Abstract
Cellular decision-making is mediated by a complex interplay of external stimuli with the intracellular environment, in particular transcription factor regulatory networks. Here we have determined the expression of a network of 18 key haematopoietic transcription factors (TFs) in 597 single primary blood stem and progenitor cells isolated from mouse bone marrow. We demonstrate that different stem/progenitor populations are characterised by distinctive TF expression states, and through comprehensive bioinformatic analysis reveal positively and negatively correlated TF pairings, including previously unrecognised relationships between Gata2, Gfi1 and Gfi1b. Validation using transcriptional and transgenic assays confirmed direct regulatory interactions consistent with a regulatory triad in immature blood stem cells, where Gata2 may function to modulate cross-inhibition between Gfi1 and Gfi1b. Single cell expression profiling therefore identifies network states and allows reconstruction of network hierarchies involved in controlling stem cell fate choices, and provides a blueprint for studying both normal development and human disease.
Haematopoiesis has long served as a model system for studying cell fate decisions during stem cell differentiation1, 2. At the molecular level, transcription factors (TFs) are major drivers of cellular identity and cell fate transitions as exemplified by their key role in reprogramming3 and lineage switching experiments4-6. TFs function within wider regulatory networks, the connectivity of which can be revealed using classical transcriptional assays or inferred from global expression and TF binding profiling studies7-12.
However, these experiments report population averages, while cell fate choices are made by individual cells. The importance of studying single cells is emphasised by the known functional heterogeneity in haematopoietic stem cells (HSCs) as well as other cell types, which manifests as relatively stable subpopulations with either balanced production of myeloid and lymphoid cells or a deficiency in lymphoid potential13-15. HSCs have also been reported to be heterogeneous in gene expression16-18, although previous studies have been limited in terms of numbers of genes, cells or populations analysed. Recent advances in microfluidics technologies have facilitated high-throughput Q-RT-PCR analysis of tens of genes in hundreds of single cells simultaneously19. This technology has recently been used to resolve cell populations in 64 cell mouse embryos20, to dissect cellular heterogeneity in human colon cancer21, and to reveal significant variation in early erythroid gene expression, which resolved upon commitment22.
Here we analysed expression of a network of 18 densely interconnected TFs in 597 single cells from five primary haematopoietic stem and progenitor cell populations, which not only revealed characteristic expression states for the different cell populations, but also identified previously unrecognized regulatory relationships. This included a putative regulatory triad consisting of Gata2, Gfi1 and Gfi1b, which was validated using cell line and transgenic mouse assays. Our 4 findings suggest that GATA2 may function in a regulatory loop to modulate Gfi1/Gfi1b cross-antagonism during entry into the myeloid/lymphoid lineages, thus demonstrating that high-throughput single cell TF expression analysis provides a powerful approach towards the identification of regulatory network links.
RESULTS
Single-cell expression analysis reveals heterogeneity in transcription factor expression in haematopoietic stem and progenitor cells
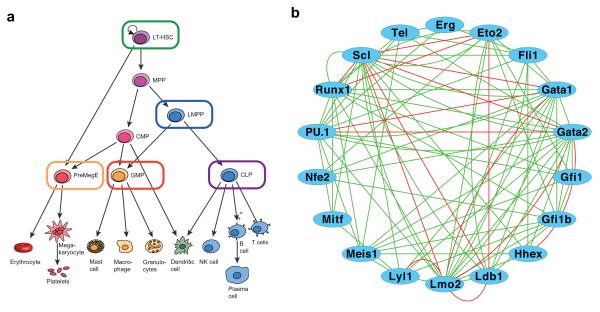
To study core regulatory circuits during early haematopoietic differentiation stages, we performed gene expression analysis for transcription factors in single primary haematopoietic stem/progenitor cells prospectively isolated from mouse bone marrow by fluorescence activated cell sorting (FACS). We analysed long-term haematopoietic stem cells (LSK CD150+CD48− HSC23), lymphoid-primed multipotent progenitors (LSK Flt3hi LMPP24), bipotential megakaryocyte/erythroid progenitors (CD16/32loCD41−CD150+CD105lo PreMegE25), granulocyte-monocyte progenitors (CD41loCD16/32hi GMP25, 26), and common lymphoid progenitor (Lin− IL7Rα+KitloSca-1lo CLP27) (Figure 1A and Supplementary Fig. 1). A total of 597 single cells (123 CLPs, 124 GMPs, 121 HSCs, 116 LMPPs, 113 PreMegEs) passed quality control measures (see Methods).
Figure 1. Single cell gene expression analysis of a core haematopoietic transcriptional regulatory network.
(a) Schematic of the haematopoietic hierarchy, with the megakaryocyte-erythroid lineage in red, the myeloid lineages in orange and the lymphoid lineage in blue. Cell types investigated in this study are outlined in the colours used to represent these populations in subsequent figures, and encompass both early multipotent stem and progenitors and committed progenitors for each of the major haematopoietic lineages. Cell surface phenotypes were LSK CD150+CD48− HSC (also gated as CD34loFlt3−), LSK Flt3hi LMPP, Lin−IL7Rα+KitloSca-1lo CLP, CD41loCD16/32hi GMP (also gated Lin−c-Kit+CD150−), CD16/32loCD41−CD150+CD105lo PreMegE (also gated Lin−c-Kit+). LT-HSC, long-term haematopoietic stem cell; MPP, multi-potent progenitor; LMPP, lymphoid-primed multi-potent progenitor; CMP, common myeloid progenitor; CLP, common lymphoid progenitor; GMP, granulocyte-monocyte progenitor; PreMegE, pre megakaryocyte erythroid progenitor; NK cell, natural killer cell. (b) Network diagram of data curated from the literature and protein interaction databases (STRING66 and FunctionalNet67) illustrating the complex interactions between 18 core haematopoietic transcription factors. Green lines indicate functional relationships and red lines indicate direct protein-protein interactions. Activating and inhibitory connections are not distinguished.
Single cell gene expression analysis was performed for 24 genes in all 597 cells (see Supplementary Table 3 for raw Ct data). Our gene set included 18 transcription factors (Figure 1B) with known key roles in haematopoiesis, as well as five housekeeping genes and the Stem Cell Factor receptor c-Kit, which is expressed on the surface of all analysed haematopoietic stem/progenitor subsets23-28. We have previously reported the potential for abundant regulatory linkages between many of the 18 TFs9, 11, 12, 29-33, and a similarly densely interconnected network was obtained using data curated from the literature and protein interaction databases (Figure 1B). Importantly, while previous studies have examined individual HSCs and commitment to the erythroid lineage, they have been limited in cell numbers16, were focussed on expression heterogeneity16, 17 or have examined a lineage-specific set of genes22. Moreover, the potential for identifying regulatory connections from single cell gene expression profiling had not been demonstrated, nor had the potential for dynamic changes of regulatory network states been studied during the differentiation of HSCs into the various multipotential blood progenitors.
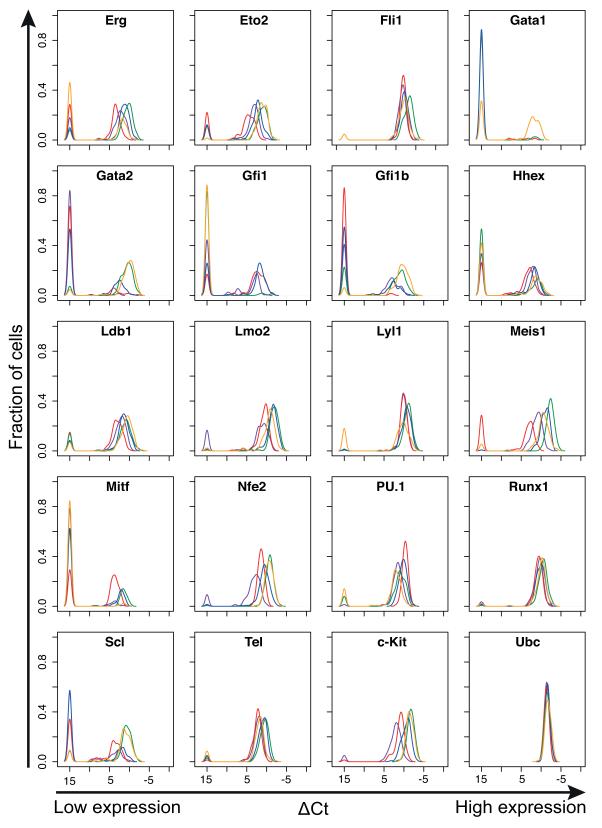
Single cell gene expression analysis recovered expected expression patterns for the 18 TFs as well as housekeepers and c-Kit (Figure 2). For example, c-Kit expression was highest in HSCs and gradually reduced in the progenitor populations, consistent with the reported downregulation in progenitors28. Gata1 is known to be expressed at high levels in erythroid and megakaryocyte lineages, but not in HSCs34, and here was expressed in around two thirds of PreMegE cells, yet absent in almost all cells of the other populations. Likewise, Gata2 is known to be expressed in HSCs and during megakaryopoiesis35, 36, and in our data was expressed in most HSCs and PreMegEs but at lower levels or not at all in LMPPs, GMPs and CLPs. GFI1B is important for the development of erythroid progenitors, while GFI1 is important for myeloid and T cell development, and the two factors are known to be mutually inhibitory37, 38. Outside of the HSC population; Gfi1 was expressed in the majority of LMPPs, CLPs and GMPs, but rarely in PreMegEs, while Gfi1b was expressed in most PreMegEs, with lower or absent expression in LMPPs, CLPs and GMPs.
Figure 2. Haematopoietic transcription factors show heterogeneous expression in haematopoietic stem and progenitor cells.
Density plots for 18 transcription factors, the stem cell factor receptor c-Kit, and the housekeeping gene Ubc, in five haematopoietic stem and progenitor populations. The density indicates the fraction of cells at each expression level, allowing direct comparison of the expression level of each gene in all five populations. Green, HSC; Blue, LMPP; Purple, CLP; Red, GMP; Orange, PreMegE.
Many genes exhibited heterogeneous expression within cell populations, with some cells expressing the gene at high levels and undetectable expression in others, in line with previous reports of expression heterogeneity in blood stem and progenitor populations16-18, 22. Several TFs, including Runx1 and Fli1, had a very similar gene expression distribution in all cell types, and these genes were expressed in almost all analysed cells. Conversely, genes including Erg, Lmo2 and Meis1, differed in expression level between cell types. Genes including Gfi1, Gfi1b and Scl (also known as Tal1) showed bimodal expression amongst the cells that expressed the gene, with the potential therefore to generate three distinct expression states (high, medium, not-expressed) within a single population that is pure based on FACS analysis. Importantly, such detailed insights into the dynamical nature of TF gene expression in primary blood stem and progenitor cells could not have been obtained from population studies.
Cell populations can be resolved by differential network activity states
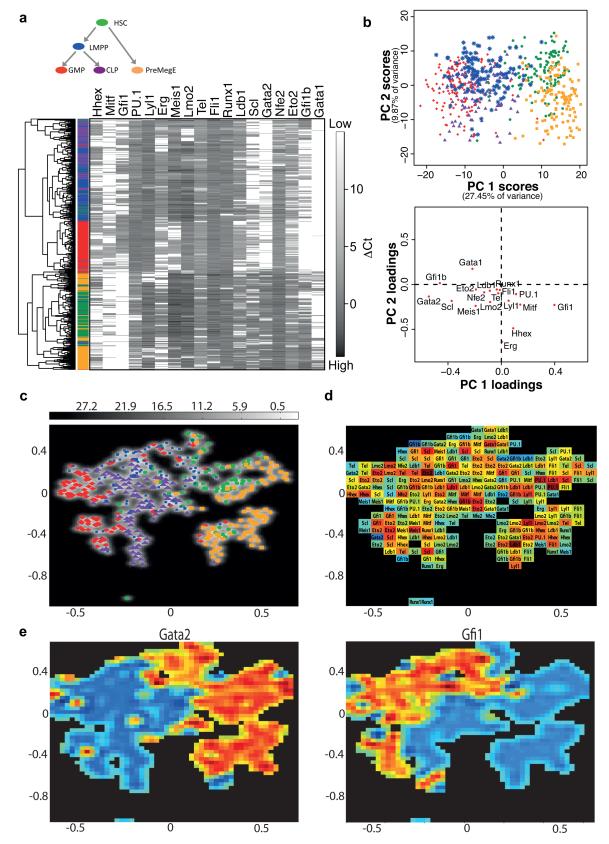
To establish cell type-specific patterns of gene expression that may aid our understanding of network activity and cell state transitions, we next performed hierarchical clustering and principal component analysis using the expression data for our TFs in all 597 haematopoietic stem/progenitor cells. The relatedness of cells is determined using only the gene expression values, without prior knowledge of which population a cell originates from. Hierarchical clustering demonstrated that mRNA levels for these 18 key TFs allow the partitioning of cells largely by sorted population (Figure 3A). This was particularly clear for the GMPs, which formed a distinct cluster. HSCs and PreMegEs formed a cluster separate from the myeloid and lymphoid lineages in which the two populations were also largely separated from each other, while LMPPs and CLPs showed significant overlap. There was some mixing of HSCs with LMPPs and PreMegEs, in line with the evidence that LMPPs and the megakaryocyte/erythroid lineage may be generated as early and alternative fates of HSCs24 and so are both closely related to HSCs but distinct from one another.
Figure 3. Single cell gene expression analysis reveals cell type-specific regulatory codes.
(a) Hierarchical clustering of 597 haematopoietic stem and progenitor cells according to the expression of the 18 TFs. Coloured bar indicates cell type of origin: Green, HSC; Blue, LMPP; Purple, CLP; Red, GMP; Orange, PreMegE. (b) Principal component projections of the 597 haematopoietic stem/progenitor cells, in the first and second components (top), from the expression of all 18 TFs. Principal component loadings (bottom) indicate the extent to which each gene contributes to the separation of cells along each component. (c) Gaussian process latent variable model (GPLVM) illustrating variations of 18 dimensional gene expression patterns between and within cell types in 2D. GPLVMs are a non-linear generalisation of PCA that allow for the analysis of more complex gene expression patterns than PCA and can thus potentially better represent variations between and within populations of different cell types. The uncertainty of the mapping from 2D to the 18 dimensional TF space is encoded in grey (white low uncertainty, grey high uncertainty). (d) Relevance map showing the most important genes across the GPLVM map. The colours correspond to the distance of the respective gene from the origin in a standard loadings plot (red far away/important, blue close to origin). (e) Expression maps for Gata2 (left) and Gfi1 (right), with high expression in red and low or absent expression in blue. Gata2 is expressed primarily in the HSC and PreMegE clusters, and Gfi1 in LMPPs, GMPs and some CLPs.
Principal component analysis confirmed the above results, where each data point represents a single cell, colour-coded according to its flow cytometric phenotype (Figure 3B, upper panel). Principal component 1, which captures the largest proportion of the variation in the data, separates the HSCs and PreMegEs from the lymphoid and myeloid populations, and partially separates the GMPs from the LMPPs and CLPs. Principal component 2 further separates the HSCs and PreMegEs. However, although individual populations can be distinguished there is also significant overlap, particularly between the LMPPs and CLPs, both of which contain lympho-myeloid-restricted progenitors39, indicating that cells at the edges of adjacent populations have similar network activity states. Gata2, Gfi1b, Scl and Gfi1 contribute to separation of the HSCs and PreMegEs from the myelolymphoid populations along component 1, while Erg, Hhex and Gata1 are important across component 2 (Figure 3B, lower panel), consistent with known expression patterns in these populations. Runx1 and Fli1 contributed little to the separation of cell types, consistent with their similar expression distributions between cell types (Figure 2).
Gaussian Process Latent Variable Models (GPLVMs) are a non-linear generalisation of PCA40 and were recently shown to be a powerful alternative, in particular for resolving non-linear differences in single-cell gene expression patterns41. GPLVM resulted in a better separation of the different cell types than PCA (Figure 3C), with the greatest improvement found for GMPs. However, LMPPs and CLPs cells could not be resolved completely. Our ability to separate populations was further confirmed by calculating the spatial median, a robust multivariate measure of the ‘centre’ of the distribution for each cell type, and then analysing distances between cell types. These GPLVM map distances reflected the differentiation hierarchy shown in Figure 3A with cell types close in the hierarchy located close together in the map (Supplementary Fig. 2).
While for standard PCA the relevance of different genes to the separation of the data (quantified by component loadings in Figure 3B) can only be found for the entire PCA map, the relevance of each gene can change across the GPLVM map, providing a greater resolution of the changes separating cell types. The GPLVM relevance map (Figure 3D) shows the most important gene at each point of the map, and illustrates for example that Gata2, Gfi1 and Gfi1b feature frequently throughout the map and particularly in the region bordering the HSC and LMPP populations. Indeed, expression maps for individual genes (Figure 3E) demonstrate that high Gata2 expression occurs mostly in the HSC and PreMegE populations, while high Gfi1 is restricted mostly to LMPPs, GMPs and a subset of CLPs. Taken together therefore, bioinformatic analysis of single cell gene expression allowed us to correlate distinct expression states of a core set of 18 key haematopoietic TFs with some of the earliest blood stem and progenitor populations, including the earliest known lineage restriction stage from HSCs resulting in distinct MegE and lympho-myeloid restricted pathways.
Single cell analysis reveals dynamic regulatory relationships
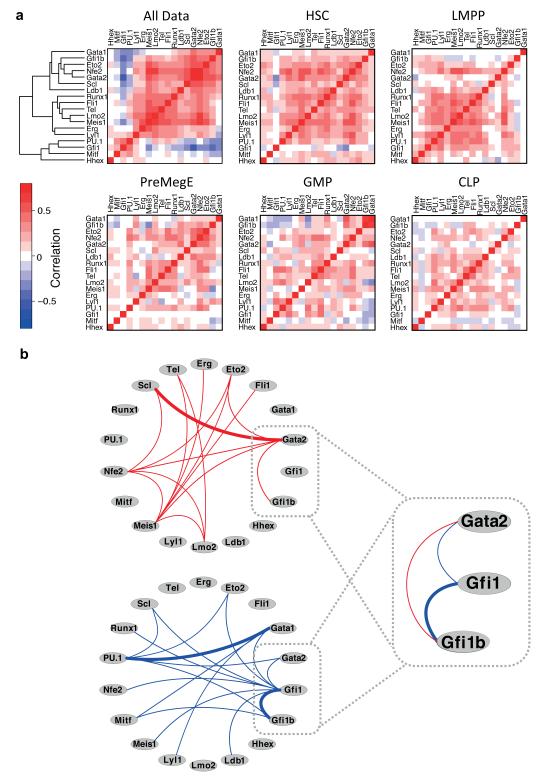
We next hypothesised that single cell expression data could be used to identify regulatory linkages by identifying pairs of factors with correlated expression, where a positive correlation suggests that one factor may activate another and a negative correlation indicates an antagonistic relationship. Correlation analysis and hierarchical clustering of the eighteen transcription factors across all 597 haematopoietic stem/progenitor cells revealed both positive and negative correlations (Figure 4A, top left panel). Among the positive correlations is a group of seven genes (Scl, Gata2, Nfe2, Eto2, Gfi1b, Gata1 and Ldb1) known to be important in the erythroid/megakaryocytic lineages35, 38, 42-49, while there was a negative correlation between PU.1 and Gata1, which are thought to function as a switch controlling erythroid and myelomonocytic fates50, 51. To establish whether such regulatory relationships were stable or dynamic during differentiation, we repeated the correlation analysis for each of the five stem/progenitor populations individually (Figure 4A). While many of the strong positive correlations identified in the whole data set remained stable between cell types, there were some clear differences, particularly in negative correlations, which could suggest that repression, or relief of repression, of some TFs by others is a vital step in cell fate transitions. For example, the strong negative correlation between Gfi1 and Gata2 present in the whole dataset is seen only in HSCs. Gfi1b and Gata1 are negatively correlated with PU.1, Mitf, Gfi1, Lyl1 and Lmo2 in GMPs, and to some extent in CLPs, but are either not correlated or positively correlated in the earlier progenitors and in the erythroid/megakaryocyte lineage. Together these results indicate that expression of the core haematopoietic transcriptional regulatory network is dynamic (Figures 2 and 3), which is presumably intimately connected to the dynamics of regulatory interactions between the components of the network (Figure 4).
Figure 4. Single cell expression analysis of haematopoietic TFs identifies previously unrecognised putative regulatory interactions between key TFs.
(a) Hierarchical clustering of Spearman Rank correlations between pairs of TFs for all 597 cells together and for the different cell types individually as indicated. Genes in all heatmaps are ordered according to the clustering performed for all data. Positive correlations (red) may result from the coordinate expression or lack of expression of pairs of factors in individual cells, while negative correlations (blue) can result either from the expression of one factor in the absence of the other, or from high expression of one factor and reciprocal low expression of the other in the same cell. (b) Network diagrams showing putative activating relationships between TFs suggested by significant positive correlations (top, red lines) and antagonistic relationships suggested by significant negative correlations (bottom, blue lines) in the whole data set. Known relationships are highlighted with bold lines. This highlights a putative transcription factor triad in which Gfi1 is negatively correlated with Gata2 and Gfi1b, but Gata2 and Gfi1b are positively correlated.
Significant positive and negative correlations between TFs were displayed as a putative interaction network (Figure 4B). Among these is the known Scl-Gata2 relationship and the PU.1-Gata151 and Gfi1-Gfi1b37, 38 inhibitory relationships, indicating that additional relationships identified may indeed signify previously unrecognised interactions. Two newly predicted regulatory links (Gata2-Gfi1 and Gata2-Gfi1b) suggested possible involvement of GATA2 in modulating the cross-inhibitory relationship between GFI1 and GFI1B (Figure 4B), which was of particular interest to us because we had seen downregulation of Gata2 and Gfi1b accompanied by reciprocal upregulation of Gfi1 in a recent transcriptomic and epigenomic analysis of leukaemia development in an MLL-ENL-driven mouse model of acute myeloid leukaemia (AML)52 (see Supplementary Fig. 3).
Direct repression of Gata2 distal enhancer elements by GFI1 provides a likely mechanism for negatively correlated expression
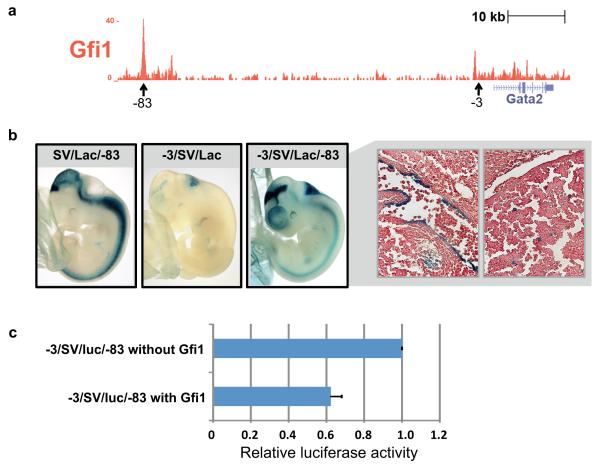
To investigate whether downregulation of Gata2 might be mediated directly through GFI1 binding to Gata2 regulatory elements, we interrogated existing ChIP-Sequencing data for GFI1 in MLL-ENL transduced cells52, 53. Across the Gata2 locus, a prominent peak was identified 83kb upstream of the Gata2 transcriptional start site (Supplementary Fig. 3B). This −83 kb region had been shown previously to loop to the Gata2 promoter54, and was bound in the HPC7 haematopoietic progenitor cell line11, 55 by multiple TFs (Supplementary Fig. 3)11. TF ChIP-Seq studies are currently not possible with the small numbers of cells that can be obtained for the highly purified blood stem cell populations used here for single cell expression analysis. We nevertheless wanted to confirm binding of GFI1 to the Gata2 gene locus in primary blood cells, and therefore performed GFI1 ChIP-Seq in primary mast cells, which like HSCs express the stem cell factor receptor c-KIT and a number of TFs important for HSCs including GFI1 and GATA2. This GFI1 ChIP-Seq experiment confirmed GFI1 binding to the Gata2 −83 kb region in primary mouse blood cells (Figure 5A).
Figure 5. Direct repression of Gata2 by GFI1 through a distal enhancer element provides a mechanism for negatively correlated expression.
(a) ChIP-seq analysis of Gfi1 in primary mast cells indicates that GFI1 binds to the Gata2 locus at the −83 kb regulatory element. (b) Representative embryos demonstrating LacZ staining for the Gata2 −83kb, −3kb and combined −3/−83kb regulatory element reporter constructs. The −83kb region alone (SV/LacZ/−83) showed consistent staining of the midbrain, hindbrain and spinal cord, but no haematopoietic staining. The −3kb enhancer (−3/SV/LacZ) had only hindbrain staining. The −83/−3kb combined element (−3/SV/LacZ/−83) showed the neural activities of both individual enhancers, but also staining in the dorsal aorta (right-hand top panel) and foetal liver haematopoietic cells (right-hand bottom panel). Images of sections taken at 40× magnification. (c) A luciferase reporter construct carrying both regulatory elements (−3/SV/luc/−83) was transfected into the HPC7 haematopoietic progenitor cell line, which expresses high levels of Gata2 but low levels of Gfi1. Co-transfection with a Gfi1 expression construct caused a 40% reduction in reporter activity. Luciferase activity is shown relative to −3/SV/luc/−83 and bars are the mean and standard deviation of three biological replicates.
As no in vivo activity has as yet been reported for the Gata2 −83 kb region, we generated a LacZ reporter construct with the −83 kb region fused to a minimal SV40 promoter/LacZ reporter cassette. Analysis of LacZ expression in E11.5 transgenic mouse embryos demonstrated consistent staining in the midbrain, hindbrain and spinal cord (Figure 5B), all known domains of endogenous Gata2 expression56. However, no haematopoietic staining was seen in any of the transgenic embryos. We had shown previously that a Gata2 −3 kb enhancer is active at E11.5 in the dorsal aorta endothelium including budding haematopoietic cells, but not the foetal liver9. Given the prominent TF binding to the −83 kb region in haematopoietic cells, we next asked whether a combination of the −83 and −3 kb enhancers was able to drive expression to foetal liver haematopoietic cells. Transgenic embryos carrying a combined enhancer construct (−3/SV/lacZ/−83) displayed the neural activities of both of the individual enhancers (Figure 5B). Moreover, staining was not only seen in the dorsal aorta but also in foetal liver haematopoietic cells. Transgenic analysis therefore confirmed the Gata2 −83 kb region as a candidate enhancer element involved in haematopoietic expression of Gata2.
To investigate whether GFI1 could repress activity of this element, we next generated a luciferase reporter construct (−3/SV/luc/−83) and performed transfection assays in the HPC7 progenitor cell line which expresses high levels of Gata2 but very low levels of Gfi133. Compared with control transfection assays, we observed that co-transfection of a Gfi1 expression construct caused a 40% reduction in reporter activity (Figure 5C). Taken together therefore, ChIP-Seq, transgenic and transfection studies validated the previously unrecognised regulatory interaction between GFI1 and Gata2.
Direct activation of Gfi1b distal enhancer elements by GATA2 provides a likely mechanism for positively correlated expression
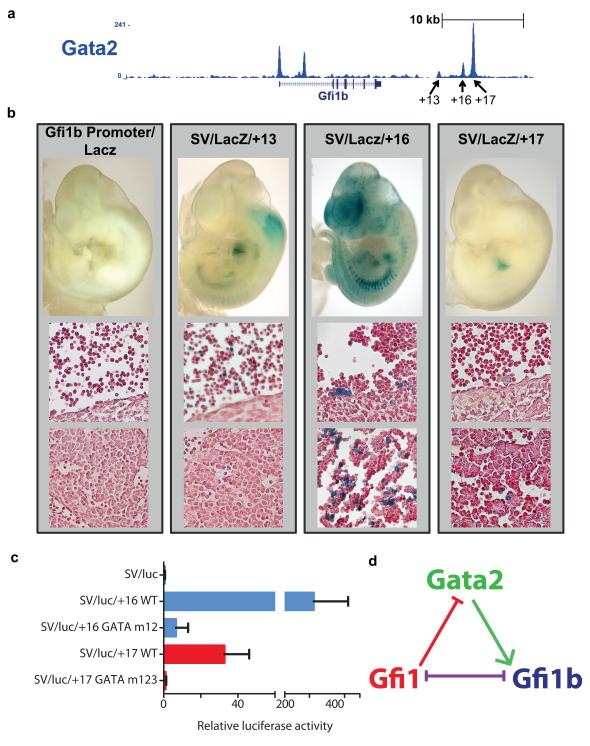
To investigate whether positively correlated expression of Gfi1b and Gata2 might be mediated directly through GATA2 binding to the Gfi1b gene locus, we interrogated existing ChIP-Seq data for the HPC7 cell line11 which demonstrated binding of GATA2 to the Gfi1b promoter as well as three 3′ candidate enhancer regions 13kb, 16kb and 17kb downstream of the start of the Gfi1b gene (Supplementary Fig. 4). To confirm binding in primary mouse blood cells, we again turned to primary mast cells, and indeed, significant GATA2 binding was observed at all four regions bound by GATA2 in HPC7 as well as a region in the first intron not bound in HPC7 (Figure 6A). Taken together, TF-binding in both mast cells and HPC7 therefore identified 4 candidate regulatory regions that might be involved in mediating control of Gfi1b expression in stem/progenitor cells by GATA2.
Figure 6. Direct activation of Gfi1b by GATA2 through distal enhancer elements.
(a) ChIP-seq analysis of GATA2 in primary mast cells indicates that GATA2 binds to the Gfi1b locus at multiple locations, including the promoter, a region in the first intron and the +13, +16 and +17 kb regulatory elements. (b) Representative embryos demonstrating LacZ staining for the Gfi1b promoter as well as the +13, +16 and +17 kb regulatory elements. The promoter shows no staining. The +13 kb region shows weak expression in a subset of circulating blood cells. The +16 kb region has strong staining in haematopoietic clusters in the dorsal aorta, and in a subset of foetal liver cells. The +17 kb region shows staining in a small subset of foetal liver haematopoietic cells. Images of sections taken at 40× magnification. (c) Luciferase reporter constructs carrying the wild type +16kb (SV/luc/+16 WT) and +17 kb (SV/luc/+17 WT) kb regulatory regions transfected in 416B cells displayed high levels of luciferase activity, particularly for the +16 kb region. Mutation of the two conserved GATA sites in the +16 kb region (SV/luc/+16 GATA m12) reduced luciferase activity by >95%. Mutation of the two conserved and one partially conserved GATA sites in the +17 kb region (SV/luc/+17 GATA m123) also reduced luciferase activity by >95%. m12 and m123 indicate that GATA sites 1, 2 and 3 were mutated. Luciferase activity is shown relative to SV/luc. Experiments were performed in biological duplicate or triplicate on two separate occasions. Shown is one representative experiment displaying the mean and standard deviation for three biological replicate transfections. (d) A putative regulatory triad including GATA2, GFI1 and GFI1B suggested by the data. In this regulatory triad, GFI1 and GFI1B are mutually inhibitory, while GATA2 can activate expression of Gfi1b and GFI1 can repress expression of Gata2.
To assess whether the four regions bound by GATA2 correspond to bona fide gene regulatory sequences, we performed transgenic assays. LacZ reporter constructs were generated for the Gfi1b promoter as well as the +13, +16 and +17 kb candidate enhancer regions and assayed in E11.5 mouse embryos. The promoter region alone did not mediate any haematopoietic expression, a phenomenon we have observed before for both Runx1 and Scl/Tal1 promoters57, 58. By contrast, all three distal regions mediated haematopoietic expression: the +13 kb region showed weak expression in a subset of circulating (likely primitive) blood cells, the +16 kb region displayed strong staining in haematopoietic clusters in the dorsal aorta as well as a subset of foetal liver cells, and the +17 kb region showed staining in a small subset of foetal liver haematopoietic cells (Figure 6B). Transgenic analysis therefore provided in vivo validation of GATA2-bound regions, with the +16 and +17 kb regions being particularly relevant due to their activity in the anatomical sites of early definitive haematopoietic development.
Since transgenic analysis had focussed our attention on the +16 and +17 kb enhancer regions as possible mediators of Gfi1b activation by GATA2, we investigated the possible presence of conserved GATA motifs. Both the +16 and +17 kb regions showed extensive sequence conservation across a wide range of mammalian species consistent with their function as gene regulatory elements (Supplementary Fig. 5). Moreover, both enhancers contained two completely conserved GATA motifs. To investigate the role of the GATA sites in enhancer activity, we generated luciferase reporter constructs with both the wild type and GATA mutant +16 and +17 kb enhancers (Figure 6C). Following stable transfection assays in the myeloid progenitor cell line 416B, both regions showed substantial enhancer activity that was almost completely lost in the GATA mutant constructs. GATA2 binding in ChIP assays, activity in transgenic assays and presence of conserved GATA motifs essential for enhancer function are therefore all consistent with a model whereby GATA2 directly activates Gfi1b expression through the +16 and +17 kb enhancer regions.
Taken together therefore, single cell gene expression analysis of primary blood stem and progenitor cells suggests the existence of a regulatory triad including Gata2, Gfi1 and Gfi1b (Figure 6D), the connectivity of which has been validated using transgenic and transcriptional assays. In this triad, the reported mutual inhibition of GFI1 and GFI1B37, 38 is retained, but is modulated by GATA2 through its activation of Gfi1b and repression by GFI1.
DISCUSSION
Cellular phenotypes are controlled by networks of interacting TFs. Development can therefore be described as a procession through multiple dynamic regulatory states, which in the case of multilineage differentiation may give rise to multiple distinct outcomes. However, classical networks derived from gene expression or ChIP-Seq data provide a population average, giving little insight into cellular heterogeneity and possible regulatory interactions likely to be critical for the lineage commitment/restriction steps of individual progenitor cells59.
In this study, we investigated a core transcriptional network of 18 TFs in single cells of five related stem and progenitor populations. Bioinformatic analyses were able to broadly distinguish sorted cell populations based only on the expression of those TFs, indicating that early haematopoietic stem/progenitor cells are characterised by related but distinct network activity states. While gross gene expression patterns were consistent with published population studies, we also found significant heterogeneity in TF expression within populations. This confirms previous observations that FACS sorted populations are molecularly and functionally heterogeneous at the single cell level16-18, and suggests either that heterogeneity is an inherent characteristic of stem cells, or that previously unresolved subpopulations may be present that could represent intermediate differentiation steps.
Importantly, heterogeneity did not confound our ability to resolve populations based on their distinct gene expression patterns. Thus, further analysis of defining TF expression patterns and interactions at important lineage restriction stages should facilitate the unravelling of critical TF interactions decisive for lineage commitment steps. Index sorting for example permits the tracing back of each cell to its position in the sorting data, and may therefore allow us in future to link heterogeneous gene expression states to novel subpopulations. Strikingly, in our study several transcription factors had similar expression levels in all 597 cells analysed, regardless of cell type of origin, including Runx1 and Fli1, while others were much more variable. This may suggest that the blood network requires, or is able to tolerate, variation in some factors but not others. Furthermore, correlation analysis revealed both stable and dynamic TF relationships across the cell types analysed, together suggesting that some TF interactions may be vital for the general stability of the network and so remain constant, while others are important for network dynamics and state transitions and are therefore more variable.
As transcriptional regulatory networks control the spatiotemporal regulation of lineage-specific genes, successful reconstruction of regulatory hierarchies represents a major step towards gaining a mechanistic understanding of cellular decision-making processes. For example, detailed experimental and computational analysis of a core circuit of Gata2, Scl and Fli1 revealed that this circuit is able to function as a bistable switch, where the internal wiring enables the network to filter noise when responding to external cues9, 60. However, the generation of large-scale experimentally validated network models is impeded by the relatively low-throughput of experiments that can provide detailed experimental information on the functionality of individual regulatory elements. Network inference, where transcriptional interactions are inferred from statistical dependencies in large expression profiling datasets, provides an alternative approach7. However, in addition to the very substantial costs of performing hundreds of expression profiling experiments, standard microarray or RNA-Seq profiling requires large cell numbers, which may not be feasible for rare stem and progenitor populations.
Here we have demonstrated that gene expression data from a large set of single cells presents an alternative approach to network reconstruction through the analysis of pairwise correlations between network components. While hundreds of samples are still required, the fact that these are single cells makes this approach applicable to rare stem cell populations. The robustness of inferred regulatory links was demonstrated by our ability to recover key known regulatory relationships such as the Gata1-PU.1 and Gfi1-Gfi1b antagonisms. Furthermore, we validated two novel putative regulatory interactions predicted from bioinformatic analysis of our single cell expression profiling data. This demonstrated that GFI1 directly represses Gata2 expression through the −83 kb regulatory region, while GATA2 activates Gfi1b through the +16 kb and +17 kb regulatory regions. The resultant connectivity resembles a type 2 coherent feed forward loop61, and suggests that GATA2 works within this regulatory triad to modulate the antagonism between GFI1 and GFI1B. Moreover, GATA2 inhibits lymphopoiesis62, and along with Gfi1b is down-regulated concurrent with Gfi1 upregulation in the myelolymphoid lineages in our data. This suggests that direct downregulation of Gata2 and Gfi1b by GFI1 may represent a key event during the specification of early lymphoid cells, similar to the role of Tif1γ in modulating the PU.1-GATA1 antagonism in the erythroid versus myeloid fate choice in zebrafish63.
Loss of function mutations in Gata2 have recently been reported to predispose carriers to developing acute myeloid leukaemia64, whereas inhibition of Gfi1 prolonged survival in leukaemia mouse models65. This suggests that identification of network states and reconstruction of network hierarchies from single cell expression profiling will not only enhance our knowledge of normal differentiation and development, but also provide a blueprint for understanding the subversion of cell fate control likely to underlie many degenerative and malignant pathologies.
METHODS
Purification of stem cells and progenitor cells
Bone marrow cells were isolated from the bones (femurs, tibiae and crista ileaca) of 9-12 week old C57BL/6 mice. For the isolation of LMPPs and HSCs, cells were enriched for CD117+ (c-Kit) cells by MACS bead separation with anti-CD117 immunomagnetic beads (Miltenyi Biotec); for the other cell populations, unenriched bone marrow was used. Cells were pre-incubated with Fc-block for the HSC, LMPP and CLP stains but not for the myeloid progenitor stain (GMP and Pre-MegE isolation). Cells were stained with antibodies to mouse antigens to allow separation of the individual populations (Supplementary Table 1). A FACSAria II (BD Biosciences) was used for all cell sorting. Fluorescence-minus-one controls and unstained populations were used as gate-setting controls. Single cells were seeded by an automated cell deposition unit directly into the Fludigm assay mixture (see below). Test sorts before and after single cell sorts verified the purity of all populations at >98% based on expression of cell-surface markers (Supplementary Fig. 1). Fluorescent beads were sorted into 96-well plates before and after samples to verify that only single events were sorted into each well.
Single cell gene expression analysis
Single-cell gene expression analysis was performed using 48.48 Dynamic Array integrated fluidics chips (M48, Fluidigm Corporation) on the BioMark HD platform (Fluidigm Corporation), which facilitates the simultaneous analysis of 48 genes in each of 48 samples. cDNA synthesis and specific target amplification (preamplification) of genes of interest were performed using the CellsDirect One-Step qRT-PCR kit (Invitrogen). Single cells were sorted by FACS directly into individual wells of 96-well plates containing 5μL CellsDirect 2× reaction mix (Invitrogen), 0.1μL SUPERase RNase inhibitor (Ambion), 2.5μL 0.2× assay mix, 1.2μL TE buffer (Invitrogen) and 1.2μL SuperscriptIII/Platinum Taq (Invitrogen). The 0.2× assay mix contained a pool of 24 TaqMan assays (Applied Biosystems; details available on request) at a 1:100 dilution of each assay in TE buffer. Reverse transcription and specific target amplification were performed in the same plates immediately after sorting as follows: 50°C for 15 minutes, 95°C for 2 minutes, 22 × (95°C for 15 seconds, 60°C for 4 minutes). cDNA was diluted 1:5 with TE prior to qPCR on the BioMark HD. cDNA was stored at −20°C before processing on the BioMark HD. 6 positive controls of 20 cells per well, 14 negative controls (no cell sorted) and 124 single cells were sorted for each population. This corresponds to 3 M48 Dynamic Arrays per population, each containing 2 positive controls, 4-6 negative controls and 40-42 single cells. For the qPCR, 3μL of each TaqMan assay was mixed with 3μL Gene Expression Assay Loading Reagent (Fluidigm). 2.7μL of diluted cDNA was mixed with 3μL 2× TaqMan Universal Mastermix (Applied Biosystems) and 0.3μL Gene Expression Sample Loading Reagent (Fluidigm). 5μL of each sample and assay were loaded into individual sample and assay inlets on the M48 Dynamic Array. Samples and assays were then loaded into the reaction chambers of the Dynamic Array using the IFC Controller MX (Fluidigm), and then transferred to the BioMark HD for qPCR (95°C for 10 minutes; 40 cycles of 95°C for 15 s and 60°C for 60 s).
Testing TaqMan Assays
TaqMan assays were tested for single cell PCR by performing standard curves on the BioMark of cDNA from a population of 100 cells of the HPC7 haematopoietic progenitor cell line55, reverse transcribed and preamplified as described above. Assays were selected based on the amplification efficiency and lack of background expression in no template controls. The linear range of all assays used was within the sensitivity of the BioMark HD (Ct 7-27).
Bioinformatic analysis of single cell gene expression data
Single cell expression data were initially analysed with the Fluidigm Data Collection software. For quality control, amplification curves were quality filtered using a threshold of 0.75 and Ct thresholds were set for each assay, with the same thresholds used across all experiments and cell populations. Data were then exported to Excel as .csv files. A table with all of the Ct values is available in the supplemental material accompanying this paper as Supplementary Table 3. Samples not expressing any genes (likely as a result of a failure of the sorter to put a cell in the well) were excluded from the analysis (n=6), as were cells not expressing the housekeeping genes Ubc (n=0) or Polr2a (n=17). Expression values over the cut off of the machine or beyond the linear range of the TaqMan assays (Ct>27) were set to 28. Each assay was performed in duplicate, and the mean of the duplicates was used for subsequent analysis. Following these quality control measures, ΔCt values were calculated as previously described20 by cell-wise normalisation to the mean expression level of Ubc and Polr2a, as the two most robustly expressed housekeeping genes. Briefly, Ct values were subtracted from the assumed no template background of the BioMark of 2820, followed by subtraction of the mean Ct value of Ubc and Polr2a for each cell. The ΔCt value for genes that were not expressed was then set to 15, representative of the detection limit of the BioMark. Hierarchical clustering and principal component analysis were performed in R (www.r-project.org). Hierarchical clustering and principal component analysis were performed only on the data for the 18 transcription factors, excluding c-Kit and housekeeping genes. Hierarchical clustering was performed on cells using Spearman Rank correlation. Positive and negative correlations between pairs of genes were tested with Spearman Rank correlation, with P-values calculated based on 10,000 permutations. Positive correlations with a Z-score above 12 (P < 3E-33) and negative correlations with a Z-score below −4 (P < 6.09E-05) were considered significant, with known antagonistic relationships recovered beyond these values. PCA was performed using the prcomp function.
A non-linear generalisation of PCA, a Gaussian Process Latent Variable Model (GPLVM), was employed to generate a nonlinear embedding of the expression data using the FGPLVM toolbox (http://staffwww.dcs.shef.ac.uk/people/N.Lawrence/fgplvm/). A probabilistic mapping was constructed from a 2 dimensional latent space to the 18 dimensional data space using Gaussian processes40. To allow for non-linear effects we used a radial basis function (rbf) kernel to construct the covariance matrix. The 2D coordinates of the latent points corresponding as well as the kernel parameters were determined by optimising the likelihood of the data-set using 1000 iterations of a scaled conjugate gradient optimiser40. A gene relevance map – corresponding to a generalised loadings plot for standard PCA – was generated; for this the mapping from latent space to data space was used to calculate the gradient of the expected value for 20×20 regularly spaced points across the GPLVM-map and the most important gene (greatest norm of gradient) was plotted41. Single-gene expression maps were generated by calculating the expected value of a single gene for 50×50 equally spaced points across the GPLVM-map. For each cell type the spatial median m in 2D was calculated by minimising the expected distance between m and the 2D coordinates of all cells of the respective cell type using an extended Weiszfeld algorithm68.
Mouse bone marrow derived mast cells (BMMC)
Bone marrow cells were collected from tibias and femurs of 3- to 5-month old adult mice. Cells were cultured in Iscove’s modified Dulbecco’s medium (IMDM) supplemented with 10% fetal bovine serum (Sigma), 1% penicillin/streptomycin (Sigma), 150 μM MTG (Sigma), 10% stem cell factor conditional media from BHK/MKL cells and 10ng/ml of recombinant mIL-3 (Peprotech). Cells were frequently transferred to new flasks to remove adherent cells and experiments were performed after 3 weeks, when cultures were homogenous. Homogeneity of culture was confirmed by presence of FcERI by FACS and toluidine blue staining of cytospins.
Chromatin Immunoprecipitation Sequencing
ChIP assays were performed as previously described11 using polyclonal antibodies against GATA2 (Santa Cruz, sc9008x) and GFI1 (Abcam, ab21061) and control nonspecific rabbit IgG (Sigma, I5006). Each sample was amplified7 and sequenced using the Illumina 2G Genome Analyzer, following manufacturer’s instructions. Sequencing reads were mapped to the mm9 mouse reference genome using Bowtie69, converted to a density plot, and displayed as UCSC genome browser custom tracks.
Transgenic Mouse Analysis and Luciferase assays
Luciferase and LacZ reporter constructs were generated using standard recombinant DNA techniques. Coordinates of chromosomal regions cloned are given in Supplementary Table 2. Luciferase7 assays were performed as described70. E11.5 transgenic mouse embryos were generated and LacZ-stained by Cyagen Biosciences. Staining patterns were analysed as described71.
Supplementary Material
Acknowledgements
We thank Sally Ann Clark for help with FACS sorting. Research in the authors’ laboratory is supported by the Medical Research Council, Leukaemia and Lymphoma Research, The Leukaemia and Lymphoma Society, Cancer Research UK, and core support grants by the Wellcome Trust to the Cambridge Institute for Medical Research and Wellcome Trust - MRC Cambridge Stem Cell Institute.
Footnotes
Author Contributions: V.M. designed and performed single cell experiments, performed analysis, and wrote the paper. B.G. conceived the study, designed experiments and wrote the paper. I.C.M. and S.E.J designed and performed FACS and wrote the paper. F.B. and F.T. analysed GPLVM data and wrote the paper. S.K. and J.S. performed transgenic and luciferase analysis. A.J. and R.H. performed bioinformatic analysis. F.J.C.-N. performed ChIP-Seq experiments. G.S. and M.d.B. designed experiments and performed preliminary studies.
Competing financial interests: The authors declare no competing financial interests.
Accession Numbers
ChIP-seq data for mast cells have been deposited into the NCBI Gene Expression Omnibus portal under the accession number GSE42518.
References
- 1.Orkin SH, Zon LI. Hematopoiesis: an evolving paradigm for stem cell biology. Cell. 2008;132:631–644. doi: 10.1016/j.cell.2008.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ottersbach K, Smith A, Wood A, Gottgens B. Ontogeny of haematopoiesis: recent advances and open questions. Br J Haematol. 2010;148:343–355. doi: 10.1111/j.1365-2141.2009.07953.x. [DOI] [PubMed] [Google Scholar]
- 3.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 4.Davis RL, Weintraub H, Lassar AB. Expression of a single transfected cDNA converts fibroblasts to myoblasts. Cell. 1987;51:987–1000. doi: 10.1016/0092-8674(87)90585-x. [DOI] [PubMed] [Google Scholar]
- 5.Gering M, Yamada Y, Rabbitts TH, Patient RK. Lmo2 and Scl/Tal1 convert non-axial mesoderm into haemangioblasts which differentiate into endothelial cells in the absence of Gata1. Development. 2003;130:6187–6199. doi: 10.1242/dev.00875. [DOI] [PubMed] [Google Scholar]
- 6.Di Tullio A, et al. CCAAT/enhancer binding protein alpha (C/EBP(alpha))-induced transdifferentiation of pre-B cells into macrophages involves no overt retrodifferentiation. Proc Natl Acad Sci U S A. 2011;108:17016–17021. doi: 10.1073/pnas.1112169108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Basso K, et al. Reverse engineering of regulatory networks in human B cells. Nat Genet. 2005;37:382–390. doi: 10.1038/ng1532. [DOI] [PubMed] [Google Scholar]
- 8.Pimanda JE, Gottgens B. Gene regulatory networks governing haematopoietic stem cell development and identity. Int J Dev Biol. 2010;54:1201–1211. doi: 10.1387/ijdb.093038jp. [DOI] [PubMed] [Google Scholar]
- 9.Pimanda JE, et al. Gata2, Fli1, and Scl form a recursively wired gene-regulatory circuit during early hematopoietic development. Proc Natl Acad Sci U S A. 2007;104:17692–17697. doi: 10.1073/pnas.0707045104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wilson NK, Calero-Nieto FJ, Ferreira R, Gottgens B. Transcriptional regulation of haematopoietic transcription factors. Stem Cell Res Ther. 2011;2:6. doi: 10.1186/scrt47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wilson NK, et al. Combinatorial transcriptional control in blood stem/progenitor cells: genome-wide analysis of ten major transcriptional regulators. Cell Stem Cell. 2010;7:532–544. doi: 10.1016/j.stem.2010.07.016. [DOI] [PubMed] [Google Scholar]
- 12.Wilson NK, et al. The transcriptional program controlled by the stem cell leukemia gene Scl/Tal1 during early embryonic hematopoietic development. Blood. 2009;113:5456–5465. doi: 10.1182/blood-2009-01-200048. [DOI] [PubMed] [Google Scholar]
- 13.Sieburg HB, et al. The hematopoietic stem compartment consists of a limited number of discrete stem cell subsets. Blood. 2006;107:2311–2316. doi: 10.1182/blood-2005-07-2970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dykstra B, et al. Long-term propagation of distinct hematopoietic differentiation programs in vivo. Cell Stem Cell. 2007;1:218–229. doi: 10.1016/j.stem.2007.05.015. [DOI] [PubMed] [Google Scholar]
- 15.Copley MR, Beer PA, Eaves CJ. Hematopoietic stem cell heterogeneity takes center stage. Cell Stem Cell. 2012;10:690–697. doi: 10.1016/j.stem.2012.05.006. [DOI] [PubMed] [Google Scholar]
- 16.Ramos CA, et al. Evidence for diversity in transcriptional profiles of single hematopoietic stem cells. PLoS Genet. 2006;2:e159. doi: 10.1371/journal.pgen.0020159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Glotzbach JP, et al. An information theoretic, microfluidic-based single cell analysis permits identification of subpopulations among putatively homogeneous stem cells. PLoS One. 2011;6:e21211. doi: 10.1371/journal.pone.0021211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hu M, et al. Multilineage gene expression precedes commitment in the hemopoietic 8 system. Genes Dev. 1997;11:774–785. doi: 10.1101/gad.11.6.774. [DOI] [PubMed] [Google Scholar]
- 19.Citri A, Pang ZP, Sudhof TC, Wernig M, Malenka RC. Comprehensive qPCR profiling of gene expression in single neuronal cells. Nat Protoc. 2012;7:118–127. doi: 10.1038/nprot.2011.430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guo G, et al. Resolution of cell fate decisions revealed by single-cell gene expression analysis from zygote to blastocyst. Dev Cell. 2010;18:675–685. doi: 10.1016/j.devcel.2010.02.012. [DOI] [PubMed] [Google Scholar]
- 21.Dalerba P, et al. Single-cell dissection of transcriptional heterogeneity in human colon tumors. Nat Biotechnol. 2011;29:1120–1127. doi: 10.1038/nbt.2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pina C, et al. Inferring rules of lineage commitment in haematopoiesis. Nat Cell Biol. 2012;14:287–294. doi: 10.1038/ncb2442. [DOI] [PubMed] [Google Scholar]
- 23.Kiel MJ, Yilmaz OH, Iwashita T, Terhorst C, Morrison SJ. SLAM family receptors distinguish hematopoietic stem and progenitor cells and reveal endothelial niches for stem cells. Cell. 2005;121:1109–1121. doi: 10.1016/j.cell.2005.05.026. [DOI] [PubMed] [Google Scholar]
- 24.Adolfsson J, et al. Identification of Flt3+ lympho-myeloid stem cells lacking erythro-megakaryocytic potential a revised road map for adult blood lineage commitment. Cell. 2005;121:295–306. doi: 10.1016/j.cell.2005.02.013. [DOI] [PubMed] [Google Scholar]
- 25.Pronk CJ, et al. Elucidation of the phenotypic, functional, and molecular topography of a myeloerythroid progenitor cell hierarchy. Cell Stem Cell. 2007;1:428–442. doi: 10.1016/j.stem.2007.07.005. [DOI] [PubMed] [Google Scholar]
- 26.Akashi K, Traver D, Miyamoto T, Weissman IL. A clonogenic common myeloid progenitor that gives rise to all myeloid lineages. Nature. 2000;404:193–197. doi: 10.1038/35004599. [DOI] [PubMed] [Google Scholar]
- 27.Kondo M, Weissman IL, Akashi K. Identification of clonogenic common lymphoid progenitors in mouse bone marrow. Cell. 1997;91:661–672. doi: 10.1016/s0092-8674(00)80453-5. [DOI] [PubMed] [Google Scholar]
- 28.Katayama N, et al. Stage-specific expression of c-kit protein by murine hematopoietic progenitors. Blood. 1993;82:2353–2360. [PubMed] [Google Scholar]
- 29.Donaldson IJ, et al. Genome-wide identification of cis-regulatory sequences controlling blood and endothelial development. Hum Mol Genet. 2005;14:595–601. doi: 10.1093/hmg/ddi056. [DOI] [PubMed] [Google Scholar]
- 30.Gottgens B, et al. Establishing the transcriptional programme for blood: the SCL stem cell enhancer is regulated by a multiprotein complex containing Ets and GATA factors. EMBO J. 2002;21:3039–3050. doi: 10.1093/emboj/cdf286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pimanda JE, et al. The SCL transcriptional network and BMP signaling pathway interact to regulate RUNX1 activity. Proc Natl Acad Sci U S A. 2007;104:840–845. doi: 10.1073/pnas.0607196104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tanaka Y, et al. The transcriptional programme controlled by Runx1 during early embryonic blood development. Dev Biol. 2012;366:404–419. doi: 10.1016/j.ydbio.2012.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wilson NK, et al. Gfi1 expression is controlled by five distinct regulatory regions spread over 100 kilobases, with Scl/Tal1, Gata2, PU.1, Erg, Meis1, and Runx1 acting as upstream regulators in early hematopoietic cells. Mol Cell Biol. 2010;30:3853–3863. doi: 10.1128/MCB.00032-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yamamoto M, Takahashi S, Onodera K, Muraosa Y, Engel JD. Upstream and downstream of erythroid transcription factor GATA-1. Genes Cells. 1997;2:107–115. doi: 10.1046/j.1365-2443.1997.1080305.x. [DOI] [PubMed] [Google Scholar]
- 35.Mouthon MA, et al. Expression of tal-1 and GATA-binding proteins during human hematopoiesis. Blood. 1993;81:647–655. [PubMed] [Google Scholar]
- 36.Orlic D, Anderson S, Biesecker LG, Sorrentino BP, Bodine DM. Pluripotent hematopoietic stem cells contain high levels of mRNA for c-kit, GATA-2, p45 NF-E2, and c-myb and low levels or no mRNA for c-fms and the receptors for granulocyte colony-stimulating factor and interleukins 5 and 7. Proc Natl Acad Sci U S A. 1995;92:4601–4605. doi: 10.1073/pnas.92.10.4601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Doan LL, et al. Targeted transcriptional repression of Gfi1 by GFI1 and GFI1B in lymphoid cells. Nucleic Acids Res. 2004;32:2508–2519. doi: 10.1093/nar/gkh570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vassen L, Okayama T, Moroy T. Gfi1b:green fluorescent protein knock-in mice reveal a dynamic expression pattern of Gfi1b during hematopoiesis that is largely complementary to Gfi1. Blood. 2007;109:2356–2364. doi: 10.1182/blood-2006-06-030031. [DOI] [PubMed] [Google Scholar]
- 39.Luc S, et al. The earliest thymic T cell progenitors sustain B cell and myeloid lineage potential. Nat Immunol. 2012;13:412–419. doi: 10.1038/ni.2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lawrence ND. Local distance preservation in the GP-LVM through back constraints. ICML Proceedings of the 23rd International Conference on Machine Learning. 2006:513–520. [Google Scholar]
- 41.Buettner F, Theis FJ. A novel approach for resolving differences in single-cell gene expression patterns from zygote to blastocyst. Bioinformatics. 2012;28:i626–i632. doi: 10.1093/bioinformatics/bts385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shivdasani RA, Orkin SH. Erythropoiesis and globin gene expression in mice lacking the transcription factor NF-E2. Proc Natl Acad Sci U S A. 1995;92:8690–8694. doi: 10.1073/pnas.92.19.8690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shivdasani RA, et al. Transcription factor NF-E2 is required for platelet formation independent of the actions of thrombopoietin/MGDF in megakaryocyte development. Cell. 1995;81:695–704. doi: 10.1016/0092-8674(95)90531-6. [DOI] [PubMed] [Google Scholar]
- 44.Hall MA, et al. The critical regulator of embryonic hematopoiesis, SCL, is vital in the adult for megakaryopoiesis, erythropoiesis, and lineage choice in CFU-S12. Proc Natl Acad Sci U S A. 2003;100:992–997. doi: 10.1073/pnas.0237324100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huang Z, et al. GATA-2 reinforces megakaryocyte development in the absence of GATA-1. Mol Cell Biol. 2009;29:5168–5180. doi: 10.1128/MCB.00482-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ikonomi P, et al. Overexpression of GATA-2 inhibits erythroid and promotes megakaryocyte differentiation. Exp Hematol. 2000;28:1423–1431. doi: 10.1016/s0301-472x(00)00553-1. [DOI] [PubMed] [Google Scholar]
- 47.Porcher C, et al. The T cell leukemia oncoprotein SCL/tal-1 is essential for development of all hematopoietic lineages. Cell. 1996;86:47–57. doi: 10.1016/s0092-8674(00)80076-8. [DOI] [PubMed] [Google Scholar]
- 48.Schuh AH, et al. ETO-2 associates with SCL in erythroid cells and megakaryocytes and provides repressor functions in erythropoiesis. Mol Cell Biol. 2005;25:10235–10250. doi: 10.1128/MCB.25.23.10235-10250.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hamlett I, et al. Characterization of megakaryocyte GATA1-interacting proteins: the corepressor ETO2 and GATA1 interact to regulate terminal megakaryocyte maturation. Blood. 2008;112:2738–2749. doi: 10.1182/blood-2008-03-146605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Huang S, Guo YP, May G, Enver T. Bifurcation dynamics in lineage-commitment in bipotent progenitor cells. Dev Biol. 2007;305:695–713. doi: 10.1016/j.ydbio.2007.02.036. [DOI] [PubMed] [Google Scholar]
- 51.Nerlov C, Graf T. PU. 1 induces myeloid lineage commitment in multipotent hematopoietic progenitors. Genes Dev. 1998;12:2403–2412. doi: 10.1101/gad.12.15.2403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bonadies N, et al. Genome-wide analysis of transcriptional reprogramming in mouse models of acute myeloid leukaemia. PLoS One. 2011;6:e16330. doi: 10.1371/journal.pone.0016330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Khandanpour C, et al. The human GFI136N variant induces epigenetic changes at the Hoxa9 locus and accelerates K-RAS driven myeloproliferative disorder in mice. Blood. 2012;120:4006–4017. doi: 10.1182/blood-2011-02-334722. [DOI] [PubMed] [Google Scholar]
- 54.Grass JA, et al. Distinct functions of dispersed GATA factor complexes at an endogenous gene locus. Mol Cell Biol. 2006;26:7056–7067. doi: 10.1128/MCB.01033-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pinto do OP, Kolterud A, Carlsson L. Expression of the LIM-homeobox gene LH2 generates immortalized steel factor-dependent multipotent hematopoietic precursors. EMBO J. 1998;17:5744–5756. doi: 10.1093/emboj/17.19.5744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nardelli J, Thiesson D, Fujiwara Y, Tsai FY, Orkin SH. Expression and genetic interaction of transcription factors GATA-2 and GATA-3 during development of the mouse central nervous system. Dev Biol. 1999;210:305–321. doi: 10.1006/dbio.1999.9278. [DOI] [PubMed] [Google Scholar]
- 57.Bee T, et al. The mouse Runx1 +23 hematopoietic stem cell enhancer confers hematopoietic specificity to both Runx1 promoters. Blood. 2009;113:5121–5124. doi: 10.1182/blood-2008-12-193003. [DOI] [PubMed] [Google Scholar]
- 58.Gottgens B, et al. cis-Regulatory remodeling of the SCL locus during vertebrate evolution. Mol Cell Biol. 2010;30:5741–5751. doi: 10.1128/MCB.00870-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Foster SD, Oram SH, Wilson NK, Gottgens B. From genes to cells to tissues-modelling the haematopoietic system. Molecular Biosystems. 2009;5:1413–1420. doi: 10.1039/B907225j. [DOI] [PubMed] [Google Scholar]
- 60.Narula J, Smith AM, Gottgens B, Igoshin OA. Modeling reveals bistability and low-pass filtering in the network module determining blood stem cell fate. PLoS Comput Biol. 2010;6:e1000771. doi: 10.1371/journal.pcbi.1000771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Alon U. Network motifs: theory and experimental approaches. Nat Rev Genet. 2007;8:450–461. doi: 10.1038/nrg2102. [DOI] [PubMed] [Google Scholar]
- 62.Tipping AJ, et al. High GATA-2 expression inhibits human hematopoietic stem and progenitor cell function by effects on cell cycle. Blood. 2009;113:2661–2672. doi: 10.1182/blood-2008-06-161117. [DOI] [PubMed] [Google Scholar]
- 63.Monteiro R, Pouget C, Patient R. The gata1/pu. 1 lineage fate paradigm varies between blood populations and is modulated by tif1gamma. EMBO J. 2011;30:1093–1103. doi: 10.1038/emboj.2011.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hahn CN, et al. Heritable GATA2 mutations associated with familial myelodysplastic syndrome and acute myeloid leukemia. Nat Genet. 2011;43:1012–1017. doi: 10.1038/ng.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Phelan JD, et al. Growth Factor Independent-1 (Gfi1) Is Critically Required for T-Cell Acute Lymphoblastic Leukemia (T-ALL) Tumor Initiation and Maintenance. Blood (ASH Annual Meeting Abstracts) 2010;116:3156. [Google Scholar]
- 66.Szklarczyk D, et al. The STRING database in 2011: functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res. 2011;39:D561–568. doi: 10.1093/nar/gkq973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kim WK, Krumpelman C, Marcotte EM. Inferring mouse gene functions from genomic-scale data using a combined functional network/classification strategy. Genome Biol 9 Suppl. 2008;1:S5. doi: 10.1186/gb-2008-9-s1-s5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Milasevic P.a.D. G.R. Uniqueness of the Spatial Median. Annals of Statistics. 1987;15:1332–1333. [Google Scholar]
- 69.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bockamp EO, et al. Transcriptional regulation of the stem cell leukemia gene by PU. 1 and Elf-1. J Biol Chem. 1998;273:29032–29042. doi: 10.1074/jbc.273.44.29032. [DOI] [PubMed] [Google Scholar]
- 71.Landry JR, et al. Fli1, Elf1, and Ets1 regulate the proximal promoter of the LMO2 gene in endothelial cells. Blood. 2005;106:2680–2687. doi: 10.1182/blood-2004-12-4755. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.