Abstract
Rice stripe virus (RSV) is the type member of the genus Tenuivirus. RSV is known to have four segmented, single-stranded RNA molecules and causes rice stripe disease in the rice fields of China, Japan, and Korea. Based on the complete genomic sequences of the determined 6 RSV isolates (from Yunnan, Jiangsu, Zhejiang, and Liaoning Provinces, China) and 27 other RSV isolates (from Yunnan, Jiangsu, Anhui, Henan, and Shandong Provinces of China, also Japan and Korea) downloaded from GenBank, we provided a genotyping profile of RSV field isolates and described the population structure of RSV. All RSV isolates, except isolate CX, could be divided into two subtypes, one including 6 isolates from Yunnan Province, and the other including 26 isolates from different parts of China, Japan, and Korea, which were referred to as subtype II and subtype I, respectively. The amino acid distances between subtypes range from 0.053 to 0.085. RSV isolates in Yunnan Province were genetically differentiated from other parts of China, Japan, and Korea and showed infrequent gene flow. The RSV populations collected from other parts of China, Japan, and Korea were only composed of subtype I and showed very low genetic diversity. We speculated that isolate CX may be the result of recombination of isolates from two subtypes. Two potential recombination events were detected in RNA4 of isolate CX.
Keywords: Rice stripe virus, Genetic variability, Genetic evolution
1. Introduction
RNA viruses have high mutation rates, mainly due to their error-prone replication and fast rates of genome replication (Domingo and Holland, 1997; Roossinck, 1997; Elena and Sanjuán, 2007; Lauring and Andino, 2010; Sanjuán et al., 2010). Thus in natural populations of RNA viruses, nucleotide variations commonly exist, leading to changes of virus host range, disease symptoms, and emergence of new viruses in nature. Analyzing the polymorphic pattern and selection pressure of these variations will help us to understand the phylogenetic relationships, epidemiological routes, population structures, and underlying evolutionary mechanisms of RNA viruses, and facilitate the development of effective control strategies for plant viral diseases.
Rice stripe virus (RSV) is one of the most damaging rice pathogens in China. It was first identified in a rice field in China in 1963 and has now been identified in 16 provinces of China, causing significant yield losses (Wei et al., 2009; Xiong et al., 2009). RSV is the type member of the genus Tenuivirus. It mainly infects rice and a few other species in the family Poaceae including maize, oats, and wheat. RSV is transmitted efficiently by the small brown planthopper (Laodelphax striatellus) in a persistent, circulative-propagative manner; it can propagate in the insect vector and is transmitted to its progenies by eggs (Koganezawa, 1975; Falk and Tsai, 1998).
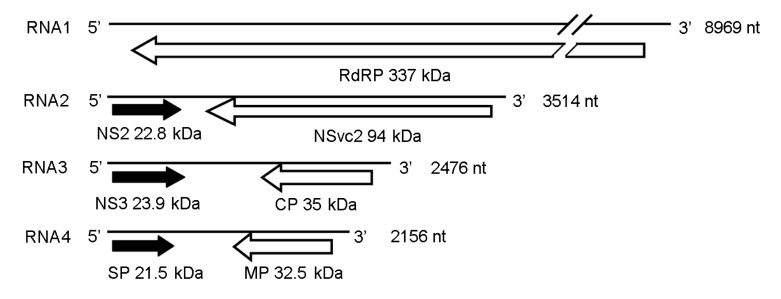
The genome of RSV is composed of four single-stranded RNAs, designated as RNA1, RNA2, RNA3, and RNA4, in the order of decreasing sizes (Fig. 1) (Zhu et al., 1991; 1992; Takahashi et al., 1993; Toriyama et al., 1994). RNA1 is negative sense and encodes a putative protein of approximately 337 kDa, which is probably the RNA-dependent RNA polymerase (RdRP) (Toriyama et al., 1994). The other three RNA segments of RSV are all ambisense. RNA2 encodes a membrane-associated protein of 22.8 kDa (NS2) from the viral sense RNA (vRNA) and a polyglycoprotein of 94.0 kDa (NSvc2) from the viral complementary sense RNA (vcRNA) (Ramirez and Haenni, 1994). RNA3 encodes a 23.9-kDa RNA silencing suppressor (NS3) from the vRNA (Xiong et al., 2009) and a nucleocapsid protein of 35.0 kDa (coat protein, CP) from the vcRNA (Kakutani et al., 1991; Zhu et al., 1991). RNA4 encodes a 21.5 kDa protein involved in disease symptom development from the vRNA (Toriyama, 1986) and a 32.5-kDa movement protein from the vcRNA (Xiong et al., 2008).
Fig. 1.
Genome organization of RSV
Open reading frames (ORFs) were indicated as black arrows on the viral sense RNAs and white arrows on the viral complementary sense RNAs; direction of arrowheads indicated the direction of translation. RdRP: RNA-dependent RNA polymerase; CP: coat protein; SP: disease-specific protein; MP: movement protein; NS2 and NS3: non-structural proteins on the viral sense RNA2 and RNA3; NSvc2: non-structural protein on the viral complementary sense RNA2
Aetiology, pathogenesis, ecology, molecular biology, and control strategies of RSV have been studied in recent decades (Falk and Tsai, 1998), and the genetic diversity and population structure of RSV have been studied based on the five gene sequences of RSV, but not on the whole genomic level (Wei et al., 2009). In this study, the sequences of 6 RSV isolates collected from rice fields in Liaoning, Jiangsu, Zhejiang, and Yunnan Provinces of China were determined. The genetic diversity and population structure of the 6 RSV isolates and other isolates with full sequences deposited in GenBank were analyzed. Our data showed that RSV isolates collected from China, Japan and Korea could be divided into two subtypes. All the subtype II isolates were collected from Yunnan Province of China, whereas subtype I isolates were from different parts of China, Japan, and Korea.
2. Materials and methods
2.1. Virus isolates
Rice infected plants with typical RSV symptoms were collected from rice fields in Liaoning, Jiangsu, Zhejiang, and Yunnan Provinces of China. RSV infection of each rice plant was confirmed by reverse transcription-polymerase chain reaction (RT-PCR) using RSV-specific primers and enzyme-linked immunosorbent assay (ELISA) using specific antibodies as previously described (Wang et al., 2004). A virus sample from an individual rice plant was considered as one isolate.
2.2. RT-PCR cloning and genomic sequencing
Total RNA of each rice plant was extracted from leaf tissues using a TRIzol reagent according to the manufacturer’s instructions (Invitrogen, Carlsbad, CA, USA). RT-PCR reactions were performed using SuperScript III reverse transcriptase (Invitrogen, Carlsbad, CA, USA) and Pfusion High-Fidelity DNA polymerase (New England Biolabs, Ipswich, MA, USA) with specific primer pairs: 5′-ACACAAAGTCCAGAGGAAAACAA-3′ and 5′-ACACATAGTCAGAGGAAAAA-3′ for RNA1; 5′-ACACAAAGTCCTGGGTATATAAGC-3′ and 5′-ACACAAAGTCTGGGTATAACTTCTT-3′ for RNA2; 5′-ACACAAAGTCCTGGGTAAAATAG-3′ and 5′-ACACAAAGTCTGGGTAATAAAAT-3′ for RNA3; 5′-ACACAAAGTCCAGGGCATTTGT-3′ and 5′-ACACAAAGTCAGGGCATATCTT-3′ for RNA4. RT-PCR products were purified by using an AxyRep PCR cleanup kit (Axygen, Union, CA, USA). The purified cDNAs were added with 3′-overhang adenine using Taq plus DNA polymerase, inserted into pMD18-T vector (TaKaRa Biotechnology, Dalian, China), and transformed into Escherichia coli DH5α. Nucleotide sequencing was performed by the Invitrogen Shanghai Sequencing Department.
2.3. Phylogenetic analysis
Neighbor-Joining (NJ) method (Saitou and Nei, 1987) was used to infer the evolutionary history of RSV. Complete genomes, noncoding sequences, and coding sequences of RSV were used as the database for phylogenetic analysis conducted in MEGA5 (Tamura et al., 2011), respectively. The maximum composite likelihood method (Tamura et al., 2004) was used to compute the evolutionary distances. The tree stability was examined by a bootstrap test of 1 000 replicates (Felsenstein, 1985).
2.4. Estimation of genetic distance
The average number of nucleotide substitutions between two randomly selected sequences in a population was referred to as genetic distance. Genetic distances of RSV isolates within and between subtypes were calculated by MEGA5 (Tamura et al., 2011). Analyses were conducted using the maximum composite likelihood model (Tamura et al., 2004).
2.5. Estimation of selection pressure
d n (the number of nonsynonymous substitutions per site) and d s (the number of synonymous substitutions per site) are used to estimate selection pressure. The P values of models of d n≠d s, d n>d s, and d n<d s were estimated separately. Values of P less than 0.05 were considered significant at the 5% level. The variance of the difference was computed using the bootstrap method (500 replicates). Analyses were conducted using the Nei-Gojobori method (Nei and Gojobori, 1986). Evolutionary analyses were conducted in MEGA5 (Tamura et al., 2011).
2.6. Genetic differentiation and gene flow
Two permutation-based statistical tests, Ks* and Z* (Hudson et al., 1992; Hudson, 2000), were used to measure genetic differentiation between RSV population, and Fst (the interpopulational component of genetic variation or the standardized variance in allele frequencies across populations) was used to measure gene flow level. The statistical tests for them were performed by DnaSP 5.0 (Librado and Rozas, 2009). Fst value <0.33 suggests frequent gene flow, while Fst value >0.33 suggests infrequent gene flow.
2.7. Recombination analysis
The recombination analysis was performed by Recombination Detection Program (RDP) 3.44, (Martin et al., 2010). The localization of potential recombination breakpoints, the possible recombination sequences, and the likely parental sequences were analyzed as previously described (Martin et al., 2010), except that the Bonferroni-corrected P value was adjusted from 0.05 to 0.01.
3. Results
3.1. Genotype profile of RSV field isolates
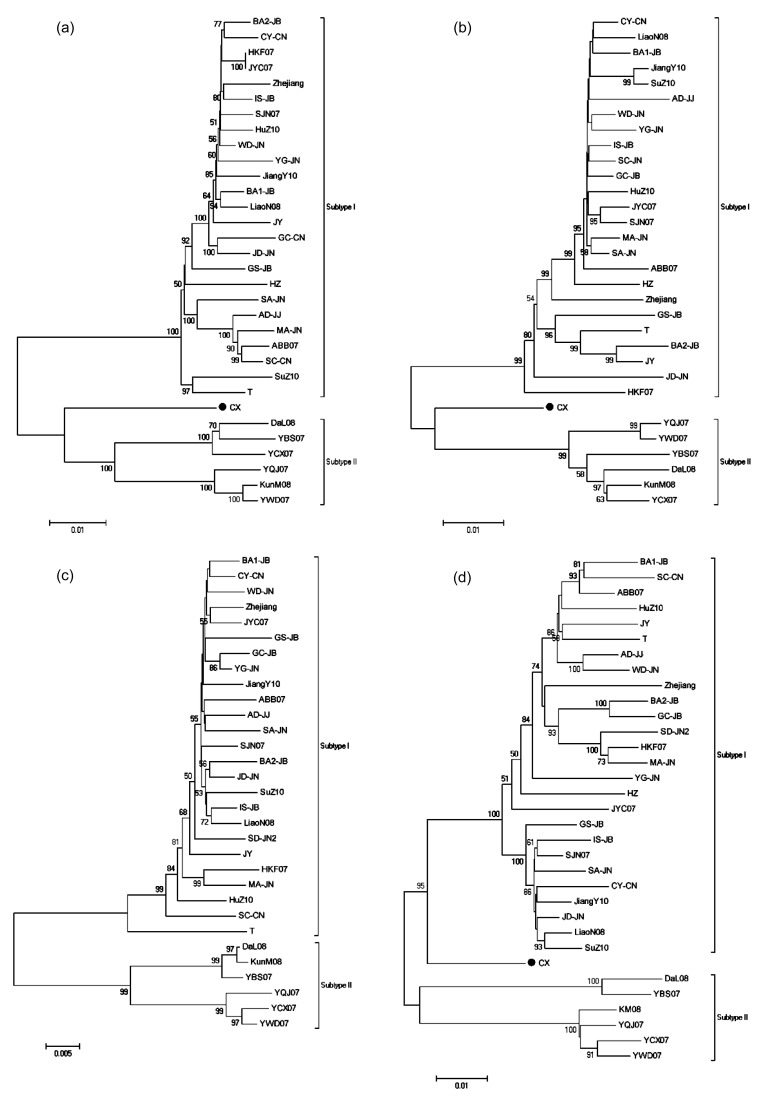
Six RSV isolates were collected from six different rice fields in four provinces of China, and the whole genomes of them were determined (Table 1). Twenty-seven isolates with full genomic sequences of more than two genomic RNAs were downloaded from GenBank (http://www.ncbi.nlm.nih.gov/) (Table 1). Note that isolates GC-JB, SC-CN, HZ, SD-JN2, and CX had one or two RNA sequences undetermined. Phylogenetic analyses of these 33 RSV isolates were performed. NJ trees were constructed by using datasets for four RSV genomic RNAs. It suggested that RSV field isolates fell into two clades except isolate CX (marked with ● in phylogenetic trees). All isolates collected from eastern China, Japan, and Korea formed one of the monophyletic clades, while all isolates collected from Yunnan Province except isolate CX formed the other, referred as subtypes I and II, respectively (Fig. 2). Isolate CX was almost equally distant from the two subtypes (Fig. 2).
Table 1.
NCBI accessions and collected locations of RSV field isolates
Six isolates sequenced by this work
Fig. 2.
Phylogenetic analyses of four RSV genomic RNAs conducted in MEGA5
(a) RNA1; (b) RNA2; (c) RNA3; (d) RNA4. Neighbor-Joining (NJ) method was used to infer the evolutionary history. The bootstrap consensus tree inferred from 1 000 replicates was taken to represent the evolutionary history of the taxa analyzed (Felsenstein, 1985). The number next to the branches represented the percentages of replicate trees, in which the associated taxa clustered together in the bootstrap test (1 000 replicates). The branch lengths were drawn corresponding to the evolutionary distances, which were computed using the maximum composite likelihood method
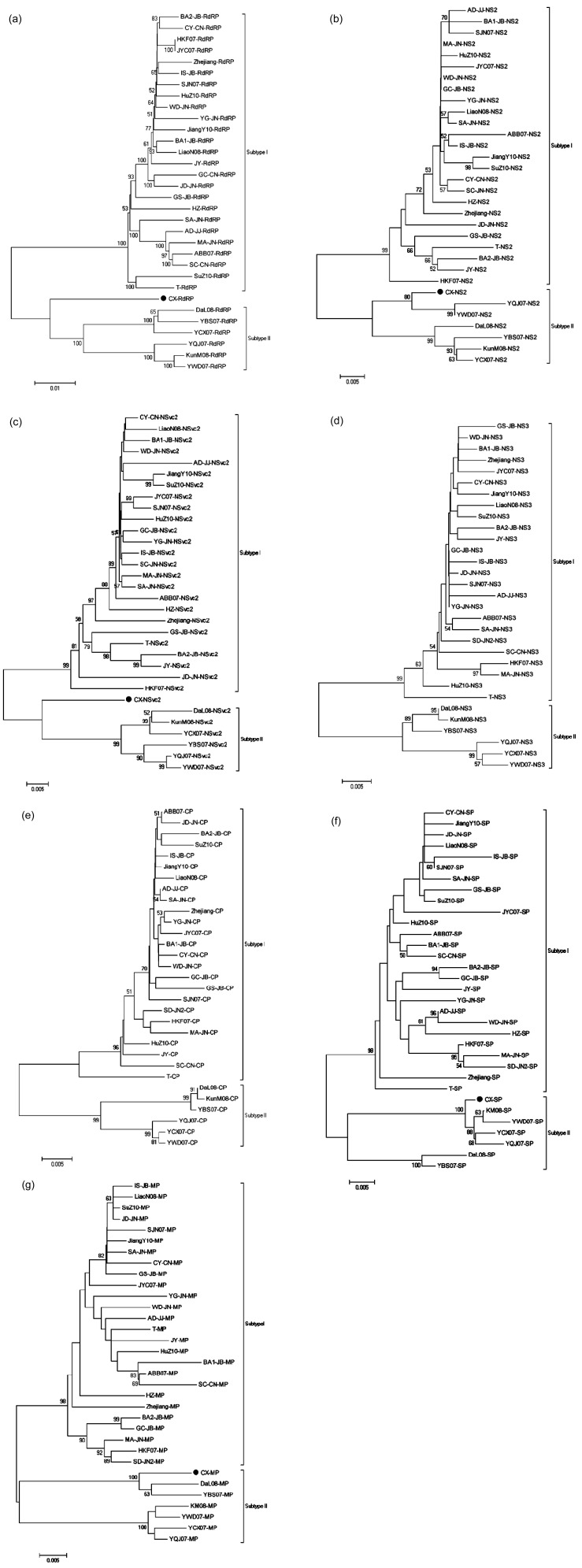
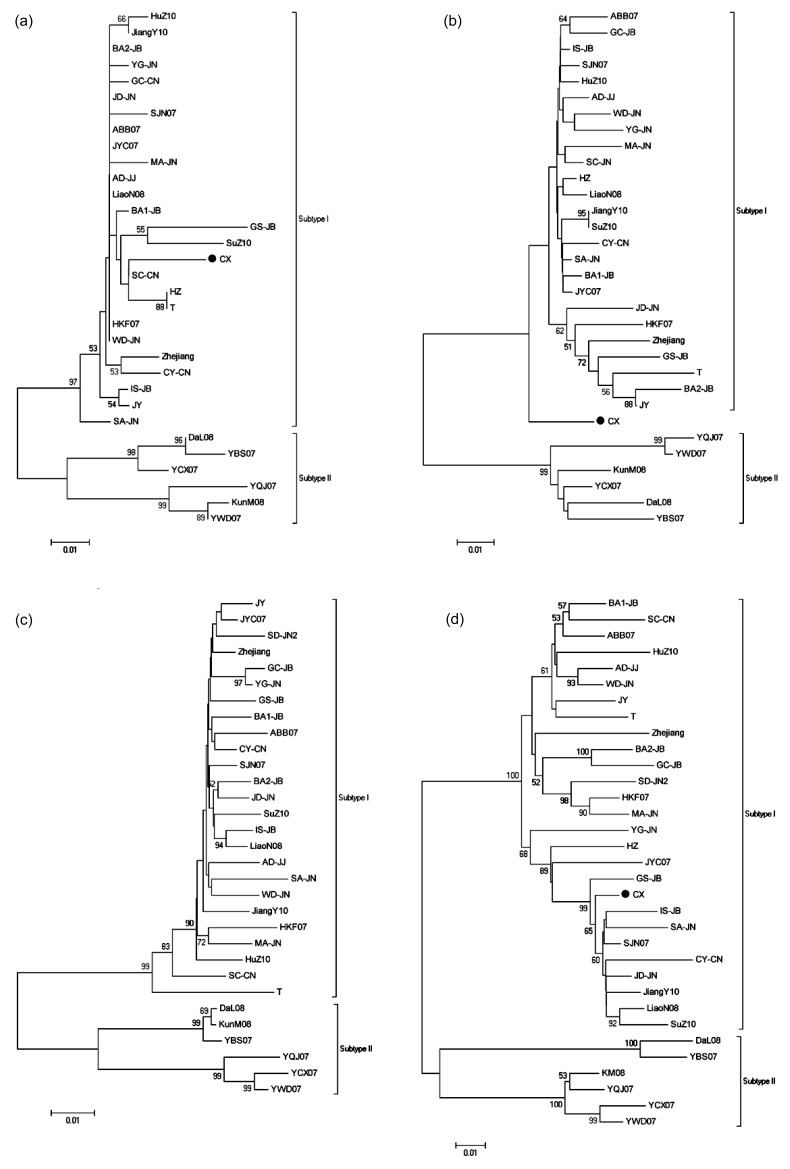
We also performed phylogenetic analyses based on sequences of seven genes and the non-coding region of these RSV isolates by using datasets (Figs. 3 and 4). The trees were similar to these constructed by using datasets for genomic RNAs, but isolates CX showed quite peculiar: it fell perfectly in subtype I when using datasets for non-coding sequences of RNA1, RNA2, RNA4, but fell perfectly in subtype II when using datasets for NS2, SP, and MP; it remained almost equally distant from two subtypes when using datasets for RdRP and NSvc2.
Fig. 3.
Phylogenetic analyses of seven RSV genes conducted in MEGA5
(a) RdRP; (b) NS2; (c) NSvc2; (d) NS3; (e) CP; (f) SP; (g) MP. Phylogenetic analyses used the statistical method as in Fig. 2
Fig. 4.
Phylogenetic analyses of non-coding sequences of four RSV genomic RNAs conducted in MEGA5
(a) RNA1; (b) RNA2; (c) RNA3; (d) RNA4. Phylogenetic analyses used the statistical method as in Fig. 2
The mean genetic distance of RSV genomic RNAs between two subtypes, within subtype I and subtype II ranged from 0.0703 to 0.0863, 0.0169 to 0.0373, and 0.0256 to 0.0536, respectively (Table 2). The average numbers of nucleotide substitutions between two randomly selected genomic RNA sequences in subtype II were generally higher than those in subtype I, suggesting that RSV isolates in subtype II were more genetically diverse than those in subtype I. The mean genetic distance of RSV genes between two subtypes, within subtype I and subtype II ranged from 0.0529 to 0.0865, 0.0123 to 0.0256, and 0.0183 to 0.0387, respectively (Table 2). The mean genetic distances of RSV genes were generally lower than those of RSV genomic RNAs whether between or within subtypes, suggesting that open reading frame (ORF) sequences were more conservative than non-coding sequences. The mean genetic distances between isolate CX and subtypes I and II ranged from 0.0537 to 0.0779 and from 0.0212 to 0.0698, respectively.
Table 2.
Genetic distances between subtype I and subtype II and within the same subtype
| Sequence | Genetic distance |
||
| Within subtype I | Within subtype II | Between subtype I and subtype II | |
| RNA1 | 0.0209±0.0007 | 0.0383±0.0013 | 0.0863±0.0027 |
| RNA2 | 0.0241±0.0012 | 0.0257±0.0016 | 0.0780±0.0043 |
| RNA3 | 0.0169±0.0010 | 0.0256±0.0023 | 0.0703±0.0052 |
| RNA4 | 0.0373±0.0021 | 0.0536±0.0037 | 0.0844±0.0054 |
| RdRP | 0.0211±0.0008 | 0.0380±0.0015 | 0.0865±0.0032 |
| NS2 | 0.0184±0.0035 | 0.0270±0.0053 | 0.0645±0.0110 |
| NSvc2 | 0.0248±0.0014 | 0.0226±0.0021 | 0.0763±0.0053 |
| NS3 | 0.0136±0.0017 | 0.0183±0.0037 | 0.0545±0.0081 |
| CP | 0.0123±0.0017 | 0.0184±0.0034 | 0.0529±0.0076 |
| SP | 0.0256±0.0033 | 0.0303±0.0054 | 0.0562±0.0076 |
| MP | 0.0220±0.0024 | 0.0387±0.0045 | 0.0545±0.0055 |
3.2. Genetic differentiation of Yunnan isolates from other isolates
To compare RSV isolates from Yunnan Province (subtype II) and other places (subtype I), the genetic differentiation of genomes between them was tested using two statistical tests, Ks* and Z* (Hudson et al., 1992; Hudson, 2000). Both P values of Ks* and Z* tests between two subtypes were <0.001, which provided significant evidence of genetic differentiation between two subtypes (Table 3). We also used Fst to estimate the extent of genetic differentiation of genomes, which is also an estimate of gene flow. The values of Fst between two subtypes were all greater than 0.33 (Table 3), suggesting infrequent gene flow between RSV population in Yunnan Province and in places other than Yunnan Province.
Table 3.
Genetic differentiation between subtype I and subtype II
| Genome |
P value#
|
Fst∆ | |
| Ks* | Z* | ||
| RNA1 | 0.0000 | 0.0000 | 0.49108 |
| RNA2 | 0.0000 | 0.0000 | 0.52272 |
| RNA3 | 0.0000 | 0.0000 | 0.51652 |
| RNA4 | 0.0000 | 0.0000 | 0.38509 |
P<0.001, which was considered as significantly rejecting the null hypothesis that there is no genetic differentiation between two subtypes
Fst is a coefficient of the extent of genetic differentiation and provides an estimate of the extent of gene flow
Value of Fst >0.33 suggested infrequent gene flow
3.3. Detection of strong negative selection pressures on all RSV genes
The selection pressure on the genes of RSV was estimated by codon-based test using the Nei-Gojobori method (Nei and Gojobori, 1986). The P values of models of d n≠d s and d n<d s of all the seven genes were <0.001 (Table 4), suggesting that negative pressures were detected on all RSV genes. In addition, the estimated d n−d s for the RdRP was −41.428 and was the lowest among all the RSV genes (Table 4), which suggested that the RdRP of RSV was under quite strong negative selection pressure.
Table 4.
Estimation of selection pressure on the RSV genes
| Gene | Unneutral H1: d
n≠d
s
|
Positive H1: d
n>d
s
|
Negative H1: d
n<d
s
|
|||
| d n−d s | P value* | d n−d s | P value | d n−d s | P value* | |
| RdRP | −41.428 | 0.000 | −40.645 | 1.000 | 40.874 | 0.000 |
| NS2 | −8.289 | 0.000 | −7.974 | 1.000 | 8.160 | 0.000 |
| NSvc2 | −19.296 | 0.000 | −19.970 | 1.000 | 19.299 | 0.000 |
| NS3 | −7.509 | 0.000 | −6.998 | 1.000 | 7.705 | 0.000 |
| CP | −9.728 | 0.000 | −9.762 | 1.000 | 9.522 | 0.000 |
| SP | −9.025 | 0.000 | −8.724 | 1.000 | 8.901 | 0.000 |
| MP | −11.444 | 0.000 | −11.814 | 1.000 | 12.646 | 0.000 |
P<0.001, which was considered as significantly rejecting the null hypothesis (H0) that there is a neutral selection, to be replaced by alternative null hypothesis (H1)
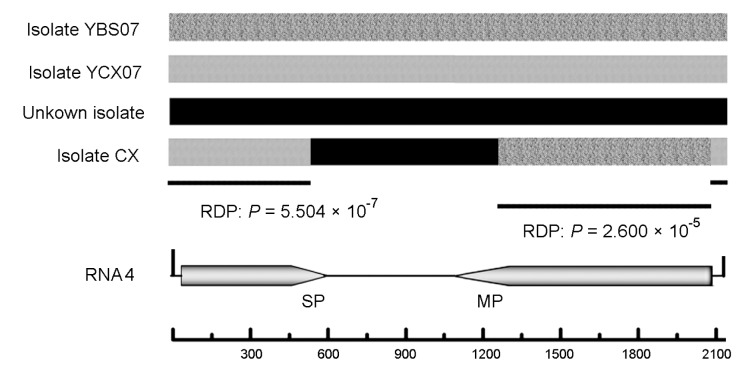
3.4. Two potential recombination events in RNA4 of isolate CX
The potential recombination events were detected by RDP3.44 (Martin et al., 2010). Among the sequences of 31 analyzed isolates, two potential recombination events were detected in RNA4 of isolate CX. The possible recombination breakpoint position and parental sequences of these two potential recombination events were detected. Potential recombination event 1 showed recombination between an unknown isolate and isolate YBS07 (1 206–2 074 nucleotides) (Fig. 5). Potential recombination event 1 was detected with a high degree of confidence by seven detection methods: GENECONV (average P value, 2.464×10−7), RDP (average P value, 2.600×10−5), Chimaera (average P value, 2.844×10−5), BOOTSCAN (average P value, 3.137×10−6), SiScan (average P value, 4.449×10−7), Max Chi (average P value, 1.117×10−5), and 3Seq (average P value, 5.882×10−16). Potential recombination event 2 showed recombination between an unknown isolate and isolate YCX07 (572 to 2 075 nucleotides) (Fig. 5). Also potential recombination event 2 showed a high degree of confidence with seven detection methods: GENECONV (average P value, 1.1315×10−7), RDP (average P value, 5.504×10−7), BOOTSCAN (average P value, 1.063×10−8), Chimaera (average P value, 2.340×10−4), SiScan (average P value, 2.876×10−8), Max Chi (average P value, 5.699×10−5), and 3Seq (average P value, 6.591×10−7) (Fig. 5). The data shown by RDP3.44 strongly suggested that isolate CX may derive from two potential recombination events among isolate YCX07, isolate YBS07, and an unknown isolate.
Fig. 5.
Schematic representation of the recombinant region of isolate CX
The possible breakpoints are presented, as are the Bonferroni-corrected P values, which indicate that the region does not have a recombinant origin
4. Discussion
In this study, we constructed the phylogenetic trees of RSV isolates collected from different areas in China, Japan, and Korea using full genomic RNA sequences as datasets. Overall, these isolates fell into two monophyletic clades, namely subtypes I and II (Fig. 2). All the isolates from Yunnan Province were in subtype II and all the isolates from other places were in subtype I. We also found that RSV isolates from Yunnan Province were more various and differed from other parts of China. We speculate that the climate and geography of Yunnan Province are not suitable for Laodelphax striatellus, the vector of RSV, to migrate long distances.
However, a peculiar isolate (CX) was found to be fluctuant between two subtypes. The isolate CX formed an individual subclade in the phylogentic trees based on RdRP and NSvc2, and fell into the same clade for NS2, SP and MP with the other isolates from Yunnan Province in subtype II (Fig. 3), but fell into the same clade with the isolates in subtype I based on RNA1 and RNA4 non-coding sequences (Fig. 4), suggesting that isolate CX may be derived from either recombination or reassortment of segments between the two subtypes. And the RDP analysis suggested that RNA4 of isolate CX may be derived from two potential recombination events among isolate YCX07, isolate YBS07 and an unknown isolate. Isolate YCX07 and isolate YBS07 were two isolates from Yunnan Province, and thus the unknown isolate was most likely from a geographical origin other than Yunnan Province.
Our data showed that the RSV population collected from Japan, Korea, and some provinces (except Yunnan) of China was only composed of subtype I and showed very low genetic diversity, indicating that the outbreak of RSV in Korea in recent years was not due to the emergence of new strong virulent isolates or invasion of isolates from other districts.
Though RSV populations in Yunnan and other districts were genetically differentiated and showed infrequent gene flow (Table 3), the genetic distance between them was still very low (0.0703 to 0.0863 for RNA segments and 0.0529 to 0.0865 for genes). And all the genes of RSV were under strong negative selection (Table 4). These findings were similar to Wei et al. (2009)’s work, in which some RSV isolates collected from Yunnan Province fell into subtype I when using gene sequences as database, while none of these were found in our work. This may be due to the fact that only seven isolates were used in this study with whole genome sequences. With more whole genome sequences of RSV isolates from Yunnan Province and other districts, the population structure and evolution of RSV will be elucidated more accurately.
Footnotes
Project supported by the National Natural Science Foundation of China (No. 31272015), the National Basic Research Program (973) of China (No. 2010CB126203), the Earmarked Fund for Modern Agro-industry Technology Research System of Ministry of Agriculture of China, and the Zhejiang Provincial Natural Science Foundation of China (No. Z3090039)
Compliance with ethics guidelines: Ling-zhe HUANG, Li-xia RAO, Xue-ping ZHOU, and Jian-xiang WU declare that they have no conflict of interest.
This article does not contain any studies with human or animal subjects performed by any of the authors.
References
- 1.Domingo E, Holland JJ. RNA virus mutations and fitness for survival. Annu Rev Microbiol. 1997;51(1):151–178. doi: 10.1146/annurev.micro.51.1.151. [DOI] [PubMed] [Google Scholar]
- 2.Elena SF, Sanjuán R. Virus evolution: insights from an experimental approach. Annu Rev Ecol Evol Syst. 2007;38(1):27–52. doi: 10.1146/annurev.ecolsys.38.091206.095637. [DOI] [Google Scholar]
- 3.Falk BW, Tsai JH. Biology and molecular biology of viruses in the genus Tenuivirus . Annu Rev Phytopathol. 1998;36(1):139–163. doi: 10.1146/annurev.phyto.36.1.139. [DOI] [PubMed] [Google Scholar]
- 4.Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39(4):783–791. doi: 10.2307/2408678. [DOI] [PubMed] [Google Scholar]
- 5.Hudson RR. A new statistic for detecting genetic differentiation. Genetics. 2000;155(4):2011–2014. doi: 10.1093/genetics/155.4.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hudson RR, Boos DD, Kaplan NL. A statistical test for detecting geographic subdivision. Mol Biol Evol. 1992;9(1):138–151. doi: 10.1093/oxfordjournals.molbev.a040703. [DOI] [PubMed] [Google Scholar]
- 7.Kakutani T, Hayano Y, Hayashi T, Minobe Y. Ambisense segment 3 of rice stripe virus: the first instance of a virus containing two ambisense segments. J Gen Virol. 1991;72(2):465–468. doi: 10.1099/0022-1317-72-2-465. [DOI] [PubMed] [Google Scholar]
- 8.Koganezawa H. Purification of rice stripe virus. Annu Phytopathol Soc Jpn. 1975;41:148–154. (in Japanese) [Google Scholar]
- 9.Lauring AS, Andino R. Quasispecies theory and the behavior of RNA viruses. PLoS Pathog. 2010;6(7):e1001005. doi: 10.1371/journal.ppat.1001005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Librado P, Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25(11):1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- 11.Martin DP, Lemey P, Lott M, Moulton V, Posada D, Lefeuvre P. RDP3: a flexible and fast computer program for analyzing recombination. Bioinformatics. 2010;26(19):2462–2463. doi: 10.1093/bioinformatics/btq467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nei M, Gojobori T. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol Biol Evol. 1986;3(5):418–426. doi: 10.1093/oxfordjournals.molbev.a040410. [DOI] [PubMed] [Google Scholar]
- 13.Ramirez BC, Haenni AL. Molecular biology of tenuiviruses, a remarkable group of plant viruses. J Gen Virol. 1994;75(3):467–475. doi: 10.1099/0022-1317-75-3-467. [DOI] [PubMed] [Google Scholar]
- 14.Roossinck MJ. Mechanisms of plant virus evolution. Annu Rev Phytopathol. 1997;35(1):191–209. doi: 10.1146/annurev.phyto.35.1.191. [DOI] [PubMed] [Google Scholar]
- 15.Saitou N, Nei M. The Neighbor-Joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4(4):406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 16.Sanjuán R, Nebot MR, Chirico N, Mansky LM, Belshaw R. Viral mutation rates. J Virol. 2010;84(19):9733–9747. doi: 10.1128/JVI.00694-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Takahashi M, Toriyama S, Hamamatsu C, Ishihama A. Nucleotide sequence and possible ambisense coding strategy of rice stripe virus RNA segment 2. J Gen Virol. 1993;74(4):769–773. doi: 10.1099/0022-1317-74-4-769. [DOI] [PubMed] [Google Scholar]
- 18.Tamura K, Nei M, Kumar S. Prospects for inferring very large phylogenies by using the Neighbor-Joining method. PNAS. 2004;101(30):11030–11035. doi: 10.1073/pnas.0404206101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28(10):2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Toriyama S. Rice stripe virus: prototype of a new group of viruses that replicate in plants and insects. Microbiol Sci. 1986;3:347–351. [PubMed] [Google Scholar]
- 21.Toriyama S, Takahashi M, Sano Y, Shimizu T, Ishihama A. Nucleotide sequence of RNA1, the largest genomic segment of rice stripe virus, the prototype of the tenuiviruses. J Gen Virol. 1994;75(12):3569–3579. doi: 10.1099/0022-1317-75-12-3569. [DOI] [PubMed] [Google Scholar]
- 22.Wang G, Zhou Y, Chen Z, Zhou X. Production of monoclonal antibodies to rice stripe virus and application in virus detection. Acta Phytopathol Sin. 2004;34:302–306. (in Chinese) [Google Scholar]
- 23.Wei TY, Yang JG, Liao FL, Gao FL, Lu LM, Zhang XT, Li F, Wu ZJ, Lin QY, Xie LH, et al. Genetic diversity and population structure of rice stripe virus in China. J Gen Virol. 2009;90(4):1025–1034. doi: 10.1099/vir.0.006858-0. [DOI] [PubMed] [Google Scholar]
- 24.Xiong R, Wu J, Zhou Y, Zhou X. Identification of a movement protein of the tenuivirus rice stripe virus. J Virol. 2008;82(24):12304–12311. doi: 10.1128/JVI.01696-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xiong R, Wu J, Zhou Y, Zhou X. Characterization and subcellular localization of an RNA silencing suppressor encoded by rice stripe tenuivirus. Virology. 2009;387(1):29–40. doi: 10.1016/j.virol.2009.01.045. [DOI] [PubMed] [Google Scholar]
- 26.Zhu Y, Hayakawa T, Toriyama S, Takahashi M. Complete nucleotide sequence of RNA3 of rice stripe virus: an ambisense coding strategy. J Gen Virol. 1991;72(4):763–767. doi: 10.1099/0022-1317-72-4-763. [DOI] [PubMed] [Google Scholar]
- 27.Zhu Y, Hayakawa T, Toriyama S. Complete nucleotide sequence of RNA4 of rice stripe virus isolate T, and comparison with another isolate and with maize stripe virus. J Gen Virol. 1992;73(5):1309–1312. doi: 10.1099/0022-1317-73-5-1309. [DOI] [PubMed] [Google Scholar]