°C than when grown at 23°C, and this difference is chiefly caused by differences in FT expression [73]. The temperature-dependent difference in flowering time is regulated by multiple factors. For example, the svp mutant flowers at the same times at both 16°C and 23°C [46]. Given that SVP mRNA levels did not change at these two temperatures [46], temperature changes may regulate SVP function. HOS1 activity significantly reduces FT gene expression through degradation of CO protein when under conditions of 4°C intermittent cold stress [74]. Genetically, phyB may control HOS1 activity under these conditions [74]. HOS1 also negatively regulates the FT expression level at 16°C, partly independent of CO activity but together with FVE and FLK [75]. HOS1 forms a protein complex with FVE; however, to date the role of the HOS1–FVE complex is unknown [75]. FVE is an Arabidopsis homolog of the retinoblastoma-associated protein, a component of a histone deacetylase complex involved in transcriptional repression, and down-regulates FLC expression [76]. The activity of FVE protein decreases under cold stress without changing its mRNA levels, resulting in elevated FLC expression; however, under lower ambient temperature conditions (16°C), FVE regulates flowering, most likely through the FLC-independent pathway [73,76]. TERMINAL FLOWER 1 (TFL1) and EARLY FLOWERING 3 (ELF3) are also involved in ambient temperature-dependent flowering regulation given that the flowering time of the tfl1 elf3 double mutant is insensitive to temperature changes [77].
In addition to these factors, several miRNAs are also involved in the temperature-dependent regulation of flowering time [52,57,78] (Figure 4). miR156 accumulates at a level several times higher at 16°C than at 23°C, whereas the level of miR172 at 16°C is about half of that at 23°C [78]. Both miRNAs concomitantly change the expression levels of FT regulators, which affect flowering time, in response to temperature changes [52,57]. The effect of miR156 overexpression on delayed flowering time is more pronounced at 16°C than at 23°C, with a corresponding reduction of FT expression in leaves [57]. Notable down-regulation of the SPL3 gene (a miR156 target) in the miR156 overexpressor is also observed specifically in leaves at 16°C. The lower levels of SPL3 mRNA are due to enhanced cleavage of the SLP3 mRNA by miR156. The SPL3 protein binds near the transcription start site of the FT promoter, where the SLP3 binding sites (GTAC motifs) are located, and the induction of miR156-resistant SPL3 transcript expression subsequently increases FT and FRUITFULL (FUL, another known target of SPL3) expression [57]. These findings support the notion that the miR156–SPL3–FT module in leaves plays an important role in flowering regulation, not only developmentally [56] but also in response to ambient temperature changes [57] (Figure 4).
Under 16°C conditions, miR172 expression is reduced and, consequently, the expression of its target genes, TOE1, TOE2 and SMZ, is increased [78]. Recent reports have revealed the mechanisms by which lower temperature reduces miR172 levels [51,52]. Post-transcriptional processing of primary miR172 (pri-miR172: miR172 transcript that has a 5′ cap and poly-adenosine tail) transcripts to mature miR172 plays a major role [52]. Even though the accumulation of miR172 is higher at 23°C, the levels of pri-miR172 and precursor-miR172 (pre-miR172: approximately 70 bp miR172 precursor that is cut out from the pri-miR172) transcripts are not drastically altered by changing temperatures (16°C and 23°C). In the pri-miR172b overexpressors, mature miR172 levels still show temperature-dependent differences, but in the pre-miR172 overexpressor, miR172 levels are similar between 16°C and 23°C. These findings indicate that the pri-miR172-to-pre-miR172 processing step is modulated by the ambient temperature. In this step, FCA, a RNA-binding protein that has a central role in the ambient temperature and autonomous pathways [73], directly binds to pri-miR172 transcripts in a non-sequence-specific manner and positively regulates the miR172 processing [52]. At 16°C FCA transcripts and FCA proteins are less abundant than at 23°C [52]. Ambient temperature may also regulate the transcription of the MIR172a gene. The miR172 levels are negatively regulated by SVP [51,52], and SVP directly binds to the CArG motifs in the MIR172a promoter [51]. Another miRNA may also be involved in flowering time regulation: miR399 is reduced at 16°C and is also regulated by FCA [52]. The miR399-target gene, PHOSPHATE2 (PHO2), which functions in the maintenance of phosphate homeostasis, modulates flowering by controlling TSF expression [79].
Responses to higher temperature
Unlike lower temperature effects, higher temperatures (27°C) promote flowering with increased FT expression [60,80]. Recent studies have indicated that PIF4 is the main regulator for higher temperature-induced morphological changes, including floral transition [81] (Figure 4). Under SD conditions the flowering time of the pif4 mutant at 27°C occurs at almost the same time as it does at 23°C, whereas the flowering of wild-type plants at 27°C is accelerated with elevated FT expression. In addition, PIF4 protein directly activates FT expression by binding to the FT promoter at 27°C under SD conditions, and PIF4 expression increases as temperature increases [60]. However, variations in PIF4 expression under different temperature conditions are not sufficient to explain the flowering phenomenon at high temperatures. The histone H2A variant H2A.Z mediates temperature signals in Arabidopsis and plays a crucial role in temperature-dependent FT expression by PIF4 through modulating the accessibility of the PIF4-binding site at the FT promoter [60,82]. Indeed, the occupancy of H2A.Z-nucleosomes on the FT promoter is decreased at high temperatures whereas the binding of PIF4 to the FT promoter is increased, indicating that the presence of H2A.Z-nucleosomes are limiting for binding of PIF4 to FT [60] (Figure 4). At 27°C, the level of miR172 is also higher than at 23°C [51], and this change may decrease the amount of transcripts of AP2-related FT repressors under these conditions. As described in this section, the same regulatory modules are used for processing different external (photoperiods and temperatures) and internal (development and hormone) information to optimize the timing of flowering.
Concluding remarks
Recently there have been large advances in our understanding of flowering time regulation, which have clarified how several exogenous and endogenous factors regulate flowering time at the molecular level, and how these signaling pathways are integrated to control the expression of a major floral regulator, FT, in leaves (Figure 1). Although not covered in this review, a complex picture has also emerged in recent years of the dynamic interactions among floral integrators, including FT, and floral homeotic genes at the shoot apex [4]. There is a missing piece to this picture. FT protein moves through the phloem from the leaves to the shoot apex [6]. However, we know of only one factor, FT-INTERACTING PROTEIN 1 (FTIP1), involved in FT protein transport, and the function of FTIP1 is not well understood [83].
Temperature regulation of flowering is another underdeveloped topic. Despite our rapidly accumulating knowledge about lower temperature-induced flowering mechanisms in Arabidopsis, the regulatory mechanisms of higher ambient temperature (i.e. 27°C)-mediated FT regulation are still not well understood. As with Arabidopsis, many crop plants respond to changes in both photoperiod and temperature. The expression of FT homologs in these crops is also regulated by photoperiodic changes impacting their flowering [3,84]. Recent studies in rice have shown that photoperiod and high temperature act synergistically on flowering time through the regulation of rice FT genes, Hd3a and RFT1 [85,86]. Further molecular and biochemical analyses are likely to focus on the interactive effects among complicated environmental conditions on the determination of flowering time. Molecular data obtained under conditions that are more similar to those observed in natural settings should give us a new insight into flowering time regulation [87]. Understanding the molecular networks by which plants incorporate photoperiod and temperature changes to generate floral signals is essential to take advantage of and offset the effects of global climate change and secure future crop production.
Highlights.
Photoperiod and developmental stages converge to regulate the expression of FT, a major component of florigen, in leaves.
The photoperiodic photoreceptor, FKF1, for time-dependent CO stabilization was revealed recently.
Lower temperature-dependent flowering regulation has been characterized recently and the temperature also regulates FT in the leaves.
The phytohormone, GA, also participates in flowering time regulation in long days by regulating FT.
Multiple external and internal factors are integrated into FT transcriptional regulation in leaves.
Acknowledgements
We thank Hannah Kinmonth-Shultz, Greg Golembeski, and Lesley Pettigrew for critical reading. This work was supported by funding from the Next-Generation BioGreen 21 Program (SSAC, PJ008109) to Y.H.S., JSPS KAKENHI Grant-in-Aid for Young Scientists (B) (25840104) to S.I., and the National Institutes of Health (GM079712) and the University of Washington Royalty Research Fund to T.I.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Thomas B, Vince-Prue D. Photoperiodism in Plants. Academic Press; 1996. [Google Scholar]
- 2.Fitter AH, Fitter RS. Rapid changes in flowering time in British plants. Science. 2002;296:1689–1691. doi: 10.1126/science.1071617. [DOI] [PubMed] [Google Scholar]
- 3.Pin PA, Nilsson O. The multifaceted roles of FLOWERING LOCUS T in plant development. Plant Cell Environ. 2012;35:1742–1755. doi: 10.1111/j.1365-3040.2012.02558.x. [DOI] [PubMed] [Google Scholar]
- 4.Pose D, et al. The end of innocence: flowering networks explode in complexity. Curr Opin Plant Biol. 2012;15:45–50. doi: 10.1016/j.pbi.2011.09.002. [DOI] [PubMed] [Google Scholar]
- 5.Andrés F, Coupland G. The genetic basis of flowering responses to seasonal cues. Nat Rev Genet. 2012;13:627–639. doi: 10.1038/nrg3291. [DOI] [PubMed] [Google Scholar]
- 6.Corbesier L, et al. FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science. 2007;316:1030–1033. doi: 10.1126/science.1141752. [DOI] [PubMed] [Google Scholar]
- 7.Srikanth A, Schmid M. Regulation of flowering time: all roads lead to Rome. Cell Mol Life Sci. 2011;68:2013–2037. doi: 10.1007/s00018-011-0673-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Imaizumi T. Arabidopsis circadian clock and photoperiodism: time to think about location. Curr Opin Plant Biol. 2010;13:83–89. doi: 10.1016/j.pbi.2009.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Andres F, Coupland G. The genetic basis of flowering responses to seasonal cues. Nat Rev Genet. 2012;13:627–639. doi: 10.1038/nrg3291. [DOI] [PubMed] [Google Scholar]
- 10.Imaizumi T, et al. FKF1 F-box protein mediates cyclic degradation of a repressor of CONSTANS in Arabidopsis. Science. 2005;309:293–297. doi: 10.1126/science.1110586. [DOI] [PubMed] [Google Scholar]
- 11.Sawa M, et al. FKF1 and GIGANTEA complex formation is required for day-length measurement in Arabidopsis. Science. 2007;318:261–265. doi: 10.1126/science.1146994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fornara F, et al. Arabidopsis DOF transcription factors act redundantly to reduce CONSTANS expression and are essential for a photoperiodic flowering response. Dev Cell. 2009;17:75–86. doi: 10.1016/j.devcel.2009.06.015. [DOI] [PubMed] [Google Scholar]
- 13.Imaizumi T, et al. FKF1 is essential for photoperiodic-specific light signalling in Arabidopsis. Nature. 2003;426:302–306. doi: 10.1038/nature02090. [DOI] [PubMed] [Google Scholar]
- 14.Mizoguchi T, et al. Distinct roles of GIGANTEA in promoting flowering and regulating circadian rhythms in Arabidopsis. Plant Cell. 2005;17:2255–2270. doi: 10.1105/tpc.105.033464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim Y, et al. ELF4 regulates GIGANTEA chromatin access through subnuclear sequestration. Cell Rep. 2013;3:671–677. doi: 10.1016/j.celrep.2013.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Morris K, et al. DAY NEUTRAL FLOWERING represses CONSTANS to prevent Arabidopsis flowering early in short days. Plant Cell. 2010;22:1118–1128. doi: 10.1105/tpc.109.066605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Koops P, et al. EDL3 is an F-box protein involved in the regulation of abscisic acid signalling in Arabidopsis thaliana. J Exp Bot. 2011;62:5547–5560. doi: 10.1093/jxb/err236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ito S, et al. FLOWERING BHLH transcriptional activators control expression of the photoperiodic flowering regulator CONSTANS in Arabidopsis. Proc Natl Acad Sci U S A. 2012;109:3582–3587. doi: 10.1073/pnas.1118876109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Robson F, et al. Functional importance of conserved domains in the flowering-time gene CONSTANS demonstrated by analysis of mutant alleles and transgenic plants. Plant J. 2001;28:619–631. doi: 10.1046/j.1365-313x.2001.01163.x. [DOI] [PubMed] [Google Scholar]
- 20.Tiwari SB, et al. The flowering time regulator CONSTANS is recruited to the FLOWERING LOCUS T promoter via a unique cis-element. New Phytol. 2010;187:57–66. doi: 10.1111/j.1469-8137.2010.03251.x. [DOI] [PubMed] [Google Scholar]
- 21.Song YH, et al. FKF1 conveys timing information for CONSTANS stabilization in photoperiodic flowering. Science. 2012;336:1045–1049. doi: 10.1126/science.1219644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wenkel S, et al. CONSTANS and the CCAAT box binding complex share a functionally important domain and interact to regulate flowering of Arabidopsis. Plant Cell. 2006;18:2971–2984. doi: 10.1105/tpc.106.043299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ben-Naim O, et al. The CCAAT binding factor can mediate interactions between CONSTANS-like proteins and DNA. Plant J. 2006;46:462–476. doi: 10.1111/j.1365-313X.2006.02706.x. [DOI] [PubMed] [Google Scholar]
- 24.Cai X, et al. A putative CCAAT-binding transcription factor is a regulator of flowering timing in Arabidopsis. Plant Physiol. 2007;145:98–105. doi: 10.1104/pp.107.102079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen NZ, et al. AtHAP3b plays a crucial role in the regulation of flowering time in Arabidopsis during osmotic stress. J Biochem Mol Biol. 2007;40:1083–1089. doi: 10.5483/bmbrep.2007.40.6.1083. [DOI] [PubMed] [Google Scholar]
- 26.Kumimoto RW, et al. The Nuclear Factor Y subunits NF-YB2 and NF-YB3 play additive roles in the promotion of flowering by inductive long-day photoperiods in Arabidopsis. Planta. 2008;228:709–723. doi: 10.1007/s00425-008-0773-6. [DOI] [PubMed] [Google Scholar]
- 27.Kumimoto RW, et al. NF-YC3, NF-YC4 and NF-YC9 are required for CONSTANS-mediated, photoperiod-dependent flowering in Arabidopsis thaliana. Plant J. 2010;63:379–391. doi: 10.1111/j.1365-313X.2010.04247.x. [DOI] [PubMed] [Google Scholar]
- 28.Song YH, et al. Isolation of CONSTANS as a TGA4/OBF4 interacting protein. Mol Cells. 2008;25:559–565. [PubMed] [Google Scholar]
- 29.Song YH, et al. CONSTANS and ASYMMETRIC LEAVES 1 complex is involved in the induction of FLOWERING LOCUS T in photoperiodic flowering in Arabidopsis. Plant J. 2012;69:332–342. doi: 10.1111/j.1365-313X.2011.04793.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Valverde F, et al. Photoreceptor regulation of CONSTANS protein in photoperiodic flowering. Science. 2004;303:1003–1006. doi: 10.1126/science.1091761. [DOI] [PubMed] [Google Scholar]
- 31.Zuo Z, et al. Blue light-dependent interaction of CRY2 with SPA1 regulates COP1 activity and floral initiation in Arabidopsis. Curr Biol. 2011;21:841–847. doi: 10.1016/j.cub.2011.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Laubinger S, et al. Arabidopsis SPA proteins regulate photoperiodic flowering and interact with the floral inducer CONSTANS to regulate its stability. Development. 2006;133:3213–3222. doi: 10.1242/dev.02481. [DOI] [PubMed] [Google Scholar]
- 33.Jang S, et al. Arabidopsis COP1 shapes the temporal pattern of CO accumulation conferring a photoperiodic flowering response. EMBO J. 2008;27:1277–1288. doi: 10.1038/emboj.2008.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu LJ, et al. COP1-mediated ubiquitination of CONSTANS is implicated in cryptochrome regulation of flowering in Arabidopsis. Plant Cell. 2008;20:292–306. doi: 10.1105/tpc.107.057281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lian HL, et al. Blue-light-dependent interaction of cryptochrome 1 with SPA1 defines a dynamic signaling mechanism. Genes Dev. 2011;25:1023–1028. doi: 10.1101/gad.2025111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu B, et al. Arabidopsis cryptochrome 1 interacts with SPA1 to suppress COP1 activity in response to blue light. Genes Dev. 2011;25:1029–1034. doi: 10.1101/gad.2025011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lazaro A, et al. The Arabidopsis E3 ubiquitin ligase HOS1 negatively regulates CONSTANS abundance in the photoperiodic control of flowering. Plant Cell. 2012;24:982–999. doi: 10.1105/tpc.110.081885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chinnusamy V, et al. Cold stress regulation of gene expression in plants. Trends Plant Sci. 2007;12:444–451. doi: 10.1016/j.tplants.2007.07.002. [DOI] [PubMed] [Google Scholar]
- 39.Mockler T, et al. Regulation of photoperiodic flowering by Arabidopsis photoreceptors. Proc Natl Acad Sci U S A. 2003;100:2140–2145. doi: 10.1073/pnas.0437826100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pittendrigh CS, Minis DH. The entrainment of circadian oscillations by light and their role as photoperiodic clocks. Am Nat. 1964:261–294. [Google Scholar]
- 41.Kim WY, et al. ZEITLUPE is a circadian photoreceptor stabilized by GIGANTEA in blue light. Nature. 2007;449:356–360. doi: 10.1038/nature06132. [DOI] [PubMed] [Google Scholar]
- 42.Baudry A, et al. F-box proteins FKF1 and LKP2 act in concert with ZEITLUPE to control Arabidopsis clock progression. Plant Cell. 2010;22:606–622. doi: 10.1105/tpc.109.072843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Takase T, et al. LOV KELCH PROTEIN2 and ZEITLUPE repress Arabidopsis photoperiodic flowering under non-inductive conditions, dependent on FLAVIN-BINDING KELCH REPEAT F-BOX1. Plant J. 2011;67:608–621. doi: 10.1111/j.1365-313X.2011.04618.x. [DOI] [PubMed] [Google Scholar]
- 44.Searle I, et al. The transcription factor FLC confers a flowering response to vernalization by repressing meristem competence and systemic signaling in Arabidopsis. Genes Dev. 2006;20:898–912. doi: 10.1101/gad.373506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Helliwell CA, et al. The Arabidopsis FLC protein interacts directly in vivo with SOC1 and FT chromatin and is part of a high-molecular-weight protein complex. Plant J. 2006;46:183–192. doi: 10.1111/j.1365-313X.2006.02686.x. [DOI] [PubMed] [Google Scholar]
- 46.Lee JH, et al. Role of SVP in the control of flowering time by ambient temperature in Arabidopsis. Genes Dev. 2007;21:397–402. doi: 10.1101/gad.1518407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Castillejo C, Pelaz S. The balance between CONSTANS and TEMPRANILLO activities determines FT expression to trigger flowering. Curr Biol. 2008;18:1338–1343. doi: 10.1016/j.cub.2008.07.075. [DOI] [PubMed] [Google Scholar]
- 48.Mathieu J, et al. Repression of flowering by the miR172 target SMZ. PLoS Biol. 2009;7:e1000148. doi: 10.1371/journal.pbio.1000148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li D, et al. A repressor complex governs the integration of flowering signals in Arabidopsis. Dev Cell. 2008;15:110–120. doi: 10.1016/j.devcel.2008.05.002. [DOI] [PubMed] [Google Scholar]
- 50.Jung JH, et al. The GIGANTEA-regulated microRNA172 mediates photoperiodic flowering independent of CONSTANS in Arabidopsis. Plant Cell. 2007;19:2736–2748. doi: 10.1105/tpc.107.054528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cho HJ, et al. SHORT VEGETATIVE PHASE (SVP) protein negatively regulates miR172 transcription via direct binding to the pri-miR172a promoter in Arabidopsis. FEBS Lett. 2012;586:2332–2337. doi: 10.1016/j.febslet.2012.05.035. [DOI] [PubMed] [Google Scholar]
- 52.Jung JH, et al. Arabidopsis RNA-binding protein FCA regulates microRNA172 processing in thermosensory flowering. J Biol Chem. 2012;287:16007–16016. doi: 10.1074/jbc.M111.337485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wu G, et al. The sequential action of miR156 and miR172 regulates developmental timing in Arabidopsis. Cell. 2009;138:750–759. doi: 10.1016/j.cell.2009.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang JW, et al. miR156-regulated SPL transcription factors define an endogenous flowering pathway in Arabidopsis thaliana. Cell. 2009;138:738–749. doi: 10.1016/j.cell.2009.06.014. [DOI] [PubMed] [Google Scholar]
- 55.Zhu QH, Helliwell CA. Regulation of flowering time and floral patterning by miR172. J Exp Bot. 2011;62:487–495. doi: 10.1093/jxb/erq295. [DOI] [PubMed] [Google Scholar]
- 56.Wu G, Poethig RS. Temporal regulation of shoot development in Arabidopsis thaliana by miR156 and its target SPL3. Development. 2006;133:3539–3547. doi: 10.1242/dev.02521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kim JJ, et al. The microRNA156-SQUAMOSA PROMOTER BINDING PROTEIN-LIKE3 module regulates ambient temperature-responsive flowering via FLOWERING LOCUS T in Arabidopsis. Plant Physiol. 2012;159:461–478. doi: 10.1104/pp.111.192369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sawa M, Kay SA. GIGANTEA directly activates Flowering Locus T in Arabidopsis thaliana. Proc Natl Acad Sci U S A. 2011;108:11698–11703. doi: 10.1073/pnas.1106771108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liu H, et al. Photoexcited CRY2 interacts with CIB1 to regulate transcription and floral initiation in Arabidopsis. Science. 2008;322:1535–1539. doi: 10.1126/science.1163927. [DOI] [PubMed] [Google Scholar]
- 60.Kumar SV, et al. Transcription factor PIF4 controls the thermosensory activation of flowering. Nature. 2012;484:242–245. doi: 10.1038/nature10928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yasui Y, et al. The phytochrome-interacting VASCULAR PLANT ONE-ZINC FINGER1 and VOZ2 redundantly regulate flowering in Arabidopsis. Plant Cell. 2012;24:3248–3263. doi: 10.1105/tpc.112.101915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Iñigo S, et al. PFT1, the MED25 subunit of the plant Mediator complex, promotes flowering through CONSTANS dependent and independent mechanisms in Arabidopsis. Plant J. 2012;69:601–612. doi: 10.1111/j.1365-313X.2011.04815.x. [DOI] [PubMed] [Google Scholar]
- 63.He Y. Chromatin regulation of flowering. Trends Plant Sci. 2012;17:556–562. doi: 10.1016/j.tplants.2012.05.001. [DOI] [PubMed] [Google Scholar]
- 64.Yamaguchi S. Gibberellin metabolism and its regulation. Annu Rev Plant Biol. 2008;59:225–251. doi: 10.1146/annurev.arplant.59.032607.092804. [DOI] [PubMed] [Google Scholar]
- 65.Byrne ME, et al. Asymmetric leaves1 mediates leaf patterning and stem cell function in Arabidopsis. Nature. 2000;408:967–971. doi: 10.1038/35050091. [DOI] [PubMed] [Google Scholar]
- 66.Osnato M, et al. TEMPRANILLO genes link photoperiod and gibberellin pathways to control flowering in Arabidopsis. Nat Commun. 2012;3:808. doi: 10.1038/ncomms1810. [DOI] [PubMed] [Google Scholar]
- 67.Porri A, et al. Spatially distinct regulatory roles for gibberellins in the promotion of flowering of Arabidopsis under long photoperiods. Development. 2012;139:2198–2209. doi: 10.1242/dev.077164. [DOI] [PubMed] [Google Scholar]
- 68.Galvão VC, et al. Spatial control of flowering by DELLA proteins in Arabidopsis thaliana. Development. 2012;139:4072–4082. doi: 10.1242/dev.080879. [DOI] [PubMed] [Google Scholar]
- 69.Yu S, et al. Gibberellin regulates the Arabidopsis floral transition through miR156-targeted SQUAMOSA PROMOTER BINDING-LIKE transcription factors. Plant Cell. 2012;24:3320–3332. doi: 10.1105/tpc.112.101014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Davière JM, Achard P. Gibberellin signaling in plants. Development. 2013;140:1147–1151. doi: 10.1242/dev.087650. [DOI] [PubMed] [Google Scholar]
- 71.de Lucas M, et al. A molecular framework for light and gibberellin control of cell elongation. Nature. 2008;451:480–484. doi: 10.1038/nature06520. [DOI] [PubMed] [Google Scholar]
- 72.Kim DH, et al. Vernalization: winter and the timing of flowering in plants. Annu Rev Cell Dev Biol. 2009;25:277–299. doi: 10.1146/annurev.cellbio.042308.113411. [DOI] [PubMed] [Google Scholar]
- 73.Blazquez MA, et al. A thermosensory pathway controlling flowering time in Arabidopsis thaliana. Nat Genet. 2003;33:168–171. doi: 10.1038/ng1085. [DOI] [PubMed] [Google Scholar]
- 74.Jung JH, et al. The E3 ubiquitin ligase HOS1 regulates Arabidopsis flowering by mediating CONSTANS degradation under cold stress. J Biol Chem. 2012;287:43277–43287. doi: 10.1074/jbc.M112.394338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lee JH, et al. The E3 ubiquitin ligase HOS1 regulates low ambient temperature-responsive flowering in Arabidopsis thaliana. Plant Cell Physiol. 2012;53:1802–1814. doi: 10.1093/pcp/pcs123. [DOI] [PubMed] [Google Scholar]
- 76.Kim HJ, et al. A genetic link between cold responses and flowering time through FVE in Arabidopsis thaliana. Nat Genet. 2004;36:167–171. doi: 10.1038/ng1298. [DOI] [PubMed] [Google Scholar]
- 77.Strasser B, et al. A complementary role for ELF3 and TFL1 in the regulation of flowering time by ambient temperature. Plant J. 2009;58:629–640. doi: 10.1111/j.1365-313X.2009.03811.x. [DOI] [PubMed] [Google Scholar]
- 78.Lee H, et al. Genetic framework for flowering-time regulation by ambient temperature-responsive miRNAs in Arabidopsis. Nucleic Acids Res. 2010;38:3081–3093. doi: 10.1093/nar/gkp1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kim W, et al. The role of the miR399-PHO2 module in the regulation of flowering time in response to different ambient temperatures in Arabidopsis thaliana. Mol Cells. 2011;32:83–88. doi: 10.1007/s10059-011-1043-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Balasubramanian S, et al. Potent induction of Arabidopsis thaliana flowering by elevated growth temperature. PLoS Genet. 2006;2:e106. doi: 10.1371/journal.pgen.0020106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Proveniers MC, van Zanten M. High temperature acclimation through PIF4 signaling. Trends Plant Sci. 2013;18:59–64. doi: 10.1016/j.tplants.2012.09.002. [DOI] [PubMed] [Google Scholar]
- 82.Kumar SV, Wigge PA. H2A.Z-containing nucleosomes mediate the thermosensory response in Arabidopsis. Cell. 2010;140:136–147. doi: 10.1016/j.cell.2009.11.006. [DOI] [PubMed] [Google Scholar]
- 83.Liu L, et al. FTIP1 is an essential regulator required for florigen transport. PLoS Biol. 2012;10:e1001313. doi: 10.1371/journal.pbio.1001313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Jung C, Muller AE. Flowering time control and applications in plant breeding. Trends Plant Sci. 2009;14:563–573. doi: 10.1016/j.tplants.2009.07.005. [DOI] [PubMed] [Google Scholar]
- 85.Luan W, et al. The effect of the crosstalk between photoperiod and temperature on the heading-date in rice. PLoS One. 2009;4:e5891. doi: 10.1371/journal.pone.0005891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Song Y, et al. Interaction between temperature and photoperiod in regulation of flowering time in rice. Sci China Life Sci. 2012;55:241–249. doi: 10.1007/s11427-012-4300-4. [DOI] [PubMed] [Google Scholar]
- 87.Nagano AJ, et al. Deciphering and prediction of transcriptome dynamics under fluctuating field conditions. Cell. 2012;151:1358–1369. doi: 10.1016/j.cell.2012.10.048. [DOI] [PubMed] [Google Scholar]