Abstract
Recent genome-wide association studies have suggested that endocytic factor, such as phosphatidylinositol-binding clathrin assembly protein (PICALM), may be implicated in the development of Alzheimer’s disease (AD). The cellular functions of PICALM are in line with this possibility: (i) PICALM is involved in regulation of amyloid-β levels and (ii) PICALM is important for a pre-synaptic function, which is diminished in AD. To facilitate the analysis of PICALM, we developed a quantitative method to assess expression level of PICALM in different biological samples. For this purpose, a stable isotope-labeled quantification concatamer (QconCAT) of PICALM was designed, expressed, purified and characterized. The PICALM QconCAT was firstly used as an internal standard in a multiple reaction monitoring assay to measure PICALM concentrations in the human frontal cortex, tissue strongly affected by AD. A second endocytic factor which is highly homologous to PICALM and also functions in clathrin-mediated endocytosis, clathrin coat assembly protein AP180 (AP180), was quantified as well. Because age-related macular degeneration (AMD) shares several clinical and pathological features with AD, the measurements were then extended to human normal neural retina. Overall, the developed method is suitable for PICALM and AP180 quantitative analysis in various biological samples of interest.
A number of genome-wide association studies (GWAS) and subsequent replication studies with meta-analysis have identified a PICALM gene as a new susceptibility gene for late-onset Alzheimer’s disease (AD) [1–3]. The identified single-nucleotide polymorphism (rs3851179) lies 88.5 kb 5′ to the actual coding region, suggesting that the expression of PICALM gene in AD may be altered. The gene product phosphatidylinositol-binding clathrin assembly protein (PICALM) is highly homologous to another endocytic factor, clathrin coat assembly protein AP180 (AP180). Both proteins play a role in clathrin-mediated endocytosis [4] and abnormalities in this pathway have been identified in cases of AD [5, 6]. Expression level studies employing immunochemical and mRNA measurements have demonstrated that levels of PICALM and AP180 can be affected in the presence of AD. For example, a significant increase of PICALM in the cortex of transgenic AD mice carrying the Swedish mutation of amyloid-β protein precursor has been reported [7]. Interestingly, other studies have shown significant decrease in AP180 protein level in brain samples from transgenic AD mice and from human AD patients [8, 9]. These findings suggest that PICALM and AP180 are linked to AD, although their exact role(s) in AD pathology remain to be established.
It is important to underline that GWAS identify variants in DNA which are associated with a disease, but this approach cannot by itself be used to specify which genes are causal, and therefore cannot be used to calculate disease risk with certainty. However, there is a hope that GWAS may elucidate disease biology and point to new targets for therapy. Thus, it would be undoubtedly beneficial to develop independent quantitative methods for validation of data obtained through GWAS and/or immunochemical and mRNA measurements.
Multiple reaction monitoring (MRM) assays use liquid chromatography/triple quadrupole mass spectrometry (LC-MS/MS) to measure the concentration of specific proteins in complex biological samples. The high specificity of MRM assays is achieved by monitoring multiple transitions (precursor ion – product ion pairs) for each target peptide derived from the protein of interest. Quantification is performed by comparing the chromatographic peak area of a transition from the (unlabeled) native peptide to that of the corresponding transition from the stable isotope-labeled internal standard. This approach has been successfully used for biomarker validation in human plasma and serum [10–12]. However, the application of MRM assays to the direct measurement of proteins in human tissues is more recent [13–15] and remains a challenge.
In this study, we first concentrated on developing an MRM assay to measure the protein levels of PICALM and AP180. It was next applied to measurements in the human frontal cortex, tissue strongly affected by AD. To validate broad applications of developed method and since there are some similarities between AD and age-related macular degeneration (AMD) [16, 17], we have extended the measurements of PICALM and AP180 to the normal neural retina. In summary, the proposed method allows quantitative assessment of PICALM and AP180 levels in various biological samples.
Material and methods
Materials
Expressway cell-free E. coli expression system was obtained from Invitrogen (Carlsbad, CA, USA). L-[13C6,15N2]-lysine and L-[13C6,15N4]-arginine (>95% purity) were obtained from Spectral Stable Isotopes (Columbia, MD, USA). The DC Protein Assay kit was obtained from Bio-Rad Laboratories (Hercules, CA, USA). Sequencing grade modified trypsin was obtained from Promega Corp. (Madison, WI, USA). All other chemicals were purchased from Sigma-Aldrich (St. Louis, MO, USA).
Design, expression and purification of PICALM QconCAT
A synthetic gene encoding amino acid sequence MVYGNERFIQYLASRNTLFNLSNFLDKSGLQGYISEFLKVAEQVGIDRGDIPDLVDEREKQAALEEEQARLKALKENPFLTKSSGDVHLSISSDVSTFTTRTPTHEM was synthesized by Integrated DNA Technologies (Coralville, IA, USA). The underlined segments represent the signature proteotypic peptides of PICALM (Q-peptides). The synthetic PICALM gene was cloned into the pEXP5CT/TOPO expression vector in-frame to the C-terminal His6-tag. Stable isotope-labeled PICALM QconCAT was expressed using an in vitro translation Expressway cell-free E. coli expression system according to the manufacture’s protocol. L-[13C6,15N2]lysine and L-[13C6,15N4]arginine were added to the amino acid mixture to replace unlabeled lysine and arginine. After in vitro translation, the labeled PICALM QconCAT was purified by nickel-nitrilotriacetic acid resin in batch mode (Qiagen, Valencia, CA, USA). Finally the purified QconCAT was loaded onto a SpinTrap G-25 spin column (GE Healthcare, Waukesha, WI, USA) to exchange buffer into 25 mmol/L NH4HCO3 with 1% SDS (mass concentration). The protein concentration of PICALM QconCAT was measured in the presence of 1% SDS (mass concentration) using the DC Protein Assay kit and bovine serum albumin as a standard. The final PICALM QconCAT was aliquoted and kept frozen at −80 °C.
To verify full-length expression of PICALM QconCAT, 1 μL of sample was mixed with 1 μL of matrix solution (20 mg/mL sinapic acid in 50% acetonitrile/0.1% trifluoroacetic acid). The mixed sample was spotted onto an ABI 01-192-6-AB target plate and allowed to dry at room temperature. Mass spectrometry analysis was performed using an AB4700 Proteomics Analyzer (Applied Biosystems, Framingham, MA, USA). Linear mode acquisition consisted of 2,000 laser shots averaged over 40 sample positions.
Human tissues
Human tissue use conformed to the Declaration of Helsinki and had been approved by Ethical Committee at the Washington University and Case Western Reserve University. Brain and retinal specimens were obtained from de-identified donors following informed consent of the respective families. Demographic information on the donors is summarized in the Supplementary Table S1. Frozen samples of frontal cortex were received from the Washington University School of Medicine Alzheimer’s Disease Research Center. Eyes were acquired through the Cleveland, Georgia or Midwest Eye Banks and dissected as described [18]. The anterior segment was removed, and fundus photos were taken to assess retinal status by a fellowship-trained retinal specialist. Thereafter, the neural retina was isolated, flash-frozen in liquid nitrogen and stored at −80 °C until analyzed.
Processing of samples
Minced human tissue was placed in 25 mmol/L NH4HCO3 with 1% SDS (mass concentration) and homogenized by sonication at 30 W using five 10 s continuous cycles (Sonicator 3000, Misonix Inc., Farmingdale, NY, USA). The homogenate was centrifuged at 2000 g for 5 min to remove tissue debris. The supernatant was used to measure total protein concentration in the presence of 1% SDS (mass concentration) using the DC Protein Assay kit and bovine serum albumin as a standard. The supernatant was then aliquoted 0.2 mg of total tissue protein per tube and kept frozen at −80 °C. During the following experiments, samples of 0.2 mg of total tissue protein were supplemented with 20 mmol/L DTT and various amounts of labeled PICALM QconCAT, ranging from 0.3 pmol to 5.0 pmol per sample. The mixtures were incubated at room temperature for 60 min to allow reduction of cysteines and were then treated with 50 mmol/L iodoacetamide for another 60 min. Alkylated samples were precipitated with chloroform/methanol [19]. This step depleted salts, SDS, and biological lipids from the samples. To remove possible trace contaminates, the protein pellets were sonicated in 1 mL of water and precipitated again by centrifugation at 20,000 g for 10 min. After washing with water, protein pellets were sonicated in 100 μL of 25 mmol/L NH4HCO3/0.1% RapiGest and treated with trypsin for 15 h at 37 °C. The substrate/trypsin ratio was 50:1 (m/m). After trypsin digestion, the peptide samples were treated with 0.5% trifluoroacetic acid for 30 min at 37 °C and centrifuged at 106,000 g for 30 min to remove RapiGest and other by-products not soluble at low pH. After centrifugation, the supernatants were dried using a vacuum centrifuge (Vacufuge, Eppendorf AG, Hamburg, Germany).
LC-MS/MS analysis
Instrumental analyses were performed on a hybrid triple quadrupole/linear ion trap mass spectrometer (4000 QTRAP, ABI/MDS-Sciex) coupled to an Eksigent nanoLC-2D system (Dublin, CA, USA). Separation of peptides was performed with an Eksigent cHiPLC- nanoflex system equipped with a nano cHiPLC column, 15 cm × 75 μm, packed with ReproSil-Pur C18-AQ, 3 μm (Dr. Maisch, Germany). Peptides were eluted over a 29 min-gradient from 15% to 35% acetonitrile, (volume concentration) containing 0.1% formic acid (volume concentration) at a flow rate of 300 nL/min. The column effluent was continuously directed into the nanospray source of the mass spectrometer. All acquisition methods used the following parameters: an ion spray voltage of 2200 V, curtain gas of 105 kPa (15 psi), source gas of 140 kPa (20 psi), interface heating temperature of 170 °C, declustering potential of 76 V for +2 precursor ions and 65 V for +3 precursor ions, collision energy of 30 V for +2 precursor ions and 22 V for +3 precursor ions, and collision cell exit potential of 16 V for +2 precursor ions and 13V for +3 precursor ions. The dwell time for all transitions was 40 ms.
Quantitative analysis and validation
Three most intensive transitions per Q-peptide used for quantification are summarized in the Supplementary Table S2. The relative ratios of these three transitions monitored in the 25 mmol/L NH4HCO3 for isotope-labeled PICALM QconCAT were similar to those observed by spiking isotope-labeled PICALM QconCAT into the biological samples. This indicated no significant interference from biological sample for the quantification based on selected transitions. The identities of the measured Q-peptides were confirmed based on the retention time of the three MRM peaks from a given Q-peptide and the ratio among the three MRM peaks. The reported value for each group was the mean with standard deviation of each biological replicate using all peptide transitions.
Results
PICALM QconCAT
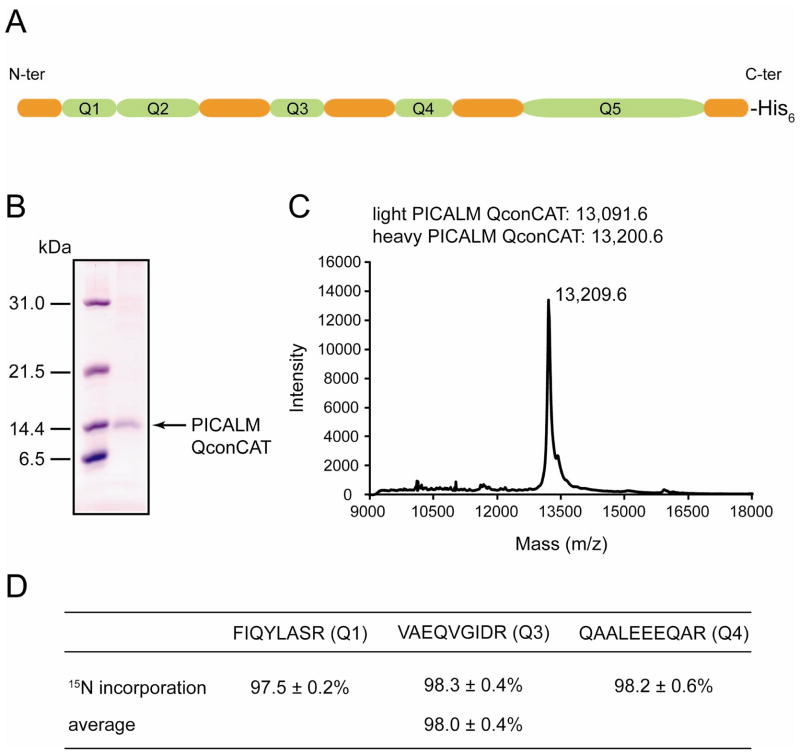
QconCATs are artificial proteins that represent concatamers of Q-peptides generated from proteins under study and used as internal standards for quantification of these proteins in MRM assay [20]. Q-peptides are usually tryptic peptides with zero missed cleavages and molecular masses between 700 Da and 2500 Da. Methionine or cysteine containing peptides are typically omitted due to the existence of various oxidation entities in nature, which could introduce variations in the quantification. Fig. 1A illustrates the design of a PICALM QconCAT with five Q-peptides (shown in green) carrying 6-amino acid long extensions from their natural sequences on both sides of the Q-peptides (shown in orange). The presence of flanking regions ensures the comparable rates of trypsinolysis for the QconCAT and analyte [21, 22]. The His6-tag necessary for purification is separated from the last Q-peptide by the flanking sequence specific for that Q-peptide and therefore will not affect the rate of proteolytic release. The uniqueness of the selected Q-peptides was verified by performing a protein blast search (http://blast.ncbi.nlm.nih.gov/Blast.cgi). Q3, Q4, and Q5 are unique PICALM peptides while Q1 and Q2 are also found in AP180, and this allows for calculation of the AP180 concentration by subtracting the PICALM concentration based on Q3, Q4, or Q5 from the protein concentration based on Q1 or Q2. The isotope-labeled PICALM QconCAT was prepared using cell-free expression with L-[13C6,15N2]-lysine and L[13C6,15N4]-arginine as the sole sources of lysine and arginine. The purified QconCAT was ≥95% pure based on SDS-PAGE analysis (Fig. 1B). Using linear mode MALDI-MS, the measured m/z [M + H]+ value for purified isotope-labeled PICALM QconCAT was 13,209.6 (Fig. 1C). This matches well with the expected m/z [M + H]+ value of 13,200.6 and confirms expression and isolation of the full-length QconCAT. The isotope incorporation was evaluated at the peptide level after digestion of the pure PICALM QconCAT with trypsin. It was calculated as the ratio of the peak area for the labeled peptides to the sum of the peak areas for the unlabeled and labeled peptides from the MRM assay and expressed as a percentage. Based on three Q-peptides, the average isotope incorporation was 98.0 ± 0.4 % (Fig. 1D). This value was interpreted as a complete labeling and so no correction for labeling efficiency was applied during data analysis.
Fig. 1.
Characterization of PICALM QconCAT. (A) Schematic representation of PICALM QconCAT which includes five concatenated Q-peptides (in green) with 6-amino acid long flanking regions (in orange). (B) 15% SDS-PAGE gel showing purified PICALM QconCAT. Molecular mass standards (in kDa) are shown on the left. (C) Linear mode MALDI spectrum of purified PICALM QconCAT. Sinapic acid was used as a matrix. The theoretical m/z [M + H]+ values for light and heavy PICALM QconCATs are indicated. (D) 15N incorporation into the heavy PICALM QconCAT was evaluated on peptide level after QconCAT digestion with trypsin and presented as mean ± SD for three transitions per peptide. For average, the data for three peptides were combined and presented as mean ± SD.
In the preliminary measurements on frontal cortex, the abundance of Q-peptides in the tryptic digest was determined in two ways: i) for observed +2 charge precursor ions using the corresponding +1 charge y fragment ions, and ii) for observed +3 charge precursor ions using the corresponding +1 and +2 charge y fragment ions. As is common, only ions with m/z values greater than the precursor ion m/z were analyzed. The intensities of the MRM transitions were recorded and the three most intense transitions per peptide were selected. It is also important to emphasize that during the measurements on real tissue samples, all signal intensities are typically lower than those observed for pure analytes and standards in buffer solutions. In our preliminary measurements on frontal cortex, the signal intensities for Q2 and Q5 were at the limit of detection and did not allow for reproducible quantification. Therefore, all subsequent measurements were performed based on Q1, Q3, and Q4 only.
To further validate the quantitative method, frontal cortex sample was supplemented with a variable amount of isotope-labeled PICALM QconCAT (from 0.3 pmol to 5.0 pmol) and standard curves were generated. The standard curves show linearity and low scattering over the QconCAT concentration range tested and indicated remarkably similar behavior of selected transitions. Fig. 2 shows standard curves for three transitions of Q4. These standard curves are representative of the other Q-peptides used for the quantification of PICALM and AP180.
Fig. 2.
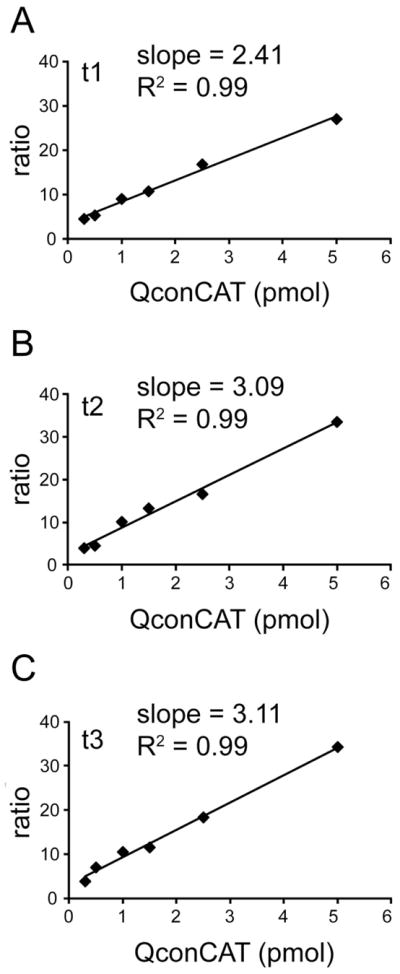
Standard curves for the PICALM QconCAT. The linearity of the optimum transitions were verified by spiking six frontal cortex samples (0.2 mg of total tissue protein each) with varying amounts of the PICALM QconCAT. The peak area ratio of a heavy peptide transition (from PICALM QconCAT) to the corresponding light peptide transition (from frontal cortex) is plotted versus the supplemented PICALM QconCAT amount. (A), (B), and (C) show three individual transitions for QAALEEEQAR (Q4); t1 is 572.8/632.3 and 577.8/642.3; t2 is 572.8/874.4 and 577.8/884.4; t3 is 572.8/945.5 and 577.8/955.5.
Measurements in frontal cortex and neural retina
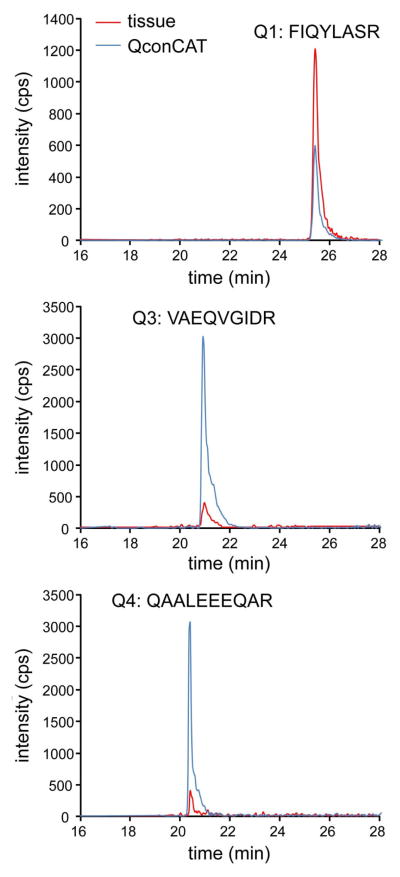
Fig. 3 shows representative extracted ion chromatograms for Q1, Q3, and Q4, with a single pair of transitions shown for each peptide. The ratio of analyte peak area (protein in frontal cortex, red line) to that of the internal standard (PICALM QconCAT, blue line) was determined, and then the analyte protein concentrations were calculated by multiplying each ratio by the known amount of PICALM QconCAT (spiked into the sample prior to digestion) and normalized to the mass of total tissue protein. Three pairs of transitions were monitored for each peptide and were treated as independent measurements. The level of each given peptide was based on the mean ± SD of the transitions from this peptide. The concentrations for individual donors were calculated for three biological replicates. Quantification data for frontal cortex are summarized in Table 1. Measurements for PICALM based on two peptides (Q3 and Q4) show small donor-to-donor variation and were combined into a consensus value of 0.62 ± 0.13 pmol/mg tissue protein. The same small donor-to-donor variation was observed for joint measurements of PICALM and AP180 based on Q1. The consensus value was 6.9 ± 0.4 pmol/mg tissue protein. Quantification data for neural retina are summarized in Table 2. Similar to quantifications in frontal cortex, the individual measurements had little donor-to-donor variation and were combined in consensus values. For PICALM, the consensus value was 0.47 ± 0.13 pmol/mg tissue protein. For joint measurements of PICALM and AP180, the consensus value was 2.2 ± 0.4 pmol/mg tissue protein.
Fig. 3.
Extracted ion chromatograms for Q1, Q3 and Q4. A pair of transitions monitored in control frontal cortex is shown for three Q-peptides: FIQYLASR (Q1), VAEQVGIDR (Q3), and QAALEEEQAR (Q4). Signals for these transitions from internal standard (blue line) and analyte (red line) are color coordinated.
Table 1.
Quantification of PICALM and AP180 in the frontal cortex
| Protein (pmol/mg tissue protein)a | ||||||
|---|---|---|---|---|---|---|
| Donor ID | #1 | #5 | #12 | #13 | #14 | #18 |
| PICALM+AP180 | ||||||
| Q1 | 7.1 ± 0.3 | 6.7 ± 0.4 | 6.2 ± 1.0 | 6.7 ± 0.2 | 7.5 ± 0.3 | 7.0 ± 0.4 |
| consensus | 6.9 ± 0.4 | |||||
| PICALM | ||||||
| Q3 | 0.82 ± 0.14 | 0.64 ± 0.17 | 0.51 ± 0.27 | 0.81 ± 0.43 | 0.63 ± 0.38 | 0.50 ± 0.20 |
| Q4 | 0.79 ± 0.40 | 0.62 ± 0.12 | 0.57 ± 0.24 | 0.61 ± 0.20 | 0.53 ± 0.30 | 0.44 ± 0.05 |
| consensus | 0.62 ± 0.13 | |||||
The concentration was calculated for three biological replicates by monitoring three transitions per individual peptide and presented as mean ± SD. For the consensus, the data for Q1 from different donors and data for Q3 and Q4 from different donors were combined and presented as mean ± SD. The monitored transitions are summarized in Supplementary Table S2.
Table 2.
Quantification of PICALM and AP180 in the neural retina
| Protein (pmol/mg tissue protein)a | ||||||
|---|---|---|---|---|---|---|
| Donor ID | #12 | #13 | #17 | #20 | #32 | #39 |
| PICALM+AP180 | ||||||
| Q1 | 2.2 ± 0.3 | 2.5 ± 0.9 | 2.4 ± 0.3 | 2.6 ± 0.3 | 1.7 ± 0.3 | 2.5 ± 0.4 |
| consensus | 2.2 ± 0.4 | |||||
| PICALM | ||||||
| Q3 | 0.73 ± 0.31 | 0.51 ± 0.09 | 0.56 ± 0.18 | 0.45 ± 0.14 | 0.39 ± 0.13 | 0.71 ± 0.12 |
| Q4 | 0.42 ± 0.15 | 0.45 ± 0.13 | 0.31 ± 0.06 | 0.51 ± 0.25 | 0.33 ± 0.06 | 0.54 ± 0.21 |
| consensus | 0.47 ± 0.13 | |||||
The concentration was calculated for three biological replicates by monitoring three transitions per individual peptide and presented as mean ± SD. For the consensus, the data for Q1 from different donors and data for Q3 and Q4 from different donors were combined and presented as mean ± SD. The monitored transitions are summarized in Supplementary Table S2.
The concentration of AP180 in each sample can be calculated by subtracting the concentration of PICALM based on Q3 and Q4 from the joint concentration of the two proteins based on Q1. The average concentration of AP180 in the frontal cortex was 6.28 pmol/mg tissue protein and the average concentration of AP180 in the neural retina was1.73 pmol/mg tissue protein, a difference of nearly 4-fold. The average concentrations of PICALM were similar in these two tissues; 0.62 pmol/mg tissue protein in the frontal cortex and 0.47 pmol/mg tissue protein in the neural retina.
Discussion
Recent GWAS of late-onset AD pointed to an association with PICALM variants [1–3]. In addition, the notion that PICALM protein might be involved in the development of neurodegenerative diseases has gained a support from previous studies, which demonstrated that endocytic factors such as PICALM and AP180 are important in the regulation of amyloid-β levels and in supporting normal pre-synaptic function (reviewed in [23]). It is therefore conceivable that expression levels of these proteins may be altered in neuronal degeneration. Several reports utilizing antibody-based approaches for proteins and real-time polymerase chain reaction for mRNA are in line with this possibility [7–9, 24]. However, because of intrinsic limitations, these approaches are not always truly quantitative. Thus, we sought to develop a truly quantitative approach to measure the protein level of PICALM and AP180, which will allow inter-laboratory comparison of data generated. For this reason, we have designed, expressed, purified, and characterized a universal internal standard for MRM assay, PICALM QconCAT. We optimized a protocol for tissue sample processing and demonstrated that it works well for such different tissues, as frontal cortex and neural retina. In addition, the PICALM and AP180 quantifications were normalized to mg of total tissue protein to support an inter-laboratory comparison of results obtained. Measurements on frontal cortex and neural retina were reproducible, showed low donor-to-donor variation and the proposed method can be recommended for quantification of PICALM and AP180 levels in various biological samples.
Supplementary Material
Acknowledgments
This work was supported in part by National Institutes of Health grant EY018383 (to I.A.P.). I.A.P. is a recipient of the Jules and Doris Stein Professorship from the Research to Prevent Blindness. We thank Dr. S. Huang for the retinal assessments, Dr. R. Reem, Dr. S. Omarova and Dr. N. Mast for the isolation of the retinal samples and Dr. A. Szakal for helpful discussions and critical reviewing the manuscript.
Footnotes
Certain commercial materials, instruments, and equipment are identified in this manuscript in order to specify the experimental procedure as completely as possible. In no case does such identification imply a recommendation or endorsement by the National Institute of Standards and Technology nor does it imply that the materials, instruments, or equipment identified are necessarily the best available for the purpose.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Harold D, Abraham R, Hollingworth P, Sims R, Gerrish A, Hamshere ML, Pahwa JS, Moskvina V, Dowzell K, Williams A, Jones N, Thomas C, Stretton A, Morgan AR, Lovestone S, Powell J, Proitsi P, Lupton MK, Brayne C, Rubinsztein DC, Gill M, Lawlor B, Lynch A, Morgan K, Brown KS, Passmore PA, Craig D, McGuinness B, Todd S, Holmes C, Mann D, Smith AD, Love S, Kehoe PG, Hardy J, Mead S, Fox N, Rossor M, Collinge J, Maier W, Jessen F, Schurmann B, van den Bussche H, Heuser I, Kornhuber J, Wiltfang J, Dichgans M, Frolich L, Hampel H, Hull M, Rujescu D, Goate AM, Kauwe JS, Cruchaga C, Nowotny P, Morris JC, Mayo K, Sleegers K, Bettens K, Engelborghs S, De Deyn PP, Van Broeckhoven C, Livingston G, Bass NJ, Gurling H, McQuillin A, Gwilliam R, Deloukas P, Al-Chalabi A, Shaw CE, Tsolaki M, Singleton AB, Guerreiro R, Muhleisen TW, Nothen MM, Moebus S, Jockel KH, Klopp N, Wichmann HE, Carrasquillo MM, Pankratz VS, Younkin SG, Holmans PA, O’Donovan M, Owen MJ, Williams J. Genome-wide association study identifies variants at CLU and PICALM associated with Alzheimer’s disease. Nat Genet. 2009;41:1088–1093. doi: 10.1038/ng.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jun G, Naj AC, Beecham GW, Wang LS, Buros J, Gallins PJ, Buxbaum JD, Ertekin-Taner N, Fallin MD, Friedland R, Inzelberg R, Kramer P, Rogaeva E, St George-Hyslop P, Cantwell LB, Dombroski BA, Saykin AJ, Reiman EM, Bennett DA, Morris JC, Lunetta KL, Martin ER, Montine TJ, Goate AM, Blacker D, Tsuang DW, Beekly D, Cupples LA, Hakonarson H, Kukull W, Foroud TM, Haines J, Mayeux R, Farrer LA, Pericak-Vance MA, Schellenberg GD. Meta-analysis confirms CR1, CLU, and PICALM as alzheimer disease risk loci and reveals interactions with APOE genotypes. Arch Neurol. 2010;67:1473–1484. doi: 10.1001/archneurol.2010.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kamboh MI, Demirci FY, Wang X, Minster RL, Carrasquillo MM, Pankratz VS, Younkin SG, Saykin AJ, Jun G, Baldwin C, Logue MW, Buros J, Farrer L, Pericak-Vance MA, Haines JL, Sweet RA, Ganguli M, Feingold E, Dekosky ST, Lopez OL, Barmada MM. Genome-wide association study of Alzheimer’s disease. Transl Psychiatry. 2012;2:e117. doi: 10.1038/tp.2012.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ford MG, Pearse BM, Higgins MK, Vallis Y, Owen DJ, Gibson A, Hopkins CR, Evans PR, McMahon HT. Simultaneous binding of PtdIns(4,5)P2 and clathrin by AP180 in the nucleation of clathrin lattices on membranes. Science. 2001;291:1051–1055. doi: 10.1126/science.291.5506.1051. [DOI] [PubMed] [Google Scholar]
- 5.Cataldo AM, Hamilton DJ, Barnett JL, Paskevich PA, Nixon RA. Abnormalities of the endosomal-lysosomal system in Alzheimer’s disease: relationship to disease pathogenesis. Adv Exp Med Biol. 1996;389:271–280. [PubMed] [Google Scholar]
- 6.Cataldo AM, Barnett JL, Pieroni C, Nixon RA. Increased neuronal endocytosis and protease delivery to early endosomes in sporadic Alzheimer’s disease: neuropathologic evidence for a mechanism of increased beta-amyloidogenesis. J Neurosci. 1997;17:6142–6151. doi: 10.1523/JNEUROSCI.17-16-06142.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thomas RS, Lelos MJ, Good MA, Kidd EJ. Clathrin-mediated endocytic proteins are upregulated in the cortex of the Tg2576 mouse model of Alzheimer’s disease-like amyloid pathology. Biochem Biophys Res Commun. 2011;415:656–661. doi: 10.1016/j.bbrc.2011.10.131. [DOI] [PubMed] [Google Scholar]
- 8.Yao PJ. Synaptic frailty and clathrin-mediated synaptic vesicle trafficking in Alzheimer’s disease. Trends Neurosci. 2004;27:24–29. doi: 10.1016/j.tins.2003.10.012. [DOI] [PubMed] [Google Scholar]
- 9.Cao Y, Xiao Y, Ravid R, Guan ZZ. Changed clathrin regulatory proteins in the brains of Alzheimer’s disease patients and animal models. J Alzheimers Dis. 2010;22:329–342. doi: 10.3233/JAD-2010-100162. [DOI] [PubMed] [Google Scholar]
- 10.Anderson L, Hunter CL. Quantitative mass spectrometric multiple reaction monitoring assays for major plasma proteins. Mol Cell Proteomics. 2006;5:573–588. doi: 10.1074/mcp.M500331-MCP200. [DOI] [PubMed] [Google Scholar]
- 11.Keshishian H, Addona T, Burgess M, Kuhn E, Carr SA. Quantitative, multiplexed assays for low abundance proteins in plasma by targeted mass spectrometry and stable isotope dilution. Mol Cell Proteomics. 2007;6:2212–2229. doi: 10.1074/mcp.M700354-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kitteringham NR, Jenkins RE, Lane CS, Elliott VL, Park BK. Multiple reaction monitoring for quantitative biomarker analysis in proteomics and metabolomics. J Chromatogr B Analyt Technol Biomed Life Sci. 2009;877:1229–1239. doi: 10.1016/j.jchromb.2008.11.013. [DOI] [PubMed] [Google Scholar]
- 13.Wang M, Heo GY, Omarova S, Pikuleva IA, Turko IV. Sample prefractionation for mass spectrometry quantification of low-abundance membrane proteins. Anal, Chem. 2012;84:5186–5191. doi: 10.1021/ac300587v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang M, Chen J, Turko IV. 15N-labeled full-length apolipoprotein E4 as an internal standard for mass spectrometry quantification of apolipoprotein E isoforms. Anal Chem. 2012;84:8340–8344. doi: 10.1021/ac3018873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen J, Wang M, Turko IV. Quantification of amyloid precursor protein isoforms using quantification concatamer internal standard. Anal Chem. 2013;85:303–307. doi: 10.1021/ac3033239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kaarniranta K, Salminen A, Haapasalo A, Soininen H, Hiltunen M. Age-related macular degeneration (AMD): Alzheimer’s disease in the eye? J Alzheimers Dis. 2011;24:615–631. doi: 10.3233/JAD-2011-101908. [DOI] [PubMed] [Google Scholar]
- 17.Ohno-Matsui K. Parallel findings in age-related macular degeneration and Alzheimer’s disease. Prog Retin Eye Res. 2011;30:217–238. doi: 10.1016/j.preteyeres.2011.02.004. [DOI] [PubMed] [Google Scholar]
- 18.Zheng W, Reem RE, Omarova S, Huang S, DiPatre PL, Charvet CD, Curcio CA, Pikuleva IA. Spatial distribution of the pathways of cholesterol homeostasis in human retina. PLoS One. 2012;7:e37926. doi: 10.1371/journal.pone.0037926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liao WL, Turko IV. Strategy combining separation of isotope-labeled unfolded proteins and matrix-assisted laser desorption/ionization mass spectrometry analysis enables quantification of a wide range of serum proteins. Anal Biochem. 2008;377:55–61. doi: 10.1016/j.ab.2008.03.016. [DOI] [PubMed] [Google Scholar]
- 20.Beynon RJ, Doherty MK, Pratt JM, Gaskell SJ. Multiplexed absolute quantification in proteomics using artificial QCAT proteins of concatenated signature peptides. Nat Methods. 2005;2:587–589. doi: 10.1038/nmeth774. [DOI] [PubMed] [Google Scholar]
- 21.Nanavati D, Gucek M, Milne JL, Subramaniam S, Markey SP. Stoichiometry and absolute quantification of proteins with mass spectrometry using fluorescent and isotope-labeled concatenated peptide standards. Mol Cell Proteomics. 2008;7:442–447. doi: 10.1074/mcp.M700345-MCP200. [DOI] [PubMed] [Google Scholar]
- 22.Chen J, Wang M, Turko IV. Mass spectrometry quantification of clusterin in the human brain. Mol Neurodegener. 2012;7:41. doi: 10.1186/1750-1326-7-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maritzen T, Koo SJ, Haucke V. Turning CALM into excitement: AP180 and CALM in endocytosis and disease. Biol Cell. 2012;104:588–602. doi: 10.1111/boc.201200008. [DOI] [PubMed] [Google Scholar]
- 24.Baig S, Joseph SA, Tayler H, Abraham R, Owen MJ, Williams J, Kehoe PG, Love S. Distribution and expression of picalm in Alzheimer disease. J Neuropathol Exp Neurol. 2010;69:1071–1077. doi: 10.1097/NEN.0b013e3181f52e01. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.