Abstract
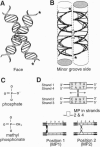
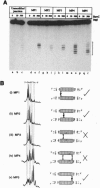
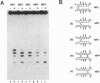
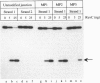
The RuvC protein of Escherichia coli catalyzes the resolution of recombination intermediates during genetic recombination and the recombinational repair of damaged DNA. Resolution involves specific recognition of the Holliday structure to form a complex that exhibits twofold symmetry with the DNA in an open configuration. Cleavage occurs when strands of like polarity are nicked at the sequence 5'-WTT decreases S-3' (where W is A or T and S is G or C). To determine whether the cleavage site needs to be located at, or close to, the point at which DNA strands exchange partners, Holliday structures were constructed with the junction points at defined sites within this sequence. We found that the efficiency of resolution was optimal when the cleavage site was coincident with the position of DNA strand exchange. In these studies, junction targeting was achieved by incorporating uncharged methyl phosphonates into the DNA backbone, providing further evidence for the importance of charge-charge repulsions in determining DNA structure.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arciszewska L., Grainge I., Sherratt D. Effects of Holliday junction position on Xer-mediated recombination in vitro. EMBO J. 1995 Jun 1;14(11):2651–2660. doi: 10.1002/j.1460-2075.1995.tb07263.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ariyoshi M., Vassylyev D. G., Iwasaki H., Nakamura H., Shinagawa H., Morikawa K. Atomic structure of the RuvC resolvase: a holliday junction-specific endonuclease from E. coli. Cell. 1994 Sep 23;78(6):1063–1072. doi: 10.1016/0092-8674(94)90280-1. [DOI] [PubMed] [Google Scholar]
- Bennett R. J., Dunderdale H. J., West S. C. Resolution of Holliday junctions by RuvC resolvase: cleavage specificity and DNA distortion. Cell. 1993 Sep 24;74(6):1021–1031. doi: 10.1016/0092-8674(93)90724-5. [DOI] [PubMed] [Google Scholar]
- Bennett R. J., West S. C. RuvC protein resolves Holliday junctions via cleavage of the continuous (noncrossover) strands. Proc Natl Acad Sci U S A. 1995 Jun 6;92(12):5635–5639. doi: 10.1073/pnas.92.12.5635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett R. J., West S. C. Structural analysis of the RuvC-Holliday junction complex reveals an unfolded junction. J Mol Biol. 1995 Sep 15;252(2):213–226. doi: 10.1006/jmbi.1995.0489. [DOI] [PubMed] [Google Scholar]
- Bhattacharyya A., Murchie A. I., von Kitzing E., Diekmann S., Kemper B., Lilley D. M. Model for the interaction of DNA junctions and resolving enzymes. J Mol Biol. 1991 Oct 20;221(4):1191–1207. doi: 10.1016/0022-2836(91)90928-y. [DOI] [PubMed] [Google Scholar]
- Crothers D. M. Upsetting the balance of forces in DNA. Science. 1994 Dec 16;266(5192):1819–1820. doi: 10.1126/science.7997876. [DOI] [PubMed] [Google Scholar]
- Dickie P., McFadden G., Morgan A. R. The site-specific cleavage of synthetic Holliday junction analogs and related branched DNA structures by bacteriophage T7 endonuclease I. J Biol Chem. 1987 Oct 25;262(30):14826–14836. [PubMed] [Google Scholar]
- Duckett D. R., Murchie A. I., Diekmann S., von Kitzing E., Kemper B., Lilley D. M. The structure of the Holliday junction, and its resolution. Cell. 1988 Oct 7;55(1):79–89. doi: 10.1016/0092-8674(88)90011-6. [DOI] [PubMed] [Google Scholar]
- Duckett D. R., Murchie A. I., Lilley D. M. The role of metal ions in the conformation of the four-way DNA junction. EMBO J. 1990 Feb;9(2):583–590. doi: 10.1002/j.1460-2075.1990.tb08146.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunderdale H. J., Benson F. E., Parsons C. A., Sharples G. J., Lloyd R. G., West S. C. Formation and resolution of recombination intermediates by E. coli RecA and RuvC proteins. Nature. 1991 Dec 19;354(6354):506–510. doi: 10.1038/354506a0. [DOI] [PubMed] [Google Scholar]
- Dunderdale H. J., Sharples G. J., Lloyd R. G., West S. C. Cloning, overexpression, purification, and characterization of the Escherichia coli RuvC Holliday junction resolvase. J Biol Chem. 1994 Feb 18;269(7):5187–5194. [PubMed] [Google Scholar]
- Iwasaki H., Takahagi M., Nakata A., Shinagawa H. Escherichia coli RuvA and RuvB proteins specifically interact with Holliday junctions and promote branch migration. Genes Dev. 1992 Nov;6(11):2214–2220. doi: 10.1101/gad.6.11.2214. [DOI] [PubMed] [Google Scholar]
- Iwasaki H., Takahagi M., Shiba T., Nakata A., Shinagawa H. Escherichia coli RuvC protein is an endonuclease that resolves the Holliday structure. EMBO J. 1991 Dec;10(13):4381–4389. doi: 10.1002/j.1460-2075.1991.tb05016.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kho S. H., Landy A. Dissecting the resolution reaction of lambda integrase using suicide Holliday junction substrates. EMBO J. 1994 Jun 1;13(11):2714–2724. doi: 10.1002/j.1460-2075.1994.tb06562.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilley D. M., Clegg R. M. The structure of the four-way junction in DNA. Annu Rev Biophys Biomol Struct. 1993;22:299–328. doi: 10.1146/annurev.bb.22.060193.001503. [DOI] [PubMed] [Google Scholar]
- Lloyd R. G., Sharples G. J. Dissociation of synthetic Holliday junctions by E. coli RecG protein. EMBO J. 1993 Jan;12(1):17–22. doi: 10.1002/j.1460-2075.1993.tb05627.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahdi A. A., Sharples G. J., Mandal T. N., Lloyd R. G. Holliday junction resolvases encoded by homologous rusA genes in Escherichia coli K-12 and phage 82. J Mol Biol. 1996 Apr 5;257(3):561–573. doi: 10.1006/jmbi.1996.0185. [DOI] [PubMed] [Google Scholar]
- Mandal T. N., Mahdi A. A., Sharples G. J., Lloyd R. G. Resolution of Holliday intermediates in recombination and DNA repair: indirect suppression of ruvA, ruvB, and ruvC mutations. J Bacteriol. 1993 Jul;175(14):4325–4334. doi: 10.1128/jb.175.14.4325-4334.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunes-Düby S. E., Azaro M. A., Landy A. Swapping DNA strands and sensing homology without branch migration in lambda site-specific recombination. Curr Biol. 1995 Feb 1;5(2):139–148. doi: 10.1016/s0960-9822(95)00035-2. [DOI] [PubMed] [Google Scholar]
- Parsons C. A., Kemper B., West S. C. Interaction of a four-way junction in DNA with T4 endonuclease VII. J Biol Chem. 1990 Jun 5;265(16):9285–9289. [PubMed] [Google Scholar]
- Parsons C. A., Stasiak A., Bennett R. J., West S. C. Structure of a multisubunit complex that promotes DNA branch migration. Nature. 1995 Mar 23;374(6520):375–378. doi: 10.1038/374375a0. [DOI] [PubMed] [Google Scholar]
- Parsons C. A., West S. C. Specificity of binding to four-way junctions in DNA by bacteriophage T7 endonuclease I. Nucleic Acids Res. 1990 Aug 11;18(15):4377–4384. doi: 10.1093/nar/18.15.4377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picksley S. M., Parsons C. A., Kemper B., West S. C. Cleavage specificity of bacteriophage T4 endonuclease VII and bacteriophage T7 endonuclease I on synthetic branch migratable Holliday junctions. J Mol Biol. 1990 Apr 20;212(4):723–735. doi: 10.1016/0022-2836(90)90233-C. [DOI] [PubMed] [Google Scholar]
- Saito A., Iwasaki H., Ariyoshi M., Morikawa K., Shinagawa H. Identification of four acidic amino acids that constitute the catalytic center of the RuvC Holliday junction resolvase. Proc Natl Acad Sci U S A. 1995 Aug 1;92(16):7470–7474. doi: 10.1073/pnas.92.16.7470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah R., Bennett R. J., West S. C. Activation of RuvC Holliday junction resolvase in vitro. Nucleic Acids Res. 1994 Jul 11;22(13):2490–2497. doi: 10.1093/nar/22.13.2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah R., Bennett R. J., West S. C. Genetic recombination in E. coli: RuvC protein cleaves Holliday junctions at resolution hotspots in vitro. Cell. 1994 Dec 2;79(5):853–864. doi: 10.1016/0092-8674(94)90074-4. [DOI] [PubMed] [Google Scholar]
- Strauss J. K., Maher L. J., 3rd DNA bending by asymmetric phosphate neutralization. Science. 1994 Dec 16;266(5192):1829–1834. doi: 10.1126/science.7997878. [DOI] [PubMed] [Google Scholar]
- Tsaneva I. R., Müller B., West S. C. ATP-dependent branch migration of Holliday junctions promoted by the RuvA and RuvB proteins of E. coli. Cell. 1992 Jun 26;69(7):1171–1180. doi: 10.1016/0092-8674(92)90638-s. [DOI] [PubMed] [Google Scholar]
- West S. C. The RuvABC proteins and Holliday junction processing in Escherichia coli. J Bacteriol. 1996 Mar;178(5):1237–1241. doi: 10.1128/jb.178.5.1237-1241.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S., Fu T. J., Seeman N. C. Symmetric immobile DNA branched junctions. Biochemistry. 1993 Aug 17;32(32):8062–8067. doi: 10.1021/bi00083a002. [DOI] [PubMed] [Google Scholar]
- Zhang S., Seeman N. C. Symmetric Holliday junction crossover isomers. J Mol Biol. 1994 May 20;238(5):658–668. doi: 10.1006/jmbi.1994.1327. [DOI] [PubMed] [Google Scholar]