Abstract
Endoglin (ENG) is a 180-kDa transmembrane glycoprotein that functions as a component of the transforming growth factor-β receptor complex. Recently, ENG promoter hypermethylation was reported in several human cancers. We examined ENG promoter hypermethylation using real-time quantitative methylation-specific PCR in 260 human esophageal tissues. ENG hypermethylation showed highly discriminative receiver-operator characteristic curve profiles, clearly distinguishing esophageal squamous cell carcinoma (ESCC) and esophageal adenocarcinoma (EAC) from normal esophagus (N) (p<0.01). Interestingly, ENG normalized methylation values were significantly higher in ESCC than in N (p<0.01) or EAC (p<0.01). ENG hypermethylation frequency was 46.2% in ESCC and 11.9% in N, but increased early and sequentially during EAC-associated neoplastic progression, to 13.3% in Barrett’s metaplasia (BE), 25% in dysplastic BE (D), and 26.9% in frank EAC. ENG hypermethylation was significantly higher in N from ESCC patients (mean = 0.0186) than in N from EAC patients (mean = 0.0117; p < 0.05). Treatment of KYSE220 ESCC cells with the demethylating agent, 5-aza-2′-deoxycytidine, reversed ENG methylation and reactivated ENG mRNA expression. We conclude that promoter hypermethylation of ENG is a frequent, tissue-specific event in human ESCC and exhibits a field defect with promising biomarker potential for the early detection of ESCC. In addition, ENG hypermethylation occurs in a subset of human EAC, and early during BE-associated esophageal neoplastic progression.
Introduction
Esophageal cancer ranks as the eighth most common cancer worldwide, with 482,000 new cases in 2008, and is the sixth most common cause of cancer death with 407,000 deaths1. This malignancy exists in two principal forms, each possessing distinct pathological characteristics: esophageal squamous cell carcinoma (ESCC), which occurs at high frequencies in many developing countries, especially Asia, and including China2; and esophageal adenocarcinoma (EAC), which is more prevalent in Western countries. These aggressive malignancies commonly present as locally advanced disease with a very poor prognosis (i.e., with approximately 17% 5-year survival)3, although significant advances have occurred in treatment. In order to improve outcome, it will be vitally important to discover early events that can serve as detection biomarkers or targets for chemoprevention and therapy.
Endoglin/CD105 (ENG) is a 180-kDa transmembrane glycoprotein that functions as a component of the transforming growth factor-β receptor complex 4, 5. ENG is expressed predominantly in proliferating vascular endothelial cells, where it plays a critical role in vascular remodeling and angiogenesis 6–10. Germline mutations in the ENG gene can lead to an autosomal dominant vascular dysplasia, hereditary hemorrhagic telangiectasia type 1 syndrome 7, 11. Its critical role in angiogenesis has prompted investigators to evaluate the role of ENG in cancer progression and metastasis. Intratumour microvessel density assessed by ENG staining strongly correlates with prognosis in different cancer patients 9, 10, 12, 13. Although ENG hypermethylation has been reported in human lung, colonrectal and breast cancers 14–16, there has been only one study that evaluated ENG hypermethylation in 2 ESCC patients and 16 ESCC cell lines 17. Therefore, to further investigate ENG hypermethylation in human esophageal carcinogenesis, we investigated whether and at which neoplastic stage promoter hypermethylation of ENG is involved in human esophageal carcinogenesis, using real-time quantitative methylation-specific PCR (qMSP) analyses of 260 endoscopic esophageal biopsy specimens of differing histologies. We also evaluated the effect of the demethylating agent, 5-aza-2′-deoxycytidine (5-Aza-dC), on reactivation of epigenetically silenced ENG in esophageal cancer cells. Our results establish that promoter hypermethylation of ENG is a common event in ESCC but not in EAC and occurs early during Barrett’s-associated esophageal neoplastic progression.
Materials and Methods
Tissue Samples
The 260 specimens examined in the current study comprised 67 normal esophageal specimens {N, including 19 obtained from non-Barrett’s/non-esophageal cancer patients (NE), 20 from ESCC patients (NEcS), and 28 from EAC patients (NEcA)}, 60 non-dysplastic Barrett’s metaplasias (BE), 40 dysplastic Barrett’s specimens (D), 67 EACs, and 26 ESCCs. All patients provided written informed consent under a protocol approved by the Institutional Review Boards at the University of Maryland and Baltimore Veterans Affairs Medical Centers, where all esophagogastroduodenoscopies were performed. Biopsies were taken using a standardized biopsy protocol, as previously described 18. Research tissues were obtained from grossly apparent Barrett’s epithelium or from mass lesions in patients manifesting these changes at endoscopic examination, and histology was confirmed using parallel aliquots taken from identical locations at endoscopy. All biopsy specimens were stored in liquid nitrogen prior to DNA extraction.
Cell Lines
The KYSE220 ESCC cell line was obtained from collaborators at Toyama University (Prof. Yutaka Shimada) and cultured in 47.5% RPMI 1640, 47.5% F-12 supplemented with 5% fetal bovine serum.
DNA and RNA Extraction
Genomic DNA and total RNA were extracted from biopsies and cultured cells using DNeasy Tissue Kits (Qiagen, Valencia, CA) and TRIzol reagent (Invitrogen, Carlsbad, CA), respectively. DNAs and RNAs were stored at −80°C before analysis.
Bisulfite Treatment and Real-Time Methylation-Specific PCR
DNA was treated with bisulfite to convert unmethylated cytosines to uracils prior to qMSP, as described previously15. Promoter methylation levels of ENG were determined by real-time qMSP with the ABI 7700 Sequence Detection System (Applied Biosystems, Foster City, CA), using primers and probes as described previously 15. Normalized methylation value (NMV) was defined as follows: NMV = (ENG-S/ENG-FM)/(ACTB-S/ACTB-FM), where ENG-S and ENG-FM represent ENG methylation levels in sample and fully methylated DNAs, respectively, while ACTB-S and ACTB-FM correspond to β-actin in sample and fully methylated DNAs, respectively.
Real-Time Quantitiative RT-PCR
To determine ENG mRNA levels, one-step real-time quantitative RT-PCR was performed using a Qiagen QuantiTect Probe RT-PCR Kit (Qiagen, Hilden, Germany) and the ABI 7700 Sequence Detection System (Applied Biosystems, Foster City, CA). β-actin was used for normalization of data. Primers and probe for ENG and β-actin were the same as previously reported (2006a). A standard curve was generated using serial dilutions of qPCR Reference Total RNA (Clontech, Mountainview, CA). Normalized mRNA value (NRV) was calculated according to the following formula for relative expression of target mRNA: NRV = (ENG-S/ENG-C)/(ACTB-S/ACTB-C, where ENG-S and ENG-C represent levels of mRNA expression for ENG in sample and control mRNAs, respectively, while ACTB-S and ACTB-C correspond to amplified ACTB levels in sample and control mRNAs, respectively.
5-Aza-dC Treatment of Esophageal Cancer Cell Lines
To determine whether ENG inactivation was due to promoter hypermethylation in esophageal cancer, KYSE220 cells were subjected to 5-Aza-dC (Sigma, St. Louis, MO) treatment as previously described19, 20. Briefly, 1 × 105 cells/ml were seeded onto a 100 mm dish and grown for 24 h. Then, 1 μl of 5mM 5-Aza-dC per ml of cells was added every 24 hours for 4 days. DNAs and RNAs were harvested on day 4.
Data Analysis and Statistics
Receiver-operator characteristic (ROC) curve analysis (1982z) was performed using NMVs for the 67 EAC, 26 ESCC and 67 N specimens by Analyse-it© software (Version 1.71, Analyse-it Software, Leeds, UK). With this approach, the area under the ROC curve (AUROC) identified optimal sensitivity and specificity levels at which to distinguish normal from malignant esophageal tissues, yielding corresponding NMV thresholds defining methylation status of ENG. The threshold NMV value determined from this ROC curve was applied to determine the status of ENG methylation in all tissue types included in the present study. For all other statistical tests, Statistica (version 6.1; StatSoft, Inc., Tulsa, OK) was used. Differences with p<0.05 were considered significant.
Results
ENG Promoter Hypermethylation in Esophageal Tissues
Promoter hypermethylation of ENG was analyzed in 67 N (including 19 NE, 20 NEcS and 28 NEcA), 60 BE, 40 D, 67 EAC, and 26 ESCC samples. ENG promoter hypermethylation showed highly discriminative ROC curve profiles and AUROCs, clearly distinguishing ESCC from both N and EAC (p<0.01 and p<0.01, respectively; Figure 1A and 1B), as well as NEcS from NEcA (p<0.01; Figure 1C), but not EAC from N (data not shown).
Figure 1. Receiver-operator characteristic (ROC) curve analysis of normalized methylation value (NMV).
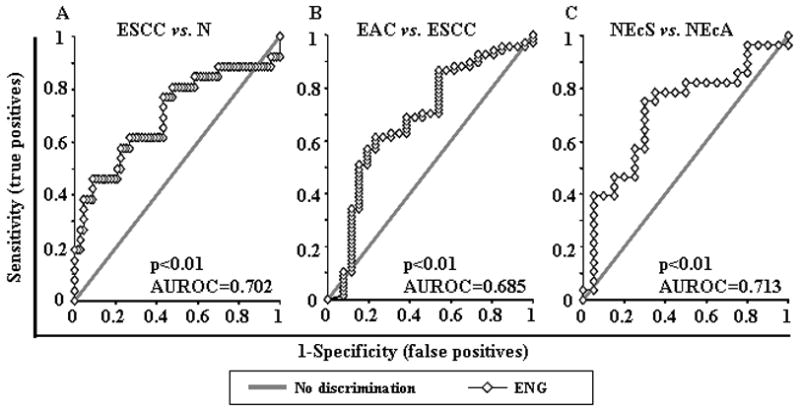
ROC curve analysis of ENG NMVs of normal esophagus (N) vs. esophageal squamous cell carcinoma (ESCC) (A), ESCC vs. esophageal adenocarcinoma (EAC) (B) and normal esophageal specimens from ESCC patients (NEcS) vs. normal esophageal specimens from EAC patients (NEcA) (C). The area under the ROC curve (AUROC) conveys this biomarker’s accuracy in distinguishing EAC from N and from ESCC in terms of its sensitivity and specificity.
The cutoff NMV for ENG (0.02) was identified from ROC curves (ESCC vs. N) as maximizing both sensitivity and specificity. Mean NMV and frequency of ENG hypermethylation for each tissue type are shown in Table 1. NMVs of ENG were significantly higher in ESCC than in N or EAC (p<0.01, Mann-Whitney U test; Table 1). Moreover, NMV of ENG was significantly higher in NEcS (mean = 0.0186) than in NEcA (mean = 0.0115 and p < 0.05, Mann-Whitney U test; Table 1). Similarly, the frequency of ENG hypermethylation was significantly higher in ESCC than in N (46.2% vs. 11.9%, p < 0.001), and was sequentially increased in BE (13.3%), D (25%), and EAC (26.9%) vs. N (11.9%; p > 0.05, p > 0.05 and p < 0.05, respectively, Chi-square for independence test). ENG hypermethylation frequency was higher in ESCC than in EAC, although these differences did not achieve statistical significance (46.2% vs. 26.9%, p = 0.074). There was no significant different for ENG hypermethylation by either NEcS vs. NE, or NEcA vs. NE. In the current study, the mean NMV in NEcS was not signigicantly higher than in NE. This finding could have resulted from differences in sample sizes between these two groups.
Table 1.
Methylation status of ENG in human esophageal tissues
| Histology | Number of samples | Age (year) mean | NMV
|
Methylation Status (cutoff 0.02)
|
||||
|---|---|---|---|---|---|---|---|---|
| mean | p$ | Frequency | UM | M | p | |||
| Normal esophagus | 67 | 64.4 | 0.0137 | 11.9% | 59 | 8 | ||
| NE | 19 | 64.1 | 0.0117 | 5.3% | 18 | 1 | ||
| NEcA | 28 | 66.9 | 0.0115 | > 0.05¶/< 0.05* | 10.7% | 25 | 3 | > 0.05¶/§/> 0.05*/§ |
| NEcS | 20 | 61.3 | 0.0186 | > 0.05¶ | 20.0% | 16 | 4 | > 0.05¶/§ |
| Barrett’s metaplasia | 60 | 63.7 | 0.0123 | > 0.05** | 13.3% | 52 | 8 | > 0.05**/† |
| Dysplasia in Barrett’s esophagus | 40 | 65.3 | 0.0141 | > 0.05** | 25.0% | 30 | 10 | > 0.05**/† |
| EAC | 67 | 65.1 | 0.0238 | > 0.05**/< 0.01# | 26.9% | 32 | 18 | < 0.05**/†/> 0.05#/† |
| ESCC | 26 | 62.5 | 0.0450 | < 0.01** | 46.2% | 14 | 12 | < 0.001**/† |
NE, normal esophagus from non-Barrett’s/cancer patients; NEcA, NE from EAC patients; NEcS, NE from ESCC patients; EAC, esophageal adenocarcinoma; ESCC, esophageal squamous cell carcinoma; NMV: normalized methylation value; UM, unmethylated; M, methylated;
Mann-Whitney U test;
Fisher’s exact test;
Chi-square for independence test;
comparisons made to NE;
comparisons made to NEcS;
comparisons made to normal esophagus;
comparisons made to ESCC.
No significant associations were observed between ENG promoter hypermethylation and patient age, survival, Barrett’s segment length, EAC tumor stage, lymph node metastasis, or smoking or alcohol consumption (data not shown).
ENG Methylation and mRNA Levels in KYSE220 Cells after 5-Aza-dC Treatment
KYSE220 cells were subjected to demethylation by 5-Aza-dC treatment. After 5-Aza-dC treatment, the NMV of ENG was diminished and ENG mRNA levels were increased (Figure 2).
Figure 2. ENG methylation and mRNA expression in KYSE220 cells after treatment with 5-aza-2′-deoxycytidine (5-Aza-dC).
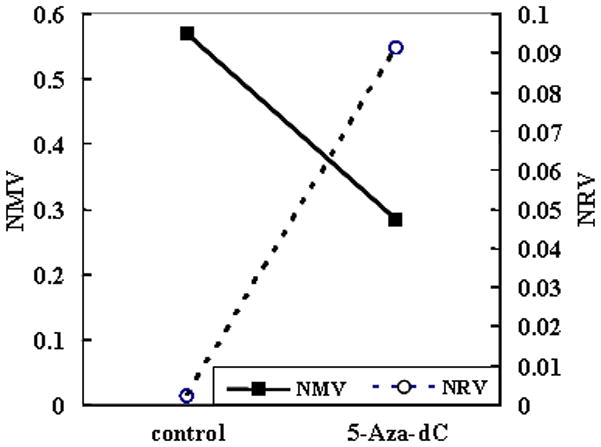
After 5-Aza-dC treatment, the NMV of ENG was diminished, while the normalized mRNA value (NRV) of ENG was increased.
Discussion
In the current study, we systematically investigated hypermethylation of the ENG gene promoter in primary human esophageal lesions of contrasting histological types. Our results demonstrate that ENG promoter hypermethylation occurs frequently in human ESCC (46.2%), but only in a smaller subset of EAC (26.9%). However, ENG hypermethylation occurs early and increases sequentially during esophageal adenocarcinogenesis, from 11.9% in N (and 10.7% in NEcA) to 13.3% in BE, 25% in D, and 26.9% in EAC. Interestingly, methylation levels of ENG were significantly higher in NEcS than in NEcA, suggesting that ENG exhibits a field defect with potential biomarker value for ESCC lurking nearby, even when analyzing non-neoplastic esophageal mucosa. In addition, ENG was hypermethylated more frequently in ESCC than in EAC. Taken together, these findings imply that hypermethylation of ENG is a common event in ESCC, occurs early in some subjects during the development of EAC, increases in frequency during Barrett’s-associated esophageal adenocarcinogenesis, and is a cell type-specific event (i.e., more common in ESCC than in EAC). Further evidence supporting this tissue specificity was provided by ROC curves, which clearly distinguished ESCC from EAC but not EAC from N. Further support for tissue specificity was evident from our finding that mean ENG NMVs were significantly higher in ESCC than in EAC. Thus, ENG hypermethylation appears to constitute a critical field-defect event in human ESCC.
Despite extensive knowledge on intratumour microvessel density assessed by ENG staining as a prognostic factor in different cancer patients 9, 10, 12, 13, limited data are available on ENG hypermethylation in tumor cells14–17. ENG was significantly downregulated in non-small cell lung cancer based on Affymetrix GeneChip assay, and its promoter was aberrantly methylated in 5 (71%) of 7 lung cancer cell lines, in 11 (69%) of 16 primary lung tumors, and in 4 (80%) of 5 normal lung tissues based on combined bisulfite restriction analysis 14. Based on qMSP assays, ENG was methylated in 3 of 34 colon cancers, but not in normal colonic mucosae15. In a large cohort of invasive breast cancers, lack of ENG expression in the tumor cell compartment correlated with ENG gene methylation and poor clinical outcome, and its expression in breast tumor cells suppressed invasion and metastasis 16. ENG was previously shown to be methylated in 2 ESCC patients and 16 ESCC cell lines by non-quantitative methylation-specific PCR, and its expression suppressed invasion in ESCC cells 17. To our knowledge, the current study is the first to quantitatively measure methylation of ENG in a large cohort of human esophageal cancers.
It has been reported that hypermethylation of gene promoters in histologically normal tissue can be related to the initiation of carcinomas 21–26. For example, methylation of the MLH1 promoter was observed in small foci of normal colonic epithelial cells from patients with colon cancer and was associated with silencing of this gene, but was not observed in sections of normal colon from healthy volunteers, suggesting that tumors with gene silencing due to epigenetic alteration may evolve from rare clones of methylated cells in normal epithelia24. Non-neoplastic epithelia from ESCC patients was significantly more methylated than in control esophageal epithelia from healthy volunteers in a panel of 14 promoter loci25. Data from our group also previously showed that AKAP12 hypermethylation was significantly higher in NEcA than in NE or NEcS26. Similarly, in the current study, ENG hypermethylation was significantly higher in NEcS than in NEcA. Thus, our highly sensitive real-time qMSP approach allowed us to show that non-neoplastic esophageal epithelia from ESCC patients already exhibit low but abnormal levels of ENG promoter methylation. It can therefore be hypothesized that increased ENG methylation in normal esophageal cells extends their lifespan enough to put them at higher risk for future malignant evolution. Furthermore, mean ENG NMVs were significantly higher in ESCC than in EAC. These results also further imply that hypermethylation of ENG is an early and unique event, constituting a potentially powerful biomarker for early ESCC detection.
5-Aza-dC and its derivatives have demonstrated effectiveness as therapeutic anti-cancer drugs 27, 28. In agreement with previous findings 16, 17, the current study found that methylation of ENG in ESCC cancer cell lines was associated with silenced or reduced expression of ENG mRNA. Treatment with 5-Aza-dC reactivated mRNA expression and reversed ENG hypermethylation in these cells. Restoration of ENG mRNA expression by demethylating agent treatment implies that DNA hypermethylation was responsible for silencing of ENG. These findings also suggest the possibility that epigenetic therapies may be useful in at least a subset of these patients. In addition, the known involvement of ENG in angiogenesis6–10, 12 suggests the possibility that anti-angiogenesis therapy may be directed toward a subset of these patients, such as those whose tumors lack ENG methylation. Further studies are needed to address this possibility.
The current findings establish that hypermethylation of the ENG promoter, leading to gene silencing, is a common event in human ESCC. In addition, these results show that ENG hypermethylation occurs early during a subset of Barrett’s-associated esophageal adenocarcinogenesis. Further large-scale prospective longitudinal validation studies of this biomarker as a potential predictive biomarker of ESCC are stimulated by these data.
Acknowledgments
Supported by NSFC grant 81172282, Natural Science Foundation of SZU grants 201108 and T201202 to Z Jin; NIH grant DK087454 to SJ Meltzer Stephen J. Meltzer is the American Cancer Society Clinical Research Professor.
Footnotes
Informed consent was obtained from the subject(s) and/or guardian(s)
Using real-time quantitative methylation-specific PCR, we examed promoter hypermethylation of Endoglin (ENG) in 260 endoscopic esophageal biopsy specimens of differing histologies. Results demonstrate that hypermethylation of ENG is a common, tissue-specific event in human esophageal squamous cell carcinomas (ESCC) and exhibits a field defect in normal mucosa with potential biomarker implications for the early detection of ESCC.
There are no financial disclosures from any authors.
References
- 1.Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127(12):2893–917. doi: 10.1002/ijc.25516. [DOI] [PubMed] [Google Scholar]
- 2.Zhang XM, Guo MZ. The value of epigenetic markers in esophageal cancer. Front Med China. 2010;4(4):378–84. doi: 10.1007/s11684-010-0230-3. [DOI] [PubMed] [Google Scholar]
- 3.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. CA Cancer J Clin. 2012;62(1):10–29. doi: 10.3322/caac.20138. [DOI] [PubMed] [Google Scholar]
- 4.Gougos A, Letarte M. Primary structure of endoglin, an RGD-containing glycoprotein of human endothelial cells. J Biol Chem. 1990;265(15):8361–4. [PubMed] [Google Scholar]
- 5.Cheifetz S, Bellon T, Cales C, Vera S, Bernabeu C, Massague J, et al. Endoglin is a component of the transforming growth factor-beta receptor system in human endothelial cells. J Biol Chem. 1992;267(27):19027–30. [PubMed] [Google Scholar]
- 6.Burrows FJ, Derbyshire EJ, Tazzari PL, Amlot P, Gazdar AF, King SW, et al. Up-regulation of endoglin on vascular endothelial cells in human solid tumors: implications for diagnosis and therapy. Clin Cancer Res. 1995;1(12):1623–34. [PubMed] [Google Scholar]
- 7.McAllister KA, Baldwin MA, Thukkani AK, Gallione CJ, Berg JN, Porteous ME, et al. Six novel mutations in the endoglin gene in hereditary hemorrhagic telangiectasia type 1 suggest a dominant-negative effect of receptor function. Hum Mol Genet. 1995;4(10):1983–5. doi: 10.1093/hmg/4.10.1983. [DOI] [PubMed] [Google Scholar]
- 8.Miller DW, Graulich W, Karges B, Stahl S, Ernst M, Ramaswamy A, et al. Elevated expression of endoglin, a component of the TGF-beta-receptor complex, correlates with proliferation of tumor endothelial cells. Int J Cancer. 1999;81(4):568–72. doi: 10.1002/(sici)1097-0215(19990517)81:4<568::aid-ijc11>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- 9.Kumar S, Ghellal A, Li C, Byrne G, Haboubi N, Wang JM, et al. Breast carcinoma: vascular density determined using CD105 antibody correlates with tumor prognosis. Cancer Res. 1999;59(4):856–61. [PubMed] [Google Scholar]
- 10.Wikstrom P, Lissbrant IF, Stattin P, Egevad L, Bergh A. Endoglin (CD105) is expressed on immature blood vessels and is a marker for survival in prostate cancer. Prostate. 2002;51(4):268–75. doi: 10.1002/pros.10083. [DOI] [PubMed] [Google Scholar]
- 11.Shovlin CL, Hughes JM, Scott J, Seidman CE, Seidman JG. Characterization of endoglin and identification of novel mutations in hereditary hemorrhagic telangiectasia. Am J Hum Genet. 1997;61(1):68–79. doi: 10.1086/513906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fonsatti E, Del Vecchio L, Altomonte M, Sigalotti L, Nicotra MR, Coral S, et al. Endoglin: An accessory component of the TGF-beta-binding receptor-complex with diagnostic, prognostic, and bioimmunotherapeutic potential in human malignancies. J Cell Physiol. 2001;188(1):1–7. doi: 10.1002/jcp.1095. [DOI] [PubMed] [Google Scholar]
- 13.Shariat SF, Karam JA, Walz J, Roehrborn CG, Montorsi F, Margulis V, et al. Improved prediction of disease relapse after radical prostatectomy through a panel of preoperative blood-based biomarkers. Clin Cancer Res. 2008;14(12):3785–91. doi: 10.1158/1078-0432.CCR-07-4969. [DOI] [PubMed] [Google Scholar]
- 14.Dammann R, Strunnikova M, Schagdarsurengin U, Rastetter M, Papritz M, Hattenhorst UE, et al. CpG island methylation and expression of tumour-associated genes in lung carcinoma. Eur J Cancer. 2005;41(8):1223–36. doi: 10.1016/j.ejca.2005.02.020. [DOI] [PubMed] [Google Scholar]
- 15.Mori Y, Cai K, Cheng Y, Wang S, Paun B, Hamilton JP, et al. A genome-wide search identifies epigenetic silencing of somatostatin, tachykinin-1, and 5 other genes in colon cancer. Gastroenterology. 2006;131(3):797–808. doi: 10.1053/j.gastro.2006.06.006. [DOI] [PubMed] [Google Scholar]
- 16.Henry LA, Johnson DA, Sarrio D, Lee S, Quinlan PR, Crook T, et al. Endoglin expression in breast tumor cells suppresses invasion and metastasis and correlates with improved clinical outcome. Oncogene. 2011;30(9):1046–58. doi: 10.1038/onc.2010.488. [DOI] [PubMed] [Google Scholar]
- 17.Wong VC, Chan PL, Bernabeu C, Law S, Wang LD, Li JL, et al. Identification of an invasion and tumor-suppressing gene, Endoglin (ENG), silenced by both epigenetic inactivation and allelic loss in esophageal squamous cell carcinoma. Int J Cancer. 2008;123(12):2816–23. doi: 10.1002/ijc.23882. [DOI] [PubMed] [Google Scholar]
- 18.Schulmann K, Sterian A, Berki A, Yin J, Sato F, Xu Y, et al. Inactivation of p16, RUNX3, and HPP1 occurs early in Barrett’s-associated neoplastic progression and predicts progression risk. Oncogene. 2005;24(25):4138–48. doi: 10.1038/sj.onc.1208598. [DOI] [PubMed] [Google Scholar]
- 19.Jin Z, Mori Y, Yang J, Sato F, Ito T, Cheng Y, et al. Hypermethylation of the nel-like 1 gene is a common and early event and is associated with poor prognosis in early-stage esophageal adenocarcinoma. Oncogene. 2007;26(43):6332–40. doi: 10.1038/sj.onc.1210461. [DOI] [PubMed] [Google Scholar]
- 20.Jin Z, Olaru A, Yang J, Sato F, Cheng Y, Kan T, et al. Hypermethylation of tachykinin-1 is a potential biomarker in human esophageal cancer. Clin Cancer Res. 2007;13(21):6293–300. doi: 10.1158/1078-0432.CCR-07-0818. [DOI] [PubMed] [Google Scholar]
- 21.Leung WK, Yu J, Ng EK, To KF, Ma PK, Lee TL, et al. Concurrent hypermethylation of multiple tumor-related genes in gastric carcinoma and adjacent normal tissues. Cancer. 2001;91(12):2294–301. [PubMed] [Google Scholar]
- 22.Kanaya T, Kyo S, Maida Y, Yatabe N, Tanaka M, Nakamura M, et al. Frequent hypermethylation of MLH1 promoter in normal endometrium of patients with endometrial cancers. Oncogene. 2003;22(15):2352–60. doi: 10.1038/sj.onc.1206365. [DOI] [PubMed] [Google Scholar]
- 23.Holst CR, Nuovo GJ, Esteller M, Chew K, Baylin SB, Herman JG, et al. Methylation of p16(INK4a) promoters occurs in vivo in histologically normal human mammary epithelia. Cancer Res. 2003;63(7):1596–601. [PubMed] [Google Scholar]
- 24.Nuovo GJ, Nakagawa H, Sotamaa K, de Chapelle AL. Hypermethylation of the MLH1 promoter with concomitant absence of transcript and protein occurs in small patches of crypt cells in unaffected mucosa from sporadic colorectal carcinoma. Diagn Mol Pathol. 2006;15(1):17–23. doi: 10.1097/00019606-200603000-00003. [DOI] [PubMed] [Google Scholar]
- 25.Ishii T, Murakami J, Notohara K, Cullings HM, Sasamoto H, Kambara T, et al. Oesophageal squamous cell carcinoma may develop within a background of accumulating DNA methylation in normal and dysplastic mucosa. Gut. 2007;56(1):13–9. doi: 10.1136/gut.2005.089813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jin Z, Hamilton JP, Yang J, Mori Y, Olaru A, Sato F, et al. Hypermethylation of the AKAP12 promoter is a biomarker of Barrett’s-associated esophageal neoplastic progression. Cancer Epidemiol Biomarkers Prev. 2008;17(1):111–7. doi: 10.1158/1055-9965.EPI-07-0407. [DOI] [PubMed] [Google Scholar]
- 27.Lemaire M, Momparler LF, Bernstein ML, Marquez VE, Momparler RL. Enhancement of antineoplastic action of 5-aza-2′-deoxycytidine by zebularine on L1210 leukemia. Anticancer Drugs. 2005;16(3):301–8. doi: 10.1097/00001813-200503000-00009. [DOI] [PubMed] [Google Scholar]
- 28.Momparler RL. Epigenetic therapy of cancer with 5-aza-2′-deoxycytidine (decitabine) Semin Oncol. 2005;32(5):443–51. doi: 10.1053/j.seminoncol.2005.07.008. [DOI] [PubMed] [Google Scholar]


