SUMMARY
Expression of miRNAs involves transcription of miRNA genes and maturation of the primary transcripts. Recent studies have shown that post-transcriptional processing of primary and precursor miRNAs are induced after DNA damage through regulatory RNA-binding proteins in the Drosha and Dicer complexes, such as DDX5 and KSRP. However, little is known about the regulation of nuclear export of pre-miRNAs in the DNA damage response, a critical step in miRNA maturation. Here we show that nuclear export of pre-miRNAs is accelerated after DNA damage in an ATM-dependent manner. The ATM-activated AKT kinase phosphorylates Nup153, a key component of the nucleopore, leading to enhanced interaction between Nup153 and Exportin-5 (XPO5) and increased nuclear export of pre-miRNAs. These findings define an important role of the DNA damage signaling in miRNA transport and maturation.
INTRODUCTION
MicroRNAs (miRNAs) are a class of small non-coding RNAs that are 18 to 24 nucleotides in length. A mature miRNA binds to its mRNA targets at their complementary sequences to downregulate gene expression by inhibiting the mRNA translation to proteins or by inducing mRNA degradation (Bartel, 2009). Whereas only 1% of the genomic transcripts in mammalian cells encode miRNA, about one-third of protein-coding genes are regulated by miRNA (Grimson et al., 2007). MiRNAs have emerged as key post-transcriptional regulators of gene expression in metazoans and plants. MiRNA biogenesis is a multi-step process that includes primary miRNA transcription and post-transcriptional maturation (Winter et al., 2009). MiRNAs are first transcribed as primary transcripts (pri-miRNAs) that contain one or more hairpin-like stem-loop structures. Pri-miRNAs are then cleaved in the nucleus by a microprocessor complex containing Drosha and DGCR8 to produce miRNA precursors (pre-miRNAs) (Lee et al., 2003). Pre-miRNAs are exported to the cytoplasm by the nuclear transport receptor XPO5, where they undergo final processing by the Dicer complex (Lund et al., 2004; Yi et al., 2003; Bohnsack et al., 2004). Mature miRNAs and Argonaute proteins form the RNA-induced silencing complex (RISC) that mediates post-transcriptional gene silencing (Diederichs and Haber, 2007; Rand et al., 2005). Recent evidence revealed that pre-miRNA processing and RISC assembly are coupled by the RISC loading complex formed by Dicer, TRBP (Tar RNA binding protein), PACT (protein activator of PKR), and Ago2 (Winter et al., 2009). Within the RISC, the single-stranded miRNA is unwound by the helicase activity of Dicer and guides target selection, causing inhibition of translation and stability of target mRNA.
In response to intrinsic and extrinsic genotoxic stresses, eukaryotic cells have evolved a sophisticated self-surveillance system, known as the DNA damage response (DDR). Ataxia Telangiectasia mutated (ATM) is one key component of the DDR that initiates DNA damage signaling by phosphorylating downstream effector proteins, leading to a dramatic change in the gene expression program (Ciccia and Elledge, 2010). Accumulating evidence has revealed important roles for miRNAs in initiation, activation and maintenance of the DDR through their target genes (Wan et al., 2011). As an example, miR-421 was found to be induced by N-myc and in turn suppresses ATM expression by targeting the 3’-UTR (untranslated region) of ATM transcripts in human neuroblastomas (Hu et al., 2010). There is also evidence indicating that miR-182 downregulates BRCA1 levels in human breast cancer cells, which leads to diminished activity of DNA double strand break repair via homologous recombination and sensitization of cells to ionizing radiation and the chemotherapeutic agent PARP (Moskwa et al., 2011).
Expression of miRNAs in the DDR is regulated transcriptionally and posttranscriptionally. The original discovery connecting the DDR to miRNA expression was the identification of the miR-34 family as a direct transcriptional target of p53 (He et al., 2007). TAp63, a major member in the p53 family, suppresses metastasis through coordinate regulation of Dicer and miRNAs. TAp63 binds to and transactivates the promoters of Dicer and miR-130b, suggesting direct control of miRNA expression by TAp63 (Su et al., 2010). In addition to transcriptional regulation, cross-talk exists between miRNA maturation and the DDR. The Persengiev group discovered that UV damage triggers a cell cycle-dependent relocalization of Argonaute 2 (Ago2) into stress granules and promoted miRNA expression (Pothof et al., 2009). Suzuki et al. defined the function of p53 as a direct interactor with the Drosha-DGCR8 microprocessor in promoting the maturation of several miRNAs (Suzuki et al., 2009). Our recent work provided direct evidence that a subset of the KSRP (KH-type regulatory protein)-associated miRNAs are induced after DNA damage in an ATM-dependent manner (Zhang et al., 2011). Following the discovery by Trabucchi and colleagues that KSRP promotes maturation of a select group of miRNA precursors (Trabucchi et al., 2009), we demonstrated that ATM directly binds to and phosphorylates KSRP, leading to enhanced interaction between KSRP and pri-miRNAs and increased KSRP activity in miRNA processing. MiRNAs complete their maturation only in the cytoplasm after their precursors are transported from the nucleus. In the present study, we show that nuclear export of pre-miRNAs is substantially induced after DNA damage in an ATM-dependent manner. The ATM-activated AKT kinase phosphorylates a Nup153, an essential component of the nuclear pore (nucleopore), enhancing the interaction between XPO5 and Nup153 and promoting nucleocytoplasmic transport of pre-miRNAs.
RESULTS
DNA Damage-induced and ATM-dependent MiRNA Expression in Human Cells
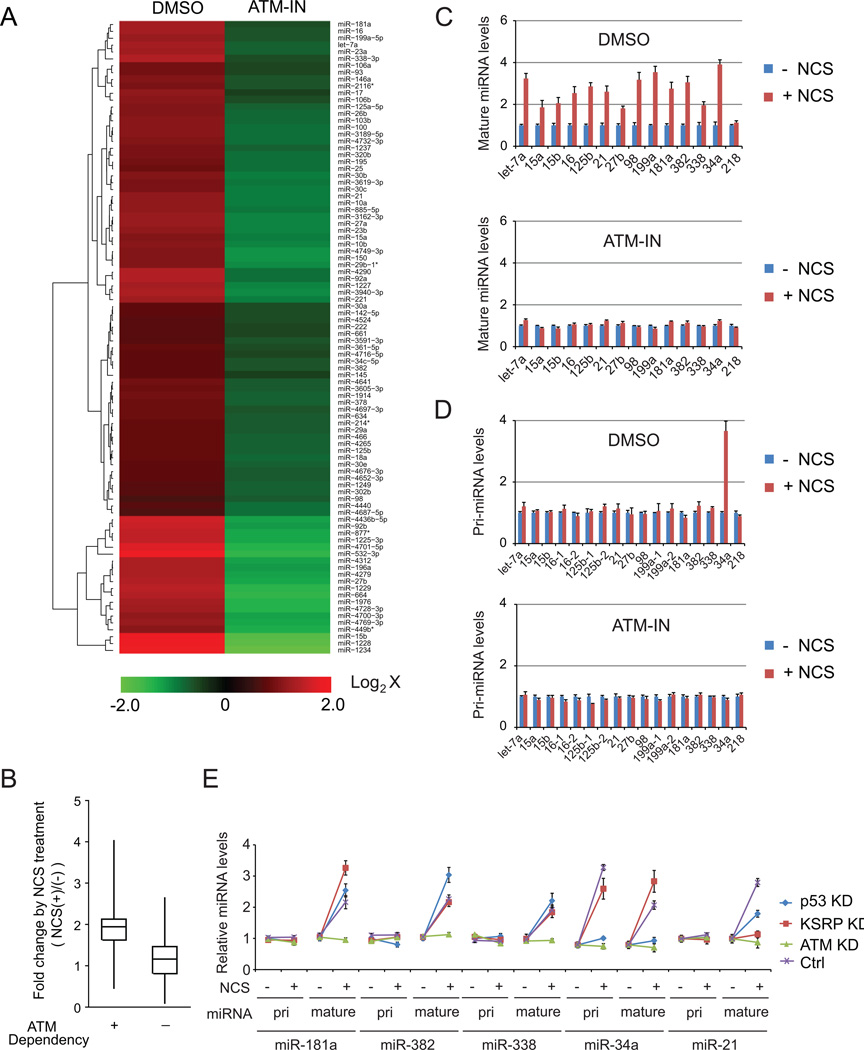
Our previous study revealed that one quarter of total identified mouse miRNAs were significantly induced in Atm+/+ mouse embryonic fibroblasts (MEFs), but not in the littermate Atm−/− MEFs, suggesting that DNA damage stress triggers wide-spectrum alterations on miRNA expression (Zhang et al., 2011). To verify the DNA damage induction of miRNAs in human cells, we examined mature miRNA expression in human fibroblast GM0637 cells treated with the radiomimetic drug, neocarzinostatin (NCS), in the presence or absence of the ATM inhibitor KU55933 (Figures 1A, 1B and S1A). A total of 331 out of 1297 human miRNAs were shown to be significantly (≥ 60%) induced after NCS treatment in the cells, but not in the cells pre-treated with the ATM inhibitor (GEO accession number GSE42248). In agreement with previous reports demonstrating that ATM-activated p53 and KSRP promotes miRNA expression (Suzuki et al., 2009; Zhang et al., 2011), we found 61 p53-dependent miRNAs and 29 KSRP-dependent miRNAs within the group of ATM-induced miRNAs.
Figure 1. MiRNAs are induced after DNA damage in an ATM-dependent manner.
(A) A set of miRNAs are induced after DNA damage in an ATM-dependent manner. Human fibroblast GM0637 (ATM-proficient) cells were pretreated with ATM inhibitor KU55933 (10 µM) or DMSO prior to NCS (500 ng/ml) treatment. Cells were harvested 4h after NCS treatment for microarray analyses. Red or green color on the heat map indicates an increase or decrease of miRNA level. Color intensity reflects relative signal levels on a logarithmic scale. (B) DNA damage induction of ATM-dependent or ATM-independent miRNAs. ATM-dependent (ATM-IN/Ctrl < 0.67) and ATM-independent (ATM-IN/Ctrl > 0.67) miRNAs were defined by the miRNA expression profile from GM0637 cells treated with DMSO or ATM inhibitor. (C) DNA damage-induced miRNAs were verified by quantitative RT-PCR. (D) DNA damage has no significant effect on pri-miRNA levels. Error bars represent the mean ± SD. (E) Five representative miRNAs are post-transcriptionally induced in an ATM-dependent manner after DNA damage. KD: knockdown.
MiRNA expression involves miRNA gene transcription and post-transcriptional maturation of primary transcripts. While transcriptional factors were identified that regulate miRNA gene transcription, increasing evidence has shown that post-transcriptional processing plays a key role in controlling miRNA expression in the DDR (Wan et al., 2011; Zhang and Lu, 2011). We examined the levels of different forms of miRNAs (pri-miRNAs, pre-miRNAs and mature miRNAs) selected from the ATM-induced miRNAs, including KSRP-dependent miRNAs (let-7a, 15a, 15b, 16, 125b, 21, 27b, 98, 199a), p53-dependent miRNAs (34a), and KSRP/p53-independent miRNAs (181a, 382, 338). The control miR-218 that is unaffected by DNA damage was also included in the examination (Figures 1C, 1D, S1B and S1C). With the exception of miR-34a that is known to be transactivated by p53, there were no significant increases in expression of primary transcripts for these miRNAs following DNA damage. These results suggest that DNA damage may promote post-transcriptional maturation of the miRNAs. MiR-181a, miR-382 and miR-338 were not dependent on either KSRP or p53. Stable knockdown of KSRP or p53 did not inhibit their induction after DNA damage, whereas knockdown of ATM abolished their induction (Figures 1E and S1D), indicating that another ATM-dependent mechanism accounts for the induced miRNAs in the DDR. As controls, miR-34a is dependent on p53, and miR-21 is dependent on KSRP as reported previously (Figure 1E) (He et al., 2007; Trabucchi et al., 2009).
While miRNA expression is induced after DNA damage, these induced miRNAs contribute to the maintenance of DDR. A number of miRNA targets have been identified in the DNA damage signaling pathways (Wan et al. 2011), one of which is Bcl2. Bcl2, an anti-apoptotic protein, is suppressed after DNA damage (Figure S2A). However, knockdown of ATM abolished its suppression. Consequently, the DNA damage-induced apoptosis was significantly inhibited (Figure S2C). Previous reports have shown that two of the DNA damage-induced miRNAs, miR-16 and miR-34a both target Bcl2 transcripts (Figure S2B) (Cimmino et al., 2005; Ji et al., 2008). Overexpression of miR-16 or miR-34a rescued apoptotic phenotypes in the ATM-knockdown U2OS cells, suggesting an important roles of the DNA damage-induced miRNAs in the DDR (Figure S2A and S2C).
Accelerated Nuclear Export of Pre-miRNAs After DNA Damage
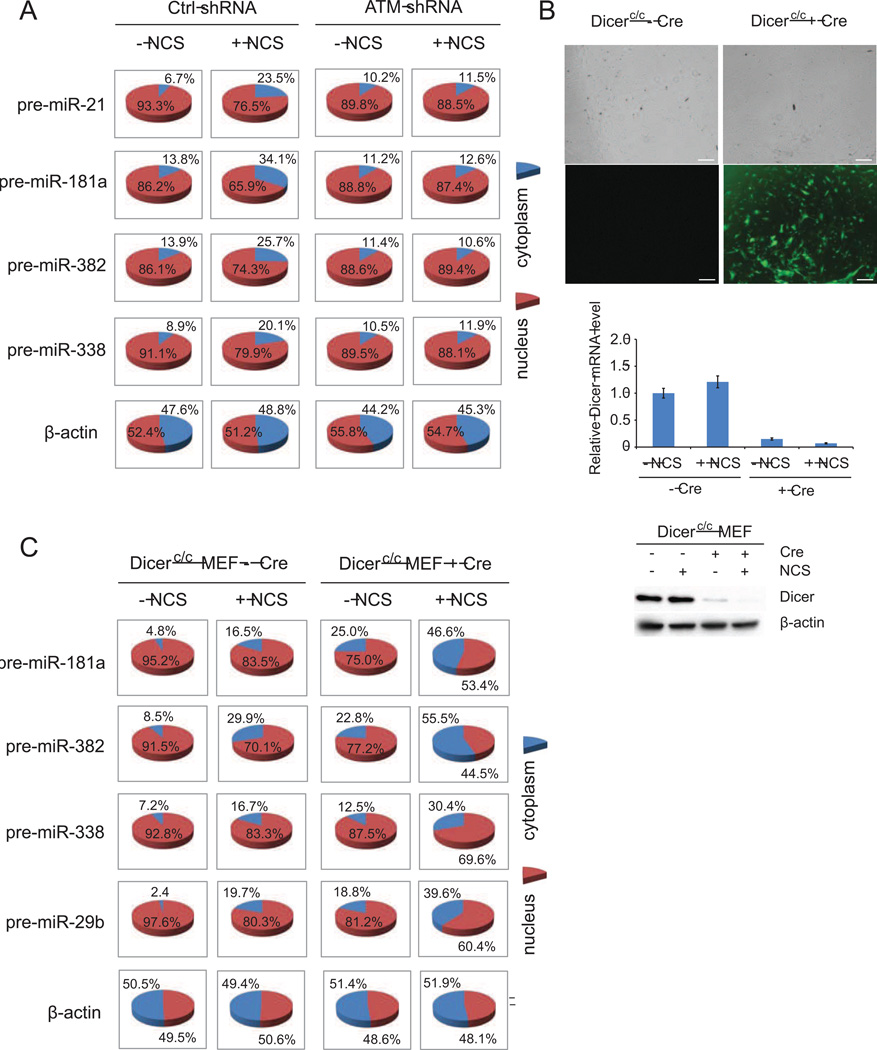
A key rate-limiting step in miRNA maturation is the transport of pre-miRNAs from the nucleus to the cytoplasm where they are further processed to mature forms by Dicer (Lund et al., 2004; Kim, 2004). To examine nuclear export of pre-miRNAs in the DDR, we determined the distribution of nuclear and cytoplasmic pre-miRNAs in control or ATM knockdown HCT116 cells (Figure 2A), and GM0637 (ATM-proficient) and GM9607 (ATM-deficient) cells (Figure S3). The tested miRNAs include KSRP-dependent miR-21, p53/KSRP-independent miR-181a, miR-382 and miR-338. Relative levels of cytoplasmic pre-miRNAs, but not pri- or mature miRNAs, were elevated significantly after NCS treatment in the ATM-proficient cells, suggesting that nucleus-to-cytoplasm transport of pre-miRNAs was accelerated following DNA damage. As a negative control, β-actin mRNA distribution had no notable change. In contrast, relative levels of cytoplasmic pre-miRNAs remained unchanged in the ATM-silenced HCT116 cells and GM9607 cells. These results indicate that accelerated nuclear export of pre-miRNAs may involve the ATM-associated signaling pathway.
Figure 2. DNA damage induces nuclear export of pre-miRNAs in an ATM-dependent manner.
(A) DNA damage induces the translocation of pre-miRNAs into the cytoplasm. Control and ATM-silenced HCT116 cells were treated with 500 ng/ml NCS. Total RNA was purified from nucleus and cytoplasm 8h after treatment and pre-miRNA levels were quantified by quantitative RT-PCR. (B) Dicer-ablated MEFs were generated by transducing adenovirus encoding Cre-GFP into Dicerc/c MEFs. The GFP signal indicates high efficiency of viral infection. Scale bar: 200 µm. Western blot and RT-PCR confirmed Dicer knockout in MEFs. (C) DNA damage promotes the distribution of pre-miRNAs in the cytoplasm in Dicer-ablated MEFs. The experiment was performed as above in Figure 2A.
Because pre-miRNAs are subject to dynamic processing by Dicer, elevated levels of pre-miRNAs in the cytoplasm may be due to blockage of Dicer activity. To exclude this possibility, we performed similar experiments in Cre-conditional Dicer knockout MEFs in which the Dicer gene is flanked by lox-P sites (Mudhasani et al., 2008). To ablate Dicer function, Cre recombinase was transiently expressed in Dicerc/c MEFs by infecting cells with recombinant adenovirus (Ad) encoding Cre and GFP (green fluorescence protein). Infection with 200 plaque-forming units of Ad-Cre did not alter the growth of wildtype MEFs, yet resulted in a greater than 95% transduction of Cre activity as determined by GFP-positive cells and mRNA and protein levels of Dicer (Figure 2B). In the control Dicerc/c MEFs, the primary transcripts of miR-181, miR-382, miR-338 and miR-29b were unchanged after NCS treatment (Figure S4). However, we noticed that the mature forms of these miRNAs were induced after DNA damage. While mature miRNAs were at extremely low levels in the Dicer knockout MEFs, total pre-miRNAs accumulated to a much higher level due to the lack of Dicer activity. In both control and Cre-expressing Dicerc/c MEFs, relative levels of pre-miRNAs in the cytoplasm were profoundly increased after DNA damage (Figure 2C), suggesting an increase in the nuclear export activity of pre-miRNAs.
DNA Damage-induced Nucleocytoplasmic Shuttling of XPO5
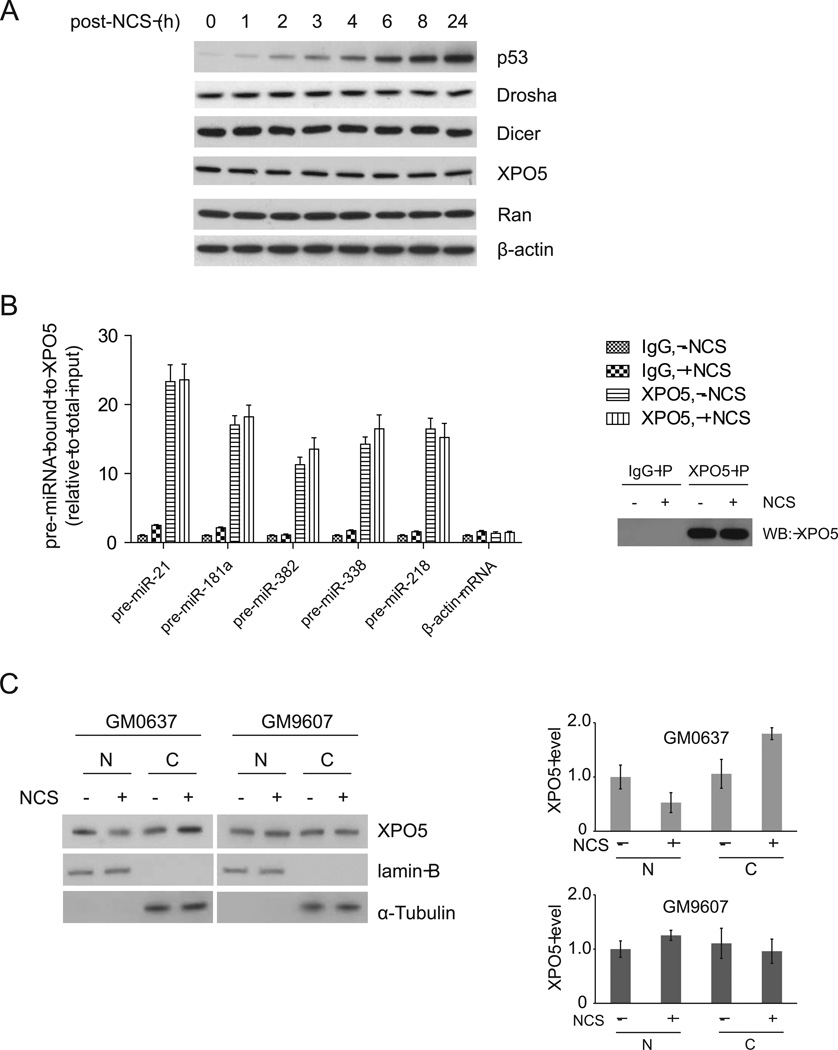
XPO5 is a RanGTP-dependent dsRNA-binding protein that mediates nuclear export of pre-miRNAs (Kim, 2004). We tested whether DNA damage affects expression of the proteins that are involved in miRNA processing and transport. In NCS-treated HCT116 cells, p53 was rapidly induced after DNA damage, indicating a functional DDR. However, total protein levels of Drosha, Dicer, XPO5 and RanGTP were not significantly changed following DNA damage (Figure 3A).
Figure 3. DNA damage enhances nucleo-cytoplasmic shuttling of XPO5.
(A) Protein levels of Drosha, Dicer, Exp5 and Ran are not changed after DNA damage. HCT116 cells were treated with NCS (500 ng/ml) and harvested at indicated time points for Western blot analyses. (B) Binding activity of XPO5 with pre-miRNAs is not increased after DNA damage. HCT116 cells were treated with NCS (500 ng/ml) for 8 h and pre-miRNA-XPO5 interaction was analyzed by RIP assay. (C) DNA damage promotes nuclear export of XPO5 in an ATM-dependent manner. XPO5 protein levels were determined by semi-quantitative immunoblotting. Error bars represent the mean ± SD. Lamin B and α-tubulin were used as nuclear and cytoplasmic markers, respectively. N: nucleus, C: cytoplasm.
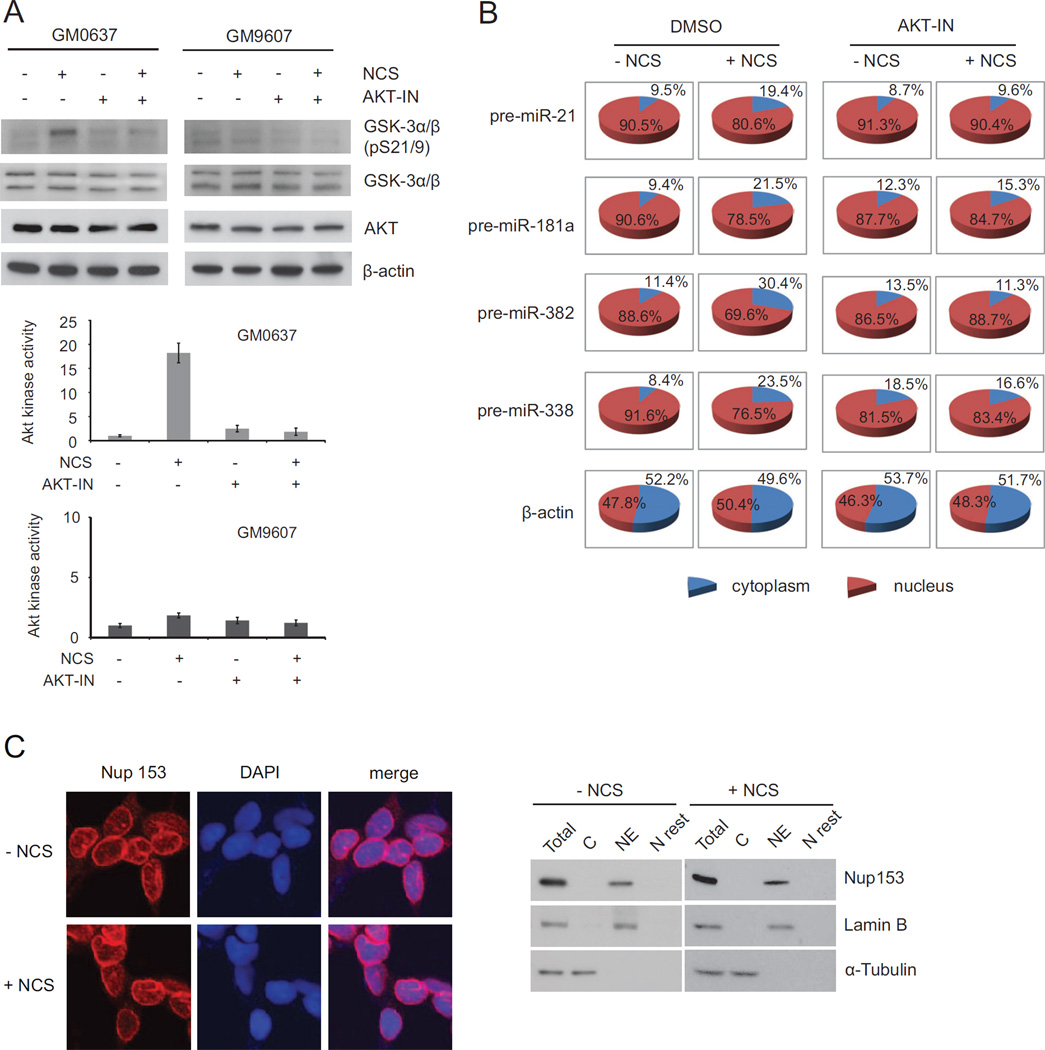
Next, we questioned how DNA damage induces nuclear export of pre-miRNAs. One possibility is that loading of pre-miRNAs to the XPO5/RanGTP complex is enhanced after DNA damage. We employed RIP (RNA Immunoprecipitation) assays to analyze the interaction of XPO5 with pre-miRNAs in cells treated with or without NCS. We measured the levels of the precursors of five DNA damage-induced miRNAs (miR-21, miR-181a, miR-382, and miR-338) and the control miR-218 in the XPO5 immunoprecipitates (Figure 3B). Although more pre-miRNAs were detected in the XPO5 complex for those DNA damage-induced miRNAs, the relative binding activity (percentage of XPO5-bound pre-miRNAs in total pre-miRNAs) was unchanged. In both NCS-treated and -untreated cells, approximately 12–23% of the total pre-miRNAs were detected in the XPO5 complex. The pre-miRNA binding activity of XPO5 was not increased after DNA damage in either the DNA damage-induced miRNAs or the control miR-218. The results suggest that accelerated nuclear export of pre-miRNAs is not due to increased binding activity of pre-miRNAs with the XPO5 transport complex. However, Drosha-mediated processing machinery is closely coupled with the XPO5-mediated transport machinery. XPO5 was detected in the Drosha complex (Figure S5A). Consistent with the previous report (Trabucchi et al., 2009), we also detected KSRP in the XPO5 complex. Knockdown of KSRP inhibited the Drosha’s processing activity for those KSRP-dependent miRNAs, but not for other DNA damage-induced miRNAs (Zhang, et al., 2011). Moreover, silencing KSRP inhibited the DNA damage-induced export of the KSRP-dependent pre-miRNAs (pre-miR-21 and pre-miR-27b), but not of other DNA damage-induced pre-miRNAs (pre-miR-382 and pre-miR-338) (Figure S5B), suggesting that induced pri-miRNA processing is a prerequisite for the induced pre-miRNA exporting activity. We next examined nuclear and cytoplasmic distributions of XPO5 proteins in cells with or without DNA damage (Figure 3C). More XPO5 proteins are transported into cytoplasm following DNA damage in the ATM-proficient GM0637 cells, but not in the ATM-deficient GM9607 cells, indicating that ATM signaling stimulates nuclear export of the pre-miRNA-loaded XPO5.
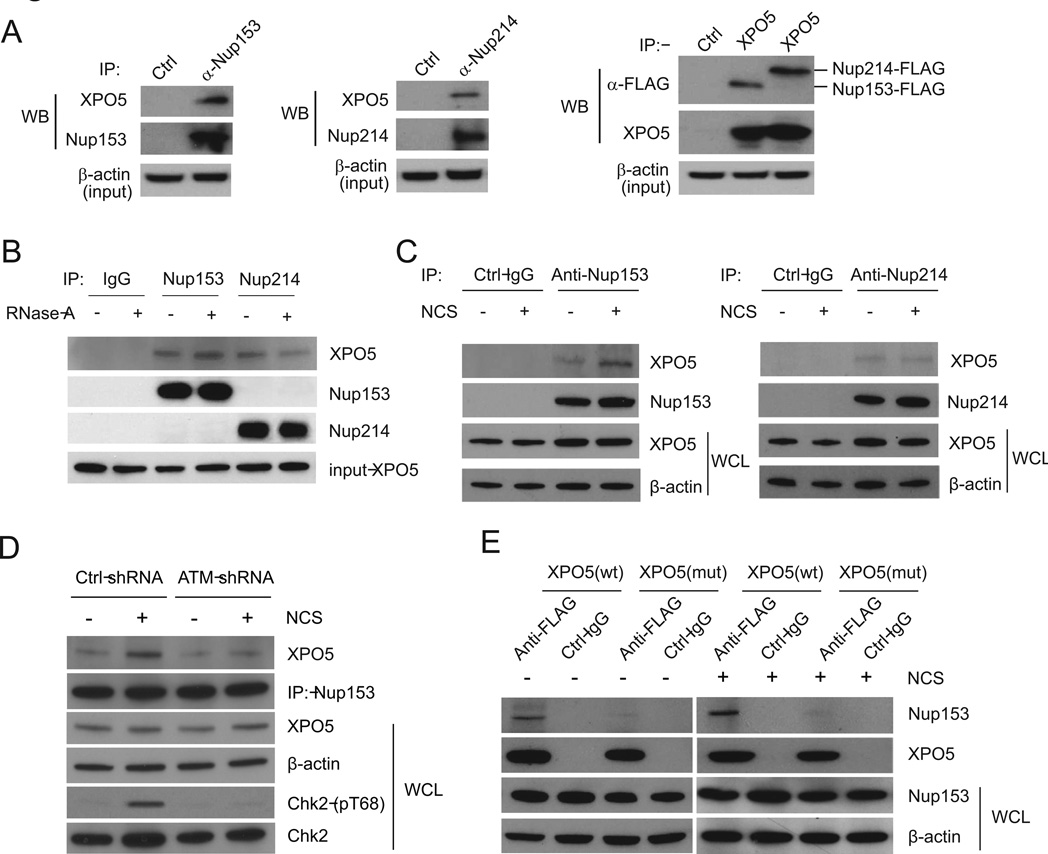
Enhanced Interaction of XPO5 with Nucleopores Following DNA Damage
Trafficking of macromolecules between the cytoplasmic and nuclear compartments in eukaryotic cells occurs through nucleopore complexes (Grunwald et al., 2011; Lim et al., 2007). Assembled from more than 30 nucleoporins, nucleopores span the nuclear envelope and form an aqueous channel with distinct cytoplasmic and nuclear faces. Whereas small molecules can diffuse freely through the pores, the translocation of larger molecules, such as pre-miRNAs, is facilitated by a family of transport receptors referred to as karyopherins. XPO5 is a member in the karyopherin β family and needs to bind nucleoporins in order to transport cargo (pre-miRNAs) from the nucleus to the cytoplasm. Consistent with previous studies showing XPO5 binds nucleoporins and other proteins (Brownawell and Macara, 2002; Chen et al., 2004), we confirmed that FLAG-XPO5 immunoprecipitates contain two nucleoporins, Nup153 and Nup214 (Figure 4A). Although XPO5 mediates pre-miRNA transport through nucleopores, the XPO5-nucleopore interaction does not require the pre-miRNA loading. In RNase A-treated cell lysates, XPO5 retained a similar binding activity with Nup153 and Nup214 (Figure 4B). We next examined XPO5-Nup153 and XPO5-Nup214 interactions in HCT116 cells treated with or without NCS (Figure 4C). We found that Nup153, but not Nup214, had an increased XPO5-binding activity after DNA damage. ATM activity appeared to be essential for the DNA damage-induced Nup153-XPO5 interaction. In the ATM-knockdown HCT116 cells and GM9607 cells, DNA damage failed to induce the interaction, which was restored by reintroduction of wildtype ATM into the cells (Figures 4D and S6).
Figure 4. Interaction between XPO5 and Nup153 is enhanced after DNA damage.
(A) XPO5 interacts with Nup153 and Nup214. The left two panels show the interaction of endogenous XPO5 with endogenous Nup153 and Nup214. The right panel shows the interaction between endogenous XPO5 and exogenous Nup153-FLAG and Nup214-FLAG. (B) The interaction between XPO5 and Nup153 or Nup214 is insensitive to RNase A treatment. Lysates from HCT116 cells were treated with RNase A prior to immunoprecipitation. (C) DNA damage induces the interaction of XPO5 with Nup153, but not with Nup214. (D) ATM is a key regulator for the XPO5-Nup153 interaction. Control or ATM-knockdown HCT116 cells were treated with NCS (500 ng/ml) for 8 h and the XPO5-Nup153 interaction was examined by immunoprecipitation-Western blotting. WCL: whole cell lysate. (E) Mutant XPO5 deficient in miRNA transport fails to bind Nup153. HCT116 cells expressing wildtype or mutant XPO5 were treated with or without NCS (500 ng/ml). The cells were harvested 4 h post-treatment and subject to immunoprecipitation-Western blotting.
Recently, a subset of human colorectal tumors were found to harbor a mutant form of XPO5 (Melo et al., 2010). This mutant XPO5 carries mutated amino acids 1181–1192 and has a truncated C-terminus (13 amino acids of 1193–1205). The XPO5 genetic defect was shown to trap pre-miRNAs in the nucleus and thus reduce miRNA processing. In the protein-binding experiments, we found that the XPO5 mutant was incapable of binding Nup153 (Figure 4E).
This finding suggests that Nup153-XPO5 interaction may facilitate nuclear export of pre-miRNA-XPO5 complex. We investigated the effect of mutant XPO5 on the pre-miRNA export and DDR using DLD-1 cells expressing mutant XPO5. Consistent with the previous report (Melo et al., 2010), we found that truncated XPO5 had a much higher expression level than wildtype XPO5 did in DLD-1 cells although the cells have one wildtype allele and one mutant allele in genome (Figure S7A). Significant induction of pre-miRNA nuclear export was observed when wildtype XPO5 was reintroduced to the cells, but not in control DLD-1 cells (Figure S7B). Wildtype XPO5 expression rebuilt up the functionality of DNA damage checkpoints (intra-S and G2/M checkpoints) that are defective in DLD-1 cells. In comparison with wildtype XPO5-expressing cells, control DLD-1 cells have less significant reduction on S-phase DNA synthesis (Figure S7C) and G2/M arrest (Figure S7D). Homologous recombination (HR) is a major error-free DNA repair for double-strand DNA breaks. We found that knockdown of XPO5 remarkably suppressed HR activity, which is rescued by the shRNA-resistance wildtype XPO5, but not mutant XPO5 (Figure S7E).
ATM-activated AKT phosphorylates Nup153
Nuclear export of pre-miRNAs and interaction of XPO5 and Nup-153 are both induced after DNA damage in an ATM-dependent manner. We postulated that ATM possibly phosphorylates and activates XPO5 or Nup153. Based on the consensus sequence for ATM phosphorylation (Matsuoka et al., 2007), Nup153 contains two putative ATM phosphorylation sites (Figure S8A, B), whereas XPO5 does not carry ATM phosphorylation sites. Using antibodies specifically against ATM/ATR-phosphorylated pS/TQ sites, we found that neither Nup153 nor XPO5 was phosphorylated by ATM or ATR in the NCS-treated human U2OS cells (data not shown).
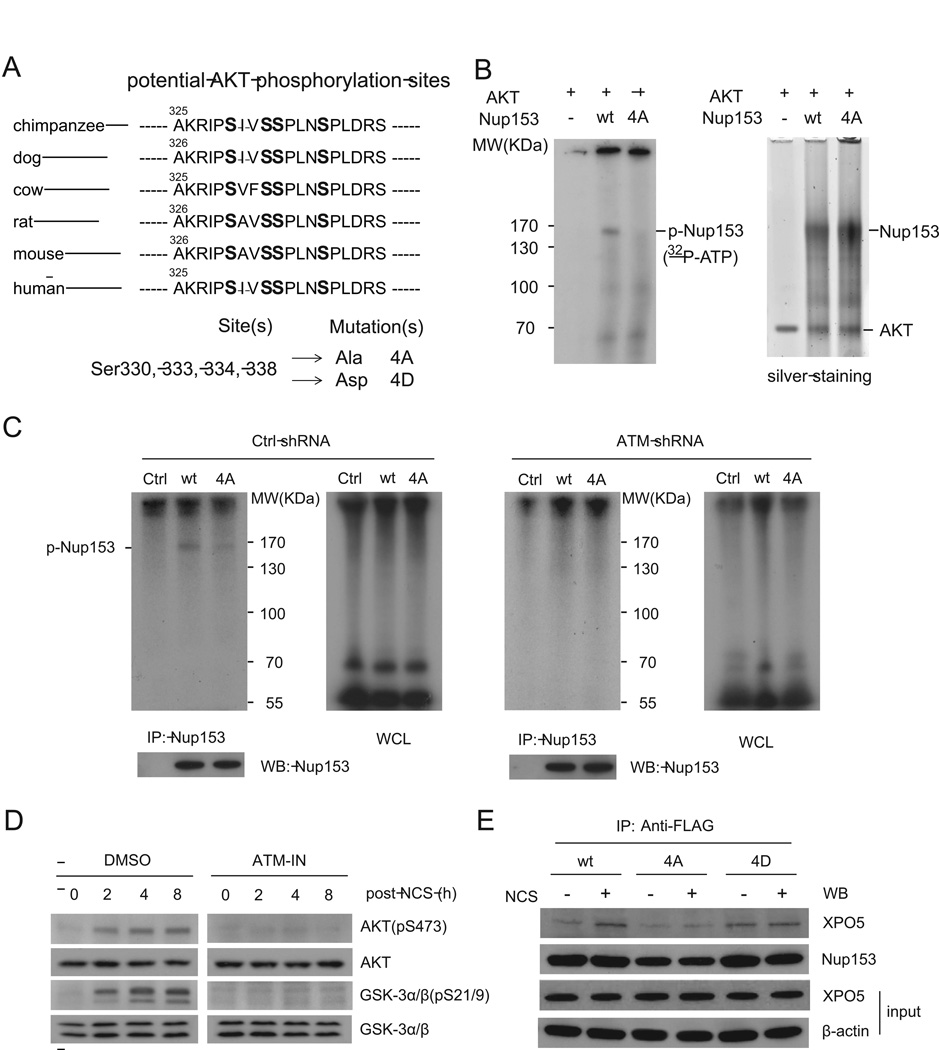
Incubating wildtype or mutant FLAG-Nup153 with immunopurified ATM kinase in vitro did not result in phosphorylation of Nup153 while the positive control, p53, was clearly phosphorylated by the ATM kinase (Figure S8C). Mutating either or both of the two putative ATM sites did not abrogate the DNA damage-induced Nup153-XPO5 interaction (Figure S8D). However, recent phosphoproteomics and kinase-motif analyses revealed a cluster of four phosphorylation sites on the 325-AKRIPSIVSSPLNSPLDRS-343 sequence of Nup153 and their phosphorylation was predicted to be mediated by AKT (Dephoure et al., 2008; Olsen et al., 2010). Protein sequence comparisons showed that these phosphorylation sites are conserved in mammals, indicating a degree of functional importance (Figure 5A).
Figure 5. AKT phosphorylates Nup153 in vitro and in vivo.
(A) Potential AKT phosphorylation sites on Nup153 are conserved in mammals. (B) Nup153 is directly phosphorylated by the AKT kinase in vitro. Immunopurified wildtype or phospho-deficient mutant Nup153 were incubated with purified AKT proteins in a kinase reaction buffer containing 32P-ATP. Immunoprecipitated Nup153 proteins were visualized by silver staining. (C) Nup153 is phosphorylated in vivo in an ATM-dependent manner. Control or ATM knockdown HCT116 cells were transfected with control vector or vector expressing FLAG-tagged wildtype or mutant form of Nup153. Cells were labeled with 32P-orthophate 24 h after transfection, and Nup153 protein was immunoprecipitated and analyzed by SDS-PAGE and Western blotting. WCL: whole cell lysate. (D) AKT is phosphorylated and activated by ATM. HCT116 cells were treated with NCS in the presence or absence of ATM inhibitor, harvested at indicated time points and analyzed by Western blotting. (E) AKT phosphorylation is critical for Nup153 to interact with XPO5. HCT116 cells overexpressing wildtype, 4A or 4D mutant forms of Nup153 were treated with or without NCS (500 ng/ml). Cell lysates were immunoprecipitated with anti-FLAG antibody. XPO5 in the immunoprecipitates was detected by Western blotting.
To study the functional role of AKT phosphorylation, we generated two mutant forms of Nup153. Serine-to-alanine mutations (4A mutant) prevent phosphorylation at the mutated site, while serine-to-aspartic acid mutations (4D) mimic constitutive phosphorylation because aspartic acid carries negative charges and structurally resembles phosphorylated serine. We first determined that wildtype Nup153, but not 4A mutant, was phosphorylated by the AKT kinase in vitro (Figure 5B). The AKT phosphorylation of Nup153 was dependent on the functionality of ATM. To verify whether ATM indirectly promotes phosphorylation of Nup153, we performed in vivo phospho-labeling assays in control and ATM-knockdown HCT116 cells (Figure 5C) and in GM0637 and GM9607 cells (Figure S8E). Radioactive orthophosphate generates DNA damage in cells without the need for additional DNA-damaging agents. In the ATM-proficient GM0637 and HCT116 cells, immunoprecipitated wildtype Nup153 was shown to be phosphorylated, while the 4A mutation resulted in a dramatically reduced phosphorylation of Nup153. Only minimal phosphorylation of Nup153 was detected in GM9607 cells and the ATM-silenced HCT116 cells due to the deficiency of ATM.
AKT has been shown to be activated in response to double-stranded DNA break damage by Ser473 phosphorylation, which is dependent on MRE11-ATM DNA damage signaling (Fraser et al., 2011). Consistent with this study, we observed that AKT was phosphorylated at Ser473 after NCS treatment, leading to an increased kinase activity of AKT (Figure 5D, left panel). Inhibiting ATM abolished the DNA damage-induced AKT phosphorylation (Figure 5D, right panel). Increased XPO5-Nup153 interaction was dependent on the AKT phosphorylation of Nup153 as the 4A mutant of Nup153 only retained a weak binding activity with XPO5 and this binding was not enhanced after DNA damage (Figure 5E). In contrast, the phospho-mimic 4D mutant exhibited much stronger binding with XPO5 compared to its wildtype form even in the absence of DNA damage, suggesting AKT phosphorylation is a functional switch for Nup153 to efficiently bind XPO5 in the DDR.
Accelerated Nuclear Export of Pre-miRNAs after DNA damage is Dependent on AKT
To determine whether the AKT kinase mediates the DNA damage-induced transport of pre-miRNAs, we used a specific AKT inhibitor to shut off AKT activity in cells. While AKT activity is extremely low in the ATM-deficient GM9607 cells, AKT was completely inhibited in the presence of 10 µM AKT inhibitor even in the ATM-proficient GM0637 cells treated with NCS (Figure 6A). We examined nuclear export of pre-miRNAs in the NCS-treated GM0637 cells with or without the AKT inhibitor. As expected, the nuclear export of pre-miRNAs had a robust increase after DNA damage as indicated by increased cytoplasmic pre-miRNA levels.
Figure 6. AKT phosphorylation facilitates Nup153 in nuclear export of pre-miRNAs.
(A) AKT activity is inhibited by an AKT inhibitor. GM0637 and GM9607 cells were pretreated with 10 µM AKT inhibitor 2 h prior to NCS treatment. Cell lysates were harvested 8 h after NCS treatment and protein levels were analyzed by Western blotting and quantified by phosphoimager. (B) DNA damage-induced nuclear export of pre-miRNAs is dependent on AKT. Levels of pre-miRNAs were measured by quantitative RT-PCR in GM0637 cells treated with or without NCS. (C) Localization of Nup153 on the nuclear envelope is unchanged after DNA damage. Cells were treated with NCS for 4 h, fixed and immunostained with anti-Nup153 antibodies. DAPI staining indicates the cell nucleus. Western blotting confirmed the localization of Nup153 on nuclear envelope. C: cytoplasm, NE: nuclear envelope, N rest: nuclear extracts without nuclear envelope.
Inhibiting AKT markedly impaired the DNA damage induction of pre-miRNA transport (Figure 6B). These results were also confirmed in AKT-knockdown HCT116 cells (Figure S9A, B). As a key component in nucleopores, Nup153 is localized on the nuclear membrane. It is essential for nucleopore architecture and macromolecular transport (Walther et al., 2001). Whereas DNA damage appears to facilitate Nup153 in pre-miRNA transport, nuclear localization and protein levels of Nup153 were unchanged after DNA damage. In both control and NCS-treated cells, Nup153 was primarily localized on the nuclear membrane (Figures 6C and S9C).
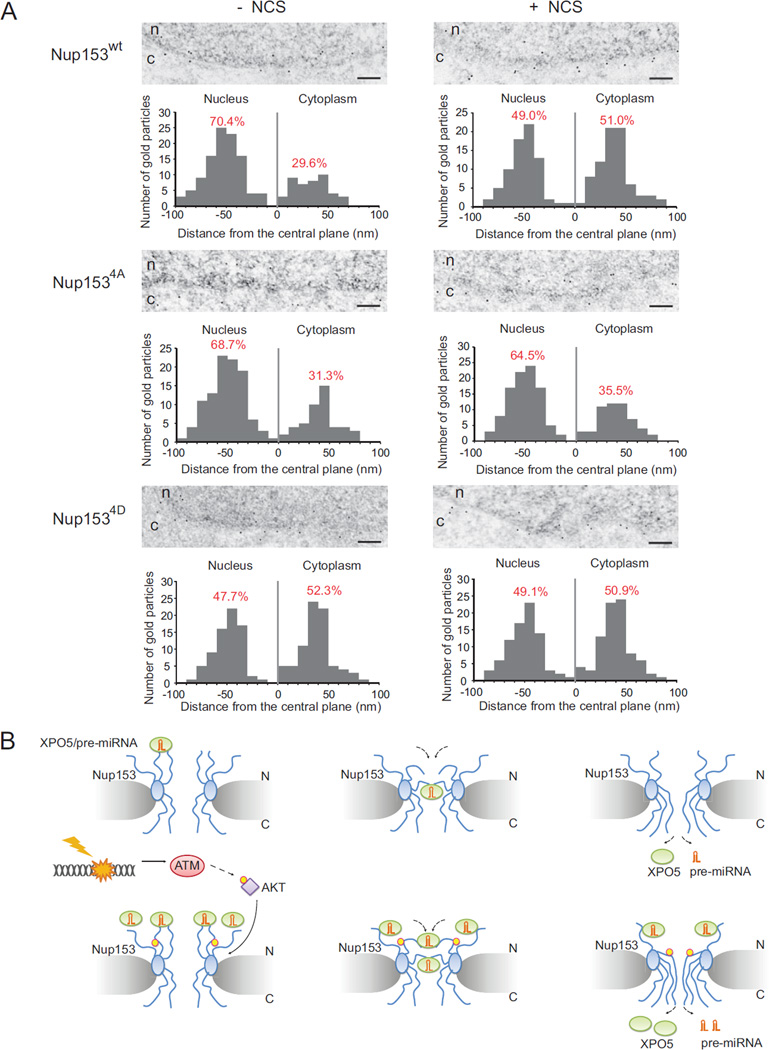
The nucleopore complex (NPC) displays an eight-fold symmetry of about 30 different nucleoporins (Grunwald et al., 2011; Lim et al., 2007). It consists of a central channel that allows the egress of cargo, a core scaffold that supports the central channel, and the cytoplasmic and nuclear ring moieties. Eight cytoplasmic filaments decorate the cytoplasmic ring moiety, whereas the nuclear ring moiety is capped by the nuclear basket. Spanning the entire nuclear basket, Nup153 has its N-terminal domain exposed at the nuclear ring of the NPC. However, the C-terminus of Nup153 is not restricted to one particular subdomain of the NPC, but rather appears to be highly flexible and mobile across nucleopores (Fahrenkrog et al., 2002). To better understand the Nup153 topology within NPC, we introduced wildtype or mutant Nup153 (4A & 4D mutants) with a C-terminal FLAG epitope into human HCT116 cells. The HCT116 cells express wildtype XPO5 and Nup153 and have functional DDR and miRNA processing machineries (data not shown). To determine the localization of the C-terminal domain of Nup153 within the 3-D architecture of the NPC, we used anti-FLAG antibodies that had been directly conjugated to 10-nm colloidal gold for immuno-electronic microscopy (EM) in the cell. As illustrated in thin-section immune-EM, a majority of Nup153 proteins had their C-termini close to the nuclear membrane in the normal unstressed state (top panels in Figure 7A and Figure S10). Quantification of the gold particle distribution with respect to the central plane of the NPC revealed that 70.4% of the gold particles were at distances between −20 and −80 nm from the central line of the nuclear membrane, with a peak between −40 and −60 nm. However, in the NCS-treated cells, a significantly higher percentage of the gold particles (51.0 %) were at the cytoplasmic face, while only 49.0 % of the gold particles were located at the nuclear face. Because the C-terminus of Nup153 dynamically binds and transports cargo through the central channel, these results suggest that pre-miRNA transport activity is at relatively low levels in unstressed cells, but increases to much higher levels by promoting the Nup153-XPO5 interaction and the “flip-out” activity of Nup153 in pre-miRNA transport. AKT phosphorylation appears to be a key switch in regulating Nup153 dynamics in the DDR. The 4A mutant had a similar pattern with its wildtype counterpart without DNA damage (middle panels in Figure 7A and Figure S10). However, 4A mutant failed to increase its mobilization to the cytoplasmic side, suggesting that the phosphorylation mutations abolish the DNA damage-induction of the Nup153 activity in pre-miRNA transport. In contrast, the 4D mutant behaved similarly to the phosphorylated form of Nup153 even in unstressed cells, and DNA damage treatment had minimal effect on its mobilization (bottom panels in Figure 7A and Figure S10). These results suggest that AKT phosphorylation plays an important role in regulating the Nup153-XPO5 interaction and the nuclear export of pre-miRNAs (Figure 7B).
Figure 7. DNA damage induces topological change of Nup153 in the nucleopore.
(A) AKT phosphorylation induces Nup153 structural change in the nucleopore. The C-terminus of wildtype Nup153 is primarily localized within the nuclear periphery of the NPC under the normal state (n=149) (upper-left EM image). Nup153 is increasingly translocalized to the cytoplasmic ring moiety after DNA damage (n=151) (upper-right EM image). The distribution of the 4A mutant of Nup153 has no significant change with (n=152) or without (n=150) DNA damage treatment (middle EM images), while the 4D mutant phenocopies the phosphorylated form of Nup153 (bottom EM images). These observations are summarized in the histograms, where 0 nm in the horizontal axis corresponds to the central plane of the NPC, and the nuclear moiety and cytoplasmic moiety in the nucleopore are located at −100 nm to 0 nm and 0 nm to 100 nm, respectively. Scale bar: 500 nm. (B) Nuclear export of pre-miRNAs is stimulated after DNA damage. Pre-miRNA is transported from nuclear to cytoplasm by XPO5. Upon DNA damage, AKT is activated by ATM, leading to the phosphorylation of Nup153 by AKT. Nuclear export of pre-miRNAs is accelerated by enhanced interaction between XPO5 and phosphorylated Nup153.
To examine the effect of Nup153 phosphorylation mutations on pre-miRNA transport, we knocked down endogenous Nup153 and re-introduced shRNA-resistant wildtype or mutant Nup153 into HCT116 cells. While wildtype Nup153 nicely rescued the nuclear export of pre-miRNAs (pre-miR-21 and pre-miR-382), phospho-deficient 4A mutant failed to restore the DNA damage-induced nuclear export (Figure S11A). In contrast, the expression of 4D mutant increased the levels of nuclear pre-miRNAs even without DNA damage. Knockdown of ATM dramatically inhibited the export activity of wildtype Nup153, but had no effect on both 4A and 4D mutants, suggesting that AKT phosphorylation is essential for the Nup153’s function on the pre-miRNA transport (Figure S11B).
DISCUSSION
DNA damage triggers a wide range of cellular responses, including altered gene expression, activation of cell cycle checkpoints and elevated DNA repair activity, to maintain the genomic integrity. Whereas DNA damage signaling seems to be well understood, non-coding RNAs are emerging as novel players in the DDR. In particular, a number of miRNAs have been shown to be part of the DDR when their mRNA targets were identified (Zhang and Lu, 2011). The miRNA-processing factors Dicer, DGCR8, Drosha, and Ago2 are essential for viability in mice. Conditional knockout of Dicer in mouse fibroblasts leads to the loss of mature miRNAs, as well as increased levels of DNA damage and premature senescence, indicating that miRNAs are a critical component of the DDR (Mudhasani et al., 2008; Bernstein et al., 2003; Wang et al., 2007; Morita et al., 2007).
Recent studies have demonstrated a wide-spectrum of alterations in miRNA expression in cells with DNA damage (Han et al., 2012). The ATM-initiated signaling pathway appears to be a major player in the process. We previously identified an ATM-dependent miRNA expression signature in mouse embryonic fibroblasts. In the current study, we verified that a significant number of human miRNAs are markedly induced in an ATM-dependent manner. The ATM dependent miRNA signatures are virtually identical in human and mouse cells except for those unique miRNAs in humans. Given a complex network of cell activities controlled by ATM, it is assumed that multiple mechanisms may be responsible for the DNA damage-induced miRNA expression. Among the ATM-dependent miRNAs, several miRNAs are modulated by the tumor suppressor p53, including miR-34, miR-16 and miR-145 (He et al., 2007; Suzuki et al., 2009). The miR-34 family was first reported to be a direct p53 transcriptional target. In addition to transcriptional regulation, p53 also promotes pri-miRNA processing. A direct interaction between p53 and DDX5 facilitates pri-miRNA processing in the Drosha complex (Suzuki et al., 2009). One of the ATM phosphorylation targets is KSRP, a RNA-binding protein that is involved in RNA splicing, localization and degradation (Chou et al., 2006). KSRP promotes maturation of a class of miRNAs as a component of both the Drosha and Dicer complexes (Trabucchi et al., 2009). Following their study, we observed that the KSRP-dependent miRNAs were induced in an ATM-dependent manner upon DNA damage (Zhang et al., 2011). ATM phosphorylation of KSRP significantly enhances recruitment of KSRP-associated pri-miRNAs to the Drosha complex and increases their processing. These findings revealed functional connections between the DDR and miRNA maturation.
Whereas Drosha and Dicer processors are the two major biochemical hubs for miRNA maturation, XPO5-mediated nuclear export of pre-miRNAs is another key step. XPO5 is an evolutionarily conserved nuclear export factor in charge of nuclear export for small noncoding RNAs such as pre-miRNAs, viral mini-helix RNAs and a subset of tRNAs in mammalian cells (Okada et al., 2009). Knockdown of XPO5 dramatically depleted mature miRNAs in cultured human cells (Lund et al., 2004; Yi et al., 2003). We found that wildtype XPO5 has a similar binding activity with pre-miRNAs regardless of DNA damage. However, XPO5-Nup153 interaction is significantly enhanced after DNA damage, suggesting that nucleopores may be a part of regulatory mechanism. This hypothesis is also supported by our findings regarding an inactivating mutant form of XPO5, which the Esteller group identified in a subset of human colorectal tumors with microsatellite instability (Melo et al., 2010). The mutant XPO5 lacks a C-terminal region and fails to form the pre-miRNA/XPO5/Ran-GTP complex, and thus traps pre-miRNAs in the nucleus. We found this mutant XPO5 also failed to bind Nup153, indicating that both miRNAs and other XPO5-exported RNA species may be affected by this genetic defect. A number of domains have been identified on Nup153, including a nuclear localization signal, a NPC-targeting sequence, an RNA-binding domain, a central zing-finger domain that interacts with RanGDP, and the C-terminal FG (phenylalanine-glycine) repeats. Previous studies demonstrated that FG repeats are essential for mediating the binding of Nup153 with transport receptors. The results in the current study show that AKT phosphorylation enhances the Nup153-XPO5 interaction, suggesting that those phosphorylation sites, although located next to the central part of Nup153, probably have profound influence on the physical state of Nup153 molecules, resulting in an increased binding with XPO5. AKT phosphorylation appears to contribute to the function of Nup153 in regulating cell activities following DNA damage. Knockdown of Nup153 in human HCT116 cells led to diminished cell proliferation and survival in response to DNA damage, which is rescued by ectopic expression of shRNA-resistant wildtype Nup153, but not phospho-deficient 4A mutant (Figure S12A, B, C). Cells stably expressing 4A mutant had weaker DNA damage checkpoint, indicated by higher S-phase DNA synthesis after DNA damage in comparison with wildtype Nup153-expressing cells (Figure S12D). Homologous recombination repair activity was also markedly inhibited by Nup153 knockdown, which was rescued by the 4D mutant, but not the 4A mutant (Figure S12E). These data suggest an important role of AKT/Nup153-mediated miRNA export in the DDR.
High-resolution structure of the pre-miRNA nuclear export machinery and experimental evidence revealed that RNA recognition by XPO5/RanGTP occurs in a sequence-independent manner, suggesting that XPO5 recognizes a variety of pre-miRNAs (Lund et al., 2004; Okada et al., 2009). Although as many as 331 human miRNAs (about a quarter of the total tested) are significantly induced after DNA damage, a large number of miRNAs are either minimally induced or repressed in the NCS-treated cells. It is interesting to know whether there is sequence specificity for the DNA damage-induced nuclear export of miRNAs. While XPO5 does not selectively bind specific pre-miRNAs, Nup153-XPO5 interaction is also independent of the pre-miRNAs. However, we showed that Drosha-mediated miRNA processing is closely coupled with the XPO5-mediated transport because XPO5 was detected in the Drosha complex. We postulate that miRNA specificity is attributed to the processing process whose induction is a prerequisite for the induced nuclear export after DNA damage, and that enhanced interaction between XPO5 and nucleopores facilitates nuclear export in the DDR. Differential readout of miRNAs after DNA damage could be a result of multiple layers of regulation, including: 1) differential processing activity by Drosha and Dicer, 2) transcriptional regulation, 3) pre-miRNA degradation pathway (an alternative destination for pre-miRNAs is degradation instead of further maturation) (Suzuki et al., 2011), 4) stability of mature miRNAs, 5) nuclear import of pre-miRNAs and mature miRNAs (CRM1 was recently shown to regulate nuclear import of miRNAs) (Castanotto et al., 2009). Each miRNA likely has its own regulatory scenario in the DDR.
In summary, we demonstrate that the ATM-AKT signaling is a key switch in accelerating nuclear export of pre-miRNAs. Our results uncover a functional interaction between the miRNA transport complex and nucleopores in the DDR. The findings in the current study provide valuable insights into the spatiotemporal regulation of miRNA biogenesis in response to DNA damage.
EXPERIMENTAL PROCEDURES
Cell Lines and Tissue Culture
The U2OS cell line is a human osteosarcoma line that was obtained from the American Type Culture Collection. GM0637 (ATM-proficient) and GM9607 (ATM-deficient) are SV40-transformed human fibroblast lines that were obtained from Coriell Cell Repositories. The HCT116 cell line was obtained from Dr. Vogelstein at Johns Hopkins University. Primary Dicerc/c MEFs were harvested and cultured as previously described (Mudhasani et al., 2008).
DNA Damaging Agents, ATM and AKT Inhibitors, RNase A and Ad-CRE-GFP
Cells were treated with 500 ng/ml neocarzinostatin (NCS, #N9162, Sigma-Aldrich) and harvested at indicated time points after treatment for RNA and protein analyses. In the assays using inhibitor, cells were pretreated with 10 µM ATM kinase inhibitor (#118500, Calbiochem) or AKT inhibitor (#124015, Calbiochem) 2 h prior to DNA damage treatment. In immunoprecipitation-Western blot assay, cell lysates were incubated at room temperature with RNase A (10 µg/ml, Ambion) for 30 min before immunoprecipitation. To generate Dicer-ablated MEFs, Dicerc/c MEFs were infected with 200 pfu of recombinant Ad-Cre-GFP adenovirus (#1700, Vector Biolabs). Adenovirus Ad-GFP (200 pfu, #1060, Vector Biolabs) was used as a control to infect Dicerc/c MEFs.
RNA Immunoprecipitation (RIP) Assay
RIP assay was performed as described previously (Zhang et al., 2011). Briefly, cells were crosslinked for 20 min with 1% formaldehyde, and cell pellets were resuspended in buffer B (1% SDS, 10 mM EDTA, 50 mM Tris-HCl [pH 8.1], 1x protease inhibitor, 50 U/ml RNase inhibitor) and disrupted by sonication. The lysates were cleared and subjected to immunoprecipitation with anti-XPO5 or control anti-IgG antibodies, followed by stringent washing, elution and reversal of crosslinking. The RNA was resuspended in TE buffer containing RNase inhibitor and incubated with DNase I to remove any remaining DNA. After extraction with phenol:chloroform:isoamyl alcohol (25:24:1), RNA was precipitated with ethanol, dissolved in DEPC-treated water, and used for cDNA synthesis reaction. Quantitative PCR reactions were then preformed on real-time PCR machine (Realplex2, Eppendorf).
Immunogold Electron Microscopy
HCT116 cells expressing wildtype and mutant Nup153-FLAG were grown on Millicell EZ slides (Millipore) and treated with NCS or DMSO alone for 4 h and then fixed with a solution containing 0.1% glutaraldehyde and 2% paraformaldehyde in PBS buffer (pH 7.3) for 1 h. After fixation, the samples were washed and treated with 0.1 M sodium borohydride for 15 min, permeabilized with 0.2% Triton X-100 for 15 min, and blocked with 2% BSA for 15 min. The samples were labeled with anti-FLAG antibody for 4 h, washed six times with PBS and incubated with 10 nm gold-conjugated secondary antibody (British Biocell) overnight at 4°C. The samples were fixed in 2% glutaraldehyde in PBS for 10 min and then were dehydrated in increasing concentrations of ethanol, infiltrated, and embedded in Spurr’s low viscosity medium. They were polymerized in a 70 °C oven for 2 days. Ultra-thin sections were processed in a Leica Ultracut microtome (Leica), stained with uranyl acetate and lead citrate in a Leica EM Stainer, and examined in a JEM-1010 transmission electron microscope (JEOL, USA) at an accelerating voltage of 80 KV. Electron micrographs were obtained using AMT Imaging System (Advanced Microscopy Techniques).
Statistical Analysis
Statistical differences were determined by analysis of variance using one-way ANOVA and Turkey’s Multiple Comparison Test on GraphPad Prism 5 software.
Supplementary Material
HIGHLIGHTS.
Pre-miRNA nuclear export is accelerated after DNA damage in an ATM-dependent manner.
Interaction between Nup 153 and XPO5 is enhanced after DNA damage.
ATM-activated AKT kinase phosphorylates Nup 153.
Nup 153 phosphorylation promotes nuclear export of pre-miRNAs.
ACKNOWLEDGEMENTS
We thank T. Michigami, M. Esteller, and M. Kastan for providing us with Nup214 expression vectors, XPO5 constructs and ATM expression vectors, respectively. We also thank K. Dunner, L. Donehower and G. Calin for their experimental assistance and intellectual contributions to various portions of this manuscript. This work is supported by grants from the National Institutes of Health (CA136549 to X.L. and CA016672 to L.M.E.), the American Cancer Society (119135-RSG-10-185-01-TBE to X.L.), the University of Texas Stars Plus award (X.L.), and the William C. Liedtke, Jr., Chair in Cancer Research (L.M.E.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein E, Kim SY, Carmell MA, Murchison EP, Alcorn H, Li MZ, Mills AA, Elledge SJ, Anderson KV, Hannon GJ. Dicer is essential for mouse development. Nat. Genet. 2003;35:215–217. doi: 10.1038/ng1253. [DOI] [PubMed] [Google Scholar]
- Bohnsack MT, Czaplinski K, Gorlich D. Exportin 5 is a RanGTP-dependent dsRNA-binding protein that mediates nuclear export of pre-miRNAs. RNA. 2004;10:185–191. doi: 10.1261/rna.5167604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brownawell AM, Macara IG. Exportin-5, a novel karyopherin, mediates nuclear export of double-stranded RNA binding proteins. J. Cell Biol. 2002;156:53–64. doi: 10.1083/jcb.200110082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castanotto D, Lingeman R, Riggs AD, Rossi JJ. CRM1 mediates nuclear-cytoplasmic shuttling of mature microRNAs. Proc. Natl. Acad. Sci. U. S. A. 2009;106:21655–21659. doi: 10.1073/pnas.0912384106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T, Brownawell AM, Macara IG. Nucleocytoplasmic shuttling of JAZ, a new cargo protein for exportin-5. Mol. Cell Biol. 2004;24:6608–6619. doi: 10.1128/MCB.24.15.6608-6619.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou CF, Mulky A, Maitra S, Lin WJ, Gherzi R, Kappes J, Chen CY. Tethering KSRP, a decay-promoting AU-rich element-binding protein, to mRNAs elicits mRNA decay. Mol. Cell Biol. 2006;26:3695–3706. doi: 10.1128/MCB.26.10.3695-3706.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciccia A, Elledge SJ. The DNA damage response: making it safe to play with knives. Mol. Cell. 2010;40:179–204. doi: 10.1016/j.molcel.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimmino A, Calin GA, Fabbri M, lorio MV, Ferracin M, Shimizu M, Wojcik SE, Ageilan RI, Zupo S, Dono M, Rassenti L, Alder H, Volinia S, Liu CG, Kipps TJ, Negrini M, Croce CM. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc. Natl. Acad. Sci. U. S. A. 2005;102:13944–13949. doi: 10.1073/pnas.0506654102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dephoure N, Zhou C, Villen J, Beausoleil SA, Bakalarski CE, Elledge SJ, Gygi SP. A quantitative atlas of mitotic phosphorylation. Proc. Natl. Acad. Sci. U. S. A. 2008;105:10762–10767. doi: 10.1073/pnas.0805139105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diederichs S, Haber DA. Dual role for argonautes in microRNA processing and posttranscriptional regulation of microRNA expression. Cell. 2007;131:1097–1108. doi: 10.1016/j.cell.2007.10.032. [DOI] [PubMed] [Google Scholar]
- Fahrenkrog B, Maco B, Fager AM, Koser J, Sauder U, Ullman KS, Aebi U. Domain-specific antibodies reveal multiple-site topology of Nup153 within the nuclear pore complex. J. Struct. Biol. 2002;140:254–267. doi: 10.1016/s1047-8477(02)00524-5. [DOI] [PubMed] [Google Scholar]
- Fraser M, Harding SM, Zhao H, Coackley C, Durocher D, Bristow RG. MRE11 promotes AKT phosphorylation in direct response to DNA double-strand breaks. Cell Cycle. 2011;10:2218–2232. doi: 10.4161/cc.10.13.16305. [DOI] [PubMed] [Google Scholar]
- Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol. Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunwald D, Singer RH, Rout M. Nuclear export dynamics of RNA-protein complexes. Nature. 2011;475:333–341. doi: 10.1038/nature10318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han C, Wan G, Langley RR, Zhang X, Lu X. Crosstalk between the DNA damage response pathway and microRNAs. Cell Mol. Life Sci. 2012 doi: 10.1007/s00018-012-0959-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L, He X, Lim LP, de SE, Xuan Z, Liang Y, Xue W, Zender L, Magnus J, Ridzon D, Jackson AL, Linsley PS, Chen C, Lowe SW, Cleary MA, Hannon GJ. A microRNA component of the p53 tumour suppressor network. Nature. 2007;447:1130–1134. doi: 10.1038/nature05939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu H, Du L, Nagabayashi G, Seeger RC, Gatti RA. ATM is down-regulated by N-Myc-regulated microRNA-421. Proc. Natl. Acad. Sci. U. S. A. 2010;107:1506–1511. doi: 10.1073/pnas.0907763107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Q, Hao X, Meng Y, Zhang M, Desano J, Fan D, Xu L. Restoration of tumor suppressor miR-34 inhibits human p53-mutant gastric cancer tumorspheres. BCM Cancer. 2008;8:266. doi: 10.1186/1471-2407-8-266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim VN. MicroRNA precursors in motion: exportin-5 mediates their nuclear export. Trends Cell Biol. 2004;14:156–159. doi: 10.1016/j.tcb.2004.02.006. [DOI] [PubMed] [Google Scholar]
- Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Radmark O, Kim S, Kim VN. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- Lim RY, Fahrenkrog B, Koser J, Schwarz-Herion K, Deng J, Aebi U. Nanomechanical basis of selective gating by the nuclear pore complex. Science. 2007;318:640–643. doi: 10.1126/science.1145980. [DOI] [PubMed] [Google Scholar]
- Lund E, Guttinger S, Calado A, Dahlberg JE, Kutay U. Nuclear export of microRNA precursors. Science. 2004;303:95–98. doi: 10.1126/science.1090599. [DOI] [PubMed] [Google Scholar]
- Matsuoka S, Ballif BA, Smogorzewska A, McDonald ER, III, Hurov KE, Luo J, Bakalarski CE, Zhao Z, Solimini N, Lerenthal Y, Shiloh Y, Gygi SP, Elledge SJ. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 2007;316:1160–1166. doi: 10.1126/science.1140321. [DOI] [PubMed] [Google Scholar]
- Melo SA, Moutinho C, Ropero S, Calin GA, Rossi S, Spizzo R, Fernandez AF, Davalos V, Villanueva A, Montoya G, Yamamoto H, Schwartz S, Jr, Esteller M. A genetic defect in exportin-5 traps precursor microRNAs in the nucleus of cancer cells. Cancer Cell. 2010;18:303–315. doi: 10.1016/j.ccr.2010.09.007. [DOI] [PubMed] [Google Scholar]
- Morita S, Horii T, Kimura M, Goto Y, Ochiya T, Hatada I. One Argonaute family member, Eif2c2 (Ago2), is essential for development and appears not to be involved in DNA methylation. Genomics. 2007;89:687–696. doi: 10.1016/j.ygeno.2007.01.004. [DOI] [PubMed] [Google Scholar]
- Moskwa P, Buffa FM, Pan Y, Panchakshari R, Gottipati P, Muschel RJ, Beech J, Kulshrestha R, Abdelmohsen K, Weinstock DM, Gorospe M, Harris AL, Helleday T, Chowdhury D. miR-182-mediated downregulation of BRCA1 impacts DNA repair and sensitivity to PARP inhibitors. Mol. Cell. 2011;41:210–220. doi: 10.1016/j.molcel.2010.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mudhasani R, Zhu Z, Hutvagner G, Eischen CM, Lyle S, Hall LL, Lawrence JB, Imbalzano AN, Jones SN. Loss of miRNA biogenesis induces p19Arf-p53 signaling and senescence in primary cells. J. Cell Biol. 2008;181:1055–1063. doi: 10.1083/jcb.200802105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada C, Yamashita E, Lee SJ, Shibata S, Katahira J, Nakagawa A, Yoneda Y, Tsukihara T. A high-resolution structure of the pre-microRNA nuclear export machinery. Science. 2009;326:1275–1279. doi: 10.1126/science.1178705. [DOI] [PubMed] [Google Scholar]
- Olsen JV, Vermeulen M, Santamaria A, Kumar C, Miller ML, Jensen LJ, Gnad F, Cox J, Jensen TS, Nigg EA, Brunak S, Mann M. Quantitative phosphoproteomics reveals widespread full phosphorylation site occupancy during mitosis. Sci. Signal. 2010;3:ra3. doi: 10.1126/scisignal.2000475. [DOI] [PubMed] [Google Scholar]
- Pothof J, Verkaik NS, van IW, Wiemer EA, Ta VT, van der Horst GT, Jaspers NG, van G, Hoeijmakers JH, Persengiev SP. MicroRNA-mediated gene silencing modulates the UV-induced DNA-damage response. EMBO J. 2009;28:2090–2099. doi: 10.1038/emboj.2009.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rand TA, Petersen S, Du F, Wang X. Argonaute2 cleaves the anti-guide strand of siRNA during RISC activation. Cell. 2005;123:621–629. doi: 10.1016/j.cell.2005.10.020. [DOI] [PubMed] [Google Scholar]
- Su X, Chakravarti D, Cho MS, Liu L, Gi YJ, Lin YL, Leung ML, El-Naggar A, Creighton CJ, Suraokar MB, Wistuba I, Flores ER. TAp63 suppresses metastasis through coordinate regulation of Dicer and miRNAs. Nature. 2010;467:986–990. doi: 10.1038/nature09459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki HI, Arase M, Matsuyama H, Choi YL, Ueno T, Mano H, Sugimoto K, Miyazono K. MCPIP1 ribonuclease antagonizes dicer and terminates microRNA biogenesis through precursor microRNA degradation. Mol. Cell. 2011;44:424–436. doi: 10.1016/j.molcel.2011.09.012. [DOI] [PubMed] [Google Scholar]
- Suzuki HI, Yamagata K, Sugimoto K, Iwamoto T, Kato S, Miyazono K. Modulation of microRNA processing by p53. Nature. 2009;460:529–533. doi: 10.1038/nature08199. [DOI] [PubMed] [Google Scholar]
- Trabucchi M, Briata P, Garcia-Mayoral M, Haase AD, Filipowicz W, Ramos A, Gherzi R, Rosenfeld MG. The RNA-binding protein KSRP promotes the biogenesis of a subset of microRNAs. Nature. 2009;459:1010–1014. doi: 10.1038/nature08025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walther TC, Fornerod M, Pickersgill H, Goldberg M, Allen TD, Mattaj IW. The nucleoporin Nup153 is required for nuclear pore basket formation, nuclear pore complex anchoring and import of a subset of nuclear proteins. EMBO J. 2001;20:5703–5714. doi: 10.1093/emboj/20.20.5703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan G, Mathur R, Hu X, Zhang X, Lu X. miRNA response to DNA damage. Trends Biochem. Sci. 2011;36:478–484. doi: 10.1016/j.tibs.2011.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Medvid R, Melton C, Jaenisch R, Blelloch R. DGCR8 is essential for microRNA biogenesis and silencing of embryonic stem cell self-renewal. Nat. Genet. 2007;39:380–385. doi: 10.1038/ng1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter J, Jung S, Keller S, Gregory RI, Diederichs S. Many roads to maturity: microRNA biogenesis pathways and their regulation. Nat. Cell Biol. 2009;11:228–234. doi: 10.1038/ncb0309-228. [DOI] [PubMed] [Google Scholar]
- Yi R, Qin Y, Macara IG, Cullen BR. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 2003;17:3011–3016. doi: 10.1101/gad.1158803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Lu X. Posttranscriptional regulation of miRNAs in the DNA damage response. RNA. Biol. 2011;8 doi: 10.4161/rna.8.6.17337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Wan G, Berger FG, He X, Lu X. The ATM Kinase Induces MicroRNA Biogenesis in the DNA Damage Response. Mol. Cell. 2011;41:371–383. doi: 10.1016/j.molcel.2011.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.