Abstract
Genetic variations resulting in a change of amino acid sequence can have a dramatic effect on stability, hydrogen bond network, conformational dynamics, activity and many other physiologically important properties of proteins. The substitutions of only one residue in a protein sequence, so-called missense mutations, can be related to many pathological conditions, and may influence susceptibility to disease and drug treatment. The plausible effects of missense mutations range from affecting the macromolecular stability to perturbing macromolecular interactions and cellular localization. Here we review the individual cases and genome-wide studies which illustrate the association between missense mutations and diseases. In addition we emphasize that the molecular mechanisms of effects of mutations should be revealed in order to understand the disease origin. Finally we report the current state-of-the-art methodologies which predict the effects of mutations on protein stability, the hydrogen bond network, pH-dependence, conformational dynamics and protein function.
Keywords: Genetic variation, single nucleotide polymorphism, SNP, rare mutations, diseases
Introduction
The differences in human DNA sequences contribute to phenotypic variations, influence an individual’s susceptibility to disease, response to the environment and drug treatment.1; 2 At the same time, the DNA differences lead to phenotypic differences between populations, for example differences in eye color.3 The genetic variations may involve several nucleotides or only one, the latter is called “single nucleotide polymorphism” or SNP. Technically a polymorphism represents a DNA variation found in more than one percent of the population.4; 5 SNPs are the most common types of genetic variations in humans5; 6; 7 and occur approximately every 1200 bases in the overall human population.6; 8 Most often, SNPs occur in the non-coding region of the genome1 whereas SNPs in coding DNA regions may result in changes of amino acid sequences either through amino acid substitutions (nsSNPs) or the introduction of nonsense/truncation mutations.5
At the opposite end of the frequency spectrum are the rare mutations (often referred to as rare variants in contrast to common variants), which are usually defined as mutations with minor allele frequency of less than 0.5 – 1%.9 Historically, effects of mutations were discussed in the context of a “Common Disease, Common Variant (CDCV)” or “Common Disease, Rare Variant (CDRV)” debate.10 The CDCV hypothesis argues that common disease mutations with low penetrance (the percent of individuals with disease mutations who exhibit disease phenotype) are the major contributors to genetic susceptibility to diseases. On the other hand, the CDRV hypothesis asserts that rare mutations with relatively high penetrance are the major contributors. The effects of common and rare mutations were extensively studied by the theoretical population genetics11 and it was found that the frequencies of susceptibility alleles for any disease ranged from rare to high and depended on the mutation rates and the strength of purifying selection.11 With the advancement of DNA sequencing technologies, many examples of rare mutations contributing to common diseases have been reported and here we list several of them. Cohen and colleagues, for example, showed that multiple rare mutations contributed significantly to the plasma levels of high-density lipoprotein (HDL)12 and low-density lipoprotein (LDL)13. In the HDL study, three HDL related genes were studied and several dozen rare mutations were identified in total. Almost all mutations were found on the HDL related genes, ABC transporter A1, and two of these mutations, N1800H and W590S, drastically changed the physicochemical properties of amino acids and affected its interaction with a partner protein.
The functional effect of rare mutations has been a topic of many recent studies which tried to understand the role of rare mutations in complex traits.14; 15 Several studies concluded that the excess of deleterious rare mutations in the human genome was due to recent fast population growth and weak purifying selection. One study, focusing on a couple of hundreds of drug target genes, reported that rare mutations were more enriched for damaging variants than common mutations.14 Deep sequencing of human exomes also confirmed the abundance and deleterious effects of rare mutations, namely that rare mutations accounted for 86% of identified single nucleotide variants and also accounted for about 96% of variants which were predicted as functionally important.15 The same study mapped rare mutations on several known protein structures and found that they were enriched in the following categories: ligand binding, active sites, and sites participating in hydrogen bonding.15
In an effort to identify a link between mutations and chronic disease susceptibility, much data was collected from the literature for disease-causing rare and common mutations. It was shown that when the frequency of variants of adenoma patients and control groups was compared, the frequency of variants was higher in patients than for controls, which suggested that disease susceptibility could be due in part to the effects of many rare variants. 16 In contrast to rare mutations contributing to Mendelian diseases, some rare mutations occur in single individuals, where the patient does not necessarily inherit the mutation from a parent. These de novo mutations are considered as the most extreme cases of rare mutations.17 Since de novo mutations have not experienced strong evolutionary selection, they tend to be more deleterious than inheritable rare mutations. Indeed, recent whole-exome sequencing studies showed that de novo mutations in a single gene contributed to many rare diseases such as Kabuki syndrome and others.18 It was also suggested that the scarcity of diseases caused by de novo mutations was related to the number of mutational target genes. Diseases caused by de novo mutations in a single target gene occur at very low population frequencies (<1/10000) whereas diseases caused by de novo mutations in more than 100 candidate genes such as intellectual disability could be relatively common (>1/100).17
The necessity to understand the effects of missense mutations becomes clear when one considers genetic diseases. It is known that the substitution of only one residue in a protein sequence can be related to a number of pathological conditions such as Alzheimer’s, Parkinson’s and Creutzfeldt-Jakob’s diseases.6 It is also well known that accumulation of autosomal mutations can lead to cancers, and that hereditary diseases are caused by one or more germline mutations.19 The analysis of the impacts of missense mutations advances our understanding of the relationships between protein structure and function, and allows us to decipher the mechanisms of the effects of disease mutations and thereby pathogenesis.20 With personalized medicine on the not-so-distant horizon, it is swiftly becoming essential to understand the process by which genetic mutations lead to disease.
The plausible effects of missense mutations21; 22 range from affecting the macromolecular stability to perturbing macromolecular interactions and cellular localization. Here we will review the current state-of-the-art methods in the area of predicting the effects of mutations on protein stability, the hydrogen bond network and pH-dependence, conformational dynamics, and activity.
Effects of mutations on macromolecular stability
Association between protein stability changes and human diseases
It is well established that protein function is closely related to stability of monomers and complexes;23;24;25 therefore in order to assess computationally the functional consequences of mutations, it is essential to identify the effect of mutations on stability and folding free energy (energy difference between folded and unfolded states). Essentially, in order for the macromolecule to carry out its function, the macromolecule, in most cases, must adopt a particular three-dimensional (3D) fold and make specific interactions with its partners. A missense mutation that affects the 3D structure and alters the stability or binding affinity of a protein complex may cause significant perturbations or complete abolishment of the function of this particular protein.
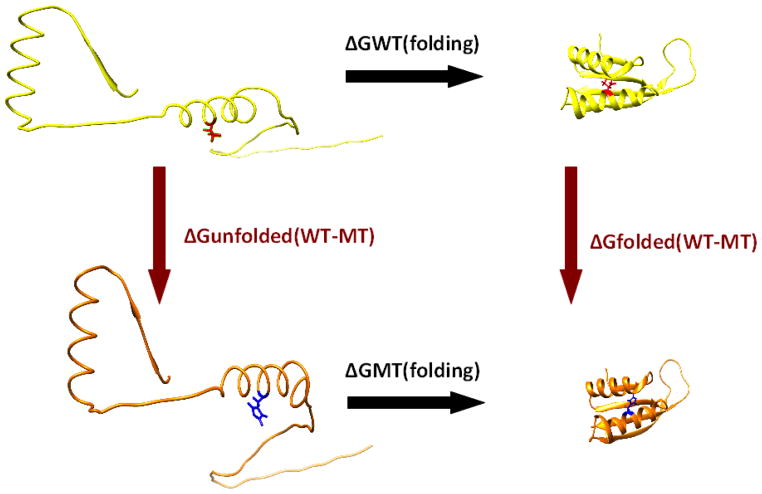
Typically the change in the folding free energy (ΔΔG) is used to quantify the magnitude of a mutation’s effect on stability. Methods using physical potential energy functions (such as those used in molecular mechanics approaches or Monte Carlo simulations) are probably the most insightful methods for predicting the details of the effects of mutations on protein stability.26 They usually are time-consuming and are frequently used for small-scale investigations.27 On the other side of the spectrum are methods utilizing machine learning techniques, which are very fast and can deliver predictions on large datasets. A typical scheme of assessing the effect of a mutation on macromolecular folding free energy is illustrated in Fig. 1. Two different approaches can be used to estimate ΔΔG: (a) in one case the folding free energy can be calculated for the wild type and mutant proteins and then the difference can be found; (b) in the second approach, the change in free energy upon mutation can be calculated for unfolded and folded states separately and then the value of the energy of the unfolded state can be subtracted from the value of the energy of the folded state. Such detailed schemes to predict ΔΔG are typically employed by methods utilizing 3D structure. However, frequently structural and sequence information are used together within the same methodological framework.
Figure 1.
The change in the folding free energy (ΔΔG) may be evaluated using two different methods: (1) using the difference in the folding free energy values calculated from the transition from an unfolded to a folded state of the wild type (ΔGWT(folding)) minus the mutant type (ΔGMT(folding)) shown with black arrows; or (2) using the difference in the folding free energy between the folded state of the wild type and mutant type (ΔGfolded(WT-MT)) minus the folding free energy between the unfolded state of the wild type and mutant type (ΔGunfolded(WT-MT)) shown with red arrows.
Before outlining the existing methods for predicting effects of missense mutations on macromolecular stability we first review recent studies showing the connection between human diseases and protein stability changes. In general the effects on protein stability can be grouped into two distinctive categories: (a) destabilizing and (b) stabilizing effects (see Table 1 for examples). Most frequently, missense mutations are found to destabilize the corresponding protein. Such cases include mutations in LMNA gene which are associated with muscular diseases,28 in the VWF A2 domain causing von Willebrand disease type,29 in retinal proteins causing retinal diseases,30 in the perforin protein resulting in hemophagocytic lymphohistiocytosis,31; 32 and mutations in prion proteins associated with prion diseases.33; 34; 35; 36 Many neurodegenerative diseases, such as Parkinson’s disease, are also associated with destabilization of the corresponding proteins.37; 38; 39 Sometimes the impact is local, affecting particular secondary structure element (SSE). For example, mutations of residues E22 and D23 in the Amyloid-β protein are associated with familial Alzheimer’s disease and are shown to destabilize an important beta-turn 40 whereas destabilizing mutations in the core of the protein lead to an inactivation of many tumor suppressors in cancer.41 Most frequently, if the effect of mutations is related to protein stability, it usually significantly destabilizes the corresponding protein.42
Table 1. The list of servers and programs to predict the effect of mutations on protein molecular characteristics. (.
| Name | Input | Description | URL |
|---|---|---|---|
| MuStab | Sequence | Predicts decrease/increase of stability upon mutation using support vector machine | http://bioinfo.ggc.org/mustab/ |
| MUPro | Sequence | Predicts decrease/increase of stability upon mutation using support vector machine | http://mupro.proteomics.ics.uci.edu/ |
| SIFT | Sequence | Estimates deleterious effect of mutation using sequence homology and site conservation | http://sift-dna.org |
| MutPred | Sequence | Estimates deleterious effect of mutation using SIFT and gain/loss of structural or functional properties predicted from sequences | http://mutpred.mutdb.org/ |
| SNPdbe | Sequence | Pre-calculated database of effects of known mutations calculated based on the neural network method and SIFT | http://www.rostlab.org/services/snpdbe/ |
| Polyphen(2) | Sequence/structure | Estimates deleterious effect of mutation using sequence homology, site conservation and structural features | http://genetics.bwh.harvard.edu/pph2/ |
| I - Mutant 2.0 | Sequence/structure | Estimates ΔΔG upon mutation using support vector machine | http://gpcr2.biocomp.unibo.it/%7Eemidio/I-Mutant/I-Mutant.htm |
| Site Directed Mutator (SDM) | Structure | Estimates ΔΔG upon mutation using statistical potential energy function | http://www-cryst.bioc.cam.ac.uk/~sdm/sdm.php |
| CUPSAT | Structure | Estimates ΔΔG upon mutation using mean force atom pair and torsion angle potentials | http://cupsat.tu-bs.de/ |
| FOLD-X | Structure | Estimates ΔΔG upon mutation using empirical force field | http://fold-x.embl-heidelberg.de |
| AUTO-MUTE | Structure | Estimates ΔΔG upon mutation using knowledge-based statistical contact potential and machine learning methods | http://proteins.gmu.edu/automute |
| ERIS | Structure | Estimates ΔΔG upon mutation using physical force field with atomic modeling | http://dokhlab.unc.edu/tools/eris/ |
| PoPMuSiC 2.0 | Structure | Estimates ΔΔG upon mutation using a combination of statistical potential and neural networks | http://babylone.ulb.ac.be/popmusic |
| CC/PBSA | Structure | Poisson-Boltzmann Calculations combined with surface accessibility term to calculate the energy on an ensemble of structures generated with Concoord | http://ccpbdsa.bioinformatik.unisaarland.de/ |
| HOPE | Structure/sequence | Uses experimentally determined or homology-based structures with conjunction with WhatIf to make predictions | http://www.cmbi.ru.nl/hope/ |
There are examples where missense mutations improve or enhance stability of the corresponding protein while still being deleterious. Recently a mutation in the CLIC2 protein, H101Q, was associated with a mental disorder and was predicted to increase the CLIC2 protein stability therefore obstructing its transport to the cell membrane.43 Later these predictions were confirmed experimentally and it was demonstrated that, indeed, this mutation makes the CLIC2 protein thermodynamically more stable, the residence time in the membrane shorter, and the mutant interacts more strongly with the ryanodine receptor.44 Similarly, the stability of a small 41-residue helical protein, the peripheral subunit-binding domain, was found to increase upon a replacement of a surface charge with a hydrophobic residue.45 In silico modeling of the effects of mutations on stability of spermine synthase (causing Snyder-Robinson Syndrome) showed that sites harboring disease-causing mutations may or may not tolerate other (different from disease-causing) amino-acid substitutions.46 This indicates that disease-causing effects on the stability may be both site and amino-acid type dependent.
Algorithms for predicting the effect of mutations on protein stability
The previous section listed several cases where missense mutations produce a very prominent effect on macromolecular stability and many efforts were invested to develop approaches and algorithms to predict the change of the folding free energy upon missense mutations. While mutations might have large effects on protein binding affinity and lead to many diseases,47; 48 here we only focus on existing approaches for predicting the effect of mutations on stability of protein monomers. In general, the existing methods can be classified into several distinct categories based on the strategy used in the calculations (see for example reviews 49; 21). Here we do not try to classify these methods because many of them utilize a mixture of different approaches. Table 2 includes several widely used methods, provides links to corresponding databases and servers and a short methods’ description. Below we summarize several methods and resources.
Table 2.
Diseases associated with changes in stability as a result of mutations
| Change In Stability | Disease | Gene/Mutation | Reference |
|---|---|---|---|
| Destabilizing | Muscular Diseases | LMNA; multiple sites | 28 |
| Von Willebrand Disease | VWF A2; R1597W, M1528V | 29 | |
| Retinal Diseases | Rhodopsin; multiple sites | 30 | |
| Hemophagocytic Lymphohistiocytosis | Perforin; multiple sites | 31; 32 | |
| Neurodegerative/Prion Diseases | Prion; D178N | 36 | |
| Autosomal recessive Parkinson’s Disease | PINK1; multiple sites | 37; 39 | |
| Familial Alzheimer’s | Amyloid protein; Glu-22, Asp-23 | 40 | |
| Inactivation of tumor suppressants | Multiple genes; multiple sites | 41 | |
| Snyder-Robinson Syndrome | Spermine Synthase; G56S, V132G, I150T, Y328C | 46; 168 | |
|
| |||
| Stabilizing | Mental Disorder | CLIC2; H101Q | 43 |
The first group of methods represents machine learning approaches which are trained on different types of data relevant to protein stability and in some cases take into account experimental conditions such as temperature, salt concentration and pH values. Taking into account such parameters is important for assessing the free energy changes upon mutations at near physiological conditions.50 Majority of these methods (MuStab51, I-Mutant and others) incorporate different physicochemical properties of amino acids and structural preferences of different sites and are trained on the experimental differences of folding free energy caused by mutations. Some methods, like I-Mutant2.0, use Support Vector Machines (SVMs), make predictions based on either structure or sequence alone;52; 19 and predict actual values of ΔΔG. Similarly, the MuPro uses SVMs leveraging both sequence and structural information.53 Other methods, such as MuStab, predict only the deleterious effects of mutations and increase or decrease (the sign) of ΔΔG values. Another recently introduced method, PoPMuSiC-2.0, uses a combination of statistical potential and neural networks to estimate the changes in stability; it exploits statistical potentials which take into account the coupling between four protein sequence and structure descriptors, and the amino acid volume variation upon mutation.54
The second group of methods exploits the evolutionary conservation data under an assumption that changes at conserved positions in the multiple sequence alignments tend to be deleterious. Although such approaches do not predict the effect of mutations on protein stability directly, they are typically used in conjunction with the above mentioned methods to achieve consensus predictions. For example, a prediction of large ΔΔG caused by a mutation will have a higher accuracy if supported by sequence based analysis. There are many sequence based methods. For example, Sorting Intolerant From Tolerant (SIFT55) method scores the normalized probabilities for all possible substitutions for a site and calculates the conditional probability that an amino acid is tolerated compared to the most frequent tolerated amino acid. PolyPhen on the other hand, predicts damaging amino acid substitutions using sequence-based as well as structure-based features such as sequence conservation, structure, and a position-specific independent counts matrix derived from the multiple sequence alignment.4; 56 Additionally, PolyPhen-2 uses eight sequence-based and three structure-based predictive features and a Naïve Bayes classifier to predict the functional significance of the mutation.57
The third group of methods relies on structural information, assuming that the ability of a protein to function properly depends on the fundamental physicochemical properties which can be derived only from structures.23 One research group, for example, investigated the relevance of combining coarse-grained structure-based stability predictions with a simple comparative modeling procedure.58 CUPSAT (Cologne University Protein Stability Analysis Tool) uses structural environment specific atom potentials and torsion angle potentials to predict ΔΔG59 whereas Site Directed Mutator (SDM) uses a statistical potential energy function;60 Some methods use empirically derived energy functions which are quite accurate in part because they are trained on the experimental data. The most prominent example is the empirical force field of FoldX which has been optimized for point mutations,19 and implemented into a computer algorithm and webserver, FOLDEF, to predict folding free energy change. 61; 56 Another method which utilizes structural information is ERIS which applies the Medusa force field while including backbone flexibility to make predictions. 62 Recently an interesting approach was introduced (MutPred) 63 which uses a broad range of different attributes based on protein sequence, structure and dynamics. This method models the changes of sequence and structural features between wild-type and mutant sequences where changes are expressed as probabilities of gain or loss of an attribute.63 Another approach for predicting changes of the folding free energy upon mutations is implemented in a method called CC/PBSA.64 It utilizes Concoord 65 to generate a structural ensemble of the target protein and calculates the folding free energy with Poisson-Boltzmann (Delphi 66) Surface Area (PBSA) method. Structural information is utilized in HOPE as will, either using experimentally available structures or structures built by homology in conjunction with energy calculations done with WhatIf.67
Several papers reported comparisons of the performance between different methods.68; 69; 70; 71 In order to assess the performance, a systematic analysis is necessary. Although there is no single measure that accurately gauges the performance, some helpful measures include ROC, AUC, sensitivity, specificity, positive and negative predictive values.72 Different factors may influence the prediction accuracy including the type of substituted amino acid, protein structural class and flexibility, structural environment of an affected site. All these factors should be considered when judging the effectiveness of a given approach.73 It is outside of the scope of this review to rank the performance of the existing tools (including Dmutant,74 MultiMutate,75 SCide,76 Scpred,77 SRide,78 nsSNPAnalyzer,79 Panther,80 PhD-SNP,6 SNAP,81 and SNPs&GO,82 among other well-known tools), because different benchmarking papers (see reviews 83;68;73;84) report conflicting results. The main conclusion is that various groups of methods complement each other and are suited for different types of tasks.
Effect of mutations on hydrogen bond network and ionization states
Protein stability is determined by many different factors and the formation of hydrogen bonds is among the most important ones. Hydrogen atoms are an essential component of the atomic structure of biological macromolecules. Among all hydrogens, those carrying significant positive partial charge, the so-called polar hydrogens, are particularly important because of their ability to form hydrogen bonds. In structures of biological macromolecules the hydrogen bond is formed between a polar hydrogen and a negatively charged hydrogen acceptor, typically an oxygen atom. In water phase, the water hydrogens and oxygens form a complex network of interactions resulting in water clusters. The arrangement of hydrogen bonds at the macromolecular surface is even more complicated involving interactions between macromolecular and water atoms. The groups of hydrogen bonds often form a cluster or a web of interactions resulting in so-called hydrogen bond network. Hydrogen bonds and hydrogen bond networks participate in biological functions of macromolecules and provide the pH-dependence associated with many biological reactions. They contribute to protein structural integrity, provide “proton wires” for proton translocation, the source of proton uptake/release and, finally, participate in many catalytic reactions. Below we briefly outline the mechanisms of how missense mutations may affect the hydrogen bond networks.
Hydrogen bond networks and macromolecular structure
As was mentioned above, the hydrogen bonds are key constituents of biomolecular structures,85 participating in the formation of SSEs,86 tertiary and quaternary structure.87 A mutation resulting in the removal or addition of a hydrogen donor or acceptor is expected to have a significant impact on the structural integrity. Even a conservative mutation which results in different donor-acceptor positions, may still be quite deleterious for the structure and may disrupt the entire hydrogen bond network (Table 3).
Table 3.
Diseases resulting from changes in the hydrogen bond network as a consequence of mutations
| Effect on | Disease | Gene/Mutation | Reference |
|---|---|---|---|
| Structure | Prolidase Deficiency | PEPD; 304insA | 169 |
| Aldosterone Synthase Deficiency | CYP11B; R374W | 88 | |
| Mitochondrial Fatty Acid Oxidation Disorder | CPT1; R595W | 89 | |
| Snyder-Robinson Syndrome | Spermine synthase; I150T | 46 | |
| Cardiovascular Disease Predisposition | Human lipoxygenase; T560M | 91 | |
| Congenital Hereditary Cataract Disease | βB1-crystallin; S129R | 92 | |
|
| |||
| Aggregation/Folding | Alzheimer’s Disease | amyloid-β peptide aggregation; multiple sites | 170 |
| Familial Alzheimer’s Disease | Fragment of amyloid beta- protein; multiple sites | 171 | |
| Amyloidosis (with severe cardiomyopathy) | Aggregation/cytotoxicity of transthyretin; S112I | 172 | |
|
| |||
| Flexibility | Aneuploidy and Solid Tumors | CEP63; L61P | 127 |
| Gaucher disease | acid-β-glucosidase; N370S | 173 | |
| Lethal Catecholaminergic Polymorphic Ventricular Tachycardia | Calsequestrin; R33Q | 174 | |
| Promotion of Aneuploidy and Tumorigenesis | Centromere-associated protein-E; Y63H | 175 | |
|
| |||
| Cellular Localization | Brain Tumors | ING1B; p33 | 176 |
| May-Hegglin anomaly, Sebastian syndrome, and Fechtner syndrome | MYH9; multiple sites | 177 | |
|
| |||
| pH | Human Amyloidosis | TTR; several residues | 178 |
| Mental Retardation | CLIC2; H101Q | 44 | |
| Alzheimer’s Disease | Apolipoprotein E, apoE4 isoform | 179 | |
|
| |||
| Activity | Pyruvate Dehydrogenase Complex | pyruvate dehydrogenase | 180 |
| Deficiency | E1α subunit protein; multiple sites | ||
| Classical Homocystinuria | cystathionine β-synthase; R266K | 181 | |
| Amyotrophic Lateral Sclerosis Disease | angiogenin protein; K17I | 182 | |
| Neonatal Epileptic Encephalopathy | pyridoxine 5′-phosphate oxidase; R229W | 183 | |
| Snyder Robinson Syndrome | spermine synthase; I150T | 90 | |
Figure 2 shows one example of mutation N550K in Isoform 3 Lactosylceramide alpha-2,3-sialyltransferase, which is associated with Autosomal Recessive Neurocutaneous Condition. As can be seen from this figure, the wild type residue, N550, is involved in several hydrogen bonds, which are deleted in the mutant. In another example of the Aldosterone synthase deficiency, it was demonstrated that molecular mechanism of this disease involved mutation R374W in CYP11B2 protein, which changed the properties of the hydrogen bond network of the wild type Arg residue.88 Similarly, in patients suffering from mitochondrial fatty acid oxidation disorder and bearing a R595W mutation in the CPT1 protein, the disease was attributed to a disruption of the wild type hydrogen bond network formed by the wild type R374 residue.89 In another case the rearrangement of the hydrogen bond network due to a missense mutation (I150T) in spermine synthase was predicted to be the main cause of the Snyder Robinson syndrome.90 Further mutational analyses of the same protein indicated that almost any mutation affecting this residue involved in a wild type hydrogen bond network will have drastic effects on the wild type properties of the hydrogen bond network and macromolecular stability.46
Figure 2.
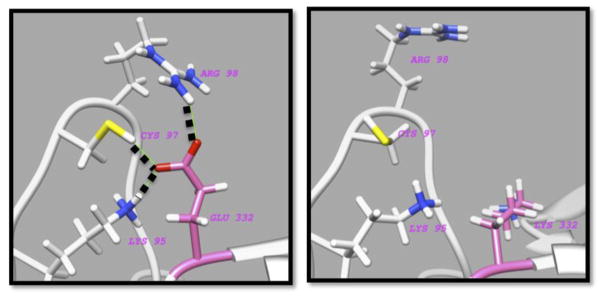
The region at the mutation site N550 of the wild type 3D structure of Isoform 3 Lactosylceramide alpha-2,3-sialyltransferase and mutant model which was built using 2wmlA.pdb as a template: (a) the wild type with N550 in the left panel and (b) the mutant N550K in the right panel, both centered at the mutation site. The hydrogen bonds between N550 and its neighbors in the wild type are shown with dashed black lines.
The effect of mutations on hydrogen bond network is especially pronounced if the network involves active site residues. For example, a recent study showed that the low catalytic efficiency of a mutant, T560M, found in human lipoxygenase is caused by the alterations of a hydrogen bond network interconnecting this residue with active sites; this in turn may lead to a predisposal to cardiovascular diseases.91 The wild type hydrogen bond network and stability of tertiary structure was also reported to be altered by missense mutations causing congenital hereditary cataract disease.92 These limited examples confirm the importance of the hydrogen bond network for structural integrity of macromolecules and point to the necessity of predicting such effects in the context of missense mutations.
Hydrogen bond network in protein aggregation and flexibility
The aggregation, or formation of amyloid fibrils is one of the primary sources of many diseases.93; 94; 95; 96 Missense mutations disrupting the native hydrogen bond(s) can destabilize the local structure and thus expose the hydrophobic core of the protein to the water phase. Such an event frequently triggers aggregation and fibril formation. Several examples are listed in Table 3. Biological reactions frequently are accompanied by small or large protein conformational changes.97 Consequently, the ability of macromolecules to retain their conformational flexibility is crucial to their function. Altering the hydrogen bond network by missense mutation can affect the conformational flexibility needed for allosteric regulation and conformational gating. Several examples of an altered protein flexibility causing diseases are listed in Table 3. At the same time, altering the wild type flexibility through mutations may not necessarily result in the pathogenic effect.98 A detailed analysis is usually required in order to attribute the change of flexibility to a particular molecular function.
Hydrogen bond networks and pH-dependence
Practically all biological processes are pH-dependent and pH is an important regulator of cellular function. Different compartments of the cell have different characteristic pH (see reviews 99,100, 24, 101). Macromolecules that shuttle between different cellular compartments, as for example, prolactin receptor, 102; 103 have their pH-dependent characteristics precisely tuned to the local pH. Thus, in order to function properly and interact with its biological partners, the protein needs maintain specific pH dependent characteristics22 which can be very sensitive to single point mutations.104 In addition, single point mutations may affect a protein’s cellular location.105 More examples are provided in Table 3.
Hydrogen bond networks and catalytic reaction
Hydrogen bonds participate in catalytic reactions in various ways: they coordinate the substrate and can be formed or disrupted during the reaction process (donating or accepting a hydrogen via general acid/base residues) (Table 3). Typically the catalytic reaction is mostly affected by a substitution of an amino acid directly involved in the catalysis. However, it should be mentioned that active site residues are rarely mutated and mutations often occur in the sites connected to the active site region via hydrogen bond networks. The most drastic change of the hydrogen bond network is caused by protonation/deprotonation of titratable groups. Thus, the pKa shifts of catalytic residues induced by a mutation will disrupt the general acid/basic reaction and will dramatically affect the catalysis.
Approaches to model effects of mutations on hydrogen bond networks
Assessing the effect of mutations on hydrogen bond networks is not a straightforward task since the positions of protons (hydrogens) are typically not resolved experimentally, but rather should be generated in silico. Even more, the protonation states of titratable groups, which are responsible for pH-dependence of biological processes including general acid/basic catalysis, are typically unknown and have to be predicted as well.
In the simplest case scenario when mutation does not involve the protonation change, the analysis of the hydrogen bond network begins with placing the missing hydrogens onto the wild type and mutant 3D structures. By doing so, one typically assumes default protonation states of titratable groups at pH equal to 7. However, many biological reactions occur at pH different from neutral and careful analysis would require obtaining the biochemical data on the optimal/characteristic pH.106; 100;24 At the next step, protons’ positions are generated with the standard Molecular Dynamics (MD) packages such as NAMD,107 Charmm,108 Amber,109 Gromos,110 Gromacs,111 or other stand-alone programs such as REDUCE112 and PDB2PQR113. Then one would compare hydrogen bonds in the minimized structures of wild type and mutant proteins.90 A more sophisticated analysis would involve the comparison of the hydrogen bonds in the snapshot structures obtained in MD simulations. All MD packages have tools for the analysis of hydrogen bond networks. A recently developed standalone program (HBonanza,114 hydrogen-bond analyzer; http://www.nbcr.net/hbonanza), allows the analysis and visualization of hydrogen-bond networks. HBonanza can be used to analyze single structures or many structures of a molecular dynamics trajectory.
Cases where mutation induces changes in the protonation state of titratable groups are much more complex. It should be mentioned that such ionization changes may occur even if mutation does not involve titratable groups.46 Predictions of protonation states can be done by calculating the pKa’s of titratable groups and then by assigning the appropriate charge states depending on the characteristic pH for a given protein. There are many approaches for computing pKa’s which are reviewed in 115. Some of them are standalone programs such as MCCE,116 ProPKA,117 while others are implemented into webservers such as H++118.
Effect of mutations on conformational dynamics
Biological macromolecules may adopt different conformations along the pathway of the corresponding biochemical reaction119; 120 and their intrinsic flexibility, the ability to sample alternative conformations is crucial for protein function.121; 122 Furthermore, a significant fraction of macromolecules is either disordered or has disordered segments at a particular stage of the biological reaction.123; 124 Missense mutations can affect the flexibility of the entire molecule or just a small region, can shift the equilibrium between different conformations or can affect the entire conformational dynamics of the molecule. In a most recent study of the NFAT5 transcription factor, different mutations from the same DNA-binding loop were analyzed.125 It was shown that even though these mutations are located very close to each other in sequence and space, their effect on protein dynamics and DNA binding is drastically different. Typically the changes of conformational dynamics are assessed computationally via monitoring the RMSD of the wild type and mutant structures and recorded via the snapshots obtained in Molecular Dynamics (MD).
Below we outline different aspects of effects of mutations on protein dynamics and provide examples of recent finding with this regard. The most commonly used approach to study protein dynamics is MD simulations, although Monte Carlo methods and Normal Mode Analysis can be utilized as well. Most widely used packages are NAMD,107 Charmm,108 Amber,109 Gromos,110 Gromacs,111 TINKER,126 and others.
Effect on protein flexibility
Molecular flexibility is reflected in the ability of a macromolecule to sample alternative conformations, as for example to open/close the gate of a channel, to mediate the recognition of the receptor or to facilitate the allosteric reactions. Typical examples are centrosomes, which are central regulators of mitosis often amplified in cancer cells. Specifically, the centrosomal protein CEP63 is associated with an aneuploidy and solid tumors in humans. When genetic alterations such as L61P occur, they increase flexibility of the protein, as seen in MD simulations, which was suggested to be the cause of the disease.127 Although mutations may cause different diseases, the common trend is the same, namely, disease can be associated with a change in the conformational or dynamical properties of the corresponding protein (Table 4).
Table 4.
Effects of disease mutations on conformational dynamics
| Effect | Disease Associated | Gene/Mutation | Reference |
|---|---|---|---|
| Conformational Flexibility | Diabetes and Cancer | PTEN; H61D | 128 |
| Multiple Diseases | cNTnC; L48Q | 184 | |
| Conformational Diseases | α(1)-antitrypsin variant; K154N | 129 | |
| Variegate Porphyria Disorder | hPPO; R59Q, R59G | 130 | |
| Colorectal Cancer | mitotic centromere-associated kinesin protein; E403K | 185 | |
| Classic Galactosemia | GALT; multiple sites | 186 | |
| Becker Muscular Dystrophy | dystrophin protein; L427P | 187 | |
|
| |||
| Disordered Regions | Parkinson disease | α-synuclein; multiple sites | 133 |
| Ovarian Cancer | HPV proteins; multiple sites | 133 | |
| Cancer | p53 protein; multiple sites | 133 | |
| Huntington’s disease | huntingtin; multiple sites | 133 | |
|
| |||
| Order-Disorder Transitions | Stargardt Disease (type 1) | Multiple mutations | 188 |
| Adrenoleukodystrophy X-linked | Multiple mutations | 188 | |
| Androgen Insensitivity Syndrome | Multiple mutations | 188 | |
| Citrullinemia (type 1) | Multiple mutations | 188 | |
|
| |||
| Aggregation | Alpha(1)-antitrypsin Deficiency and Cystic Fibrosis | human serine protease inhibitor (serpin) α-1 antitrypsin; E342K | 140; 189 |
| Creutzfeldt-Jakob disease and Gerstmann-Straussler-Scheinker Disease | prion protein; multiple sites | 141 | |
|
| |||
| Local Motion | Cancers | KLK3; multiple sites | 190; 191; 192 |
Another case is phosphatase and tensin homolog (PTEN) protein which plays essential roles in cellular processes including survival, proliferation, energy metabolism, and cellular architecture. Mutations in PTEN are implicated in diabetes and cancer. A particular mutation (H61D) was studied using MD simulations and it was shown that it increases the flexibility, radius of gyration, and solvent accessibility of PTEN.128 The list of examples would be incomplete without mentioning the “conformational diseases”, where native protein conformers convert to pathological intermediates that can polymerize. Recently a forme fruste deficiency variant of α(1)-antitrypsin (K154N) was investigated by the Nuclear Magnetic Resonance spectroscopy and it was found that this mutation alters the wild type interaction of the side chain of K154 with the backbone carbonyl oxygen of K174, affecting protein flexibility and resulting in polymerization. 129
Disease mutations might not only lead to increased flexibility, on the contrary, they can restrict the conformational transitions. One example includes mutations in the human protoporphyrinogen oxidase (hPPO) gene which are responsible for the dominantly inherited disorder variegate porphyria (VP). Two missense mutations (R59Q and R59G) were investigated in a recent work130 where MD modeling revealed that these mutations affect the catalytic activity of hPPO by changing its ability to sample different conformations. Analysis of mutation H101Q in the CLIC2 protein demonstrated that this mutation restricted the mobility of the N-terminal domain and prevented the conformational change presumed to be required for entering of CLIC2 into the membrane.43; 44 In this regard, channels and pores are very interesting objects to study since their selectivity is regulated by structural fragments. For instance, aquaporins play physiological roles in several organs and tissues, and their alteration is associated with disorders of water regulation. A mutant, D184E, was shown to affect the mobility of the aquaporin D-loop, which acquires a higher propensity to equilibrate in a “closed conformation”, thus affecting the rate of water flux 131.
Protein intrinsic disorder and disease mutations
Disordered proteins, existing in dynamic equilibrium between various conformers, may provide a way to tolerate many mutations due to the loosely packed cores. However, the relationship between disorder, stability, and function is not well understood. According to some studies, about 10% of inherited disease mutations from HGMD database affect disordered regions,63 and cancer-associated proteins are especially enriched in intrinsically disordered regions.132 In particular, disease-related genes encoding disordered proteins or proteins with the extended disordered regions, include α-synuclein (Parkinson’s disease), BRCA1 (breast and/or ovarian cancer), p53 (cancer), huntingtin (Huntington’s disease).133; 134
There are many ways in which mutations might impact disordered regions and lead to dysfunctional proteins. If a residue that promotes disorder is mutated into a residue that favors structural regions, such a mutation can have a drastic effect on disorder-order transition, post-translational modifications, binding, and other functions relevant to intrinsic disorder. Examples of such substitutions are abundant among substitutions of disorder-promoting arginine into other residues types. Overall, about a quarter of disease mutations in disordered regions were predicted to disrupt disorder-promoting properties. Therefore the prediction and prioritization of disease mutations should account for the disorder-promoting tendency of the region where mutations occur.135; 136 Several such examples are provided in Table 4.
Protein aggregation, misfolding and dynamics
Protein aggregation is driven by the exposure of the hydrophobic core of the macromolecule to the water phase due to the changes in native structure and dynamics. It was shown that mutations in peripheral myelin protein 22 resulted in the common peripheral neuropathy Charcot-Marie-Tooth disease. One of them, L16P, caused misfolding which led to a loss of function and toxic accumulation of aggregates.137 Another recent work studied the effects of several mutations (V180I, F198S, V203I and V210I) known to cause prion disease and showed that these mutations induced the misfolding and aggregation of the prion protein.138 Another case involved mutations or deletions in the FMRP protein, which participates in the regulation of mRNA metabolism in brain, leading to the Fragile X syndrome. A severe manifestation of the disease has been associated with the I304N mutation, located on the KH2 domain of the protein. This mutation was found to destabilize the hydrophobic core causing a partial unfolding and a displacement of alpha-helices.139
Similarly, mutation E342K in human serine protease inhibitor (serpin) α-1 antitrypsin causes polymerization in the endoplasmic retuculim of hepatocytes and is associated with a lack of secretion into the circulation. It was shown that this mutation increases local flexibility, favors polymerization and promotes aggregation.140 Another case involves mutations in the human prion protein resulting in Creutzfeldt-Jakob or Gerstmann-Straussler-Scheinker diseases. The molecular dynamics modeling suggested that these mutations promote amyloid formation.141 Other examples of disease causing mutations in relation to aggregation and misfolding are shown in Table 4.
Effect of mutations on protein activity
Challenges of modeling of mutation effects on protein function
Protein structure-function relationships are complex and crucial for understanding and predicting the effects of benign or disease-causing mutations on the fitness. Proteins largely evolve through the acquisition of new mutations, the majority of them are destabilizing but neutral. At the same time some mutations can be deleterious or damaging or may result in advantageous novel functions. Interestingly, mutations which modify and produce novel binding specificities were shown to have larger destabilizing effects compared to mutations occurring on protein surfaces.142 On the other hand, since protein functional regions can be energetically unfavorable, mutations of functionally important residues, especially of polar and charged residues, may often result in more stable structures.143; 144; 145 Protein stability is necessary but not sufficient for protein functioning and proteins are not necessarily optimized to maximize their stability 146; 147. Therefore in order to assess the damaging effect of mutations, it would be crucial to understand how they will impact functionally important sites. Functional site prediction methods can be subdivided into several categories: those that use evolutionary conservation of binding site motifs, those that use information about a structure of a complex and docking methods. Some of them include PHUNCTIONER,148 FIRESTAR,149 IBIS,150 ConCavity,151 and others.
The effects of missense mutations on proteins and their function can be understood and modeled within the framework of the energy landscape theory which describes the potential energy of a protein as a function of conformational coordinates 127. Different conformational states might be characterized by different functional specificities and structural differences. The equilibrium between conformational states can be shifted by binding of different ligands, post-translational modifications, by changing the environmental conditions, and, finally, by mutations. Recently, human spermine synthase activity was enhanced by engineered novel mutations152 and it was demonstrated that the activity depended on electrostatic field distribution, the intrinsic flexibility of protein domains, and the overall protein stability. This indicates the complexity of biochemical functions involving various factors which in turn may be affecting each other. Table 5 lists exemplary mutations and their effect on protein activity.
Table 5.
Examples of disease-related mutations that affect protein activity and function.
| Gene/Protein | Disease | Mutation | Effect | Reference |
|---|---|---|---|---|
| ATP-binding cassette transporter (ABCA1) | Tangier disease(reduction of HDL Cholesterol in plasma) | N1800H | Changes the localization of protein, resulting in intracellular accumulation of ABCA1 and loss of interaction with ApoA-I. | 193 |
| W590S | Impairs dissociation of apoA-I from ABCA1, which may affect ATP-dependent lipid translocation. | 194 | ||
| Niemann–Pick C1-Like protein 1 (NPC1L1) | Hypocholesterolemia(reduction of LDL Cholesterol in the plasma) | T61M, S881L | Severely dysfunctional mutations; affect cholesterol uptake function, cholesterol-regulated recycling, glycosylation and stability of the protein. | 195 |
| G402S, R1268H | Partially dysfunctional mutations; affect cholesterol-regulated recycling. | 195 | ||
| Thiazide-sensitive sodium-chloride cotransporter (SLC12A3) | Gitelman syndrome (salt wasting and low blood pressure) | G439S, G741R | Abolish sodium uptake function due to loss of localization at plasma membrane, which may be caused by incomplete glycosylation. | 196 |
| Chloride intracellular channel protein 2(CLIC2) | X-linked intellectual disability | H101Q | Stabilizes the protein, which in turn may increase ryanodine receptor activity. | 44 |
| Histone-lysine N- methyltransferase (MLL2) | Kabuki syndrome | W5065X, R5179H | May affect the epigenetic control of active chromatin states via methylation of histones. | 18 |
Case study: Receptor Tyrosine Kinases
Here we analyze the mechanisms of cancer mutations on protein stability, dynamics, and activity using an example of the well-studied human receptor tyrosine kinase (RTK) family that is frequently mutated in cancer. The connection between cancer and kinase activation was found fairly recently153 and an increased RTK activity in tumor tissues was attributed to gene amplifications, enhanced transcription, translation, and to mutations. Several cancer mutation hotspots were identified in several RTKs, most of them were located in the activation loop, P-loop, and DFG loops. Moreover, driver mutations were found to be more often associated with the functionally important regions in kinases than passenger mutations.154 The activation of certain RTKs is tightly linked with their dimerization and high RTK activity is sometimes achieved by promoting dimerization. According to long-timescale molecular dynamics simulations, it was suggested that cancer mutations may suppress the intrinsic disorder on the dimer interface and stimulate the EGFR dimerization.155
Structural studies of kinases revealed different structural perturbations in response to cancer mutations. In particular, the mechanisms of kinase activation in cancer is probably linked to transitions between the active and inactive states.156; 157 Indeed, the thorough analysis of the effect of cancer mutations on kinase structure and activity showed that some mutations disturbed autoinhibitory interactions and considerably accelerated the catalysis.156 At the same time the crystal structure of the EGFR L858R mutant revealed that this mutation prevented the activation loop from adopting the inactive conformation157 thereby activating the kinase. On the other hand, the secondary EGFR T790M mutation facilitated the transition between inactive and active conformations and increased the stability of the active conformation.158 Enhanced mobility was observed near cancer mutations sites and modeling of autoinhibited conformations revealed that these mutations disrupted the local stabilizing interactions and destabilized the inactive form.159 Recently the effect of cancer mutations was analyzed using the dataset of all available structural pairs of active in inactive states. It was shown that cancer mutations destabilized active states, but to a lesser degree than the random mutations; moreover, to a lesser degree than mutations destabilized inactive states.160 This led to kinase activation. The same study found a relationship between the statistics-based estimate of oncogenic potential of mutation and its activation effect calculated based on thermodynamics principles. Namely, more frequent mutations had a higher activating effect.
Multiple mutations
A more comprehensive understanding of molecular mechanisms of disease mutations may come from the analysis of the effects of multiple mutations occurring in the same gene. It was demonstrated previously that the number of cases where multiple mutations may occur simultaneously in one gene is higher than the number predicted from the random mutation distribution.161 Such multiple mutations may be the result of an accumulation of single mutations in sequential cell replications or in the same cell cycle. The latter synchronous mutations are usually characterized by non-random proximal spacing in higher eukaryotes.162 At the same time, a recent study showed that the extent of the clustering of cancer mutations might differ between oncogenes and tumor suppressors and the former are found to be clustered while latter are not.41 A significant fraction of cancer-associated mutations comes in doublets or triplets where overall about 6–8% of all cancer mutations represent double cancer mutations.160; 162; 163 Similarly to single mutations, the majority of cancer multiple mutations occur in non-synonymous codon sites implying that doublets are under positive selection.160; 164 The observed and simulated spectra of double mutations generated from the spectra of single mutations were found to be significantly different for receptor tyrosine kinase (RTK) genes, which pointed to different mechanisms underlying single and double mutation spectra.160
Moreover, multiple disease mutations might have a synergistic phenotypic effect manifested in enhanced or decreased protein stability or activity. For example, a double mutation was found in β-amyloid precursor protein which resulted in an increased production and secretion of amyloid- β-peptide causing early-onset Alzheimer’s disease.165 On the other hand double mutations in the α-galactosidase encoding gene, which cause Fabry disease, resulted in reduced or non-detectable activity compared to single mutants.166 A positive epistasis was found for many double cancer mutants in RTK genes,160 namely, the effect of multiple mutations on kinase activity was higher than a total of individual mutations. This trend was especially pronounced for double mutations observed in more than one tumor sample.
Concluding remarks
The effects of human missense mutations on various biophysical characteristics, stability, hydrogen bond network, dynamics and activity, were reviewed in this work. It was demonstrated that mutations can frequently affect several biophysical characteristics simultaneously and may or may not cause diseases (see also 167). There is no clear threshold of how large the change of the wild type characteristics should be in order to alter protein function and result in disease. Predictions of damaging effects of mutations are further complicated by the observations that enhanced activity 152, higher stability or binding affinity,43; 44 are not necessarily advantageous for the cell and protein and can be disease causing.
Missense mutations affect protein stability, hydrogen bonds, dynamics and activity and cause diseases
Any deviation away from wild type characteristics can be deleterious
Changes in stability, dynamics, hydrogen bonds and activity upon mutations are interconnected
Rare mutations are typically more deleterious than common variants
Acknowledgments
S.S., M.P, and E.A. acknowledge the support from NIH, grant number R01GM093937. H.N. and A.R.P. were supported by the Intramural Research Program of the National Library of Medicine at the U.S. National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Taillon-Miller P, Gu Z, Li Q, Hillier L, Kwok PY. Overlapping genomic sequences: a treasure trove of single-nucleotide polymorphisms. Genome Res. 1998;8:748–54. doi: 10.1101/gr.8.7.748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.De Baets G, Van Durme J, Reumers J, Maurer-Stroh S, Vanhee P, Dopazo J, Schymkowitz J, Rousseau F. SNPeffect 4.0: on-line prediction of molecular and structural effects of protein-coding variants. Nucleic Acids Research. 2012;40:D935–D939. doi: 10.1093/nar/gkr996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sturm RA, Frudakis TN. Eye colour: portals into pigmentation genes and ancestry. Trends Genet. 2004;20:327–32. doi: 10.1016/j.tig.2004.06.010. [DOI] [PubMed] [Google Scholar]
- 4.Fernald GH, Capriotti E, Daneshjou R, Karczewski KJ, Altman RB. Bioinformatics challenges for personalized medicine. Bioinformatics. 2011;27:1741–8. doi: 10.1093/bioinformatics/btr295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mooney S. Bioinformatics approaches and resources for single nucleotide polymorphism functional analysis. Brief Bioinform. 2005;6:44–56. doi: 10.1093/bib/6.1.44. [DOI] [PubMed] [Google Scholar]
- 6.Capriotti E, Calabrese R, Casadio R. Predicting the insurgence of human genetic diseases associated to single point protein mutations with support vector machines and evolutionary information. Bioinformatics. 2006;22:2729–34. doi: 10.1093/bioinformatics/btl423. [DOI] [PubMed] [Google Scholar]
- 7.Collins FS, Guyer MS, Chakravarti A. Variations on a Theme: Cataloging Human DNA Sequence Variation. Science. 1997;278:1580–1581. doi: 10.1126/science.278.5343.1580. [DOI] [PubMed] [Google Scholar]
- 8.Sherry ST, Ward MH, Kholodov M, Baker J, Phan L, Smigielski EM, Sirotkin K. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–11. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Raychaudhuri S. Mapping rare and common causal alleles for complex human diseases. Cell. 2011;147:57–69. doi: 10.1016/j.cell.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schork NJ, Murray SS, Frazer KA, Topol EJ. Common vs. rare allele hypotheses for complex diseases. Curr Opin Genet Dev. 2009;19:212–9. doi: 10.1016/j.gde.2009.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pritchard JK. Are rare variants responsible for susceptibility to complex diseases? Am J Hum Genet. 2001;69:124–37. doi: 10.1086/321272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cohen JC, Kiss RS, Pertsemlidis A, Marcel YL, McPherson R, Hobbs HH. Multiple rare alleles contribute to low plasma levels of HDL cholesterol. Science. 2004;305:869–72. doi: 10.1126/science.1099870. [DOI] [PubMed] [Google Scholar]
- 13.Cohen JC, Pertsemlidis A, Fahmi S, Esmail S, Vega GL, Grundy SM, Hobbs HH. Multiple rare variants in NPC1L1 associated with reduced sterol absorption and plasma low-density lipoprotein levels. Proc Natl Acad Sci U S A. 2006;103:1810–5. doi: 10.1073/pnas.0508483103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nelson MR, Wegmann D, Ehm MG, Kessner D, St Jean P, Verzilli C, Shen J, Tang Z, Bacanu SA, Fraser D, Warren L, Aponte J, Zawistowski M, Liu X, Zhang H, Zhang Y, Li J, Li Y, Li L, Woollard P, Topp S, Hall MD, Nangle K, Wang J, Abecasis G, Cardon LR, Zollner S, Whittaker JC, Chissoe SL, Novembre J, Mooser V. An abundance of rare functional variants in 202 drug target genes sequenced in 14,002 people. Science. 2012;337:100–4. doi: 10.1126/science.1217876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tennessen JA, Bigham AW, O’Connor TD, Fu W, Kenny EE, Gravel S, McGee S, Do R, Liu X, Jun G, Kang HM, Jordan D, Leal SM, Gabriel S, Rieder MJ, Abecasis G, Altshuler D, Nickerson DA, Boerwinkle E, Sunyaev S, Bustamante CD, Bamshad MJ, Akey JM, Broad GO, Seattle GO, Project NES. Evolution and functional impact of rare coding variation from deep sequencing of human exomes. Science. 2012;337:64–9. doi: 10.1126/science.1219240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bodmer W, Bonilla C. Common and rare variants in multifactorial susceptibility to common diseases. Nat Genet. 2008;40:695–701. doi: 10.1038/ng.f.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Veltman JA, Brunner HG. De novo mutations in human genetic disease. Nat Rev Genet. 2012;13:565–75. doi: 10.1038/nrg3241. [DOI] [PubMed] [Google Scholar]
- 18.Ng SB, Bigham AW, Buckingham KJ, Hannibal MC, McMillin MJ, Gildersleeve HI, Beck AE, Tabor HK, Cooper GM, Mefford HC, Lee C, Turner EH, Smith JD, Rieder MJ, Yoshiura K, Matsumoto N, Ohta T, Niikawa N, Nickerson DA, Bamshad MJ, Shendure J. Exome sequencing identifies MLL2 mutations as a cause of Kabuki syndrome. Nat Genet. 2010;42:790–3. doi: 10.1038/ng.646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shen B, Bai J, Vihinen M. Physicochemical feature-based classification of amino acid mutations. Protein Engineering Design and Selection. 2008;21:37–44. doi: 10.1093/protein/gzm084. [DOI] [PubMed] [Google Scholar]
- 20.Kato S, Han SY, Liu W, Otsuka K, Shibata H, Kanamaru R, Ishioka C. Understanding the function–structure and function–mutation relationships of p53 tumor suppressor protein by high-resolution missense mutation analysis. Proceedings of the National Academy of Sciences. 2003;100:8424–8429. doi: 10.1073/pnas.1431692100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Teng S, Michonova-Alexova E, Alexov E. Approaches and resources for prediction of the effects of non-synonymous single nucleotide polymorphism on protein function and interactions. Curr Pharm Biotechnol. 2008;9:123–33. doi: 10.2174/138920108783955164. [DOI] [PubMed] [Google Scholar]
- 22.Zhang Z, Miteva MA, Wang L, Alexov E. Analyzing Effects of Naturally Occurring Missense Mutations. Computational and Mathematical Methods in Medicine. 2012;2012:15. doi: 10.1155/2012/805827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Araya CL, Fowler DM, Chen W, Muniez I, Kelly JW, Fields S. A fundamental protein property, thermodynamic stability, revealed solely from large-scale measurements of protein function. Proceedings of the National Academy of Sciences. 2012;109:16858–16863. doi: 10.1073/pnas.1209751109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Talley K, Alexov E. On the pH-optimum of activity and stability of proteins. Proteins. 2010;78:2699–706. doi: 10.1002/prot.22786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang Z, Wang L, Gao Y, Zhang J, Zhenirovskyy M, Alexov E. Predicting folding free energy changes upon single point mutations. Bioinformatics. 2012;28:664–71. doi: 10.1093/bioinformatics/bts005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Worth CL, Preissner R, Blundell TL. SDM—a server for predicting effects of mutations on protein stability and malfunction. Nucleic Acids Research. 2011;39:W215–W222. doi: 10.1093/nar/gkr363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kang S, Chen G, Xiao G. Robust prediction of mutation-induced protein stability change by property encoding of amino acids. Protein Engineering Design and Selection. 2009;22:75–83. doi: 10.1093/protein/gzn063. [DOI] [PubMed] [Google Scholar]
- 28.Zwerger M, Jaalouk DE, Lombardi ML, Isermann P, Mauermann M, Dialynas G, Herrmann H, Wallrath LL, Lammerding J. Myopathic lamin mutations impair nuclear stability in cells and tissue and disrupt nucleo-cytoskeletal coupling. Human Molecular Genetics. 2013 doi: 10.1093/hmg/ddt079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xu AJ, Springer TA. Mechanisms by which von Willebrand disease mutations destabilize the A2 domain. J Biol Chem. 2013;288:6317–24. doi: 10.1074/jbc.M112.422618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rakoczy EP, Kiel C, McKeone R, Stricher F, Serrano L. Analysis of disease-linked rhodopsin mutations based on structure, function, and protein stability calculations. J Mol Biol. 2011;405:584–606. doi: 10.1016/j.jmb.2010.11.003. [DOI] [PubMed] [Google Scholar]
- 31.An O, Gursoy A, Gurgey A, Keskin O. Structural and functional analysis of perforin mutations in association with clinical data of familial hemophagocytic lymphohistiocytosis type 2 (FHL2) patients. Protein Sci. 2013;22:823–839. doi: 10.1002/pro.2265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Goransdotter Ericson K, Fadeel B, Nilsson-Ardnor S, Soderhall C, Samuelsson A, Janka G, Schneider M, Gurgey A, Yalman N, Revesz T, Egeler R, Jahnukainen K, Storm-Mathiesen I, Haraldsson A, Poole J, de Saint Basile G, Nordenskjold M, Henter J. Spectrum of perforin gene mutations in familial hemophagocytic lymphohistiocytosis. Am J Hum Genet. 2001;68:590–7. doi: 10.1086/318796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Prusiner S. Molecular biology of prion diseases. Science. 1991;252:1515–1522. doi: 10.1126/science.1675487. [DOI] [PubMed] [Google Scholar]
- 34.Collinge J. Prion diseases of humans and animals: their causes and molecular basis. Annu Rev Neurosci. 2001;24:519–50. doi: 10.1146/annurev.neuro.24.1.519. [DOI] [PubMed] [Google Scholar]
- 35.Kretzschmar H, Tatzelt J. Prion disease: a tale of folds and strains. Brain Pathol. 2013;23:321–32. doi: 10.1111/bpa.12045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jetha NN, Semenchenko V, Wishart DS, Cashman NR, Marziali A. Nanopore analysis of wild-type and mutant prion protein (PrP(C)): single molecule discrimination and PrP(C) kinetics. PLoS One. 2013;8:e54982. doi: 10.1371/journal.pone.0054982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lin W, Kang UJ. Characterization of PINK1 processing, stability, and subcellular localization. J Neurochem. 2008;106:464–74. doi: 10.1111/j.1471-4159.2008.05398.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nuytemans K, Theuns J, Cruts M, Van Broeckhoven C. Genetic etiology of Parkinson disease associated with mutations in the SNCA, PARK2, PINK1, PARK7, and LRRK2 genes: a mutation update. Hum Mutat. 2010;31:763–80. doi: 10.1002/humu.21277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Morais VA, Verstreken P, Roethig A, Smet J, Snellinx A, Vanbrabant M, Haddad D, Frezza C, Mandemakers W, Vogt-Weisenhorn D, Van Coster R, Wurst W, Scorrano L, De Strooper B. Parkinson’s disease mutations in PINK1 result in decreased Complex I activity and deficient synaptic function. EMBO Mol Med. 2009;1:99–111. doi: 10.1002/emmm.200900006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Grant MA, Lazo ND, Lomakin A, Condron MM, Arai H, Yamin G, Rigby AC, Teplow DB. Familial Alzheimer’s disease mutations alter the stability of the amyloid beta-protein monomer folding nucleus. Proc Natl Acad Sci U S A. 2007;104:16522–7. doi: 10.1073/pnas.0705197104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stehr H, Jang SH, Duarte JM, Wierling C, Lehrach H, Lappe M, Lange BM. The structural impact of cancer-associated missense mutations in oncogenes and tumor suppressors. Mol Cancer. 2011;10:54. doi: 10.1186/1476-4598-10-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shi Z, Moult J. Structural and functional impact of cancer-related missense somatic mutations. J Mol Biol. 2011;413:495–512. doi: 10.1016/j.jmb.2011.06.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Witham S, Takano K, Schwartz C, Alexov E. A missense mutation in CLIC2 associated with intellectual disability is predicted by in silico modeling to affect protein stability and dynamics. Proteins. 2011;79:2444–54. doi: 10.1002/prot.23065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Takano K, Liu D, Tarpey P, Gallant E, Lam A, Witham S, Alexov E, Chaubey A, Stevenson RE, Schwartz CE, Board PG, Dulhunty AF. An X-linked channelopathy with cardiomegaly due to a CLIC2 mutation enhancing ryanodine receptor channel activity. Hum Mol Genet. 2012;21:4497–507. doi: 10.1093/hmg/dds292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Spector S, Wang M, Carp SA, Robblee J, Hendsch ZS, Fairman R, Tidor B, Raleigh DP. Rational Modification of Protein Stability by the Mutation of Charged Surface Residues†. Biochemistry. 2000;39:872–879. doi: 10.1021/bi992091m. [DOI] [PubMed] [Google Scholar]
- 46.Zhang Z, Norris J, Schwartz C, Alexov E. In silico and in vitro investigations of the mutability of disease-causing missense mutation sites in spermine synthase. PLoS One. 2011;6:e20373. doi: 10.1371/journal.pone.0020373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Teng S, Madej T, Panchenko A, Alexov E. Modeling effects of human single nucleotide polymorphisms on protein-protein interactions. Biophys J. 2009;96:2178–88. doi: 10.1016/j.bpj.2008.12.3904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nishi H, Tyagi M, Teng S, Shoemaker BA, Hashimoto K, Alexov E, Wuchty S, Panchenko AR. Cancer missense mutations alter binding properties of proteins and their interaction networks. PLoS One. 2013;8:e66273. doi: 10.1371/journal.pone.0066273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang Z, Miteva MA, Wang L, Alexov E. Analyzing effects of naturally occurring missense mutations. Comput Math Methods Med. 2012;2012:805827. doi: 10.1155/2012/805827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kang S, Chen G, Xiao G. Robust prediction of mutation-induced protein stability change by property encoding of amino acids. Protein Eng Des Sel. 2009;22:75–83. doi: 10.1093/protein/gzn063. [DOI] [PubMed] [Google Scholar]
- 51.Teng S, Srivastava AK, Wang L. Sequence feature-based prediction of protein stability changes upon amino acid substitutions. BMC Genomics. 2010;11(Suppl 2):S5. doi: 10.1186/1471-2164-11-S2-S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Capriotti E, Fariselli P, Casadio R. I-Mutant2.0: predicting stability changes upon mutation from the protein sequence or structure. Nucleic Acids Research. 2005;33:W306–W310. doi: 10.1093/nar/gki375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cheng J, Randall A, Baldi P. Prediction of protein stability changes for single-site mutations using support vector machines. Proteins: Structure, Function, and Bioinformatics. 2006;62:1125–1132. doi: 10.1002/prot.20810. [DOI] [PubMed] [Google Scholar]
- 54.Dehouck Y, Grosfils A, Folch B, Gilis D, Bogaerts P, Rooman M. Fast and accurate predictions of protein stability changes upon mutations using statistical potentials and neural networks: PoPMuSiC-2.0. Bioinformatics. 2009;25:2537–2543. doi: 10.1093/bioinformatics/btp445. [DOI] [PubMed] [Google Scholar]
- 55.Ng PC, Henikoff S. SIFT: predicting amino acid changes that affect protein function. Nucleic Acids Research. 2003;31:3812–3814. doi: 10.1093/nar/gkg509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ramensky V, Bork P, Sunyaev S. Human non-synonymous SNPs: server and survey. Nucleic Acids Research. 2002;30:3894–3900. doi: 10.1093/nar/gkf493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, Kondrashov AS, Sunyaev SR. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–9. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gonnelli G, Rooman M, Dehouck Y. Structure-based mutant stability predictions on proteins of unknown structure. J Biotechnol. 2012;161:287–93. doi: 10.1016/j.jbiotec.2012.06.020. [DOI] [PubMed] [Google Scholar]
- 59.Parthiban V, Gromiha MM, Schomburg D. CUPSAT: prediction of protein stability upon point mutations. Nucleic Acids Research. 2006;34:W239–W242. doi: 10.1093/nar/gkl190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Worth CL, Preissner R, Blundell TL. SDM--a server for predicting effects of mutations on protein stability and malfunction. Nucleic Acids Res. 2011;39:W215–22. doi: 10.1093/nar/gkr363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Guerois R, Nielsen JE, Serrano L. Predicting Changes in the Stability of Proteins and Protein Complexes: A Study of More Than 1000 Mutations. Journal of Molecular Biology. 2002;320:369–387. doi: 10.1016/S0022-2836(02)00442-4. [DOI] [PubMed] [Google Scholar]
- 62.Ding F, Dokholyan NV. Emergence of Protein Fold Families through Rational Design. PLoS Comput Biol. 2006;2:e85. doi: 10.1371/journal.pcbi.0020085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li B, Krishnan VG, Mort ME, Xin F, Kamati KK, Cooper DN, Mooney SD, Radivojac P. Automated inference of molecular mechanisms of disease from amino acid substitutions. Bioinformatics. 2009;25:2744–2750. doi: 10.1093/bioinformatics/btp528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Benedix A, Becker CM, de Groot BL, Caflisch A, Bockmann RA. Predicting free energy changes using structural ensembles. Nat Methods. 2009;6:3–4. doi: 10.1038/nmeth0109-3. [DOI] [PubMed] [Google Scholar]
- 65.de Groot BL, van Aalten DM, Scheek RM, Amadei A, Vriend G, Berendsen HJ. Prediction of protein conformational freedom from distance constraints. Proteins. 1997;29:240–51. doi: 10.1002/(sici)1097-0134(199710)29:2<240::aid-prot11>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- 66.Li L, Li C, Sarkar S, Zhang J, Witham S, Zhang Z, Wang L, Smith N, Petukh M, Alexov E. DelPhi: a comprehensive suite for DelPhi software and associated resources. BMC Biophys. 2012;5:9. doi: 10.1186/2046-1682-5-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Venselaar H, Te Beek TA, Kuipers RK, Hekkelman ML, Vriend G. Protein structure analysis of mutations causing inheritable diseases. An e-Science approach with life scientist friendly interfaces. BMC Bioinformatics. 2010;11:548. doi: 10.1186/1471-2105-11-548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Khan S, Vihinen M. Performance of protein stability predictors. Hum Mutat. 2010;31:675–84. doi: 10.1002/humu.21242. [DOI] [PubMed] [Google Scholar]
- 69.Masso M, Vaisman II. Accurate prediction of stability changes in protein mutants by combining machine learning with structure based computational mutagenesis. Bioinformatics. 2008;24:2002–2009. doi: 10.1093/bioinformatics/btn353. [DOI] [PubMed] [Google Scholar]
- 70.Potapov V, Cohen M, Schreiber G. Assessing computational methods for predicting protein stability upon mutation: good on average but not in the details. Protein Engineering Design and Selection. 2009;22:553–560. doi: 10.1093/protein/gzp030. [DOI] [PubMed] [Google Scholar]
- 71.Thiltgen G, Goldstein RA. Assessing predictors of changes in protein stability upon mutation using self-consistency. PLoS One. 2012;7:e46084. doi: 10.1371/journal.pone.0046084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Vihinen M. How to evaluate performance of prediction methods? Measures and their interpretation in variation effect analysis. BMC Genomics. 2012;13(Suppl 4):S2. doi: 10.1186/1471-2164-13-S4-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Thusberg J, Olatubosun A, Vihinen M. Performance of mutation pathogenicity prediction methods on missense variants. Hum Mutat. 2011;32:358–68. doi: 10.1002/humu.21445. [DOI] [PubMed] [Google Scholar]
- 74.Zhou H, Zhou Y. Distance-scaled, finite ideal-gas reference state improves structure-derived potentials of mean force for structure selection and stability prediction. Protein Sci. 2002;11:2714–26. doi: 10.1110/ps.0217002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Deutsch C, Krishnamoorthy B. Four-body scoring function for mutagenesis. Bioinformatics. 2007;23:3009–15. doi: 10.1093/bioinformatics/btm481. [DOI] [PubMed] [Google Scholar]
- 76.Dosztanyi Z, Magyar C, Tusnady G, Simon I. SCide: identification of stabilization centers in proteins. Bioinformatics. 2003;19:899–900. doi: 10.1093/bioinformatics/btg110. [DOI] [PubMed] [Google Scholar]
- 77.Kurgan L, Cios K, Chen K. SCPRED: accurate prediction of protein structural class for sequences of twilight-zone similarity with predicting sequences. BMC Bioinformatics. 2008;9:226. doi: 10.1186/1471-2105-9-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Magyar C, Gromiha MM, Pujadas G, Tusnady GE, Simon I. SRide: a server for identifying stabilizing residues in proteins. Nucleic Acids Res. 2005;33:W303–5. doi: 10.1093/nar/gki409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bao L, Zhou M, Cui Y. nsSNPAnalyzer: identifying disease-associated nonsynonymous single nucleotide polymorphisms. Nucleic Acids Res. 2005;33:W480–2. doi: 10.1093/nar/gki372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Thomas PD, Campbell MJ, Kejariwal A, Mi H, Karlak B, Daverman R, Diemer K, Muruganujan A, Narechania A. PANTHER: a library of protein families and subfamilies indexed by function. Genome Res. 2003;13:2129–41. doi: 10.1101/gr.772403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bromberg Y, Rost B. SNAP: predict effect of non-synonymous polymorphisms on function. Nucleic Acids Res. 2007;35:3823–35. doi: 10.1093/nar/gkm238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Calabrese R, Capriotti E, Fariselli P, Martelli PL, Casadio R. Functional annotations improve the predictive score of human disease-related mutations in proteins. Hum Mutat. 2009;30:1237–44. doi: 10.1002/humu.21047. [DOI] [PubMed] [Google Scholar]
- 83.Gromiha MM, Huang LT. Machine learning algorithms for predicting protein folding rates and stability of mutant proteins: comparison with statistical methods. Curr Protein Pept Sci. 2011;12:490–502. doi: 10.2174/138920311796957630. [DOI] [PubMed] [Google Scholar]
- 84.Kumar S, Suleski MP, Markov GJ, Lawrence S, Marco A, Filipski AJ. Positional conservation and amino acids shape the correct diagnosis and population frequencies of benign and damaging personal amino acid mutations. Genome Res. 2009;19:1562–9. doi: 10.1101/gr.091991.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Nisius L, Grzesiek S. Key stabilizing elements of protein structure identified through pressure and temperature perturbation of its hydrogen bond network. Nat Chem. 2012;4:711–7. doi: 10.1038/nchem.1396. [DOI] [PubMed] [Google Scholar]
- 86.Gong S, Worth CL, Bickerton GR, Lee S, Tanramluk D, Blundell TL. Structural and functional restraints in the evolution of protein families and superfamilies. Biochem Soc Trans. 2009;37:727–33. doi: 10.1042/BST0370727. [DOI] [PubMed] [Google Scholar]
- 87.Horowitz S, Trievel RC. Carbon-oxygen hydrogen bonding in biological structure and function. J Biol Chem. 2012;287:41576–82. doi: 10.1074/jbc.R112.418574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Nguyen HH, Hannemann F, Hartmann MF, Malunowicz EM, Wudy SA, Bernhardt R. Five novel mutations in CYP11B2 gene detected in patients with aldosterone synthase deficiency type I: Functional characterization and structural analyses. Mol Genet Metab. 2010;100:357–64. doi: 10.1016/j.ymgme.2010.04.016. [DOI] [PubMed] [Google Scholar]
- 89.Fontaine M, Dessein AF, Douillard C, Dobbelaere D, Brivet M, Boutron A, Zater M, Mention-Mulliez K, Martin-Ponthieu A, Vianey-Saban C, Briand G, Porchet N, Vamecq J. A Novel Mutation in CPT1A Resulting in Hepatic CPT Deficiency. JIMD Rep. 2012;6:7–14. doi: 10.1007/8904_2011_94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zhang Z, Teng S, Wang L, Schwartz CE, Alexov E. Computational analysis of missense mutations causing Snyder-Robinson syndrome. Hum Mutat. 2010;31:1043–9. doi: 10.1002/humu.21310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Schurmann K, Anton M, Ivanov I, Richter C, Kuhn H, Walther M. Molecular basis for the reduced catalytic activity of the naturally occurring T560M mutant of human 12/15-lipoxygenase that has been implicated in coronary artery disease. J Biol Chem. 2011;286:23920–7. doi: 10.1074/jbc.M110.211821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wang S, Zhao WJ, Liu H, Gong H, Yan YB. Increasing betaB1-crystallin sensitivity to proteolysis caused by the congenital cataract-microcornea syndrome mutation S129R. Biochim Biophys Acta. 2013;1832:302–11. doi: 10.1016/j.bbadis.2012.11.005. [DOI] [PubMed] [Google Scholar]
- 93.Gorbenko G, Trusova V. Protein aggregation in a membrane environment. Adv Protein Chem Struct Biol. 2011;84:113–42. doi: 10.1016/B978-0-12-386483-3.00002-1. [DOI] [PubMed] [Google Scholar]
- 94.Goto Y, Yagi H, Yamaguchi K, Chatani E, Ban T. Structure, formation and propagation of amyloid fibrils. Curr Pharm Des. 2008;14:3205–18. doi: 10.2174/138161208786404146. [DOI] [PubMed] [Google Scholar]
- 95.Khare SD, Dokholyan NV. Molecular mechanisms of polypeptide aggregation in human diseases. Curr Protein Pept Sci. 2007;8:573–9. doi: 10.2174/138920307783018703. [DOI] [PubMed] [Google Scholar]
- 96.Naeem A, Fazili NA. Defective protein folding and aggregation as the basis of neurodegenerative diseases: the darker aspect of proteins. Cell Biochem Biophys. 2011;61:237–50. doi: 10.1007/s12013-011-9200-x. [DOI] [PubMed] [Google Scholar]
- 97.Wilbur JD, Hwang PK, Brodsky FM, Fletterick RJ. Accommodation of structural rearrangements in the huntingtin-interacting protein 1 coiled-coil domain. Acta Crystallogr D Biol Crystallogr. 2010;66:314–8. doi: 10.1107/S0907444909054535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Mason AB, Halbrooks PJ, James NG, Byrne SL, Grady JK, Chasteen ND, Bobst CE, Kaltashov IA, Smith VC, MacGillivray RT, Everse SJ. Structural and functional consequences of the substitution of glycine 65 with arginine in the N-lobe of human transferrin. Biochemistry. 2009;48:1945–53. doi: 10.1021/bi802254x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Garcia-Moreno B. Adaptations of proteins to cellular and subcellular pH. J Biol. 2009;8:98. doi: 10.1186/jbiol199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Mitra RC, Zhang Z, Alexov E. In silico modeling of pH-optimum of protein-protein binding. Proteins. 2011;79:925–36. doi: 10.1002/prot.22931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Chan P, Warwicker J. Evidence for the adaptation of protein pH-dependence to subcellular pH. BMC Biol. 2009;7:69. doi: 10.1186/1741-7007-7-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kulkarni MV, Tettamanzi MC, Murphy JW, Keeler C, Myszka DG, Chayen NE, Lolis EJ, Hodsdon ME. Two independent histidines, one in human prolactin and one in its receptor, are critical for pH-dependent receptor recognition and activation. J Biol Chem. 2010;285:38524–33. doi: 10.1074/jbc.M110.172072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Wang L, Witham S, Zhang Z, Li L, Hodsdon M, Alexov E. In Silico Investigation of pH-Dependence of Prolactin and Human Growth Hormone Binding to Human Prolactin Receptor. 2013 doi: 10.4208/cicp.170911.131011s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Reichold M, Zdebik AA, Lieberer E, Rapedius M, Schmidt K, Bandulik S, Sterner C, Tegtmeier I, Penton D, Baukrowitz T, Hulton SA, Witzgall R, Ben-Zeev B, Howie AJ, Kleta R, Bockenhauer D, Warth R. KCNJ10 gene mutations causing EAST syndrome (epilepsy, ataxia, sensorineural deafness, and tubulopathy) disrupt channel function. Proc Natl Acad Sci U S A. 2010;107:14490–5. doi: 10.1073/pnas.1003072107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Hanemann CO, D’Urso D, Gabreels-Festen AA, Muller HW. Mutation-dependent alteration in cellular distribution of peripheral myelin protein 22 in nerve biopsies from Charcot-Marie-Tooth type 1A. Brain. 2000;123 ( Pt 5):1001–6. doi: 10.1093/brain/123.5.1001. [DOI] [PubMed] [Google Scholar]
- 106.Petukh M, Stefl S, Alexov E. The Role of Protonation States in Ligand-Receptor Recognition and Binding. Curr Pharm Des. 2012 doi: 10.2174/1381612811319230004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Nelson MT, Humphrey W, Gursoy A, Dalke A, Kalé LV, Skeel RD, Schulten K. NAMD: a Parallel, Object-Oriented Molecular Dynamics Program. International Journal of High Performance Computing Applications. 1996;10:251–268. [Google Scholar]
- 108.Brooks BR, Bruccoleri RE, Olafson BD, States DJ, Swaminathan S, Karplus M. CHARMM: A program for macromolecular energy, minimization, and dynamics calculations. Journal of Computational Chemistry. 1983;4:187–217. [Google Scholar]
- 109.Case DA, Cheatham TE, 3rd, Darden T, Gohlke H, Luo R, Merz KM, Jr, Onufriev A, Simmerling C, Wang B, Woods RJ. The Amber biomolecular simulation programs. J Comput Chem. 2005;26:1668–88. doi: 10.1002/jcc.20290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Christen M, Hunenberger PH, Bakowies D, Baron R, Burgi R, Geerke DP, Heinz TN, Kastenholz MA, Krautler V, Oostenbrink C, Peter C, Trzesniak D, van Gunsteren WF. The GROMOS software for biomolecular simulation: GROMOS05. J Comput Chem. 2005;26:1719–51. doi: 10.1002/jcc.20303. [DOI] [PubMed] [Google Scholar]
- 111.Berendsen HJC, van der Spoel D, van Drunen R. GROMACS: A message-passing parallel molecular dynamics implementation. Computer Physics Communications. 1995;91:43–56. [Google Scholar]
- 112.Word JM, Lovell SC, Richardson JS, Richardson DC. Asparagine and glutamine: using hydrogen atom contacts in the choice of side-chain amide orientation. Journal of Molecular Biology. 1999;285:1735–1747. doi: 10.1006/jmbi.1998.2401. [DOI] [PubMed] [Google Scholar]
- 113.Dolinsky TJ, Czodrowski P, Li H, Nielsen JE, Jensen JH, Klebe G, Baker NA. PDB2PQR: expanding and upgrading automated preparation of biomolecular structures for molecular simulations. Nucleic Acids Research. 2007;35:W522–W525. doi: 10.1093/nar/gkm276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Durrant JD, McCammon JA. HBonanza: a computer algorithm for molecular-dynamics-trajectory hydrogen-bond analysis. J Mol Graph Model. 2011;31:5–9. doi: 10.1016/j.jmgm.2011.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Murray AE. A routine method for the quantification of physical change in melanocytic naevi using digital image processing. J Audiov Media Med. 1988;11:52–7. doi: 10.3109/17453058809051356. [DOI] [PubMed] [Google Scholar]
- 116.Georgescu RE, Alexov EG, Gunner MR. Combining conformational flexibility and continuum electrostatics for calculating pK(a)s in proteins. Biophys J. 2002;83:1731–48. doi: 10.1016/S0006-3495(02)73940-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Li H, Robertson AD, Jensen JH. Very fast empirical prediction and rationalization of protein pKa values. Proteins. 2005;61:704–21. doi: 10.1002/prot.20660. [DOI] [PubMed] [Google Scholar]
- 118.Gordon JC, Myers JB, Folta T, Shoja V, Heath LS, Onufriev A. H++: a server for estimating pKas and adding missing hydrogens to macromolecules. Nucleic Acids Res. 2005;33:W368–71. doi: 10.1093/nar/gki464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Maier T, Leibundgut M, Boehringer D, Ban N. Structure and function of eukaryotic fatty acid synthases. Q Rev Biophys. 2010;43:373–422. doi: 10.1017/S0033583510000156. [DOI] [PubMed] [Google Scholar]
- 120.Svetlov V, Nudler E. Macromolecular micromovements: how RNA polymerase translocates. Curr Opin Struct Biol. 2009;19:701–7. doi: 10.1016/j.sbi.2009.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Kokkinidis M, Glykos NM, Fadouloglou VE. Protein flexibility and enzymatic catalysis. Adv Protein Chem Struct Biol. 2012;87:181–218. doi: 10.1016/B978-0-12-398312-1.00007-X. [DOI] [PubMed] [Google Scholar]
- 122.Chow MK, Lomas DA, Bottomley SP. Promiscuous beta-strand interactions and the conformational diseases. Curr Med Chem. 2004;11:491–9. doi: 10.2174/0929867043455936. [DOI] [PubMed] [Google Scholar]
- 123.Oldfield CJ, Xue B, Van YY, Ulrich EL, Markley JL, Dunker AK, Uversky VN. Utilization of protein intrinsic disorder knowledge in structural proteomics. Biochim Biophys Acta. 2013;1834:487–98. doi: 10.1016/j.bbapap.2012.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Xue B, Dunker AK, Uversky VN. Orderly order in protein intrinsic disorder distribution: disorder in 3500 proteomes from viruses and the three domains of life. J Biomol Struct Dyn. 2012;30:137–49. doi: 10.1080/07391102.2012.675145. [DOI] [PubMed] [Google Scholar]
- 125.Li M, Shoemaker BA, Thangudu RR, Ferraris JD, Burg MB, Panchenko AR. Mutations in DNA-Binding Loop of NFAT5 Transcription Factor Produce Unique Outcomes on Protein-DNA Binding and Dynamics. J Phys Chem B. doi: 10.1021/jp403310a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Ponder J, Richards F. TINKER molecular modeling package. J Comput Chem. 1987;8:1016–1024. [Google Scholar]
- 127.Kumar A, Purohit R. Computational investigation of pathogenic nsSNPs in CEP63 protein. Gene. 2012;503:75–82. doi: 10.1016/j.gene.2012.04.032. [DOI] [PubMed] [Google Scholar]
- 128.George Priya Doss C, Rajith B. A New Insight into Structural and Functional Impact of Single-Nucleotide Polymorphisms in PTEN Gene. Cell Biochem Biophys. 2012 doi: 10.1007/s12013-012-9472-9. [DOI] [PubMed] [Google Scholar]
- 129.Nyon MP, Segu L, Cabrita LD, Levy GR, Kirkpatrick J, Roussel BD, Patschull AO, Barrett TE, Ekeowa UI, Kerr R, Waudby CA, Kalsheker N, Hill M, Thalassinos K, Lomas DA, Christodoulou J, Gooptu B. Structural dynamics associated with intermediate formation in an archetypal conformational disease. Structure. 2012;20:504–12. doi: 10.1016/j.str.2012.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Wang B, Wen X, Qin X, Wang Z, Tan Y, Shen Y, Xi Z. Quantitative Structural Insight into Human Variegate Porphyria Disease. J Biol Chem. 2013;17:11731–40. doi: 10.1074/jbc.M113.459768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Nicchia GP, Ficarella R, Rossi A, Giangreco I, Nicolotti O, Carotti A, Pisani F, Estivill X, Gasparini P, Svelto M, Frigeri A. D184E mutation in aquaporin-4 gene impairs water permeability and links to deafness. Neuroscience. 2011;197:80–8. doi: 10.1016/j.neuroscience.2011.09.023. [DOI] [PubMed] [Google Scholar]
- 132.Iakoucheva LM, Brown CJ, Lawson JD, Obradovic Z, Dunker AK. Intrinsic disorder in cell-signaling and cancer-associated proteins. J Mol Biol. 2002;323:573–84. doi: 10.1016/s0022-2836(02)00969-5. [DOI] [PubMed] [Google Scholar]
- 133.Uversky VN, Oldfield CJ, Midic U, Xie H, Xue B, Vucetic S, Iakoucheva LM, Obradovic Z, Dunker AK. Unfoldomics of human diseases: linking protein intrinsic disorder with diseases. BMC Genomics. 2009; 10(Suppl 1):S7. doi: 10.1186/1471-2164-10-S1-S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Bugg CW, Isas JM, Fischer T, Patterson PH, Langen R. Structural features and domain organization of huntingtin fibrils. J Biol Chem. 2012;287:31739–46. doi: 10.1074/jbc.M112.353839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Hu Y, Liu Y, Jung J, Dunker AK, Wang Y. Changes in predicted protein disorder tendency may contribute to disease risk. BMC Genomics. 2011;12(Suppl 5):S2. doi: 10.1186/1471-2164-12-S5-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Vacic V, Iakoucheva LM. Disease mutations in disordered regions--exception to the rule? Mol Biosyst. 2012;8:27–32. doi: 10.1039/c1mb05251a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Sakakura M, Hadziselimovic A, Wang Z, Schey KL, Sanders CR. Structural basis for the Trembler-J phenotype of Charcot-Marie-Tooth disease. Structure. 2011;19:1160–9. doi: 10.1016/j.str.2011.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.van der Kamp MW, Daggett V. Pathogenic mutations in the hydrophobic core of the human prion protein can promote structural instability and misfolding. J Mol Biol. 2010;404:732–48. doi: 10.1016/j.jmb.2010.09.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Di Marino D, Achsel T, Lacoux C, Falconi M, Bagni C. Molecular dynamics simulations show how the FMRP Ile304Asn mutation destabilizes the KH2 domain structure and affects its function. J Biomol Struct Dyn. 2013 doi: 10.1080/07391102.2013.768552. [DOI] [PubMed] [Google Scholar]
- 140.Kass I, Knaupp AS, Bottomley SP, Buckle AM. Conformational properties of the disease-causing Z variant of alpha1-antitrypsin revealed by theory and experiment. Biophys J. 2012;102:2856–65. doi: 10.1016/j.bpj.2012.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Jahandideh S, Zhi D. Systematic investigation of predicted effect of nonsynonymous SNPs in human prion protein gene: a molecular modeling and molecular dynamics study. J Biomol Struct Dyn. 2013 doi: 10.1080/07391102.2012.763216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Tokuriki N, Stricher F, Serrano L, Tawfik DS. How protein stability and new functions trade off. PLoS Comput Biol. 2008;4:e1000002. doi: 10.1371/journal.pcbi.1000002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Dessailly BH, Lensink MF, Wodak SJ. Relating destabilizing regions to known functional sites in proteins. BMC Bioinformatics. 2007;8:141. doi: 10.1186/1471-2105-8-141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Elcock AH. Prediction of functionally important residues based solely on the computed energetics of protein structure. J Mol Biol. 2001;312:885–96. doi: 10.1006/jmbi.2001.5009. [DOI] [PubMed] [Google Scholar]
- 145.Studer RA, Dessailly BH, Orengo CA. Residue mutations and their impact on protein structure and function: detecting beneficial and pathogenic changes. Biochem J. 2013;449:581–94. doi: 10.1042/BJ20121221. [DOI] [PubMed] [Google Scholar]
- 146.Meiering EM, Serrano L, Fersht AR. Effect of active site residues in barnase on activity and stability. J Mol Biol. 1992;225:585–9. doi: 10.1016/0022-2836(92)90387-y. [DOI] [PubMed] [Google Scholar]
- 147.Shoichet BK, Baase WA, Kuroki R, Matthews BW. A relationship between protein stability and protein function. Proc Natl Acad Sci U S A. 1995;92:452–6. doi: 10.1073/pnas.92.2.452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Pazos F, Sternberg MJ. Automated prediction of protein function and detection of functional sites from structure. Proc Natl Acad Sci U S A. 2004;101:14754–9. doi: 10.1073/pnas.0404569101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Lopez G, Valencia A, Tress ML. firestar--prediction of functionally important residues using structural templates and alignment reliability. Nucleic Acids Res. 2007;35:W573–7. doi: 10.1093/nar/gkm297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Shoemaker BA, Zhang D, Tyagi M, Thangudu RR, Fong JH, Marchler-Bauer A, Bryant SH, Madej T, Panchenko AR. IBIS (Inferred Biomolecular Interaction Server) reports, predicts and integrates multiple types of conserved interactions for proteins. Nucleic Acids Res. 2012;40:D834–40. doi: 10.1093/nar/gkr997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Capra JA, Laskowski RA, Thornton JM, Singh M, Funkhouser TA. Predicting protein ligand binding sites by combining evolutionary sequence conservation and 3D structure. PLoS Comput Biol. 2009;5:e1000585. doi: 10.1371/journal.pcbi.1000585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Zhang Z, Zheng Y, Petukh M, Pegg A, Ikeguchi Y, Alexov E. Enhancing human spermine synthase activity by engineered mutations. PLoS Comput Biol. 2013;9:e1002924. doi: 10.1371/journal.pcbi.1002924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Martin GS. The road to Src. Oncogene. 2004;23:7910–7. doi: 10.1038/sj.onc.1208077. [DOI] [PubMed] [Google Scholar]
- 154.Izarzugaza JM, Redfern OC, Orengo CA, Valencia A. Cancer-associated mutations are preferentially distributed in protein kinase functional sites. Proteins. 2009;77:892–903. doi: 10.1002/prot.22512. [DOI] [PubMed] [Google Scholar]
- 155.Shan Y, Eastwood MP, Zhang X, Kim ET, Arkhipov A, Dror RO, Jumper J, Kuriyan J, Shaw DE. Oncogenic mutations counteract intrinsic disorder in the EGFR kinase and promote receptor dimerization. Cell. 2012;149:860–70. doi: 10.1016/j.cell.2012.02.063. [DOI] [PubMed] [Google Scholar]
- 156.Yun CH, Boggon TJ, Li Y, Woo MS, Greulich H, Meyerson M, Eck MJ. Structures of lung cancer-derived EGFR mutants and inhibitor complexes: mechanism of activation and insights into differential inhibitor sensitivity. Cancer Cell. 2007;11:217–27. doi: 10.1016/j.ccr.2006.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Zhang X, Gureasko J, Shen K, Cole PA, Kuriyan J. An allosteric mechanism for activation of the kinase domain of epidermal growth factor receptor. Cell. 2006;125:1137–49. doi: 10.1016/j.cell.2006.05.013. [DOI] [PubMed] [Google Scholar]
- 158.Yun CH, Mengwasser KE, Toms AV, Woo MS, Greulich H, Wong KK, Meyerson M, Eck MJ. The T790M mutation in EGFR kinase causes drug resistance by increasing the affinity for ATP. Proc Natl Acad Sci U S A. 2008;105:2070–5. doi: 10.1073/pnas.0709662105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Dixit A, Verkhivker GM. Hierarchical modeling of activation mechanisms in the ABL and EGFR kinase domains: thermodynamic and mechanistic catalysts of kinase activation by cancer mutations. PLoS Comput Biol. 2009;5:e1000487. doi: 10.1371/journal.pcbi.1000487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Hashimoto K, Rogozin IB, Panchenko AR. Oncogenic potential is related to activating effect of cancer single and double somatic mutations in receptor tyrosine kinases. Hum Mutat. 2012;33:1566–75. doi: 10.1002/humu.22145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Colgin LM, Hackmann AF, Emond MJ, Monnat RJ., Jr The unexpected landscape of in vivo somatic mutation in a human epithelial cell lineage. Proc Natl Acad Sci U S A. 2002;99:1437–42. doi: 10.1073/pnas.032655699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Chen JM, Ferec C, Cooper DN. Closely spaced multiple mutations as potential signatures of transient hypermutability in human genes. Hum Mutat. 2009;30:1435–48. doi: 10.1002/humu.21088. [DOI] [PubMed] [Google Scholar]
- 163.Chen Z, Feng J, Buzin CH, Sommer SS. Epidemiology of doublet/multiplet mutations in lung cancers: evidence that a subset arises by chronocoordinate events. PLoS One. 2008;3:e3714. doi: 10.1371/journal.pone.0003714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Meng L, Lin L, Zhang H, Nassiri M, Morales AR, Nadji M. Multiple mutations of the p53 gene in human mammary carcinoma. Mutat Res. 1999;435:263–9. doi: 10.1016/s0921-8777(99)00053-1. [DOI] [PubMed] [Google Scholar]
- 165.Haass C, Lemere CA, Capell A, Citron M, Seubert P, Schenk D, Lannfelt L, Selkoe DJ. The Swedish mutation causes early-onset Alzheimer’s disease by beta-secretase cleavage within the secretory pathway. Nat Med. 1995;1:1291–6. doi: 10.1038/nm1295-1291. [DOI] [PubMed] [Google Scholar]
- 166.Yasuda M, Shabbeer J, Benson SD, Maire I, Burnett RM, Desnick RJ. Fabry disease: characterization of alpha-galactosidase A double mutations and the D313Y plasma enzyme pseudodeficiency allele. Hum Mutat. 2003;22:486–92. doi: 10.1002/humu.10275. [DOI] [PubMed] [Google Scholar]
- 167.Gong S, Blundell TL. Structural and functional restraints on the occurrence of single amino acid variations in human proteins. PLoS One. 2010;5:e9186. doi: 10.1371/journal.pone.0009186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Zhang Z, Norris J, Kalscheuer V, Wood T, Wang L, Schwartz C, Alexov E, Van Esch H. A Y328C missense mutation in spermine synthase causes a mild form of Snyder-Robinson syndrome. Hum Mol Genet. 2013 doi: 10.1093/hmg/ddt229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Pandit RA, Chen CJ, Butt TA, Islam N. Identification and analysis of a novel mutation in PEPD gene in two Kashmiri siblings with prolidase enzyme deficiency. Gene. 2013;516:316–9. doi: 10.1016/j.gene.2012.12.070. [DOI] [PubMed] [Google Scholar]
- 170.Attanasio F, Convertino M, Magno A, Caflisch A, Corazza A, Haridas H, Esposito G, Cataldo S, Pignataro B, Milardi D, Rizzarelli E. Carnosine Inhibits Abeta42 Aggregation by Perturbing the H-Bond Network in and around the Central Hydrophobic Cluster. Chembiochem. 2013;14:583–92. doi: 10.1002/cbic.201200704. [DOI] [PubMed] [Google Scholar]
- 171.Murray MM, Krone MG, Bernstein SL, Baumketner A, Condron MM, Lazo ND, Teplow DB, Wyttenbach T, Shea JE, Bowers MT. Amyloid beta-protein: experiment and theory on the 21–30 fragment. J Phys Chem B. 2009;113:6041–6. doi: 10.1021/jp808384x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Mizuguchi M, Yokoyama T, Nabeshima Y, Kawano K, Tanaka I, Niimura N. Quaternary structure, aggregation and cytotoxicity of transthyretin. Amyloid. 2012;19(Suppl 1):5–7. doi: 10.3109/13506129.2012.666510. [DOI] [PubMed] [Google Scholar]
- 173.Liou B, Kazimierczuk A, Zhang M, Scott CR, Hegde RS, Grabowski GA. Analyses of Variant Acid β-Glucosidases: EFFECTS OF GAUCHER DISEASE MUTATIONS. Journal of Biological Chemistry. 2006;281:4242–4253. doi: 10.1074/jbc.M511110200. [DOI] [PubMed] [Google Scholar]
- 174.Bal NC, Sharon A, Gupta SC, Jena N, Shaikh S, Gyorke S, Periasamy M. The catecholaminergic polymorphic ventricular tachycardia mutation R33Q disrupts the N-terminal structural motif that regulates reversible calsequestrin polymerization. J Biol Chem. 2010;285:17188–96. doi: 10.1074/jbc.M109.096354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Kumar A, Purohit R. Computational screening and molecular dynamics simulation of disease associated nsSNPs in CENP-E. Mutat Res. 2012;738–739:28–37. doi: 10.1016/j.mrfmmm.2012.08.005. [DOI] [PubMed] [Google Scholar]
- 176.Vieyra D, Senger DL, Toyama T, Muzik H, Brasher PM, Johnston RN, Riabowol K, Forsyth PA. Altered subcellular localization and low frequency of mutations of ING1 in human brain tumors. Clin Cancer Res. 2003;9:5952–61. [PubMed] [Google Scholar]
- 177.Kunishima S, Matsushita T, Kojima T, Sako M, Kimura F, Jo EK, Inoue C, Kamiya T, Saito H. Immunofluorescence analysis of neutrophil nonmuscle myosin heavy chain-A in MYH9 disorders: association of subcellular localization with MYH9 mutations. Lab Invest. 2003;83:115–22. doi: 10.1097/01.lab.0000050960.48774.17. [DOI] [PubMed] [Google Scholar]
- 178.Yokoyama T, Mizuguchi M, Nabeshima Y, Kusaka K, Yamada T, Hosoya T, Ohhara T, Kurihara K, Tomoyori K, Tanaka I, Niimura N. Hydrogen-bond network and pH sensitivity in transthyretin: Neutron crystal structure of human transthyretin. J Struct Biol. 2012;177:283–90. doi: 10.1016/j.jsb.2011.12.022. [DOI] [PubMed] [Google Scholar]
- 179.Garai K, Baban B, Frieden C. Self-association and stability of the ApoE isoforms at low pH: implications for ApoE-lipid interactions. Biochemistry. 2011;50:6356–64. doi: 10.1021/bi2006702. [DOI] [PubMed] [Google Scholar]
- 180.Imbard A, Boutron A, Vequaud C, Zater M, de Lonlay P, de Baulny HO, Barnerias C, Mine M, Marsac C, Saudubray JM, Brivet M. Molecular characterization of 82 patients with pyruvate dehydrogenase complex deficiency. Structural implications of novel amino acid substitutions in E1 protein. Mol Genet Metab. 2011;104:507–16. doi: 10.1016/j.ymgme.2011.08.008. [DOI] [PubMed] [Google Scholar]
- 181.Smith AT, Su Y, Stevens DJ, Majtan T, Kraus JP, Burstyn JN. Effect of the disease-causing R266K mutation on the heme and PLP environments of human cystathionine beta-synthase. Biochemistry. 2012;51:6360–70. doi: 10.1021/bi300421z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Padhi AK, Kumar H, Vasaikar SV, Jayaram B, Gomes J. Mechanisms of loss of functions of human angiogenin variants implicated in amyotrophic lateral sclerosis. PLoS One. 2012;7:e32479. doi: 10.1371/journal.pone.0032479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Musayev FN, Di Salvo ML, Saavedra MA, Contestabile R, Ghatge MS, Haynes A, Schirch V, Safo MK. Molecular basis of reduced pyridoxine 5′-phosphate oxidase catalytic activity in neonatal epileptic encephalopathy disorder. J Biol Chem. 2009;284:30949–56. doi: 10.1074/jbc.M109.038372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Wang D, Robertson IM, Li MX, McCully ME, Crane ML, Luo Z, Tu AY, Daggett V, Sykes BD, Regnier M. Structural and functional consequences of the cardiac troponin C L48Q Ca(2+)-sensitizing mutation. Biochemistry. 2012;51:4473–87. doi: 10.1021/bi3003007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Kumar A, Rajendran V, Sethumadhavan R, Purohit R. Evidence of Colorectal Cancer-Associated Mutation in MCAK: A Computational Report. Cell Biochem Biophys. 2013 doi: 10.1007/s12013-013-9572-1. [DOI] [PubMed] [Google Scholar]
- 186.Tang M, Facchiano A, Rachamadugu R, Calderon F, Mao R, Milanesi L, Marabotti A, Lai K. Correlation assessment among clinical phenotypes, expression analysis and molecular modeling of 14 novel variations in the human galactose-1-phosphate uridylyltransferase gene. Hum Mutat. 2012;33:1107–15. doi: 10.1002/humu.22093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Acsadi G, Moore SA, Cheron A, Delalande O, Bennett L, Kupsky W, El-Baba M, Le Rumeur E, Hubert JF. Novel mutation in spectrin-like repeat 1 of dystrophin central domain causes protein misfolding and mild Becker muscular dystrophy. J Biol Chem. 2012;287:18153–62. doi: 10.1074/jbc.M111.284521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Uversky VN, Iakoucheva LM, Dunker AK. eLS. John Wiley & Sons, Ltd; 2001. Protein Disorder and Human Genetic Disease. [Google Scholar]
- 189.Doring G. Serine proteinase inhibitor therapy in alpha(1)-antitrypsin inhibitor deficiency and cystic fibrosis. Pediatr Pulmonol. 1999;28:363–75. doi: 10.1002/(sici)1099-0496(199911)28:5<363::aid-ppul9>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- 190.Zhang Z, Liu M, Li B, Wang Y, Yue J, Liang L, Sun J. Exploring the mechanism of a regulatory SNP of KLK3 by molecular dynamics simulation. J Biomol Struct Dyn. 2013;31:426–40. doi: 10.1080/07391102.2012.703067. [DOI] [PubMed] [Google Scholar]
- 191.Kote-Jarai Z, Amin Al Olama A, Leongamornlert D, Tymrakiewicz M, Saunders E, Guy M, Giles GG, Severi G, Southey M, Hopper JL, Sit KC, Harris JM, Batra J, Spurdle AB, Clements JA, Hamdy F, Neal D, Donovan J, Muir K, Pharoah PD, Chanock SJ, Brown N, Benlloch S, Castro E, Mahmud N, O’Brien L, Hall A, Sawyer E, Wilkinson R, Easton DF, Eeles RA. Identification of a novel prostate cancer susceptibility variant in the KLK3 gene transcript. Hum Genet. 2011;129:687–94. doi: 10.1007/s00439-011-0981-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.O’Mara TA, Nagle CM, Batra J, Kedda MA, Clements JA, Spurdle AB. Kallikrein-related peptidase 3 (KLK3/PSA) single nucleotide polymorphisms and ovarian cancer survival. Twin Res Hum Genet. 2011;14:323–7. doi: 10.1375/twin.14.4.323. [DOI] [PubMed] [Google Scholar]
- 193.Singaraja RR, Visscher H, James ER, Chroni A, Coutinho JM, Brunham LR, Kang MH, Zannis VI, Chimini G, Hayden MR. Specific mutations in ABCA1 have discrete effects on ABCA1 function and lipid phenotypes both in vivo and in vitro. Circ Res. 2006;99:389–97. doi: 10.1161/01.RES.0000237920.70451.ad. [DOI] [PubMed] [Google Scholar]
- 194.Nagao K, Zhao Y, Takahashi K, Kimura Y, Ueda K. Sodium taurocholate-dependent lipid efflux by ABCA1: effects of W590S mutation on lipid translocation and apolipoprotein A-I dissociation. J Lipid Res. 2009;50:1165–72. doi: 10.1194/jlr.M800597-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Wang LJ, Wang J, Li N, Ge L, Li BL, Song BL. Molecular characterization of the NPC1L1 variants identified from cholesterol low absorbers. J Biol Chem. 2011;286:7397–408. doi: 10.1074/jbc.M110.178368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.De Jong JC, Van Der Vliet WA, Van Den Heuvel LP, Willems PH, Knoers NV, Bindels RJ. Functional expression of mutations in the human NaCl cotransporter: evidence for impaired routing mechanisms in Gitelman’s syndrome. J Am Soc Nephrol. 2002;13:1442–8. doi: 10.1097/01.asn.0000017904.77985.03. [DOI] [PubMed] [Google Scholar]