Abstract
Non-coding RNAs (ncRNAs) recently were discovered to outnumber their protein-coding counterparts, yet their diverse functions are still poorly understood. Here we report on a method for the intracellular Single-molecule High Resolution Localization and Counting (iSHiRLoC) of microRNAs (miRNAs), a conserved, ubiquitous class of regulatory ncRNAs that controls the expression of over 60% of all mammalian protein coding genes post-transcriptionally, by a mechanism shrouded by seemingly contradictory observations. We present protocols to execute single particle tracking (SPT) and single-molecule counting of functional microinjected, fluorophore-labeled miRNAs and thereby extract diffusion coefficients and molecular stoichiometries of micro-ribonucleoprotein (miRNP) complexes from living and fixed cells, respectively. This probing of miRNAs at the single molecule level sheds new light on the intracellular assembly/disassembly of miRNPs, thus beginning to unravel the dynamic nature of this important gene regulatory pathway and facilitating the development of a parsimonious model for their obscured mechanism of action.
Introduction
Current estimates place the fraction of the mammalian genome that is transcribed into RNA at up to ~75% [1,2]. Considering that only ~1.2% of the entire genome is protein coding [1], non-coding RNAs (ncRNAs) have been predicted to be one of the most abundant molecular species within the eukaryotic cell. Although ncRNAs were initially assumed to be transcriptional noise, i.e., a basal amount of erroneous transcription accompanying the fruitful synthesis of protein-coding messenger RNAs (mRNAs), recent findings suggest that a majority of them exhibit profound structural, catalytic and regulatory roles [2–5], thus forming the basis of cellular homeostasis. One such class of endogenous regulatory RNAs is microRNA (miRNA).
miRNAs are endogenously expressed and processed small ncRNAs, ~22 nucleotides (nt) in length, which in association with a cohort of proteins, collectively termed the miRNA induced silencing complex (miRISC), bind to short (6–8 nt) so-called seed sequences in the 3’ untranslated regions (UTRs) of target mRNAs to typically downregulate gene expression at the post-transcriptional level [6,7]. As abundant high-level gene regulators they play key roles in almost all cellular processes [6,7], and their aberrant expression has been associated with several human pathologies, including cancer [5]. About 1,500 different mammalian miRNAs have been discovered so far and the expression of ~60% of all human mRNAs are computationally predicted to be controlled by miRNAs, while the rest appears to have evolved to avoid such control [6,8]. As an additional layer of complexity, a single miRNA is thought to typically regulate several hundred mRNAs, whereas a single mRNA is predicted to often be regulated by multiple miRNAs of the same or different sequence. A picture thus emerges of a complex regulatory network that allows for rapid adaptation of the whole transcriptome to changing environmental cues during, for example, cell development or cancer onset. However, tools to experimentally dissect and verify this regulatory network are emerging only slowly.
A number of competing hypothesis regarding the mechanism of miRNA-induced gene repression have been proposed [9–11], and when we developed the method described here it was not yet clear whether miRNAs repress their target in a specific cell either by a single mechanism or by several parallel mechanisms. Moreover, the nature and extent of miRNA-mRNA interactions and the spatial organization of miRNAs as they mediate repression are still a mystery. The functionally pervasive nature of miRNAs and lack of an incisive tool to quantitatively differentiate the intracellular miRNA pathways prompted us to utilize single molecule fluorescence microscopy (SMFM); an unparalleled tool that has exposed molecular dispersion and short-lived and/or rare intermediates otherwise masked in the ensemble average [12].
SMFM has been extensively applied to living cells in the form of single particle tracking (SPT) or single molecule counting to understand diffusion and oligomerization states of biomolecular complexes [13]. The existing strategies for SPT in living cells either decorate the target molecule with endogenously expressed fluorescent protein(s) [14] or endocytosed nanoparticles [15], leading to high-molecular weight appendices that may impede function; or they employ microinjection of target particles externally labeled with multiple fluorophores [16]. These approaches so far have been applied to proteins, protein assemblies and large RNA:protein complexes. Complementary methods have recently been developed to assess the number and location of single intracellular molecules by fixing and permeabilizing a cell and incubating it with probes that are labeled with multiple fluorophores or nanoparticles [17], limiting this approach to the detection of relatively large protein and RNA molecules. Small RNAs, such as miRNAs have also been previously counted both in vitro [18] and in situ [19], but neither in vivo nor in a form that allowed them to still be functional. All prior methods required the hybridization of microscopically detectable complementary oligonucleotides to miRNAs, after cell lysis or fixation, and were used predominantly to quantify endogenous miRNAs. Although efficient in understanding miRNA expression patterns, even at the single cell level [19], these methods could not visualize the intracellular assembly and transport of miRNA-associated complexes, the processes that are most fundamental to their biological function. Here we report a method to visualize single, functional, singly-fluorophore labeled miRNA molecules inside live as well as formaldehyde-fixed human cells, a technique laying the foundation for understanding their intracellular localization, assembly and diffusion. We have termed our tool intracellular Single-molecule High Resolution Localization and Counting, or iSHiRLoC.
1. iSHiRLoC – a detailed description of the materials and methods
1.1. Outline of the method
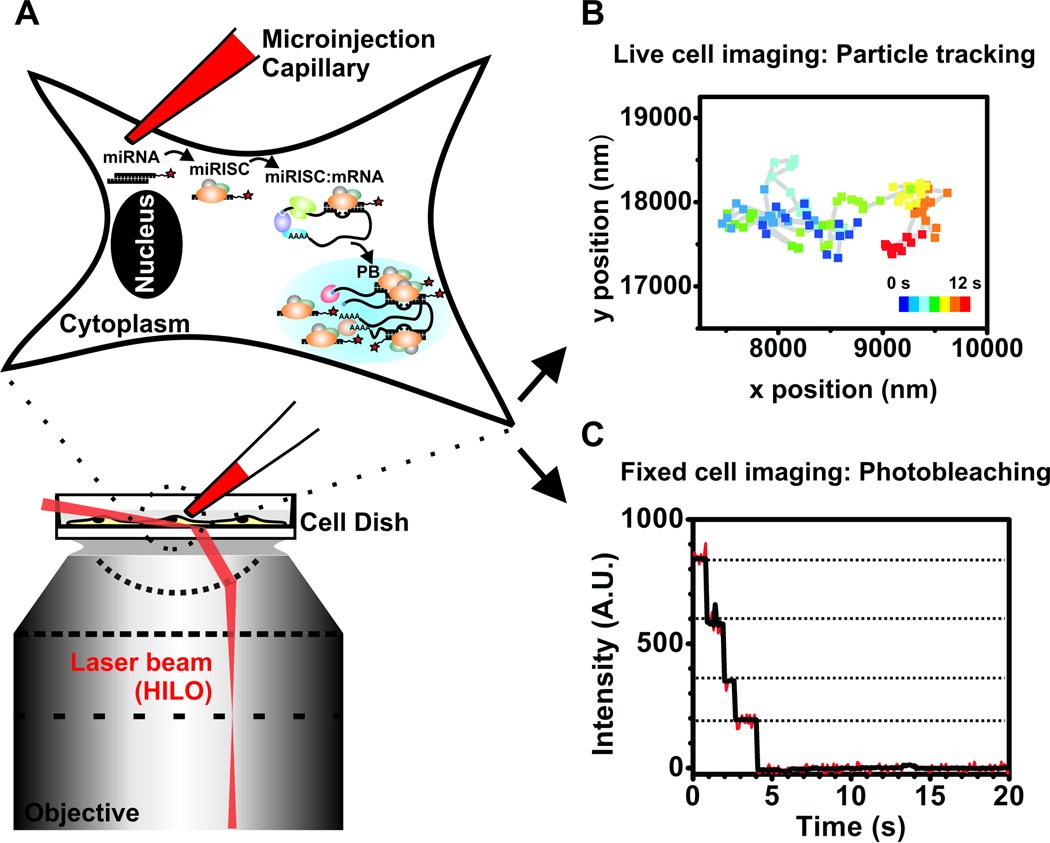
Three previously reported, disparate methods – intracellular single molecule detection utilizing an optimized illumination strategy, single particle tracking microscopy and single molecule counting by stepwise photobleaching of fluorescent probes – were carefully combined with microinjection into a widely applicable single molecule tool to investigate the intracellular diffusion and distribution of miRNAs (Fig. 1) [20]. More specifically, adherent mammalian cells maintained under their natural growth conditions (typically 37 °C supplemented with 5% CO2 in air) were first microinjected in their cytoplasm with fluorophore labeled functional miRNAs (1.2. Validating iSHiRLoC as a biologically relevant method). We chose microinjection over other methods of intracellular delivery for two main reasons: (a) it allowed us to precisely control the number of molecules that were delivered to each cell and, (b) it defined a clear starting point in our assays. By controlling the number of molecules injected, we were able to introduce close to endogenous levels of the miRNA, thereby minimizing cell perturbation and achieving sufficiently low densities of miRNA-containing particles to discern them as distinct point spread functions, facilitating single molecule detection. As microinjected materials are immediately exposed to the cellular content, we could clearly define a starting point in our assays – seldom achievable using methods like transfection, where the introduced materials, at least initially, are trapped in endocytic vesicles. After a certain period of incubation, the cells were imaged by highly inclined laminar optical sheet (HILO) microscopy to achieve deeper penetration into cells without compromising on signal-to-noise ratio [21]. Single particle tracking (SPT) fluorescence microscopy was then used to localize and track individual miRNA particles inside living cells with a temporal resolution of 100 ms and a spatial accuracy of ~30 nm, effectively breaking the theoretical limit of resolution that conventional light microscopy suffers. Diffusion coefficients and diffusive patterns of individual particles thus obtained shed light on the molecular mass of complexes and their intracellular behavior. In a complementary method, cells were formaldehyde-fixed post-microinjection to immobilize all particles and then continuously illuminated until all particles photobleached. The quantized stochastic, and hence stepwise, photobleaching of single fluorophores quantifies the number of miRNA molecules per particle and thus the oligomerization state of micro-ribonucleoprotein (miRNP) complexes. Put together, iSHiRLoC illuminates the intracellular assembly of miRNP complexes and advances the understanding of their biological mechanism of action.
Figure 1.
Schematic of iSHiRLoC.
1.2. Instrumentation
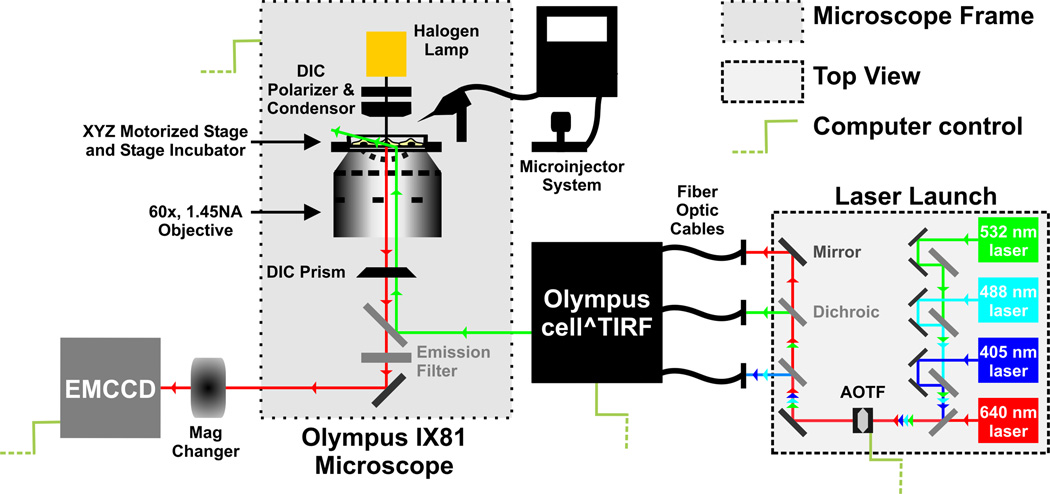
Fluorescence microscopy was performed on an Olympus IX81 microscope (Fig. 2) that was equipped with 3 different objectives; 10× and 20× low magnification objectives, and a 60× 1.45 NA oil-immersion objective (Olympus). The microscope featured an internal 1.6× magnification and was additionally equipped with a 1–4× magnification changer (Olympus). We routinely performed ensemble fluorescence microscopy using the 20× objective and single-molecule imaging using the 60× objective with additional 2× magnification to a total of 120× magnification. Samples were positioned on a nanometer-precision motorized stage (ASI Imaging) with xyz-position motor control and z-position piezo control. The microscope also contains an infrared laser coupled zero-drift control module (Olympus) to correct for focal drift (which was not used in our experiments since drift was minor over the imaging period). Images were acquired using a 512 × 512 pixel area of an EVOLVE EM-CCD camera (Photometrics) at 100 ms exposure time. Solid state lasers with wavelengths of 405 nm (20 mW), 488 nm (25 mW), 532 nm (100 mW) and 640 nm (100 mW), were directed through an acousto-optical tunable filter (AOTF) and split into separate fiber-optic cables within a home-built laser launch system. The AOTF allowed for computer mediated selection of the appropriate laser wavelength for illumination and modulation of laser intensity. AOTF coupling also enabled sub-millisecond switching between multiple laser wavelengths, forming the basis for alternating or interlaced excitation schemes. The fiber-coupled laser beams were directed into a cell-TIRF laser-combining module (Olympus). All laser lines were optically filtered using 10 nm bandwidth clean-up filters to ensure narrow bandwidth illumination. A maximum of ~ 0.8 mW, 1.2 mW, 7 mW and 8 mW of laser power were achieved at the objective for the 405 nm, 488 nm, 532 nm and 640 nm laser lines, respectively. All laser beams were focused on the back-focal plane of the objective and aligned to travel parallel to the optical axis such that the incident angle of illumination at the dish-medium interface can be controlled electronically by changing the distance of the beam from the optical axis at ~0.01° angular resolution using the cell-TIRF module. GFP and Cy5 were excited by the 488 nm and 640 nm lasers, respectively, and their fluorescence was detected using a dual-band filter cube consisting of a z491/639rpc dichroic filter (Chroma) and a z491/639m emission filter (Chroma). mCherry and Cy3 were excited using the 532 nm laser and their emission was detected using a Q570LP dichroic filter (Chroma) and HQ610/75m emission filter (Chroma). Cells grown on DeltaT dishes (Bioptechs) were maintained at 37 °C on the microscope stage while imaging using the DeltaT open dish system (Bioptechs) and a heated lid (Bioptechs). 5% CO2 in air was supplemented to cells using the side port of the heated lid. Microinjections were performed using a Femtojet pump (Eppendorf) and an Injectman NI2 micromanipulator (Eppendorf), which was mounted on the microscope via the back adapter, just adjacent to the stage. Capillaries (500-nm tip diameter Femtotips, Eppendorf), positioned at an angle of 45° with respect to the plane of the sample, were loaded with the appropriate concentration of injection solution just prior to microinjection.
Figure 2.
Schematic of the microscope used for iSHiRLoC.
1.3. Cell culture, media formulations, injection sample and microinjection
Selecting an appropriate cell line with favorable physical and morphological characteristics is critical to iSHiRLoC’s experimental design. For instance, it is preferable to use adherent cells that are large yet flat. The use of adherent cells prevents the accidental removal of cells during microinjection. Optimal cell size and flatness ensures favorable spatial distribution of fluorescent particles and reduced likelihood for particles to diffuse out of the imaging plane, both of which are ideal for single particle tracking and single molecule imaging. Moreover, the cell lines should be capable of accommodating the fluorophore labeled molecule of interest in its appropriate cellular function, even after microinjection. U2OS and HeLa cells are two examples of human cell lines that meet the above criteria and have been successfully used in iSHiRLoC experiments.
Cells are typically maintained in their regular growth medium in an incubator set at 37 °C, supplemented with 5% CO2 and 95% relative humidity in air. We use DMEM (GIBCO, Cat# 11995) supplemented with 10% FBS and 1× NEAA to culture HeLa cells and McCoy’s 5A basal medium (GIBCO, Cat# 16600) supplemented with 10% FBS for U2OS cells. Prior to microinjection, the cell medium is exchanged for a phenol red-free basal medium supplemented with reduced FBS (minimal tolerated FBS media, MTF) to reduce autofluorescence and stabilize the dish for the addition of the microinjection medium. We use HEPES buffered phenol red-free DMEM (GIBCO, Cat# 20163) supplemented with 2% FBS, 1× NEAA and 1 mM sodium pyruvate for HeLa cells and phenol red-free McCoy’s 5A basal medium (Promocell GmbH, C-73227) supplemented with 2% FBS for U2OS cells.
Cell medium is a viscous solution of high osmolarity that can easily clog or reduce the flow of injectate from the narrow opening of a microinjector capillary and thus can influence the number of successful injections. To reduce capillary clogging, cells are soaked in a HEPES buffered saline (HBS) minimal medium (20 mM HEPES-KOH, pH 7.4, 135 mM NaCl, 5 mM KCl, 1 mM MgCl2, 1.8 mM CaCl2 and 5.6 mM glucose) of lower viscosity and osmolarity than the injection solution. These characteristics, together with the constant compensation pressure applied during microinjection, prevents clogging and backflow of solution into the capillary. HBS is also used as an imaging medium for live cell imaging because it contains neither serum nor vitamins, both of which significantly contribute to autofluorescence [22]. However, due to its minimalistic nature, it is essential to test the compatibility of both MTF and HBS with each cell line to ensure minimal perturbation to cell health, especially cell division time, during and after microinjection.
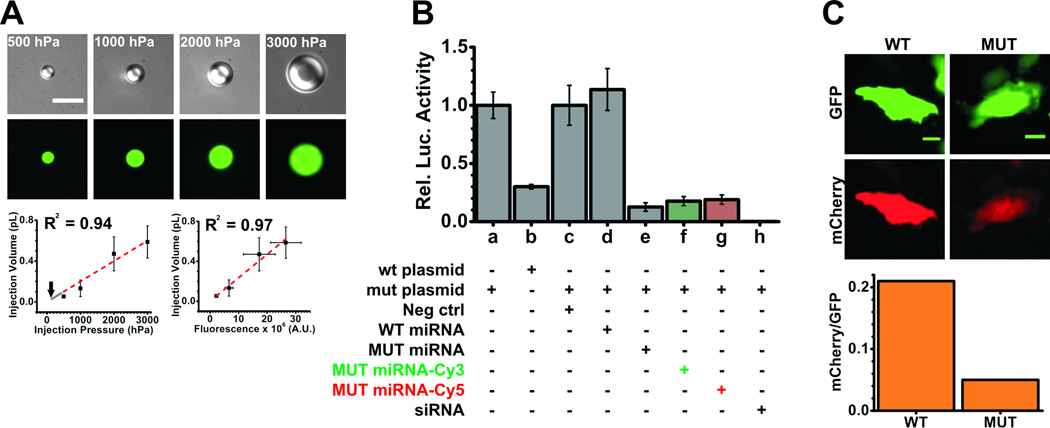
Selection of injection pressure is based on the extent to which the cells are perturbed by the act of microinjection and the amount of material that is required to be injected into the cell. Beyond a certain pressure threshold cells may simply explode upon injection, hence it is best to start at an injection pressure recommended by the manufacturer for adherent cells. For instance, Eppendorf recommends using an injection pressure of 50 – 500 hPa, 0.3 s injection time and 30 – 300 hPa compensation pressure. The injection pressure is the pressure applied by the pump to the solution during each injection, whereas the compensation pressure is the constant pressure that is applied to the capillary to prevent back-flow of solution into the capillary. The injection time corresponds to the length of time the capillary remains in the cell during injection while the set injection pressure is applied. Selection of compensation pressure is dependent on the osmolarity of the medium and is set by finding the lowest value at which there is a constant flow of materials from the capillary. Typically, the injection pressure is set higher than the compensation pressure. In practice, microinjections for all SPT and single-molecule counting experiments [20] were performed at 100 hPa injection pressure for 0.5 s with 20 hPa compensation pressure. Based on our calibration (Fig. 3A), the microinjection volume was ~0.02 pL or ~0.5% – 5% of the total cell volume, which translates to ~18,000 molecules of miRNA (using a 1.5 µM microinjection concentration) at these settings.
Figure 3.
Control experiments. (A) Microinjector calibration. Fluorescein dextran (0.5 mg/ml, 10 kDa) reconstituted in PBS was injected into cargille type FF oil at injection pressures ranging from 500–3000 hPa. Top, representative DIC (gray scale) and fluorescence images (green) of fluorescein dextran droplets (top) at different injection pressure. Scale bar, 10 µm. Bottom left, injection volume as calculated from the volume of dextran droplets was plotted as a function of injection pressure and fit by linear regression (y = 0.0002×, red dotted line). Grey line shows extrapolation of fit to 100 hPa, marked by an arrow, which corresponds to an injection volume of 0.02 pL and translates to ~18,000 molecules of miRNAs injected at a concentration of 1.5 µM. Error bars, standard deviation of at least 3 independent microinjections. Bottom right, plot of injection (or dextran droplet) volume versus fluorescence, also fit by linear regression (y = 0.0235×106x, red dotted line). In either case, the linearity was not maintained beyond 3,000 hPa injection pressure. (B) Functional analyses of miRNAs using luciferase assays. HeLa cells were co-transfected with luciferase reporter plasmids and double-stranded (ds) small RNAs. The plasmids either contained the wild-type (wt) or mutant (mut) 3`UTR of HMGA2. Ds-RNA (50 nM) was a negative control siRNA (neg ctrl), wt-let-7-a1 miRNA (WT miRNA), mut-let-7-a1 miRNA (MUT miRNA, with or without fluorophore) or a positive control luciferase siRNA (siRNA). Firefly luciferase activity was normalized with Renilla luciferase activity within each sample. wt (a) and mut (b) plasmid samples were normalized with respect to (a) to show the expression of the wt plasmid relative to the mut plasmid. The reporter bearing the wt 3`UTR is strongly repressed by endogenous let-7-a1 miRNA. All other samples were normalized with respect to the neg ctrl (c) sample. (C) Functional analyses of miRNAs using fluorescence reporter assays. Representative pseudocolored images (top) of HeLa cells expressing GFP (green) and mCherry (red) from samples that were injected with the mut 3`UTR containing mCherry reporter plasmid, GFP control plasmid and either the WT (seed-mismatch) or MUT (seed-match) miRNA. Scale bar, 20 µm. The bottom panel represents quantification of the mCherry fluorescence relative to the GFP fluorescence in the cells above.
The miRNAs used for microinjection were synthesized with a 5’ phosphate (P) and, in the case of amine modified RNA, a 3’ amino group on a C7 carbon linker (Howard Hughes Medical Institute Biopolymer/Keck Foundation Biotechnology Resource RNA Laboratory at the Yale University School of Medicine). RNA was purified as described [23] and the 3’ amine group was labeled with Cy3 or Cy5 NHS ester as described [23,24]. The absorbance of labeled miRNAs was measured to ensure the presence of one fluorophore label per miRNA, as calculated from the molar extinction coefficients of label and RNA [23,24]. Size homogeneity of purified and labeled RNA strands were tested on a 20% Urea-PAGE. Guide and passenger strands were heat-annealed in a 1:1 ratio in water at 10 – 20 µM concentrations, and the resulting duplex miRNAs were kept at −20 °C for further use. The extent of duplex formation was tested by electrophoretic mobility shift assay (EMSA) on a 15% non-denaturing polyacrylamide gel. Sequences of miRNAs used in our assays are listed in Table 1.
Table 1.
RNA/DNA oligonucleotide sequences
| Oligo name | RNA/DNA | Sequence (5` - 3`) |
|---|---|---|
| let-7-a1 guide | RNA | P-UGAGGUAGUAGGUUGUAUAGUU |
| let-7-a1 passenger | RNA | P-CUAUACAAUCUACUGUCUUUCC |
| let-7-a1 mutant guide | RNA | P-UGCGUUAGUAGGUUGUAUAGUU |
| let-7-a1 mutant passenger | RNA | P-CUAUACAAUCUACUGUCGUUCC |
| siluc2 siRNA guide | RNA | GAAGUGCUCGUCCUCGUCCUU |
| siluc2 siRNA passenger | RNA | GGACGAGGACGAGCACUUCUU |
| MS2 | DNA | AXGTCGACCTGCAGACAXGGGTGATCCTCAXGTTTTCTAGGCAATXA |
| C-MS2 | DNA | P-TAATTGCCTAGAAAACATGAGGATCACCCATGTCTGCAGGTCGACAT |
P – 5` Phosphate, X - amine modified deoxy-uridine, underlined bases represent differences between wild-type and mutant sequences
The injection solution typically contains just the fluorophore tagged miRNA or an additional injection marker diluted in a buffered solution at pH ~7. It is critical to prepare the microinjection sample in a solution with an ionic concentration and osmolarity similar to that found inside cells, mainly because the cell volume increases upon microinjection proportional to the injection pressure, and it is preferable not to perturb the cell’s ionic equilibria; microinjection of samples diluted in water results in cell swelling and death. We typically used PBS (phosphate buffered saline, GIBCO) as an injection buffer because it is isotonic with the intracellular milieu, it has been previously used for a wide range of injection experiments [25,26], and cellular functions were not impaired upon injecting this buffer. Although mammalian cells may not contain more than typically ~12 mM and ~5 mM of sodium and chloride ions, respectively, PBS also contains potassium and phosphate ions that are present at levels that are more appreciable. More elaborate close-to physiological injection buffers, such as those containing acetate ions, HEPES or glutamate ions, can also be used [27,28]. It is noteworthy that we found Tris-based injection buffers, including those containing Tris-acetate, to be detrimental to cell health. A majority of our experiments were performed using samples that contained both the fluorophore labeled miRNA and a comparably blue shifted, and thus spectrally distinct, 10 kDa fluorophore labeled dextran, used as an injection marker. In addition, we have successfully used cascade blue, fluorescein, Alexa555 or Alexa647 conjugated dextran (Invitrogen), in some cases of higher molecular weight to reduce leakage between cell compartments, for injector calibration (1.3.1. Microinjector calibration) and as a marker during miRNA microinjections. We found the usage of such injection markers to neither adversely affect cell growth and function nor to contribute to background signal while imaging the miRNA if chosen sufficiently blue-shifted [20]. Moreover, the capillary can be tested for clogging by visualizing the flow of injection marker from the tip rather than the fluorophore labeled miRNA, thereby preventing photobleaching of the latter.
Our miRNA concentrations were based on the desired particle density and the biologically required intracellular concentration. If the biologically required concentration and hence the intracellular particle density is refractory to single molecule detection, non-fluorescent RNA can be titrated into the injection mixture to achieve the appropriate concentration without oversaturating the fluorescent particle density. Based on the ratio of fluorescent to non-fluorescent RNA in the cell, the distribution of molecular stoichiometry or aggregation state of the RNA will vary, i.e., the fraction of higher order complexes will be more pronounced if the ratio is high. However, we typically probed the relative time evolution of the miRNA assembly state so that the results were not affected as this ratio was maintained through the time course and between experiments. Moreover, a single target mRNP may bind multiple miRNAs of the same or different sequence and miRNA bound mRNPs can be targeted to common degradation sites like processing bodies (PBs) [8], thus it is likely that the fluorescent particles we observe also contain endogenous “dark” miRNAs. In essence, the assembly states we measured were those of the fluorophore labeled miRNAs only. We typically microinjected 1.5 µM Cy5 labeled duplex miRNAs and 0.5 mg/ml 10 kDa fluorescein-dextran (Invitrogen) in PBS. As Cy5 photobleaches relatively quickly, fluorescein served as a marker to locate microinjected cells. More specifically, cells were first brought into focus and imaged using 488 nm (fluorescein) excitation and then quickly imaged using 640 nm (Cy5) excitation. The injection marker can be omitted from experiments when it is detrimental to cell health or imaging, by simply increasing to 2 µM Cy3 or Cy5 labeled miRNA in PBS. Although Cy3 labeled particles were visible for a longer time than those containing Cy5, the majority of experiments were performed with Cy5 labeled miRNAs, mainly because cellular autofluorescence was higher upon 532 nm (Cy3) excitation than 640 nm excitation. Co-microinjecting let-7-a1-Cy3 along with an injection marker, either fluorescein-dextran or Alexa-647-dextran, resulted in high background fluorescence due to spectral overlap and strong Cy3 fluorescence quenching by energy transfer, respectively, when illuminated at 532 nm.
1.3.1. Microinjector calibration
To ensure that the intracellular concentration of the RNA microinjected reflects endogenous levels, it is essential to know the exact amount of sample that is introduced into each cell. Based on the injection volume and the exact concentration of the injection sample in the capillary, one can calculate the number of molecules injected per cell (Fig. 3A). The microinjector is calibrated as described [29]. We used the following protocol:
- Prepare siliconized coverslips
- Sonicate coverslips in 1 M KOH for 10 min.
- Rinse three times each with water, methanol, acetone, water and dry coverslips under N2.
- Add 1 ml sigmacote (Sigma) on the coverslip and incubate at RT for 10 min.
- Rinse three times with water.
- Bake coverslip at 100 °C for 30 min.
- Rinse three times with water and dry under N2.
Attach the coverslip onto a slide with an ~ 2 cm (diameter) hole in the middle such that the coverslip and the slide create a small well. Ensure that the siliconized side faces into the well.
Fill the well with Type FF immersion oil (Cargille).
Fill capillaries with 0.01 – 0.5 mg/ml injection marker in 1× PBS.
Inject into the oil at various injection pressures varying from 100 hPa – 7,000 hPa.
Take bright-field and fluorescence z-stacks of the droplet that settles on the coverslip surface. Based on these images measure the diameter and calculate the volume of the droplet.
Plot the integrated density of fluorescence (IDF) of the droplet versus volume to create a standard curve. This curve is used to gauge the average injection volume at specific injection pressures. It can also be used to calculate the number of miRNA molecules microinjected into individual cells based on the intensity of fluorescent injection marker. For image analysis, we selected cells with up to 30% variation in IDF from the average IDF at the chosen microinjection pressure.
Notes, tips and troubleshooting
If glass bottom plastic dishes are used for siliconization, then the baking step can be omitted.
Siliconization and Type FF oil are absolutely essential to execute this protocol. The former creates a water repellant layer that preserves the spherical nature of the droplet formed upon injection with very little or no flattening. The latter has the optimal density to make the injected droplet sink to the bottom of the well.
This method can be used to calculate the number of miRNA molecules injected per cell, by reading cellular fluorescence values off the standard curve. To compare cellular fluorescence with the above standard curve the fluorescent probe used here need to be similar in identity and concentration to one used in cellular microinjection experiment as an injection marker. Moreover, the excitation laser intensity and camera acquisition conditions ought to be similar to that used for cellular imaging.
Occasionally, the droplet remains attached to the capillary after injection. Quickly remove the capillary out of the oil (using the “HOME” function on the Eppendorf micromanilpulator) to dislodge the droplet. Surface tension at the oil surface causes the droplet to dislodge into the oil and sink once the tip moves beyond the air-oil interface.
Recombinant luciferase proteins (Promega) or radioactive tracers can also be used for calibration purposes. However, it is difficult to assay for these reagents microscopically and hence they are used only to provide an average injection volume.
Other methods for calibration can be employed as described [30].
Methods such as those described above seldom account for the exact volume of the cell and only provide an average number of molecules injected per cell.
1.3.2. Cell culture and microinjection protocol
Seed cells at a density of 1–1.25 × 105 cells per 4.2 cm2 of plate surface.
Incubate cells overnight at 37 °C in a 5% CO2 and 95% relative humidity atmosphere.
Replace the growth medium with MTF 4 h prior to microinjection.
Remove cells from incubator, remove MTF, wash cells three times with pre-warmed (37 °C) HBS, add 1.25 ml pre-warmed (37 °C) HBS and place cells on the on-stage cell incubator to allow the system to come to thermal equilibrium. This step can be performed under non-sterile conditions.
Prepare the microinjection sample in 1× PBS and centrifuge the sample at 16,000 × g for 15 min at 4 °C and store sample on ice.
- Microinject up to three different regions of the plate for a total of 100–200 cells injected within 15–20 min.
- Focus on a patch of cells on the dish using the 10× objective and the eyepiece.
- Slowly lower the injector capillary in ‘coarse’ mode such that the tip of the capillary is close to the center of the bright-field illumination spot, until it comes in contact with the medium. Switch to ‘fine’ mode.
- If present near the field of view, the capillary can be easily found as it scatters the incident light, i.e., a bright area within the darker field of cells. Focus on the tip of the capillary and slowly lower it until the cells are barely in focus.
- Switch to the 20× objective and use the additional 1.6× magnification. Bring the capillary tip into focus.
- Check for microinjector clogging by visualizing the flow of injection marker from the capillary upon illumination with the appropriate excitation light. Application of a constant compensation pressure results in a perennial flow of solution from the capillary.
- Lower the tip until it comes in contact with a cell and creates a light depression on the cell membrane. Set the z-limit on the injector and begin injecting cells.
- To move to the next region, raise the capillary by ~100 µm, raise it to the non-operational position (press the ‘HOME’ button on the injector), move to a second region and return the capillary to the dish (press the ‘HOME’ button again). Repeat step f.
Remove HBS, wash cells 3 times with pre-warmed (37 °C) HBS, replace with pre-warmed (37 °C) MTF and incubate cells for the desired time at 37 °C in 5% CO2 and 95% relative humidity atmosphere. The cells are now ready to image.
Notes, tips and troubleshooting
Samples should be centrifuged to pellet any large debris that may clog the microinjector capillary. Alternatively, the sample can be filtered using 0.2/0.45 µm Ultrafree MC spin filtration units (Millipore), centrifuged at 12,000 × g for 4 min at RT.
If the microinjector is clogged or if an air bubble is present in the capillary, apply maximum pressure (“CLEAN” functionality on Eppendorf Femtojet pumps) to remove the obstacle.
It is important to limit the microinjection time and cell exposure to HBS. Long exposure of sample to white light, the preferred source of illumination for injections, can result in fluorophore photobleaching. In some cases, long exposure to HBS may be detrimental to cells. Certain cell lines, such as HeLa, are not affected by exposure to HBS of greater than 1 h, whereas with other cell lines, such as U2OS, it causes massive cell death, potentially due to reduced buffering capacity or the absence of vitamins and serum in the medium.
If MTF affects cell growth, phenol-red free regular growth medium can be used; however, increased background may mar single molecule detection.
Microinjected cells are a potential source of contamination of a laboratory incubator due to their exposure to the atmosphere. Therefore, using a separate incubator for them is preferred. Alternatively, a make-shift incubator can be created by placing the dish in one half of a divided plate, filling the second half with a small volume of water, enclosing it in an airtight sandwich bag inflated with 5% CO2 air and incubating this bag in an oven preset at 37 °C. Cells are stable in such an incubator for up to 36 h.
1.4. Validating iSHiRLoC as a biologically relevant method
iSHiRLoC requires the injection of fluorophore labeled molecules, whose activity should be preserved inside cells to extract biologically relevant information. Thus, for every cell line used, it is essential to test if fluorophore conjugation or the act of microinjection affect the expected biomolecular function; in the case of miRNAs, the reduction in protein expression by sequence-specific binding to their cognate mRNA targets. To this end, ensemble luciferase and fluorescence reporter assays should be performed. We found that both labeled and unlabeled miRNAs are functional to similar extents (Fig. 3B) whether they were transfected or microinjected (Fig. 3C).
1.4.1. Luciferase assay to validate functionality of fluorophore tagged microRNAs
Our reporter plasmids were based on the pmiRGLO dual luciferase expression vector (Promega). The plasmid contained both the firefly luciferase (FL, luc2) reporter gene and the Renilla luciferase (RL, rl-neo) control gene, thereby reducing the number of plasmids required for transfection. Furthermore, the RL gene is fused with the neomycin resistance gene as an antibiotic selection marker for stable cell line creation; such a fusion effectively reduces the size of the plasmid compared to one that contains both genes under separate promoters. As the FL reporter gene is under the control of the human phosphor-glycerate kinase (hPGK) promoter, expression levels are close to endogenous amounts and thus over-expression related artifacts are reduced. To create reporter plasmids, 3`UTRs that are typically responsive to miRNA binding, for instance the 3`UTR of mouse HMGA2, are cloned downstream of the FL gene.
As let-7 miRNAs are abundant in HeLa cells, reporter plasmids with the 3`UTR of HMGA2 were strongly repressed even in the absence of additional miRNA (Fig. 3B). To increase the sensitivity of our assays, we used a reporter plasmid that contained two base mutations in all let-7 seed sequences within the 3’UTR of HMGA2, which could be repressed efficiently only by a mutant let-7-a1 miRNA with complementary mutations. We created luciferase reporter plasmids pmG-mH3U (wt) and pmG-mH3UM (mut) by inserting PCR amplified wild-type and mutant mouse HMGA2 3`UTR sequences, respectively, downstream of the FL gene in the pmiRGLO vector. Negative control siRNA (Ambion) and siLuc2 siRNA (Dharmacon, Table 1), a positive control siRNA designed against the coding sequence of the firefly luciferase gene (luc2), were purchased as duplexes without 5`P on either strand. These siRNAs are rendered functional inside cells by cellular kinases that phosphorylate the 5` ends of both strands of the duplex [31].
Seed 15,000 cells with 100 µl of growth medium per well of a 96 well plate, such that the cells will be ~70% confluent after 24 h.
Replace with fresh growth medium just before transfection.
Prepare transfection complexes by combining 0.4 µl of lipofectamine 2000 (Invitrogen), 60 ng of luciferase reporter plasmid and 10–100 nM dsRNAs in 50 µl of OptiMEM (GIBCO). Follow the manufacturer’s transfection protocol and add the nucleic acid–transfection reagent complexes to each well.
Replace old medium with 100 µl fresh growth medium 6 h after transfection.
- Assay for luciferase expression 24 h after transfection using the Dual-Glo luciferase assay system (Promega). Measure luminescence using a Genios Pro (Tecan) or GloMAX plate reader (Promega).
- Replace growth medium with 40 µl phenol-red free DMEM.
- Add 40 µl of Dual-Glo luciferase assay reagents. Incubate on a rocker for 15 min at RT. Measure firefly luminescence.
- Add 40 µl stop and Dual-Glo Stop & Glo reagent. Incubate on a rocker for 15 min at RT. Measure Renilla luciferase luminescence.
- Compare the ratio of FL to RL expression between all samples.
Notes, tips and troubleshooting
The above protocol is optimal for HeLa and U2OS cells. Standardize the ratio of nucleic acid to transfection reagent for each cell type. If plasmids and siRNAs with markedly different molecular weights are to be compared, include a carrier plasmid such as pUC19 or pGEM-3z in the lower molecular weight siRNA sample. Carrier plasmids are chosen such that they do not express any gene in mammalian cells. Ideally, it should be attempted to maintain the nucleic acid amount over all samples such that the net mass of nucleic acids (in µg) and the net moles of the specific plasmid or siRNA remain the same.
Titrate siRNA or miRNA amount to identify the minimal concentration at which maximal repression is achieved.
It is required that the relative concentration of the luciferase substrate is much higher than the enzyme for such reporter assays. Thus, assays can also be performed as per manufacturer’s (Promega) protocol with 100 µl of all reagents, although it suffices to use 40 µl.
To ensure that measured luminescence readings are directly comparable, it is important for them to fall within the linear range of luminescence detection. To this end, luminescence of lysates at various concentrations is measured, the range of concentrations over which the readings are linear is identified, and the amount of reporter plasmid added is titrated such that measured luminescence values of experimental samples are within that range.
It is important to note that Dual-Glo luciferase assay reagents, unlike dual-luciferase assay reagents, are not designed for use with automated injectors as excessive reagent foaming may occur.
1.4.2. Fluorescence reporter assay to validate functionality of microinjected microRNAs
Microinjection is the act of introducing molecules into one cell at a time. We typically inject ~300–400 cells in a dish that contains over 105 cells, thus, performing ensemble assays with lysates created from all cells in the dish, such as that required for luciferase assays, are not preferred because they dilute the reporter several fold, thus obstructing sensitive and accurate quantification. To overcome this problem, a reporter gene assay that measures specific output from individual cells was required. As microscopes are typically not equipped to measure luminescence, we resorted to fluorescence. To this end, we microinjected a solution containing three components: 1) an mCherry reporter plasmid that contained a miRNA sensitive 3`UTR; 2) a GFP control plasmid that was used for normalizing variation in injection amount and protein expression; and 3) a 3’UTR specific or non-specific miRNA. Fluorescence from individual cells was then used to measure the extent of repression. For our assays [20] we created a fluorescence reporter plasmid pEF6-mCherry-mH3UM by inserting the mCherry gene and the mutant 3`UTR of HMGA2 into the pEF6 (Invitrogen) vector backbone. pEGFP-C1 was purchased from Clontech. The injection solution contained 0.1 µg/µl of each plasmid and 1.5 µM of the respective miRNA diluted in PBS. Injections were performed at 100 hPa for 0.5 s with a compensation pressure of 40 hPa. Using the above experimental conditions, we found that the normalized fluorescence in cells microinjected with the mut-let-7-a1 miRNA were, on average, much lower than those injected with the let-7-a1 miRNA, strongly suggesting that the act of microinjection does not affect miRNA function (Fig. 3C).
Culture and microinject cells as described above (1.3.2. Cell culture and microinjection protocol).
Incubate cells at 37 °C supplemented with 5% CO2 for 24 h.
Transfer cells into HBS and image at 20 × magnification with 488 nm and 532 nm excitation to image GFP and mCherry fluorescence, respectively.
- Measure background-subtracted, bleed-through corrected IDF of GFP and mCherry from each cell. Normalize mCherry fluorescence with that of GFP, average over several cells and compare between different samples.
- Compute the cell boundary by setting an absolute threshold of GFP signal at which only cellular signal has non-zero values.
- Compute background intensity by first averaging signal intensity from a 25 × 25 pixel area in the vicinity of each cell, calculating the product of this value and the area of each cell (in pixels) and subtracting this from the IDF within the corresponding cell (IDFBS).
- Measure fluorescence from cells injected with just GFP and mCherry at both 488 nm and 532 nm excitation to compute the contribution of GFP signal to the mCherry signal and vice-versa, i.e., the bleed-through. Find the ratio of background subtracted IDF at non-optimal (NO) excitation (532 nm for GFP and 488 nm for mCherry) to optimal (O) excitation and average over several cells; this is the fraction of bleed-through (BT) signal (rIDFBT = average of IDFBS,NO / IDFBS,O over all cells). Calculate the product of this value and the IDF of individual cells and subtract this bleed-through signal from the IDF (IDFfinal = IDFBS,sample* (1 - rIDFBT)).
Notes, tips and troubleshooting
Both labeled and unlabeled miRNAs can be used in these assays. However, bleed-through contributed by the fluorophore on the miRNA should also be considered for final IDF calculations.
Although fluorescence reporter assays are valuable in differentiating repressed cells from those that are not, they should be complemented with luciferase reporter gene assays mainly because the latter are more sensitive over a much larger range of reporter concentrations.
1.5 Single particle tracking in live cells
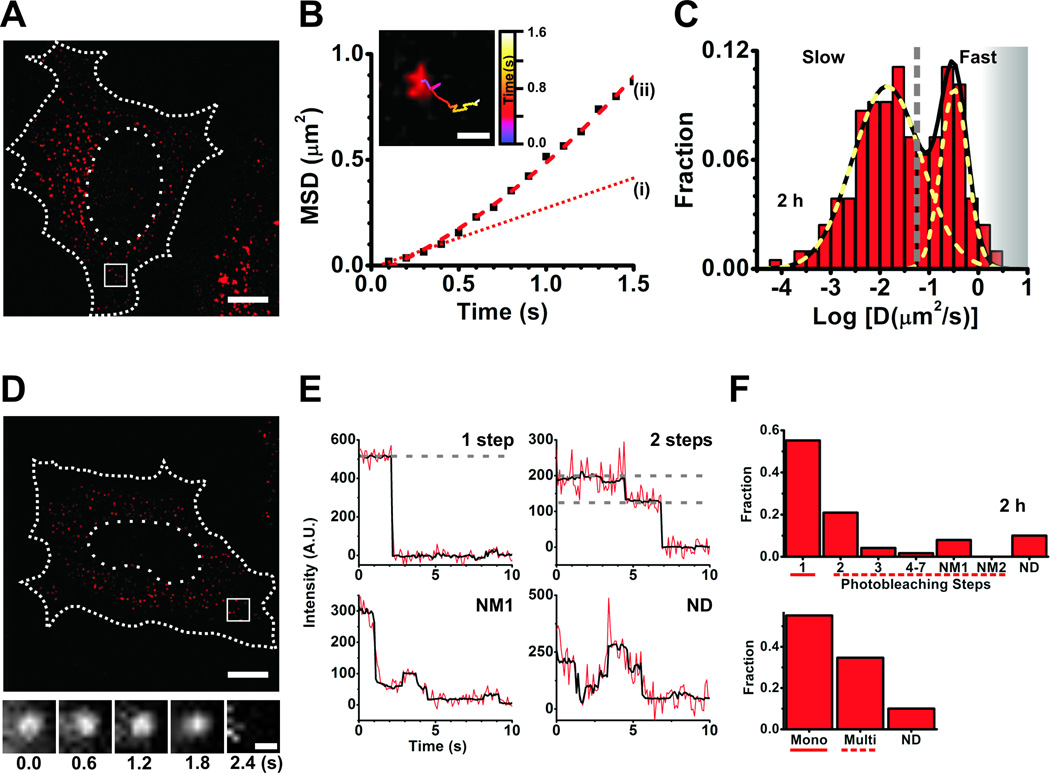
The live-cell SPT component of the iSHiRLoC assay is designed to capture the dynamic information of biomolecular interactions and intracellular localization. Through monitoring the diffusive motion of particles, various physical parameters such as particle number, size, velocity, motion type, and diffusion coefficients can be obtained, which sheds light on the nature of the particle itself and its immediate surroundings. Population analyses of the calculated particle parameters typically show multiple distributions owing to differences in their behavior, which in some cases may illuminate the diversity in their mechanism of action (Fig. 4A–D). This property is especially true for miRNAs since they have the tendency to form complexes of various sizes through the course of their assembly and repressive pathways [20, 26]. We used 1.5 µM Cy5 labeled duplex miRNAs and 0.5 mg/ml 10 kDa Fluorescein dextran (Invitrogen) in PBS for injections. Microinjections were performed at 100 hPa injection pressure for 0.5 s with 20 hPa compensation pressure. Based on these experimental conditions we found that intracellular miRNA assembly is temporally regulated [20]. Briefly, at initial time points (up to 1 h post injection) miRNAs were either free or bound by miRISC complexes that diffused faster than the time resolution of our camera and presented as a blur, thus spreading the signal over the entire span of the cell. As they assembled into larger complexes (after ~1 h and beyond), localization of individual particles was enhanced and diffusion coefficients of all such particles were distributed over two Gaussian populations resembling those of mRNPs (fast population) and PBs (slow population), as expected if miRNAs are target bound and miRNA-mRNA complexes localize to the cellular mRNA degradation machinery, respectively. We further found that the fraction of the slowly moving population decreased with time, signifying increased degradation at later time points and the release of miRNPs from such large complexes, i.e., disassembly. The protocol for live cell particle tracking was as follows.
Culture and microinject cells as described above (1.3.2. Cell culture and microinjection protocol).
Incubate the microinjected cells for desired time intervals, then wash the cells five times with pre-warmed (37 °C) HBS.
Replace the cell medium with 1 ml pre-warmed (37 °C) HBS containing an oxygen scavenger system (OSS)-antixoidant mix (HBS-OSS; 0.3 U/ml Oxyfluor, 20 mM sodium succinate, 2 mg/ml ascorbic acid and 200 µM trolox). Incubate the cells on a 37 °C stage incubator for ~5 min.
Set the imaging plane by first visualizing fluorescein by WF-epi illumination using 488 nm excitation.
Quickly switch to 640 nm excitation. Use HILO illumination to image cells at 120× magnification with 100 ms camera exposure at 10 MHz image transfer rate and a gain of 1×, 100 A.U. on an EVOLVE camera. Record 50–400 frame videos.
Track particles using an appropriate analysis routine, see below.
Figure 4.
Single particle tracking and single molecule counting. (A–C) Data from single particle tracking experiments in live HeLa cells. (A) A representative background-subtracted pseudocolored image of a cell injected with 1.5 µM let-7-a1-Cy5 miRNA and fluorescein dextran (injection marker), imaged 2 h post-microinjection. Scale bar, 10 µm. Dashed and dotted lines represent cell and nuclear boundaries, respectively. (B) Plot of mean squared displacement (MSD) versus time of the track within the inset. Scale bar within inset, 500 nm. The track corresponds to the particle within the white box in (A). Brownian micro-diffusion coefficient (D(i)), calculated from the first three data points of the MSD versus time plot, is 0.0561 µm2/s, whereas the diffusion coefficient calculated by best-fitting all data points with a parabolic curve, the canonical equation for a particle exhibiting biased motion, yields D(ii) = 0.085 µm2/s (velocity = 0.43 µm/s). The line fitted to the first three data points is extrapolated for ease of viewing (dotted red line (i)). Based on RD analysis, we do not find a significant difference in the number of particles exhibiting non-Brownian diffusion between sample sets. As we are interested only in the change in diffusion coefficient and not its actual magnitude, such differences do not significantly affect our analyses. (C) Distribution of Brownian micro-diffusion coefficients calculated from individual let-7-a-Cy5 particles in (A), that were visible for at least 9 consecutive frames. Particles distributed into at least two distinct Gaussian populations of diffusion coefficients, with an average of diffusion coefficients of 0.322 µm2/s and 0.014 µm2/s. The grey dashed line represents a crude demarcation of the fast and slow moving particles that resemble mRNPs and PBs, respectively. The grey shaded region represents diffusion coefficients of fast moving particles that we increasingly lose either due to time resolution limitations or due to our stringent track length threshold for calculating diffusion coefficient. (D–F) Data from single molecule counting experiments in formaldehyde-fixed HeLa cells. (D) A representative background subtracted, pseudocolored image of a cell injected with 0.25 µM let-7-a1-Cy5 miRNA and 10 kDa fluorescein-dextran, imaged 2 h after microinjection. Scale bar, 10 µm. Dashed and dotted lines represent cell and nuclear boundaries, respectively. Cells were imaged immediately after fixation. The bottom panel contains a set of frames that shows the photobleaching of a miRNA particle, within the white box in (D), over the indicated time. Scale bar, 500 nm. (E) Representative photobleaching trajectories of particles in (D). Grey dashed lines are drawn to guide the eye to discrete intensity levels. (F) Distribution of photobleaching steps corresponding to particles in (D). The bottom panel represents particles either grouped as monomers (photobleaching steps spanning solid line in top panel), multimers (photobleaching steps spanning dashed line in top panel) or non-discernible (ND).
Notes, Tips and Troubleshooting
OSS-antioxidant mix depletes oxygen from the medium, thereby reducing photooxidation induced photobleaching and quenches unwanted dark (triplet) states that prevents fluorophore “blinking”.
Cells should be imaged immediately after HBS-OSS addition to prevent stress induced by hypoxia. The concentration of OSS, however, is much lower than that required for complete oxygen depletion. We typically image cells for a maximum of 10–15 min under these conditions. If OSS affects cell health, it can be removed from the imaging medium. However, this might increase the propensity for photobleaching.
Increasing laser intensity and integration times are a great way to maximize the signal-to-noise/background ratio (SNR). However, the rate of photobleaching increases with increasing laser intensity and particle blurring may be a problem upon long exposure time. Optimal imaging conditions are set by identifying the least laser intensity and exposure time at which a maximum number of distinct particles is visible.
Stroboscopic illumination techniques [32] can be used to image particles at high laser intensity with reduced average exposure of fluorophore to excitation light.
Prior to tracking, SNR can be artificially improved by applying certain mathematical filters to the image that markedly increase localization accuracy and decrease uncertainty in diffusion coefficient calculations. 2D deconvolution (using AutoQuant X2 or Huygens) can be used to decrease the width of the spatial intensity distribution (or point-spread function, PSF) arising from diffraction limited particles and additionally aid in de-blurring, i.e., increasing the sharpness of images.
1.5.1. Selection of a Tracking Program
Tracking programs have two major functions, particle detection and trajectory reconstruction, and the mathematical relations used to execute these functions distinguish one software from another. However, there is a common link between all software programs; they assume that the intensity of the particle of interest must be greater than the background. Particle localization can be performed using one of several methods, as described [33]. Based on localization of particles in every frame of a video, trajectory reconstruction can be achieved using either multiple hypothesis tracking (MHT) [34] or a nearest neighbor approach. As the former is computationally expensive, mainly because tracks are evaluated every frame for the largest connected patch to identify the most probable path within a distribution of tracks, the latter is employed predominantly. We utilized ImarisTrack (Bitplane), mainly due to its automated tracking program and its intuitive visualization tool to assess tracks in real-time while selecting filters and thresholds. Particle localization is achieved by localizing the centroid of a particle’s intensity distribution after applying a Gaussian low-pass filter, whereas tracking is accomplished by the nearest neighbor method. The Gaussian filter helps to remove high-frequency noise and approximates the PSF to a Gaussian function without the need for actual fitting, thereby increasing the accuracy of localization at reduced computational cost [35]. Localization is further enhanced by correcting for the local background around a particle of preset dimensions in every frame of the video. Put together, ImarisTrack and Imaris MeasurementPro (Bitplane) provide a statistical readout of all parameters from the assigned tracks that are necessary for data processing.
Open image in Imaris. Videos open as a 3D stack, so changing Z position with time.
Select an intensity threshold at which a majority of the particles are visible. Select the “spots” option in Imaris to track particles. Track particles over a small (100 × 100 pixel) region within the cell and then extend to the entire field of view.
Choose an average diameter (~6–10 pixels) for the particles to be localized and include adaptive background subtraction.
Select localized particles based on the “quality” parameter that is directly proportional to the SNR. The magnitude of this value is a function of the intensity histogram range.
Track particles using the nearest neighbor “Brownian” method and setting a maximal inter-frame jump distance of a magnitude that is not higher than the diameter of the particle. Set the “gap size”, i.e., the maximal duration (in frames) for which a particle may disappear from the field of view, to zero.
Extend these settings to the entire field of view.
Notes, Tips and Troubleshooting
Choose a small region that contains the most number of least bright particles during particle tracking within the 100 × 100 pixel region.
The apparent diameters of particles depend on the magnification used and should be chosen by analyzing several particles over multiple videos.
To increase the confidence in trajectory reconstruction, the jump distance was set to the particle diameter. Based on the density of particles and the above criteria, it was certain that a position on the adjacent frame belonged to the track of the same particle.
Tracks should be inspected for merging, separating and disappearing/reappearing particles to ensure that they arose from well-behaved and isolated particles.
Particle disappearance may occur due to fluorophore blinking or a particles moving out of the imaging plane; we typically do not account for these effects.
1.5.2. SPT data processing
We only tracked particles that are observed for at least 9 frames. Shorter tracks resulted in diffusion coefficients with ~50% error or more and were discarded from the analyses [36]. Micro-diffusion coefficients are calculated as the slope of the line fit over first three data points (D0–3) of the mean squared displacement (MSD) versus time plot, using in-house MATLAB routines (Fig. 4B, C). More specifically, we consider all tracks to be Brownian in nature, a valid assumption given the very short analysis window [37]. The intercept of the line represents the error in particle localization at the initial time point, the distribution of which closely mirrors the experimentally verified localization precision (1.6.3. Precision of particle localization).
Due to the limited length of tracks, a relative deviation (RD) measurement is used to characterize variations in diffusive pattern between different trajectories [37]. An RD value of ~1 suggests that the data fit a Brownian diffusion model, thereby providing increased confidence in the calculated diffusion coefficients. Based on the track length and the time lag at which RD was calculated, deviations from unity suggest the presence of additional types of diffusion: anomalous, directed, and corralled motions. In such instances, the diffusion coefficients were also calculated using the canonical equation for the appropriate diffusive pattern [38].
1.6. Single molecule counting in fixed cells
Biomolecules function as monomeric or multimeric units either individually or within complexes inside cells. Precisely understanding such oligomerization states of biomolecular complexes and their associated heterogeneity or diversity is one of the main aims of our iSHiRLoC method. This second aspect of iSHiRLoC entails single molecule counting in fixed cells through the stochastic, stepwise photobleaching of single fluorescent probes. Although photobleaching analysis has been extensively employed to understand molecular stoichiometry [13], it has been limited to in vitro assays [39] or the membrane of live or fixed mammalian cells. The majority of such investigations employed total internal reflection fluorescence (TIRF) microscopy to selectively illuminate a thin lamina at the interface of glass and cell medium, hence limiting visualization to the vicinity of cell membranes. To overcome this caveat, we developed a method that employs HILO illumination to investigate the distribution and assembly of singly fluorophore-labeled, functional miRNAs in the cytoplasm of fixed cells after microinjection [20]. Our study revealed that most microinjected miRNAs exist as monomers, with a significant fraction of multimers (Fig. 4E – H), suggesting the existence of two distinct populations, i.e., those assembled into individual miRNAs/miRNPs and others assembled into even larger RNP aggregates, the distribution of which changed over time [20]. Microinjection sample and conditions were the same as described above (1.5. Single particle tracking in live cells). The protocol for photobleaching was as follows.
Culture cells and microinject them as described above (1.3.2. Cell culture and microinjection protocol).
After the microinjected cells are incubated for specific time intervals, wash the cells five times with pre-warmed (37 °C) PBS.
Fix cells using pre-warmed (37 °C) 4% (w/v) paraformaldehyde in PBS for 20 min at RT.
Wash the cells five times with PBS after fixing.
Transfer cells to PBS containing OSS-antioxidant mix as mentioned above (PBS-OSS).
Image as described above (1.5. Single particle tracking in live cells), record 200–400 frame videos, or until all signal has disappeared due to photobleaching.
Determine the number of photobleaching steps using an appropriate analysis routine.
Notes, Tips and Troubleshooting
The incubation time for fixing cells can be varied from 15–30 min at RT or 37 °C. The cells can also be fixed with a solution of 4% formaldehyde and 0.5% glutaraldehyde for 25 min if the above fixing protocol does not work well.
Increase concentrations of OSS if photobleaching is too rapid.
As mentioned above, imaging conditions may vary with different cameras. Increase gain values if particles are not clearly visible. To choose an appropriate gain setting, image cells at various gain values, use any software to detect the average number of particles per cell maintaining the same particle identification sensitivity, i.e., the threshold constant, plot the number of particles detected over gain, and choose the lowest gain value that results in the maximal number of particles.
The density of the spots within the imaged cells should be between ~0.1–0.4 spots/µm2 to ensure that individual spots can be analyzed.
1.6.1. Photobleaching data analysis
The photobleaching data obtained from continuous illumination of fluorescent particles can be analyzed using a custom LabView (National Instruments) code, as described [40]. This code incorporates noise-filtering and photon-counting histogram (PCH) techniques effective for use in photobleaching trajectory analysis. The non-linear Chung-Kennedy (CK) noise-filter [41] was used as it is designed to preserve the fast and sudden transitions in the signal with minimal distortions and yet average out instrument noise effectively with minimum edge blurring based on the parameters chosen. The PCH approach is based on the premise that the number of photons collected per sampling time and molecule follows a Poisson distribution, which can help reveal the intensity distributions of the different species that bear distinct peak brightness values. The area under such a curve can then be used to calculate dwell times at individual states as well. Procedures for data analysis using this software were as follows.
Compute cell boundary by setting an absolute threshold of fluorescein signal at which only the intracellular pixels bear non-zero values.
Average the first 5 frames of the video for particle recognition.
Set the software to detect particles that are spread over an area of 11 × 11 pixels at most, but larger than a pixel.
Select particles whose maximal intensity value is at least 1.2-fold higher than the average local background. Particle centers are chosen based on the pixel with the maximum intensity.
Use the coordinates of chosen particles to locate particles in the video where the intensity of each particle per frame is calculated by subtracting the average intensity values of a 1 pixel strip around the 11 × 11 particle area from the average intensity within.
Process the intensity trajectory thus created by the CK non-linear filter with K (bank of predictors) = 4 predictors, moving average window length of 32 points, M (predictor window size) = 10, p (weighting factor or exponent) = 100 over two iterations.
Overlay original trajectory with the filtered trajectory and count the number of photobleaching steps for all particles. Use the filtered trajectory and the PCH to identify the number of steps.
Notes, Tips and Troubleshooting
All parameters used above are assay and software dependent. The above settings are valid only for samples imaged using our specific microscope, microinjection settings and software [20].
Particle size is chosen such that the largest particle is at least one pixel strip smaller than our setting.
Parameters for the CK non-linear filter should be chosen carefully. The size of the analysis window is represented by M and, based on the data, typically 4 ≤ M ≤ 20. The weighting factor assigned to each forward and backward predictor, p, can be chosen independently and typically 1 ≤ p ≤ 100. The higher the value the better (e.g., p = 100). Although there is a drastic reduction in noise with lower p values, the amplitude and the edges of the signal get distorted [41]. Perform 2–3 iterations at most, as increasing iterations are accompanied by a distortion of the original signal as when using lower p values. Additionally, the number and length of predictors, K (in frames), can be varied based on the average dwell times at each state. It is safe to choose a value that is larger than the smallest dwell time. A short pair of forward and backward predictors helps extract exponentially decaying signals without much effect on reducing background noise.
We distinguish particles by the number of photobleaching steps. However, a few traces (~20– 25%) will contain photobleaching trajectories that do not bleach at finite steps. These traces were labeled as NM1, NM2, ND or noPB based on certain characteristic features. NM1 included those traces that contained multiple photobleaching steps, but the exact number was indiscernible. By contrast, NM2 also bleached with multiple steps, but the maximal intensity was higher than ~1750 A.U. Such intensity values are expected for particles with more than seven molecules, considering that the average maximal intensity per single photobleaching event was ~250 A.U. ND (~2–10%) were traces where it was difficult to determine if the particle bleached in one or many steps. Lastly, noPB included those non-zero traces that did not contain any stepwise photobleaching. Finally, the distribution was binned into monomers, i.e., single step photobleaching particles, or multimers, i.e., those that bleached in multiple steps including NM1s and NM2s (Fig. 4H).
Other software packages that use hidden Markov modeling (HMM) to identify photobleaching steps are also available [42].
1.6.2. Photobleaching control
It is critical to reduce pre-bleaching of fluorophores, i.e., photobleaching prior to imaging, to accurately count molecules. To ensure minimal pre-bleaching in our samples, we performed photobleaching assays on cells injected with the same sample or ones that were prepared and stored in a similar fashion, immediately after microinjection. We then counted the number of photobleaching steps and correlated them with the number of independently determined fluorophores per particle. We used a DNA oligonucleotide, MS2 (Table 1), with four internal amine modifications, purified and labeled it similar to the miRNAs [23,24] for this purpose. MS2 and its 5`P containing complementary strand (C-MS2) were annealed in a 1:1 ratio in water, at a concentration of 10–20 µM and frozen for further use.. We used 0.5 µM Cy5 labeled duplex DNA and 0.5 mg/ml 10 kDa Fluorescein dextran (Invitrogen) in PBS for microinjection. Microinjections were performed at 100 hPa injection pressure for 0.5 s with 20 hPa compensation pressure. Under these conditions, we found that the distribution of photobleaching steps closely matched the theoretical binomial distribution of the number of dyes per DNA for our labeling efficiency of 3.45 labels per DNA, suggesting that pre-bleaching was minimal [20]. Moreover, we could clearly visualize multi-step photobleaching particles with high confidence, thus further validating our method of analysis.
Notes, Tips and Troubleshooting
This assay can alternatively be performed in vitro as follows. Using a modified protocol, a DNA containing both biotin and a fluorophore (Cy3) is annealed to a complementary miRNA with a spectrally distinct fluorophore label (Cy5). The amount of miRNA should be in a large excess over the DNA, for instance a 20:1 ratio of miRNA:DNA. This duplex is bound to a streptavidin functionalized surface, excess miRNAs are washed away, and the fraction of colocalized spots is then calculated. The non-colocalized spots that only contain the DNA represent duplexes with pre-bleached miRNAs.
1.6.3. Precision of particle localization
Localization accuracy is the average radius of a circle within which a particle can be confidently located, i.e., the error associated with its location. This uncertainty plays a significant role in SPT by negatively affecting particle detection, tracking and accurate diffusion coefficient calculations; the greater the uncertainty, the blurrier the particle and the greater the error in diffusion coefficient. Reduced precision typically can be attributed to small movements in the stage position or low SNR. To ensure that the localization accuracy is much smaller that the width of the particles (in pixels) and their corresponding inter-frame jump distances, we experimentally quantified the former using the following protocol.
Culture cells, microinject (1.3.2. Cell culture and microinjection protocol), fix and image cells as described above (1.6. Single molecule counting in fixed cells).
Track the fixed particles using the tracking software.
Plot a histogram of particle displacement over the duration of their track.
Fit the histogram with a Gaussian function, and predict the mean and standard deviation of the measured inter-frame displacements. The mean value of the Gaussian represents the average uncertainty in the particle position.
2. Conclusions and future directions
In conclusion, here we report the detailed protocol for the iSHiRLoC analysis of miRNAs, designed to probe their intracellular assembly and hence their mechanism of action. It is only a matter of time before this method is extended to probing the intracellular function of other ncRNAs.
Microinjection allowed for the homogeneous, controlled delivery of miRNAs directly into cells in an efficient and non-invasive fashion. Ensemble reporter gene assays showed that fluorophore modification and microinjection do not affect miRNA function (Fig. 3), attesting to the low invasiveness of the method. In this fashion, each cell functions as an individual reaction vessel into which functional fluorophore labeled miRNAs are introduced. SPT and single molecule counting methods complement each other in understanding assembly states of miRNAs; the former sheds light on the molecular mass and spatiotemporal distribution of miRNP complexes and the latter predicts the exact number of labeled miRNA molecules within each complex. Based on SPT data (Fig. 4A–D), diffusion coefficients of miRNPs typically distributed into (at least) two distinct Gaussian populations that were indicative of miRNA-bound mRNAs [43] and PBs [44], respectively, both functional intermediates of miRNA mediated gene regulation. Particle counting (Fig. 4E–H) showed that a majority of such particles contained individual miRNA molecules; however, significant fractions of particles also held multiple molecules (up to seven), thus representing higher-order complexes. Put together, we mapped the spatiotemporal evolution of miRNA assembly with single molecule sensitivity [20].
Ensemble reporter assays and distribution of diffusion coefficients strongly suggested that a significant proportion of the particles we observed are functional. However, further studies using dual-color tracking and colocalization, are required to pinpoint with single molecule precision the functional relevance of each miRNA. For instance, we can microinject miRNAs with fluorescent labels on both strands of the duplex and quantify the ratio of double-stranded to single-stranded miRNA. As only one strand of the miRNA duplex (guide) is preferentially retained in active RISC [6,7], this experiment would measure the extent of duplex unwinding and strand selection. As we can now also label and visualize single mRNA molecules [43,45] and PBs [44], it would be relatively straightforward to determine the exact percentage of target bound and PB-localized miRNAs. In this fashion, we will be able to monitor the progress of miRNAs though every step of RNA silencing, as well as other, competing cellular RNA pathways [46] at single molecule sensitivity, with the ultimate goal of understanding the RNA silencing mechanism through a novel single molecule systems biology approach.
Highlights.
Method to probe the activity of microRNAs (miRNAs, a class of regulatory non-coding RNAs) in cellulo with single-molecule sensitivity.
Microinjection and HILO microscopy to deliver and image, respectively, repression-competent, fluorophore labeled miRNAs.
Single particle tracking in live cells to obtain diffusion coefficients of micro-ribonucleoprotein (miRNP) complexes.
Single molecule counting by stepwise photobleaching in fixed cells to obtain the stoichiometry of miRNAs within miRNPs.
Mobility and assembly changes enable the understanding of intracellular miRNA function.
Acknowledgments
We thank D. Bartel, C. Mayr, R. Tsien, N. Kedersha, and R. Singer for generous gifts of plasmids containing the 3’ UTR of HMGA2, mCherry ORF, Dcp1a ORF, and the MS2 system of plasmids, A. Manzo for MSD software development, H. Ding, A. Gafni and D. Steel for sharing their intensity analysis routine, the Microscopy and Image Analysis Laboratory at the University of Michigan for access to Imaris, Sunney Xie for important input at the onset of the project, A. Mapp for access to her plate reader, J. Androsavich for help with the reporter gene assays, and Laurie Heinicke for helpful comments on the manuscript. This work was supported by NIH grants GM081025 and GM098023 to N.G.W.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Carninci P. Nat. Cell Biol. 2008;10:1023–1024. doi: 10.1038/ncb0908-1023. [DOI] [PubMed] [Google Scholar]
- 2.Bernstein BE, Birney E, Dunham I, Green ED, Gunter C, Snyder M. Nature. 2012;489:57–74. [Google Scholar]
- 3.Schmeing TM, Ramakrishnan V. Nature. 2009;461:1234–1242. doi: 10.1038/nature08403. [DOI] [PubMed] [Google Scholar]
- 4.Kruger K, Grabowski PJ, Zaug AJ, Sands J, Gottschling DE, Cech TR. Cell. 1982;31:147–157. doi: 10.1016/0092-8674(82)90414-7. [DOI] [PubMed] [Google Scholar]
- 5.Esteller M. Nat. Rev. Genet. 2011;12:861–874. doi: 10.1038/nrg3074. [DOI] [PubMed] [Google Scholar]
- 6.Bartel DP. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Czech B, Hannon GJ. Nat. Rev. Genet. 2011;12:19–31. doi: 10.1038/nrg2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Krol J, Loedige I, Filipowicz W. Nat. Rev. Genet. 2010;11:597–610. doi: 10.1038/nrg2843. [DOI] [PubMed] [Google Scholar]
- 9.Filipowicz W, Bhattacharyya SN, Sonenberg N. Nat. Rev. Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- 10.Djuranovic S, Nahvi A, Green R. Science. 2011;331:550–553. doi: 10.1126/science.1191138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guo H, Ingolia NT, Weissman JS, Bartel DP. Nature. 2010;466:835–840. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Walter NG, Huang CY, Manzo AJ, Sobhy MA. Nat. Methods. 2008;5:475–489. doi: 10.1038/nmeth.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Leake MC, Chandler JH, Wadhams GH, Bai F, Berry RM, Armitage JP. Nature. 2006;443:355–358. doi: 10.1038/nature05135. [DOI] [PubMed] [Google Scholar]
- 14.Grunwald D, Singer RH. Nature. 2010;467:604–607. doi: 10.1038/nature09438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pinaud F, Clarke S, Sittner A, Dahan M. Nat. Methods. 2010;7:275–285. doi: 10.1038/nmeth.1444. [DOI] [PubMed] [Google Scholar]
- 16.Babcock HP, Chen C, Zhuang X. Biophys. J. 2004;87:2749–2758. doi: 10.1529/biophysj.104.042234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Itzkovitz S, van Oudenaarden A. Nat. Methods. 2011;8:S12–S19. doi: 10.1038/nmeth.1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Neely LA, Patel S, Garver J, Gallo M, Hackett M, McLaughlin S, Nadel M, Harris J, Gullans S, Rooke J. Nat. Methods. 2006;3:41–46. doi: 10.1038/nmeth825. [DOI] [PubMed] [Google Scholar]
- 19.Lu J, Tsourkas A. Nucleic Acids Res. 2009;37:e100. doi: 10.1093/nar/gkp482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pitchiaya S, Androsavich JR, Walter NG. EMBO rep. 2012;13:709–715. doi: 10.1038/embor.2012.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tokunaga M, Imamoto N, Sakata-Sogawa K. Nat. Methods. 2008;5:159–161. doi: 10.1038/nmeth1171. [DOI] [PubMed] [Google Scholar]
- 22.Bogdanov AM, Bogdanova EA, Chudakov DM, Gorodnicheva TV, Lukyanov S, Lukyanov KA. Nat. Methods. 2009;6:859–860. doi: 10.1038/nmeth1209-859. [DOI] [PubMed] [Google Scholar]
- 23.Walter NG. Curr. Protoc. Nucleic Acid Chem. Chapter. 2003;11 doi: 10.1002/0471142700.nc1110s11. Unit 11 10. [DOI] [PubMed] [Google Scholar]
- 24.Walter NG, Burke JM. Methods Enzymol. 2000;317:409–440. doi: 10.1016/s0076-6879(00)17027-2. [DOI] [PubMed] [Google Scholar]
- 25.Lukacs GL, Haggie P, Seksek O, Lechardeur D, Freedman N, Verkman AS. J. Biol. Chem. 2000;275:1625–1629. doi: 10.1074/jbc.275.3.1625. [DOI] [PubMed] [Google Scholar]
- 26.Liu X, Fernandes R, Gertsenstein M, Perumalsamy A, Lai I, Chi M, Moley KH, Greenblatt E, Jurisica I, Casper RF, Sun Y, Jurisicova A. PLoS One. 2011;6:e21687. doi: 10.1371/journal.pone.0021687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grunwald D, Spottke B, Buschmann V, Kubitscheck U. Mol. Biol. Cell. 2006;17:5017–5027. doi: 10.1091/mbc.E06-06-0559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ohrt T, Mutze J, Staroske W, Weinmann L, Hock J, Crell K, Meister G, Schwille P. Nucleic Acids Res. 2008;36:6439–6449. doi: 10.1093/nar/gkn693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee GM. J. Cell Sci. 1989;94(Pt 3):443–447. doi: 10.1242/jcs.94.3.443. [DOI] [PubMed] [Google Scholar]
- 30.Minaschek G, Bereiter-Hahn J, Bertholdt G. Exp. Cell Res. 1989;183:434–442. doi: 10.1016/0014-4827(89)90402-3. [DOI] [PubMed] [Google Scholar]
- 31.Nykanen A, Haley B, Zamore PD. Cell. 2001;107:309–321. doi: 10.1016/s0092-8674(01)00547-5. [DOI] [PubMed] [Google Scholar]
- 32.Elf J, Li GW, Xie XS. Science. 2007;316:1191–1194. doi: 10.1126/science.1141967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cheezum MK, Walker WF, Guilford WH. Biophys. J. 2001;81:2378–2388. doi: 10.1016/S0006-3495(01)75884-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jaqaman K, Loerke D, Mettlen M, Kuwata H, Grinstein S, Schmid SL, Danuser G. Nat. Methods. 2008;5:695–702. doi: 10.1038/nmeth.1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Park HY, Buxbaum AR, Singer RH. Methods Enzymol. 2010;472:387–406. doi: 10.1016/S0076-6879(10)72003-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pinaud F, Michalet X, Iyer G, Margeat E, Moore HP, Weiss S. Traffic. 2009;10:691–712. doi: 10.1111/j.1600-0854.2009.00902.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kusumi A, Sako Y, Yamamoto M. Biophys. J. 1993;65:2021–2040. doi: 10.1016/S0006-3495(93)81253-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Saxton MJ, Jacobson K. Annu. Rev. Biophys. Biomol. Struct. 1997;26:373–399. doi: 10.1146/annurev.biophys.26.1.373. [DOI] [PubMed] [Google Scholar]
- 39.Kuszak AJ, Pitchiaya S, Anand JP, Mosberg HI, Walter NG, Sunahara RK. J. Biol. Chem. 2009;284:26732–26741. doi: 10.1074/jbc.M109.026922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ding H, Wong PT, Lee EL, Gafni A, Steel DG. Biophys. J. 2009;97:912–921. doi: 10.1016/j.bpj.2009.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chung SH, Kennedy RA. J. Neurosci. Methods. 1991;40:71–86. doi: 10.1016/0165-0270(91)90118-j. [DOI] [PubMed] [Google Scholar]
- 42.Simonson PD, Deberg HA, Ge P, Alexander JK, Jeyifous O, Green WN, Selvin PR. Biophys J. 2010;99:L81–L83. doi: 10.1016/j.bpj.2010.08.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fusco D, Accornero N, Lavoie B, Shenoy SM, Blanchard JM, Singer RH, Bertrand E. Curr. Biol. 2003;13:161–167. doi: 10.1016/s0960-9822(02)01436-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Aizer A, Brody Y, Ler LW, Sonenberg N, Singer RH, Shav-Tal Y. Mol. Biol. Cell. 2008;19:4154–4166. doi: 10.1091/mbc.E08-05-0513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Winz ML, Samanta A, Benzinger D, Jaschke A. Nucleic Acids Res. 2012;40:e78. doi: 10.1093/nar/gks062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ding XC, Weiler J, Grosshans H. Trends Biotechnol. 2009;27:27–36. doi: 10.1016/j.tibtech.2008.09.006. [DOI] [PubMed] [Google Scholar]