Abstract
Objective: To investigate the inhibitory effect of plasmid-based survivin-specific short hairpin RNA and GRIM-19 on the growth of Hep-2 laryngeal cancer cells. Methods: The plasmid expressing survivin-specific short hairpin RNA (shRNA) and GRIM-19 (p-siRNA survivin/GRIM-19) was prepared and transfected into Hep-2 cells with Lipofectamine 2000. The mRNA and protein expression of surviving and GRIM-19 were measured with RT-PCR and western blot assay, respectively. MTT assay was employed to detect the proliferation of Hep-2 cells, and flow cytometry and AO/EB assay were done to determine the apoptosis of Hep-2 cells. Results: In the p-siRNA survivin/GRIM-19, the mRNA and protein expression of survivin was markedly reduced by 54.4% and 42.2%, and the reduction in protein expression of surviving was more obvious than that in the p-siRNA survivin group (37%) (P<0.05). The protein expression of GRIM-19 was markedly enhanced when compared with the control group (P<0.01). MTT assay revealed the proliferation of Hep-2 cells undergoing transfection with p-siRNA survivin/GRIM-19 was markedly inhibited, and the inhibition rate was as high as 79%, which was higher than that in the psi-survivin group (45%) and p-GRIM-19 group (35%). AO/EB assay and flow cytometry indicated that the apoptotic cells in the p-siRNA survivin/GRIM-19 group were dramatically increased as compared to the psi-survivin group and p-GRIM-19 group. Conclusion: The p-siRNA survivin/GRIM-19 has marked decrease in survivin expression and dramatic increase in GRIM-19 expression. Moreover, silencing of survivin and over-expression of GRIM-19 can significantly inhibit the growth and induce the apoptosis of Hep-2 in vitro and in vivo.
Keywords: Co-expression plasmid, gene silencing, survivin, laryngeal cancer
Introduction
Cancer is caused by multiple factors, which makes the single gene therapy difficult. Therefore, numerous studies focus on blocking several signaling pathways involving in the proliferation and survival of cancer cells. In recent years, great progress has been achieved in the gene therapy of genetic disease, metabolic diseases and cancer. However, the safety and effectiveness of gene therapy are still controversial in clinical practice, which significantly limits the wide application of gene therapy. Therefore, many studies focus on developing new strategies targeting only cancer cells in gene therapy.
The hypoxia in solid tumors is a major cause of resistance to anti-cancer drugs and radiotherapy, which contributes to the poor prognosis in these patients. On the other hand, the hypoxia and necrosis in solid tumors provide a good environment for the growth of anaerobic bacteria, which provide us a possibility of selective treatment. The possible mechanism about tumor targeting of anaerobic bacteria: 1) The hypoxic environment in the tumor provides a suitable place for the growth and reproduction of anaerobic bacteria; 2) Abnormal vascularization and high pressure in the tumors influence the level of immune substances and suppress their functions, forming a so-called relative immune vacuum area. Under this condition, the removal of bacteria is harder than that under normal condition [1]; (3) Some auxotrophic bio-engineering anaerobes can use the intermediate products produced in the tumors due to active metabolism. Thus, these anaerobes grow and proliferate rapidly in tumors. Thus, the anaerobic bacteria can be used as vectors to destroy the hypoxia in the tumor, or anaerobic bacteria as vectors may enhance the anti-tumor effect of other treatments.
Survivin gene is an apoptosis suppressor gene isolated and identified in 1997. It is a member of the inhibitor of apoptosis protein (IAP) family and can inhibit the apoptosis and regulate the cell proliferation [2]. Among apoptosis suppressor genes, survivin gene is the most potent one close to the initiation of cancers. Different from the other members of IAP family, survivin is lowly or not expressed in normal tissues while has a high expression in a majority of malignant tumors in humans [3-6]. This makes it a new target in the gene therapy of cancers. Studies have developed several methods to lower the survivin expression in cancer cells to induce apoptosis and inhibit proliferation [7-13].
Gene associated with retinoid-IFN-induced mortality 19 (GRIM-19 gene) is a new member of GRIM family and a new tumor suppressor gene. Studies have shown that GRIM-19 is expressed in many normal tissues, but has a low or no expression in several cancers [14,15]. The incidence of cancer is higher in subjects with low or negative GRIM-19 expression. On the other hand, GRIM-19 can enhance the cell sensitivity to IFN-β-RA-induced apoptosis, gaining itself a name of Death Activator [16].
In this study, the plasmid expressing GRIM-19 and survivin-specific short hairpin RNA (p-si RNA Survivin/GRIM-19) was constructed and transfected into Hep-2 cells using the attenuated Salmonella as a vector. Our results showed that silencing of survivin could significantly inhibit the growth of human laryngeal cancer cells and induce their apoptosis in vitro and in vivo.
Materials and methods
Materials
Survivin small interfering RNA plasmid pGCsilencer-siRNA-survivin (psi-survivin), negative control plasmid pGCsi-shRNA-scramble (psi-scramble) and survivin small interfering RNA and GRIM-19 co-expression plasmid pCDNA3.1-siRNA survivin/GRIM-19 (p-siRNA survivin/GRIM-19) were constructed by our group, and pCDNA3.1-GRIM-19 (p-GRIM-19) was kindly provided by the Center for Prostate Disease Research, Department of Pathophysiology, Jilin University. Attenuated Salmonella mutant Ty21a was a gift from Professor Xu DQ in FDA of U.S.; mouse anti-human GRIM-19 monoclonal antibody was kindly provided by the Dr. Hu JD in the University of Maryland. Plasmid extraction kit, BamH I and Hind III restriction enzymes, and T4 DNA were purchased from Takara. Gel extraction kits (Ding Guo Biochemical Reagent Company of Beijing), rabbit anti-human survivin monoclonal antibody (Santa Cruz), alkaline phosphatase (ALP) conjugated goat anti-rabbit IgG polyclonal antibody (Zhong San company), lipofectamine2000 and Trizol reagent (Invitrogen), TUNEL kit (Roche, Germany) were used in this study.
Cell culture and transfection of recombinant plasmid
Laryngeal cancer Hep-2 cells (Shanghai Institute of Biological Sciences and Bio-chemistry and Cell Biology) were grown in RPMI-1640 medium containing 10% fetal calf serum (FCS) at 37°C and 5% CO2. By using plasmid extraction kit, plasmids (psi-survivin, P-GRIM-19, p-siRNA survivin/GRIM-19) and pCDNA3.1 blank plasmid and psi-scramble negative control plasmid were extracted and λDNA quantified. According to manufacturer’s instructions, Hep-2 cells were transfected with the plasmids in the presence of lipofetamine2000. After 72 h, cells were harvested and named as Hpsi-survivin, Hpsi-scramble, HP-GRIM-19, Hp-siRNA survivin/GRIM-19 and HpCDNA3.1, respectively. Cells underwent lysis for western blot assay, and total RNA was extracted using Trizol reagent.
Detection of survivin and GRIM-19 mRNA expression in Hep-2 cells by semi-quantitative RT-PCR
In brief, 5 μg of total RNA was used for reverse transcription into template cDNA. According to the cDNA sequences of survivin and GRIM-19 gene in Genbank database, corresponding primers were designed and synthesized by Shanghai Biological Engineering Company. For Survivin, the forward primer was 5’-GAATTCATGGGTGCCCCGACGTTGCC-3’, reverse primer was 5’-AGATCTTTCTTCTTATTGTTGGTTTCC-3’. The mixture for PCR included cDNA (2 μl), 25 mM dNTP (0.5 μl), 10×PCR Buffer (2.5 μl), forward and reverse primer (50 pmol) (1 μl), Taq Enzyme (1 U) (1.0 μl), and ddH2O (1 u) (17 μl). The conditions for PCR were as follows: 94°C for 3 min; 30 cycles of 94°C for 30 s, 59°C for 30 s, 72°C for 1 min, and a final 72°C for 5 min. GRIM-19 primers were as follows: forward primer: 5’-GAGTCACGCACTGTCTGGG-3’, reverse primer: 5’-CGGTCGGTTTCTGCCTGTA-3’. The components of mixture for PCR were the same to those above. The conditions for PCR were as follows: 30 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 1 min, and a final 72°C for 5 min. The PCR products were subjected to 1% agarose gel electrophoresis. β-actin served as an internal reference to normalize the expression of target proteins. The experiments were repeated thrice and t-test was employed for analysis.
Survivin and GRIM-19 protein expression in Hep-2 cells by western blot assay
Total protein was extracted from Hep-2 cells at 72 h after transfection. The protein quantity was determined with Bio-rad method. Then, 100 μg of proteins were mixed with 4×SDS buffer (1/4 volume), and this mixture was boiled for 5 min. Subsequently, 28 μl of proteins and protein Marker were loaded independently for gel electrophoresis at 100 V for 1.5 h. After electrophoresis, the membrane was balanced in methanol for 30 s, and the membrane, gel and Whatman filter paper were immersed in blotting buffer for 10 min. Proteins were then transferred onto the PVDF membranes for 45 min. The membrane was blocked in 5% non-fat milk overnight at 40°C, then at room temperature for 1 h, and washed thrice with TBST. The membrane was incubated with primary antibody (survivin or GRIM-19) at room temperature for 3 h. After washing in TBST thrice, the membrane was incubated with secondary antibody for 1 h. Following washing in TBST thrice, visualization was performed, and representative photographs were captured. The optical density of each band was determined with analysis system. Because the molecular weight of survivin (16.5 kD) and GRIM-19 (16 kD) was similar, the detection of survivin and GRIM-19 expression in transfected Hep-2 cells were detected separately by western blot assay. The expression of target proteins was normalized with that of β-Actin. The experiments were repeated thrice and t-test was employed for analysis.
Cell growth inhibition test by MTT assay
When the cell confluence reached 80%, Hep-2 cells were trypsinized, washed twice in PBS, and seeded into 96-well plates at a density of 2×105/ml followed incubation in serum-free RPMI 1640 (100 μl/well; 2×104 cells/well). These cells were divided into 5 groups: blank plasmid control, psi-scramble, psi-survivin, p-GRIM-19 and p-siRNA survivin/GRIM-19. Transfection was performed with 6 μg of plasmid and 5 μl of Lipofectamine 2000 followed by incubation for 24 h, 72 h and 96 h. Then, 5 g/L MTT solution (20 μl) was added to each well followed by incubation for 4 h. The supernatant was removed and 100 μl of DMSO was added. After micro-vibration for 10 min, the absorbance (A) was measured at 570 nm with a microplate reader. The inhibition rate was calculated formulas follow: Inhibition rate (%) = (Acontrol group–Aexperiment group)/Acontrol group×100%. The inhibition rate of >30% suggested the sensitivity to the drugs. Experiment was repeated thrice.
Detection of apoptosis by flow cytometry
Hep-2 cells in the logarithmic growth phase were transfected with psi-survivin, p-GRIM-19, p-siRNA survivin/GRIM-19, psi-scramble and blank vector, respectively. After 72 h, cells were digested with 0.25% trypsin and single cell suspension was prepared. After treatment with 70% ethanol for more than 12 h, cells were washed with PBS twice. Then, cell density was adjusted to 1×106 cells/ml. Following RNAase digestion, cells were incubated in 1.5 ml of propidium iodide (PI) at 5 mg/100 ml for 1 h. Then, flow cytometry was performed. A total of 1×104 cells were detected for each sample. The cells in the G1 phase were determined as apoptotic cells.
Acridine orange staining
Hep-2 cells in the logarithmic growth phase were transfected with psi-survivin, p-GRIM-19, p-siRNA survivin/GRIM-19 and psi-scramble, respectively. After 72 h, the cells were treated with 0.25% trypsin and single cell suspension was prepared. After washing in PBS twice, the cell density was adjusted into 1×107/ml. Then, 95 μl of cell suspension was mixed with 5 μl of acridine orange solution. A drop of this mixture was put on clean glass slides and covered with coverslips before detection by fluorescent microscopy. Under the fluorescence microscope, the normal nuclear DNA has uniformly yellow or yellow-green fluorescence, while cytoplasmic and nucleolar RNA presents with orange or orange-red fluorescence. For apoptotic cells, the nucleus or cytoplasm shows intensely yellow-green fluorescence, or yellow-green or orange pieces are found. For necrotic cells, the cytoplasmic yellow-green or orange fluorescence is reduced or lost.
Establishment of mouse laryngeal cancer model and gene therapy
A total of 30 (15 males and 15 females) BALB/c-nu/nu nude mice aged 4-6 weeks (18-20 g) were purchased from the SLAC Laboratory Animal, Then, 2×106 (100 μl) Hep-2 cells were subcutaneous injected to the left abdominal wall. The tumor size was measured daily since 15 days after injection. The tumor volume was calculated as follow: m1 2×m2×0.5236 (m1: width, m2: length). About 22 days after inoculation of Hep-2 cells, the average tumor volume was up to 111.18±9.99 mm3. These cancer bearing nude mice were randomly divided into six groups: (1) control group, (2) psi-scramble group, (3) psi-scramble group, (4) psi-survivin group, (5) p-GRIM-19 group and (6) p-siRNA survivin/GRIM-19 group. In the control group, nude mice were injected with saline. In the psi-scramble group, 20 μg of psi-scramble plasmid were injected into the tumor at several sites. The electric transfection was done to increase the transfection efficiency. Injection was performed once every 3 days. Attenuated Salmonella typhi y21 were treated with calcium chloride to prepare the competent cells, and then transfected with p-GRIM-19, psi-survivin, psi-scramble and p-siRNA survivin/GRIM-19 plasmid, independently. The Salmonella transfected with different plasmids were injected into nude mice in the corresponding groups, once every 6 days. The general conditions of these mice were observed, including body weight and tumor size. One month later, the animals were killed for measuring the tumor size. The tumors were collected, paraffin-embedded, sliced, H&E stained and Tunel stained. In addition, protein was extracted for further testing. The liver, spleen, kidney, lung, heart and tumor were collected from mice in the psi-survivin group and the psi-scramble group and weighed, and their size was measured. The Salmonella also was checked to see if there is more in tumor tissues.
Immunohistochemistry
A fraction of tumor tissues was fixed in 10% formalin, embedded in paraffin, and cut into 4-μm sections. The chain-avidin-biotin-peroxidase complex (SP) method was used for immunohistochemistry according to the manufacture’s instructions. The pathologically proven gastric cancer tissues served as a positive control; the primary antibody was replaced with PBS in the negative control. Cells with brown nucleus and cytoplasm were considered as positive for target proteins. A total of 500 cells were counted under a microscope at a magnification of 400× (positive: ≥10%; negative: <10%).
Western blot assay
Total protein was extracted from the tumors, and western blot assay was done with Bio-Rad method. In brief, 80 μg of proteins were mixed with 4×SDS sample buffer (1/4 volume) and this mixture was boiled water for 5 min followed by electrophoresis for 1.5 h at 100 V and being transferred onto PVDF membrane. The membrane were sequentially incubated with primary antibody and secondary followed by visualization. The optical density of each band was normalized to that of β-actin. The experiment was done thrice.
Tunnel staining
The above sections were washed in PBS, and treated with 0.1% TritonX-100 and 0.1% sodium citrate on ice. Following washing in PBS, 50 L Tunnel reaction mixture (5’ end transferase and 45l fluorescently labeled nucleic acid) was added to each sections. After washing with PBS thrice, DAPI was added for double staining followed by mounting with 1:1 glycerol-PBS. Representative photographs were captured under a fluorescence microscope.
Statistical analysis
Data are expressed as mean ± standard deviation. SPSS 14.0 for Window was employed for statistical analysis. Data from three independent experiments were averaged and compared among different groups with analysis of variance. A value of P<0.05 was considered statistically significant.
Results
mRNA and protein expression of survivin and GRIM-19 levels in Hep-2 cells
Our results showed that the mRNA expression of survivin was significantly reduced in the Hep-2 cells transfected with psi-survivin and p-siRNA survivin/GRIM-19 plasmid when compared with blank control group and the negative control group. The inhibition rates were 54.4% and 53.4%, respectively (P<0.01), indicating that psi-survivin and p-siRNA survivin/GRIM-19 transfected Hep-2 cells are able to silence the survivin expression at mRNA level. However, the survivin expression was comparable between p-siRNA survivin/GRIM-19 transfected cells and psi-survivin transfected cells (P>0.05). When compared with blank control group, the survivin expression in the p-GRIM-19 plasmid transfected cells was significantly lower (P<0.01), indicating that GRIM-19 can decrease the survivin expression. The GRIM-19 expression was increased in the p-GRIM-19 transfected cells and p-siRNA survivin/GRIM-19 transfected cells; but no significant difference was observed between the later two groups (P>0.05), showing that the silencing of survivin has no influence on the GRIM-19 expression in Hep-2 cells.
Western blot assay showed that no significant difference in the expression of survivin and GRIM19 between the negative control group and the blank control group. However, in the psi-survivin and p-siRNA survivin/GRIM-19 groups, the survivin expression was significantly reduced (P<0.01), and the inhibition rate was 37% and 42.2%, respectively. When compared with psi-survivin group, the survivin expression was lower in the p-siRNA survivin/GRIM-19 group (P<0.05). In addition, the survivin expression was also reduced in p-GRIM-19 transfected cells (P<0.05), indicating that p-GRIM-19 transfection reduced the survivin expression at both mRNA and protein levels. The GRIM-19 expression was increased in the p-GRIM-19 group and p-siRNA survivin/GRIM-19 group, when compared with the blank control group (P<0.01). As shown in the Figure 1, the survivin expression was reduced while GRIM-19 expression increased in the p-siRNA survivin/GRIM-19 transfected cells.
Figure 1.
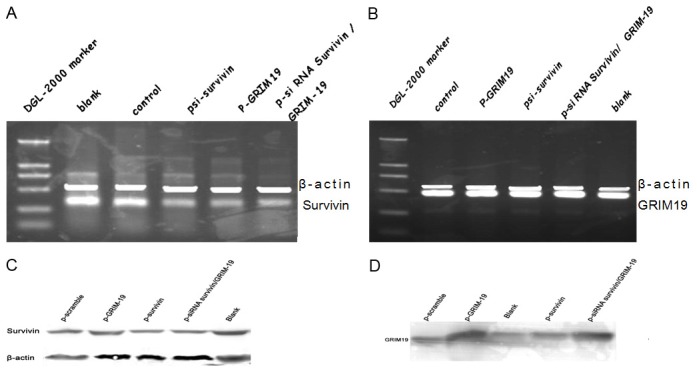
Expression of Survivin and GRIM-19 in Hep-2 cells transfected with various plasmids. A. RT-PCR analysis of Survivin mRNA. B. RT-PCR analysis of GRIM-19 mRNA. C. Western blot analyses for Survivin. D. Western blot analyses for GRIM-19. E. Quantification of Survivin and GRIM-19 protein by densitometric analysis.
Detection of cell apoptosis and effect of plasmid transfection on growth of Hep-2 cells
Flow cytometry showed, 72 h after transfection with psi-survivin, p-GRIM-19, or p-siRNA survivin/GRIM-19, the apoptotic Hep-2 cells were significantly increased. No or few apoptotic cells were found in the blank control group and the negative control group. In addition, the apoptotics cells significantly increased in the plasmid group as compared to the negative control group (P<0.01) (Table 1); while there was no significant difference between negative control group and transfection control group (P>0.05), indicating no influence of negative control plasmid on the apoptosis of Hep-2 cells. Moreover, more apoptosis cells were found in the p-siRNA survivin/GRIM-19 group than in the psi-survivin group (P<0.05) and the p-GRIM-19 group (P<0.05), indicating that transfection with p-siRNA survivin/GRIM-19 has more potent promotive effect on apoptosis than the transfection with psi-survivin or p-GRIM-19.
Table 1.
Percentage of apoptosis cells 72 hours after transfection with psi-scramble, psi-survivin, p-GRIM-19, p-siRNA Survivin/GRIM-19
| Group | Apoptotic cells (%) |
|---|---|
| Untreated | 0.753±0.23 |
| psi-scramble | 1.277±0.369 |
| psi-survivin | 13.047±0.561* |
| p-GRIM-19 | 10.313±0.561* |
| p-siRNA survivin/GRIM-19 | 13.933±0.244* |
n=3 Mean±SD
P<0.05 vs psi-scramble and untreated groups.
In the acridine orange staining, normal viable cells showed uniform yellow-green fluorescence. Hep-2 cells undergoing transfection with p-GRIM-19, psi-survivin or p-siRNA survivin/GRIM-19 showed massive orange fluorescence in the apoptotic cells (Figure 2C-E), while cells in the blank control group and negative control group had no the apoptosis related fluorescence (Figure 2A, 2B).
Figure 2.
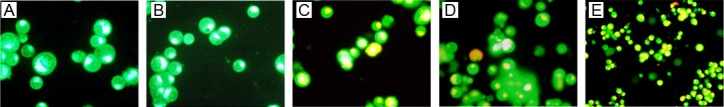
Effects of coexpressed siRNA-Survivin and GRIM-19 on cell growth and apoptosis: A-E. Acridine orange staining; A. Hep-2 cells 200×; B. psi-scramble Hep-2 cells 200×; C. P-GRIM-19 Hep-2 cells 200×; D. psi-survivin Hep-2 cells 200×; E. Growth inhibitory effects of various plasmids on Hep-2 cells.
The transfection with psi-survivin, p-GRIM-19, p-siRNA survivin/GRIM-19 could significantly inhibit the proliferation of Hep-2 cells, and the inhibition rate was 70.4%, 60.7% and 83.4%, respectively. These results suggest that transfection with these plasmids may inhibit the proliferation of Hep-2 cells. No significantly difference was found in the proliferation between negative control and the blank control cells in vitro (P>0.05); while the growth of Hep-2 cells was inhibited by after transfection with psi-survivin, p-GRIM-19 or p-siRNA survivin/GRIM-19. When compared with the control group, significant difference was found (P<0.01). Of interest, in the p-siRNA survivin/GRIM-19 group, the inhibition of survivin expression was more obvious than that in the psi-survivin group and p-GRIM-19 group (P<0.01).
Effects of recombinant plasmids on tumor growth in vivo
When compared with the negative control group (psi-scramble), the tumor growth in the psi-survivin group, p-GRIM-19 group and p-siRNA survivin/GRIM-19 group was significantly inhibited. Moreover, the inhibition of cancer group in the p-siRNA survivin/GRIM-19 group was more evident that that in the former two groups. At 2 weeks after inoculation, the xenografts disappeared in 1 mouse; at 4 weeks, the tumors disappeared in other 3 mice. The inhibition of cancer growth in the psi-survivin group was stronger than that in the p-GRIM-19 group. The mice were generally in good condition as shown in daily observations, and no failure and death were observed during the experiment. The tumor size was comparable between nude mice injected with saline and psi-scramble, indicating the negative control plasmid has no inhibitory effect on the tumor growth in nude mice. As compared to the plasmid-negative group, the anti-tumor effect was more obvious in the Salmonella control plasmid group, suggesting that Salmonella itself has anti-tumor effect.
Salmonella in organs of nude mice
One mouse from the psi-survivin group and psi-scramble group was collected and sacrificed. The liver, spleen, kidney, lung, heart and tumor were harvested and weighted, and their size was measured. The tissues were mashed and seeded on plates. The numbers of colonies were detected on the next day, which was divided by the tissue weight as the number of colonies per gram of tissues. Results showed that the bacteria were most in the liver, followed by the tumor and kidney, and the heart had the least bacteria.
Immunohistochemistry for survivin and GRIM-19
The survivin expression in the saline control (mock) group, negative plasmid group (scramble-injected) and Salmonella-negative plasmid group (scramble) was higher than that in the p-GRIM-19 group, psi-survivin group and p-siRNA survivin/GRIM-19 group. However, for the GRIM-19 expression, the saline control (mock) group, negative plasmid group (scramble-injected) and Salmonella-negative plasmid group (scramble) was lower than that in the p-GRIM-19 group and psi-survivin group. In the psi-survivin group, GRIM-19 was negative (Figure 3).
Figure 3.
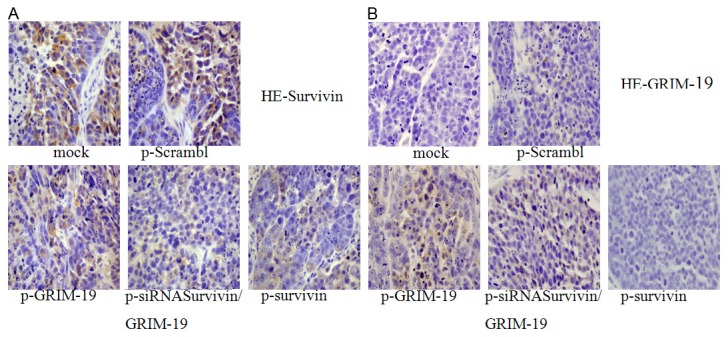
Immunohistochemistry for survivin and GRIM-19. HE (400×): A. Immunohistochemistry for surviving, mock-saline control, p-Scramble-negative control; B. Immunohistochemistry for GRIM-19.
Western blot assay and tunel staining of survivin and GRIM-19
Figure 4A showed the survivin expression decreased in the p-siRNA survivin/GRIM-19 group and psi-survivin group, and the inhibition rate was 50.3% and 41.3%, respectively. The inhibition of survivin expression was more evident in the p-siRNA survivin/GRIM-19 group than that in the psi-survivin group. The expression of survivin in the p-GRIM-19 group was also lowered (inhibition rate: 34.5%); but the inhibition was weaker than that in the p-siRNA survivin/GRIM-19 group and psi-survivin group. The GRIM-19 expression increased in the p-siRNA survivin/GRIM-19 group and p-GRIM-19 group.
Figure 4.
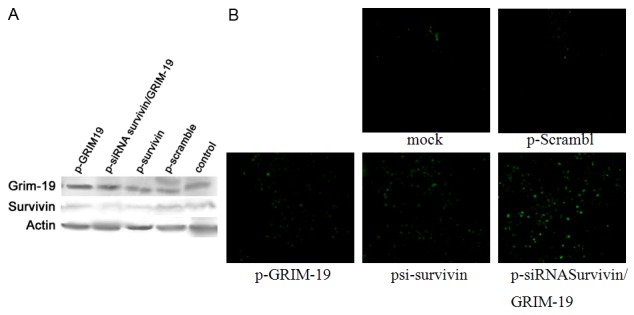
Western blot assay and Tunnel staining of survivin and GRIM-19: A. Western blot assay of survivin and GRIM-19. B. Tunnel staining: mock-saline control, p-scramble-negative control.
Results of Tunel staining are shown in Figure 4B. In the saline control group, negative plasmid control group and Salmonella negative plasmid group, few green apoptotic cells were found. However, in the p-GRIM-19 group, psi-survivin group and p-siRNA survivin/GRIM-19 group, more green apoptotic cells were observed, especially in the p-siRNA survivin/GRIM-19 group, indicating that transfection of these plasmids can induce the apoptosis of laryngeal cancer cells.
Discussion
Abnormal apoptosis plays an important role in the occurrence of cancer [17], and therefore, controlling the apoptosis of cancer cells has biological and clinical significance [18,19]. In this study, plasmid expression GRIM-19 and survivin specific siRNA wasconstructed and transfected into laryngeal cancer cells (Hep-2 cells). Then, the expression of target genes was detected in vivo and in vitro. We hypothesized that the transfection with p-siRNA survivin/GRIM-19 induce the apoptosis of Hep-2 cells and inhibit their growth, which were more potent than the transfection with p-surviving or p-GRIM-19.
There are several strategies for the combined gene therapy. For example, vectors carrying different genes are transfected into target cells concomitantly. The design of vectors is simple and it is easy to adjust the ratio of each vector. However, it is difficult to ensure that a variety of target genes are introduced into the target cells at an appropriate ratio. In addition, we can prepare vectors carrying multiple genes. The design of this system is a challenge. The single vector may carry a limited number of genes, and whether these genes can be expressed efficiently in the target cells is still unclear. Theoretically, p-siRNA survivin/GRIM-19 co-expressing siRNA survivin and GRIM-19 has both U6 promoter and CMV promoter. p-siRNA survivin and GRIM-19 have the corresponding promoters and may exert effect independently in cells after transfection. They can silence survivin and increase the GRIM-19 expression. Our results showed that, p-siRNA survivin/GRIM-19 could be transfected into Hep-2 cells and influence the expression of target genes. In this study, the survivin expression was reduced, while the GRIM-19 expression increased at mRNA and protein levels. In addition, western blot assay showed that, as compared to single psi-survivin group, transfection with p-siRNA survivin/GRIM-19 more effectively silenced the survivin expression, suggesting that the p-siRNA survivin/GRIM-19 was successfully constructed. At the mRNA level, expression of survivin was similar after transfection with p-siRNA survivin/GRIM-19 and psi-survivin. This may be attributed to the limited sensitivity of semi-quantitative RT-PCR which did not reflect the small changes in the quantity of mRNA.
It has been shown that GRIM-19 can enhance the cell sensitivity to IFN-β-RA-induced apoptosis. GRIM-19 can interact with many cellular factors involving in the apoptosis. GRIM-19 can specifically inhibit the activity of signal transduction and activator of transcription 3 (Stat3) [20]; and Stat3 is activated in many malignant tumors, including the head and neck squamous cell carcinoma [21,22]. In addition, survivin gene is a direct downstream gene of Stat3. Inhibition of STAT3 can reduce the decrease in survivin expression, and induce the apoptosis of tumor cells [23]. As shown in semi-quantitative RT-PCR, p-GRIM-19 transfected Hep-2 cells had higher survivin expression when compared with blank control; while the psi-survivin transfected cells had compareble GRIM-19 expression to the negative control group. These results suggested that high expression of GRIM-19, possibly through specific inhibition of its downstream gene Stat3, indirectly reduces the expression of survivin in Hep-2 cells. However, the reduction in survivin expression following transfection with survivin siRNA had no influence on the GRIM-19 expression.
Combined gene therapy is to kill tumor cells by synergistically targeting several genes involved in the occurrence and/or development of cancers. Theoretically, it is more effective to inhibit the cancer growth, and improve the prognosis. In the present study, flow cytometry showed that, in the psi-survivin, p-GRIM-19 and p-siRNA survivin/GRIM-19, the apoptotic cells were markedly increased, and the apoptosis rate was 13.047%, 10.313% and 13.933%, respectively. When compared with the control group and negative plasmid group, the apoptosis rate was significantly elevated. However, the apoptotic peak was absent in the control group and the negative control group, and the apoptosis rate was 0.753% and 1.277%, respectively showing no marked difference between them. These findings suggested that the induction of apoptosis in Hep-2 cells could be specific following transfection. In addition, the apoptosis rate was higher in the p-siRNA survivin/GRIM-19 group than in the psi-survivin group and p-GRIM-19 group. This suggested that, in the p-siRNA survivin/GRIM-19 group, the survivin expression was reduced not only by silencing the surviving gene but also through increasing the expression of GRIM-19 which inhibits the Stat3. MTT assay showed that, in the psi-survivin group and p-GRIM-19 group, the growth of Hep-2 cells was inhibited; the inhibition rate was 48% and 35%, respectively. In the p-siRNA survivin/GRIM-19 group, the growth inhibition rate was 71%, which was higher than that in the psi-survivin group and p-GRIM-19 group. These results indicated that p-siRNA survivin/GRIM-19 is more effective to inhibit the in vitro Hep-2 cell growth than psi-survivin and p-GRIM-19.
In order to detect the in vivo effect of plasmids on the growth of laryngeal cancer, nude mice xenograft model was established. How can the plasmid carrying exogenous genes be delivered to the tumors? Anaerobic bacteria with anaerobic characteristics were chosen, and the center of solid tumors has a hypoxic environment which provides possibility for the survival of anaerobic bacteria. In this study, the attenuated Salmonella were used. At 32 days after inoculation, the nude mice were killed, and the tumor, heart, liver, lung, kidney and spleen were collection for the detection of bacterial colonies. Results showed that the bacterial colonies were the most in liver, followed by tumor, indicating that attenuated Salmonella tends to localize in tumors. As shown in the growth curve of tumors in nude mice, in the psi-survivin group, p-GRIM-19 group and p-siRNA survivin/GRIM-19 group, the tumor growth in nude mice were reduced when compared with the saline group and negative control group, and the inhibition was the most evident in the p-siRNA survivin/GRIM-19 group. Among the 5 nude mice, tumor disappeared in 1 mouse in the second week and in 3 mice in the fourth week. All the nude mice were in good condition generally; and death was not observed during the experiment, indicating the plasmid and Salmonella had no side effects. In addition, the tumor growth was also significantly reduced in the Salmonella negative control group when compared with the control group, indicating that Salmonella itself has anti-tumor effect. To further explore the mechanism underlying the inhibition of tumor growth in nude mice, immunohistochemistry and western blot assay were employed to detect the survivin and GRIM-19 expression. Results showed that, in the p-siRNA survivin/GRIM-19 group and psi-survivin group, the survivin expression was significantly lowered (inhibition rate: 53.6% and 35.4%, respectively). Although survivin expression was also reduced in the p-GRIM-19 group, the reduction was small. p-siRNA survivin/GRIM-19 was more effective than psi-survivin to reduce the survivin expression in vivo. GRIM-19 expression increased in the p-siRNA survivin/GRIM-19 group and p-GRIM-19 group, indicating that the Salmonella carrying siRNA survivin/GRIM-19 was successfully constructed.
Survivin can inhibit cell apoptosis. Thus, to reduce the survivin expression in tumor cells may promote their apoptosis. GRIM-19 can specifically inhibit the Stat3 activity. High expression of GRIM-19 further enhances the apoptosis by inhibiting survivin expression via Stat3. Tunel staining showed that, in the psi-survivin group, p-GRIM-19 group and p-siRNA survivin/GRIM-19 group, apoptotic cells were seen, and the number of apoptotic cells was highest in the p-siRNA survivin/GRIM-19 group. Few apoptotic cells were noted in the saline control group and the two negative control groups. These results indicated that all these plasmids can induce the apoptosis of laryngeal cancer cells.
In summary, our study demonstrate that psi-survivin, p-GRIM-19 and p-siRNA survivin/GRIM-19 can promote the apoptosis of laryngeal cancer cells and inhibit their proliferation, and p-siRNA survivin/GRIM-19 is more effective than psi-survivin and p-GRIM-19. In addition, attenuated Salmonella carrying plasmid can be delivered to the tumors in nude mice, and then exhibit an inhibitory effect. Moreover, the attenuated Salmonella itself has anti-tumor effect.
Acknowledgements
This study was supported by Science and Technology Fund of Jilin Province (200705213) and National Natural Science Foundation of China (81072210).
References
- 1.Theys J, Barbe S, Landuyt W, Nuyts S, Van Mellaert L, Wouters B, Anne J, Lambin P. Tumor-specific gene delivery using genetically engineered bacteria. Curr Gene Ther. 2003;3:207–21. doi: 10.2174/1566523034578357. [DOI] [PubMed] [Google Scholar]
- 2.Ambrosini G, Adida C, Altieri DC. A novel anti-apoptosis gene, survivin, expressed in cancer and lymphoma. Nat Med. 1997;3:917–21. doi: 10.1038/nm0897-917. [DOI] [PubMed] [Google Scholar]
- 3.Wen LJ, Li CQ, Sun YX, Jin CS, Gao HW. Expression and implication of survivin in laryngeal carcinoma. Chinese Archives of Otolaryngology-head and Neck Surgery. 2004;11:327–329. [Google Scholar]
- 4.Ko YH, Roh SY, Won HS, Jeon EK, Hong SH, Lee MA, Kang JH, Hong YS, Kim MS, Jung CK. Prognostic significance of nuclear survivin expression in resected adenoid cystic carcinoma of the head and neck. Head Neck Oncol. 2010;2:30. doi: 10.1186/1758-3284-2-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Atlasi Y, Mowla SJ, Ziaee SA. Differential expression of survivin and its splice variants, survivin-DeltaEx3 and survivin-2B, in bladder cancer. Cancer Detect Prev. 2009;32:308–13. doi: 10.1016/j.cdp.2008.12.001. [DOI] [PubMed] [Google Scholar]
- 6.Taubert H, Heidenreich C, Holzhausen HJ, Schulz A, Bache M, Kappler M, Eckert AW, Würl P, Melcher I, Hauptmann K, Hauptmann S, Schaser KD. Expression of survivin detected by immunohistochemistry in the cytoplasm and in the nucleus is associated with prognosis of leiomyosarcoma and synovial sarcoma patients. BMC Cancer. 2010;10:65. doi: 10.1186/1471-2407-10-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guan HT, Xue XH, Wang XJ, Li A, Qin ZY. Knockdown of survivin expression by small interfering RNA suppresses proliferation of two human cancer cell lines. Chin Med Sci J. 2006;21:115–9. [PubMed] [Google Scholar]
- 8.Hou JQ, He J, Wang XL, Wen DG, Chen ZX. Effect of small interfering RNA targeting survivin gene on biological behaviour of bladder cancer. Chin Med J (Engl) 2006;119:1734–9. [PubMed] [Google Scholar]
- 9.Hou JQ, He J, Wen DG, Wang XL. [Experimental study on bladder cancer by the small interfering RNA targeting survivin] . Zhonghua Yi Xue Yi Chuan Xue Za Zhi. 2007;24:401–4. [PubMed] [Google Scholar]
- 10.Yang JH, Zhang YC, Qian HQ. Survivin antisense oligodeoxynucleotide inhibits growth of gastric cancer cells. World J Gastroenterol. 2004;10:1121–4. doi: 10.3748/wjg.v10.i8.1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zaffaroni N, Pennati M, Daidone MG. Survivin as a target for new anticancer interventions. J Cell Mol Med. 2005;9:360–72. doi: 10.1111/j.1582-4934.2005.tb00361.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu X, Gao R, Dong Y, Gao L, Zhao Y, Zhao L, Zhao X, Zhang H. Survivin gene silencing sensitizes prostate cancer cells to selenium growth inhibition. BMC Cancer. 2010;10:418. doi: 10.1186/1471-2407-10-418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nakao K, Hamasaki K, Ichikawa T, Arima K, Eguchi K, Ishii N. Survivin downregulation by siRNA sensitizes human hepatoma cells to TRAIL-induced apoptosis. Oncol Rep. 2006;16:389–92. [PubMed] [Google Scholar]
- 14.Shulga N, Pastorino JG. GRIM-19-mediated translocation of STAT3 to mitochondria is necessary for TNF-induced necroptosis. J Cell Sci. 2012;125:2995–3003. doi: 10.1242/jcs.103093. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 15.Alchanati I, Nallar SC, Sun P, Gao L, Hu J, Stein A, Yakirevich E, Konforty D, Alroy I, Zhao X, Reddy SP, Resnick MB, Kalvakolanu DV. A proteomic analysis reveals the loss of expression of the cell death regulatory gene GRIM-19 in human renal cell carcinomas. Oncogene. 2006;25:7138–47. doi: 10.1038/sj.onc.1209708. [DOI] [PubMed] [Google Scholar]
- 16.Angell JE, Lindner DJ, Shapiro PS, Hofmann ER, Kalvakolanu DV. Identification of GRIM-19, a novel cell death-regulatory gene induced by the interferon-beta and retinoic acid combination, using a genetic approach. J Biol Chem. 2000;275:33416–26. doi: 10.1074/jbc.M003929200. [DOI] [PubMed] [Google Scholar]
- 17.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 18.Chau BN, Wang JY. Coordinated regulation of life and death by RB. Nat Rev Cancer. 2003;3:130–8. doi: 10.1038/nrc993. [DOI] [PubMed] [Google Scholar]
- 19.Chene P. Inhibiting the p53-MDM2 interaction: an important target for cancer therapy. Nat Rev Cancer. 2003;3:102–9. doi: 10.1038/nrc991. [DOI] [PubMed] [Google Scholar]
- 20.Nikitakis NG, Siavash H, Sauk JJ. Targeting the STAT pathway in head and neck cancer: recent advances and future prospects. Curr Cancer Drug Targets. 2004;4:637–51. doi: 10.2174/1568009043332736. [DOI] [PubMed] [Google Scholar]
- 21.Gao LF, Wen LJ, Yu H, Zhang L, Meng Y, Shao YT, Xu DQ, Zhao XJ. Knockdown of Stat3 expression using RNAi inhibits growth of laryngeal tumors in vivo. Acta Pharmacol Sin. 2006;27:347–52. doi: 10.1111/j.1745-7254.2006.00277.x. [DOI] [PubMed] [Google Scholar]
- 22.Xi S, Zhang Q, Dyer KF, Lerner EC, Smithgall TE, Gooding WE, Kamens J, Grandis JR. Src kinases mediate STAT growth pathways in squamous cell carcinoma of the head and neck. J Biol Chem. 2003;278:31574–83. doi: 10.1074/jbc.M303499200. [DOI] [PubMed] [Google Scholar]
- 23.Gritsko T, Williams A, Turkson J, Kaneko S, Bowman T, Huang M, Nam S, Eweis I, Diaz N, Sullivan D, Yoder S, Enkemann S, Eschrich S, Lee JH, Beam CA, Cheng J, Minton S, Muro-Cacho CA, Jove R. Persistent activation of stat3 signaling induces survivin gene expression and confers resistance to apoptosis in human breast cancer cells. Clin Cancer Res. 2006;12:11–19. doi: 10.1158/1078-0432.CCR-04-1752. [DOI] [PubMed] [Google Scholar]


