Abstract
Maternal undernutrition increases the risk of perinatal complications and predisposes offspring to obesity, diabetes, and cardiovascular disease later in life. Emerging evidence suggests that changes in placental function play a role in linking altered maternal nutrition in pregnancy to the subsequent development of adult disease. The susceptibility for disease in response to an adverse intrauterine environment differs distinctly between boys and girls, with girls typically having better outcomes. Here, we tested the hypothesis that regulation of the placental transcriptome by maternal nutrient reduction (NR) is dependent on fetal sex. We used a nonhuman primate model of NR in which maternal global food intake was reduced by 30% in baboons starting at gestational day (GD) 30. At GD 165 (term = GD 183), placental genome expression profiling of 6 control (n = 3 females, 3 males) and 6 nutrient restricted (n = 3 females, 3 males) fetuses was carried out followed by bioinformatic analysis. Surprisingly, there was no coordinated placental molecular response to decreased nutrient availability when analyzing the data without accounting for fetal sex. In contrast, female placentas exhibited a highly coordinated response that included upregulation of genes in networks, pathways, and functional groups related to programmed cell death and downregulation of genes in networks, pathways, and functional groups associated with cell proliferation. These changes were not apparent in the male placentas. Our data support the concept that female placentas initiate complex adaptive responses to an adverse intrauterine environment, which may contribute to increased survival and better pregnancy outcomes in girls.
Introduction
Genetic variants and adult lifestyle factors have traditionally been regarded as the primary determinants for risk of developing metabolic and cardiovascular diseases. However, it is now established that adult disease also has fetal origins, because an adverse intrauterine environment, such as decreased nutrient availability, is strongly associated with the development of obesity, diabetes, and cardiovascular disease later in life (1–3). Limitations in fetal nutrient availability sometimes result in fetal growth restriction (FGR)7, which is associated with increased perinatal morbidity and risk of neurodevelopmental impairment. Emerging evidence suggests that changes in placental function may link altered maternal nutrition to changes in fetal growth and the development of adult disease (4–6).
Animal experiments have demonstrated a sex difference in the response to an adverse intrauterine environment and the subsequent risk to develop disease later in life (7). Clinically, girls have better outcomes than boys in pregnancies complicated by preeclampsia, preterm delivery, or FGR (8–11), and boys are overrepresented among stillborn babies (12). This difference in perinatal morbidity and mortality between girls and boys has been suggested to be due to sex-specific differences in placental function (13, 14). Indeed, the female placenta is less “efficient” than its male counterpart and girls have slower intrauterine growth and lower birth weights (15, 16). This more conservative strategy for fetal growth may explain why female placentas show more extensive and varied changes in gene and protein expression and function than male placentas in response to alterations in the maternal environment (17). However, the molecular mechanisms underlying sex dependent in utero growth strategies are largely unknown.
A subset of placental genes is differentially expressed according to fetal sex, including genes involved in immune regulation, which are expressed at higher levels in female placentas (18). Furthermore, recent reports show sex-specific responses in placental function and gene expression to various stresses and perturbations in the maternal environment. For example, Osei-Kumah et al. (19) reported that in pregnancies complicated by asthma, the expression of 65 placental genes was altered in female fetuses, whereas only 6 genes were differentially expressed in male placentas. Male placentas have a higher expression of toll-like receptor-4 (TLR4) and an exaggerated release of tumor necrosis factor-α (TNF-α) in response to endotoxin (20). In mice, females show much more extensive changes in placental gene expression than males in response to maternal diets with differing fat contents (21). To the best of our knowledge, the regulation of the placental transcriptome in response to the challenge of maternal nutrient reduction (NR) in the primate is not known.
We hypothesized that there is a difference in the expression of key genes and molecular pathways in male and female baboon placenta when nutrition is moderately reduced. To this effect, pregnant baboons were fed either a control (CTR) diet or a diet reduced in nutrients (70% of diet consumed by CTR) from gestational day (GD) 30. At GD 165 (term = GD 183), we isolated placental RNA and analyzed and compared gene expression in male and female placentas.
Methods
Animal care and maintenance.
All procedures were approved by the Texas Biomedical Research Institute and University of Texas Health Science Center Institutional Animal Care and Use Committees and conducted in Association for Assessment and Accreditation of Laboratory Animal Care-approved facilities. Animals were fed in individual feeding cages. The feeding system for controlling animal diet and details of housing and environmental enrichment have been published elsewhere (22–24). Animals were fed Purina Monkey Diet 5038, which is a complete life-cycle diet for Old World primates. Biscuits contained stabilized vitamin C as well as all other required vitamins. Its basic composition was crude protein not <15%, crude fat not <5%, crude fiber not >6%, ash not >5%, and added minerals not >3%. At the start of the feeding period, ad libitum-fed CTR baboons were given 60 biscuits in their feeding tray. At the end of the 2-h feeding period, baboons were returned to the group cage. The biscuits remaining in the tray, on the cage floor, and in the pan beneath the cage were counted. Females in the maternal NR group were fed 70% of the CTR diet eaten by contemporaneous CTR animals on a per-kilogram basis. Animal food consumption, weights, and health status were recorded daily.
Study design.
Healthy female baboons of similar body weights (10–15 kg) were initially placed into 2 group cages. Each cage contained a social group of 10–16 randomly assigned nonpregnant females with a vasectomized male. After acclimation to eating in the feeding cages (30 d), a fertile male was substituted into each breeding cage. Pregnancy was initially dated by timing of ovulation and changes in sex skin color and confirmed at GD 30 by ultrasonography. Over a period of 6 mo, 6 CTR females and 6 NR females became pregnant. Data from these 12 baboons are presented in the current study.
Morphometric data collection.
Maternal weight was collected at the beginning of the NR challenge and at the time of cesarean section. Placental and fetal weights were collected at cesarean section and weight gain and fetal/placental weights were calculated.
Cesarean sections.
Cesarean sections were performed at GD 165 as described in detail (24). In brief, baboons were premedicated with ketamine hydrochloride (10 mg/kg, i.m.) and anesthetized using isoflurane (2%). Following hysterotomy, the umbilical cord was identified and elevated to the surgical opening to enable the fetus to be exsanguinated (25). The placenta and fetus were removed, fetal and placental weights recorded, and tissue was rapidly collected. Postoperative maternal analgesia was provided [buprenorphine hydrochloride, 0.015 mg/(kg · d)] for 3 d (Buprenex Injectable, Reckitt Benckiser Health Care) (24).
Processing of placental tissue samples.
The placenta was trimmed of fetal membranes, umbilical cord and decidua, and weighed. Five to seven samples, ~1 cm × 1 cm × 0.1–0.5 cm, extending from the chorionic to the basal plate were taken in a systematic, random manner using a transparent sheet with quadratic patterns (grid system) described by Burton and Jauniaux (26). These samples were then used to prepare samples for RNA isolation and histology. For RNA isolation, small tissue pieces were rapidly dissected from the 5–7 larger tissue samples and snap-frozen for later use. For histology, to ensure randomness of tissue orientation, the samples were processed as described by Mayhew (27). Briefly, each sample was cut to produce a cube, fixed in formalin (10% buffered formalin), and embedded in paraffin in a random direction.
RNA isolation from tissue.
RNA was isolated from ~100-mg sections of frozen placenta using TRIzol Reagent (Invitrogen) as previously described (24). RNA quality was determined using an Agilent 2100 Bioanalyzer (Agilent Technologies) and RNA concentrations confirmed by quantitation using a NanoDrop 8000 spectrophotometer (Thermo Fisher Scientific).
Whole placental genome expression profiling.
Whole genome expression profiling was performed using gene arrays (Human-6 BeadChips, Illumina) as described (28). Individual cRNA samples were used to interrogate each BeadChip (CTR female placentas, n = 3; CTR male placentas, n = 3; NR female placentas, n = 3; NR male placentas, n = 3). Gene expression data were processed and analyzed using GenomeStudio (Illumina) and GeneSifter (VizX Labs) as detailed in (28, 29) (GEO accession no: GSE40878).
Pathway and network analysis.
Array data for differently expressed genes were overlaid onto gene ontological (30) pathways and Kyoto Encyclopedia of Genes and Genomes (KEGG) (31) pathways using GeneSifter. Network analysis [Ingenuity Pathway Analysis (IPA), Ingenuity Systems] was performed using differentially expressed genes (P < 0.05) from each pairwise comparison. Networks were built with the IPA Knowledge Base by using expression profiles from this dataset and requiring direct connections between molecules based on experimental evidence.
Validation of gene expression for targeted genes.
We performed a 2-step qRT-PCR protocol using Assay on Demand kits (Applied Biosystems) to determine gene expression of peroxisome proliferator-activated receptor γ (PPARγ; cat. no. Hs00234592_m1), proteoglycan 2 (PRG2; cat. no. Hs00794928_m1), tumor protein 53 (TP53; cat. no. Hs99999147_m1), and histone cluster 3 (HIST3H3; cat. no. Hs00605762_s1) according to the manufacturer’s instructions. In brief, total RNA (50 ng) was reverse transcribed in a 100-μL reaction using the High-Capacity cDNA Archive kit (Applied Biosystems). Complimentary DNA synthesis was followed by qPCR using gene-specific primers provided by the manufacturer, TaqMan Universal PCR master mix (Applied Biosystems), and the target cDNA. Human 18s rRNA (cat. no. Hs99999901_s1) and mitochondrial ribosomal protein L48 (MRPL48; cat. no. Hs00740658_m1) were used as endogenous CTR. All probe sets spanned introns to ensure specificity amplifying cDNA. All samples were assayed in triplicate and a no-template negative CTR was included.
Immunohistochemistry.
Immunohistochemistry (IHC) was performed using standard avidin-biotin histochemical techniques as previously described (32). The primary antibodies and blocking peptides were as follows: caspase 3 (CASP3; Cell Signaling Technology, cat no. 9661, dilution 1:50) and blocking peptide (for negative CTR) (Cell Signaling Technology. cat no.1050, dilution 1:10); BCL2-associated X protein (BAX; Cell Signaling Technology, cat no. 2772, dilution 1:150), and blocking peptide (Cell Signaling Technology, cat no. 2772, dilution 1:30). The primary antibody in question was preabsorbed. Images were acquired digitally and analyzed with a 5.1 mega-pixel SPOT (Diagnostics Instruments) color cooled CCD camera (2650 × 1920 pixels) and ImagePro Plus 4.5 software (Media Cybernetics). Quantification was performed on randomly chosen slides from each animal and 3 pictures were taken at 3, 6, and 9 o'clock on the slide. Each picture covered 64,900 square microns. The fraction of the slide stained positive was recorded as the fraction stained relative to the total area interrogated.
Statistical analysis.
Data were analyzed using the 2-tailed Student’s t test to compare CTR and NR groups, females of the 2 groups, and males of the 2 groups. Results are presented as means ± SEMs. Significance was set at <0.05. For pathway analysis of differentially expressed genes, significance was determined by Z-score as previously described (24, 28, 33). Pathways were considered significantly different between groups if the Z-score for that pathway was >2.00 or < −2.00. Network significance was calculated in IPA using Fisher's exact t test.
Results
Morphometrics.
Morphometric data including maternal weight, maternal weight gain, fetal weight, placenta weight, and fetal:placenta weight ratio for CTR and NR animals are shown in Table 1. Maternal weight was less in NR than CTR but not when male and female offspring were analyzed separately. Weight gain was less in the NR than in the CTR dams and in dams with NR males than in CTR males. There were no significant differences in fetal weight. Placenta weight was significantly less in NR than in CTR and in NR females than in CTR females.
TABLE 1.
Morphometric data for CTR and NR female baboons and their offspring1
| Maternal weight | Weight gain | Fetal weight | Placenta weight | Fetal:placenta ratio | |
| kg | kg/165 d | g | g | ||
| CTR | |||||
| F | 17.75 | 0.78 | 693.00 | 208.00 | 3.33 |
| F | 15.64 | 1.43 | 648.00 | 160.00 | 4.05 |
| F | 19.70 | 0.82 | 836.00 | 181.00 | 4.62 |
| M | 16.75 | 2.02 | 694.00 | 165.00 | 4.21 |
| M | 17.02 | 3.36 | 813.00 | 200.00 | 4.07 |
| M | 13.95 | 1.33 | 846.00 | 176.00 | 4.81 |
| Mean | 16.80 ± 0.79 | 1.62 ± 0.39 | 755.00 ± 35.22 | 181.67 ± 7.77 | 4.18 ± 0.21 |
| F Mean | 17.70 ± 1.17 | 1.01 ± 0.21 | 726.00 ± 56.68 | 183.00 ± 13.89 | 4.00 ± 0.37 |
| M Mean | 15.91 ± 0.98 | 2.24 ± 0.60 | 784.33 ± 46.16 | 180.33 ± 10.33 | 4.36 ± 0.23 |
| NR | |||||
| F | 15.30 | −0.62 | 600.00 | 150.00 | 4.00 |
| F | 16.14 | 1.28 | 680.00 | 146.00 | 4.66 |
| F | 12.44 | −0.94 | 598.00 | 131.00 | 4.56 |
| M | 15.90 | −0.08 | 809.00 | 164.00 | 4.93 |
| M | 12.90 | −1.75 | 706.00 | 118.00 | 5.98 |
| M | 11.96 | −2.82 | 618.00 | 161.00 | 3.84 |
| Mean | 14.11 ± 0.77* | −0.82 ± 0.57* | 668.50 ± 33.41 | 145.00 ± 7.23* | 4.66 ± 0.31 |
| F Mean | 14.63 ± 1.12 | −0.09 ± 0.69 | 626.00 ± 27.01 | 142.33 ± 5.78* | 4.41 ± 0.21 |
| M Mean | 13.59 ± 1.19 | −1.55 ± 0.80* | 711.00 ± 55.19 | 148.00 ±14.86 | 4.92 ± 0.62 |
Results are means ± SEMs. *Different from corresponding CTR, P ≤ 0.05. CTR, control; NR, nutrient reduction.
Whole genome expression profiling.
Analysis of gene array data comparing NR with CTR without taking fetal sex into account showed that of 8928 expressed placental genes, 120 were upregulated, and 195 were downregulated (3.5% differentially expressed) (Supplemental Table 1). When data were analyzed separately for placentas of female and male fetuses, we found 9906 expressed genes in female placentas with 190 upregulated and 503 downregulated when comparing NR with CTR (7% differentially expressed) (Supplemental Table 2). In male placentas, we found 9757 expressed genes with 166 upregulated and 267 downregulated in NR compared with CTR (4% differentially expressed) (Supplemental Table 3). Comparing the list of differentially expressed genes for females and males, we found only 6 genes with a similar response to NR: hypothetical protein LOC283867 (LOC283867), kelch repeat and BTB (POZ) domain containing 3 (KBTBD3), NOP56 ribonucleoprotein homolog (NOP56), and solute carrier family 25, member 1 (SLC25A1) were downregulated, and cell death-inducing DFFA-like effector b (CIDEB) and Rho GTPase activating protein 30 (ARHGAP30) were upregulated in both sexes. An additional 10 placental genes were discordant between females and males: hypothetical protein LOC284531 (LOC284531), four and a half LIM domains 3 (FHL3), eukaryotic translation initiation factor 4E binding protein 1 (EIF4EBP1), CUE domain containing 1 (CUEDC1), hydroxypyruvate isomerase homolog (E. coli) (HYI), and KIAA1228 protein (KIAA1228) were downregulated in females and upregulated in males, and polybromo 1 (PBRM1), cyclin-dependent kinase inhibitor 2C (CDKN2C), non-SMC condensin II complex, subunit D3 (NCAPD3), and zinc family member 4 (ZIC4) were upregulated in females and downregulated in males (Supplemental Table 4).
Pathway analysis.
KEGG pathway and IPA network analyses included 428 mappable genes of the 693 differentially expressed genes in response to NR in female placentas (Supplemental Table 5) and 239 mappable genes of the 433 differentially expressed genes in placentas of male NR compared with CTR fetuses (Supplemental Table 6). Genes not included were hypothetical genes or proteins deficient in functional information necessary for inclusion in the analyses.
KEGG pathway analysis comparing NR against CTR revealed 15 upregulated and 25 downregulated pathways. When data were separately analyzed by fetal sex, we found 14 upregulated and 22 downregulated pathways in females and 11 upregulated and 13 downregulated in males in response to NR (Tables 2 and 3). A comparison of the differential KEGG pathways showed only 2 upregulated pathways (dilated cardiomyopathy and hypertrophic cardiomyopathy), which were common to both female and male placentas. Cell cycle, aminoacyl-tRNA synthesis, and renin-angiotensin system pathways were upregulated in NR female placentas but not in male NR placentas; endocytosis, transforming growth factor-beta (TGF-β) signaling, and tight junction pathways were downregulated in response to NR in female but not in male placentas. Furthermore, glycolysis/gluconeogenesis and protein digestion and absorption pathways were upregulated in male but not in female placentas. Pathways involving aldosterone-regulated sodium reabsorption, oxidative phosphorylation, and wingless/integration family signaling were downregulated in response to NR in male but not in female placentas.
TABLE 2.
KEGG pathways upregulated in female and male placentas of NR female baboons1
| Total differentially expressed | Genes upregulated | Genes downregulated | Z-score | |
| KEGG pathways up in NR females compared with CTR females | ||||
| Aminoacyl-tRNA biosynthesis | 4 | 2 | 2 | 2.65 |
| Arrhythmogenic right ventricular cardiomyopathy | 7 | 3 | 4 | 2.74 |
| Cell cycle | 8 | 4 | 4 | 2.62 |
| Dilated cardiomyopathy | 7 | 3 | 4 | 2.32 |
| Glycosylphosphatidylinositol-anchor biosynthesis | 2 | 2 | 0 | 3.67 |
| Histidine metabolism | 2 | 2 | 0 | 3.25 |
| Hypertrophic cardiomyopathy | 7 | 3 | 4 | 2.39 |
| Mucin type O-glycan biosynthesis | 3 | 2 | 1 | 3.41 |
| One carbon pool by folate | 1 | 1 | 0 | 2.06 |
| Phagosome | 8 | 4 | 4 | 2.20 |
| Phenylalanine metabolism | 1 | 1 | 0 | 2.15 |
| Renin-angiotensin system | 1 | 1 | 0 | 2.06 |
| Sphingolipid metabolism | 2 | 2 | 0 | 2.65 |
| Tyrosine metabolism | 2 | 2 | 0 | 2.60 |
| KEGG pathways up in NR males compared with CTR males | ||||
| Cardiac muscle contraction | 2 | 2 | 0 | 2.55 |
| Dilated cardiomyopathy | 2 | 2 | 0 | 2.21 |
| Glycolysis/gluconeogenesis | 2 | 2 | 0 | 2.83 |
| Glyoxylate and dicarboxylate metabolism | 1 | 1 | 0 | 3.00 |
| Hypertrophic cardiomyopathy | 2 | 2 | 0 | 2.27 |
| Maturity onset diabetes of the young | 1 | 1 | 0 | 2.42 |
| Protein digestion and absorption | 2 | 2 | 0 | 2.45 |
| Protein export | 2 | 1 | 1 | 2.49 |
| RNA degradation | 2 | 2 | 0 | 2.77 |
| Small cell lung cancer | 2 | 2 | 0 | 2.31 |
| Thyroid cancer | 1 | 1 | 0 | 2.19 |
| KEGG pathways up in NR compared with CTR | ||||
| Antigen processing and presentation | 4 | 2 | 2 | 2.39 |
| Galactose metabolism | 1 | 1 | 0 | 2.03 |
| Glycolysis/gluconeogenesis | 2 | 2 | 0 | 2.48 |
| Leishmaniasis | 3 | 2 | 1 | 2.34 |
| Maturity onset diabetes of the young | 1 | 1 | 0 | 2.15 |
| mRNA surveillance pathway | 2 | 2 | 0 | 2.16 |
| Mucin type O-glycan biosynthesis | 2 | 2 | 0 | 4.38 |
| Pentose phosphate pathway | 1 | 1 | 0 | 2.09 |
| Peroxisome | 3 | 2 | 1 | 2.12 |
| RNA degradation | 3 | 3 | 0 | 3.96 |
| Selenoamino acid metabolism | 1 | 1 | 0 | 2.09 |
| Synthesis and degradation of ketone bodies | 1 | 1 | 0 | 3.91 |
| Type II diabetes mellitus | 2 | 2 | 0 | 3.09 |
| Ubiquitin mediated proteolysis | 5 | 3 | 2 | 2.30 |
| Viral myocarditis | 2 | 2 | 0 | 2.39 |
The significance of identified KEGG pathways was determined by Z-score. Pathways were considered significantly different between CTR and NR groups if the Z-score for that pathway was >2.00 or < −2.00. CTR, control; KEGG, Kyoto Encyclopedia of Genes and Genomes; NR, nutrient reduction; tRNA, transfer RNA.
TABLE 3.
KEGG pathways downregulated in female and male placentas of NR female baboons1
| Total differentially expressed | Genes upregulated | Genes downregulated | Z-score | |
| KEGG pathways down in NR females compared with CTR females | ||||
| Acute myeloid leukemia | 6 | 2 | 4 | 2.55 |
| Adherens junction | 6 | 1 | 5 | 2.74 |
| Bacterial invasion of epithelial cells | 7 | 0 | 7 | 4.60 |
| d-Glutamine and d-glutamate metabolism | 2 | 0 | 2 | 6.54 |
| ECM-receptor interaction | 9 | 3 | 6 | 3.20 |
| Endocytosis | 11 | 2 | 9 | 2.34 |
| ErbB signaling pathway | 5 | 0 | 5 | 2.35 |
| Fatty acid elongation in mitochondria | 1 | 0 | 1 | 2.19 |
| Fc gamma R-mediated phagocytosis | 6 | 1 | 5 | 2.23 |
| Focal adhesion | 13 | 3 | 10 | 2.81 |
| Glycosaminoglycan biosynthesis: chondroitin sulfate | 2 | 0 | 2 | 2.22 |
| Glycosphingolipid biosynthesis: ganglio series | 2 | 0 | 2 | 2.96 |
| Nitrogen metabolism | 2 | 0 | 2 | 2.14 |
| Pathogenic Escherichia coli infection | 4 | 0 | 4 | 2.73 |
| Pathways in cancer | 15 | 1 | 14 | 2.69 |
| Proximal tubule bicarbonate reclamation | 2 | 0 | 2 | 2.14 |
| Regulation of actin cytoskeleton | 14 | 2 | 12 | 3.64 |
| Shigellosis | 5 | 0 | 5 | 3.32 |
| Small cell lung cancer | 5 | 0 | 5 | 2.38 |
| TGF-β signaling pathway | 8 | 0 | 8 | 4.68 |
| Thyroid cancer | 3 | 0 | 3 | 3.09 |
| Tight junction | 9 | 2 | 7 | 2.55 |
| KEGG pathways down in NR males compared with CTR males | ||||
| Aldosterone-regulated sodium reabsorption | 2 | 0 | 2 | 3.00 |
| β-Alanine metabolism | 1 | 0 | 1 | 2.02 |
| Cell cycle | 4 | 0 | 4 | 3.18 |
| Colorectal cancer | 2 | 0 | 2 | 2.24 |
| Glioma | 2 | 0 | 2 | 2.15 |
| Long-term depression | 2 | 0 | 2 | 2.07 |
| Long-term potentiation | 3 | 0 | 3 | 3.36 |
| Natural killer cell mediated cytotoxicity | 3 | 0 | 3 | 2.00 |
| Non-small cell lung cancer | 2 | 0 | 2 | 2.46 |
| Oxidative phosphorylation | 3 | 0 | 3 | 2.04 |
| Sulfur relay system | 1 | 0 | 1 | 3.53 |
| Vibrio cholerae infection | 3 | 1 | 2 | 2.46 |
| Wnt signaling pathway | 4 | 0 | 4 | 2.69 |
| KEGG pathways down in NR compared with CTR | ||||
| Adipocytokine signaling pathway | 2 | 0 | 2 | 2.42 |
| Antigen processing and presentation | 4 | 2 | 2 | 2.44 |
| β-Alanine metabolism | 1 | 0 | 1 | 2.32 |
| Cell cycle | 3 | 0 | 3 | 2.57 |
| Chronic myeloid leukemia | 2 | 0 | 2 | 2.34 |
| Circadian rhythm: mammal | 1 | 0 | 1 | 2.32 |
| Colorectal cancer | 2 | 0 | 2 | 2.62 |
| ErbB signaling pathway | 2 | 0 | 2 | 2.02 |
| Ether lipid metabolism | 2 | 0 | 2 | 4.10 |
| Fatty acid elongation in mitochondria | 1 | 0 | 1 | 4.56 |
| Fc gamma R-mediated phagocytosis | 3 | 0 | 3 | 3.28 |
| Fructose and mannose metabolism | 3 | 1 | 2 | 3.80 |
| Glycosaminoglycan biosynthesis: chondroitin sulfate | 1 | 0 | 1 | 2.32 |
| Glycosaminoglycan biosynthesis: heparan sulfate | 1 | 0 | 1 | 2.07 |
| Glycosphingolipid biosynthesis: globo series | 1 | 0 | 1 | 3.08 |
| Nitrogen metabolism | 1 | 0 | 1 | 2.25 |
| Non-small cell lung cancer | 2 | 0 | 2 | 2.86 |
| Pathways in cancer | 6 | 0 | 6 | 2.84 |
| PPAR signaling pathway | 3 | 0 | 3 | 3.92 |
| Prostate cancer | 3 | 0 | 3 | 3.31 |
| Shigellosis | 2 | 0 | 2 | 2.69 |
| Small cell lung cancer | 2 | 0 | 2 | 2.04 |
| Thyroid cancer | 3 | 0 | 3 | 6.75 |
| Vibrio cholerae infection | 2 | 0 | 2 | 2.86 |
| Wnt signaling pathway | 3 | 0 | 3 | 2.16 |
The significance of identified KEGG pathways was determined by Z-score. Pathways were considered significantly different between CTR and NR groups if the Z-score for that pathway was >2.00 or < −2.00. CTR, control; ECM, extracellular matrix; ErbB, v-erb-b erythroblastic leukemia viral oncogene; Fc, fragment, crystallizable; KEGG, Kyoto Encyclopedia of Genes and Genomes; NR, nutrient reduction; PPAR, peroxisome proliferator-activated receptor; R, receptor; TGF-β, transforming growth factor-beta; Wnt, wingless/integration family.
Network analysis.
Network analysis of placental RNA expression from female CTR placentas compared with NR revealed 5 networks with >20 molecules and a significant P value (<10−20). The top annotated functions of the genes composing the networks included: cellular function and maintenance, cell-to-cell signaling and interaction, cellular assembly and organization, cellular compromise, cell morphology, gene expression, reproductive system disease, cellular movement, and cellular growth and proliferation. Evaluation of these networks showed a coordinated response to NR with the majority of genes downregulated (e.g., Fig. 1A; Supplemental Table 7). Merging the overlapping networks and counting edges to identify likely major hub genes (>35 edges) in the coordinated network showed 13 genes central to the merged network and includes PPARγ and TP53. TP53 was not detected on the gene array (likely due to human to baboon sequence differences in the probe site of the gene) (Supplemental Table 8).
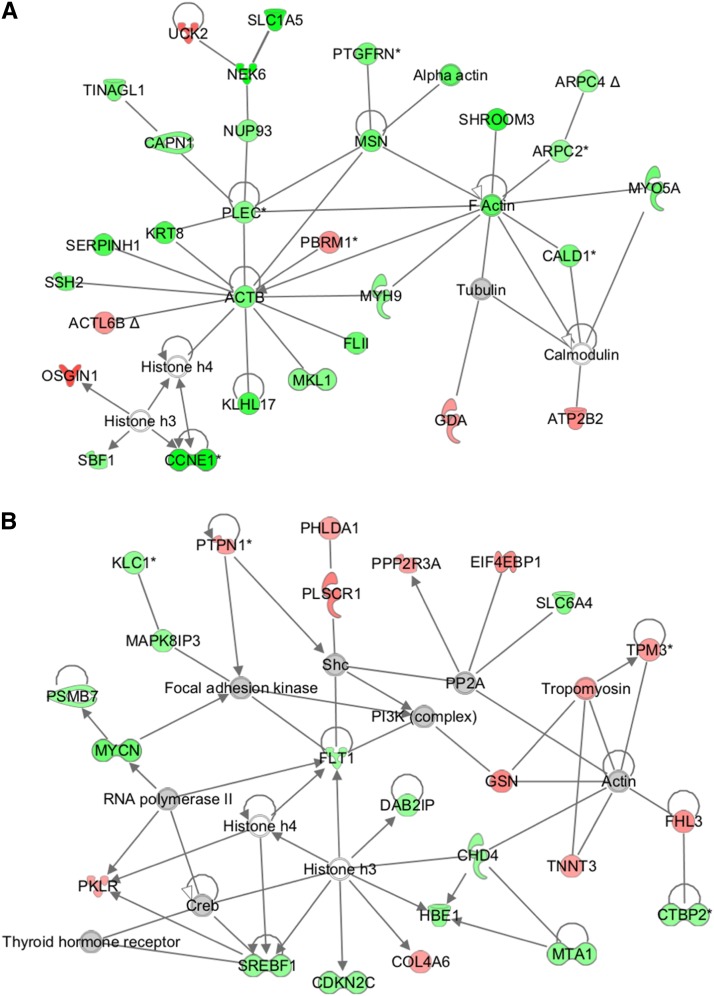
FIGURE 1.
Top-ranking networks for gene expression differences in female (A) and male (B) offspring of CTR and NR female baboons. Genes are denoted by gene IDs, green indicates downregulated, red indicates upregulated, gray indicates no difference in gene expression in NR compared with CTR, and white denotes the lack of quality signal on the array. ACTB, actin, beta; ACTL6B, actin-like protein 6B; ARPC2, actin related protein 2/3 complex, subunit 2, 34kDa; ARPC4, actin related protein 2/3 complex, subunit 4, 20kDa; ATP2B2, ATPase, Ca++ transporting, plasma membrane 2; CALD1, caldesmon 1; CAPN1, calpain 1, (mu/I) large subunit; CCNE1, cyclin E1; CDKN2C, cyclin-dependent kinase inhibitor 2C (p18, inhibits CDK4); CHD4, chromodomain helicase DNA binding protein 4; COL4A6, collagen, type IV, alpha 6; Creb, c-AMP response element-binding protein; CTBP2, C-terminal binding protein 2; CTR, control; DAB2IP, DAB2 interacting protein; EIF4EBP1, eukaryotic translation initiation factor 4E binding protein 1; F actin, fibrous actin; FHL3, four and a half LIM domains 3; FLII, flightless I homolog (Drosophila); FLT1, fms-related tyrosine kinase 1; GDA, guanine deaminase; GSN, gelsolin; HBE1, hemoglobin, epsilon 1; KLC1, kinesin light chain 1; KLHL17, kelch-like family member 17; KRT8, keratin 8; MAPK8IP3, mitogen-activated protein kinase 8 interacting protein 3; MKL1, megakaryoblastic leukemia (translocation) 1; MSN, moesin; MTA1, metastasis associated 1; MYCN, v-myc myelocytomatosis viral related oncogene, neuroblastoma derived (avian); MYH9, myosin, heavy chain 9, non-muscle; MYO5A, myosin VA (heavy chain 12, myoxin); NEK6, NIMA-related kinase 6; NR, nutrient reduction; NUP93, nucleoporin 93kDa; OSGIN1, oxidative stress induced growth inhibitor 1; PBRM1, polybromo 1; PHLDA1, pleckstrin homology-like domain, family A, member 1; PI3K, phosphoinositide 3-kinase; PKLR, pyruvate kinase, liver and RBC; PLEC, plectin; PLSCR1, phospholipid scramblase 1; PP2A, protein phosphatase 2; PPP2R3A, protein phosphatase 2, regulatory subunit B'', alpha; PSMB7, proteasome (prosome, macropain) subunit, beta type, 7; PTGFRN, prostaglandin F2 receptor inhibitor; PTPN1, protein tyrosine phosphatase, non-receptor type 1; SBF1, SET binding factor 1; SERPINH1, serpin peptidase inhibitor, clade H (heat shock protein 47), member 1, (collagen binding protein 1); Shc, Src (sarcoma) homology 2 domain-containing transforming protein 1; SHROOM3, shroom family member 3; SLC1A5, solute carrier family 1 (neutral amino acid transporter), member 5; SLC6A4, solute carrier family 6 (neurotransmitter transporter, serotonin), member 4; SREBF1, sterol regulatory element binding transcription factor 1; SSH2, slingshot protein phosphatase 2; TINAGL1, tubulointerstitial nephritis antigen-like 1; TNNT3, troponin T type 3 (skeletal, fast); TPM3, tropomyosin 3; UCK2, uridine-cytidine kinase 2.
Network analysis of placental RNA expression from placentas of CTR males compared with NR revealed one network with >20 molecules and a significant P value (<10−20). The top annotated functions of the genes in this network included cellular assembly and organization and cellular compromise. There were 12 upregulated genes and 13 downregulated genes in this network with no coordinated activation or repressive effects of NR (Figure 1B; Supplemental Table 9).
Analysis of functional groups.
Analysis of top-ranking functional groups for all differentially expressed genes (including those not built into networks) for CTR females compared with NR indicated that genes related to cell death were upregulated and cell proliferation, tissue development, and cellular assembly and organization were downregulated in response to NR (Supplemental Table 10).
Analysis of top-ranking functional groups in placentas from CTR male fetuses compared with NR showed differential expression of genes related to endocrine system development, organ development, and cardiovascular disease risk. Expression profiles of genes included in these functional groups were mixed with some genes upregulated and some downregulated with no coordinated response to NR, i.e., we did not find upregulation of activators and downregulation of repressors or vice versa (Supplemental Table 11).
Validation of gene expression.
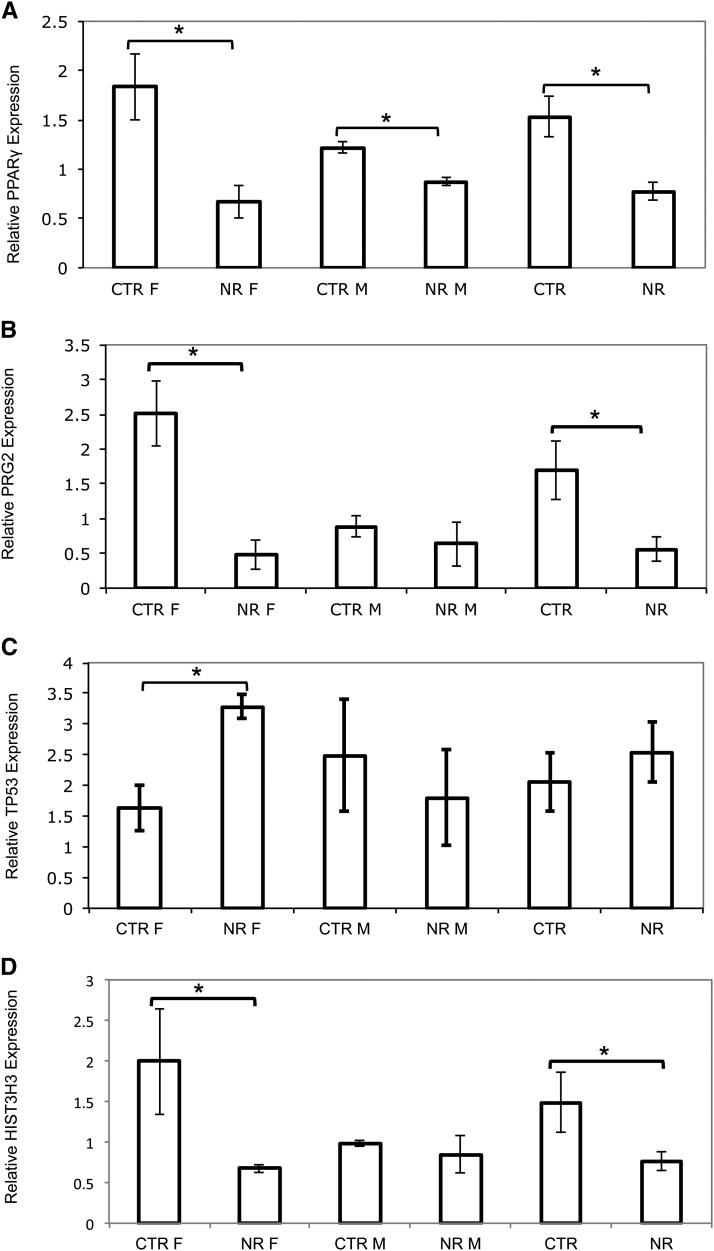
Quantification of PPARγ mRNA expression using qRT-PCR was consistent with our gene array results. PPARγ was significantly downregulated in both NR female and male placentas compared with CTR (Fig. 2A). PRG2 gene expression by qRT-PCR analysis was also consistent with our gene array results showing significant downregulation in the NR female placentas compared with CTR and no change in PRG2 expression in response to NR in male placentas (Fig. 2B). TP53 expression was not detected on the gene arrays. qRT-PCR analysis showed significant upregulation in NR female placentas compared with CTR with no change in male placentas (Fig. 2C). These findings are consistent with inclusion of TP53 in a top-ranked, downregulated, merged network in NR female placentas but not present in male placenta networks. Similarly, HIST3H3 was consistent with our gene array results showing significant downregulation in the NR female placentas compared with CTR and no expression differences in CTR and NR male placentas (Fig. 2D).
FIGURE 2.
qRT-PCR validation of PPARγ (A), PRG2 (B), TP53 (C), and HIST3H3 (D) gene expression in female and male offspring of CTR and NR female baboons. The y-axis shows mRNA expression relative to both human 18S and MRPL48 endogenous CTR. Bars denote SEM. *Different from CTR, P < 0.05. CTR, control; HIST3H3, histone cluster 3 H3; MRPL48, mitochondrial ribosomal protein L48; NR, nutrient reduction; PPARγ, peroxisome proliferator-activated receptor γ PRG2, proteoglycan 2; TP53, tumor protein 53.
IHC.
BAX protein expression using IHC showed the majority of positive staining in the syncytiotrophoblast. Quantification of staining showed no significant difference between NR male (Fig. 3B) and CTR male (Fig. 3A) placentas (summarized in Fig. 3F). In contrast, in female placentas, BAX protein expression was upregulated in response to NR (Fig. 3E) as compared with CTR (Fig. 3D; summarized in Fig. 3F). Quantification of CASP3 protein expression using IHC showed no differences between NR male (Fig. 3H) and CTR male (Fig. 3G) placentas (summarized in Fig. 3L). However, the female placentas responded differently in that CASP3 protein expression was upregulated in NR (Fig. 3K) compared with CTR placentas (Fig. 3J; summarized in Fig. 3L).
FIGURE 3.
BAX/CASP3 immunoreactivity in male placenta of CTR female baboons, n = 3 (A,G); male placenta of NR female baboon, n = 3 (B,H); preabsorbed negative CTR (C,I); female placenta of CTR female baboon, n = 4 (D,J); female placenta of NR female baboon, n = 3 (E,K); and summary of results expressed as fraction percent of area stained (F,L). Green staining, nuclei; black staining, BAX/CASP3. Data are means ± SEMs. *Different from CTR, P < 0.05. Microbar applies to all images. BAX, BCL2-associated X protein; CASP3, caspase 3; CTR, control; NR, nutrient reduction.
Discussion
To the best of our knowledge, this is the first report exploring regulation of the transcriptome of the primate placenta in response to maternal NR. Analysis of the term placental transcriptome in the baboon showed a striking sex difference in responsiveness. Gene expression profiling indicated that almost twice as many genes were differentially regulated in the female compared with the male placenta in response to NR. Pathway and network analysis revealed that the female placental transcriptome exhibited a coordinated response to decreased nutrient availability, including upregulation of genes related to programmed cell death and downregulation of genes in networks, pathways, and functional groups related to cell proliferation. These changes were not apparent in male placentas. Our data support the concept that female placentas initiate complex adaptive responses to an adverse intrauterine environment, which may contribute to increased survival and better pregnancy outcomes in girls.
Maternal under-nutrition during pregnancy remains a daunting problem worldwide and also constitutes a significant problem in the United States, because >50 million Americans live in households experiencing food insecurity or hunger at least some time during the year (34). Thus, our nonhuman primate model involving a 30% maternal calorie reduction throughout most of pregnancy is highly relevant for human health and disease. Babies who were in utero during the wartime famine in the winter of 1944–1945 in Holland (the Dutch famine cohort) have an increased incidence of obesity and metabolic and cardiovascular disease in adult age (13, 35), demonstrating that maternal under-nutrition during gestation has important effects on health in later life in humans.
The concept of sex differences in placental responses to altered maternal environment in pregnant women has been pioneered by Clifton et al. (36), who have explored sexual dimorphic effects of maternal asthma on placental gene expression and function. Asthma typically becomes worse in pregnancy, but asthmatic symptoms are much more aggravated if the fetus is female. This phenomenon has been suggested to be related to sex differences in placental cytokine expression in the presence of maternal asthma (37) or differences in cortisol responsiveness between male and female placentas (38). Specifically, the male placenta appears to be glucocorticoid resistant, because pathways typically responsive to cortisol, such as cytokine expression and the insulin-like growth factor (IGF) axis, remained unaffected in the presence of increasing cortisol (17). In addition, using global gene expression analysis, it was shown that the expression of 65 placental genes was altered in female fetuses in pregnancies complicated by asthma, whereas only 6 genes were differentially expressed in male placentas (19). Based on these observations and reports in the literature, it has been proposed that the male strategy for responding to an adverse maternal environment is a minimalistic approach involving differential regulation of only a very limited number of genes, allowing continued growth in a suboptimal environment (17). In contrast, the female placenta responds to an environmental challenge by regulating multiple genes, resulting in a modestly decreased growth (19). Indeed, our data are in general agreement with this model, because the activation of pathways involved in programmed cell death and downregulation of genes related to cell proliferation in female placentas may represent a conservative strategy to adapt placental function to decreased nutrient availability. The male placental transcriptome, on the other hand, appeared less responsive to maternal NR. Recent studies in mice have demonstrated that female placentas respond with more extensive changes in gene expression to diets with different fat content than male placentas (21). Because these observations have been made in species with distinct placentation and using different experimental paradigms (asthma, maternal NR, high-fat diet), they strongly suggest that there are fundamental molecular differences in environmental responsiveness between female and male placentas.
Gene expression profiling demonstrated that almost twice as many genes were differentially regulated in the female (7%) compared with the male placenta (4%) in response to NR in baboons. We proceeded with a more in-depth investigation of sex differences using network analysis, which does not rely on annotated pathways and may provide additional insights into the repertoire of placental genes that are differentially expressed in response to decreased nutrient availability.
Network and functional category analyses of the female placentas showed a coordinated molecular genetic response to reduced maternal nutrition. Network analysis revealed 5 significant, coordinated networks with upregulation of genes related to programmed cell death and downregulation of genes involved with cell proliferation. On the other hand, analyses of the transcriptome in male placentas revealed only one network that met the cutoff criteria that was regulated by NR. Importantly, the responses in this network lacked coordination, because it included 11 upregulated genes resulting in activation of the network and 12 downregulated genes causing repression. We validated gene expression profiles from gene array data and for genes highlighted from the network analysis that did not give a signal on the gene arrays and genes known to be relevant to pregnancy outcomes.
TP53 is a tumor suppressor gene, which inhibits cell proliferation and glycolysis and promotes apoptosis in response to various stress signals, including hypoxia and DNA damage. We did not detect the gene on our arrays (due to baboon/human sequence differences in the TP53 gene array probe); however, network analysis revealed TP53 as a major hub in female offspring overlapping networks with the 98 edges, the greatest number of functional connections to other molecules than any other hubs. qRT-PCR showed this gene significantly upregulated in female placentas with no change in males in response to NR. TP53 is known to activate BAX, which in turn activates apoptosis. IHC results for BAX and another apoptotic factor, CASP3, provide additional evidence that programmed cell death is observed in female NR placentas compared with CTR but not males. Our results show sex-specific expression of TP53 gene (upregulated in females but not males) and sex-specific expression of BAX (upregulated in females but not males). Thus, TP53 activation in female placentas may act to adapt placental metabolism and growth to the reduced nutrient availability.
PPARγ was identified as a major hub in the merged network of female, but not in male, placentas and was therefore analyzed further. PPARγ is a member of the nuclear receptor superfamily of ligand-dependent transcription factors activated by nutrients such as fatty acids and their metabolites. The transcriptional activator PPARγ and apoptosis activator TP53 should be expressed in opposite directions for a coordinated response to NR. PPARγ is essential for adipose tissue development and has well-established insulin-sensitizing effects, and this transcription factor has also been shown to be critical for normal placental development (39), mediated by regulation of placental angiogenesis (40), trophoblast invasion (41), and proliferation (42). Both gene array and qRT-PCR results showed a marked downregulation in female placentas in response to NR, inverse to TP53, with only a modest reduction in placental PPARγ gene expression in males and no difference in TP53. PPARγ downregulation is expected to stimulate trophoblast invasion (41), decrease trophoblast lipid accumulation (43), and inhibit villous trophoblast proliferation and differentiation (42), which may represent an appropriate adaptive strategy to nutrient restriction.
HIST3H3 encodes the core component of the nucleosome and has been shown to be associated with transcriptionally active chromatin (44). DNA accessibility is regulated via a complex set of post-translational modifications of histones and nucleosome remodeling. Our gene array results showed that HIST3H3 expression was less in female placentas, with no difference in males, in response to maternal NR. Furthermore, HIST3H3 was identified as a central hub in the female placenta networks. These targeted results support our network findings with coordinated response to NR via enhanced apoptosis and reduced transcriptional activation in females but not males. In addition, the results suggest that the response of the female placental transcriptome to NR may be mediated in part by epigenetic regulation. Additional studies are required to validate this.
PRG2 was chosen for validation, because it has been linked to pregnancy outcomes. The PRG2 gene encodes proform of eosinophil major basic protein (proMBP). MBP is toxic to bacteria and parasites and is important for the defense against infection. proMBP is also highly expressed in the placenta and has been reported to bind and inhibit the activity of pregnancy-associated plasma protein A (PAPPA) and angiotensinogen (AGT) (45). PAPPA is a protease regulating local IGF bioavailability by degrading IGF binding proteins. Thus, the pronounced reduction in PRG2 expression in female placentas could lead to greater local IGF levels. Because IGFs stimulate placental function, including nutrient transport, these alterations may constitute a compensatory response to NR. Maternal NR in pregnant baboons leads to complex modifications in the IGF system (32). The serum IGF-I:IGFBP-3 ratio is less in both mothers and fetuses, whereas maternal serum IGF-II is unaffected by maternal NR. Placental IGF-I mRNA and protein abundance are reduced, whereas placental IGF-II mRNA is greater. Systemic (maternal) and local (placental) IGFBP-1 and IGFBP-3 mRNA and protein abundance were unaffected by maternal NR. Thus, moderate maternal NR alters the maternal and placental IGF-IGFBP axis in many interactive ways. We have proposed that these alterations are directed toward preserving placental growth (32) and the current data suggest these mechanisms may be more effective in the female than the male placenta.
As mentioned previously, apoptosis was identified by network analyses as one of 2 key functions that were different between male and female placentas. To further validate this finding, we performed IHC for BAX and CASP3, players in programmed cell death. TP53, upon activation by BAX, activates apoptosis (46). Studies in humans have shown TP53-mediated apoptosis in FGR placentas (47). CASP3 is one of 3 executioner caspases that cleaves downstream products in the apoptotic process (48). Increased CASP3 protein expression has been observed in FGR human placentas (49) and associated with increased placental apoptosis in our baboon model of maternal NR (50). In addition, using stereology, we have shown that NR in the baboon impairs placental structure (23). Our results show sex-specific expression of TP53 gene (upregulated in females but not males) and concordant sex-specific expression of BAX protein and CASP3 protein (upregulated in females but not males). These results are consistent with our gene network analysis showing coordinated programmed cell death in female placentas but not male placentas.
The mechanisms underlying sex differences in the placental response to maternal NR remain to be established. In addition to the obvious candidates (sex hormones, in particular testosterone), the sex-dependent differences in responsiveness to corticosteroids, with the male placenta glucocorticoid resistant (19), may be involved. In conclusion, our data show that the regulation of the placental transcriptome in maternal NR shows differences that are fetal sex dependent. Our findings support the concept that female placentas initiate complex adaptive responses to an adverse intrauterine environment, which may contribute to enhanced survival and better pregnancy outcomes in girls.
Supplementary Material
Acknowledgments
Dr. Natalia Schlabritz-Loutsevitch contributed to animal husbandry and collection of placental samples. L.A.C. designed research, provided essential reagents, analyzed data, wrote the paper, and had primary responsibility for final content; C.L., J.P.G., and K.L. conducted research; K.D.S. conducted research and edited the paper; P.W.N. provided essential reagents and wrote the paper; and T.J. provided essential reagents, analyzed data, and wrote the paper. All authors read and approved the final manuscript.
Footnotes
Abbreviations used: AGT, angiotensinogen; ARHGAP30, Rho GTPase activating protein 30; BAX, BCL2-associated X protein; CASP3, caspase 3; CDKN2C, cyclin-dependent kinase inhibitor 2C; CIDEB, cell death-inducing DFFA-like effector b; CTR, control; CUEDC1, CUE domain containing 1; EIF4EBP1, eukaryotic translation initiation factor 4E binding protein 1; FGR, fetal growth restriction; FHL3, four and a half LIM domains 3; GD, gestational day; HIST3H3, histone cluster 3 H3; HYI, hydroxypyruvate isomerase homolog (E. coli); IGF, insulin-like growth factor; IHC, Immunohistochemistry; IPA, Ingenuity Pathway Analysis; KBTBD3, kelch repeat and BTB (POZ) domain containing 3; KEGG, Kyoto Encyclopedia of Genes and Genomes; KIAA1228, KIAA1228 protein; LOC283867, hypothetical protein LOC283867; LOC284531, hypothetical protein LOC284531; MRPL48, mitochondrial ribosomal protein L48; NCAPD3, non-SMC condensin II complex, subunit D3; NOP56, NOP56 ribonucleoprotein homolog; NR, nutrient reduction; PAPPA, pregnancy-associated plasma protein A; PBRM1, polybromo 1; PPARγ, peroxisome proliferator-activated receptor γ PRG2, proteoglycan 2; proMBP, proform of eosinophil major basic protein; SLC25A1, solute carrier family 25, member 1; TGF-β, transforming growth factor-β TLR4, toll-like receptor-4; TNF-α, tumor necrosis factor-α TP53, tumor protein 53; ZIC4, zinc family member 4.
Literature Cited
- 1.Osmond C, Barker DJ. Fetal, infant, and childhood growth are predictors of coronary heart disease, diabetes, and hypertension in adult men and women. Environ Health Perspect. 2000;108 Suppl 3:545–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gluckman PD, Hanson MA. Living with the past: evolution, development, and patterns of disease. Science. 2004;305:1733–6. [DOI] [PubMed] [Google Scholar]
- 3.Barker DJP. The developmental origins of insulin resistance. Horm Res. 2005;64 Suppl 3:2–7. [DOI] [PubMed] [Google Scholar]
- 4.Thornburg KL, O'Tierney PF, Louey S. Review: the placenta is a programming agent for cardiovascular disease. Placenta. 2010;31 Suppl:S54–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jansson T, Powell TL. Role of the placenta in fetal programming: Underlying mechanisms and potential interventional approaches. Clin Sci (Lond). 2007;113:1–13. [DOI] [PubMed] [Google Scholar]
- 6.Godfrey KM. The role of the placenta in fetal programming-a review. Placenta. 2002;23 Suppl A:S20–7. [DOI] [PubMed] [Google Scholar]
- 7.Gilbert JS, Nijland MJ. Sex differences in the developmental origins of hypertension and cardiorenal disease. Am J Physiol Regul Integr Comp Physiol. 2008;295:R1941–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Papageorgiou AN, Colle E, Farri-Kostopoulos E, Gelfand MM. Incidence of respiratory distress syndrome following antenatal betamethasone: Role of sex, type of delivery, and prolonged rupture of membranes. Pediatrics. 1981;67:614–7. [PubMed] [Google Scholar]
- 9.Stevenson DK, Verter J, Fanaroff AA, Oh W, Ehrenkranz RA, Shankaran S, Donovan EF, Wright LL, Lemons JA, Tyson J, et al. Sex differences in outcomes of very low birthweight infants: the newborn male disadvantage. Arch Dis Child Fetal Neonatal Ed. 2000;83:F182–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Di Renzo GC, Rosati A, Sarti RD, Cruciani L, Cutuli AM. Does fetal sex affect pregnancy outcome? Gend Med. 2007;4:19–30. [DOI] [PubMed] [Google Scholar]
- 11.Vatten LJ, Skjaerven R. Offspring sex and pregnancy outcome by length of gestation. Early Hum Dev. 2004;76:47–54. [DOI] [PubMed] [Google Scholar]
- 12.Engel PJ, Smith R, Brinsmead MW, Bowe SJ, Clifton VL. Male sex and pre-existing diabetes are independent risk factors for stillbirth. Aust N Z J Obstet Gynaecol. 2008;48:375–83. [DOI] [PubMed] [Google Scholar]
- 13.Roseboom TJ, Painter RC, de Rooij SR, van Abeelen AFM, Veenendaal MVE, Osmond C, Barker DJP. Effects of famine on placental size and efficiency. Placenta. 2011;32:395–9. [DOI] [PubMed] [Google Scholar]
- 14.Stark MJ, Wright IMR, Clifton VL. Sex-specific alterations in placental 11beta-hydroxysteroid dehydrogenase 2 activity and early postnatal clinical course following antenatal betamethasone. Am J Physiol Regul Integr Comp Physiol. 2009;297:R510–4. [DOI] [PubMed] [Google Scholar]
- 15.Tanner JM. Fetus into man: physical growth from conception to maturity. 2nd revised, illustrated ed. Cambridge (MA): Harvard University Press; 1990.
- 16.Pedersen JF. Ultrasound evidence of sexual difference in fetal size in first trimester. BMJ. 1980;281:1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Clifton VL. Review: sex and the human placenta: mediating differential strategies of fetal growth and survival. Placenta. 2010;31 Suppl:S33–9. [DOI] [PubMed] [Google Scholar]
- 18.Sood R, Zehnder JL, Druzin ML, Brown PO. Gene expression patterns in human placenta. Proc Natl Acad Sci USA. 2006;103:5478–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Osei-Kumah A, Smith R, Jurisica I, Caniggia I, Clifton VL. Sex-specific differences in placental global gene expression in pregnancies complicated by asthma. Placenta. 2011;32:570–8. [DOI] [PubMed] [Google Scholar]
- 20.Yeganegi M, Watson CS, Martins A, Kim SO, Reid G, Challis JRG, Bocking AD. Effect of lactobacillus rhamnosus GR-1 supernatant and fetal sex on lipopolysaccharide-induced cytokine and prostaglandin-regulating enzymes in human placental trophoblast cells: Implications for treatment of bacterial vaginosis and prevention of preterm labor. Am J Obstet Gynecol. 2009;200:532.e1–8. [DOI] [PubMed] [Google Scholar]
- 21.Mao J, Zhang X, Sieli PT, Falduto MT, Torres KE, Rosenfeld CS. Contrasting effects of different maternal diets on sexually dimorphic gene expression in the murine placenta. Proc Natl Acad Sci USA. 2010;107:5557–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schlabritz-Loutsevitch NE, Howell K, Rice K, Glover EJ, Nevill CH, Jenkins SL, Bill Cummins L, Frost PA, McDonald TJ, Nathanielsz PW. Development of a system for individual feeding of baboons maintained in an outdoor group social environment. J Med Primatol. 2004;33:117–26. [DOI] [PubMed] [Google Scholar]
- 23.Schlabritz-Loutsevitch N, Ballesteros B, Dudley C, Jenkins S, Hubbard G, Burton GJ, Nathanielsz P. Moderate maternal nutrient restriction, but not glucocorticoid administration, leads to placental morphological changes in the baboon (papio sp.). Placenta. 2007;28:783–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cox LA, Nijland MJ, Gilbert JS, Schlabritz-Loutsevitch NE, Hubbard GB, McDonald TJ, Shade RE, Nathanielsz PW. Effect of 30 per cent maternal nutrient restriction from 0.16 to 0.5 gestation on fetal baboon kidney gene expression. J Physiol. 2006;572:67–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grieves JL, Dick EJ, Schlabritz-Loutsevich NE, Butler SD, Leland MM, Price SE, Schmidt CR, Nathanielsz PW, Hubbard GB. Barbiturate euthanasia solution-induced tissue artifact in nonhuman primates. J Med Primatol. 2008;37:154–61. [DOI] [PubMed] [Google Scholar]
- 26.Burton GJ, Jauniaux E. Sonographic, stereological and doppler flow velocimetric assessments of placental maturity. Br J Obstet Gynaecol. 1995;102:818–25. [DOI] [PubMed] [Google Scholar]
- 27.Mayhew TM. Recent applications of the new stereology have thrown fresh light on how the human placenta grows and develops its form. J Microsc. 1997;186:153–63. [DOI] [PubMed] [Google Scholar]
- 28.Shi Q, Cox LA, Hodara V, Wang XL, VandeBerg JL. Repertoire of endothelial progenitor cells mobilized by femoral artery ligation: a nonhuman primate study. J Cell Mol Med. 2012;16:2060–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kerr MK, Martin M, Churchill GA. Analysis of variance for gene expression microarray data. J Comput Biol. 2000;7:819–37. [DOI] [PubMed] [Google Scholar]
- 30.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kanehisa M, Goto S, Kawashima S, Okuno Y, Hattori M. The KEGG resource for deciphering the genome. Nucleic Acids Res. 2004;32 Database issue:D277–80. [DOI] [PMC free article] [PubMed]
- 32.Li C, Levitz M, Hubbard GB, Jenkins SL, Han V, Ferry RJ, McDonald TJ, Nathanielsz PW, Nathanielsz PW, Schlabritz-Loutsevitch NE. The IGF axis in baboon pregnancy: placental and systemic responses to feeding 70% global ad libitum diet. Placenta. 2007;28:1200–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Doniger SW, Salomonis N, Dahlquist KD, Vranizan K, Lawlor SC, Conklin BR. MAPPFinder: using Gene Ontology and GenMAPP to create a global gene-expression profile from microarray data. Genome Biol. 2003;4:R7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nord M. Household food security in the United States, 2009. Darby (PA): DIANE Publishing; 2010.
- 35.Roseboom TJ, Painter RC, van Abeelen AFM, Veenendaal MVE, de Rooij SR. Hungry in the womb: what are the consequences? Lessons from the Dutch famine. Maturitas. 2011;70:141–5. [DOI] [PubMed] [Google Scholar]
- 36.Clifton VL, Stark MJ, Osei-Kumah A, Hodyl NA. Review: the feto-placental unit, pregnancy pathology and impact on long term maternal health. Placenta. 2012;33 Suppl:S37–41. [DOI] [PubMed] [Google Scholar]
- 37.Scott NM, Hodyl NA, Murphy VE, Osei-Kumah A, Wyper H, Hodgson DM, Smith R, Clifton VL. Placental cytokine expression covaries with maternal asthma severity and fetal sex. J Immunol. 2009;182:1411–20. [DOI] [PubMed] [Google Scholar]
- 38.Scott NM, Hodyl NA, Osei-Kumah A, Stark MJ, Smith R, Clifton VL. The presence of maternal asthma during pregnancy suppresses the placental pro-inflammatory response to an immune challenge in vitro. Placenta. 2011;32:454–61. [DOI] [PubMed] [Google Scholar]
- 39.Barak Y, Nelson MC, Ong ES, Jones YZ, Ruiz-Lozano P, Chien KR, Koder A, Evans RM. PPAR [gamma] is required for placental, cardiac, and adipose tissue development. Mol Cell. 1999;4:585–95. [DOI] [PubMed] [Google Scholar]
- 40.Nadra K, Quignodon L, Sardella C, Joye E, Mucciolo A, Chrast R, Desvergne B. PPARγ in placental angiogenesis. Endocrinology. 2010;151:4969–81. [DOI] [PubMed] [Google Scholar]
- 41.Tarrade A, Schoonjans K, Pavan L, Auwerx J, Rochette-Egly C, Evain-Brion D, Fournier T. PPARγ/rxrα heterodimers control human trophoblast invasion. J Clin Endocrinol Metab. 2001;86:5017–24. [DOI] [PubMed] [Google Scholar]
- 42.Parast MM, Yu H, Ciric A, Salata MW, Davis V, Milstone DS. PPARgamma regulates trophoblast proliferation and promotes labyrinthine trilineage differentiation. PLoS ONE. 2009;4:e8055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fournier T, Guibourdenche J, Handschuh K, Tsatsaris V. PPAR [gamma] and human trophoblast differentiation. J Reprod Immunol. 2011;90:44–9. [DOI] [PubMed] [Google Scholar]
- 44.Hake SB, Garcia BA, Duncan EM, Kauer M, Dellaire G, Shabanowitz J, Bazett-Jones DP, Allis CD, Hunt DF. Expression patterns and post-translational modifications associated with mammalian histone H3 variants. J Biol Chem. 2006;281:559–68. [DOI] [PubMed] [Google Scholar]
- 45.Weyer K, Glerup S. Placental regulation of peptide hormone and growth factor activity by prombp. Biol Reprod. 2011;84:1077–86. [DOI] [PubMed] [Google Scholar]
- 46.Chipuk JE, Maurer U, Green DR, Schuler M. Pharmacologic activation of p53 elicits bax-dependent apoptosis in the absence of transcription. Cancer Cell. 2003;4:371–81. [DOI] [PubMed] [Google Scholar]
- 47.Nishizawa H, Ota S, Suzuki M, Kato T, Sekiya T, Kurahashi H, Udagawa Y. Comparative gene expression profiling of placentas from patients with severe pre-eclampsia and unexplained fetal growth restriction. Reprod Biol Endocrinol. 2011;9:107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wilson MR. Apoptosis: unmasking the executioner. Cell Death Differ. 1998;5:646–52. [DOI] [PubMed] [Google Scholar]
- 49.Agata KB, Anita S, Urszula KK, Agnieszka NK, Grzegorz B. Expression of caspase-3, bax nad bcl-2 in placentas from pregnancies complicated by treated and non-treated fetal growth restriction. Ginekol Pol. 2009;80:652–6. [PubMed] [Google Scholar]
- 50.Arroyo JA, Li C, Schlabritz-Loutsevitch N, McDonald T, Nathanielsz P, Galan HL. Increased placental XIAP and caspase 3 is associated with increased placental apoptosis in a baboon model of maternal nutrient reduction. Am J Obstet Gynecol. 2010;203:364e13–. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.