Summary
Background
When standard therapies fail, clinical trials provide experimental treatment opportunities for patients with drug-resistant illnesses or terminal diseases. Clinical Trials can also provide free treatment and education for individuals who otherwise may not have access to such care. To find relevant clinical trials, patients often search online; however, they often encounter a significant barrier due to the large number of trials and in-effective indexing methods for reducing the trial search space.
Objectives
This study explores the feasibility of feature-based indexing, clustering, and search of clinical trials and informs designs to automate these processes.
Methods
We decomposed 80 randomly selected stage III breast cancer clinical trials into a vector of eligibility features, which were organized into a hierarchy. We clustered trials based on their eligibility feature similarities. In a simulated search process, manually selected features were used to generate specific eligibility questions to filter trials iteratively.
Results
We extracted 1,437 distinct eligibility features and achieved an inter-rater agreement of 0.73 for feature extraction for 37 frequent features occurring in more than 20 trials. Using all the 1,437 features we stratified the 80 trials into six clusters containing trials recruiting similar patients by patient-characteristic features, five clusters by disease-characteristic features, and two clusters by mixed features. Most of the features were mapped to one or more Unified Medical Language System (UMLS) concepts, demonstrating the utility of named entity recognition prior to mapping with the UMLS for automatic feature extraction.
Conclusions
It is feasible to develop feature-based indexing and clustering methods for clinical trials to identify trials with similar target populations and to improve trial search efficiency.
Keywords: medical informatics, search engine, clinical trials, knowledge representation, eligibility determination
1. Introduction
Randomized Controlled Trials (RCTs) provide high-quality evidence for clinical practice. Many RCTs, especially those that test hopeful therapies for terminal diseases or offer free medications to patients, also represent valuable treatment opportunities for patients. Various informatics techniques have been developed to boost RCT recruitment (1) including methods that help trial sponsors or investigators to use electronic data to prescreen potentially eligible patients (2–4), as well as methods that aid patients in identifying applicable trial opportunities (5–20). Several trial search engines have been developed, including ASCOT (15), TrialX (6, 10), ResearchMatch (16, 17), caMatch (18), Corengi (19) and a special search engine developed by the University of Florida (20). The focus of this research was to assess the feasibility of a feature-based indexing method to facilitate clinical trial indexing, clustering, and search to better engage patients in finding relevant trials.
At the point of writing, there were more than 130,000 trials for about 5,000 diseases on ClinicalTrials.gov with a few thousand trials each for common diseases (21, 22). However, searching through a large collection of clinical trials using the existing technology is a daunting task (23). Trial eligibility criteria are not structured to facilitate fine-grained search because they are listed in free-text form so that existing trial search engines routinely return results with unsatisfactory specificity and hence require laborious manual reviews to identify a relevant study from dozens, hundreds, or thousands of trials. Therefore, efficient indexing of clinical trials using eligibility criteria is greatly needed to facilitate user-friendly clinical trial search.
Moreover, the existing state-of-the-art search engines tend to use term-based information retrieval methods that measure the similarity between each document and an input query (24–26). For example, in the medical domain the cosine similarity metric was used to generate tag clouds (27) and rank clinical trial summaries (28). Other methods use Boolean operators to indicate the presence of search terms in a set of documents, which are then retrieved unordered (29, 30). To improve accuracy, a user is often required to specify relevant disease-related terms or synonyms in queries, which can be difficult for patients (31), who may not be familiar with such synonyms without support (32–34).
Medical Subject Heading (MeSH) terms (35) enable users to easily locate pertinent articles indexed by these terms in MEDLINE. Observing that MeSH terms are effective at indexing MEDLINE documents, we were inspired to develop MeSH’s counterpart for ClinicalTrials.gov. Noticing that researchers in nursing informatics have developed concept-oriented data dictionaries to meet the context-specific information needs of users (36), we hypothesized that concept-oriented eligibility features could facilitate meaningful and efficient indexing for clinical trials to facilitate effective user search. In the context of this paper, a concept-oriented eligibility feature is a clinically meaningful atomic patient state, which can be a diagnosis, a symptom, or a demographic characteristic, for ascertaining a volunteer’s eligibility for a trial, and is derived from eligibility criteria. Feature identification, whereby distinctive characteristics associated with a digital file are identified and extracted (37), has been an established technique for indexing both textual documents or images (38–41).
Given that there is no prior feature-based clinical trial indexing system, this study aims to set a baseline for feature-based trial indexing and clustering methods through manual review. To make the scope manageable, this study focuses on clinical trial eligibility criteria for studies of one disease, stage III breast cancer. We explore the feasibility of feature-based methods for indexing clinical trials using two use cases: clustering of clinical trials with similar eligibility criteria (42) and indexing of clinical trials to improve the specificity of trial search results.
The objective of this paper is four-fold: 1.) manual identification of concept-oriented eligibility features for 80 randomly selected clinical trials on stage III breast cancer; 2.) feature-based trial clustering to identify trials with similar recruitment populations; 3.) feature-based trial search to find trials matching volunteer conditions; and 4.) analysis of Unified Medical Language System (UMLS) terms (43) in eligibility features in order to assess the eligibility feature complexity and to inform the design of automated methods for feature discovery from eligibility criteria.
2. Methods
2.1 Data and feature annotation
We used the keyword “breast cancer stage 3” in the search form on Clinicaltrials.gov to retrieve stage III breast cancer clinical trials (22). From the result of 1,853 relevant trials, we randomly selected 80 and downloaded their free-text eligibility criteria for further analyses.
Due to the complexity in free-text eligibility criteria (44, 45) and the limitations of current natural language understanding techniques, the first author used the competency decomposition method developed by Sim et al.(46) to manually decompose each eligibility criterion into a vector of discrete eligibility features (47). Using a set of heuristics, we identified atomic eligibility data elements, which were sometimes implicit, and used sentence segments to determine their boundaries. Sentence fragments were iteratively decomposed until the lowest level clinical concept was reached. If a conjunction was reached, the two concepts directly involved in the conjunction were treated as separate features. For example, “synchronous bilateral invasive or non-invasive breast cancer” was decomposed into four eligibility features - synchronous bilateral breast cancer, invasive breast cancer, non-invasive breast cancer, and breast cancer. Hence if one trial contained “synchronous bilateral invasive breast cancer” and another contained “synchronous bilateral non-invasive breast cancer” the two trials would share three out of the four features. This process was iterated for each criterion, generating a hierarchical representation whose leaf nodes were eligibility features.
Next, we annotated feature modifiers (e.g., positive, negative) when available for all categorical features (e.g., hormone estrogen receptor 2 or HER-2). For eligibility features with continuous values (e.g., laboratory test values), we annotated the value attributes only for age and life expectancy. In addition, in feature specifications we did not include temporal constraints, e.g., “within the past 6 months”, or negation except for one standalone feature, i.e., no evidence of disease (ned), as these constraints are simply modifiers. An example of typical feature annotations is shown in Figure 1.
Figure 1. Example criteria with feature annotations.

Criteria are denoted by italics and features follow after the arrows
In order to assess the features’ reliability, three raters – two informaticians and one informatics-trained physician – evaluated a small subset of the extracted features, each being present in more than 20 trials in our sample of 80 trials. We evaluated the inter-rater reliability on feature extraction using these features by asking each rater to annotate the presence and absence of each feature in all the 80 trials (Table 2). The raters were provided with a list of feature synonyms to help recognize equivalent semantic representations of the features. If a rater determined a feature to be present in a trial they denoted its presence with a ‘1’ otherwise they coded a ‘0’. We then computed Fleiss’s Kappa coefficient (1 = perfect agreement; 0 = chance agreement) between the three annotators to assess the strength of the raters’ agreement for feature presence and absence (48, 49).
Table 2.
Characteristics of 37 common eligibility features each present in > 20 trials
| Feature Class Path | Feature Name | Frequency |
|---|---|---|
| Patient-Characteristics, Accepts Healthy Volunteers | accepts healthy volunteers | 80 |
| Patient-Characteristics, Gender | Gender | 80 |
| Patient-Characteristics, Age | Age | 71 |
| Disease-Characteristics, Disease | breast cancer | breast carcinoma | 58 |
| Patient-Characteristics, Other | Pregnant | 57 |
| Disease-Characteristics, Disease, Treatment, Therapy, Chemotherapy | Chemotherapy | 52 |
| Disease-Characteristics, Laboratory Variable | ecog performance status | zubrod performance status | 52 |
| Disease-Characteristics, Laboratory Variable | Bilirubin | 48 |
| Disease-Characteristics, Laboratory Variable | platelet count | 47 |
| Disease-Characteristics, Laboratory Variable | Creatinine | 46 |
| Patient-Characteristics, Other | effective contraception | adequate contraception | 46 |
| Disease-Characteristics, Disease, Diagnosis, Staging Information, TNM stage | stage iii | stage 3 | stage three | 43 |
| Patient-Characteristics, Other | Nursing | 41 |
| Disease-Characteristics, Disease, Treatment, Therapy, Radiotherapy | Radiotherapy | 41 |
| Patient-Characteristics, Menopausal Status | menopause | menopausal status | 36 |
| Disease-Characteristics, Laboratory Variable | negative pregnancy test | 35 |
| Disease-Characteristics, Disease | carcinoma in situ of the cervix | in situ cervical cancer | 34 |
| Disease-Characteristics, Laboratory Variable | anc | absolute neutrophil count | 33 |
| Disease-Characteristics, Disease, Diagnosis, Staging Information, TNM stage | stage ii | stage 2 | stage two | 33 |
| Patient-Characteristics, Other | fertile patients must use effective contraception | willing to use medically acceptable form of contraception | 32 |
| Disease-Characteristics, Disease, Diagnosis, Staging Information, Receptor | hormone receptor status: not specified | 31 |
| Disease-Characteristics, Disease | secondary malignancy | other malignancy | 30 |
| Disease-Characteristics, Laboratory Variable | Ast | 28 |
| Disease-Characteristics, Disease | Infection | 28 |
| Disease-Characteristics, Disease | psychiatric illness | 28 |
| Disease-Characteristics, Disease, Treatment, Procedure, Surgery | Surgery | 27 |
| Disease-Characteristics, Disease | Tumor | 27 |
| Disease-Characteristics, Disease | basal cell skin cancer | 26 |
| Disease-Characteristics, Disease | congestive heart failure | chf | 26 |
| Disease-Characteristics, Disease | Metastatic | 26 |
| Disease-Characteristics, Disease | basal cell carcinoma | 25 |
| Disease-Characteristics, Disease | angina pectoris | 24 |
| Disease-Characteristics, Laboratory Variable | creatinine clearance | 23 |
| Disease-Characteristics, Laboratory Variable | lvef | left ventricular ejection fraction | 23 |
| Disease-Characteristics, Laboratory Variable | Wbc | 22 |
| Disease-Characteristics, Laboratory Variable | Alt | 21 |
| Disease-Characteristics, Disease, Diagnosis, Biopsy | Biopsy | 21 |
We then organized eligibility features into two main classes:
disease-characteristic: characteristics indicative of the patient’s disease status (e.g., medications, disease diagnoses, laboratory variables, and diagnostic tests)
patient-characteristic: other characteristics that are not disease specific and are mainly demographic (e.g., age, gender, menopausal status, life expectancy)
We generated the feature hierarchy in Protégé OWL 4.2, which will be made publically available on Bioportal (http://bioportal.bioontology.org/). In addition, we performed automatic post-annotation processing for features with subsumption dependencies. For example, all trials containing the feature breast cancer also contain the feature cancer. Therefore, the higher-level feature – cancer – was annotated as present whenever any of its lower level features – e.g., breast cancer, lung cancer – were present. Since the post-annotation processing was performed automatically and to avoid introducing errors at this step, all annotations were also manually reviewed and counterchecked. Table 1 illustrates an example of post-annotation processing results, where * denotes the implicit inferred features.
Table 1.
Post-annotation processing of feature subsumptions
| Human Annotation | Post-Annotation | |||
|---|---|---|---|---|
| Trial 1 | Trial 2 | Trial 1 | Trial 2 | |
| Cancer | 1 | 0 | 1 | 1* |
| Breast cancer | 0 | 1 | 0 | 1 |
| Age: 21 years to 65 years | 1 | 0 | 1 | 1* |
| Age: 21 years to 75 years | 0 | 1 | 0 | 1 |
| Infection | 1 | 0 | 1 | 1* |
| Viral infection | 0 | 1 | 0 | 1 |
| Effective contraception | 1 | 0 | 1 | 1* |
| Fertile patients must use effective contraception | 0 | 1 | 0 | 1 |
| CT scan | 1 | 0 | 1 | 1* |
| CT scan of the breast | 0 | 1 | 0 | 1 |
| Skin cancer | 1 | 0 | 1 | 1* |
| Basal cell skin cancer | 0 | 1 | 0 | 1 |
indicates an implicit inferred feature
2.2 Trial clustering using the identified eligibility features
Each trial is represented by an eligibility feature vector. We implemented Ward’s hierarchical clustering method, using the Euclidean metric in order to avoid clusters with a high within-cluster variance (i.e., between trials in the same cluster) that could result when correlation is used for clustering (50, 51). Initially each of the 80 trials was in a separate cluster (i.e., 80 clusters each containing one trial). Then clusters were progressively merged one at a time to form new clusters that generate the smallest increase in variance thereby minimizing the total within-cluster variance. The algorithm stopped when all trials were grouped into a single cluster. During clustering, the trials were compared to each other using binary feature vectors (1=presence, 0=absence). The Euclidean metric was used to define the distance between two vectors X and Y as follows:
| (1) |
where N is the number of features (i.e., N = 1,437). The distance is calculated per element in the feature vector. For example, x1 and y1 both indicate whether or not a trial contains the feature at position 1 in the feature vector, e.g., bilirubin. Therefore, if trial X contains bilirubin (x1=1) and trial Y does not (y1=0) then the quantity (y1 − x1)2 = 1. This is calculated for each of the 1,437 features and then the quantities are summed together and the square root is calculated.
The feature hierarchy was used for segmenting the feature set into smaller feature subsets for trial clustering. In total, clustering was performed for three times: first using all feature classes, second using only features from disease-characteristic classes and third using only features from patient-characteristic classes (t-test, p ≤ 0.05) (52). No maximum number of clusters was set a priori and cluster selection was based purely on observing a statistically significant difference (50–52). Trials found in significant clusters were reported and analyzed.
2.3 Search-space partitioning using eligibility features
Using a set of heuristics, we explored how features may be used to reduce the trial search-space for the general public searching for relevant clinical trials. First, we selected a few eligibility features from within the 1,437 feature set that contained information that an average user is likely to have readily available. For example, an average person is likely to know their age and gender but may not know details regarding docetaxel (a breast cancer medication). Therefore, we partitioned the trial search space using these features: gender, age, contraception status, and pregnancy test, and assessed the trial filtering efficiency using this method.
2.4 Mapping features to the UMLS
Using a previously developed UMLS semantic annotator, we mapped eligibility features to the UMLS (53). This annotator uses the UMLS Metathesaurus and SPECIALIST Lexical tools combined with a rule-based algorithm to meaningfully map eligibility criteria to the UMLS while minimizing ambiguity (53). Compared to MetaMap Transfer (MMTx), our annotator benefits from a specialized lexicon for clinical research eligibility criteria (53) and provides more fine-grained annotation. An example criterion sentence fragment, serious cardiac, renal and hepatic disorders, is used to illustrate the differences in mapping between the previously developed semantic annotator and MMTx 2.4C (UMLS concepts are shown in braces []). Our annotator resulted in the following annotation: serious [Qualitative Concept] cardiac, [Body Part, Organ or Organ Component] renal [Body Part, Organ or Organ Component] and hepatic [Body Location or Region] disorders [Disease or Syndrome]. In contrast MMTx 2.4C produced the following annotation: serious cardiac, renal [Idea or Concept] and hepatic disorders [Disease or Syndrome].
We then used a classification algorithm to tag the entire feature with one UMLS semantic type. First, each term in the feature was labeled with its corresponding UMLS concept from the Metathesaurus (53). Second, each sentence was classified with one semantic class. For example, the feature fertile patients must use effective contraception was first labeled with each of its UMLS concepts shown in braces []: Fertile [Organism Function] patients [Patient or Disabled Group] must use [Functional Concept] effective [Qualitative Concept] contraception [Contraception]. Then the sentence was automatically classified with the semantic class “Pregnancy Related Activities” using our previously developed algorithm (54). The algorithm uses a hierarchical clustering method whereby each sentence or feature is classified based on its set of UMLS concepts. We then analyzed the distribution of UMLS concepts and semantic class labels throughout our eligibility feature corpus.
3. Results
3.1 Hierarchical organization of eligibility features
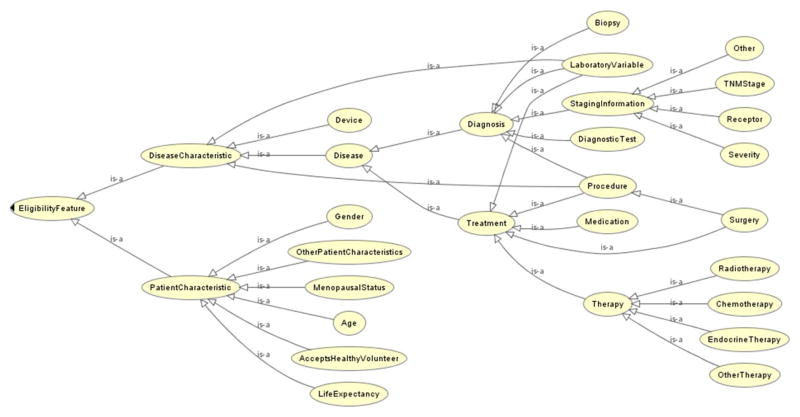
We identified 1,437 eligibility features with unique value ranges including 1,406 distinctive data elements for the 80 sample trials. Figure 2 shows the class hierarchy of the features, including six leaf node classes under the patient-characteristic class (i.e., 271 features) and eighteen classes under the disease-characteristic class (i.e., 1,166 features). We visualized the class hierarchy using Protégé’s OWLViz (55). Many eligibility features, e.g., gender and age, were shared by all trials. However, we found at least one novel feature per trial.
Figure 2.
Feature class hierarchy
The inter-rater agreement for the 37 eligibility features with trial frequency > 20 was substantial with a Kappa coefficient of 0.73 (56) (Table 2). The disagreement was resolved so that some equivalent features were merged and additional synonyms were identified.
3.2 Trial clustering by eligibility features
We arrived at six clusters by patient-characteristic features only (P), five clusters by disease-characteristic features only (D), and two clusters by combined features (C). Appendix Table A.1 denotes the trials of each cluster type and their corresponding p-values. Table 3 shows the clusters obtained using disease-characteristic features and patient-characteristic features respectively. Tumor-related features accounted for 24 shared features across all trials in clusters D1 and D2, while infection-related features accounted for 6 shared features across all trials in clusters D1 and D3. This indicates that the trials in these clusters contain extensive tumor or infection related information in their eligibility criteria.
Table 3.
Number of features shared across all trials in that cluster by class
| Cluster | Class - Information | No. of Shared Features Present |
|---|---|---|
| D11 | Disease - Tumor | 24 |
| Disease – Infection | 6 | |
| Laboratory Variable | 4 | |
| Disease – General | 3 | |
| D2 | Disease – Tumor | 24 |
| D3 | Disease – Infection | 6 |
| Chemotherapy | 1 | |
| D4 | No features shared completely by all 14 trials | |
| D5 | No features shared completely by all 22 trials | |
| P12 | Age | 17 |
| Life Expectancy | 10 | |
| Other Patient Characteristics | 4 | |
| Accepts Healthy Volunteers | 1 | |
| Gender | 1 | |
| Menopause | 1 | |
| P2 | Age | 7 |
| Other Patient Characteristics | 4 | |
| Accepts Healthy Volunteers | 1 | |
| Gender | 1 | |
| P3 | Other Patient Characteristics | 4 |
| Age | 2 | |
| Accepts Healthy Volunteers | 1 | |
| Gender | 1 | |
| P4 | Age | 17 |
| Accepts Healthy Volunteers | 1 | |
| Gender | 1 | |
| Other Patient Characteristics | 1 | |
| P5 | Age | 12 |
| Other Patient Characteristics | 4 | |
| Accepts Healthy Volunteers | 1 | |
| Gender | 1 | |
| Menopause | 1 | |
| P6 | Age | 12 |
| Accepts Healthy Volunteers | 1 | |
| Gender | 1 | |
| Menopause | 1 |
For disease-characteristic clusters only the disease-characteristic features are shown since these are the features used in the clustering.
For patient-characteristic clusters only the patient-characteristic features are shown since these are the features used in the clustering.
Next, we analyzed the clusters with patient-characteristic features. Accordingly, cluster P5 contained two trials with highly similar patient-characteristic eligibility requirements but very different study interventions. For example, the first trial studied the effectiveness of sunitinib for patients with stage I, II, or III breast cancers that have tumor cells in the bone marrow; however, the other trial focused on studying the effect of combining two treatments, paclitaxel and radiation therapy, on surgery patients with stage II or III breast cancer. In order to demonstrate the similarity between the two trials, Table 4 provides the alignment between the trials’ eligibility criteria from the patient characteristics section.
Table 4.
Similarity of patient characteristics for trials in cluster P5 (p<0.01)
| Trial NCT00824538 | Trial NCT00647218 |
|---|---|
| Menopausal status not specified | Menopausal status not specified |
| ECOG performance status 0–1 | ECOG performance status 0–1 |
| WBC count normal (3.4–10 × 10∧9/L) | WBC ≥ 3,000/mm∧3 |
| Platelet count normal (140–450 × 10∧9/L) | Platelet count ≥ 100,000/mm∧3 |
| Serum creatinine ≤ 1.5 times upper limit of normal (ULN) | Creatinine ≤ 1.5 times upper limit of normal (ULN) |
| Total bilirubin ≤ 1.5 times ULN | Bilirubin ≤ 1.5 times ULN |
| LVEF > 50% | Left ventricular ejection fraction ≥ 45% |
| Not pregnant or nursing | Not pregnant or nursing |
| Negative pregnancy test | Negative pregnancy test |
| Fertile patients must use effective contraception | Fertile patients must use effective contraception |
| No other malignancy within the past 5 years except basal cell carcinoma of the skin | No other malignancies within the past 5 years, except curatively treated nonmelanomatous skin cancer |
| No concurrent severe illness that would likely preclude study compliance | No serious medical illness that, in the judgment of the treating physician, places the patient at risk |
|
| |
| Not Aligned (from trial: NCT00824538) | |
|
| |
| TSH and T4 levels normal | |
| Absolute Neutrophil Count (ANC) normal (1.8–6.8 × 10∧9/L) | |
| Alkaline phosphatase ≤ 1.5 times ULN | |
| Aspartate Aminotransferase (AST) and Alanine Aminotransferase (ALT) ≤ 2.5 times ULN | |
| Hemoglobin > 9.0 g/dL | |
| Systolic blood pressure (BP) < 140 mm Hg | |
| Diastolic BP < 90 mm Hg | |
| No history of HIV infection | |
|
| |
| Not Aligned (from trial: NCT00647218) | |
|
| |
| No other malignancies within the past 5 years, except curatively treated carcinoma in situ of the cervix | |
| No history of hypersensitivity reaction to products containing polysorbate 80 (Tween 80) | |
| No peripheral neuropathy ≥ grade 2 | |
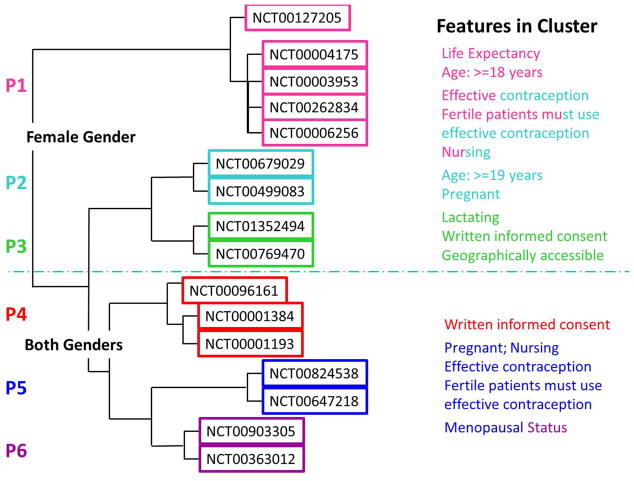
Looking at the cluster dendrogram (Figure 3), one can see that the trials split into two groups based on gender. This was not set a priori but rather a result of the clustering. Some features were present in multiple clusters when gender is ignored such as written informed consent found in P3 and P4 and effective contraception found in P1, P2 and P5. In general, trials in the same cluster contained similar features and information. For example, all trials contained in P3 shared gender=female, lactating and geographically accessible. Also trials in P5 contained: gender=both, pregnant, nursing, effective contraception, menopausal status and fertile patients must use effective contraception. On the other hand, some clusters contained features that defined that cluster, such as lactating, which was only found in P3.
Figure 3. Related trials cluster together using patient-characteristic features.
Cluster dendrogram with labels P1–P6 are consistent with Table A.1. The aqua colored dashed line shows how the clusters can be partitioned using gender (i.e., both genders allowed on bottom and females only on top). The other features shared within each cluster are displayed on the far right. They are colored with the same color as their cluster label (P1: pink; P2: sky blue; P3: green; P4: red; P5: navy blue; P6: purple). Dual color labels mean that a particular feature is shared across two related clusters on one side of the gender partition, applied a posteriori. For example, P1, P2 and P5 all share the feature effective contraception but it is shown twice: once on the top part of the dendrogram when gender=female and once on the bottom when gender=both. Similarly, written informed consent is found in P3 on the female only side of the gender partition and it also appears in P4 on the gender=both side of the gender partition. On the other hand, some features were only found on one side of the partition - for example, menopausal status is a unique feature when compared to the other trial clusters, and it is shared only by P5 and P6.
Because of the number of disease-characteristic features, i.e., 1,166 features, the clusters formed using only these features contained trials pertaining to certain stages of breast cancer or shared comorbidity information, e.g., neuropathy. Table 5 shows an alignment between two trials in D3, namely NCT00203372 and NCT00769470. These two trials share many eligibility criteria, including “history of any other malignancy within the past 5 years, with the exception of non-melanoma skin cancer or carcinoma-in-situ of the cervix,” which includes four features other malignancy, non-melanoma skin cancer, skin cancer and carcinoma in situ of the cervix. This illustrates that trials contained in the same disease cluster have similar target populations.
Table 5.
Similarity of disease characteristics for two trials in cluster D3 (p=0.04)
| Trial NCT00203372 | Trial NCT00769470 |
|---|---|
| Histologically or cytologically proven adenocarcinoma of the breast | Histologically or cytologically confirmed adenocarcinoma of the breast |
| Inflammatory Breast Cancer, clinically defined as the presence of erythema or induration involving one-third or more of the breast | Inflammatory breast cancer, defined as the presence of erythema or induration involving > 1/3 of the breast |
| Stage II or Stage III disease | Stage II or III disease |
| HER2-negative disease (as defined by fluorescence in situ hybridization [FISH]) | HER2/neu-positivity by fluorescence in situ hybridization (FISH) |
| New York Heart Association (NYHA) Grade II or greater congestive heart failure | New York Heart Association class II-IV congestive heart failure |
| A history of a severe hypersensitivity reaction to bevacizumab, or docetaxel or other drugs formulated with polysorbate 80 | Known hypersensitivity to Chinese hamster ovary products or other recombinant human or humanized antibodies and/or known hypersensitivity to any of the study drugs or their ingredients (e.g., polysorbate 80 in docetaxel) |
| Ejection fraction > lower limit of normal as determined by MUGA or echocardiogram | Ejection fraction>- lower limit of normal as determined by MUGA or echocardiogram. |
| Bilateral invasive breast cancer | Bilateral invasive breast cancer |
| Chemotherapy, radiotherapy, or endocrine therapy | Chemotherapy, radiotherapy, or endocrine therapy |
| Adequate organ function | Adequate organ function |
| Active, uncontrolled infection requiring parenteral antimicrobials | Active, uncontrolled infection requiring parenteral antimicrobials |
| Other medical or psychiatric disorder that, in the opinion of the treating physician, would contraindicate the use of the drugs in this protocol or place the subject at undue risk for treatment complications | Other medical or psychiatric disorder that, in the opinion of the treating physician, would contraindicate the use of study drugs or place the subject at undue risk for treatment complications |
| History of any other malignancy within the past 5 years, with the exception of non- melanoma skin cancer or carcinoma-in-situ of the cervix | History of any other malignancy within the past 5 years, with the exception of nonmelanoma skin cancer or carcinoma in situ of the cervix |
| Invasive or noninvasive breast cancer | Invasive or noninvasive breast cancer |
| Ipsilateral radiation therapy | Ipsilateral radiotherapy |
| Concurrent therapy with any other non- protocol anti-cancer therapy | Concurrent therapy with any other non- protocol anti-cancer therapy |
| Hormonal agent such as raloxifene, tamoxifen, or other selective estrogen receptor modulators | Hormonal agent (e.g., raloxifene, tamoxifen citrate, or other selective estrogen receptor modulators) |
| Hormone replacement therapy | Hormonal replacement therapy |
| Unstable angina | Unstable angina |
| Myocardial infarction | Myocardial infarction |
| Cardiovascular disease | Cardiac disease |
| Inflammatory bowel disease | Inflammatory bowel disease |
|
| |
| Not Aligned (from trial: NCT00203372) | |
|
| |
| Normal cardiac function | |
| Prior treatment with an anti-angiogenic agent | |
| Presence of neuropathy > grade 2 (NCI-CTC version 3.0) at baseline | |
| Presence of any non-healing wound, bone fracture, or ulcer, or the presence of clinically significant (> grade 2) peripheral vascular disease | |
| Hypertension [BP > 150/100], | |
| Stroke | |
| Cardiac arrhythmia requiring medication | |
| Active peptic ulcer disease or other gastrointestinal condition increasing the risk of perforation; history of abdominal fistula, gastrointestinal perforation, or intra-abdominal abscess within 6 months prior to beginning therapy | |
| Evidence of bleeding diathesis or coagulopathy | |
| Major surgical procedure, open biopsy, or significant traumatic injury within 28 days prior to beginning therapy, or anticipation of the need for a major surgical procedure during the course of the study; minor surgical procedure, fine needle aspiration, or core biopsy within 7 days prior to beginning therapy | |
|
| |
| Not Aligned (from trial: Trial NCT00769470) | |
|
| |
| Stage I | |
| Gilbert’s syndrome confirmed by genotyping or Invader UGTIA1 molecular assay | |
| Tumor measuring ≥ 1 cm | |
| Tumor Grade | |
| Estrogen and progesterone receptor status known prior to study entry | |
| Metastatic disease | |
| Pre-existing motor or sensory neurotoxicity ≥ grade 2 by NCI NTCAE version 3.0 | |
3.3 Feature-based trial search
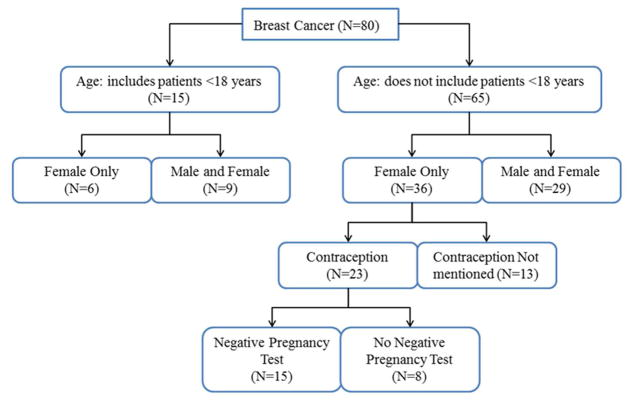
In our hypothetical use case, an adult female is searching for breast cancer trials. Figure 4 illustrates how the search space can be partitioned into smaller groups, whose sizes range from 5 to 15, using a few features. Each branch in Figure 4 represents a possible search path. Each branch-point in the path represents a place where additional information is required from the user to further narrow down the search results. We envision a question-answer style trial search system where features can be used to generate questions to elicit additional input from the user and guide them to filter search results iteratively. For example, if a user responds that their “age < 18” and “gender = female”, the system would return a set of 6 trials out of 80. Therefore, using only four eligibility features a hypothetical user can arrive at a much smaller (i.e., an order of magnitude reduction) set of trials for manual review.
Figure 4. Feature-based clinical trial search.
The initial group of 80 trials was stratified into multiple smaller groups after 3 questions; each group has a size between 6 and 15.
3.4 Semantic complexity of eligibility features
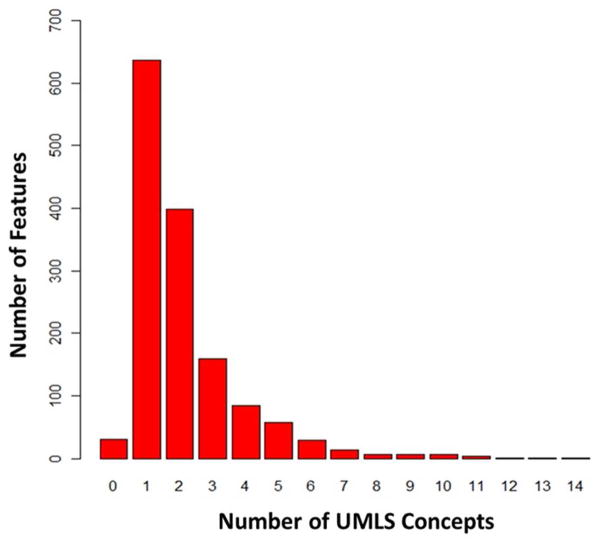
We analyzed the feature complexity by mapping each feature to its corresponding UMLS concepts. The number of UMLS concepts per feature is shown in Figure 5. Table 6 shows the mapping results broken down by patient-characteristic and disease-characteristic feature classes. Most of the unmatched terms were modifiers or common words, e.g., that, in, the, of, would, this, or, at, must, by, undue. As it can be seen, using the UMLS led to better term coverage for disease-characteristic features (87.01%) than patient-characteristic features (36.56%). The UMLS coverage among all features was 81.14% with the Medication (94.42%) and Laboratory Variable (90.80%) features achieving the highest coverage.
Figure 5.
Distribution of features (N = 1,437) by number of UMLS terms
Table 6.
Distribution of the number of UMLS terms in the 1,437 eligibility features
| Class/Category (M=Number of features per category) | % of exact UMLS match1 | %Contain unmatched terms2 |
|---|---|---|
|
Disease-Characteristic Features
| ||
| Biopsy (M=10) | 100% | 0% |
| Device (M=11) | 81.82% | 18.18% |
| Diagnostic Test (M=34) | 64.71% | 35.29% |
| Disease (M=550) | 87.27% | 12.73% |
| Laboratory Variable (M=87) | 90.80% | 9.20% |
| Medication (M=251) | 94.42% | 5.58% |
| Procedure (M=70) | 87.14% | 12.86% |
| Staging Info (M=102) | 87.25% | 12.75% |
| Surgery (M=55) | 74.55% | 25.45% |
| Therapy (M=100) | 77.00% | 23.00% |
| Total within Disease-Characteristic | 87.01% (M=1,105) | 12.99% (M=165) |
|
| ||
| Patient-Characteristic Features | ||
|
| ||
| Accepts Healthy Volunteers (M=1) | 0% | 100% |
| Age (M=17) | 0% | 100% |
| Gender (M=2) | 0% | 100% |
| Life Expectancy (M=10) | 100% | 0% |
| Menopause (M=6) | 100% | 0% |
| Other Patient Characteristic (M=131) | 34.35% | 65.65% |
| Total within Patient-Characteristic | 36.53% (M=61) | 63.47% (M=106) |
|
| ||
| Grand Total (M=1437) | 81.14% (M=1,166) | 18.86% (M=271) |
Exact UMLS match indicates that all terms in the feature were mapped to UMLS concepts.
‘% contain unmatched terms’ indicates the percentage of features per category that were not exactly matched to the UMLS.
Yet we noted that six menopausal status-related features were mapped exactly to the UMLS, though to different UMLS semantic types: Organ or Tissue Function, Organism Function, Temporal Concept, and Physiologic Function. For example, postmenopause has semantic type Organism Function while premenopausal and perimenopausal are Temporal Concepts. This result reveals some ambiguities in the UMLS semantic type assignments and potential challenges for automating this process using natural language processing tools.
4. Discussion
Inspired by the effectiveness of using MeSH terms to index MEDLINE documents, we demonstrated the feasibility of using feature-based indexing for trial clustering and search. We used a manual approach to extract concept-oriented features from clinical trial eligibility criteria and to construct a hierarchy of eligibility features. Through feature-based indexing, we were able to successfully cluster trials of similar eligibility criteria and illustrate how the clinical trial search space can be stratified to a small subset of relevant trials amenable for manual review. The results also inform a conceptual design for automating feature identification.
4.1 Benefits of trial clustering
We used eligibility features to effectively cluster trials recruiting similar volunteers. Typically, clustering results are evaluated using external validation measures, e.g., F-measure (57), Jaccard-index (58), or Rand measure (59), to name a few. Since we did not have a predefined “gold-standard” regarding which trials belong to each cluster, we evaluated our clustering results manually to determine the trial relatedness within each cluster. We determined success based on whether or not the trials within each cluster shared common themes or features (Figure 3). For example, a sub-cluster within D3 contained trials recruiting volunteers for bevacizumab (i.e., NCT00203372) and trastuzumab (i.e., NCT00769470), which are different medications and yet the trials share similar patient disease characteristics. The two medications are also similar in that both are monoclonal antibodies and are often given in combination with docetaxel (60, 61). We conclude that the identification of similar trials can be useful for individuals who are interested in participating in a particular trial but then cannot for some reason (e.g., the trial was discontinued). In fact, these individuals can then look into the possibility of enrolling in one of the other trials in the same cluster, as they are likely to be eligible for these trials as well. We observed that some features defined a particular trial cluster. For example, lactating was only found in P3. Clusters with defining features may be useful to users looking for a trial that is related to a previous trial of interest. From the researchers’ perspective, these trial clusters may also be useful for identifying trials with similar target populations.
4.2 Trial search space reduction
Furthermore, we also successfully used this feature-based index to optimize a hypothetical individual’s query for relevant clinical trials by reducing the search-space up to 90% within four questions, which were for gender, age, contraception status, and pregnancy status. If a clinical trial search engine were developed using a feature-based indexing method then patients would retrieve a subset of trials representing the most relevant trials that is also small enough to be amenable for manual review. Furthermore, a feature-based clinical trial search engine designed specifically to engage users’ interaction would be useful not only to patients with their complete medical history but also for patients with only a few key data elements.
4.3 A conceptual design for automating eligibility feature extraction
Our method used 80 breast cancer trials to derive a set of 1,437 eligibility features that we organized into a feature hierarchy. If our method is applied to another disease, e.g., liver cancer, it is likely that additional features will be identified. Therefore, automating the feature extraction process is necessary to extend the method for all diseases listed on ClinicalTrials.gov. Our manual feature extraction process contributes design ideas for automating this process in the future. The feature hierarchy proves to be useful for organizing features into a meaningful class structure. Next, we outline a few considerations for automating eligibility feature extraction.
We observed that patient-characteristic features tended to contain multiple UMLS terms, which are not easily identifiable by regular named entity recognition methods. For example, one patient-characteristic feature - fertile patients must use effective contraception - contains five UMLS concepts. By focusing on extracting concept-oriented eligibility features and not individual UMLS terms, we were able to identify more meaningful eligibility features that contain multiple UMLS terms. Prior research on this topic only supported UMLS-based term recognition in eligibility criteria text without multi-term named entity recognition, which was confirmed to be insufficient by this study (53, 54). Therefore, our conceptual design calls for an automatic feature extraction method that supports multi-term recognition. N-gram named entity recognition is one such Natural Language Processing (NLP) method that presents a viable solution for automatically extracting features that could contain multiple terms. Semantic methods that take into account the grammatical structure of the text, including its phrasal (62, 63) and clausal (64) structures, may also be needed to derive meaningful features. Once the features are extracted, they can then be mapped to the UMLS to avoid performing complex synonymous term grouping and multi-term named entity recognition with UMLS concepts.
Other methods for automatically extracting eligibility features could be used as well. Some machine learning methods used for classifying medical text rely heavily on the availability of labeled training data (65). However, others did not use labeled training data but instead they exploited unsupervised techniques, e.g., latent Dirichlet (66). Our proposed method of combining a NLP method, e.g., N-gram named entity recognition, with a conceptual feature hierarchy has literature support as others have successfully used ontologies to automatically map patients to relevant clinical trials (67). As mentioned previously, many NLP methods require the use of a “gold-standard” for evaluation; therefore, we propose using our manually derived concept-oriented eligibility feature hierarchy resulting from this study as a gold-standard to gauge the “success” of any future automatic feature extraction approach that we develop.
4.4 Limitations and future work
This study has two main limitations. First, as a case study, we limited our analysis to one disease, stage III breast cancer. Therefore, future studies should extend and apply our methods to other diseases. The second limitation is that we did not process all synonyms during feature extraction so that some features with different semantic representations were mistakenly treated as separate features. For example, features basal cell skin cancer and basal cell carcinoma should be combined into one feature. Whenever the annotator was in doubt as to whether or not two features should be merged, the two features were kept separate. Synonym identification was difficult in other instances as well, such as breast-feeding, lactating and nursing. Breast-feeding and nursing should be merged to form the more frequently occurring feature (i.e., nursing); while lactating should be kept separate since this has a different clinical meaning.
Our future work includes automating the feature extraction process using NLP methods, e.g., n-gram named entity recognition, followed by normalization of the terms in features using the UMLS. Our other future work includes assessing the meaningfulness of the trial clusters by incorporating feedback from health care consumers. This would allow us to determine the usefulness of the trial clusters to the general public for finding relevant clinical trials.
5. Conclusion
This study demonstrates the usefulness of concept-oriented feature-based indexing of clinical trial eligibility criteria using two use cases: trial similarity clustering and efficient trial search. We also present a conceptual design for automating the feature discovery process. The eligibility feature class hierarchy resulting from this study could serve as a “gold-standard” for evaluating future automatic feature-extraction methods. This hierarchy will be made publically available on BioPortal (http://bioportal.bioontology.org/).
Acknowledgments
The research was supported by grants R01LM009886 and R01LM010815 from the National Library of Medicine, grant R01 HQ 1R01HS019853-01 from Agency for Healthcare Research Quality, and grant UL1 TR000040 from the National Center for Advancing Translational Sciences. We thank Dr. Nanfang Xu for using his medical expertise to help with feature evaluation.
Appendix
Table A.1.
Cluster membership by type (D: disease-characteristic; P: patient-characteristic; C: combined-all-classes)
The trials in this cluster contained very few eligibility criteria requirements
References
- 1.Weng C, Embi P. Informatics Approaches to Participant Recruitment. In: Richesson R, Andrews J, editors. Clinical Research Informatics. Springer; 2012. p. 428. [Google Scholar]
- 2.Thadani SR, Weng C, Bigger JT, Ennever JF, Wajngurt D. Electronic Screening Improves Efficiency in Clinical Trial Recruitment. Journal of the American Medical Informatics Association. 2009 Nov 1;16(6):869–73. doi: 10.1197/jamia.M3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Herasevich V, Pieper MS, Pulido J, Gajic O. Enrollment into a time sensitive clinical study in the critical care setting: results from computerized septic shock sniffer implementation. Journal of the American Medical Informatics Association. 2011 Sep 1;18(5):639–44. doi: 10.1136/amiajnl-2011-000228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yamamoto K, Sumi E, Yamazaki T, Asai K, Yamori M, Teramukai S, et al. A pragmatic method for electronic medical record-based observational studies: developing an electronic medical records retrieval system for clinical research. BMJ Open. 2012 Jan 1;2(6) doi: 10.1136/bmjopen-2012-001622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Niland J. Integration of Clinical Research and EHR: Eligibility Coding Standards: ASPIRE (Agreement on Standardized Protocol Inclusion Requirements for Eligibility) 2007 http://crisummit2010amiaorg/files/symposium2008/S14_Nilandpdf.
- 6.Patel C, Khan S, Gomadam K. TrialX: Using Semantic Technologies to Match Patients to Relevant Clinical Trials Based on Their Personal Health Records. Proceedings of the 8th International Semantic Web Conference; 2009. pp. 1–7. [Google Scholar]
- 7.Weng C, Tu SW, Sim I, Richesson R. Formal representation of eligibility criteria: a literature review. J Biomed Inform. 2010 Jun;43(3):451–67. doi: 10.1016/j.jbi.2009.12.004. Epub 2009/12/26. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Penberthy L, Brown R, Puma F, Dahman B. Automated matching software for clinical trials eligibility: Measuring efficiency and flexibility. Contemporary Clinical Trials. 2010;31(3):207–17. doi: 10.1016/j.cct.2010.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heiney SP, Adams SA, Drake BF, Bryant LH, Bridges L, Hebert JR. Successful subject recruitment for a prostate cancer behavioral intervention trial. Clinical Trials. 2010 Aug 1;7(4):411–7. doi: 10.1177/1740774510373491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Patel C, Gomadam K, Khan S, Garg V. TrialX: Using semantic technologies to match patients to relevant clinical trials based on their Personal Health Records. J Web Sem. 2010;8(4):342–7. [Google Scholar]
- 11.Kernan W, Viscoli C, Brass L, Amatangelo M, Birch A, Clark W, et al. Boosting enrolment in clinical trials: validation of a regional network model. Clinical Trials. 2011 Oct 1;8(5):645–53. doi: 10.1177/1740774511414925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weng C, Wu X, Luo Z, Boland MR, Theodoratos D, Johnson SB. EliXR: an approach to eligibility criteria extraction and representation. Journal of the American Medical Informatics Association. 2011 Dec 1;18(Suppl 1):i116–i24. doi: 10.1136/amiajnl-2011-000321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heinemann S, Thuring S, Wedeken S, Schafer T, Scheidt-Nave C, Ketterer M, et al. A clinical trial alert tool to recruit large patient samples and assess selection bias in general practice research. BMC Med Res Methodol. 2011;11(16):1–10. doi: 10.1186/1471-2288-11-16. Epub 2011/02/16. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Beauharnais CC, Larkin ME, Zai AH, Boykin EC, Luttrell J, Wexler DJ. Efficacy and cost-effectiveness of an automated screening algorithm in an inpatient clinical trial. Clinical Trials. 2012 Apr 1;9(2):198–203. doi: 10.1177/1740774511434844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Korkontzelos I, Mu T, Ananiadou S. ASCOT: a text mining-based web-service for efficient search and assisted creation of clinical trials. BMC Medical Informatics and Decision Making. 2012;12(Suppl 1):S3. doi: 10.1186/1472-6947-12-S1-S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harris PA, Scott KW, Lebo L, Hassan N, Lightner C, Pulley J. ResearchMatch: a national registry to recruit volunteers for clinical research. Academic medicine: journal of the Association of American Medical Colleges. 2012 Jan;87(1):66–73. doi: 10.1097/ACM.0b013e31823ab7d2. Epub 2011/11/23. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. [Accessed on August 9, 2012.];ResearchMatch. www.researchmatch.org.
- 18. [Accessed on January 7, 2013.];caMATCH. https://cabigncinihgov/community/tools/caMATCH.
- 19. [Accessed on January 7, 2013.];Corengi. https://wwwcorengicom/
- 20. [Accessed on January 7, 2013.];University of Florida Research Affairs Clinical Trials. http://wwwhscjufledu/research/SearchClinicalTrialsaspx.
- 21.McCray A. Better access to information about clinical trials. Annals of Internal Medicine. 2000;133(8):609–14. doi: 10.7326/0003-4819-133-8-200010170-00013. [DOI] [PubMed] [Google Scholar]
- 22.NIH. [Accessed on February 10, 2012 and October 2, 2012.]; www.clinicaltrials.gov.
- 23.Muller H, Hanbury A, Al Shorbaji N. Health information search to deal with the exploding amount of health information produced. Methods of information in medicine. 2012 Dec 4;51(6):516–8. Epub 2012/12/06. eng. [PubMed] [Google Scholar]
- 24.Tan P-N, Steinbach M, Kumar V. Introduction to Data Mining. Addison-Wesley; 2005. [Google Scholar]
- 25.Tata S, Patel JM. Estimating the selectivity of tf-idf based cosine similarity predicates. SIGMOD Rec. 2007;36(2):7–12. [Google Scholar]
- 26.Manning CD, Raghavan P, Schütze H. Introduction to information retrieval. New York: Cambridge University Press; 2008. p. 482. [Google Scholar]
- 27.Durao F, Dolog P, Leginus M, Lage R. SimSpectrum: A Similarity Based Spectral Clustering Approach to Generate a Tag Cloud. In: Harth A, Koch N, editors. Current Trends in Web Engineering. Lecture Notes in Computer Science 7059. Springer; Berlin Heidelberg: 2012. pp. 145–54. [Google Scholar]
- 28.Korkontzelos I, Mu T, Ananiadou S. ASCOT: a text mining-based web-service for efficient search and assisted creation of clinical trials. BMC Medical Informatics and Decision Making. 2012;12(Suppl 1):S3. doi: 10.1186/1472-6947-12-S1-S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Salton G, Fox EA, Wu H. Extended Boolean information retrieval. Commun ACM. 1983;26(11):1022–36. [Google Scholar]
- 30.Salton G. Developments in Automatic Text Retrieval. Science. 1991 Aug 30;253(5023):974–80. doi: 10.1126/science.253.5023.974. [DOI] [PubMed] [Google Scholar]
- 31.Denecke K. An Architecture for Diversity-aware Search for Medical Web Content. Methods of information in medicine. 2012 Dec 4;51(6):549–56. doi: 10.3414/ME11-02-0022. Epub 2012/10/20. eng. [DOI] [PubMed] [Google Scholar]
- 32.Turney P. Mining the web for synonyms: PMI-IR versus LSA on TOEFL. Proceedings of the Twelfth European Conference on Machine Learning (ECML-2001) 2001:1–12. [Google Scholar]
- 33.Aula A. Proceedings of IADIS international conference WWW/Internet. Lisboa(IADIS Press); 2003. Query formulation in web information search; pp. 403–10. [Google Scholar]
- 34.Steinbrook R. Searching for the Right Search — Reaching the Medical Literature. New England Journal of Medicine. 2006;354(1):4–7. doi: 10.1056/NEJMp058128. [DOI] [PubMed] [Google Scholar]
- 35.Rogers FB. Medical subject headings. Bulletin of the Medical Library Association. 1963 Jan;51:114–6. Epub 1963/01/01. eng. [PMC free article] [PubMed] [Google Scholar]
- 36.Bakken S, Currie LM, Lee N-J, Roberts WD, Collins SA, Cimino JJ. Integrating evidence into clinical information systems for nursing decision support. International Journal of Medical Informatics. 2008;77(6):413–20. doi: 10.1016/j.ijmedinf.2007.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Burstein J, Kukich K, Wolff S, Lu C, Chodorow M, Braden-Harder L, et al. Automated scoring using a hybrid feature identification technique. Proceedings of the 36th Annual Meeting of the Association for Computational Linguistics and 17th International Conference on Computational Linguistics; Montreal, Quebec, Canada. Association for Computational Linguistics; 1998. pp. 206–10. 980879. [Google Scholar]
- 38.Forman G, Kirshenbaum E. Extremely fast text feature extraction for classification and indexing. Proceedings of the 17th ACM conference on Information and knowledge management; Napa Valley, California, USA. ACM; 2008. pp. 1458243pp. 1221–30. [Google Scholar]
- 39.Lowe D, Webb AR. Optimized Feature Extraction and the Bayes Decision in Feed-Forward Classifier Networks. IEEE Transations on Pattern Analysis and Machine Intelligence. 1991;13(4):355–64. [Google Scholar]
- 40.Clausen M, Korner H, Kurth F. An Efficient Indexing and Search Technique for Multimedia Databases. SIGIR Multimedia Information Retrieval Workshop. 2003:1–12. [Google Scholar]
- 41.Lewis DD. Feature selection and feature extraction for text categorization. Proceedings of the workshop on Speech and Natural Language; Harriman, New York. Association for Computational Linguistics; 1992. pp. 1075574pp. 212–7. [Google Scholar]
- 42.Similarity trials. Nat Biotech. 2011;29(1):1. doi: 10.1038/nbt.1760. [DOI] [PubMed] [Google Scholar]
- 43.Bodenreider O. The Unified Medical Language System (UMLS): integrating biomedical terminology. Nucleic Acids Research. 2004 Jan 1;32(suppl 1):D267–D70. doi: 10.1093/nar/gkh061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ross J, Tu S, Carini S, Sim I. Analysis of eligibility criteria complexity in clinical trials. AMIA Summits Transl Sci Proc. 2010 Mar 1;:46–50. [PMC free article] [PubMed] [Google Scholar]
- 45.George S. Reducing patient eligibility criteria in cancer clinical trials. J Clin Oncol. 1996;14(4):1364–70. doi: 10.1200/JCO.1996.14.4.1364. [DOI] [PubMed] [Google Scholar]
- 46.Sim I, Olasov B, Carini S. An ontology of randomized controlled trials for evidence-based practice: content specification and evaluation using the competency decomposition method. Journal of Biomedical Informatics. 2004;37:108–19. doi: 10.1016/j.jbi.2004.03.001. [DOI] [PubMed] [Google Scholar]
- 47.Sarkar IN. A vector space model approach to identify genetically related diseases. Journal of the American Medical Informatics Association. 2012;19(2):249–54. doi: 10.1136/amiajnl-2011-000480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Geertzen J. [Accessed on August 15, 2012.];Cohen’s Kappa for more than two annotators with multiple classes. http://cosmion.net/jeroen/software/kappao/
- 49.Fleiss JL. Measuring nominal scale agreement among many raters. Psychological Bulletin. 1971;76(5):378–82. [Google Scholar]
- 50.Wishart D. 256. Note: An Algorithm for Hierarchical Classifications. Biometrics. 1969;25(1):165–70. [Google Scholar]
- 51.Ward JH., Jr Hierarchical Grouping to Optimize an Objective Function. Journal of the American Statistical Association. 1963;58(301):236–44. [Google Scholar]
- 52.Suzuki R, Shimodaira H. Pvclust: an R package for assessing the uncertainty in hierarchical clustering. Bioinformatics. 2006 Jun 15;22(12):1540–2. doi: 10.1093/bioinformatics/btl117. [DOI] [PubMed] [Google Scholar]
- 53.Luo Z, Duffy R, Johnson SB, Weng C. Corpus-based Approach to Creating a Semantic Lexicon for Clinical Research Eligibility Criteria from UMLS. AMIA Summits Transl Sci Proc. 2010 Mar 1;:26–30. [PMC free article] [PubMed] [Google Scholar]
- 54.Luo Z, Johnson SB, Weng C. Semi-Automatically Inducing Semantic Classes of Clinical Research Eligibility Criteria Using UMLS and Hierarchical Clustering; AMIA Annu Symp Proc; 2010. Nov 13, pp. 487–91. [PMC free article] [PubMed] [Google Scholar]
- 55.Horridge M. [Accessed on September 24, 2012];OWLViz - A visualisation plugin for the Protege OWL Plugin. http://www.co-ode.orgldownloads/owlvizl.
- 56.Landis JR, Koch GG. The Measurement of Observer Agreement for Categorical Data. Biometrics. 1977;33(1):159–74. [PubMed] [Google Scholar]
- 57.Hripcsak G, Rothschild AS. Agreement, the F-Measure, and Reliability in Information Retrieval. Journal of the American Medical Informatics Association. 2005 May 1;12(3):296–8. doi: 10.1197/jamia.M1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hamers L, Hemeryck Y, Herweyers G, Janssen M, Keters H, Rousseau R, et al. Similarity measures in scientometric research: The Jaccard index versus Salton’s cosine formula. Information Processing & Management. 1989;25(3):315–8. [Google Scholar]
- 59.Krieger AM, Green PE. A Generalized Rand-Index Method for Consensus Clustering of Separate Partitions of the Same Data Base. Journal of Classification. 1999;16(1):63. [Google Scholar]
- 60.Meriggi F, Abeni C, Di Biasi B, Zaniboni A. The use of bevacizumab and trastuzumab beyond tumor progression: a new avenue in cancer treatment? Rev Recent Clin Trials. 2009;4(3):163–7. doi: 10.2174/157488709789957682. [DOI] [PubMed] [Google Scholar]
- 61.Martín M, Makhson A, Gligorov J, Lichinitser M, Lluch A, Semiglazov V, et al. Phase II Study of Bevacizumab in Combination with Trastuzumab and Capecitabine as First-Line Treatment for HER-2-positive Locally Recurrent or Metastatic Breast Cancer. The Oncologist. 2012 Apr 1;17(4):469–75. doi: 10.1634/theoncologist.2011-0344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Evans DA, Zhai C. Noun-phrase analysis in unrestricted text for information retrieval. Proceedings of the 34th annual meeting on Association for Computational Linguistics; Santa Cruz, California. Association for Computational Linguistics; 1996. pp. 981866pp. 17–24. [Google Scholar]
- 63.Novichkova S, Egorov S, Daraselia N. MedScan, a natural language processing engine for MEDLINE abstracts. Bioinformatics. 2003 Sep 1;19(13):1699–706. doi: 10.1093/bioinformatics/btg207. [DOI] [PubMed] [Google Scholar]
- 64.Molina A, Pla F. Clause detection using HMM. Proceedings of the 2001 workshop on Computational Natural Language Learning; Toulouse, France. Association for Computational Linguistics; 2001. p. 1455688.p. 1. [Google Scholar]
- 65.Pakhomov S, Buntrock J, Duffy P. High throughput modularized NLP system for clinical text. Proceedings of the ACL 2005 on Interactive poster and demonstration sessions; Ann Arbor, Michigan. Association for Computational Linguistics; 2005. pp. 1225760pp. 25–8. [Google Scholar]
- 66.Restificar A, Ananiadou S. Inferring appropriate eligibility criteria in clinical trial protocols without labeled data. Proceedings of the ACM sixth international workshop on Data and text mining in biomedical informatics; Maui, Hawaii, USA. ACM; 2012. pp. 2390074pp. 21–8. [Google Scholar]
- 67.Patel C, Cimino J, Dolby J, Fokoue A, Kalyanpur A, Kershenbaum A, et al. Matching Patient Records to Clinical Trials Using Ontologies. In: Aberer K, Choi K-S, Noy N, Allemang D, Lee K-I, Nixon L, et al., editors. The Semantic Web. Lecture Notes in Computer Science. Springer; Berlin Heidelberg: 2007. pp. 4825pp. 816–29. [Google Scholar]