SUMMARY
Protein•protein interactions, which often involve interactions between an acyl carrier protein (ACP) and its partner enzymes, are important for coordinating polyketide biosynthesis. However, the nature of such interactions is not well understood, especially in the fungal non-reducing polyketide synthases (NR-PKSs) that biosynthesize toxic and pharmaceutically important polyketides. Here, we employ a mechanism-based crosslinker to successfully probe ACP and ketosynthase (KS) domain interactions in NR-PKSs. We found that crosslinking efficiency is closely correlated with the strength of ACP•KS interactions, and that KS demonstrates strong starter unit selectivity. We further identified positively charged surface residues by KS mutagenesis, which mediate key interactions with the negatively-charged ACP surface. Such complementary/matching contact pairs can serve as “adapter surfaces” for future efforts to generate new polyketides using NR-PKSs.
INTRODUCTION
Polyketide natural products from filamentous fungi are highly diverse in both chemical structures and bioactivities, and they include current top-selling drugs such as lovastatin (for cholesterol lowering), as well as potent toxins such as aflatoxin and cercosporin (Crawford and Townsend, 2010; Hopwood, 1997). At the center of polyketide biosynthesis is the polyketide synthase (PKS), a multi-domain enzyme complex. PKSs produce a huge variety of possible products via the controlled variation of building blocks, chain lengths, and modification reactions such as cyclizations. There are at least three types of PKSs in fungi (Shen, 2003). The non-reducing PKS (NR-PKS) subclass is an important, yet relatively less well-understood family (Crawford and Townsend, 2010). NR-PKSs are involved in the synthesis of a wide spectrum of aromatic polyketides, including mycotoxins such as aflatoxin (Fig. 1D) (Watanabe et al., 1996) and zearalenone (Kim et al., 2005) that have a profound influence on agriculture (Huffman et al., 2010), as well as current therapeutics such as griseofulvin (Rhodes et al., 1961) (antifungal) and potential future therapeutics such as bikaverin (anti-cancer) (Linnemannstons et al., 2002; Ma et al., 2007) (Fig. 1D). Similar to bacterial-derived tetracycline or daunorubicin metabolites (Otten et al., 1990; Perlman et al., 1960; Pickens and Tang, 2009), products from the fungal NR-PKSs have conjugated aromatic rings. These aromatic polyketides display an enormously rich spectrum of bioactivities (Hopwood, 1997); therefore, understanding how NR-PKSs generate polyketide diversity is significant with respect to (1) the identification and prediction of new polyketides based on the proteins responsible for their biosynthesis, which are encoded in the vast number of emerging sequenced microbial genomes, and (2) the generation of new bioactive polyketides by PKS engineering.
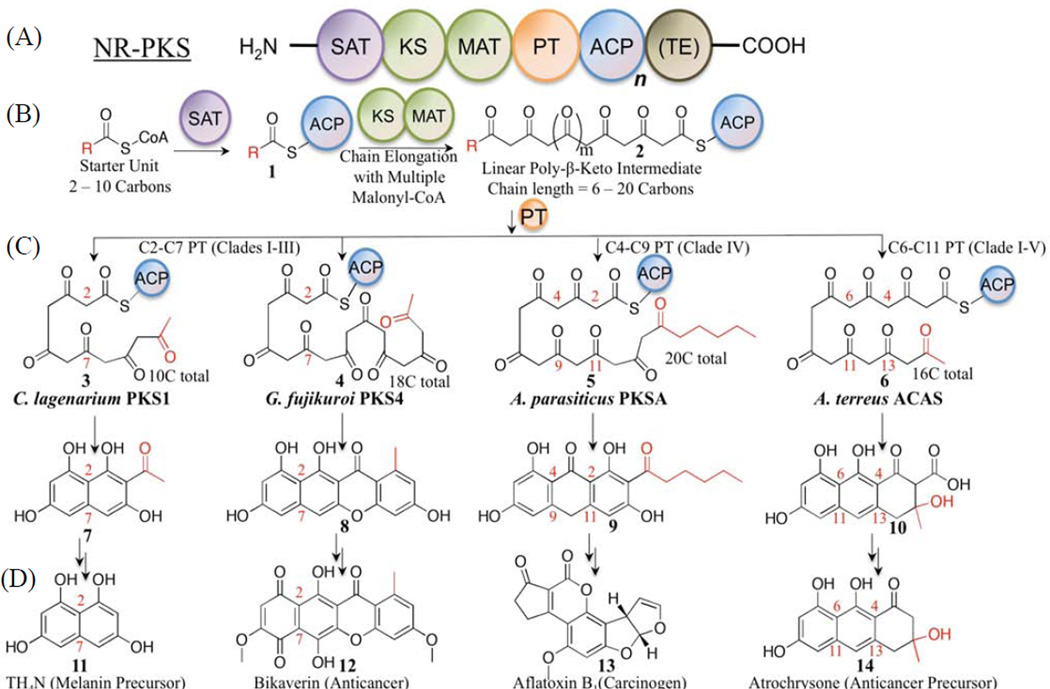
Figure 1.
(A) Domain architecture of a typical NR-PKS: there may be multiple ACPs (n=1–3), and TE is optional (in brackets). (B) NR-PKS enzymatic steps: chain initiation by SAT results in 1 (starter unit in red), followed by chain elongation by KS-AT (2); (C) cyclization is promoted by PT in 3 patterns: C2–C7 (Clades I–III, 3–4), C4–C9 (Clade IV, 5), and C6–C11 (Clade V, 6). Target PKSs are indicated in (C) and their final products in (D) (11–14).
A typical NR-PKS contains a core set of six covalently linked catalytic domains (Crawford and Townsend, 2010) that execute the following generalized biosynthetic scheme (Fig. 1B) (Cox, 2007; Crawford et al., 2008): the starter ACP transacylase (SAT) domain selects and loads the starter unit onto the acyl carrier protein (ACP). The chain is then transferred from ACP onto the ketosynthase (KS). Meanwhile, the malonyl-CoA:ACP transacylase (MAT) loads the extension unit building block from malonyl-CoA onto the ACP. Iterative rounds of decarboxylative Claisen condensations by KS domains synthesize a linear poly-β-keto intermediate, which is in turn cyclized and aromatized to form substituted aromatic rings by the product template (PT) domain (Fig. 1C). The growing polyketide intermediate remains covalently attached to the ACP via the phosphopantetheine (PPT) prosthetic group during chain elongations and PT-catalyzed cyclizations. The final intermediate is transferred to the thioesterase (TE) domain, which catalyzes product release.
Over the past decade, many NR-PKSs have been cloned, heterologously expressed, and their products characterized (Crawford and Townsend, 2010; Huffman et al., 2010; Zhou et al., 2010). How one enzyme domain interacts with the other domains within the PKS mega-synthase, however, remains largely unknown (Crawford et al., 2009; Korman et al., 2010). Prior engineering attempts indicate that protein•protein interactions are important in transporting the starter unit from SAT to KS, stabilizing the poly-β-keto intermediate, sequestering the linear chain in the KS and PT, and releasing the final cyclized product (Fig. 1B) (Crawford and Townsend, 2010). The ACP mediates all enzymatic reactions that occur within catalytic domains, and, presumably, protein•protein interactions are essential for coordinating polyketide assembly. Specifically, docking interactions between KS and ACP domains should be important for starter unit selection, chain length determination, and sequestration of the linear intermediate prior to cyclization. At this time, practically nothing is understood about ACP•KS interactions in NR-PKSs. Without such information, random domain shuffling has often resulted in abolished enzyme activity in microbial systems (Petkovic et al., 2002).
Although structures have been obtained for some individual NR-PKS domains (Crawford et al., 2009; Korman et al., 2010; Wattana-amorn et al., 2010), to date, the only related mega-synthase structure reported is the 3.2 Å crystal structure of the porcine fatty acid synthase (FAS) (Maier et al., 2008), which bears little (< 12 %) sequence homology to NR-PKSs (Crawford et al., 2009). The mammalian FAS exists as a homo-dimer, both in the crystal structure (Maier et al., 2008) and in solution (Fig. 2A) (Smith et al., 1985). The dimeric KS, dehydratase (DH) and enoyl reductase (ER) form the central “torso”, and the MAT and ketoreductase (KR) monomers (the “legs” and “arms”) are located on both sides of the torso. Our previous protein-sizing experiments showed that the SAT-KS-MAT tridomain of PksA exists as a dimer in solution (Crawford et al., 2009), which is architecturally similar to the KS-AT dimer found in porcine FAS (Fig. 2B). As is the case for porcine FAS, the KS exists as a major part of the dimer interface in the overall PksA architecture, while the ACP of each monomer, may interact alternately with each KS monomer during polyketide biosynthesis (Witkowski et al., 2004). Whereas in FAS, ACP presents the growing chain sequentially to the KR, DH, and ER domains following each extension to form a reduced methylene at the β-carbon position, in NR-PKSs the unaltered poly-β-ketide intermediate is fully extended in the KS before being transported to the PT domain for cyclization; and the nature of ACP•KS interactions in NR-PKS remains a mystery.
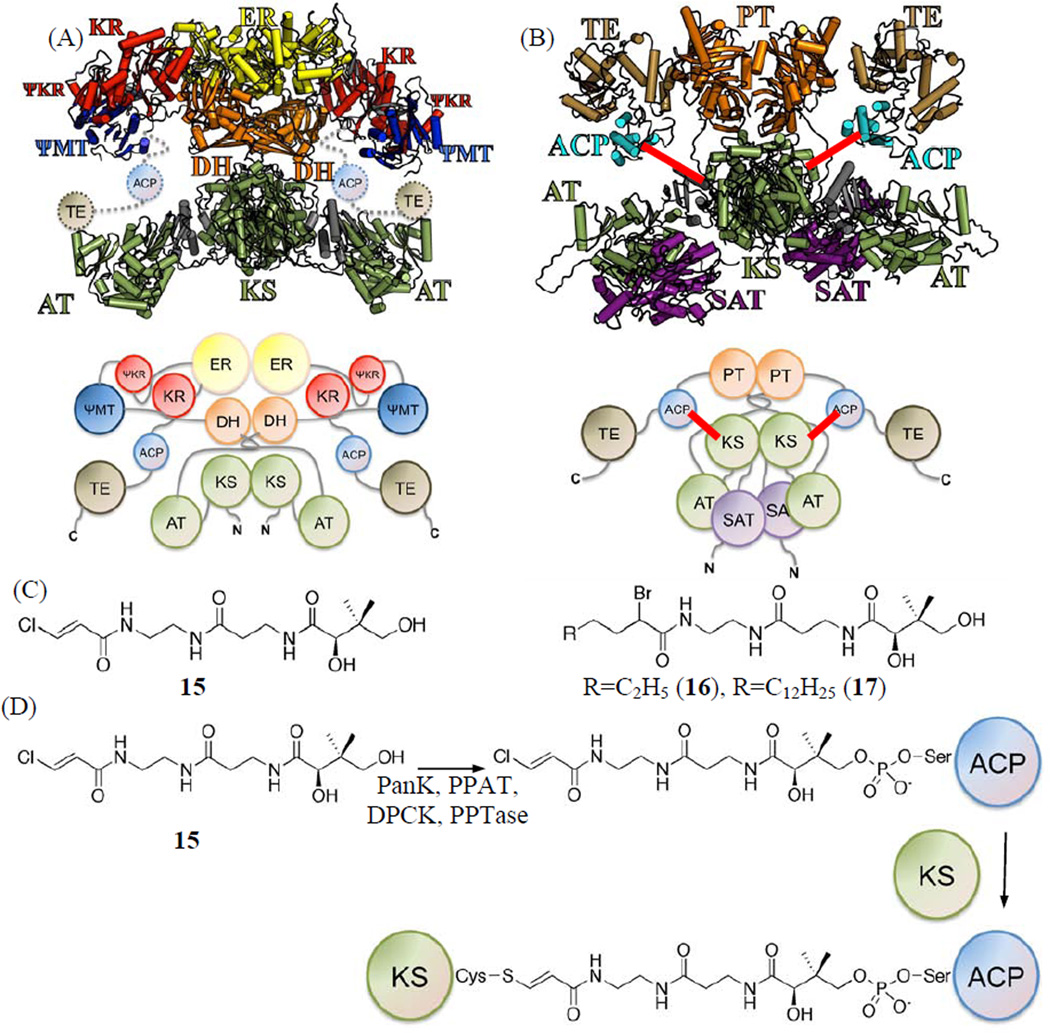
Figure 2.
(A) Crystal structure and cartoon of porcine FAS model as a homo-dimer, where KS is located at the center as a dimer. (B) Protein sizing studies of PksA supports an overall architecture similar to that of porcine FAS, where KS also exists as a dimer, and two ACPs on either side of the torso as shown in this homology model and cartoon. The cross-linking between KS and ACP is shown as red lines. (C) Pantetheinylated probes with a chloroacryl 15, 2-bromo hexyl 16 or 2-bromo palmitoyl 17 chemical groups that target the nucleophilic cysteine of the KS domain. (D) Chemo-enzymatic attachment of 15, 16 or 17 to ACP is followed by KS crosslinking the nucleophilic cysteine of the KS domain.
In order to probe for protein•protein interactions between ACP and the catalytic domains within the PKS mega-synthase, we have developed mechanism-based crosslinkers that can specifically bond the ACP to the KS (Fig. 2C) (Haushalter et al., 2008; Kapur et al., 2008; Meier et al., 2010; Meier et al., 2006; Worthington and Burkart, 2006; Worthington et al., 2008; Worthington et al., 2010; Worthington et al., 2006). To attach the synthetic crosslinkers to ACP, a chemo-enzymatic method using four enzymes— PanK, PPAT, DPCK, and a phosphopantetheinyl transferase (PPTase)—converts the pantetheine crosslinker analogue to the coenzyme A-like species and appends it to the conserved serine residue of ACP (Fig. 2D). When ACP forms a functional complex with the KS domain, the “warhead” group is delivered to the KS active site and crosslinks to the Cys nucleophile (Haushalter et al., 2008; Kapur et al., 2008; Meier et al., 2010; Meier et al., 2006; Worthington and Burkart, 2006; Worthington et al., 2008; Worthington et al., 2010; Worthington et al., 2006). Our previous crosslinking studies showed that the degree of crosslinking is reliant on the strength of direct protein•protein interactions between ACP and its target partner (Meier et al., 2010; Worthington et al., 2010) and closely tracks with standard kinetic and thermodynamic techniques to reveal binding specificity (Worthington 2006). Three crosslinkers (15, 16 and 17, Fig. 2C) were developed, which can be covalently attached to the ACP active site Ser chemo-enzymatically by the aforementioned four enzymes. They contain halide leaving groups to enable nucleophilic attack by the KS active site Cys resulting in formation of a covalent link between ACP and KS (Fig. 2D). The different chain lengths of the acyl moieties in 15–17 further enabled us to probe the substrate selectivity of KS.
In this work, ACP•KS interactions were probed by examining the crosslinking activity and compatibility between different domain combinations from PksA and Pks4, two six-domain NR-PKSs (Fig. 1A). The chemistries of PksA and Pks4 are shown in Fig. 1B–D; they are involved in the biosynthesis of aflatoxin and bikaverin, respectively. PksA extends and cyclizes a hexanoyl, a short fatty acid, starter unit to generate the 20-carbon product norsolorinic acid anthrone, while Pks4 uses the more common acetyl starter unit in the synthesis of the 18-carbon product pre-bikaverin/SMA76a (Fig. 1). Two central hypotheses were tested for this study: (1) Cys-specific crosslinkers, such as 15–17, will crosslink the ACP to the KS of an NR-PKS; and (2) crosslinking efficiency will correlate with the strength of domain•domain interactions. Here we show that the first hypothesis is operable and that the starter unit specificity of KSs from different NR-PKSs is correlated with crosslinker specificity. We also demonstrate that only ACPs from NR-PKSs, but not from other PKS types tested, can be crosslinked effectively to the KSs. Furthermore, structure-based mutants of KS were prepared and studied to support the second hypothesis. The crosslinking studies between ACP and KS of PksA and Pks4 NR-PKSs elucidate the degree of protein•protein interactions between a matched or mixed pair of KS and ACP. The results identify structural features important for inter-domain interactions that may be useful for future combinatorial biosynthesis efforts among mixed NR-PKS domain pairs.
RESULTS AND DISCUSSION
Chemoenzymatic loading of 15–17 to NR-PKS ACPs
To ensure that the crosslinkers 15–17 (Fig. 2C) can be functionally loaded onto the ACP domain of an NR-PKS, we cloned the stand-alone ACP domains from PksA and Pks4 as strep-tag fusions for affinity purification. The loading of pantetheine analogues 15–17 to ACP was monitored by MALDI, and the loading efficiency for one hour, four hours, or overnight incubations of the NR-PKS ACPs was compared to that of other ACPs; including the ACPs from modular type I PKS DEBS2, the type I FAS mycocerosic acid synthase, the type II E. coli FAS, and the type II actinorhodin PKS (examples of loading as detected by MALDI are shown in Fig. S2–S4). The ACPs were selected to reflect the diversity of ACPs from both type I and II FASs and PKSs, with a sequence identity of 10 – 24% to that of the PksA ACP. The loading efficiency of 15–17 was 100% for all apo ACPs tested within an hour, indicating that under these conditions, the loading of the crosslinker is independent of the affinity tag or the source of ACP (data not shown).
Crosslinking is probe-dependent based on the starter unit selectivity of KS
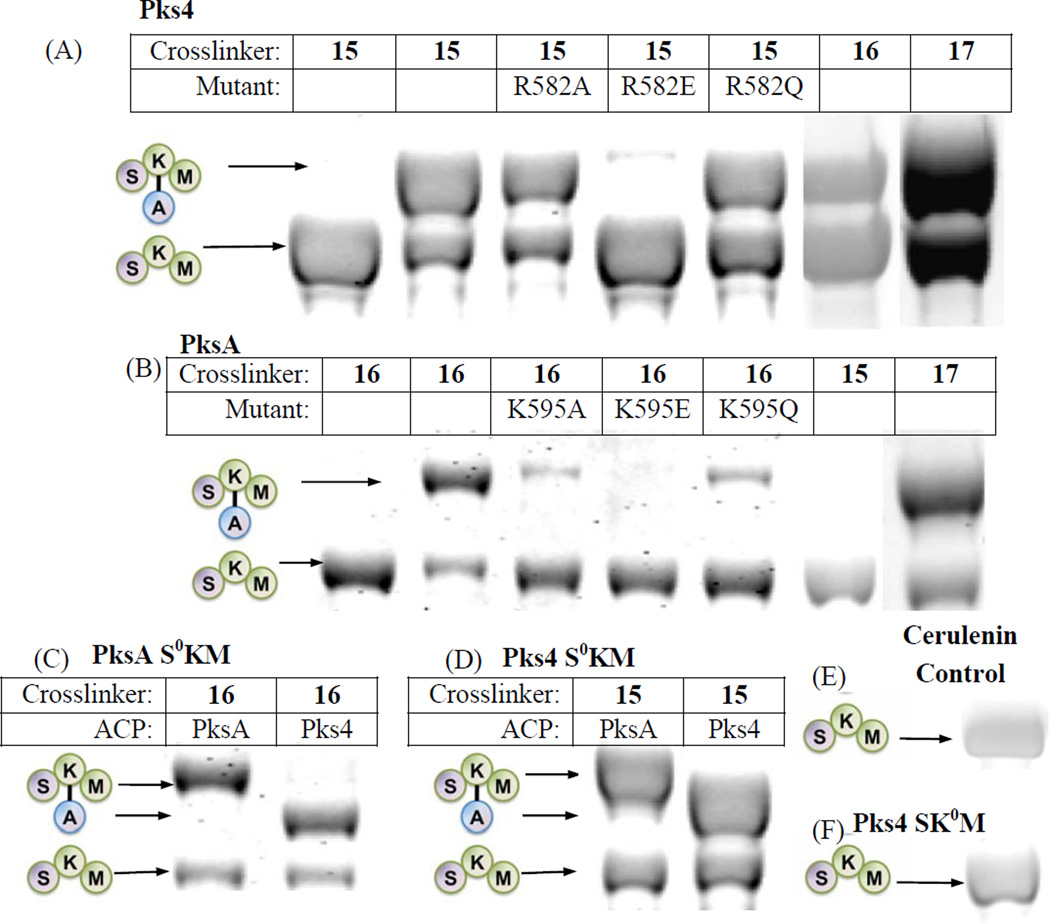
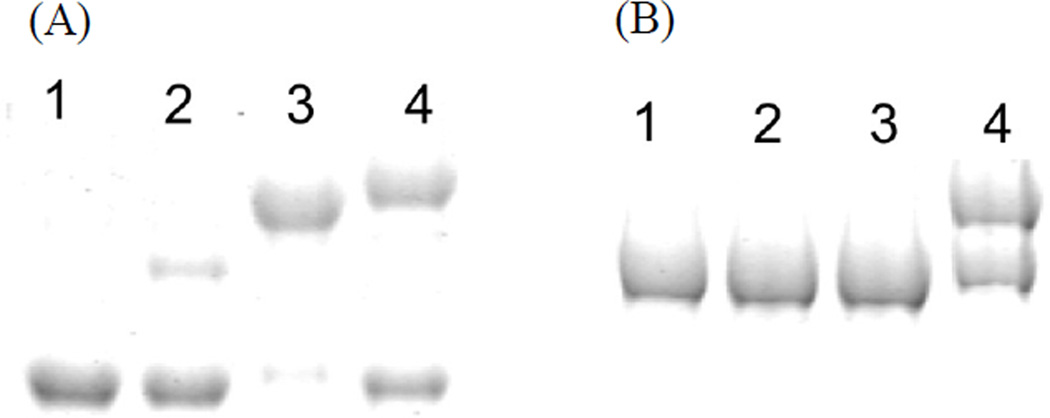
To ensure that the crosslinking between KS and ACP is specific, with no side reactions, we generated the N-terminal tridomain constructs, SAT(C117A)-KS-MAT (referred to as “S0KM” throughout the text) of both PksA and Pks4, where the active site Cys nucleophile of the SAT domain has been inactivated by mutation to Ala, ensuring that SAT would not compete with KS for crosslinking. We used the S0KM tridomain construct due to insoluble expression of the KS monodomain. For the ease of downstream purification, we also cloned the S0KM construct with an N-terminal 6-His tag cleavable by thrombin. Covalent crosslinking between the stand-alone ACP and the KS domain of S0KM was monitored by an SDS-PAGE gel shift assay in which successful crosslinking results in an increase in the protein mass (Fig. 3)(Haushalter et al., 2008; Kapur et al., 2008; Worthington et al., 2010; Worthington et al., 2006). The 2-carbon substrate mimic, chloroacrylic pantetheine (15), crosslinks the ACP to Pks4 S0KM but not to PksA S0KM (Fig. 3A – B). In contrast, the 6-carbon mimic (16) and 16-carbon mimic (17) crosslink the ACP to both Pks4 and PksA S0KM constructs (Fig. 3A–B). Further, when 16 or 17 was used as the crosslinker, the ACP from Pks4 or PksA could be crosslinked to the S0KM of either Pks4 or PksA, demonstrating that crosslinking between noncognate fungal NR-PKS domains occurred with nearly equal efficiency (Fig. 3C–D) under the conditions tested. As a control experiment to confirm that the crosslinker is interacting exclusively with the KS, but not with SAT or MAT, the KS-specific inhibitor cerulenin was pre-incubated with S0KM for an hour on ice before initiating the crosslinking reaction (Fig. 3E). The addition of cerulenin resulted in the abolishment of crosslinking, confirming that the KS active site Cys is essential for crosslinking. Additionally, when using an SKM construct where the KS active site Cys (C543 or C541 for PksA and Pks4, respectively) was mutated to Ala, no crosslinking occured for either construct (Fig. 3F). Therefore, for NR-PKSs, the crosslinking (and thereby protein•protein interactions between ACP and KS) is probe-dependent, but not ACP-dependent.
Figure 3.
see also Figures S1–6. SKM crosslinking gels. (A) Pks4 ACP crosslinked to Pks4 S0KM and KS surface mutants. Pks4 can crosslink by crosslinkers with acyl chains ranging from 2 – 16 carbons (15 – 17). Surface mutations of Pks4 R582 (and K593 in the SI, Fig. S1A) abolished crosslinking when the side chain charge is reversed (R582E), which can be rescued by mutation to a polar amino acid (R582Q). (B) PksA ACP crosslinked to PksA S0KM and additional KS surface mutants. PksA requires a minimum of 6 carbons in the acyl chain of the crosslinkers (16–17). Reversing the charge of K595 (K595E) abolishes crosslinking, which can be rescued by mutation to a polar amino acid (K595Q). Similar results were found for K584 mutants (detailed in the SI, Fig. S1C) When comparing (A) and (B) or PksA vs Pks4, the alanine mutation do not necessarily abolish protein-protein interactions. Rather, the reversal of charge (the mutation to glutamate) effectively abolishes the KS-ACP interactions. The difference is likely due to the distinct character at the protein interface of different PKSs. (C) A comparison of PksA S0KM crosslinked to PksA and Pks4 ACP, showing comparable crosslinking efficiency for the two ACPs from NR-PKSs. (D) A comparison of Pks4 S0KM crosslinked to PksA and Pks4 ACP, showing comparable crosslinking efficiency for the two ACPs from NR-PKSs. (E) The negative control of cerulenin, which binds KS and prevents crosslinking. The result supports that the KS active site must be accessible for crosslinking to occur. (F) As an additional control, the KS0 mutant abolishes crosslinking, supporting that KS active site must be present for crosslinking to occur.
The above observation can be rationalized based on the starter unit selectivity of the KS domains. The PksA KS natively accepts a 6-carbon starter unit for chain elongation (Crawford et al., 2006), but not an acetyl unit, which is consistent with its specificity for 16 (the 6-carbon mimic) but not 15 (the 2-carbon mimic). In comparison, the Pks4 KS accepts a 2-carbon starter unit and subsequently conducts chain elongation until the intermediate reaches 18 carbons (Crawford et al., 2006; Ma et al., 2007). The 2-carbon mimic 15 is thus accepted by the Pks4 KS as the starter unit, while the 6-carbon mimic 16 is likely accepted by the Pks4 KS as a surrogate elongation intermediate. Similarly, the 16-carbon mimic 17 is likely accepted by both PksA and Pks4 KS domains as a pseudo-elongation intermediate. The above result is significant, not only because it offers strong support that both SAT and KS domains (as opposed to only SAT) of PksA are selective for the hexanoyl starter unit, but also that probe specificity directly affects protein•protein interactions (as detected by crosslinking) between KS and ACP.
Crosslinking ratio is correlated with observed PKS activity
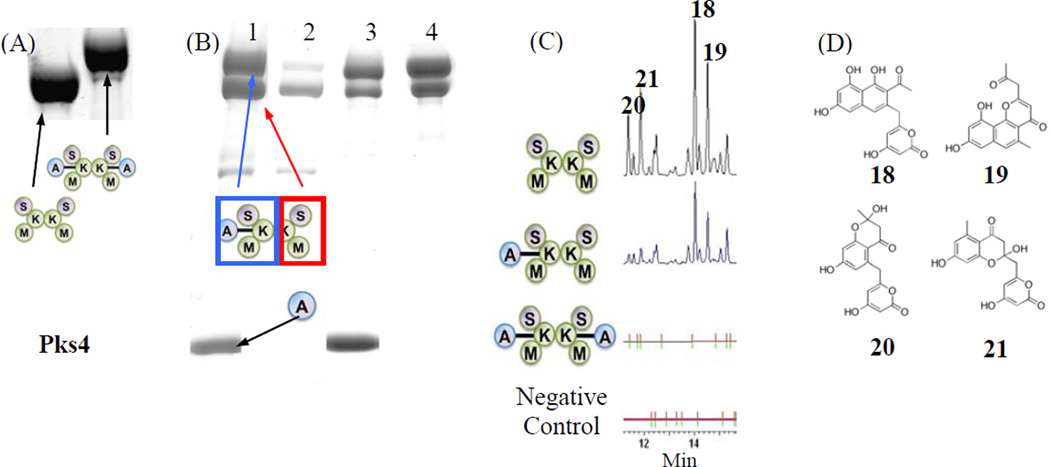
The intact NR-PKS (six domains), as well as the tridomain S0KM construct, exist as dimers in solution (Crawford et al., 2009). For either PksA or Pks4, the extent of crosslinking is dependent on the reaction conditions. Under our standard conditions (detailed in the Methods) with a one hour incubation, only one subunit of the S0KM dimer crosslinks to a single ACP monomer (Fig. 3 and also detected by size exclusion chromotography). The ~2:1 (SKM:ACP) crosslinking ratio persists despite individually lengthening the incubation time, increasing the concentration of S0KM and ACP, or changing the temperature, buffer, or the concentrations of the coupling enzymes CoaA, CoaD, CoaE and Sfp. In contrast—when the crosslinker and ATP concentrations are doubled, ACP concentration is tripled, 10% DMSO is added, and incubation time is extended to overnight—the S0KM dimer is crosslinked to two monomers of ACP (Fig. 4A). The addition of DMSO improves overall protein solubility, while increasing ATP concentration may help increase the crosslinking efficiency, thus increasing the population of the doubly crosslinked species.
Figure 4.
Composite gels. (A) In comparison with Fig 3 (one hour incubation), overnight incubation with doubling concentration of crosslinker and ATP, and triple concentration of ACP resulted in 100% crosslinking. (B) Purification of Strep-tagged Pks4 ACP crosslinked to His-tagged Pks4 S0KM. Lane 1: Crosslinking reaction before purification steps, lane 2: StrepTrap flow-through indicates that excess S0KM purified away from the crosslinked S0KM=ACP, lane 3: StrepTrap elution shows the resulting cross-linked S0KM=ACP and its co-purifying un-crosslinked S0KM, plus contaminating Strep-tagged ACP, lane 4: gel filtration-purified Pks4 S0KM crosslinked to ACP. (C) Pks4 in vitro reconstitution. The major products are PK8 (18), naphthopyrone (19), Sek4 (20) and Sek4b (21). All reactions are incubated with ACP and malonyl-CoA. In addition, reaction 1 contains pure SKM, reaction 2 contains pure, partially-crosslinked SKM to ACP, reaction 3 contains fully crosslinked SKM to ACP, and reaction 4 has no protein present. The result supports that crosslinking abolishes enzyme activity. (D) Chemical structures of 18–21.
To ensure that the observed ~2:1 (SKM:ACP) ratio is not a result of the second KS being blocked by excess unloaded crosslinker or the coupling enzymes, we separated the crosslinker-loaded ACP (referred to as crypto-ACP) by fast protein liquid chromatography (FPLC, data not shown). Addition of purified crypto-ACP to S0KM yielded a similar crosslinking result to the in situ reaction: the S0KM:ACP ratio remains ~2:1 with similar overall efficiency. This control experiment confirms that the ~2:1 ratio is not a result of chemical interference, but rather an inherent property of the ACP•KS complex.
To evaluate the enzymatic activity of the crosslinked S0KM=ACP species (“=” denotes chemical crosslinking throughout the text), we conducted in vitro reconstitution assays using Pks4. S0KM=ACP was generated and isolated by an orthogonal purification scheme (Fig. 4B) (Haushalter et al., 2008). We crosslinked His-tagged Pks4 S0KM to strep-tagged ACP. The reaction mixture was then passed through a StrepTrap FPLC column so that S0KM=ACP and ACP were purified from the excess S0KM and coupling enzymes CoaA, CoaD, CoaE and Sfp (Fig. 4B, lane 2–3). The S0KM=ACP and ACP mixture was then passed through a Superdex 200 size exclusion column to separate ACP (MW = 16 kD) from crosslinked S0KM=ACP (MW = 298 kD as the 2:1 complex). As expected, the purified S0KM=ACP ran as a single band by native-PAGE and analysis under denaturing conditions by SDS-PAGE showed that 50% of the S0KM was the uncrosslinked species, which formed a dimer with crosslinked S0KM and was carried through the purification process (Fig. 4B, lane 4). The above result confirms the 2:1 S0KM:ACP stoichiometry. We then conducted in vitro reconstitution assays to compare the product output of the 100% (2:2 S0KM:ACP) and 50% (2:1 S0KM:ACP) crosslinked S0KM=ACP to an un-crosslinked S0KM + ACP reaction (Fig. 4C–D). The ACP concentration was standardized for each reaction. Because the SAT domain is mutated, starter units were provided by self-acylation of the KS domain under the high substrate conditions (Vagstad et al., 2012). We found that relative to the un-crosslinked reaction, only approximately 20% production (measured by HPLC peak area) was detected for the 50% crosslinked S0KM=ACP, supporting the view that (1) only one monomer of the S0KM dimer is crosslinked to one ACP, and (2) the free S0KM monomer is still capable of interacting with additional ACP in the solution. In comparison, the 100% crosslinked S0KM=ACP completely lost its activity, supporting that both S0KM monomers have been blocked by the crosslinked ACPs.
Why can we only crosslink one ACP to one monomer of the S0KM dimer unless the reaction conditions are pushed? Previously, crystal structures of porcine FAS and yeast FAS showed that while mega-synthases such as PKSs or FASs exist as dimers in solution (Fig. 2A), the two reaction chambers are in fact asymmetric (Maier et al., 2008; Rangan et al., 2001). Namely, as suggested by Smith et al (Rangan et al., 2001), the conformation of one subunit may trigger a corresponding change in its dimeric partner. Therefore, it is possible in NR-PKSs, that crosslinking of one monomeric ACP to one KS within the S0KM dimer triggers a conformational change in the second S0KM, resulting in a slower rate of complex formation with the second ACP. The free S0KM only forms a stable complex with a second ACP after overnight incubation with a high concentration of crosslinker, ATP and ACP. This is also supported by the result where 50% crosslinked S0KM produces just 20% of product output compared to non-crosslinked S0KM during in vitro reconstitution, rather than an expected 50% if no differences in the reactivity of the second monomer existed.
KS mutations identify surface residues important for protein•protein affinity
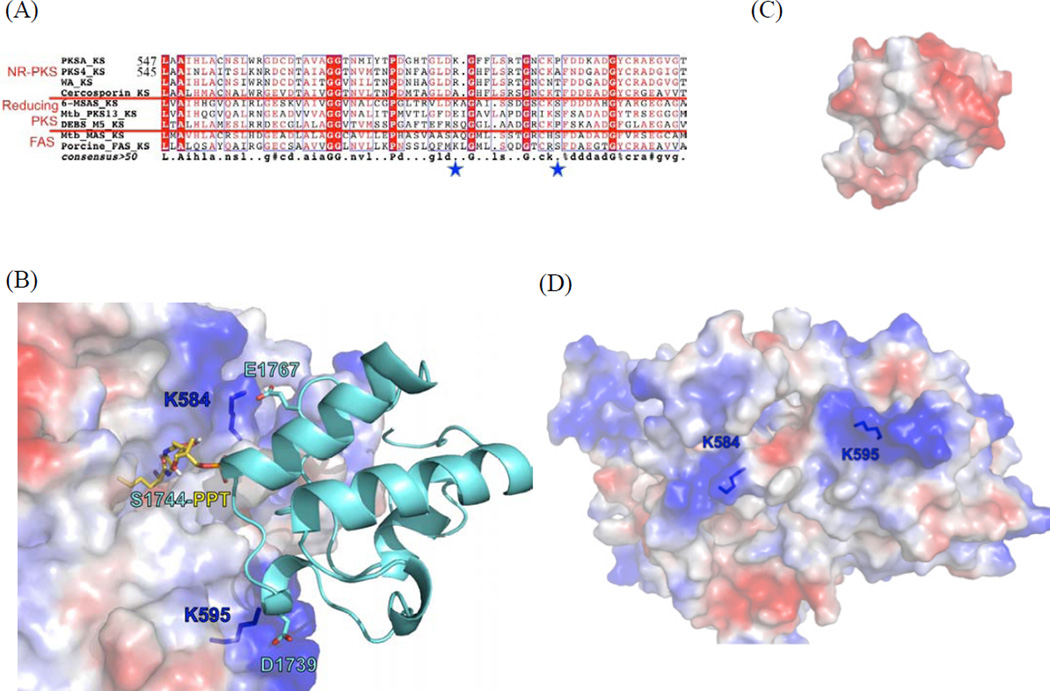
In order to visualize potential protein•protein interactions mediated by surface residues of KS and ACP, we generated homology models of Pks4 and PksA KS domains with Modeller (Fiser and Šali, 2003) using DEBS KS5 as a template (Tang et al., 2006). The sequence identity between the KS domains of Pks4 and PksA is 57%, and the sequence identities between Pks4 KS and PksA A and DEBS KS5 are 31% and 34%, respectively. A >30% sequence identity is typically required for reliable homology modeling by Modeller (Fiser and Šali, 2003). We additionally conducted docking simulations of the 4’-phosphopantetheine analogues 15 and 16 to the KS active site models using the program GOLD (Verdonk et al., 2003). We found that the putative PPT binding pockets of the Pks4 and PksA KSs are highly conserved. Furthermore, when we compared the KS homology models, docking simulations, and sequence alignments (Fig. 5A), we found that the 4’-phosphate of 15 and 16 consistently docked near residue K584 of PksA (corresponding to R582 of Pks4). Docking simulations between the PksA KS model and the PksA ACP structure (Wattana-amorn et al., 2010) further support that the positively-charged surface created by K584 and K595 may be docked with the negatively charged surface of ACP (Fig. 5 B-D). The PksA ACP residues D1739 and E1767 are each situated in close proximity to the KS residues, K595 and K584, respectively, and they may be involved in hydrogen bonding that contributes to ACP•KS interactions. Sequence alignment of NR-PKS ACP’s indicate that D1739 is highly conserved among NR-PKS and the negative charge at residue 1767 is conserved in most NR-PKSs (Fig. S7A). We hypothesize that the specificity between KS and ACP may require both electrostatic interactions between specific residues and overall protein conformation. Both lysines are located at the proposed substrate pocket entrance of KS (Fig. 5D), and residues with positive polarity are relatively conserved across KS domains of different NR-PKSs at these positions (Fig. 5A). The fact that the lysines are conserved among the KSs of NR-PKSs but not other types of FASs and PKSs supports the hypothesis that different types of mega-synthases may have different residues that interact with their cognate ACPs. The above analysis leads to the hypothesis that K584 and K595 of PksA (R582 and K593 of Pks4, respectively) are important for protein•protein association between ACP and KS in NR-PKS.
Figure 5.
see also Figure S7. Probing surface residues important for KS-ACP interactions. (A) Sequence alignment identified two relatively conserved, positively charged residues (star). The boxed residues are conserved ones. (B) Homology modeling and docking simulation of KS domains of PksA with PksA ACP suggests that K584 and K595 of PksA are located at the protein surface, where positively-charged lysines are docked with acid residues of ACP, while active site Ser tethered 2-bromohexyl pantetheine docks into the active site. D1739 and E1767 of PksA ACP were identified based on protein-protein docking simulation (Fig. S8). (C) The negatively-charged ACP surface that is docked to KS. (D) The positively-charged KS surface that is docked to ACP
To evaluate the importance of these surface lysines and arginine on ACP•KS interactions, we generated Ala, Glu, and Gln mutations for residues K584 and K595 of the PksA S0KM and residues R582 and K593 of the Pks4 S0KM. Circular dichroism experiments that compare the mutants to wild type of both PKSA and PKS4 S0KM indicate that the mutations do not significantly change the overall protein fold (Fig. S5–S6). The extent of crosslinking between these mutant S0KMs and ACP was evaluated (Fig. 3). We found that the charge reversing Glu mutations abolished crosslinking. In comparison, the neutral Ala mutants showed significantly diminished crosslinking, while the Gln mutants that preserve polarity maintained crosslinking. According to our model, polar residues at these positions appears to be important, such that a reversal of charge (the Lys or Arg to Glu mutants) abolished the interaction between the negatively charged ACP and the 4’-phosphate of pantetheine with the positively charged KS surface. However, if we maintain the polar character of the KS surface, in the case of the Lys or Arg to Gln mutation, a high degree of protein•protein affinity is maintained, resulting in high crosslinking efficiency. In support of the importance of polar (positively-charged or non-charged) residues at these positions, eliminating the charge (the Lys to Ala mutants) does not eliminate crosslinking, but it severely decreases the crosslinking efficiency for PksA.
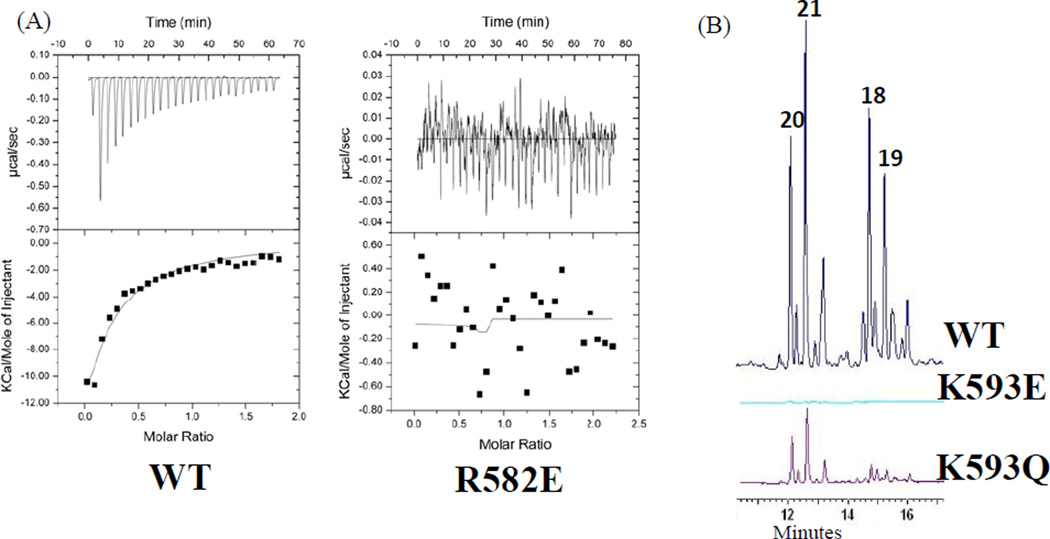
To validate that the observed crosslinking efficiency is, indeed, correlated with domain•domain association between ACP•KS and the PKS activity, we conducted isothermal titration calorimetry (ITC) and product analyses of in vitro reconstitution reactions from wild-type and mutant S0KMs (Fig. 6). By varying the concentration of S0KM and ACP, we found that wild-type Pks4 S0KM has an estimated KD of 15 µM for Pks4 apo-ACP and 30 µM for PksA apo-ACP. In comparison, the Pks4 R582E mutant completely lost protein•protein interactions, and we could not measure its KD (the dissociation constant between Pks4 ACP and S0KM) by ITC (Fig. 6A). Furthermore, the in vitro reconstitution assays with wild-type Pks4 S0KM and Pks4 ACP resulted in the normal shunt products, 18, 19, 20 and 21 (Fig. 6B) (Ma et al., 2007), indicating that chain elongation is functional. In comparison, Pks4 K593E completely lost the ability to produce 18–21 (Fig. 6B). This absence of polyketide extension and the loss of affinity by ITC for a single KS point mutant allow us to deduce that the ACP domain does not measurably bind either the SAT or MAT domains, and the NR-PKS resting state may be with the ACP docked to KS. Such an observation was made in the crystal structure of fungal FAS, where the holo-ACP and PPT were found to be “stalled” in the KS domain (Leibundgut et al., 2007). Alternatively, ACP may interact with SAT and MAT if it were in either the holo or acylated state. A third possibility is that certain conformational changes may need to occur via the holo or acylated form of ACP so that SAT or MAT may be allowed to interact with ACP. The above results offer strong support that the observed difference in crosslinking efficiency between wild-type and mutant S0KM is correlated with the strength of protein•protein interactions, which in turn is crucial for the biosynthesis of full-length polyketides. In addition, these results further reinforce the utility of crosslinking probes to provide a rapid readout for detecting docking interactions.
Figure 6.
ITC and in vitro reconstitution results that compare Pks4 wild type (wild type refers to C117A SAT inactive mutant) with KS surface mutants. (A) ITC results show that interactions between Pks4 S0KM WT and ACP are disrupted with the KS surface mutant R582E. (B) Similarly, in vitro reconstitution shows that the surface mutant K593E disrupts interactions with ACP, whose in vitro activity is rescued by the K593Q mutant.
As further control experiments, in addition to the above mutants, we also generated Pks4 R546A, R546E and R546Q, since R546 is also near the surface of the Pks4 KS active site pocket. We found that these mutants do not change the crosslinking efficiency to the same degree as the other two mutants in Pks4 S0KM (Fig. S1). In summary, the above results confirm the importance of K584 and K595 of PksA (R582 and K593 of Pks4, respectively) to protein•protein interactions between KS and ACP domains, as well as correlating the ACP•KS interactions to polyketide biosynthetic activity.
Implications for directing biosynthesis: the ACP specificity of NR-PKSs
A major goal of contemporary PKS research efforts is to enable combinatorial biosynthesis, the process of mixing different enzyme domains to synthesize new products. To do so, the maintenance of protein•protein interactions between ACP and other catalytic domains is essential (Mercer and Burkart, 2007). Although we showed that ACPs can be mixed with SKMs from different NR-PKSs, it is of significant interest to evaluate if such ACP promiscuity also extends to different types of PKSs and FASs. To more comprehensively evaluate ACP•KS interactions, we incubated ACP domains from a type II PKS (Act ACP) and a FAS (type II E.coli AcpP) with the S0KM of either Pks4 or PksA and a crosslinker, either 15 for Pks4 or in the case of PksA S0KM, 16 (Fig. 7). As a positive control, we also included the matched pair, the E. coli type II ACP (AcpP) and KS (FabF). As expected, both type II ACPs, including the AcpP positive control, crosslink to FabF (Fig. 7A); however, neither Act ACP nor AcpP crosslinks to the S0KM of Pks4 or PksA (Fig. 7B). Sequence alignment of ACP also shows that D1739 is conserved in both type I and II PKS ACPs, while E1767 is only conserved for NR-PKS ACPs. D1739 is located in the loop between helices I and II of ACP, while E1767 is located in helix III, a key region for ACP structural integrity and specificity with its partner proteins (Crosby and Crump, 2012). In the NMR structure by Crump et al (Wattana-amorn et al., 2010), helix III of an NR-PKS ACP is significantly different in orientation relative to ACPs of type II FASs and PKSs. In type II PKS and FAS ACPs, the residue aligned to PksA E1767 is uncharged (Fig. S7A); therefore, both the specific charge-charge interactions and overall shape at the ACP-partner interface may be different between type I and type II ACPs. As Crump et al pointed out (Wattana-amorn et al., 2010), the helix III region of NR-PKS is unusually hydrophobic when compared to ACPs from type II PKSs. Our docking simulation also supports that hydrophobic residues of helix III from PksA ACP (such as F1771 and I1772) may form a hydrophobic interface with F586, F587 and L588 of the KS domain (Fig. S7B). These hydrophobic residues are not conserved in type II PKSs (Fig. S7A). A recent critical review by Crosby and Crump compared ACPs from type I and II FASs and PKSs in details (Crosby and Crump, 2012). The differences of charged residues and overall shape between the ACPs of NR-PKS and type II PKS may help explain how the NR-PKS KS•ACP partners are exchangeable among NR-PKSs but not with type II PKSs (Fig. 7). The above observation is also consistent with the report by Simpson et al that substituting the NR-PKS ACP into type II PKS results in significantly reduced PKS activity (Ma et al., 2006).
Figure 7.
Crosslinking gel with Type II ACP using pantetheine analogue 15. (A) FabF crosslinked to the following ACPs: lane 1: negative control, lane 2: Act ACP, lane 3: AcpP, lane 4: Pks4 ACP. (B) Pks4 S0KM that is crosslinked to the following ACPs: lane 1: negative control, lane 2: Act ACP, lane 3: AcpP, lane 4: Pks4 ACP. The result showed that KS-ACP pair is specific within the similar type of FAS or PKS.
The above result has implications for downstream bioengineering efforts, that NR-PKS ACPs can be mixed-and-matched with enzymes from other NR-PKSs, but not efficiently with type II FAS and PKS components. Previously, the fungal Pks1 and WA ACPs have also been reported to be interchangeable (Watanabe and Ebizuka, 2004). Similarly, Du et al. have reported successful swapping of fungal KS domains for the reducing fungal PKSs (Zhu et al., 2007). These observations are in contrast to the previous report by Simpson and Cox et al that PksA ACP can be mixed with the type II actinorhodin ketosynthase/chain length factor to produce the type II shunt products, albeit with much reduced PKS activity (Ma et al., 2006). One likely explanation for such a difference is the nature of the architectures of type I and type II PKSs; whereas an NR-PKS needs to maintain a comparatively fixed architecture (Fig. 2B) similar to the mammalian FAS (Fig. 2A), a type II PKS consists of 5–10 stand-alone enzymes and does not have a stable organization. Consequently, the loosely-associated type II PKS elongation machinery can accept a fungal ACP, but the organized NR-PKS machinery cannot accept the type II ACP, which presumably does not have a matching protein interface present in PksA and Pks4 ACPs. We conclude that for a successful project of combinatorial biosynthesis, the selection of a competent ACP is key to the maintenance of protein•protein interactions and intermediate transit between ACP and other enzyme domains.
Significance
It is well recognized that protein•protein interactions must be important in transporting the starter unit, stabilizing and sequestering the poly-β-keto intermediate, and releasing the final cyclization product (Crawford and Townsend, 2010). However, only until our recent structural work on the NR-PKS PT (Crawford et al., 2009) and TE (Korman et al., 2010) and other’s work on the ACP (Wattana-amorn et al., 2010) was it possible to appreciate the molecular interactions that result in orderly polyketide biosynthesis in detail. Further, although there are more than 150 NR-PKS-containing gene clusters annotated (Li et al., 2010) and extensive phylogenetic analyses of the NR-PKSs (Kroken et al., 2003), these correlations do not identify residues important for protein•protein interactions essential for orchestrated domain function. Recognizing the need to probe for such interactions in the polyketide mega-synthases, we applied mechanism-based crosslinkers to probe the ACP•KS interactions in NR-PKSs for the first time in this work. We found that ACPs from fungal NR-PKSs can be interchanged, but that their KS domains do not accept ACPs from other types of PKSs or FASs, indicating the need of NR-PKSs to maintain their architecture (Fig. 2B). We further found that the crosslinking efficiency is highly dependent on the chain length of the probe, which reflects the starter unit selectivity of the corresponding NR-PKSs (Fig. 3A–B). Surface mutagenesis, in combination with crosslinking, ITC, and in vitro reconstitution experiments, further identified some residues important for ACP•KS interactions and established a strong correlation between domain docking and PKS activity. These results confirm the importance of protein•protein interaction in the NR-PKS complex. In the long run, knowledge about inter-domain interactions in NR-PKSs can be applied to guide future biosynthetic efforts in the development of new “non-native natural products” that can be screened for new biomedical activities.
EXPERIMENTAL PROCEDURES
Cloning and mutagenesis
Plasmid pENKA-C117A containing the PksA SAT-KS-MAT (S0KM) C117A gene was cloned into pET28a(+), and pEPKS4-NKA-C117A containing the Pks4 S0KM C117A gene was cloned into pET24a(+) (Vagstad et al, 2012). Finnzymes Phasing High-Fidelity DNA Polymerase was used in polymerase chain reaction (PCR) to generate K582A, K582E, K582Q, K593A, K593E and K593Q for pEPKS4-NKA-C117A, and R584A, R584E, R584Q, K595A, K595E and K595Q for pENKA-C117A. E. coli strain NovaBlue (Novae) was used for plasmid amplification. All mutations were confirmed by sequencing. Primers were obtained from Integrated DNA Technologies. Specific primer sequences used are:
PKS4 SKM K582A Forward: 5’- GCCTCGATGCGGGGCATTTCCTTTCCCGTAC
PKS4 SKM K582A Reverse: 5’- ATGCCCCGCATCGAGGCCTGCAAAGTTGTCC
PKS4 SKM K582E Forward: 5’- GCCTCGATGAAGGGCATTTCCTTTCCCGTAC
PKS4 SKM K582E Reverse: 5’- ATGCCCTTCATCGAGGCCTGCAAAGTTGTCC
PKS4 SKM K582Q Forward: 5’- CCTCGATCAGGGGCATTTCCTTTCCCG
PKS4 SKM K582Q Reverse: 5’- GAAATGCCCCTGATCGAGGCCTGCAAAG
PKS4 SKM K593A Forward: 5’- GAAACTGCGCGGCTTTTAACGACGGTGCGG
PKS4 SKM K593A Reverse: 5’- CGTTAAAAGCCGCGCAGTTTCCCGTACGGG
PKS4 SKM K593E Forward: 5’- GGGAAACTGCGAAGCTTTTAACGACGGTGCGG
PKS4 SKM K593E Reverse: 5’- CGTTAAAAGCTTCGCAGTTTCCCGTACGGGAAAGG
PKS4 SKM K593Q Forward: 5’- GAAACTGCCAGGCTTTTAACGACGGTGCG
PKS4 SKM K593Q Reverse: 5’- CGTTAAAAGCCTGGCAGTTTCCCGTACGG
PKSA SKM K584A Forward: 5’- GATTGGACGCGGGGTTCTTTCTTTCCCGGAC
PKSA SKM K584A Reverse: 5’- GAACCCCGCGTCCAATCCTGTGTGACCATC
PKSA SKM K584E Forward: 5’- GATTGGACGAAGGGTTCTTTCTTTCCCGG
PKSA SKM K584E Reverse: 5’- GAACCCTTCGTCCAATCCTGTGTGACC
PKSA SKM K584Q Forward: 5’- GATTGGACCAGGGGTTCTTTCTTTCCCGGAC
PKSA SKM K584Q Reverse: 5’- GAACCCCTGGTCCAATCCTGTGTGACCATC
PKSA SKM K595A Forward: 5’- CAACTGCGCGCCCTACGACGACAAGGCCG
PKSA SKM K595A Reverse: 5’- GTAGGGCGCGCAGTTGCCAGTCCGGGAAAG
PKSA SKM K595E Forward: 5’- GCAACTGCGAACCCTACGACGACAAGGCC
PKSA SKM K595E Reverse: 5’- GTAGGGTTCGCAGTTGCCAGTCCGGGAAAG
PKSA SKM K595Q Forward: 5’- GCAACTGCCAGCCCTACGACGACAAGGCCG
PKSA SKM K595Q Reverse: 5’- CGTCGTAGGGCTGGCAGTTGCCAGTCCGG
PKSA SKM R546A Forward: 5’- CACCGCGCCTCTCGCAGACGCCCAGG
PKSA SKM R546A Reverse: 5’- GCGAGAGGCGCGGTGATGGAGACGGAC
PKSA SKM R546E Forward: 5’- CACCGAACCTCTCGCAGACGCCCAGG
PKSA SKM R546E Reverse: 5’- GCGAGAGGTTCGGTGATGGAGACGGAC
PKSA SKM R546Q Forward: 5’- ACCCAGCCTCTCGCAGACGCCCAGG
PKSA SKM R546Q Reverse: 5’- GCGAGAGGCTGGGTGATGGAGACGGAC
Expression and purification of His-tagged Sfp, FabF, apo- ACP and S0KM mutants
Plasmids pEACP41 and pEPKS4-ACP that contain the PKSA ACP and PKS4 ACP genes, respectively, were cloned into pET28a(+) (Crawford et al., 2006). All plasmids were transformed into BL21 (DE3) cells. Transformed cells were grown by being shaken at 37°C in 2 1-L of Luria-Bertani medium supplemented with 50 µg/mL kanamycin (or 50 µg/mL carbenecillin for Sfp) to an OD600 of 0.4–0.6. Protein expression was induced by the addition of 0.5 mM IPTG at 18°C overnight. The cells were harvested by centrifugation (5000 rpm for 10 min at 4°C) and resuspended in 25 mM Tris-HCl, pH 8.0 at 4°C. The cells were pelleted, flash frozen in liquid nitrogen and stored at −80°C until purification. Frozen cell pellets were defrosted on ice and resuspended in 40 mL lysis buffer (50 mM Tris-HCl, pH 7.5 at room temperature, 300 mM NaCl, 10% glycerol, 20 mM Imidazole) and lysed by sonication (5 × 30 sec pulses). The cell debris was removed by centrifugation (14000 rpm for 1 h at 4°C). Protein was bound to Ni-IMAC resin (Bio-Rad) by batch binding at 4°C for 1 h. The lysis slurry was loaded onto a gravity column where the lysate was eluted followed by 2 × 25-mL washes with lysis buffer. Protein was eluted at 40, 100, 150, 250 and 450 mM imidazole in lysis buffer to at least 95% purity. One mM 1,4-dithiothreitol (DTT) was added as elutions were collected. The purified protein was buffer exchanged overnight in 3-L dialysis buffer at 4°C with a 3 kDa (ACP) or 10 kDa (S0KM and Sfp) Thermo Scientific Snakeskin membrane into 50 mM Tris-HCl, pH 8.0 at 4°C, 50 mM NaCl, 10% glycerol, and 1 mM DTT. The protein samples were concentrated to between 4 and 10 mg/mL as determined by Bradford assay (Bio-Rad), flash frozen in liquid nitrogen and stored at −80°C.
Expression and purification of MBP-tagged E. coli CoA enzymes
The plasmids that contain MBP-tagged E. coli constructs CoaA (PanK), CoaD (PPAT) and CoaE (DPCK) were transformed into BL21 (DE3) cells (Haushalter et al., 2008). Transformed cells were grown by being shaken at 37°C in 2 1-L of Luria-Bertani medium supplemented with 50 µg/mL carbenicillin and 0.2% (w/v) glucose to an OD600 of 0.4–0.6. Protein expression was induced by the addition of 0.5 mM IPTG at 18°C overnight. The cells were harvested by centrifugation (5000 rpm for 10 min at 4°C) and resuspended in 25 mM Tris-HCl, pH 8.0 at 4°C. The cells were pelleted, flash frozen in LiN2 and stored at −80°C until purification. Frozen cell pellets were defrosted on ice and resuspended in 40 mL lysis buffer (50 mM Tris-HCl, pH 7.5 at room temp, 300 mM NaCl, 10% glycerol, 1 mM EDTA and 1 mM DTT) and lysed by sonication (5 × 30 sec pulses). The cell debris was removed by centrifugation (14000 rpm for 1 h at 4°C). Protein was bound to the amylose resin (NEB) by batch binding at 4°C for 1 h. The lysis slurry was loaded onto a gravity column where the lysate was eluted followed by 2 × 25-mL washes with lysis buffer. The protein was eluted at 1, 10 and 100 mM maltose in lysis buffer to at least 95% purity. The purified protein was buffer exchanged overnight in 3-L dialysis buffer at 4°C with a 10 kDa Thermo Scientific Snakeskin membrane into 50 mM Tris-HCl, pH 8.0 at 4°C, 10% glycerol, and 1 mM DTT. Protein samples were concentrated to 3 mg/mL as determined by Bradford assay (Bio-Rad), flash frozen in liquid nitrogen and stored at −80°C.
Expression and purification of Strep(II)-tagged ACP proteins
Plasmids pEACP41-SB and pEPKS4-SB contain the PKSA ACP and PKS4 ACP genes, respectively, cloned into a modified pET-28a construct in which the N-terminal 6×–His tag was replaced by an N-terminal Strep(II) tag. All plasmids were transformed into BL21 (DE3) cells. Transformed cells were grown by being shaken at 37°C in 2 1-L of Luria-Bertani medium supplemented with 50 µg/mL kanamycin to an OD600 of 0.4–0.6. Protein expression was induced by the addition of 0.5 mM IPTG at 18°C overnight. The cells were harvested by centrifugation (5000 rpm for 10 min at 4°C) and resuspended in 25 mM Tris-HCl, pH 8.0 at 4°C. The cells were pelleted, flash frozen in liquid nitrogen and stored at −80°C until purification. Frozen cell pellets defrosted on ice and resuspended in 40 mL lysis buffer (100 mM Tris-HCl, pH 7.5 at RT, 200 mM NaCl, 1 mM DTT and 1 mM EDTA) and lysed by sonication (5 × 30 sec pulses). The cell debris was removed by centrifugation (14000 rpm for 1 h at 4°C), and protein was bound to a 5 mL StrepTrap FF column (GE), washed with 30 to 40 mL lysis buffer and eluted with 2.5 mM d-desthiobiotin (Sigma-Aldrich) in lysis buffer. Column flow through and wash were combined and loaded on the column again, washed and eluted from the StrepTrap column repeatedly until no protein was eluted from the column. The purified protein was buffer-exchanged overnight in 3-L dialysis at 4°C with a 3 kDa Thermo Scientific Snakeskin membrane into 50 mM Tris-HCl, pH 8.0 at 4°C, 50 mM NaCl, 10% glycerol, and 1 mM DTT. Protein samples were concentrated to between 3 and 5 mg/mL as determined by Bradford assay (Bio-Rad), flash frozen in liquid nitrogen and stored at −80°C.
Crosslinking of ACP and S0KM Resulting in Partial Crosslinking
Crosslinking probes 15–17 were prepared as previously described (Worthington 2006, Worthington 2008, Blatti 2012). The loading of pantetheine analogues 15–17 to ACP was accomplished by combining a one-pot chemo-enzyme reaction containing 50 mM Na Phosphate, pH 7.0, 12.5 mM MgCl2, 2 mM ATP, 15 µg/mL each of CoaA (250 nM), CoaD (200 nM), CoaE (300 nM) and Sfp (500 nM), 0.8 µg/µL ACP (50 µM), and 200 µM pantetheine analogue (Haushalter et al., 2008). This reaction was incubated at 37°C for 1 hr. Pantetheine loading results were determined by MALDI-TOF. One µg/µL S0KM (7 µM) was added to the one-pot reaction and incubated an additional 1 hr at 37°C. Calculations were made so that the final volume after S0KM addition was 50 µL. Samples were run on SDS-PAGE using 7.5% Mini-PROTEAN TGX gels (Bio-Rad) to determine the extent of crosslinking.
Purification of Crosslinked S0KM-ACP
Pure strep-tagged ACP and His-tagged S0KM from PksA or Pks4 were crosslinked and purified as follows. A scaled up crosslinking reaction to purify crosslinked protein was accomplished by combining a one-pot chemo-enzyme reaction with 100 mM Na Phosphate, pH 7.0, 12.5 mM MgCl2, 2 mM ATP, 15 µg/mL each of CoaA (250 µM), CoaD (200 µM), CoaE (300 µM) and Sfp (500 µM), 0.8 mg/mL ACP (50 µM), and 200 µM pantetheine analogue (Haushalter et al., 2008). This reaction was incubated at 37°C for 1 h. 1 mg/mL S0KM (7 µM) was added to the one-pot reaction and incubated an additional 1 h at 37°C. Calculations were made so the final volume after S0KM was added was between 5 and 20 mL. Protein was bound to a 5 mL StrepTrap FF column (GE), washed with 30 to 40 mL lysis buffer and eluted with 2.5 mM d-desthiobiotin (Sigma-Aldrich) in lysis buffer. The StrepTrap column separated crosslinked S0KM=ACP and un-crosslinked ACP from un-crosslinked S0KM and chemo-enzymes. The fractions were combined and concentrated to 10 mg/mL as determined by Bradford assay. The crosslinked S0KM=ACP was separated from un-crosslinked ACP by gel filtration (Superdex 200, GE) in 25 mM Tris-HCl, pH 8.0 at 4°C, and 1 mM DTT, and concentrated to 5 mg/mL, where it was flash frozen using liquid nitrogen and stored at −80°C.
Crosslinking of ACP and S0KM Resulting in Complete Crosslinking
Crosslinking that resulted in 100% crosslinking utilized the same conditions as partial crosslinking with the following exceptions: 4 mM ATP, 2.5 µg/µL ACP (150 µM) and 500 µM pantetheine analogue were used instead of the concentrations listed above. In addition, a final concentration of 10% DMSO was present during the reaction and the reaction was carried out overnight at 37°C once S0KM was added to the solution.
In vitro reconstitution
The in vitro reconstitution assays were prepared with 100 mM Tris-HCl, pH 7.5 at room temp, 20 mM ATP (Sigma-Aldrich), 5 mM MgCl2, 20 uM MatB, 5 mM CoA (Sigma-Aldrich), 50 µM holo PKS4 ACP, 10 µM PKS4 S0KM or crosslinked protein as determined by Bradford assay, and 100 mM Na Malonate (Fisher), with a total final volume of 250 µL. The reaction tubes were left overnight in the dark. 300 µL extraction solution (94:5:1 Ethyl Acetate Methanol:Acetic Acid) was added to the reactions, vortexed and centrifuged for 3 min at 14k rpm. The extraction layer was transferred to a fresh tube where it was speed-vacuumed until dry. 100 µL DMSO was added to each tube which was stored at −20°C until analysis. Product analysis was performed by reverse phase HPLC on a C18 analytical column (Phenomenex, 4 µm, 150 mm × 4.6 mm); 20 µL of sample was loaded and separated with a 5 to 95% acetonitrile/0.1% formic acid gradient over 20 min with absorbance monitored by a UV-vis diode array detector tuned to 280 nm.
Docking in silico
PKS4 and PKSA KS homology models were generated using DEBS KS5 (PDB ID code 2HG4) from amino acids P40 to D457 as a template with the program Modeller (Fiser and Šali, 2003). GOLD was used to dock compounds to KS homology models (Verdonk et al., 2003).
Isothermal Calorimetry
Pure apo ACP and S0KM or the S0KM KS surface mutant proteins were buffer exchanged into the ITC buffer 50 mM Tris-HCl, 10% glycerol and 50 mM NaCl with a PD-10 buffer exchange column (GE Healthcare). The ACP protein was diluted to 200 µM and S0KM or the S0KM KS surface mutants were diluted to 20 µM in ITC buffer. The ITC sample chamber was loaded with S0KM or the KS surface mutants and the sample syringe was loaded with ACP. The ITC was run with 25–10 µL injections of ACP with 150 seconds between injections. The Kd were calculated by VPViewer 2000 (Micro Cal Inc).
Circular Dichroism
Samples for circular dichroism (CD) experiments were prepared by diluting protein to 0.05–0.1 mg/mL in 20 mM Tris-HCl, pH 7.5, and the CD data were collected using a Jasco J810 CD spectro-polarimeter.
Supplementary Material
HIGHLIGHTS.
An activity-based crosslinker successfully detects PKS inter-domain interactions
The crosslinking efficiency is correlated with starter unit specificity of KSs
The ACPs and KSs from NR-PKSs are interchangeable for crosslinking
Mutations identify KS surface residues important for ACP•KS interactions
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Cox RJ. Polyketides, proteins and genes in fungi: programmed nano-machines begin to reveal their secrets. Org Biomol Chem. 2007;5:2010–2026. doi: 10.1039/b704420h. [DOI] [PubMed] [Google Scholar]
- Crawford JM, Dancy BCR, Hill EA, Udwary DW, Townsend CA. Identification of a starter unit acyl-carrier protein transacylase domain in an iterative type I polyketide synthase. Proc Natl Acad Sci USA. 2006;103:16728–16733. doi: 10.1073/pnas.0604112103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford JM, Korman TP, Labonte JW, Vagstad AL, Hill EA, Kamari-Bidkorpeh O, Tsai SC, Townsend CA. Structural basis for biosynthetic programming of fungal aromatic polyketide cyclization. Nature. 2009;461:1139–1143. doi: 10.1038/nature08475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford JM, Thomas PM, Scheerer JR, Vagstad AL, Kelleher NL, Townsend CA. Deconstruction of iterative multidomain polyketide synthase function. Science. 2008;320:243–246. doi: 10.1126/science.1154711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford JM, Townsend CA. New insights into the formation of fungal aromatic polyketides. Nat Rev Microbiol. 2010;8:879–889. doi: 10.1038/nrmicro2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crosby J, Crump MP. The structural role of the carrier protein--active controller or passive carrier. Nat Prod Rep. 2012;29:1111–1137. doi: 10.1039/c2np20062g. [DOI] [PubMed] [Google Scholar]
- Fiser A, Šali A. Modeller: Generation and refinement of homology-based protein structure models. Method Enzymol. 2003;374:461–491. doi: 10.1016/S0076-6879(03)74020-8. [DOI] [PubMed] [Google Scholar]
- Haushalter RW, Worthington AS, Hur GH, Burkart MD. An orthogonal purification strategy for isolating crosslinked domains of modular synthases. Bioorg Med Chem Lett. 2008;18:3039–3042. doi: 10.1016/j.bmcl.2008.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopwood D. Genetic Contributions to Understanding Polyketide Synthases. Chemical Reviews. 1997;97:2465–2497. doi: 10.1021/cr960034i. [DOI] [PubMed] [Google Scholar]
- Huffman J, Gerber R, Du L. Recent advancements in the biosynthetic mechanisms for polyketide-derived mycotoxins. Biopolymers. 2010;93:764–776. doi: 10.1002/bip.21483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapur S, Worthington A, Tang Y, Cane D, Burkart M, Khosla C. Mechanism based protein crosslinking of domains from the 6-deoxyerythronolide B synthase. Bioorg Med Chem Lett. 2008;18:3034–3038. doi: 10.1016/j.bmcl.2008.01.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YT, Lee YR, Jin J, Han KH, Kim H, Kim JC, Lee T, Yun SH, Lee YW. Two different polyketide synthase genes are required for synthesis of zearalenone in Gibberella zeae. Mol Microbiol. 2005;58:1102–1113. doi: 10.1111/j.1365-2958.2005.04884.x. [DOI] [PubMed] [Google Scholar]
- Korman TP, Crawford JM, Labonte JW, Newman AG, Wong J, Townsend CA, Tsai SC. Structure and function of an iterative polyketide synthase thioesterase domain catalyzing Claisen cyclization in aflatoxin biosynthesis. Proc Natl Acad Sci U S A. 2010;107:6246–6251. doi: 10.1073/pnas.0913531107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroken S, Glass NL, Taylor JW, Yoder OC, Turgeon BG. Phylogenomic analysis of type I polyketide synthase genes in pathogenic and saprobic ascomycetes. Proc Natl Acad Sci U S A. 2003;100:15670–15675. doi: 10.1073/pnas.2532165100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leibundgut M, Jenni S, Frick C, Ban N. Structural basis for substrate delivery by acyl carrier protein in the yeast fatty acid synthase. Science. 2007;316:288–290. doi: 10.1126/science.1138249. [DOI] [PubMed] [Google Scholar]
- Li Y, Image II, Xu W, Image I, Tang Y. Classification, prediction, and verification of the regioselectivity of fungal polyketide synthase product template domains. The Journal of Biological Chemistry. 2010;285:22764–22773. doi: 10.1074/jbc.M110.128504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linnemannstons P, Schulte J, del Mar Prado M, Proctor RH, Avalos J, Tudzynski B. The polyketide synthase gene pks4 from Gibberella fujikuroi encodes a key enzyme in the biosynthesis of the red pigment bikaverin. Fungal Genet Biol. 2002;37:134–148. doi: 10.1016/s1087-1845(02)00501-7. [DOI] [PubMed] [Google Scholar]
- Ma SM, Zhan J, Watanabe K, Xie X, Zhang W, Wang CC, Tang Y. Enzymatic synthesis of aromatic polyketides using PKS4 from Gibberella fujikuroi. Journal of the American Chemical Society. 2007;129:10642–10643. doi: 10.1021/ja074865p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y, Smith LH, Cox RJ, Beltran-Alvarez P, Arthur CJ, Simpson FRST. Catalytic relationships between type I and type II iterative polyketide synthases: The Aspergillus parasiticus norsolorinic acid synthase. Chembiochem. 2006;7:1951–1958. doi: 10.1002/cbic.200600341. [DOI] [PubMed] [Google Scholar]
- Maier T, Leibundgut M, Ban N. The crystal structure of a mammalian fatty acid synthase. Science. 2008:1315–1322. doi: 10.1126/science.1161269. [DOI] [PubMed] [Google Scholar]
- Meier JL, Haushalter RW, Burkart MD. A mechanism based protein crosslinker for acyl carrier protein dehydratases. Bioorg Med Chem Lett. 2010:4936–4939. doi: 10.1016/j.bmcl.2010.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier JL, Mercer AC, Rivera H, Burkart MD. Synthesis and evaluation of bioorthogonal pantetheine analogues for in vivo protein modification. Journal of the American Chemical Society. 2006:12174–12184. doi: 10.1021/ja063217n. [DOI] [PubMed] [Google Scholar]
- Mercer AC, Burkart MD. The ubiquitous carrier protein--a window to metabolite biosynthesis. Nat Prod Rep. 2007;24:750–773. doi: 10.1039/b603921a. [DOI] [PubMed] [Google Scholar]
- Otten SL, Stutzman-Engwall KJ, Hutchinson CR. Cloning and expression of daunorubicin biosynthesis genes from Streptomyces peucetius and S. peucetius subsp. caesius. J Bacteriol. 1990;172:3427–3434. doi: 10.1128/jb.172.6.3427-3434.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlman D, Heuser LJ, Dutcher JD, Barrett JM, Boska JA. Biosynthesis of tetracycline by 5-hydroxy-tetracycline-producing cultures of Streptomyces rimosus. J Bacteriol. 1960;80:419–420. doi: 10.1128/jb.80.3.419-420.1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petkovic H, Hunter I, P R. Engineering of polyketide synthases: how close are we to reality? Microbiol Immunol Hung. 2002;49:493–500. doi: 10.1556/AMicr.49.2002.4.7. [DOI] [PubMed] [Google Scholar]
- Pickens LB, Tang Y. Decoding and engineering tetracycline biosynthesis. Metab Eng. 2009;11:69–75. doi: 10.1016/j.ymben.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangan VS, Joshi AK, Smith S. Mapping the functional topology of the animal fatty acid synthase by mutant complementation in vitro. Biochemistry. 2001;40:10792–10799. doi: 10.1021/bi015535z. [DOI] [PubMed] [Google Scholar]
- Rhodes A, Boothroyd B, Mc GP, Somerfield GA. Biosynthesis of griseofulvin: the methylated benzophenone intermediates. Biochem J. 1961;81:28–37. doi: 10.1042/bj0810028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen B. Polyketide biosynthesis beyond the type I, II and III polyketide synthase paradigms. Curr Opin Chem Biol. 2003;7:285–295. doi: 10.1016/s1367-5931(03)00020-6. [DOI] [PubMed] [Google Scholar]
- Smith S, Stern A, Randhawa ZI, Knudsen J. Mammalian fatty acid synthetase is a structurally and functionally symmetrical dimer. FEBS. 1985;152:547–555. doi: 10.1111/j.1432-1033.1985.tb09230.x. [DOI] [PubMed] [Google Scholar]
- Tang Y, Kim CY, Mathews II, Cane DE, Khosla C. The 2.7-Angstrom crystal structure of a 194-kDa homodimeric fragment of the 6-deoxyerythronolide B synthase. Proc Natl Acad Sci U S A. 2006;103:11124–11129. doi: 10.1073/pnas.0601924103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagstad AL, Bumpus SB, Belecki K, Kelleher NL, Townsend CA. Interrogation of global active site occupancy of a fungal iterative polyketide synthase reveals strategies for maintaining biosynthetic fidelity. Journal of the American Chemical Society. 2012;134:6865–6877. doi: 10.1021/ja3016389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdonk ML, Cole JC, Hartshorn MJ, Murray CW, Taylor RD. Improved protein-ligand docking using GOLD. Proteins. 2003;52:609–623. doi: 10.1002/prot.10465. [DOI] [PubMed] [Google Scholar]
- Watanabe A, Ebizuka Y. Unprecedented mechanism of chain length determination in fungal aromatic polyketide synthases. Chem Biol. 2004;11:1101–1106. doi: 10.1016/j.chembiol.2004.05.015. [DOI] [PubMed] [Google Scholar]
- Watanabe CM, Wilson D, Linz JE, Townsend CA. Demonstration of the catalytic roles and evidence for the physical association of type I fatty acid synthases and a polyketide synthase in the biosynthesis of aflatoxin B1. Chem Biol. 1996;3:463–469. doi: 10.1016/s1074-5521(96)90094-0. [DOI] [PubMed] [Google Scholar]
- Wattana-amorn P, Williams C, Ploskon E, Cox RJ, Simpson TJ, Crosby J, Crump MP. Solution structure of an acyl carrier protein domain from a fungal type I polyketide synthase. Biochemistry. 2010;49:2186–2193. doi: 10.1021/bi902176v. [DOI] [PubMed] [Google Scholar]
- Witkowski A, Joshi AK, Smith S. Characterization of the beta-carbon processing reactions of the mammalian cytosolic fatty acid synthase: role of the central core. Biochemistry. 2004;43:10458–10466. doi: 10.1021/bi048988n. [DOI] [PubMed] [Google Scholar]
- Worthington AS, Burkart MD. One-pot chemo-enzymatic synthesis of reporter-modified proteins. Org Biomol Chem. 2006;4:44–46. doi: 10.1039/b512735a. [DOI] [PubMed] [Google Scholar]
- Worthington AS, Hur GH, Meier JL, Cheng Q, Moore BS, Burkart MD. Probing the compatibility of type II ketosynthase-carrier protein partners. Chembiochem. 2008;9:2096–2103. doi: 10.1002/cbic.200800198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worthington AS, Porter DF, Burkart MD. Mechanism-based crosslinking as a gauge for functional interaction of modular synthases. Org Biomol Chem. 2010;8:1769–1772. doi: 10.1039/b925966j. [DOI] [PubMed] [Google Scholar]
- Worthington AS, Rivera H, Torpey JW, Alexander MD, Burkart MD. Mechanism-based protein cross-linking probes to investigate carrier protein-mediated biosynthesis. ACS Chemical Biology. 2006;1:687–691. doi: 10.1021/cb6003965. [DOI] [PubMed] [Google Scholar]
- Zhou H, Li Y, Tang Y. Cyclization of aromatic polyketides from bacteria and fungi. Nat Prod Rep. 2010;27:839–868. doi: 10.1039/b911518h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X, Yu F, Li XC, Du L. Production of dihydroisocoumarins in Fusarium verticillioides by swapping ketosynthase domain of the fungal iterative polyketide synthase Fum1p with that of lovastatin diketide synthase. Journal of the American Chemical Society. 2007;129:36–37. doi: 10.1021/ja0672122. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.