Abstract
Robotic and automation technologies have played a huge role in in vitro biological science, having proved critical for scientific endeavors such as genome sequencing and high-throughput screening. Robotic and automation strategies are beginning to play a greater role in in vivo and in situ sciences, especially when it comes to the difficult in vivo experiments required for understanding the neural mechanisms of behavior and disease. In this perspective, we discuss the prospects for robotics and automation to impact neuroscientific and intact-system biology fields. We discuss how robotic innovations might be created to open up new frontiers in basic and applied neuroscience, and present a concrete example with our recent automation of in vivo whole cell patch clamp electrophysiology of neurons in the living mouse brain.
Keywords: robotics, neuroscience, patch clamping
Introduction: automation in biology
Robotics has played a major role in the advancement of biological research in the past few decades. Semi-autonomous machines integrate hardware, wetware, and software from precision engineered or microfabricated parts to nimbly load, manipulate, and measure thousands to millions of biological samples simultaneously, more rapidly, more sensitively, more accurately, or in a more repeatable manner than manual approaches. Their applications span the research space from automated phenotyping to high-throughput screening to imaging to genome sequencing. Examples abound for how these tools have opened the door to essential, comprehensive biological studies. In the race to sequence the human genome in the 1990s, robots capable of high-throughput analysis of microliter volumes of liquid were developed and deployed massively (Fig. 1). As just a few examples, high-throughput microfluidics1, 2 can now be used to perform nearly 10,000 independent real time PCR reactions for genotyping3 and transcriptome profiling applications.4–8 Fluid handling robots have revolutionized synthetic biology by enabling the efficient, rapid transfer of reagents from one set of plates to another.9,10 Automated plate readers and microscopy methods enable time-lapse imaging of physiological changes in cultured cells, and in vitro patch clamping enables automated electrophysiology in cell lines.11–15 These innovations are very widely used, and sometimes ubiquitous, in major research institutions and industrial settings such as in pharmaceutical and biotechnology companies.
Figure 1.

One of many rows of ABI 3730xl automated DNA Analyzers for shotgun sequencing of the human genome in months (30 billion bp/year) in 2005. (Courtesy: Steve Jurvetson)
Value and principles of in vivo and in situ automation
Despite this progress and the pervasive presence of automation in molecular biology today, there remain many tedious and repetitive manual tasks, as well as more complex tasks which defy straightforward automation, and that are more akin to art forms than scientific processes. Often they have not been systematically analyzed but are passed down through generations of researchers as best practices. This practice sometimes limits use to a few highly-skilled laboratories, especially when living organs or organisms are involved, as in in vivo neuroscience experiments. This frontier represents an opportunity because automation would not only make things simpler, increasing the rate of adoption and progress of specific approaches, but broadens the number of individuals and laboratories that can contribute innovations, since they no longer have to improve an art form, but rather can iteratively improve the protocols performed by a robotic agent. Further, higher throughput automation can enable scaling of the number of parallel samples being analyzed, or the number of analyses being performed per sample, or the sampling rate. It can also lead to increased standardization of procedures across laboratories, important for improving the ability of different parts of the literature to be integrated, and for results from different groups to be compared.
In many cases, increasing the number of observations that can be made in parallel is important not only for augmenting the amount of data that can be collected, but can also open up fundamentally new kinds of investigation. For example, making many simultaneous observations on different parts of an intact system – an organ, such as the brain, or even an entire organism – might reveal correlated, and perhaps coordinated, physiological processes taking place in different parts of the system, which would never be revealed by investigation of single sites one at a time. As another example, the ability to perform tasks often done in vitro – from pharmacological assessment to biochemical analysis – in the living organism, would enable detailed understanding of how specific processes of basic or applied scientific interest take place, in the full context of an intact organism, including its baseline activity, awake or behaving state-dependent modulation, or disease states.
Rethinking in vivo procedures for automation
In order to automate a complex in vivo or in situ procedure, it is very important to understand how humans perform the procedure, not only analyzing the procedure at face value but also delving deeply into the parameters that govern success or failure of the procedure. This often means that the engineer seeking to automate a procedure must not simply take requests from biologists, simply attempting to automate their stated methods, but instead must herself master the procedure, so that it is possible for the engineer to understand the best possible way for the procedure to be automated. Very often, the way that a human performs will not necessarily be the easiest way to perform the task in an automated fashion. Humans often use complex sets of cues—visual and auditory, for example—to perform in vivo experiments. But for automation, it may be important to rely upon more straightforward and less complex modalities, such as electrical impedance (which can nevertheless indicate important properties of a tissue that a robotic device is exploring). In addition, humans conduct in vivo experiments with their very-high-degree-of-freedom hands and synergistic muscular systems, but the range of inexpensive actuators that are reliable and inexpensive enough to become commonly used in messy and complex biology lab environments, may require a greater reliance on good software and rethinking of the procedure to minimize the number of expensive actuators required, or even a rethinking of the modality of actuation (e.g., replacing a complex robot arm with a scanning laser beam). Below, we explore these two arenas of endeavor – how to find the most easily automated methodology for performing a complex in vivo procedure, and how to devise the simplest and most robust modality of actuation and style of robot –in the context of an area that we have recently pioneered, the automation of intracellular neural recording in the living brain.
In vivo neuroscience and past automation efforts
The vertebrate brain is a complex organ consisting of billions of neurons,16 each of which is interconnected with thousands of other neurons through synapses.17 Each neuron receives information via synaptic transmission, computes an electrical signal within it, and transmits information to downstream neurons. They express different sets of genes,18 have myriad morphologies, and undergo plasticity in different ways during performance of cognitive tasks and learning. Thus, one of the fundamental challenges for neuroscientists has been the difficulty of linking the knowledge we have on cellular level phenomena, such as synaptic transmission, often gained by manual in vitro experimental preparations; to emergent properties of the intact living system such as learning and memory. Technologies including electrical neural recording, the generation of transgenic animals, the use of optogenetic neural control,19,20 optics to image intact neural systems, and cell- and circuit-resolution molecular and biochemical analyses, are all important. However, many of these techniques are art forms, requiring extensive effort to learn, typically time consuming to perform, and lacking in scale, without standardization across groups, and with innovations typically driven in different directions by different laboratories. Arguably, many of these areas are ripe for robotic innovation.
In these in vivo and in situ (i.e., intact tissue) spaces of endeavor, robotics and automation have already begun to make inroads. The use of laser capture microdissection to automatically isolate cellular contents from tissues is enabling new kinds of systems biology,21 and also supporting a diversity of histopathological studies. Microfluidic devices for whole-organism imaging and sorting are having great impact on the study of organisms, such as C. elegans,22–25 Drosophila,26 and zebrafish,27 enabling rapid imaging, sorting, and adaptive control of these organisms for both advancement of basic biology as well as accelerated pharmacological screening. Another area where robotics has played a crucial role for in situ analysis is in the field of intact tissue imaging. Automated serial block-face scanning electron microscopy, as well as automated histology systems,28 have driven progress in the nascent field of connectomics.29 High-throughput, automated in situ hybridization along with automated imaging platforms were indispensible for charting the mouse brain gene expression maps of the Allen Brain Atlas.30 Intact tissue analysis has benefited from automated sectioning, in situ hybridization, and imaging of tissue samples.28,31–33 Motorized devices have been devised to support the lowering of tetrodes into the living rat brain,34 and to enable automated stabilization of extracellular recording electrodes for maintaining optimal recording quality.35 In addition, automated electrode recording stabilization techniques have been explored for stabilizing sharp recordings against brain movement in awake, behaving zebra finches.36 We have recently explored the automation of multisite viral injection, using precisely-timed fluidic delivery of viruses to three-dimensional structures in the brain, in an easily user-customizable fashion37 (a process that one can imagine would easily be extended to stem cell or pharmacological injection in many sites at once). Microfabricated strategies for adaptively moving many extracellular microelectrodes could lead to improvements in the development of reliable and stable interfaces with single neurons, important for basic neurophysiological studies and emerging cortical prosthetic technologies.38 Beyond the analysis of live animals and preserved animal and human tissues, robotic actuators are now routinely used in clinical settings to enhance the ability of humans to perform complex surgical procedures.39 For example, the Amadeus and Da Vinci robotic are used to perform minimally invasive laproscopic procedures, as well as robot-assisted telesurgery.40 Robots are increasingly playing a role in even delicate neurosurgeries, as well as in cardiac surgery,41 and being incorporated into operating room systems that enable noninvasive visualization as well (e.g., the neuroArm, a MRI-guided robotic actuator42). These advances illustrate the broad and deep impacts already stemming from automation technologies on the study or manipulation of living or intact biological systems in basic biology and medicine.
Case study: in vivo patch clamp neural recording
To explore in depth a specific avenue of in vivo robotic engineering, we discuss a technology we recently developed that assists in the mechanistic understanding of how cellular level activities of neuronal networks give rise to higher level cognitive abilities, and how they go awry in brain disorders. To study cellular level activity, ideally one would be able to observe electrical activities in neurons with intracellular, synaptic resolution, and ideally in a fashion capable of linking this physiological information to the genetic and morphological information associated with the cellular identity. Such integrated network-wide studies will require new technologies that can access these single cells efficiently and in way that is able to be scaled. One method for doing this, which works even in vivo, is whole-cell patch clamp neural recording. In this technique, a glass micropipette establishes an electrical and molecular connection to the insides of an individual cell embedded in intact tissue. Invented in 1981,43 and winning Neher and Sakmann the Nobel Prize in 1991, whole-cell patch clamping enables recording of the electrical activity of neurons in vivo that exhibit signal quality and temporal fidelity sufficient to report synaptic and ion channel–mediated subthreshold events of importance for understanding not only how neurons compute during behavior, but how their physiology changes in disease states or in response to drug administration. Further, it enables dye infusion for morphological visualization, and extraction of cell contents for transcriptomic analysis.44–46 Potentially, in vivo patch clamping could have clinical impact, being used in neurosurgical settings to do integrative measurements on single cells in, for example, the epileptic brain. This technique could also be used to analyze biopsy samples (e.g., from tumors) or from surgical resections of the brain, at single-cell resolution, potentially revealing tissue heterogeneity and thus new principles of personalized medicine. Such studies that link molecular, cellular, and anatomical properties of individual cells to their behavioral or disease circuit context are difficult in vitro.
Despite these compelling opportunities, in vivo patching requires skill, and the hardware required is specialized and expensive. Thus, in vivo patching has been utilized by a relatively small number of labs, and is usually regarded as a difficult technique, performed manually by highly skilled operators trained by masters, in the anesthetized brain, and in very limited applications in the awake brain.47–52 Accordingly, we considered it as an ideal example of a neuroscience technique to automate.
We began by analyzing what humans do while they perform in vivo patch clamping. This required examination of humans in the laboratory as they performed patch clamping, to analyze the actual methodology they used to perform this task. Importantly, we then examined the physics and mechanics of what was being done, in order to isolate parameters most amenable to automation (e.g., focusing on a time-series electrical impedance analysis for our automation, rather than relying on visual detection of stereotyped patterns of electrical signal, such as heartbeat-rhythm modulation of the recording).
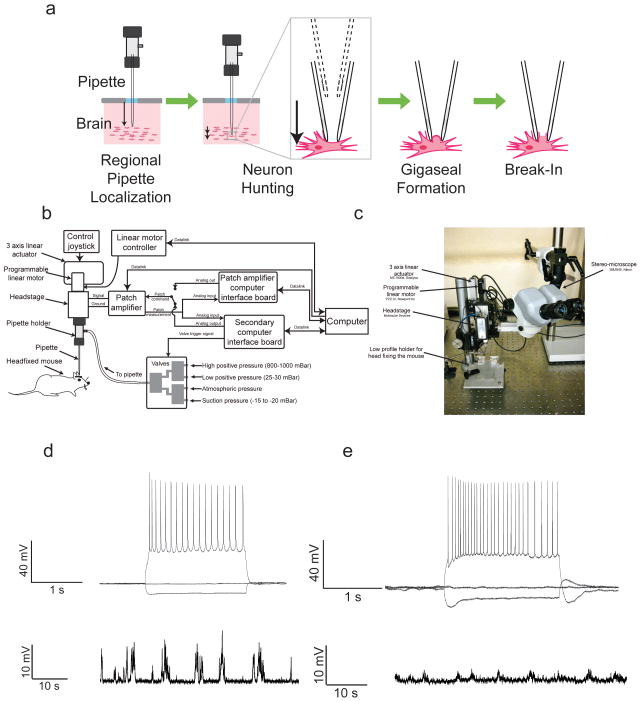
By doing this analysis, we discovered that single cells could be accurately detected by analyzing the temporal sequence of micropipette impedance changes as the micropipette is lowered into the brain (Fig. 2A), looking for particular signatures of temporal change in pipette resistance. Building from this observation, we found that blind in vivo whole-cell patching of neurons, in which micropipettes were lowered until a cell is detected and then recorded, could be reduced to a reliable algorithm, in which cells are detected with > 90% yield, and the whole-cell state established in 40–60% of detected cells,53 with the yields for whole-cell state establishment exceeding 60–70% at the beginning of a session when the brain was intact, and declining to 30–50% over time as multiple brain penetrations occurred.
Figure 2.
The autopatcher: a robot for in vivo patch clamping. (Ai) The four stages of the automated in vivo patch algorithm, discovered through iterative exploration of the parameters governing successful patch clamping: regional pipette localization, in which the pipette is lowered to a target zone in the brain; neuron hunting, in which the pipette is advanced until a neuron is detected via a change in pipette resistance; gigaseal formation, in which a gigaseal state is achieved (if cell-attached patching is desired, the algorithm can end here); break-in, in which the whole cell state is achieved. (Aii) Yields and durations of each of the four stages, when executed by the robot shown in B, running the autopatching algorithm in the living mouse brain, aiming for targets in cortex and hippocampus. (B) Schematic of a simple robotic system capable of performing the autopatching algorithm. The system consists of a conventional in vivo patch setup (i.e., pipette, headstage, three axis linear actuator, patch amplifier plus computer interface board, and computer), equipped with a few additional modules: a programmable linear motor (to move the pipette up and down in a temporally precise fashion), a controllable bank of pneumatic valves for pressure control, and a secondary computer interface board to enable closed-loop control of the motor based upon sequences of pipette resistance measurements. (C) Photograph of the setup, focusing on three axis linear actuator (with additional programmable linear motor) and the holder for head fixing the mouse. (D) Current clamp traces during current injection for a cortical neuron for which whole-cell state was established via autopatcher. (E) Current clamp traces during current injection for a hippocampal neuron for which whole-cell state was established via autopatcher. Adapted, with permission, from Kodandaramaiah et al.53
The next step was to devise the simplest robotic invention that could perform this algorithm. The algorithm could be realized by a simple robot (Figs. 2B and 2C), which actuates a set of motors and valves rapidly upon recognition of specific temporal sequences of micropipette impedance changes, achieving whole-cell patch clamp recordings in 3–7 min each in the anesthetized and awake mouse brain. The robot we designed is relatively inexpensive, is made out of easily accessible, commercially available parts, and thus can easily be appended to an existing patch clamp electrophysiology rig, by adding a few valves, a computer-controlled linear motor, and a digital interface board for analyzing micropipette impedance changes, and actuating the motor and valves.
The robot can obtain very high quality intracellular electrical recordings of neurons, even millimeters deep, in living mouse brain (Fig. 2D), and works in multiple brain regions, suggesting that our algorithm has a degree of generality. The fact that a robot performs it makes exploration of new algorithm variants a simple task. We are starting to explore the capabilities of our robot in the awake rodent brain54 and its compatibility with optogenetics, which could enable on-the-fly cell type identification.54 We are currently aiming to derive new algorithms for procedures which humans do not perform at all, such as the simultaneous patch clamp of many neurons at once,55,56 which requires study of how multiple independent patch pipettes might interact in the dense tissue of the living brain.
Potential pitfalls in in vivo robotics, and how they might be addressed
In vivo neuroscience is a combination of multiple experimental procedures, often required to be performed sequentially. For fully realizing the potential of robotics, it is not necessary just to automate one aspect of the experimental workflow, but develop platforms for automating upstream and downstream processes as well. These systems will need to be built modularly, so as to be easily integrated into the work flow and be reconfigurable to be broadly applicable to many experimental protocols, while taking into consideration inherent variability in the in vivo biological milieu. For example, in the case study of our automated patch clamp robot, true high throughput will only be achieved if we can scale the control algorithms and the hardware to control large arrays of recording electrodes in the fashion of tetrodes and silicon probes, and additionally automated accessory tasks, such as animal surgery, the fabrication of electrodes, the swapping in of fresh electrodes after each experiment, and real-time logging and analyzing of the acquired data. In particular, as automation of other procedures such as animal neurosurgeries emerge, ethical considerations will be important in the engineering as well – for example, if the robot were to fail, it should fail in a safe mode that does not jeopardize the health or well-being of the animal subject. Also, if a robot must halt a procedure that is partially completed, it must be able to either bring the experimental episode to a conclusion in a way that preserves animal welfare, or promptly alert a nearby human attendant to intervene. One possibility is that in vivo automation will enable scientists to perform experiments much more efficiently and effectively than before, enabling higher success rates in animal experimentation, and more data acquired per animal used, important for ethical, scientific, and financial reasons.
Endeavors in in vivo robotics may benefit from multi-lab collaborations that collectively contain deep domain knowledge in multiple aspects of neuroscience as well as multiple aspects of the engineering. These efforts will require significant investments for innovating technologies and scaling up infrastructure as technologies mature. Recent work by the Allen Institute for Brain Science,30 the emerging European Human Brain Project,57 and the excitement generated by recent proposals in the United States, such as the Brain Activity Map (BAM) initiative,58–60 suggest that the time may be ripe for the automation and scaling up of neuroscientific procedures.
This also brings into discussion the role that will have to be played by entities outside of academia. Although many entities have launched to foster the distribution of DNA (e.g., http://addgene.org) and viruses (e.g., various university viral core facilities), there are no comparable methodologies for distributing robots. Entrepreneurial and commercialization endeavors may be the de facto path, but open access and open source models may well be as important (if not more important, in the early days when many groups may seek to customize robots for specific kinds of application). Ensuring broad dissemination means increasing usability (i.e., through the creation of simple yet powerful user interfaces, working to make devices fault tolerant, and connecting devices to existing laboratory hardware), while working to keep costs and prices down to maximize impact. To enable rapid dissemination to the scientific community, it may be important to pursue such simplification and robustness activities in commercial entities, while simultaneously enabling immediate free access to non-profit and academic researchers who seek to try it out right away (e.g., by making all parts lists, computer-aided design (CAD) drawings, and software available on the internet, or even by setting up core facilities within universities to manufacture components for their communities)).
There may well be great demand in the future for innovation in the field of in vivo robotics, particularly in neuroscience. In vivo stem cell biology, in vivo imaging (especially over long periods of time), stereotactic surgery (to insert drugs and devices, or to make measurements), and ex vivo analyses of tissues, are all in need of automation that powerfully enables everyday art forms to become simple and inexpensive, and to later enable these tasks to be performed at such scales, and in such integrated fashions, as to reveal new kinds of integrated patterns and principles of biological operation. As such in vivo automation tools find uses driven by biological discovery, it is likely that they, like in vitro automation tools, will find clinical uses, perhaps in contexts such as diagnostics, neurosurgery, or other fields.
Acknowledgments
E.S.B. and C.R.F. acknowledge NIH Single Cell Grant 1 R01 EY023173. E.S.B. acknowledges Human Frontiers Science Program; IET A. F. Harvey Prize; MIT McGovern Institute and McGovern Institute Neurotechnology (MINT) Program; MIT Media Lab and Media Lab Consortia; New York Stem Cell Foundation-Robertson Investigator Award; NIH Director’s New Innovator Award 1DP2OD002002, NIH EUREKA Award 1R01NS075421, NIH Transformative R01 1R01GM104948, and NIH Grants 1R01DA029639, and 1R01NS067199; NSF CAREER Award CBET 1053233 and DMS1042134 (the Cognitive Rhythms Collaborative); Paul Allen Distinguished Investigator in Neuroscience Award; Shelly Razin; and SkTech. C.R.F. acknowledges funding from NSF (EHR 0965945 and CISE 1110947), NIH Computational Neuroscience Training grant (DA032466-02), Georgia Tech Translational Research Institute for Biomedical Engineering & Science (TRIBES) Seed Grant Awards Program, Wallace H. Coulter Translational/Clinical Research Grant Program, and support from Georgia Institute of Technology through the Institute for Bioengineering and Biosciences Junior Faculty Award, Technology Fee Fund, Invention Studio, and the George W. Woodruff School of Mechanical Engineering.
Footnotes
Conflicts of interest
C.R.F., E.S.B., and S.B.K. are co-inventors on a patent owned by MIT and Georgia Institute of Technology. C.R.F. and S.B.K. are financially affiliated with Neuromatic Devices, which is seeking to manufacture and sell autopatching robots.
References
- 1.Thorsen T, Maerkl SJ, Quake SR. Microfluidic Large-Scale Integration. Science. 2002;298:580–584. doi: 10.1126/science.1076996. [DOI] [PubMed] [Google Scholar]
- 2.White AK, et al. High-throughput microfluidic single-cell RT-qPCR. Proceedings of the National Academy of Sciences. 2011 doi: 10.1073/pnas.1019446108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fan HC, Wang J, Potanina A, Quake SR. Whole-genome molecular haplotyping of single cells. Nat Biotech. 2011;29:51–57. doi: 10.1038/nbt.1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sanchez-Freire V, Ebert AD, Kalisky T, Quake SR, Wu JC. Microfluidic single-cell real-time PCR for comparative analysis of gene expression patterns. Nat Protocols. 2012;7:829–838. doi: 10.1038/nprot.2012.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Warren L, Bryder D, Weissman IL, Quake SR. Transcription factor profiling in individual hematopoietic progenitors by digital RT-PCR. Proceedings of the National Academy of Sciences. 2006;103:17807–17812. doi: 10.1073/pnas.0608512103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dalerba P, et al. Single-cell dissection of transcriptional heterogeneity in human colon tumors. Nat Biotech. 2011;29:1120–1127. doi: 10.1038/nbt.2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Citri A, Pang ZP, Sudhof TC, Wernig M, Malenka RC. Comprehensive qPCR profiling of gene expression in single neuronal cells. Nat Protocols. 2012;7:118–127. doi: 10.1038/nprot.2011.430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Teh SY, Lin R, Hung LH, Lee AP. Droplet microfluidics. Lab Chip. 2008;8:198–220. doi: 10.1039/b715524g. [DOI] [PubMed] [Google Scholar]
- 9.Wang HH, et al. Programming cells by multiplex genome engineering and accelerated evolution. Nature. 2009;460:894–898. doi: 10.1038/nature08187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tian J. Automation in Proteomics and Genomics. John Wiley & Sons, Ltd; 2009. pp. 175–191. [Google Scholar]
- 11.Dunlop J, Bowlby M, Peri R, Vasilyev D, Arias R. High-throughput electrophysiology: an emerging paradigm for ion-channel screening and physiology. Nat Rev Drug Discov. 2008;7:358–368. doi: 10.1038/nrd2552. [DOI] [PubMed] [Google Scholar]
- 12.Estacion M, et al. Can robots patch-clamp as well as humans? Characterization of a novel sodium channel mutation. J Physiol. 2010;588:1915–1927. doi: 10.1113/jphysiol.2009.186114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Milligan CJ, et al. Robotic multiwell planar patch-clamp for native and primary mammalian cells. Nat Protocols. 2009;4:244–255. doi: 10.1038/nprot.2008.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.wang X, Li M. Automated Electrophysiology: High Throughput of Art. ASSAY and Drug Development Technologies. 2004;1:695–708. doi: 10.1089/154065803770381057. [DOI] [PubMed] [Google Scholar]
- 15.Brueggemann A, et al. Current Drug Discovery Technologies. Vol. 1. Bentham Science; 2004. pp. 91–96. [DOI] [PubMed] [Google Scholar]
- 16.Nowakowski RS. Stable neuron numbers from cradle to grave. Proceedings of the National Academy of Sciences. 2006;103:12219–12220. doi: 10.1073/pnas.0605605103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Drachman DA. Do we have brain to spare? Neurology. 2005;64:2004–2005. doi: 10.1212/01.WNL.0000166914.38327.BB. [DOI] [PubMed] [Google Scholar]
- 18.Markram H, et al. Interneurons of the neocortical inhibitory system. Nature reviews. Neuroscience. 2004;5:793–807. doi: 10.1038/nrn1519. [DOI] [PubMed] [Google Scholar]
- 19.Boyden ES. A history of optogenetics: the development of tools for controlling brain circuits with light. F1000 Biology Reports. 2011;3 doi: 10.3410/B3-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bernstein JG, Garrity PA, Boyden ES. Optogenetics and thermogenetics: technologies for controlling the activity of targeted cells within intact neural circuits. Curr Opin Neurobiol. 2011 doi: 10.1016/j.conb.2011.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Boyden ES, et al. Selective engagement of plasticity mechanisms for motor memory storage. Neuron. 2006;51:823–834. doi: 10.1016/j.neuron.2006.08.026. [DOI] [PubMed] [Google Scholar]
- 22.Kocabas A, Shen CH, Guo ZV, Ramanathan S. Controlling interneuron activity in Caenorhabditis elegans to evoke chemotactic behaviour. Nature. 2012;490:273–277. doi: 10.1038/nature11431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leifer AM, Fang-Yen C, Gershow M, Alkema MJ, Samuel AD. Optogenetic manipulation of neural activity in freely moving Caenorhabditis elegans. Nat Methods. 2011;8:147–152. doi: 10.1038/nmeth.1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Samara C, et al. Large-scale in vivo femtosecond laser neurosurgery screen reveals small-molecule enhancer of regeneration. Proc Natl Acad Sci U S A. 2010;107:18342–18347. doi: 10.1073/pnas.1005372107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Crane MM, et al. Autonomous screening of C. elegans identifies genes implicated in synaptogenesis. Nat Methods. 2012;9:977–980. doi: 10.1038/nmeth.2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Furlong EE, Profitt D, Scott MP. Automated sorting of live transgenic embryos. Nat Biotechnol. 2001;19:153–156. doi: 10.1038/84422. [DOI] [PubMed] [Google Scholar]
- 27.Pardo-Martin C, et al. High-throughput in vivo vertebrate screening. Nat Methods. 2010;7:634–636. doi: 10.1038/nmeth.1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tsai PS, et al. All-Optical Histology Using Ultrashort Laser Pulses. Neuron. 2003;39:27–41. doi: 10.1016/s0896-6273(03)00370-2. [DOI] [PubMed] [Google Scholar]
- 29.Briggman KL, Helmstaedter M, Denk W. Wiring specificity in the direction-selectivity circuit of the retina. Nature. 2011;471:183–188. doi: 10.1038/nature09818. [DOI] [PubMed] [Google Scholar]
- 30.Lein ES, et al. Genome-wide atlas of gene expression in the adult mouse brain. Nature. 2007;445:168–176. doi: 10.1038/nature05453. [DOI] [PubMed] [Google Scholar]
- 31.Denk W, Horstmann H. Serial block-face scanning electron microscopy to reconstruct three-dimensional tissue nanostructure. PLoS Biol. 2004;2:e329. doi: 10.1371/journal.pbio.0020329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lein ES, et al. Genome-wide atlas of gene expression in the adult mouse brain. Nature. 2007;445:168–176. doi: 10.1038/nature05453. [DOI] [PubMed] [Google Scholar]
- 33.Ragan T, et al. Serial two-photon tomography for automated ex vivo mouse brain imaging. Nat Methods. 2012;9:255–258. doi: 10.1038/nmeth.1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yamamoto J, Wilson MA. Large-scale chronically implantable precision motorized microdrive array for freely behaving animals. J Neurophysiol. 2008;100:2430–2440. doi: 10.1152/jn.90687.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cham JG, et al. Semi-Chronic Motorized Microdrive and Control Algorithm for Autonomously Isolating and Maintaining Optimal Extracellular Action Potentials. J Neurophysiol. 2005;93:570–579. doi: 10.1152/jn.00369.2004. [DOI] [PubMed] [Google Scholar]
- 36.Fee MS. Active stabilization of electrodes for intracellular recording in awake behaving animals. Neuron. 2000;27:461–468. doi: 10.1016/s0896-6273(00)00057-x. [DOI] [PubMed] [Google Scholar]
- 37.Chan S, Bernstein J, Boyden E. Scalable fluidic injector arrays for viral targeting of intact 3-D brain circuits. J Vis Exp. 2010 doi: 10.3791/1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Muthuswamy J, Anand S, Sridharan A. Adaptive movable neural interfaces for monitoring single neurons in the brain. Front Neurosci. 2011;5:94. doi: 10.3389/fnins.2011.00094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Beasley RA. Medical Robots: Current Systems and Research Directions. Journal of Robotics. 2012;2012:14. [Google Scholar]
- 40.Marescaux J, et al. Transatlantic robot-assisted telesurgery. Nature. 2001;413:379–380. doi: 10.1038/35096636. [DOI] [PubMed] [Google Scholar]
- 41.Yuen SG, Perrin DP, Vasilyev NV, del Nido PJ, Howe RD. Force Tracking With Feed-Forward Motion Estimation for Beating Heart Surgery. Robotics, IEEE Transactions on. 2010;26:888–896. doi: 10.1109/TRO.2010.2053734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sutherland GR, Wolfsberger S, Lama S, Zarei-nia K. The Evolution of neuroArm. Neurosurgery. 2013;72:A27–A32. doi: 10.1227/NEU.1220b1013e318270da318219. [DOI] [PubMed] [Google Scholar]
- 43.Hamill OP, Marty A, Neher E, Sakmann B, Sigworth FJ. Improved patch-clamp techniques for high-resolution current recording from cells and cell-free membrane patches. Pflugers Arch. 1981;391:85–100. doi: 10.1007/BF00656997. [DOI] [PubMed] [Google Scholar]
- 44.Eberwine J, et al. Analysis of gene expression in single live neurons. Proceedings of the National Academy of Sciences of the United States of America. 1992;89:3010–3014. doi: 10.1073/pnas.89.7.3010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Van Gelder RN, et al. Amplified RNA synthesized from limited quantities of heterogeneous cDNA. Proceedings of the National Academy of Sciences of the United States of America. 1990;87:1663–1667. doi: 10.1073/pnas.87.5.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sucher NJ, Deitcher DL. PCR and patch-clamp analysis of single neurons. Neuron. 1995;14:1095–1100. doi: 10.1016/0896-6273(95)90257-0. [DOI] [PubMed] [Google Scholar]
- 47.Crochet S, Poulet JF, Kremer Y, Petersen CC. Synaptic mechanisms underlying sparse coding of active touch. Neuron. 2011;69:1160–1175. doi: 10.1016/j.neuron.2011.02.022. [DOI] [PubMed] [Google Scholar]
- 48.Gentet LJ, Avermann M, Matyas F, Staiger JF, Petersen CC. Membrane potential dynamics of GABAergic neurons in the barrel cortex of behaving mice. Neuron. 2010;65:422–435. doi: 10.1016/j.neuron.2010.01.006. [DOI] [PubMed] [Google Scholar]
- 49.Lee AK, Epsztein J, Brecht M. Head-anchored whole-cell recordings in freely moving rats. Nat Protocols. 2009;4:385–392. doi: 10.1038/nprot.2009.5. [DOI] [PubMed] [Google Scholar]
- 50.Lee AK, Manns ID, Sakmann B, Brecht M. Whole-cell recordings in freely moving rats. Neuron. 2006;51:399–407. doi: 10.1016/j.neuron.2006.07.004. [DOI] [PubMed] [Google Scholar]
- 51.Lee D, Lin BJ, Lee AK. Hippocampal Place Fields Emerge upon Single-Cell Manipulation of Excitability During Behavior. Science. 2012;337:849–853. doi: 10.1126/science.1221489. [DOI] [PubMed] [Google Scholar]
- 52.Harvey CD, Collman F, Dombeck DA, Tank DW. Intracellular dynamics of hippocampal place cells during virtual navigation. Nature. 2009;461:941–946. doi: 10.1038/nature08499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kodandaramaiah SB, Franzesi GT, Chow BY, Boyden ES, Forest CR. Automated whole-cell patch-clamp electrophysiology of neurons in vivo. Nat Meth. 2012;9:585–587. doi: 10.1038/nmeth.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kodandaramaiah SB, et al. Society for NeuroscienceOnline. 2012 [Google Scholar]
- 55.Kodandaramaiah SB, et al. Society for NeuroscienceOnline. 2012. [Google Scholar]
- 56.Kodandaramaiah SB. Mechanical Engineering, Vol PhD 56. Georgia Institute of Technology; Atlanta: 2012. [Google Scholar]
- 57.Markram H. The Blue Brain Project. Nat Rev Neurosci. 2006;7:153–160. doi: 10.1038/nrn1848. [DOI] [PubMed] [Google Scholar]
- 58.Alivisatos AP, et al. Nanotools for Neuroscience and Brain Activity Mapping. ACS Nano. 2013;7:1850–1866. doi: 10.1021/nn4012847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Alivisatos AP, et al. The Brain Activity Map. Science. 2013;339:1284–1285. doi: 10.1126/science.1236939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Alivisatos AP, et al. The Brain Activity Map Project and the Challenge of Functional Connectomics. Neuron. 2012;74:970–974. doi: 10.1016/j.neuron.2012.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]