Abstract
Chronic lymphocytic leukemia (CLL) is a malignancy of mature lymphocytes that is manifest by the progressive accumulation of transformed cells, mostly due to their decreased apoptosis. CD84 belongs to the Signaling Lymphocyte Activating Molecule (SLAM) family of immunoreceptors and has an as yet unknown function in normal B cells and CLL lymphocytes. We show that CD84 is over-expressed in CLL cells. Activation of cell surface CD84 initiates a signaling cascade, which enhances cell survival. Both immunoneutralization or blockade of CD84 induce cell death in vitro and in vivo. Thus, overexpression of CD84 from an early stage may be critical for the survival of CLL. These findings suggest novel therapeutic strategies based on the blockade of a CD84 dependent survival pathway.
Introduction
Chronic lymphocytic leukemia is the most common leukemia in the Western world and is characterized by the progressive accumulation of small mature CD5+ lymphocytes, in the peripheral blood, lymphoid organs and bone marrow (BM). The main feature of the disease is decreased apoptosis, resulting in the pathologic accumulation of these malignant cells 1. Despite major progress in the last few years in the understanding of the biology and pathophysiology of this disease, as well as the development of better treatment modalities, CLL remains incurable in most patients and even control of the disease requires aggressive treatment with significant side effects. A better understanding of the cellular events involved in the pathogenesis and progression of CLL should lead to more specific and less toxic therapies, with early treatment in patients at risk, and possibly enabling cure.
Adaptive and innate immune responses are orchestrated by the dynamic interactions between cell-surface molecules. The Signaling Lymphocyte Activation Molecule (SLAM) family includes homophilic and heterophilic receptors that modulate both adaptive and innate immune responses 2. These receptors share a common ectodomain organization: a membrane-proximal immunoglobulin (Ig)-like constant domain and a membrane-distal Ig variable domain that is responsible for ligand recognition (with the exception of CD229, which consists of a tandem repeat of two V-Ig/C2 sets of domains). The SLAM family modulates immune cell function 3–6, most likely because of the ability of these receptors to interact with SLAM-associated protein (SAP)-related molecules, which are a group of SRC homology 2 (SH2) domain adaptors. The SAP family comprises three members: SAP, Ewing’s sarcoma-associated transcript-2 (EAT2), and in rodents, EAT2-related transducer (ERT) 4,7. In humans, the gene encoding ERT has evolved into a non-functional pseudo-gene. SAP does not appear to be expressed in B cells; rather, EAT2 was proposed to be the functional homologue in these cells 8,9. The interaction of the SLAM family receptors with SAP family members is mediated by tyrosine-based motifs in the cytoplasmic region of SLAM-related receptors, and by the SH2 domain of SAP-related adaptors 9.
CD84 is a member of this SLAM immunoglobulin superfamily. It is a single chain cell-surface protein with a 199 aa extracellular portion containing four potential N-glycosylation sites. The transmembrane region consists of 25 aa, and the 83 aa cytoplasmic tail contains four tyrosines 10. CD84 strongly self-associates 11,12 with a Kd in the submicromolar range; the association is driven by the Ig-V domain, forming an orthogonal homophilic dimer 13. CD84 is predominantly expressed by B cells, T cells, platelets, monocytes, and dendritic cells (DCs). CD84 also is expressed early in hematopoiesis, where its function is being to be versatile but not fully understood 2. Although CD84 was originally cloned from a human B cell line cDNA library, very little is known regarding its biology. B cells can be subdivided into CD84hi and CD84lo populations. The CD84hi population represents a subset of memory B cells, which are characterized by co-expression of CD27, somatically mutated Ig variable region genes, and vigorous proliferation in response to CD40L and IL-4, compared to CD84lo B cells 8,11. It was recently shown that CD84 is required for prolonged T cell:B cell contact and for optimal T follicular helper function and germinal center formation 14. A striking feature of human CD84 is the expression in tumor cells of a complex series of isoforms with several cytoplasmic tails 15. Nevertheless, both the expression and the function of CD84 in the biology of normal and malignant B cells remains to be fully explored.
Macrophage migration inhibitory factor (MIF) is an upstream activator of innate and adaptive immune responses 16,17. It was shown previously that MIF binds to the CD74 extracellular domain, a process that results in the initiation of a signaling pathway in a CD44-dependent manner 18–20. In addition, over-expression of CD74 was shown to be an important survival mechanism in CLL starting from the very early disease stages 21. Activation of cell surface CD74 by MIF on CLL cells initiates a signaling cascade that induces NF-κB activation, TAp63 expression, and secretion of interleukin-8 (IL-8), which together promote cell survival 21,22.
In the current study, we examined the expression and function of CD84 in CLL cells. Our results show an elevation in CD84 expression in CLL cells from early disease stages, in a MIF-CD74 dependent manner. CD84 expression in turn regulates the survival of CLL cells in vitro and in vivo. Thus, blocking CD84 activity may be a novel therapeutic strategy for interfering with tumorigenic survival pathways.
Materials and Methods
Patient population
B lymphocytes obtained from the peripheral blood of both healthy subjects (control) and CLL patients at varying stages of the disease were provided by the Hematology Institute of Kaplan Medical Center and the Sourasky Medical Center in accordance with the IRB of the hospitals, as previously described 23,24. The diagnosis of CLL was based on standard criteria 25. Patients were staged according to the Rai staging system 26.
Reagents
MIF: Recombinant human MIF was purified from an expression system as previously described, and the contaminating endotoxin removed by C8 chromatography 27. The final endotoxin concentration was <0.032 EU μg−1 MIF.
hLL1 antibody: The hLL1 mAb (IMMU-115; milatuzumab) was provided by Immunomedics, Inc. (Morris Plains, NJ). Development and properties of hLL1 were described previously 28,29.
Cell purification
B-lymphocytes were purified using a RosettSep antibody cocktail (StemCell), as previously described 23. Briefly, cells were incubated for 20 min with the cocktail. An equal amount of PBS then was added to the mixture, and the cells were placed on a Ficoll gradient. Cells were used fresh, or viably frozen in fetal calf serum (FCS) plus 10% dimethyl sulfoxide (DMSO) for storage in liquid nitrogen. Frozen cells were thawed and cultured overnight in 5% CO2 in RPMI medium with 10% FCS and antibiotics. We have previously shown that the freezing and thawing process had no effect on protein expression or function of the cells 23.
CD84 stimulation and blocking
CD84 stimulation was performed as previously described 8. Briefly, 1×107 CLL cells were suspended in 1 ml of RPMI medium containing 10% FCS in the presence of 0.25 μg of antibody specific for the luminal domain of CD84 (152–1D5, ab-3202 Abcam) followed by 0.5 μg anti Fab (Jackson) or an isotype control antibody, IgG (MOPC-21 BioLegend). Immediately following incubation, the cells were washed and fast-frozen in liquid N2.
CD84 blocking was performed as previously described 11. Briefly, 1×107 CLL cells were incubated in RPMI medium containing 0.1% (v/v) FCS at 37°C for 2 h. Next, cells were resuspended in medium containing 200 ng/ml of MIF, or MIF with 2.5 μg of anti CD84 (CD84.1.21 BioLegend) and incubated at 37°C for 18–24 h.
Cell lysis
Lysis of cells was performed as previously described 30. Briefly, cell pellets were resuspended in preboiled 0.5% SDS, 50 mM triethanolamine pH 7.4, 0.1 mM NaCl, and 2 mM EDTA, and boiled at 99°C for 2 minutes, frozen in liquid nitrogen, boiled again for 2 minutes, and sonicated.
For detection of phosphorylated proteins, cells were lysed as previously described 31.
DNA chip arrays
The Affymetrix genechip expression analysis system was used for differential expression analysis. The Weizmann Institute has an Affymetrix-based service facility, which routinely performs RNA labeling, hybridization, and data analysis. RNA from MIF treated or unterated cells were reverse transcribed to cDNA in the presence of biotinylated nucleotides. The target cDNA was then hybridized to the genechip under stringent conditions. The hybridized probe array was stained with a streptavidin-phycoerythrin conjugate and scanned by using the GeneArray scanner.
Western blot analysis
To detect changes in protein levels, lysates were separated by 8–12% SDS-PAGE. The proteins were then transferred onto a nitrocellulose membrane and probed with anti-CD84 (152–1D5:sc-23899 Santa Cruz), anti-Bcl-2 (C-2; Santa Cruz), anti-pAKT (PY99: sc-7020 Santa Cruz), or anti-myc followed by horseradish peroxidase-conjugated anti-mouse (Jackson Laboratories). The membrane was then stripped and reprobed with anti-tubulin antibody (Sigma), followed by peroxidase-conjugated anti-mouse (Jackson Labs).
Immunoprecipitation and western blot
Protein-G Sepharose beads (Pharmacia) were conjugated to Tyr(P) (PY99:sc-7020 Santa Cruz) for 2 hours at 4°C, followed by three washes in PBS. Beads were added to the cell lysates and tyr(p) proteins were immunoprecipitated overnight. The protein G bound material was washed three times with PBS containing 0.1% SDS and 0.5% NP40. Immunoprecipitates were separated by 8–15% (w/v) SDS-PAGE. The protein bands were transferred onto a nitrocellulose membrane and probed with anti-CD84 (152–1D5:sc-23899 Santa Cruz), or anti-EAT-2 (N-14:sc-21572 Santa Cruz Biotechnology) followed by horseradish peroxidase-conjugated anti mouse IgG (Jackson ImmunoResearch Laboratories).
Flow cytometry
Staining of CLL cells was performed as previously described 23. The following antibodies were used: anti-CD84 (Ab-2, 152–1D5 Labvision), followed by PE anti-mouse secondary antibody (Jackson ImmunoResearch Laboratories).
Cell death detection
Annexin and PI staining
Purified CLL cells were cultured in 24-well plates at 1×107 cells/well in RPMI medium supplemented with 10% FCS, 2 mM glutamate, 100 U/ml penicillin, 100 μg/ml streptomycin, with or without 0.25 μg of anti-CD84 (ab-3202 Abcam) followed by 0.5 μg of anti Fab (Jackson) or an IgG isotype control antibody (MOPC-21 BioLegend) for 24h. Cells were centrifuged, washed, and stained with Annexin (BD Biosciences), and propidium iodide (PIl; 25μg/ml) (Sigma) was added for 15 min at room temp. The extent of Annexin+PI staining was analyzed by FACS.
Magic Red Apoptosis Detection Kit
Cells were incubated with Magic Red (Immunochemistry Technology) according to the manufacturer’s instructions, at 37 °C for 1 h. Magic Red staining then was measured by FACS analysis.
Cell transfection
HEK 293 cells (a human embryonic kidney cell line) were seeded in a 24 well dish. Transfections were performed using the standard CaPO4 method, as previously described 32. A total of 1 μg of DNA was used per well.
CD74 stimulation and blocking
MIF stimulation was performed as previously described 31. Briefly, 1×107 CLL cells were incubated in RPMI medium containing 0.1% (v/v) FCS at 37°C for 2 h. Next, cells were resuspended in medium containing 200 ng/ml of human recombinant MIF and incubated at 37°C for 18–24 h.
For MIF blocking, 1× 107 CLL cells were incubated for 2 h in 1 ml RPMI medium containing 0.1% (v/v) FCS. Next, cells were resuspended in medium containing 20 μM of the MIF antagonist, ISO-1 33 and incubated at 37°C for 18–24 h.
Blocking the CD74 pathway: CLL (1×107) cells were incubated in the presence of hLL1 (50 mg/ml) or ISO-1 (20 mM; CalBiochem, San Diego, CA) at 37°C for 18 h, as described previously 21.
Hybridoma activation/blocking
Hybridoma supernatant was added to CLL cells for 18 or 24 hours, with or without MIF stimulation.
RNA isolation and reverse transcription
Total RNA was isolated from cells by using the Tri Reagent kit (Molecular Research Center, Cincinnati, OH), according to the manufacturer’s instructions. Reverse transcription was carried out using SuperScript II RT (Invitrogen, Carlsbad, CA). Primers that were used in the PCR reactions included:
Bcl-2: 5′ AGATCTCTGGTTGGGATTC 3′ CACCGAACACTTGATTCTG
Actin: 5′ TGAAGTGTGACGTGGACATCCG 3′ GCTGTCACCTTCACCGTTCCAG
CD84a-c: 5′ATGTCCTTCAAATCTTCCAGACTCCT 3′ GTTTACTGTCCTTGTTGCTGGCTTTC
CD84d: 5′ TACATGCCTTAGGTCCGA 3′ GAGGGAAGCACCTTGT
CD84e:5′ ATGTCCTTCAAATCTTCCAGACTCCT 3′ GGTGAGTTTCCACATTTTACCTTCTG
CD84all: 5′ TGCCTGCAAACCTGGCCGGAAGCA 3′ TGCAGGTTGTAGCGCTTGGTGGTGGT
Real-time reverse transcription–PCR analysis
mRNA levels of Rp2, CD84 and Bcl-2 were analyzed by quantitative real-time RT-PCR using a Light-Cycler instrument (Roche Diagnostics, Mannheim, Germany). The reaction volume (10 ml) contained 3 mM MgCl2, LightCycler HotStart DNA SYBR Green I mix (Roche Diagnostics), specific primer pairs, and 2.5 ml of cDNA. Conditions for PCR were as follows: 10 minutes at 95°C followed by 40–55 cycles of 15 seconds at 95°C, 15 seconds at 60°C, and 15 seconds at 72°C. PCR was performed in duplicates as previously described 34. Primer sequences were as follows:
Bcl-2: 5′ GGATCAGGGAGTTGGAAG 3′ GCACTGCCAAACGGAG
RP2: 5′ GCACACGTCCAATGACAT 3′ GTGCGGCTGCTTCCATAA
CD84: 5′ TTGTTCCGTTTGTTCAAGAG 3′ CGGAATAAACTGTGTTCACTG
RP-2 levels were used to normalize samples for calculation of the relative expression levels of other genes.
Constructs
The full-length human CD84 (25–1010) and the ECD-CD84 (25–685) constructs in the pEF4/Myc-His vector (Invitrogen) vector were designed as was previously described 18.
Primers used for cloning:
BamH1CD84: 5′CGTCGGATCCATGGCTCAGCACCAC
EcoR1CD84: 3′TAGCGAATTCACGGAAGCCCATTGC
Site directed mutagenesis
The Stratagene QuickChange Site-directed Mutagenesis Kit was used, following the manufacturer’s instructions. The following tyrosine mutants were derived from the CD84-full construct:
Y262F
Y262F, Y299F
Y279F
Y279F, Y324F
Primers used for cloning are listed below
-
Y262:5′GATGCTGCCTCAAAGAAAACCATATTCACATATATCATGGCTTC
3′GAAGCCATGATATATGTGAATATGGTTTTCTTTGAGGCAGCATC
-
Y299:5′GAGCCAGTGAACACAGTTTTTTCCGAAGTGCAGT
3′ACTGCACTTCGGAAAAAACTGTGTTCACTGGCTC
-
Y279:5′GCCAGCAGAGTCCAGAATCTTTGATGAAATCCTG
3′CAGGATTTCATCAAAGATTCTGGACTCTGCTGGC
-
Y324:5′ACCTCCTGGGACTTCAAGCTTTGAAATTGTGATCG
3′CGATCACAATTTCAAAGCTTGAAGTCCCAGGAGGT
Transfection and protein expression of secreted CD84-ECD
For expression and secretion of CD84-ECD protein, HEK 293T cells were plated in 175cm2 cell culture flasks to a final confluence of 75% at 16 to 24 hr before transfection. For transfection, 30mL of complete DMEM medium and 3mL of DNA precipitate-containing solution were used. Cultures were incubated an additional 16 to 24 hours after transfection, and the medium then was changed to serum-free DMEM for 3 to 4 days. The conditioned medium was collected and protein purification from conditioned medium was performed using FLPC. After binding of the protein to the Ni2+ column, it was washed and eluted with high concentrations of imidazole, which competes with the His6-tag and displaces the protein.
After purification, concentrated samples from each purification step and from the different peaks were analyzed by SDS-PAGE followed by western blotting and Coomassie staining of the gel.
Size-exclusion chromatography (gel filtration)
The pooled, concentrated fractions of the Ni2+ affinity chromatography were loaded onto a 16/60 Superdex 200 column (preparative grade) by injection into the FPLC system. The proteins were eluted at a flow rate of 5 mL/min with PBS. Fractions were collected according to the UV absorption, and changes in pH of the eluates. After pooling and concentrating the different peak fractions, samples were analyzed by SDS-PAGE, followed by western blotting and Coomassie staining.
Hybridoma protocol
CD84-ECD protein was purified from conditioned medium derived from 293 cells transfected with the CD84-ECD construct. Mice were immunized with CD84-ECD over a period of 5 months. Following a positive ELISA test of blood for antibodies against CD84-ECD, the spleens were removed and the lymphocytes isolated and mixed with NSO cells. Hybridomas were selected and their supernatants were analyzed for recognition of CD84-ECD using an ELISA assay 35. Hybridomas were shown to be of the IgM class with a kappa light chain by commercial ELISA kit.
In vivo cell tracking
CLL cells were labeled with 5 μM CFSE (Molecular Probes) for 15 min at room temperature. CFSE labeling had no effect on cell survival throughout the experiment. Following labeling, 1 × 107 cells were i.v. injected into control C57BL/6 mice. After 1h, anti-CD84 (CD84.1.21 BioLegend) or an isotype control (Biolegend) were i.v. injected. Survival of CLL cells in the spleen was determined 3 h later by FACS analysis of isolated spleens. All animal procedures were approved by the Animal Research Committee at the Weizmann Institute.
Statistics
Statistical analysis was performed using SPSS. The data were tested for normal distribution, and t-tests were performed on normally distributed data sets, as described previously 21.
Results
CD84 expression is elevated in CLL cells in a MIF /CD74 dependent manner
To better understand the biologic function of CD74 in CLL cells, we sought to identify target genes induced by CD74 activation. Thus, CLL cells were incubated in the presence or absence of the CD74 ligand, MIF (100 ng/ml) for 18 hrs, and RNA from MIF-stimulated cells was compared to RNA derived from unstimulated cells using the Affymetrix GeneChip® expression analysis system (Gene expression data is available upon publication on http://www.biochemmcb.rwth-aachen.de/mif_consortium). Many genes were found to be differentially expressed in these populations; one striking example was CD84, whose expression was markedly elevated in the MIF-induced CLL cells.
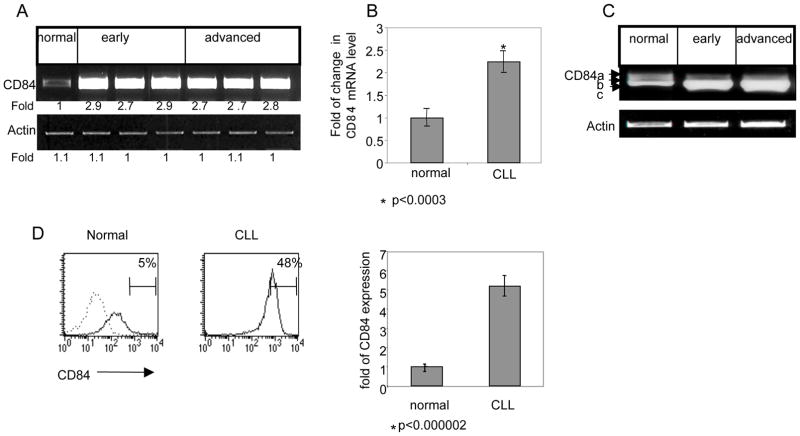
Since the expression of CD84 was not previously characterized in CLL, we next wished to determine its expression levels in CLL cells from patients at various disease stages. Purified B cells from healthy subjects as well as early- and advanced-stage CLL cells were analyzed for the presence of CD84 mRNA (a segment common to all isoforms). As shown in Fig. 1A–B, low levels of CD84 message were detected in normal B cells, while elevated levels of CD84 mRNA were observed in all the CLL patients, regardless of the stage of disease (Table 2).
Figure 1.
Elevated expression of CD84 in CLL cells. B cells derived from healthy subjects (normal), as well as early- and advanced-stage CLL patients were purified and examined for CD84 expression. (A) CD84 and actin mRNA were analyzed by RT-PCR. The results presented are representative of two normal, six early-stage, and six advanced-stage CLL patients. (B) Quantitative Real Time PCR (qPCR) was performed using primers for CD84 and RP-2. Results are expressed as a fold of change in CD84 mRNA in CLL cells compared to normal B cells, which was defined as 1. The graph summarizes results of three normal and seven CLL patients. (C) Expression of several CD84 isoforms and actin mRNA were analyzed by RT-PCR, as described in Methods. The results presented are representative of three normal, six early-stage, and five advanced-stage CLL patients. (D) Cells were stained with anti-CD84 followed by a secondary (goat anti-mouse) antibody. Histograms show CD84 expression (grey line) or staining with secondary Ab alone (dotted line) in normal B and CLL cells. The graph summarizes the results of two normal and 18 CLL patients.
Table 2.
CD84 expression
| patient | age | m/f | zap70 | doubling time (month) | previous chemotherapy | RAI stage | autoimmune phenomena | lymphocyte count (103/ml) | serum IgG(mg/dl) | Fold of CD84 (PCR) | Fold of CD84 (FCS) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 82 | f | positive | 7 month | none | i | no | 30 | <700 | 2.9 | |
| 2 | 34 | m | negative | none | I | no | 40 | >700 | 2.7 | 6.1 | |
| 3 | 75 | f | nd | 12 month | none | 0 | no | 30 | >700 | 2.9 | |
| 4 | 55 | f | nd | 6 month | 0 | no | 20 | 700 | 2.1 | ||
| 5 | 70 | f | negative | not reached | none | 0 | no | 12.5 | <700 | 2.24 | |
| 6 | 80 | f | negative | 12 month | none | 0/I | no | 140 | <700 | 2.1 | |
| 7 | m | positive | 3 month | FCR | II | no | 40 | <700 | 2.7 | 4 | |
| 8 | 78 | m | negative | 1 month | FCR, L/P | IV | no | 46 | <700 | 2.83 | 5.24 |
| 9 | 58 | f | positive | not reached | COP | IV | coombs pos 150 | >700 | 2.64 | ||
| 10 | 72 | m | positive | 2 month | CHOP-likeIV | IV | no | 80 | <700 | 3.12 | |
| 11 | 86 | f | positive | not reached | none | II | no | 57 | >700 | 1.4 | |
| 12 | 52 | m | positive | 6 month | none | 0 | no | 12 | >700 | 1.48 | |
| 13 | 53 | m | negative | not reached | FC/R | IV | yes | 45 | <700 | 1.16 | |
| 14 | 72 | f | 6 month | none | 0 | no | 40 | >700 | 1.57 | ||
| 15 | 76 | f | positive | 2 month | CP, FC/R | IV | no | 120 | <700 | 1.6 | 4.89 |
| 16 | 72 | m | negative | 6 month | L/P | III–IV | no | 40 | >700 | 1.6 | |
| 17 | 70 | m | positive | not reached | L/P | IV | no | 15 | 1500 | 1.48 | |
| 18 | 77 | m | positive | 3 month | COP | III | AIHA | 80 | 700 | 6.68 | |
| 19 | 72 | f | not reached | none | 0 | no | 40 | >700 | 5.24 | ||
| 20 | 34 | m | negative | none | I | no | 40 | >700 | 5.44 | ||
| 21 | 80 | m | positive | 5 month | COP, FCR | IV | no | 6 | <700 | 5.72 | |
| 22 | 40 | f | negative | none | 0 | no | 6.34 | ||||
| 23 | 54 | m | positive | 2 month | FCR | I | no | 30 | >700 | 4.55 | |
| 24 | 70 | f | not reached | none | I | no | 12 | >700 | 6 | ||
| 25 | 74 | m | positive | FC | I | no | 5.58 | ||||
| 26 | 75 | f | positive | 12 month | none | 0 | no | 40 | <700 | 4.9 | |
| 27 | 80 | f | negative | 6 month | CP | IV | no | 80 | <700 | 5.65 | |
| 28 | 61 | m | FC | III | no | 5.8 | |||||
| 29 | 66 | f | positive | FCR | III–IV | no | 4.96 | ||||
| 30 | 65 | m | negative | 1 year | none | I | no | 20 | 700 | 6 |
Genomic characterization of CD84 has revealed the existence of five isoforms differing in their cytoplasmic domains 15. In order to determine which isoforms are expressed and function in CLL cells, the expression pattern of the various isoforms (a,b,c; Fig 1c; d,e; data not shown) in healthy donors and CLL cells were analyzed by semi-quantitative RT-PCR, as previously described 15. As shown in Fig. 1C, in all tested samples (13 patients), CD84c was the dominant isoform in healthy, early and advanced stage CLL patients.
We next compared by flow cytometry the cell surface expression levels of CD84 in normal and CLL cells. As shown in Fig. 1D, CD84 cell surface levels were significantly higher in all CLL cells when compared to normal B cells. These results were observed uniformly in all the samples examined, regardless of the clinical parameters of the patients, including stage of the disease (Table 2).
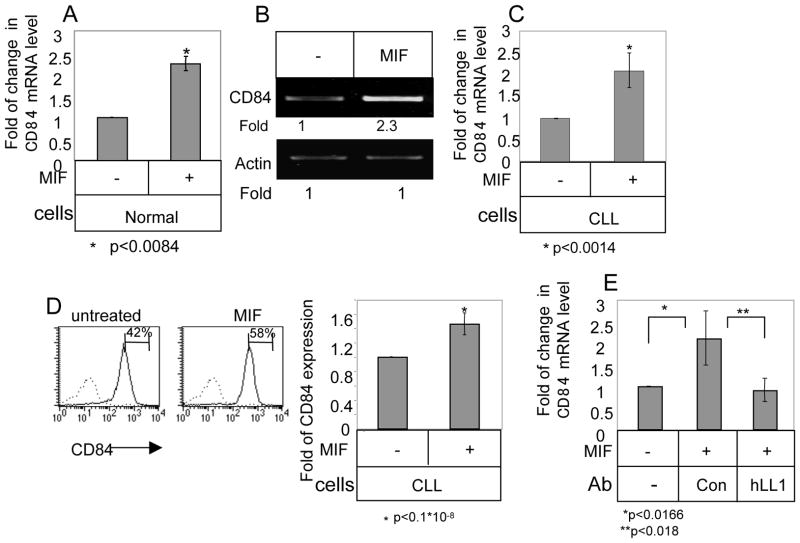
To verify that CD84 expression is indeed modulated by CD74 and its ligand, MIF, we analyzed its mRNA levels in MIF-stimulated and unstimulated normal (Fig. 2A) and CLL (Fig. 2B–C; Table 3) cells. MIF stimulation elevated CD84 mRNA levels in both normal and CLL cells. This elevation resulted in a significant increase in CD84 cell surface expression on CLL cells (Fig. 2D; Table 3). ISO-1 is a non-toxic inhibitor of MIF that binds to bioactive MIF at its N-terminal tautomerase site and interferes with tis biologic activity 36. Blocking MIF by ISO-1 downregulated CD84 mRNA levels and cell surface expression levels (Supplementary Fig. 1), further demonstrating that MIF regulates CD84 expression levels. In addition, blocking CD74 using the humanized anti-CD74 blocking antibody (hLL1), specifically downregulated CD84 mRNA levels (Fig. 2E). Thus, MIF and its receptor, CD74, regulate CD84 expression in CLL cells.
Figure 2.
CD84 expression is elevated in CLL cells in a MIF/CD74 dependent manner. (A) Normal B cells were purified and incubated in the presence or absence of MIF (100 ng/ml). After 18 h, RNA was purified and Quantitative Real Time PCR was performed using primers for CD84 and RP-2. Results are expressed as fold of change in CD84 expression following MIF stimulation; untreated cells were defined as 1. The graph summarizes results of B cells from two normal donors. (B–D) CLL cells were purified and incubated in the presence or absence of MIF. (B) After 18 h, RNA was purified, and CD84 and actin mRNA levels were analyzed by RT-PCR. The results presented are representative of nine CLL patients. (C) Following 18h, the RNA was purified, and qPCR was performed using primers for CD84 and RP-2. Results are expressed as fold of change in CD84 expression in MIF stimulated relative to untreated cells, which were defined as 1. The graph summarizes results of six CLL patients. (D) Following 24h, cells were stained with anti-CD84 followed by a secondary (goat anti-mouse) antibody. Histograms show CD84 expression (grey line) or secondary Ab (dotted line) on CLL cells. The graph summarizes results of 10 CLL patients. (E) CLL cells were incubated in the presence or absence of MIF (100 ng/ml), hLL-1 (50 μM) or an isotype control Ab (50 mM). Following 18 h, RNA was purified and qPCR was performed using primers for CD84 and RP-2. Results are expressed as a fold change in CD84 following the various treatments. Untreated cells were defined as 1. The graph summarizes results from three CLL patients.
Table 3.
CD84 expression following MIF stimulation
| patient | age | m/f | zap70 | doubling time (month) | previous chemotherapy | RAI stage | autoimmune phenomena | lymphocyte count (103/ml) | serum IgG(mg/dl) | Fold of CD84 (PCR) | Fold of CD84 (FCS) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 78 | m | negative | 1 month | FCR, L/P | IV | no | 46 | <700 | 1.71 | |
| 2 | 56 | f | 6 month | L/P | I | sjorgen | 30 | >700 | 1.3 | ||
| 3 | 74 | m | positive | 6 month | none | II | no | 50 | <700 | 1.97 | |
| 4 | 53 | m | negative | not reached | FC/R | IV | yes | 45 | <700 | 2.8 | |
| 5 | 72 | f | 6 month | none | 0 | no | 40 | >700 | 1.33 | ||
| 6 | 60 | m | 6 month | none | 0 | no | 19 | >700 | 2.53 | ||
| 7 | 80 | f | negative | 12 month | none | 0/I | no | 140 | <700 | 2.2 | |
| 8 | 52 | m | positive | 6 month | none | 0 | no | 12 | >700 | 1.47 | |
| 9 | 84 | f | 6 month | none | 0 | no | 42 | >700 | 1.5 | ||
| 10 | 72 | f | 6 month | none | 0 | no | 40 | >700 | 1.7 | ||
| 11 | 70 | m | negative | 6 month | none | I | no | 15 | >700 | 1.72 | |
| 12 | 58 | f | positive | not reached | COP | IV | coombs pos150 | >700 | 1.2 | ||
| 13 | 75 | m | negative | not reached | none | 0 | no | 10 | <700 | 2.36 | |
| 14 | 59 | f | positive | 3 month | L/P | II | no | 100 | >700 | 2.6 | |
| 15 | 72 | m | positive | 2 month | CHOP-likeIV | IV | no | 80 | <700 | 1.41 | |
| 16 | 76 | f | positive | 2 month | CP, FC/R | IV | no | 120 | <700 | 1.6 | |
| 17 | 72 | f | not reached | none | 0 | no | 40 | >700 | 1.24 | ||
| 18 | m | positive | 3 month | FCR | II | no | 40 | <700 | 1.8 | ||
| 19 | 75 | f | positive | 12 month | none | 0 | no | 40 | <700 | 1.5 | |
| 20 | 34 | m | negative | none | I | no | 40 | >700 | 1.5 | ||
| 21 | 80 | m | positive | 5 month | COP, FCR | IV | no | 6 | <700 | 1.37 | |
| 22 | 54 | m | positive | 2 month | FCR | I | no | 30 | >700 | 1.52 | |
| 23 | 55 | f | nd | 6 month | 0 | no | 20 | 700 | 1.33 |
Activation of cell surface CD84 in CLL cells initiates a survival cascade
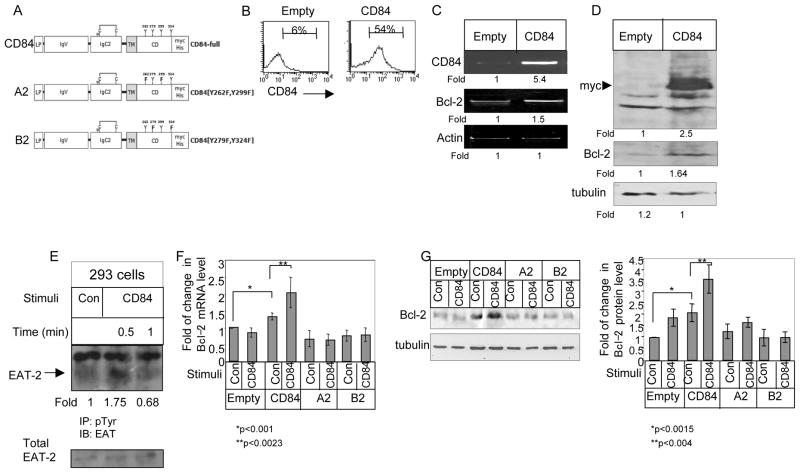
To characterize the signaling cascade induced by CD84 in CLL cells, CD84 was activated by an agonistic anti-CD84 mAb 8. The cells then were washed and bound mAb was cross-linked with F(ab′)2 goat anti-mouse Ig. While CD84 stimulation did not augment its own expression levels (data not show), it induced CD84 tyrosine phosphorylation (Fig. 3A). We next followed the CD84 signaling cascade in CLL cells. As shown in Fig 3B, increased Akt phosphorylation was detected following CD84 stimulation, peaking at 0.5–1 minutes following stimulation (Fig. 3B). To determine which SAP molecule is recruited following CD84 activation, cells were incubated in the presence or absence of anti-CD84, and phosphorylated SAP or EAT-2 levels were analyzed. SAP expression could not be detected at the mRNA level in CLL cells (data not shown). We therefore analyzed EAT-2 phosphorylation. As shown in Fig 3C, CD84 stimulation augmented EAT-2 phosphorylation, suggesting that activation of CD84 initiates a cascade that results in EAT-2 recruitment to the phosphorylated cytoplasmic tail of CD84.
Figure 3.
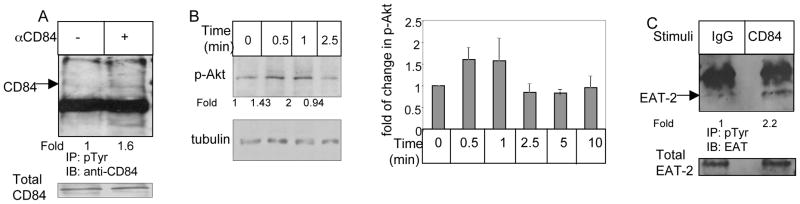
Activation of cell surface CD84 in CLL cells initiates a signaling cascade. CLL cells were incubated in the presence of anti-CD84 (0.25 μg/ml) or a control antibody (IgG) for 30 minutes. The cells then were washed and bound mAb was cross-linked with F(ab′)2 goat anti mouse Ig (0.5 μg). (A) After 5 minutes, cells were lysed and phosphorylated proteins were immunoprecipitated, separated on 8% (wt/vol) SDS/PAGE and blotted with anti-CD84 antibody. Results presented are representative of four separate experiments. (B) Cells were lysed at different time points following stimulation, and p-AKT and tubulin expression were analyzed by western blot analysis. Graph summarizes the results of five different experiments. (C) After 30 sec of stimulation, cells were lysed and phosphorylated proteins were immunoprecipitated, separated on 15% (wt/vol) SDS/PAGE, and blotted with anti-EAT-2 antibody. Results presented are representative of three separate experiments.
We next wished to determine whether the CD84 induced cascade regulates CLL survival. Cells were incubated in the presence or absence of anti-CD84, and Bcl-2 mRNA levels were analyzed. CD84 stimulation specifically elevated Bcl-2 mRNA (Fig. 4A–B; Table 4) and protein (Fig. 4C; Table 4) levels, suggesting that activation of CD84 initiates a survival cascade. We next followed CLL survival following CD84 stimulation by Annexin staining (Fig. 4D) and their caspase 3 and 7 activity was additionally analyzed as a marker of apoptosis by magic red staining (Fig. 4E). CD84 stimulation specifically reduced the Annexin/Magic Red-positive populations, showing that CD84 activation augments CLL survival.
Figure 4.
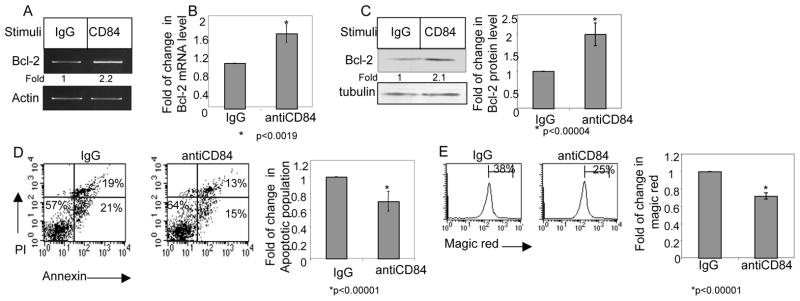
Stimulation of CD84 induces a survival cascade in CLL cells. CLL cells were incubated in the presence of agonistic anti-CD84 (1 ng/ml) or a control antibody (IgG) for 30 minutes. The cells then were washed and bound mAb was cross-linked with F(ab′)2 goat anti mouse Ig. (A) Following 18 h, the RNA was purified, and Bcl-2 and actin mRNA levels were analyzed. The results presented are representative of seven CLL patients. (B) Following 18 h, RNA was purified, and qPCR was performed using primers for Bcl-2 and RP-2. Results are expressed as fold of change in Bcl-2 mRNA in stimulated and non-stimulated cells; untreated cells were defined as 1. The graph summarizes results from four CLL patients. (C) Following 24 hr, cells were lysed and Bcl-2 and tubulin proteins were analyzed by western blot analysis. Graph summarizes the results of four different experiments. (D) Following 26h, cells were stained with Annexin V and PI, and analyzed by FACS. The graph summarizes the results of seven CLL patients. (E) Following 26h, cells were stained with Magic Red and analyzed by FACS. The graph summarizes the results of four CLL patients.
These results were further confirmed using another agonistic antibody, derived from the anti-CD84 agonistic hybridoma D1, that was generated against the CD84 extracellular domain (Materials and Methods). This stimulation resulted in a similar elevation of Bcl-2 mRNA levels (Supplementary Fig. 2A) and cell survival (Supplementary Fig. 2B). Together, these results show that CD84 functions as a survival receptor on CLL cells, and its activation initiates a cascade that regulates CLL survival.
CD84 tyrosine signaling motifs as mediators of the survival signal in HEK 293 cells
We next wished to determine whether the survival cascade induced by CD84 is specific to CLL cells, or whether this molecule can trigger a similar cascade in additional cell types. Therefore, a human CD84 isoform c (CD84-full) myc construct (Fig. 5A) was transfected to HEK-293 cells, which express low levels of endogenous CD84 (Fig. 5B–D). Expression of CD84 and its downstream cascade were then followed. CD84 cell surface expression levels were significantly elevated in HEK-293 transfected cells (Fig 5B). Overexpression of hCD84 elevated Bcl-2 mRNA (Fig. 5C) and protein (Fig. 5D) levels in the transfected cells. This signaling cascade resulted in EAT-2 recruitment to the phosphorylated cytoplasmic tail of CD84 (Fig 5E).
Figure 5.
Intracytoplasmic tyrosines are mediators of the CD84-induced survival signal in HEK 239 cells. (A) Schematic representation of the CD84-full construct and tyrosine-mutated versions. LP-leader peptide, IgV-immunoglobulin domain type V, IgC2- Immunoglobulin type C2, TM-transmembrane domain, CD-cytoplasmic domain. N indicates predicted site of N-glycosylation, C indicates disulfide bonds. Construct length is 367 aa. (B–D) HEK-293 cells transfected with full length CD84 (CD84) or empty constructs. (B) 24 h following transfection, cells were stained with anti-CD84, followed by secondary (goat anti-mouse) antibody, and analyzed by FACS. The results presented are representative of eight independent experiments with similar results. (C) RNA was purified 8h after transfection and levels of CD84, Bcl-2, and actin mRNA were analyzed. The results presented are representative of 17 independent experiments with similar results. (D) Cells were lysed by hot SDS, 24 h after transfection, and the lysates were separated on 12% (wt/vol) SDS/PAGE and blotted with anti-Myc, anti-Bcl-2 or anti-tubulin Abs, followed by anti-mouse HRP antibodies. The results shown represent 25 independent experiments with similar results. (E) HEK-293 cells transfected with FL-CD84. CD84 was cross-linked 24 h following transfection as described in Methods for 30 sec. Cells were then lysed, and phosphorylated proteins were immunoprecipitated, separated on 15% (wt/vol) SDS/PAGE and blotted with anti-EAT-2 antibody. Results presented are representative of three separate experiments. (F–G) HEK-293 cells were transfected with an empty vector (empty), full-length, native CD84 (CD84), mutated CD84 (A2), or mutated CD84 (B2) plasmid constructs. CD84 was cross-linked 6 h following transfection as described in Methods. (F) Following 18 h, RNA was purified, and Quantitative Real Time PCR was performed using primers for Bcl-2 and RP-2. Results are expressed as a fold of change in Bcl-2 levels in stimulated and non-stimulated cells; leves in untreated cells were defined as 1. The graph summarizes results of four independent experiments with similar results. (G) Following 24 hr, cells were lysed with hot SDS and the lysates were separated on 12% (wt/vol) SDS/PAGE and blotted with anti-Bcl-2 and tubulin antibodies, followed by anti-mouse HRP antibodies. The graph summarizes results of four independent experiments giving similar results.
In order to further dissect the mechanism of action of CD84 that results in induction of cell survival, we wished to determine the role of the tyrosine residues in the CD84 intra-cytoplasmic domain. The cytoplasmic tail of CD84c contains four tyrosine motifs (Y262, Y279, Y299, Y324). Two of them (Y262 and Y299) were characterized as consensus motifs for SH2-interactions with SAP/EAT2 8,37. Little is known about the other two SH2-interacting motifs (Y279 and Y324), besides the resemblance of Y324 to an ITIM motif 38. Thus, two different tyrosine substituted mutants were generated, one with Y262F and Y299F (A2) and the other with Y279F and Y324F (B2).
To determine which motifs regulate Bcl-2 expression, HEK-293 cells were transiently transfected with native hCD84, the two different tyrosine substituted CD84s (A2, B2), or empty vector as a control. After 6h, the cells were stimulated with an activating anti-CD84 antibody or control antibody. The cells then were harvested the expression of the constructs was verified by probing for the myc-tagged recognition sequence (not shown), and the Bcl-2 mRNA (Fig. 5F) and protein (Fig. 5G) levels were analyzed. Transfection of hCD84 elevated Bcl-2 expression. Moreover, hCD84 ligation with an agonistic anti-CD84 antibody further upregulated the levels of Bcl-2 mRNA (Fig. 5F) and protein (Fig. 5G). However, CD84 induced elevation of Bcl-2 mRNA was not observed in the HEK-293 cells transfected with the A2 or B2 mutated constructs. Thus, both pairs of tyrosine residues are essential for the CD84-induced survival cascade.
Blocking cell surface CD84 results in apoptosis of CLL cells
Since CD84 is a survival receptor for CLL cells, we next wished to determine whether blocking CD84 could induce cell death.
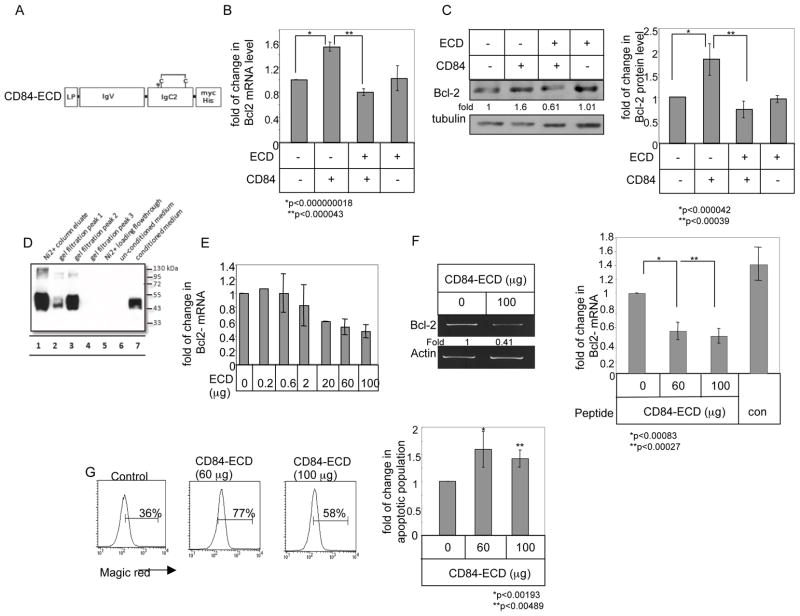
Cell-cell contacts and homophilic interactions were previously shown to regulate CD84 function 39. We therefore hypothesized that inhibiting these interactions using a truncated CD84 fragment that can bind to CD84 and thus blocks the receptor from further interactions. Thus, a myc/his6 tagged CD84 construct was generated, expressing CD84 extracellular domain, without its membranal and cytosolic parts (CD84-ECD; Fig. 6A). We first determined whether CD84-ECD can inhibit hCD84 activity. Therefore, HEK-293 cells were transfected with hCD84 and CD84-ECD constructs and the effect of CD84-ECD on CD84 activity was analyzed. Bcl-2 mRNA (Fig. 6B) and protein (Fig. 6C) were elevated in hCD84-transfected cells. This increase was abolished in cells co-transfected with CD84-ECD (Fig. 6B–C), while transfection with CD84-ECD alone had no significant effect on Bcl-2 expression levels. Finally we wished to determine whether CD84-ECD could inhibit the CD84 induced survival cascade in CLL cells. To that end, the secreted CD84-ECD protein was purified as described in Materials and Methods. The purified molecule was detected by western blot analysis as a prominent band between 43 kDa and 50 kDa (Fig. 6D). ECD-CD84 was then added to CLL cells and Bcl-2 levels were measured. As seen in Fig. 6E–F incubation with ECD-CD84 specifically down regulated Bcl-2 mRNA levels, and cell survival (Fig. 6G). Thus, homophilic interactions regulate CLL survival; interfering with these CD84 interactions induces cell death.
Figure 6.
CD84-ECD blocks the CD84–induced survival cascade in CLL cells. (A) Schematic view of the structure of CD84-ECD. LP-leader peptide, IgV-immunoglobulin domain type V, IgC2- Immunoglobulin domain type C2, TM-transmembrane domain, CD-cytoplasmic domain. N indicates predicted site of N-glycosylation; C indicates disulfide bonds. Length of construct is 330 aa. (B–C) HEK 293 cells were transfected in the presence or absence of empty vector, CD84, or CD84-ECD. (B) Following 18 hours, RNA was purified. Quantitative Real Time PCR was performed using primers for Bcl-2 and RP-2. Results are expressed as the fold change in Bcl-2 expression following the various treatments relative to cells transfected with empty construct, which was defined as 1. The graph summarizes results of four independent experiments with similar results. (C) Cells were lysed 24 hr after transfection, and Bcl-2 and tubulin expression were analyzed by western blot analysis. The graph summarizes results of four independent experiments with similar results. (D) Samples from different purification stages were separated on a 12% gel, and myc expression was analyzed by western blot analysis. Conditioned medium (lane 7) was purified using Ni2+ affinity chromatography (lanes 1 and 5), followed by size exclusion chromatography. Lanes 2–4 shows the peak fraction of the size exclusion chromatography. (E–F) CLL cells were incubated with different amounts of CD84-ECD (E. F) or control (F) proteins. Following 18 h, RNA was purified. RT-PCR and qPCR were performed using primers for Bcl-2 and RP-2. Results are expressed as fold change in Bcl-2 mRNA levels in treated compared to untreated cells, which was defined as 1. The graph summarizes results from three independent experiments with similar results (E), and six (CD84-ECD) and three (Control) independent experiments with similar results (F). (G) CLL cells were incubated with CD84-ECD protein. Following 24h, the cells were stained with Magic Red and analyzed by FACS. The graph summarizes results of five CLL patients.
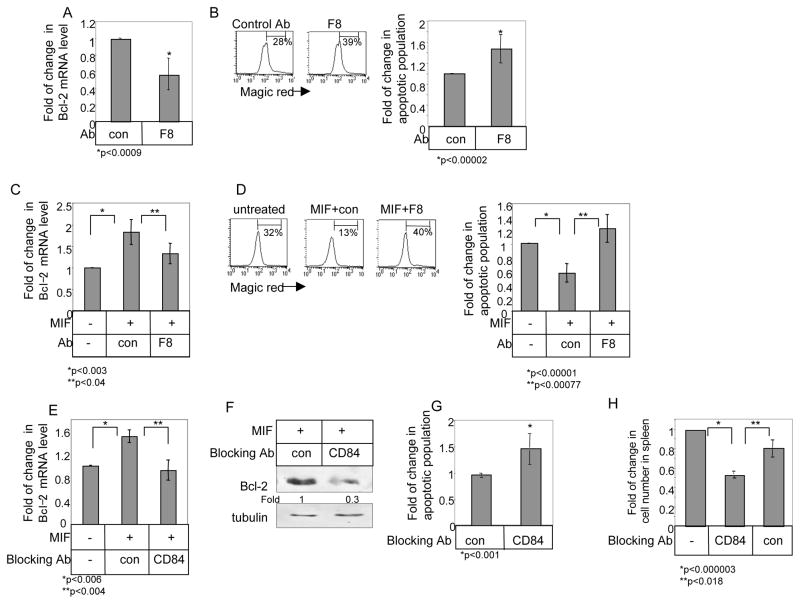
We next wished to determine whether blocking CD84 activity with antibodies induces cell death. CLL cells were incubated with supernatant derived from an antagonistic hybridoma (F8) that recognizes the extracellular domain fragment of CD84 (Materials and Methods) and survival followed over time. Supernatant derived from hybridoma F8 reduced Bcl-2 mRNA levels (Fig. 7A) and CLL survival (Fig. 7B). Thus, blocking of CD84 regulates CLL survival. Moreover, blocking CD84 with the F8 hybridoma inhibited the MIF-induced elevation of Bcl-2 mRNA (Fig. 7C), and cell survival (Fig. 7D). Together, our results show that CD84 antagonistic antibodies induce cell death.
Figure 7.
Blocking cell surface CD84 induces CLL cell death. (A–B) CLL cells were incubated in the presence of an anti-CD84 containing supernatant of hybridoma F8 or supernatant of control hybridoma. (A) Following 18h, the RNA was purified, and Quantitative Real Time PCR was performed using primers for Bcl-2 and RP-2. Results are expressed as fold of change in Bcl-2 expression in treated or control cells; control cells were defined as 1. The graph summarizes results from three CLL patients. (B) Following 26h, cells were stained with Magic Red and analyzed by FACS. The graph summarizes results of eight CLL patients. (C–D) CLL cells were incubated in the presence or absence of MIF, and supernatant of hybridoma F8 or supernatant of control hybridoma. (C) Following 18h, the RNA was purified and qPCR was performed using primers for Bcl-2 and RP-2. Results are expressed as a fold of change in Bcl-2 expression in the presence or absence of the hybridoma supernatant; control cells were defined as 1. The graph summarizes results of four CLL patients. (D) Following 26h, cells were stained with Magic Red and analyzed by FACS. The graph summarizes results of six CLL patients. (E–F) CLL cells were incubated in the presence or absence of MIF (100 ng/ml) and CD84 blocking (Biolegend; 2.5 μg) antibody or a control antibody at 37°C for 18–24 h. (E) Following 18h, the RNA was purified and qPCR was performed using primers for Bcl-2 and RP-2. Results are expressed as fold change in Bcl-2 mRNA levels in stimulated cells compared to unstimulated cells, which were defined as 1. The graph summarizes the results from three patients. (F) After 24 hr, cells were lysed and Bcl-2 and tubulin proteins were analyzed by western blot analysis. The results presented are representative of cells from three CLL patients. (G) CLL cells were incubated in the presence or absence of CD84 blocking (Biolegend; 4 μg) antibody or a control antibody at 37°C. Following 26h, cells were stained with Magic Red and analyzed by FACS. The graph summarizes results of five CLL patients. (H) CLL cells were stained with CFSE and were injected i.v. into C57BL/6 mice. After 1h, mice were i.v. injected with anti-CD84 blocking or isotype control antibodies (Biolegend; 2.5 μg). After an additional 3 h, spleen cells were analyzed by FACS. The graph shows the numbers of labeled cells recovered from the spleens of five mice in each group.
In addition, CLL cells were incubated with a commercially available antagonistic anti-CD84 Ab (blocking Ab), that recognizes the extracellular domain fragment of CD84 and survival was followed over time. Similarly to the results with the supernatant of the hybridoma F8, incubation with the blocking antibody reduced Bcl-2 mRNA levels (Fig 7E), Bcl-2 protein levels (Fig 7F) and cell survival (Fig 7G).
Finally, we wished to determine whether blocking CD84 activity might have a therapeutic effect by regulating the in vivo survival of CLL cells. It was recently shown in a CLL xenograft model that the highest absolute numbers of recovered human cells were consistently observed in the murine spleens, especially when the tumor cells were administered i.v. 40; therefore, CLL survival following CD84 blockade was analyzed in the spleen. CLL cells were stained with CFSE and i.v. injected to C57BL/6 mice to allow their homing to the spleen, an event that was shown to be almost integrin independent 22. After, 1 h, mice were injected with anti-CD84 blocking antibody or an isotype control antibody, and after an additional 3 hours, CLL numbers in the spleen were analyzed by FACS. As shown in Fig 7H, blocking CD84 resulted in a significant reduction in the CLL cell population. Thus, CD84 has a significant in vivo effect in regulating CLL survival.
Discussion
Chronic lymphocytic leukemia is a malignant disease characterized by the progressive accumulation of small mature B-lymphocytes in peripheral blood, BM and secondary lymphoid organs. The accumulation of tumor cells in patients results primarily from a defect in apoptosis. Several mechanisms were previously suggested to regulate CLL survival. CLL cells are endowed with a functional B-cell receptor (BCR) that allows interaction with antigen (Ag). The nature of the Ag together with BCR affinity promote malignant cell survival and growth. In addition, the CLL microenvironment was found to control CLL cell survival and growth 41. Despite these insights into the nature of these survival pathways and steady improvements in patient outcomes over the last decade, there is still a need for more targeted and curative therapy in CLL.
We have previously shown that CLL cells express high levels of CD74, which upon stimulation with its natural ligand, MIF, initiates a signaling cascade leading to cell survival. We further demonstrated that the humanizd anti-CD74 mAb, hLL-1 (milatuzumab), blocks the signaling cascade initiated by MIF 21. In addition, MIF stimulation was shown to induce the expression of TAp63, resulting in augmented expression of the integrin, VLA-4, particularly during the advanced stage of CLL. In vivo blockade of CD74, TAp63 or VLA-4 inhibits the homing of CLL cells to the bone marrow. Thus, CD74 and its downstream target genes, TAp63 and VLA-4, facilitate the migration of CLL cells back to the bone marrow, where they interact with a supportive marrow environment that rescues them from apoptosis 22.
In the current study, we searched for novel MIF/CD74 target genes in CLL cells. We show that the expression of the SLAM family member, CD84, whose expression levels are significantly elevated on CLL cells from the early stages of the disease, is regulated by MIF and its receptor, CD74.
We further show that CD84 isoform c is the predominant isoform in both cells from healthy controls, and in early and advanced stage CLL patients, and that its expression is significantly upregulated in the CLL cells. Homophilic interactions, or activation (cross-linking) of CD84 in CLL cells induce a signaling cascade that involves CD84 tyrosine phosphorylation, EAT-2 recruitment, and increased Akt phosphorylation, resulting in augmented Bcl-2 expression and CLL survival. A similar survival cascade was observed in HEK-293 cells transfected with hCD84, suggesting that CD84 survival activity is not restricted to CLL cells, and that this receptor may serve as a survival receptor in various cell types.
The cytoplasmic tail of CD84 isoform c contains both ITSM and non-ITSM phosphotyrosine motifs: Y262, Y279, Y299 and Y324. While it is known that Y262 and Y299 interact with SH2-domain containing proteins, such as SAP and EAT-2, the characteristics of Y279 and Y324 are less well-established 38. Our results show that the two pairs of tyrosines in CD84 are essential for the CD84-induced survival cascade (a model summarizing our results is presented in Supplementary Fig. 3). Together, these results suggest that CD84 is a survival receptor and therefore might play a major role in survival of tumor cells (Supplementary Fig. 3).
EAT-2 transcripts have been detected in murine NK cells, macrophages and B cells, and in human B cell lines 37,42 and activated T cells 43, whereas the EAT-2 protein has been detected in human NK cells and CD8+ T cells but not in B cells 44. Here, we show expression of EAT-2 protein in CLL cells. SAP and EAT-2 were found to have a high degree of homology; consequently, it was proposed that SAP and EAT-2 might have functional similarities. However, it was demonstrated that SAP and EAT-2 have opposing roles in lymphocyte activation 42. SAP was recently shown to have a pro-apoptotic function in T cells and high levels of SAP are associated with increased death rated in response to activation in human and mice T cells 45,46. We show herein that EAT-2 recruitment induces cell survival, further confirming the opposing roles of these molecules.
Homophilic interactions of SLAM family members lead to clustering and activation of the receptor 39,47. It was recently shown that CD84 is involved in T cell:B cell contact, optimal T follicular helper function, and germinal center formation in vivo. Moreover, both CD84 and another SLAM member, Ly108, mediate T cell adhesion and participate in stable T cell:B cell interactions 14. In the bone marrow and lymph nodes, CLL cells interact with a variety of accessory and bystander cells that support CLL cell survival and are believed to promote resistance to therapy. It is plausible that these interactions are CD84 dependent as well. It will be important therefore to analyze the peripheral cells that are involved in the CD84 dependent interaction.
In an attempt to block CD84 activity, we used a commercially available antibody and generated the F8 hybridoma, which recognizes the CD84 extracellular domain, thereby inhibiting the CD84 induced downstream signaling cascade. In addition, we prepared and purified a recombinant CD84 extracellular domain (CD84-ECD), which most likely binds cellular CD84 and serves as a competitive inhibitor. Our results show that inhibition of CD84 leads to reduced Bcl-2 expression and elevation of CLL cell death in vitro. In addition, the significant effect of the anti-CD84 in vivo treatment suggests a novel target and new potential modalities for the therapy of CLL. The clinical potential of monoclonal antibodies in oncology has been fully realized only recently. The use of anti-CD20 monoclonal antibody (Rituximab) in CLL set the stage for the development of novel agents to target B cell malignancies. Anti-CD52 (Alemtuzumab) has also shown remarkable activity in patients with CLL, and a series of additional monoclonal antibodies with specificity against a variety of antigens on the surface of B cells (CD22,23,40,70 HLA and others) are currently undergoing evaluation. Our results suggest that antibody-mediated blocking of the MIF/CD74/CD84 pathway also could lead to better treatment of CLL, resulting in decreased cell survival from the early stage of the disease.
In conclusion, our data show that overexpression of CD84 in CLL is an important survival mechanism that appears to be an early event in the pathogenesis of the disease. These findings could pave the way toward unique therapeutic strategies aimed at interrupting this survival pathway.
Table 1.
Patients’ characteristics
| Summary of 71 patients | ||
|---|---|---|
| Age (mean) | 67 (range-34–94) | |
| Male/female | 29/34 | |
| Lymphocyte count at time of study (103/μl) (average) | 58 (range-6–300) | |
| Doubling time | Less then six month | 11 |
| More then six month | 24 | |
| NA* | 36 | |
| RAI stage | 0–I | 36 |
| II | 15 | |
| III | 7 | |
| IV | 13 | |
| ZAP-70 (PCR) | positive | 22 |
| negative | 22 | |
| NA | 27 | |
| Serum IgG | <700 mg/dl | 21 |
| >700 mg/dl | 25 | |
| NA | 25 | |
| Autoimmune phenomena** | 7 | |
| Previous chemotherapy | Leukeran/prednisone | 9 |
| FCR | 17 | |
| chlorambucil | 3 | |
| none | 47 |
Due to recurrent courses of chemotherapy.
Includes Coomb’s positive hemolytic anemia, Evan’s syndrome, immune thryroiditis and Sjögren’s disease.
L/P-Leukeran/Prednisone, FC/R-Fludarabine and Endoxan with or without Rituximab.
Acknowledgments
The authors gratefully acknowledge members of the Shachar laboratory team for helpful discussions. This research was supported by the Minerva foundation, Israel Science Foudnation, The Gurwin Foundation, the NIH, and the Alliance for Lupus Research (RB, LL), and in part by USPHS grant PO1-CA103985 from the National Cancer Institute, NIH (DMG), and by the United Israel Appeal of Canada. I.S. is the incumbent of the Dr. Morton and Ann Kleiman Professorial Chair.
Footnotes
Conflict of interest statement: D.M.G. is a director and stockholder of Immunomedics, Inc., which is developing the hLL1 mAb.
Author contribution:
I.B.-E.- Designed and performed most of the experiments, analyzed results, wrote the paper.
A.M.- Designed and performed some of the experiments, analyzed results, participate in writing the paper.
M.C.S.- Designed and performed some of the experiments, analyzed results, participate in writing the paper.
F.L.- Designed and performed some of the experiments, analyzed results.
N.H.- Provided reagents.
L.S.- Provided reagents.
A.B.- Suprevised some of the experiments.
I.H.-H.- Provided reagents.
M.H.- Provided reagents.
Y.H.- Provided reagents, discussed some of the results.
D.M.G.- Provided reagents and participate in writing the paper.
A.A.- Purified reagent.
I.S.- Provided reagent, discussed some of the results.
L.L.- Provided reagents.
R.B.- Provided reagents and participate in writing the paper.
I.S.- Designed and supervised the experiments, analyzed the results and wrote the paper.
References
- 1.Caligaris-Cappio F, Hamblin TJ. B-cell chronic lymphocytic leukemia: a bird of a different feather. J Clin Oncol. 1999;17:399–408. doi: 10.1200/JCO.1999.17.1.399. [DOI] [PubMed] [Google Scholar]
- 2.Calpe S, Wang NH, Romero X, et al. The SLAM and SAP gene families control innate and adaptive immune responses. Advances in Immunology. 2008;97:177–250. doi: 10.1016/S0065-2776(08)00004-7. [DOI] [PubMed] [Google Scholar]
- 3.Cocks BG, Chang CCJ, Carballido JM, Yssel H, Devries JE, Aversa G. A Novel Receptor Involved in T-Cell Activation. Nature. 1995;376:260–263. doi: 10.1038/376260a0. [DOI] [PubMed] [Google Scholar]
- 4.Engel P, Eck MJ, Terhorst C. The SAP and SLAM families in immune responses and X-linked lymphoproliferative disease. Nature Reviews Immunology. 2003;3:813–821. doi: 10.1038/nri1202. [DOI] [PubMed] [Google Scholar]
- 5.Veillette A, Latour S. The SLAM family of immune-cell receptors. Current Opinion in Immunology. 2003;15:277–285. doi: 10.1016/s0952-7915(03)00041-4. [DOI] [PubMed] [Google Scholar]
- 6.Nichols KE, Ma CS, Cannons JL, Schwartzberg PL, Tangye SG. Molecular and cellular pathogenesis of X-linked lymphoproliferative disease. Immunological Reviews. 2005;203:180–199. doi: 10.1111/j.0105-2896.2005.00230.x. [DOI] [PubMed] [Google Scholar]
- 7.Latour S, Veillette A. The SAP family of adaptors in immune regulation. Seminars in Immunology. 2004;16:409–419. doi: 10.1016/j.smim.2004.08.020. [DOI] [PubMed] [Google Scholar]
- 8.Tangye SG, van de Weerdt BCM, Avery DT, Hodgkin PD. CD84 is up-regulated on a major population of human memory B cells and recruits the SH2 domain containing proteins SAP and EAT-2. European Journal of Immunology. 2002;32:1640–1649. doi: 10.1002/1521-4141(200206)32:6<1640::AID-IMMU1640>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 9.Veillette A. Immune regulation by SLAM family receptors and SAP-related adaptors. Nature Reviews Immunology. 2006;6:56–66. doi: 10.1038/nri1761. [DOI] [PubMed] [Google Scholar]
- 10.delaFuente MA, Pizcueta P, Nadal M, Bosch J, Engel P. CD84 leukocyte antigen is a new member of the Ig superfamily. Blood. 1997;90:2398–2405. [PubMed] [Google Scholar]
- 11.Martin M, Romero X, de la Fuente MA, et al. CD84 functions as a homophilic adhesion molecule and enhances IFN-gamma secretion: Adhesion is mediated by Ig-like domain 1. Journal of Immunology. 2001;167:3668–3676. doi: 10.4049/jimmunol.167.7.3668. [DOI] [PubMed] [Google Scholar]
- 12.Romero X, Zapater N, Calvo M, et al. CD229 (Ly9) lymphocyte cell surface receptor interacts homophilically through its N-terminal domain and relocalizes to the immunological synapse. Journal of Immunology. 2005;174:7033–7042. doi: 10.4049/jimmunol.174.11.7033. [DOI] [PubMed] [Google Scholar]
- 13.Yan QR, Malashkevich VN, Fedorov A, et al. Structure of CD84 provides insight into SLAM family function. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:10583–10588. doi: 10.1073/pnas.0703893104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cannons JL, Qi H, Lu KT, et al. Optimal Germinal Center Responses Require a Multistage T Cell:B Cell Adhesion Process Involving Integrins, SLAM-Associated Protein, and CD84. Immunity. 2010;32:253–265. doi: 10.1016/j.immuni.2010.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Palou E, Pirotto F, Sole J, et al. Genomic characterization of CD84 reveals the existence of five isoforms differing in their cytoplasmic domains. Tissue Antigens. 2000;55:118–127. doi: 10.1034/j.1399-0039.2000.550203.x. [DOI] [PubMed] [Google Scholar]
- 16.Calandra T, Bernhagen j, Mitchell RA, Bucala R. The macrophage is an important and previously unrecognized source of macrophage migration inhibitory factor. J Exp Med. 1994;179:1895–1902. doi: 10.1084/jem.179.6.1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bernhagen J, Bacher M, Calandra T, et al. An essential role for macrophage migration inhibitory factor in the tuberculin delayed-type hypersensitivity reaction. J Exp Med. 1996;183:277–282. doi: 10.1084/jem.183.1.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leng L, Metz CN, Fang Y, et al. MIF Signal Transduction Initiated by Binding to CD74. J Exp Med. 2003;197:1467–1476. doi: 10.1084/jem.20030286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gore Y, Starlets D, Maharshak N, et al. Macrophage migration inhibitory factor (MIF) induces B cell survival by activation of a CD74/CD44 receptor complex. J Biol Chem. 2008;283:2784–2792. doi: 10.1074/jbc.M703265200. [DOI] [PubMed] [Google Scholar]
- 20.Shi X, Leng L, Wang T, et al. CD44 Is the Signaling Component of the Macrophage Migration Inhibitory Factor-CD74 Receptor Complex. Immunity. 2006;25:595–606. doi: 10.1016/j.immuni.2006.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Binsky I, Haran M, Starlets D, et al. IL-8 secreted in a macrophage migration-inhibitory factor- and CD74-dependent manner regulates B cell chronic lymphocytic leukemia survival. Proc Natl Acad Sci U S A. 2007;104:13408–13413. doi: 10.1073/pnas.0701553104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Binsky I, Lantner F, Grabovsky V, et al. TAp63 regulates VLA-4 expression and CLL cell migration to the BM in a CD74 dependent manner. J Immunol. 2010;184:4761–4769. doi: 10.4049/jimmunol.0904149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Haran M, Chebatco S, Flaishon L, et al. Grb7 expression and cellular migration in chronic lymphocytic leukemia: a comparative study of early and advanced stage disease. Leukemia. 2004;18:1948–1950. doi: 10.1038/sj.leu.2403512. [DOI] [PubMed] [Google Scholar]
- 24.Kay S, Herishanu Y, Pick M, et al. Quantitative flow cytometry of ZAP-70 levels in chronic lymphocytic leukemia using molecules of equivalent soluble fluorochrome. Cytometry Part B-Clinical Cytometry. 2006;70B:218–226. doi: 10.1002/cyto.b.20078. [DOI] [PubMed] [Google Scholar]
- 25.Hallek M, Cheson BD, Catovsky D, et al. Guidelines for the diagnosis and treatment of chronic lymphocytic leukemia: a report from the International Workshop on Chronic Lymphocytic Leukemia updating the National Cancer Institute-Working Group 1996 guidelines. Blood. 2008;111:5446–5456. doi: 10.1182/blood-2007-06-093906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cheson BD, Bennett JM, Grever M, et al. National Cancer Institute-sponsored Working Group guidelines for chronic lymphocytic leukemia: revised guidelines for diagnosis and treatment. Blood. 1996;87:4990–4997. [PubMed] [Google Scholar]
- 27.Bernhagen J, Mitchell RA, Calandra T, Voelter W, Cerami A, Bucala R. Purification, bioactivity, and secondary structure analysis of mouse and human macrophage migration inhibitory factor (MIF) Biochemistry. 1994;33:14144–14155. doi: 10.1021/bi00251a025. [DOI] [PubMed] [Google Scholar]
- 28.Stein R, Qu Z, Cardillo TM, et al. Antiproliferative activity of a humanized anti-CD74 monoclonal antibody, hLL1, on B-cell malignancies. Blood. 2004;104:3705–3711. doi: 10.1182/blood-2004-03-0890. [DOI] [PubMed] [Google Scholar]
- 29.Stein R, Mattes MJ, Cardillo TM, et al. CD74: A new candidate target for the immunotherapy of B-Cell neoplasms. Clinical Cancer Research. 2007;13:5556S–5563S. doi: 10.1158/1078-0432.CCR-07-1167. [DOI] [PubMed] [Google Scholar]
- 30.Matza D, Kerem A, Lantner F, Shachar I. Invariant chain induced B cell differentiation requires intramembrane - proteolytic release of the cytosolic domain. Immunity. 2002;17:549–560. doi: 10.1016/s1074-7613(02)00455-7. [DOI] [PubMed] [Google Scholar]
- 31.Starlets D, Gore Y, Binsky I, et al. Cell Surface CD74 initiates a signaling cascade leading to cell proliferation and survival. Blood. 2006;107:4807–4816. doi: 10.1182/blood-2005-11-4334. [DOI] [PubMed] [Google Scholar]
- 32.Matza D, Wolstein O, Dikstein R, Shachar I. Invariant Chain Induces B Cell Maturation by Activating TAFII105-NF-kB Dependent Transcription Program. J Biol Chem. 2001;276:27203–27206. doi: 10.1074/jbc.M104684200. [DOI] [PubMed] [Google Scholar]
- 33.Arjona a, Foellmer HG, Town T, et al. Abrogation of macrophage migration inhibitory factor decreases West Nile virus lethality by limiting viral neuroinvasion. Journal of Clinical Investigation. 2007;117:3059–3066. doi: 10.1172/JCI32218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Luger D, Dayan M, Zinger H, Liu JP, Mozes E. A peptide based on the complementarity determining region 1 of a human monoclonal autoantibody ameliorates spontaneous and induced lupus manifestations in correlation with cytokine immunomodulation. J Clin Immunol. 2004;24:579–590. doi: 10.1007/s10875-004-6245-2. [DOI] [PubMed] [Google Scholar]
- 35.Ho MK, Springer TA. Preparation and use of monoclonal antimacrophage antibodies. Methods Enzymol. 1984;108:313–324. doi: 10.1016/s0076-6879(84)08098-8. [DOI] [PubMed] [Google Scholar]
- 36.Dios A, Mitchell RA, Aljabari B, et al. Inhibition of MIF bioactivity by rational design of pharmacological inhibitors of MIF tautomerase activity. J Med Chem. 2002;45:2410–2416. doi: 10.1021/jm010534q. [DOI] [PubMed] [Google Scholar]
- 37.Morra M, Lu J, Poy F, et al. Structural basis for the interaction of the free SH2 domain EAT-2 with SLAM receptors in hematopoietic cells. Embo Journal. 2001;20:5840–5852. doi: 10.1093/emboj/20.21.5840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Oliver-Vila I, Saborit-Villarroya I, Engel P, Martin M. The leukocyte receptor CD84 inhibits Fc epsilon RI-mediated signaling through homophilic interaction in transfected RBL-2H3 cells. Molecular Immunology. 2008;45:2138–2149. doi: 10.1016/j.molimm.2007.12.006. [DOI] [PubMed] [Google Scholar]
- 39.Nanda N, Andre P, Bao M, et al. Platelet aggregation induces platelet aggregate stability via SLAM family receptor signaling. Blood. 2005;106:3028–3034. doi: 10.1182/blood-2005-01-0333. [DOI] [PubMed] [Google Scholar]
- 40.Durig J, Ebeling P, Grabellus F, et al. A novel nonobese diabetic/severe combined immunodeficient xenograft model for chronic lymphocytic leukemia reflects important clinical characteristics of the disease. Cancer Research. 2007;67:8653–8661. doi: 10.1158/0008-5472.CAN-07-1198. [DOI] [PubMed] [Google Scholar]
- 41.Caligaris-Cappio F, Ghia P. Novel insights in chronic lymphocytic leukemia: Are we getting closer to understanding the pathogenesis of the disease? Journal of Clinical Oncology. 2008;26:4497–4503. doi: 10.1200/JCO.2007.15.4393. [DOI] [PubMed] [Google Scholar]
- 42.Roncagalli R, Taylor JER, Zhang SH, et al. Negative regulation of natural killer cell function by EAT-2, a SAP-related adaptor. Nature Immunology. 2005;6:1002–1010. doi: 10.1038/ni1242. [DOI] [PubMed] [Google Scholar]
- 43.Tangye SG, Nichols KE, Hare NJ, van de Weerdt BCM. Functional requirements for interactions between CD84 and Src homology 2 domain-containing proteins and their contribution to human T cell activation. Journal of Immunology. 2003;171:2485–2495. doi: 10.4049/jimmunol.171.5.2485. [DOI] [PubMed] [Google Scholar]
- 44.Tassi I, Colonna M. The cytotoxicity receptor CRACC (CS-1) recruits EAT-2 and activates the PI3K and phospholipase C gamma signaling pathways in human NK cells. Journal of Immunology. 2005;175:7996–8002. doi: 10.4049/jimmunol.175.12.7996. [DOI] [PubMed] [Google Scholar]
- 45.Nagy N, Matskova L, Kis LL, Hellman U, Klein G, Klein E. The proapoptotic function of SAP provides a clue to the clinical picture of X-linked lymphoproliferative disease. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:11966–11971. doi: 10.1073/pnas.0905691106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen G, Tai AK, Lin M, Chang F, Terhorst C, Huber BT. Increased proliferation of CD8(+) T cells in SAP-deficient mice is associated with impaired activation-induced cell death. European Journal of Immunology. 2007;37:663–674. doi: 10.1002/eji.200636417. [DOI] [PubMed] [Google Scholar]
- 47.Howie D, Simarro M, Sayos J, Guirado M, Sancho J, Terhorst C. Molecular dissection of the signaling and costimulatory functions of CD150 (SLAM): CD150/SAP binding and CD150-mediated costimulation. Blood. 2002;99:957–965. doi: 10.1182/blood.v99.3.957. [DOI] [PubMed] [Google Scholar]