SUMMARY
The tumor-suppressive Hippo pathway controls tissue homeostasis through balancing cell proliferation and apoptosis. Activation of the kinases Mst1/2 is a key upstream event in this pathway and remains poorly understood. Mst1/2 and their critical regulators RASSFs contain SARAH domains that can homo- and hetero-dimerize. Here, we report the crystal structures of human Mst2 alone and bound to RASSF5. Mst2 undergoes activation through trans-autophosphorylation at its activation loop, which requires SARAH-mediated homodimerization. RASSF5 disrupts Mst2 homodimer and blocks Mst2 autoactivation. Binding of RASSF5 to already activated Mst2, however, does not inhibit its kinase activity. Thus, RASSF5 can act as an inhibitor or a potential positive regulator of Mst2, depending on whether it binds to Mst2 before or after activation-loop phosphorylation. We propose that these temporally sensitive functions of RASSFs enable the Hippo pathway to respond to and integrate diverse cellular signals.
Keywords: Hippo, Mst autoactivation, RASSF, SARAH domain, X-ray, dimerization
INTRODUCTION
The Hippo pathway controls organ size and tissue homeostasis through coordinately regulating cell growth, proliferation, and death. This tumor suppressive pathway was first characterized in Drosophila through genetic screens for mutations that caused tissue overgrowth, and was later shown to be conserved in mammals (Badouel et al., 2009; Edgar, 2006; Halder and Johnson, 2011; Harvey and Tapon, 2007; Harvey et al., 2013; Pan, 2010; Staley and Irvine, 2012; Zhao et al., 2010a). The core components of the mammalian Hippo pathway include the Ste20 family kinases Mst1/2, the scaffolding protein Salvador (Sav1), the NDR family kinases Lats1/2, and the adaptor protein Mob1. They form a central kinase cascade to transduce signals from cell-surface receptors (Avruch et al., 2012; Hergovich, 2012).
In the canonical Hippo kinase cascade, Mst1/2 in complex with Sav1 phosphorylate and activate the Lats1/2-Mob1 complexes, which then phosphorylate the transcriptional co-activator YAP (Yes-associated protein), a major downstream target of the Hippo pathway (Dong et al., 2007; Hao et al., 2008; Hong and Guan, 2012; Huang et al., 2005; Zhao et al., 2007). Lats1/2-mediated phosphorylation inhibits YAP in two ways. Phosphorylation of YAP at S127 by Lats1/2 creates a docking site for 14-3-3 proteins. Binding of 14-3-3 causes the cytoplasmic sequestration and inactivation of YAP (Dong et al., 2007; Hao et al., 2008; Zhao et al., 2007). Phosphorylation of YAP at S381 by Lats1/2 promotes its ubiquitination and degradation (Zhao et al., 2010b).
When the Hippo pathway is turned off, YAP is dephosphorylated and translocates into the nucleus. Although YAP does not contain a DNA-binding domain, it binds to the TEAD family of transcription factors (which contains a sequence-specific DNA-binding domain) to form a functional hybrid transcription factor (Luo, 2010; Sudol et al., 2012; Zhao et al., 2008). The YAP–TEAD hybrid then activates the transcription of Hippo-responsive genes that promote cell growth and proliferation and inhibit apoptosis.
Tremendous progress has been made towards the dissection of the molecular circuitry of the Hippo pathway and towards the understanding of the pathophysiology of this pathway in multiple organisms. By contrast, mechanistic and structural studies in this area have lagged behind. In particular, the activation mechanisms of the core Mst1/2-Lats1/2 kinase cascade remain elusive. The upstream kinases Mst1/2 contain an N-terminal kinase domain and a C-terminal SARAH (Salvador/RASSF1A/Hippo) domain (Figure 1A). Mst1 and Mst2 can each form a constitutive homodimer through the SARAH domain, and kinase activation requires autophosphorylation of the activation loop (T183 for Mst1 and T180 for Mst2) (Avruch et al., 2012; Creasy et al., 1996). The Mst1/2 regulators, Sav1 and RASSF proteins, also contain SARAH domains (Figure 1A). The Mst1/2 SARAH domain can form a heterodimer with RASSF SARAH (Hwang et al., 2007) and a heterotetramer with Sav1 SARAH (data not shown). RASSF binding and Sav1 binding to Mst1/2 are mutually exclusive. How RASSFs and Sav1 regulate Mst1/2 activation by forming different SARAH domain-dependent complexes is not understood.
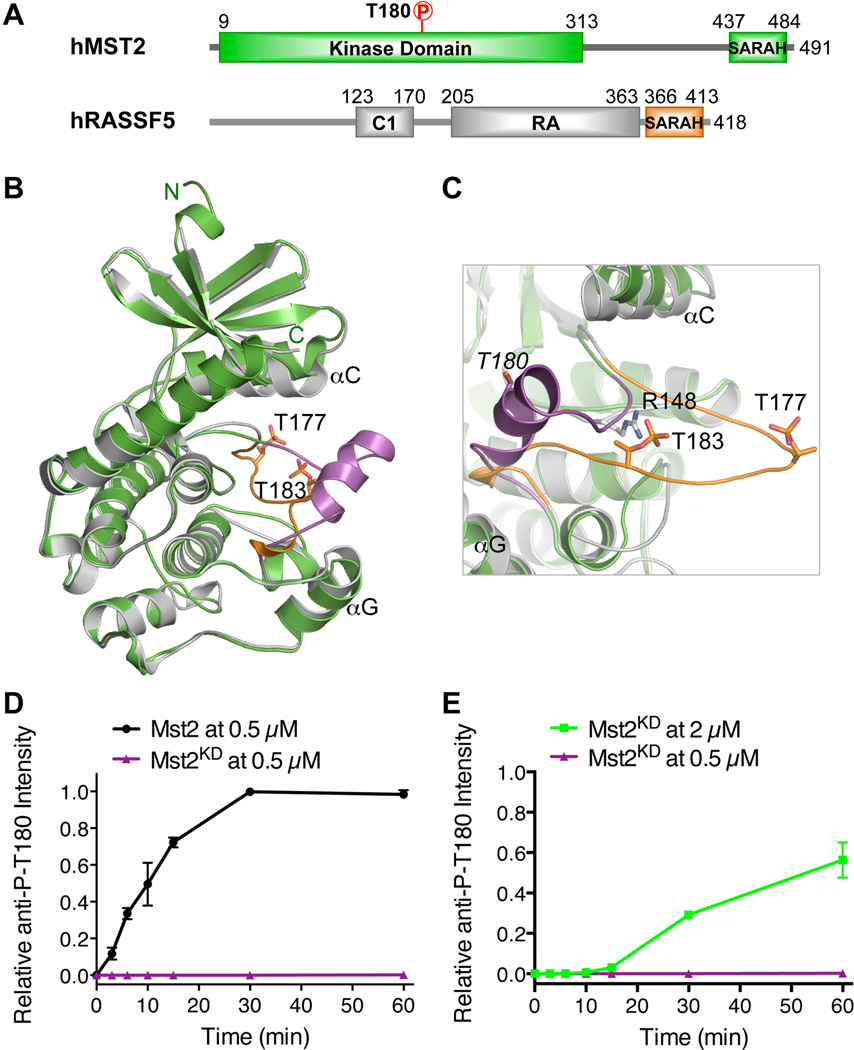
Figure 1. Structural Basis for Mst2 Autoactivation.
(A) Domain organization of human Mst2 and RASSF5. RA, Ras-association domain. Residue numbers for the boundaries of corresponding domains are labeled.
(B) Structure superposition of the kinase-active Mst1 kinase domain (PDB code: 3COM) and the kinase-inactive Mst2 kinase domain (this study). Mst1 is colored in gray and Mst2 is colored in green. The activation loop from Mst1 is colored in orange with residues phospho-T177 and phospho-T183 shown in sticks. The activation loop from Mst2 is colored in magenta.
(C) Close-up view of the activation loops from Mst1 and Mst2. The color scheme is the same as in B. R148 and phospho-T183 of Mst1 are shown as sticks. T180 in Mst2 is shown as sticks and labeled in italic. All structural figures were generated with PyMOL (http://www.pymol.org).
(D) SARAH-mediated homodimerization is critical for Mst2 auto-activation at T180. The kinetic profiles of T180 autophosphorylation of full-length Mst2 (0.5 µM) and Mst2 kinase domain (KD) (0.5 µM) were shown. The relative α-Mst2 P-T180 intensities were normalized against Mst2 reaction at 60 minute (100%). See also Figure S1.
(E) Mst2 kinase domain alone is inefficient in T180 autophosphorylation. The kinetic profiles of T180 autophosphorylation of Mst2 kinase domain (KD) at two different concentrations (2 µM and 0.5 µM) were shown. The relative α-Mst2 P-T180 intensities were normalized against Mst2KD without λ phosphatase treatment (100%). Data are representative of at least two independent experiments.
RASSFs are important tumor suppressors (Avruch et al., 2009; Richter et al., 2009). Their expression is frequently silenced in human cancers through promoter methylation, and reintroduced expression of RASSF1A or RASSF5 inhibits human tumor cell growth (Aoyama et al., 2004). In addition, RASSF1A knockout mice have increased spontaneous and chemical-induced tumor susceptibility (Tommasi et al., 2005). The roles of RASSFs in the tumor-suppressive Hippo pathway are far from clear, however. In Drosophila, the only RASSF protein (dRASSF) forms a heterodimer with Hippo through SARAH heterodimerization and prevents Salvador from binding to Hippo, thereby inhibiting the Hippo pathway (Polesello et al., 2006).Paradoxically, dRASSF also antagonizes Ras signaling and, in this capacity, has tumor-suppressive function. In mammals, RASSF1, RASSF5, and RASSF6 block Mst1/2 autophosphorylation and activation, possibly through SARAH heterodimerization (Ikeda et al.,2009; Khokhlatchev et al., 2002; Praskova et al., 2004). On the other hand, RASSFs can positively regulate Mst1/2 (Guo et al., 2007; Oh et al., 2006). For example, co-expression of RASSFs with Ras enhances the Mst1 kinase activity. RASSF1A disrupts the interaction between proto-oncogene Raf1 and Mst2, and enhances the Mst2-Lats1 interaction to promote apoptosis through the Hippo pathway (Matallanas et al., 2007). RASSF2 associates with Mst1/2 through its SARAH domain to stabilize and activate Mst1/2 (Cooper et al., 2009; Song et al., 2010).Thus, in both Drosophila and mammals, RASSFs appear to have both negative and positive regulatory functions in the Hippo pathway.
Here, we report the crystal structures of the human Mst2 kinase domain and Mst2 in complex with the SARAH domain of RASSF5. SARAH-mediated homodimerization of Mst2 is critical for its trans-autophosphorylation and activation. RASSF5 disrupts this dimer interface and blocks Mst2 autoactivation. Interestingly, binding of RASSF5 to Mst2 that has already undergone autoactivation does not inhibit the kinase activity of Mst2 towards the downstream substrate Mob1. This lack of inhibition of active Mst2 might permit RASSF5 to have a positive regulatory role in the Hippo signaling. Thus, the order of RASSF5 binding and activation-loop phosphorylation determines whether RASSF5 acts as an inhibitor of Mst2. We speculate that the temporal regulation of the binding between Mst1/2 and RASSFs might enable RASSFs to perform dual functions in the Hippo pathway.
RESULTS and DISCUSSION
Structure of the Mst2 Kinase Domain in Its Inactive State
The crystal structure of the activated Mst1 kinase domain (Mst1KD) with two residues (T177 and T183) of its activation loop phosphorylated has recently been determined in a structural genomics effort. The structure of the Mst1/2 kinase domain in its inactive, unphosphorylated state is unknown, however. We thus created a catalytically inactive mutant of the human Mst2 kinase domain (Mst2KD/D146N) with its catalytic residue D146 mutated to an asparagine and determined its crystal structure (Figure 1B). The structure was solved by molecular replacement with the Mst1KD structure (PDB code 3COM) as the search model (Table 1). The kinase domain of Mst2 in our structure adopts a canonical kinase fold, without Mg2+-ATP bound to the active site located at the interface of the N- and C-lobes. The salt bridge between K56 of strand β3 and E70 of helix αC (which is critical for ATP binding) is not present in our unphosphorylated Mst2 structure. The overall structures of the Mst1 and Mst2 kinase domains are very similar, with an RMSD of 1.0 Å excluding the activation loop, as expected from their 90% amino acid identity (Figure 1B). Comparison of the inactive and active states of Mst1/2, however, reveals phosphorylation-dependent conformational changes in its activation loop. The phosphorylated activation loop in the active Mst1 adopts an extended conformation suitable for substrate binding (Figure 1C). As in other kinases that require activation-loop phosphorylation for activation, the phospho-residue in the activation loop (T183 in Mst1) makes favorable electrostatic interactions with the arginine (R148 in Mst1) in the catalytic loop. By contrast, the unphosphorylated activation loop in the inactive Mst2 structure folds into an α helix that blocks the active site of the kinase. Phosphorylation-mediated conformational rearrangement of its activation loop thus underlies Mst1/2 activation.
Table 1.
Data collection, phasing and refinement statistics for Mst2KD and Mst2-RASSF5
| Data collection | |||
|---|---|---|---|
| Crystal | Mst2KD | Mst2-RASSF5 | Mst2-RASSF5 SeMet |
| Space group | C2 | C2 | C2 |
| Energy (eV) | 12,683.7 | 12,683.7 | 12,683.7 |
| Resolution range (Å) | 46.0 – 2.42 (2.46 – 2.42) | 47.1 – 3.05 (3.10 –3.05) | 45.6 – 3.55 (3.61 – 3.55) |
| Unique reflections | 83,949 (3,787) | 65,746 (2,170) | 31,896 (1,617) |
| Multiplicity | 4.5 (3.5) | 4.1 (3.8) | 4.5 (4.4) |
| Data completeness (%) | 99.5 (91.2) | 99.1 (90.7) | 99.9 (100.0) |
| Rmerge (%)a | 5.2 (53.7) | 10.9 (76.2) | 14.9 (76.5) |
| I/σ(I) | 27.4 (1.7) | 14.4 (1.6) | 11.7 (1.7) |
| Wilson B-value (Å2) | 56.4 | 80.2 | 82.7 |
| Refinement statistics | |||
| Crystal | Mst2KD | Mst2-RASSF5 | |
| Resolution range (Å) | 29.8 – 2.42 (2.48 – 2.42) | 29.6 – 3.05 (3.11 – 3.05) | |
| No. of reflections Rwork/Rfree | 83,826/1,997 (5,491/134) | 50,374/2,524 (2,308/124) | |
| Data completeness (%) | 99.5 (94.0) | 98.6 (88.9) | |
| Atoms (non-H protein/nucleotide/solvent/) | 13,179/NA/279 | 12,609/124/NA | |
| Rwork (%) | 19.3 (26.4) | 19.9 (29.9) | |
| Rfree (%) | 23.1 (32.4) | 24.4 (36.9) | |
| R.m.s.d. bond length (Å) | 0.002 | 0.006 | |
| R.m.s.d. bond angle (°) | 0.64 | 0.61 | |
| Mean B-value (Å2) (protein/nucleotide/solvent) | 79.7/NA/60.0 | 135.8/142.9/NA | |
| Ramachandran plot (%) (favored/additional/disallo wed)b | 96.4/3.4/0.2 | 95.4/4.3/0.3 | |
| Maximum likelihood coordinate error | 0.30 | 0.29 | |
| Missing residues, by chain | A: 16–17, 36–37, 309–313; B: 16–18, 36–38, 165–170, 306–313; C: 16–17, 308–313; D: 16–25, 48–51, 58–68, 92–95, 165–182, 309–313; E: 16–17, 35–38, 59–63, 174–179, 303–313; F: 16–17, 166–182, 309–313. | A: 9–15, 428–436, 489–491; B: 9, 428–442, 489–491; C: 9–11, 33–39, 428–436, 491; D: 9–10, 428–454, 490–491; E: 366; F: 366–369, 399–413; H: 406–413. | |
Data for the outermost shell are given in parentheses.
Rmerge = 100 ΣhΣi|Ih, i— 〈Ih〉|/ΣhΣiIh, i, where the outer sum (h) is over the unique reflections and the inner sum (i) is over the set of independent observations of each unique reflection.
As defined by the validation suite MolProbity
Mst2 Undergoes Activation through Trans-autophosphorylation of T180
The full-length Mst1/2 or their kinase domains purified from bacteria are phosphorylated at their respective activation loops, indicating that Mst1/2 can undergo autophosphorylation-dependent activation and does not require upstream kinases for activation. To monitor the kinetics of Mst2 activation in vitro, we treated purified, active Mst2 with λ phosphatase to remove its activation-loop phosphorylation, incubated the dephosphorylated Mst2 with cold ATP for 1 hour, and blotted Mst2 with an antibody against phospho-T180 Mst2 (α-Mst2 pT180). The full-length Mst2 indeed underwent efficient autophosphorylation at T180 in vitro (Figure 1D). By contrast, the Mst2 kinase domain alone was inefficient in T180 autophosphorylation, even at a higher concentration (Figure 1E). Consistent with previous findings, the full-length Mst2 eluted as a dimer during gel filtration whereas the kinase domain of Mst2 migrated predominantly as a monomer (Figure S1). Our results suggest that the C-terminal SARAH domain of Mst2 is critical for the homodimerization of Mst2 and for T180 autophosphorylation and Mst2 activation.
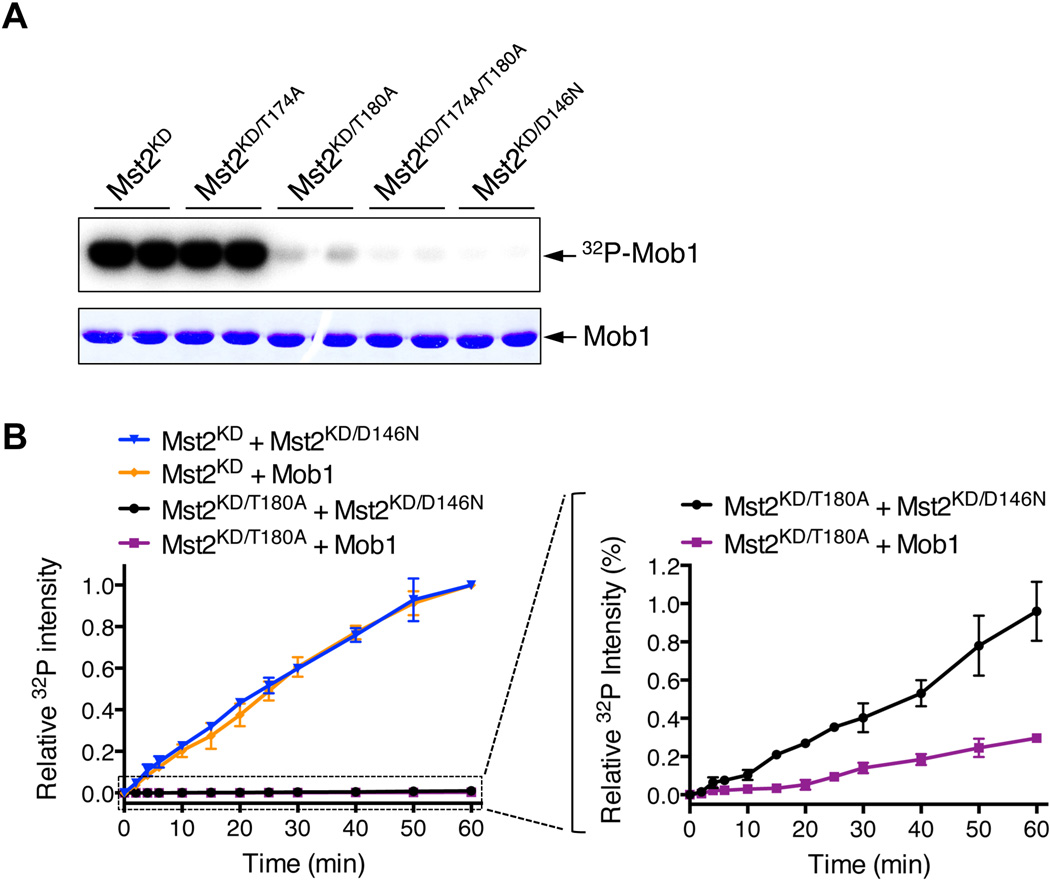
In the conventional model of the initial activation of kinases that undergo activation-loop trans-autophosphorylation, the activation loop of an unphosphorylated kinase molecule transiently adopts an active conformation and phosphorylates the activation loop of a neighboring kinase molecule to fully activate it (Pirruccello et al., 2006). We tested whether Mst2 used a similar trans-autophosphorylation mechanism. Mst2 undergoes autophosphorylation on two residues: T174 and T180 in its activation loop. The T174A mutant of the Mst2 kinase domain (Mst2KD/T174A) is fully active, whereas the T180A mutant is largely inactive towards a key downstream substrate Mob1 (Figure 2A). Thus, T180 phosphorylation,but not T174 phosphorylation, is critical for Mst2 activation. Mst2KD phosphorylated the catalytically inactive Mst2KD/D146N, indicating that Mst2 could undergo trans-autophosphorylation (Figures 2B and S2). Interestingly, despite not being able to permanently reach the phosphorylated active state, the phosphorylation-deficient Mst2KD/T180A mutant phosphorylated Mst2KD/D146N to some extent and, in fact, more efficiently than it phosphorylated Mob1 (Figure 2B). Thus, the unphosphorylated activation loop of Mst2 might transiently adopt a conformation that is more amenable for trans-autophosphorylation than for phosphorylation of a bona fide substrate. Our results support the conventional model for Mst2 trans-autoactivation.
Figure 2. The Conventional Model for Mst2 Trans-autophosphorylation.
(A) T180 phosphorylation is critical for the Mst2 kinase activity. In vitro kinase assay of Mst2 kinase domain (KD) and its mutants using Mob1 as substrate and in the presence of γ-32P-ATP. The reaction mixtures were separated on SDS-PAGE and analyzed by a phosphorimager (top panel) and Coomassie blue staining (bottom panel). Each reaction was done in duplicate.
(B) The phosphorylation-defective mutant T180A of Mst2 kinase domain (KD) phosphorylates kinase-inactive Mst2KD/D146N more efficiently than it phosphorylates Mob1. The kinetic profiles of Mst2KD/D146N and Mob1 phosphorylation by either Mst2KD or Mst2KD/T180A were shown on the left. An enlarged view of the boxed region on the left graph was shown on the right. The 32P intensities were normalized against Mst2KD wild-type reactions at 60 minute (100%) with substrates Mst2KD/D146N and Mob1, respectively. Data are representative of at least two independent experiments. See also Figure S2.
Distinct Residues Mediate Mst2 Autophosphorylation and Phosphorylation of Its Downstream Substrates
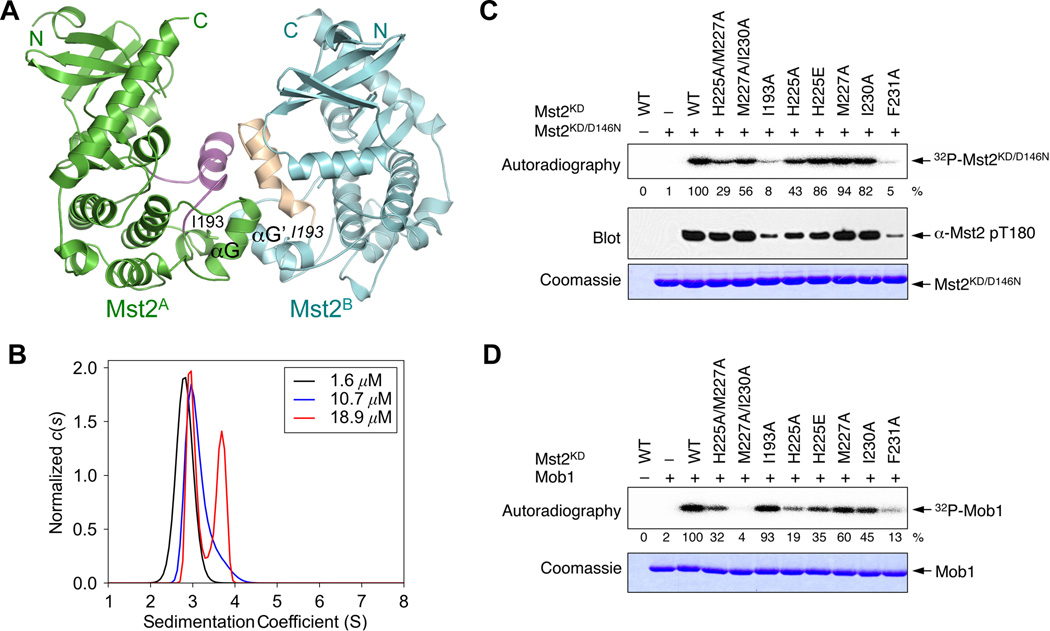
Despite being predominantly a monomer on gel filtration columns (Figure S1B), the Mst2 kinase domain exists as a homodimer in the crystal (Figure 3A). The Mst2 kinase domain in isolation indeed formed a weak dimer in solution with a Kd of 36 µM as measured by analytical ultracentrifugation (Figure 3B, Figure S3 and Table S1). To test whether the weak dimerization of the Mst2 kinase domain involved the interface observed in the crystal, we introduced mutations at this interface and measured the dimerization affinity. Unexpectedly, none of the mutations appreciably altered the dimerization of the Mst2 kinase domain in its active or inactive states (Table S1). Therefore, this interface might not mediate the dimerization seen in solution.
Figure 3. Identification of Key Residues Required for Mst2 Autophosphorylation.
(A) Ribbon drawing of the Mst2 crystallographic dimer. Monomer A is colored in green with the activation loop in magenta. Monomer B is colored in cyan with the activation loop in wheat. I193 is shown in sticks.
(B) Sedimentation velocity analytical ultracentrifugation analysis of Mst2KD/D146N. Three c(s) size distributions are shown, normalized by the total amount of signal in each. The distributions represent three separate sedimentation velocity experiments performed at differing concentrations. Populations of monomer (2.9 S) and dimer (4.1 S) change as a function of concentration, a hallmark of a monomer-dimer equilibrium. This part was made using GUSSI (http://biophysics.swmed.edu/MBR/software.html). See also Figure S3 and Table S1.
(C) In vitro kinase assay of the interface mutants of Mst2 kinase domain based on the structure shown in (A). Mst2KD/D146N was used as the substrate. The autoradiography of 32P incorporation was shown in the top panel. The 32P intensities of each mutant normalized against wild-type Mst2KD (100%) were quantified and indicated below. The Western blot of phospho-T180 Mst2 was shown in the middle panel. The Coomassie-blue staining of Mst2KD/D146N was shown in the bottom panel.
(D) In vitro kinase assay of the interface mutants of Mst2 kinase domain using Mob1 as the substrate. The autoradiography of 32P incorporation was shown in the upper panel. The 32P intensities of each mutant normalized against wild-type Mst2KD (100%) were quantified and indicated below. The Coomassie-blue staining of Mob1 was shown in the lower panel.
Because our mutations targeted residues close to the active site, we tested the ability of these mutants to phosphorylate the kinase-inactive Mst2KD/D146N or its downstream substrate Mob1. Based on both 32P incorporation and anti-Mst2 pT180 blot, two mutations, I193A and F231A, severely inhibited Mst2 autophosphorylation at T180 (Figure 3C). Whereas the F231A mutant was also defective in Mob1 phosphorylation, the I193A mutant was fully functional in Mob1 phosphorylation (Figure 3D). On the other hand, the M227A/I230A mutant was capable of undergoing autophosphorylation at T180, but was inactive towards Mob1. Therefore, Mst2 appears to use overlapping but distinct surfaces to perform trans-autophosphorylation or phosphorylation of other substrates. I193 is specifically involved in autophosphorylation. We note that the I193A mutant purified from bacteria still underwent autophosphorylation at T180, indicating that this residue is not absolutely required for Mst2 autoactivation.
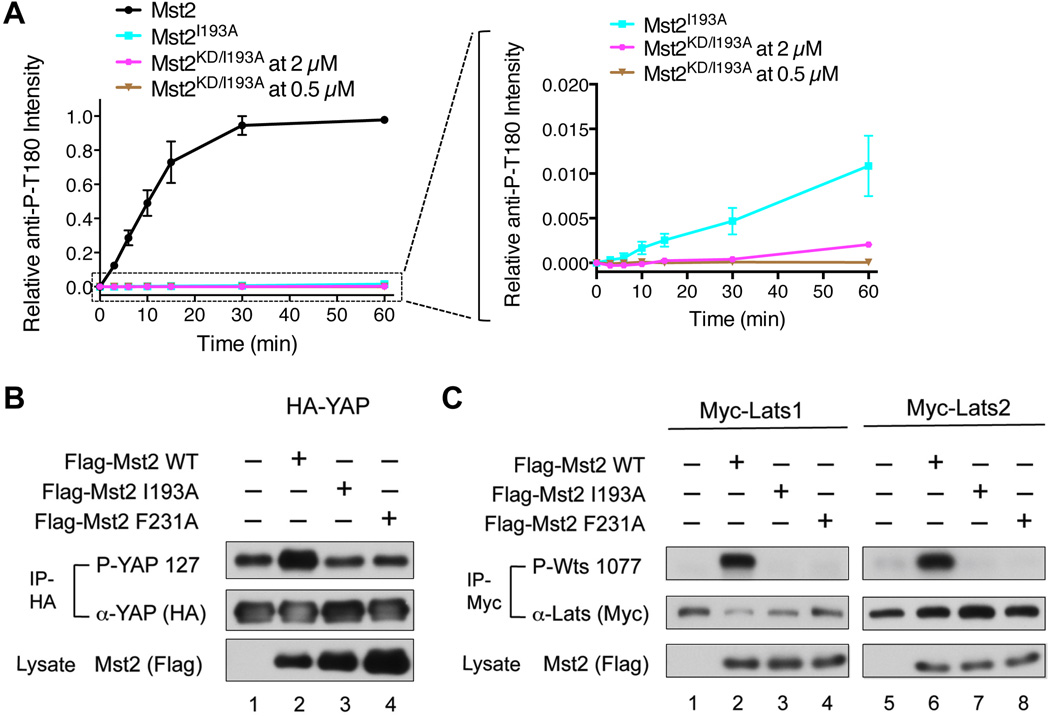
Importantly, the I193A mutation inhibited Mst2 autophosphorylation at T180, even in the context of the full-length Mst2 (Figure 4A). Furthermore, ectopic expression of Flag-Mst2 WT, but not the I193A and F231A mutants, enhanced the phosphorylation of HA-YAP and Myc-Lats1/2 in human cells (Figures 4B and 4C), indicating that I193 was required for Mst2 activity and function in vivo.
Figure 4. Mst2 Autoactivation Is Required for Lats1/2 and YAP Phosphorylation in Human Cells.
(A) I193A inhibits Mst2 autophosphorylation at T180 even in the context of the full-length Mst2. The kinetics profiles of T180 autophosphorylation of full-length Mst2 (0.5 µM), full-length point mutant Mst2I193A (0.5 µM) and Mst2KD/I193A (2 µM and 0.5 µM) were shown on the left. An enlarged view of the boxed region on the left graph was shown on the right. The full-length Mst2I193A was slightly more efficient than Mst2KD/I193A in autophosphorylation. Data are representative of at least two independent experiments. See also Figure S4.
(B) Overexpression of Flag-Mst2 WT, but not the I193A and F231A mutants, enhanced the phosphorylation of HA-YAP. HA-IP from HEK293 cells expressing the indicated constructs was probed with α-phospho-YAP and α-YAP (HA). A fraction of the cell lysate was probed with α-Mst2 (Flag) to evaluate protein expression levels.
(C) Overexpression of Flag-Mst2 WT, but not the I193A and F231A mutants, enhanced the phosphorylation of Myc-Lats1 (left panel) and Myc-Lats2 (right panel). Myc-IP from HEK293 cells expressing the indicated constructs was probed with α-phospho-Wts and α-Myc. A fraction of the cell lysate was probed with α-Mst2 (Flag) to evaluate protein expression levels.
The underlying reason for why Mst2 I193A undergoes autophosphorylation in bacteria, but not in human cells, is unclear at present, but this difference could be due to the presence of phosphatases in human cells. A similar mutation in the Hippo kinase domain has been shown to impair Hippo autophosphorylation and activation in Drosophila, suggesting that the activation mechanism of Mst1/2 is conserved (Jin et al., 2012). Finally, our results indicate that SARAH-mediated dimerization of Mst2 is critical for Mst2 autophosphorylation and activation. Even though the Mst2 kinase domain in isolation forms a weak dimer, there is no evidence supporting the importance of this dimerization in Mst2 activation.
Structure of the Mst2-RASSF5 Complex
We failed to obtain crystals of full-length Mst1/2. We thus analyzed the overall structural architecture of Mst2 by nuclear magnetic resonance (NMR) spectroscopy. Due to the large molecular weight of Mst2, the transverse relaxation optimized spectroscopy (TROSY) version of heteronuclear single quantum correlation (HSQC) experiments was performed (Pervushin, 2000). The 1H/15N TROSY-HSQC spectrum of Mst2 contained many sharp peaks between 7.5–8.5 ppm in the 1H dimension, indicating that many residues of Mst2 were disordered (Figure S4A). By contrast, the 1H/15N TROSY-HSQC spectrum of the Mst2 mutant with its linker between the kinase and SARAH domains deleted (Mst2ΔL) had few sharp peaks, indicating that the Mst2 linker was natively unfolded (Figure S4B). Therefore, Mst2 existed as a homodimer with two juxtaposed kinase-domain heads and a SARAH coiled-coil domain connected by flexible linkers (Figure S4C). Mst2ΔL was fully functional and capable of autophosphorylation (Figure S4D). Similar to Mst2WT, Mst2ΔL formed a 1:1 heterodimer with RASSF5 on a gel filtration column (Figures S5A and S5B). We thus used Mst2ΔL in subsequent structural studies.
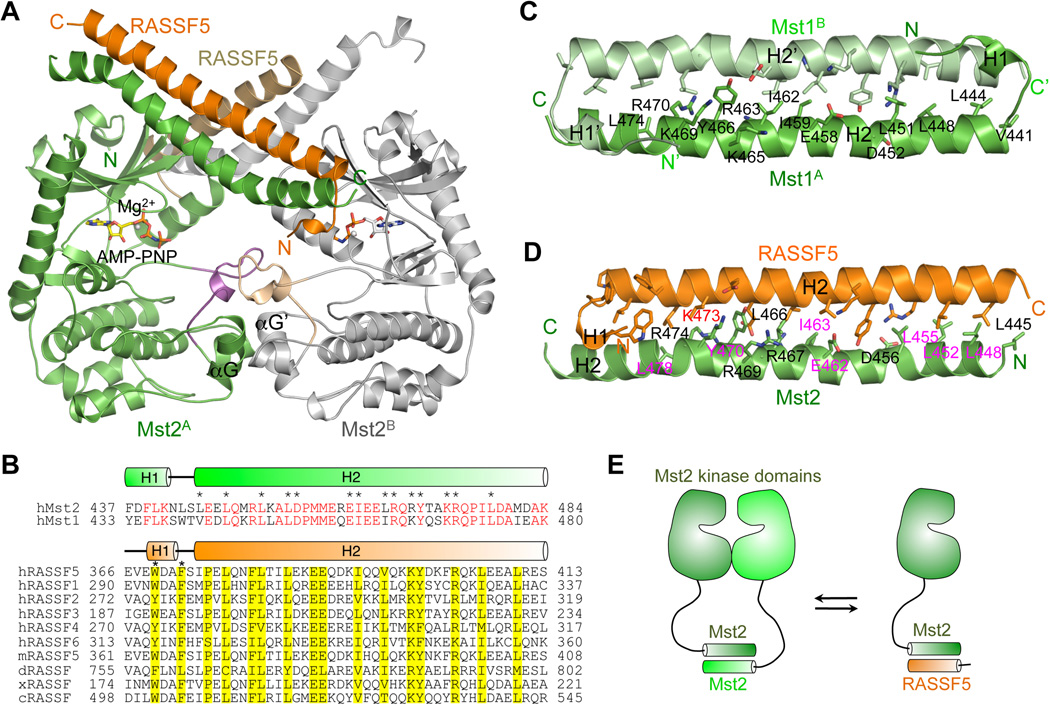
We determined the structure of kinase-dead Mst2ΔL/D146N bound to AMP-PNP and the SARAH domain of RASSF5, which we referred to as Mst2-RASSF5 hereafter for simplicity. Mst2-RASSF5 again formed a dimer in crystal through the Mst2 kinase domain (Figure 5A). Intriguingly, although the dimer interface of the Mst2 kinase domains also mainly involves αG, the relative orientation of the two kinase domains and their specific contacts are different from those observed in the active Mst1KD (Figure S5C) or inactive Mst2KD structures, suggesting that the dimer interfaces of Mst1/2 kinase heads in crystals might not be specific.
Figure 5. Structure of the Mst2-RASSF5 Complex.
(A) Ribbon diagram of the crystallographic dimer of the Mst2-RASSF5 complex. In monomer A, Mst2 is colored in green, RASSF5 is in orange and the activation loop in magenta. In monomer B, Mst2 is colored in gray, RASSF5 is in sand and the activation loop is in wheat. AMP-PNP is shown in sticks and Mg2+ is shown as gray sphere.
(B) Sequence alignment of SARAH domains from Mst1/2 and RASSF proteins. The sequence alignment of Mst1 and Mst2 SARAH domains is shown in the top panel with identical residues colored red and the conserved interface residues marked by asterisks. The secondary structure elements are shown above the sequences and colored in green. The sequence alignment of RASSF SARAH domains is shown in the bottom panel. h, Homo sapiens; m, mouse; d, Drosophila; x, Xenopus; c, C. elegans. The conserved residues are shaded in yellow. W369 and F372 in the H1 helix are marked by asterisks. The secondary structure elements are shown above the sequences and colored in orange.
(C) The solution structure of Mst1 SARAH homodimer. Protomer A is colored in green with the conserved Mst1 interface residues shown in sticks and labeled. Protomer B is colored in light green.
(D) The crystal structure of Mst2-RASSF5 SARAH heterodimer. The same color scheme as in (A) was used. The conserved interface residues were shown in sticks, and residues from Mst2 were labeled. The homodimerization-defective mutants are labeled in magenta and K473 in red.
(E) A schematic drawing of SARAH domain exchange between the Mst2 homodimer and Mst2-RASSF5 heterodimer. See also Figure S5.
The active site of Mst2-RASSF5 is well defined, as the Mg2+ ion and AMP-PNP are clearly visible in the electron density map. The kinase domain in Mst2-RASSF5 is very similar to the Mst2 kinase alone, except that the activation-loop helix observed in the Mst2 kinase structure has partially melted in the Mst2-RASSF5 structure (Figure S5D). The K56–E70 salt bridge between strand β3 and helix αC is also not formed in the Mst2-RASSF5 structure. The residual linker between the kinase domain and SARAH domain is invisible and presumably disordered. The SARAH domains of Mst2 and RASSF5 form a long anti-parallel coiled coil. Because the SARAH domains of Mst1 and Mst2 share 70% sequence identity (Figure 5B), they most likely share the same fold. The solution structure of the Mst1 SARAH homodimer has been previously determined (Figure 5C) (Hwang et al., 2007). The surfaces of Mst1/2 SARAH for binding to RASSF5 SARAH (heterodimerization) or to another Mst1/2 SARAH (homodimerization) are virtually identical (Figures 5C and 5D), readily explaining the disruption of SARAH-mediated Mst2 homodimerization by RASSF5 binding (Figure 5E).
Structural Determinants of Mst2 SARAH Homodimerization and Mst2-RASSF5 SARAH Heterodimerization
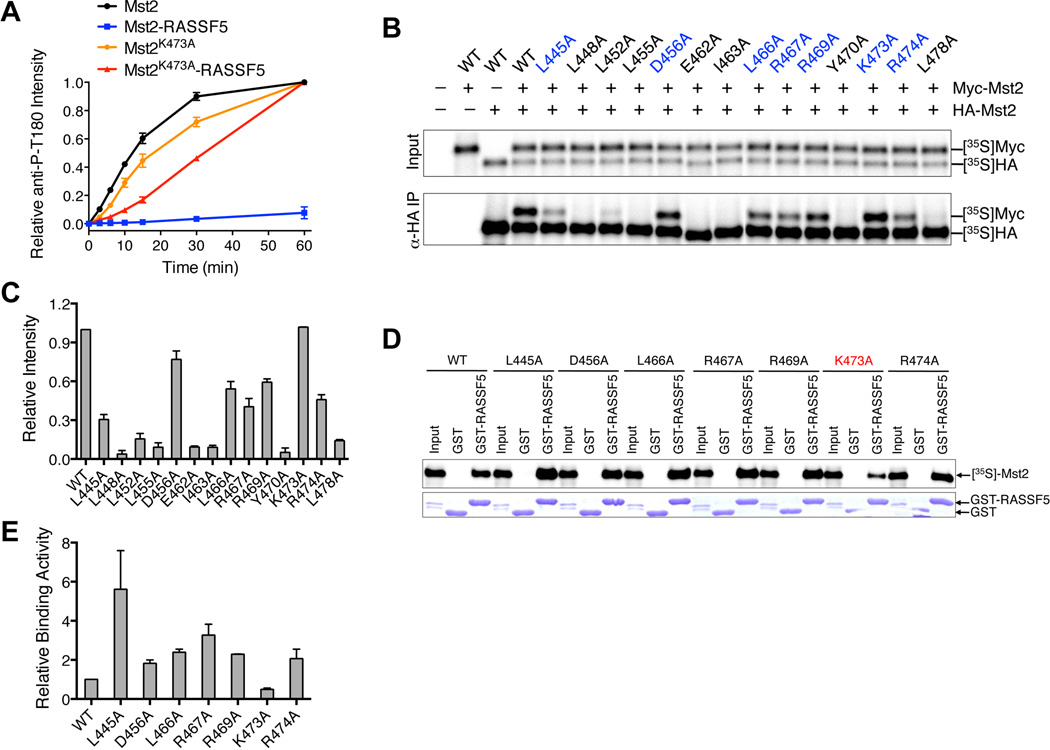
Addition of purified RASSF5 to the full-length Mst2 abolished its autophosphorylation and activation in vitro (Figure 6A). To test whether RASSF5 blocked Mst2 activation through disrupting Mst2 homodimerization, we sought to obtain Mst2 mutants that retained homodimerization but lost their ability to heterodimerize with RASSF5. Based on the Mst1 SARAH homodimer structure, we designed the point mutations of all conserved interface residues in the Mst2 SARAH domain (Figures 5B–D). We systematically mutated these interface residues in the context of the full-length Mst2 and tested their dimerization in an in vitro binding assay (Figures 6B and 6C). Briefly, we co-translated HA- or Myc-Mst2 in rabbit reticulocyte lysate in the presence of 35S-methionine and performed immunoprecipitation with anti-HA beads. The wild-type HA-Mst2 efficiently pulled down Myc-Mst2, confirming that Mst2 indeed formed dimers. Among the 14 mutants, Mst2 L448A, L452A, L455A, E462A, I463A, Y470A, and L478A displayed minimal dimerization (Figure 6C), indicating that the SARAH domain of Mst2 was the major dimerization interface. The weak dimerization of the Mst2 kinase domain was not detectable by this assay.
Figure 6. RASSF5 Blocks Mst2 Autoactivation through Heterodimerization.
(A) The kinetic profiles of T180 autophosphorylation of full-length Mst2, the full-length Mst2 and RASSF5-SARAH complex, the full-length point mutant Mst2K473A, and the full-length point mutant Mst2K473A and RASSF5-SARAH complex were shown. The relative α-Mst2 P-T180 intensities were normalized against Mst2 reaction at 60 minute (100%). Data are representative of at least two independent experiments.
(B) In vitro dimerization assay of Mst2 mutants. HA- and Myc-tagged Mst2 proteins were co-translated in vitro in the presence of 35S-methionine. The input and anti-HA IP were separated by SDS-PAGE and analyzed with a phosphor imager. Mutants that retain the dimerization ability were colored in blue.
(C) Quantification of the homodimerization of Mst2 mutants in (B).
(D) Binding between GST-RASSF5-SARAH and in vitro translated Mst2 proteins. Mst2K473A has a weakened interaction with GST-RASSF5 and is labeled in red.
(E) Quantification of GST-RASSF5 binding of Mst2 mutants in (D).
Compared to the wild-type (WT) Mst2, HA-Mst2 L445A, D456A, L466A, R467A, R469A, K473A and R474A still retained at least 30% binding to their Myc-tagged counterparts, (Figures 6B and 6C). We thus tested the binding of these 35S-labeled Mst2 mutants to GST-RASSF5 SARAH. As expected, Mst2 WT bound specifically to GST-RASSF5-SARAH, but not to GST (Figures 6D and 6E). Six out of the seven Mst2 mutants had stronger binding to RASSF5, presumably because these mutations disrupted the Mst2 homodimerization more severely than they disrupted Mst2-RASSF5 heterodimerization. Only Mst2 K473A exhibited reduced RASSF5 binding, as compared to Mst2 WT. Mst2 K473A still underwent efficient autophosphorylation and activation, which were not effectively blocked by RASSF5 (Figure 6A). This result confirms that RASSF5 blocks Mst2 activation through disrupting SARAH-mediated Mst2 homodimerization.
Functions of RASSFs in Mst1/2 Regulation
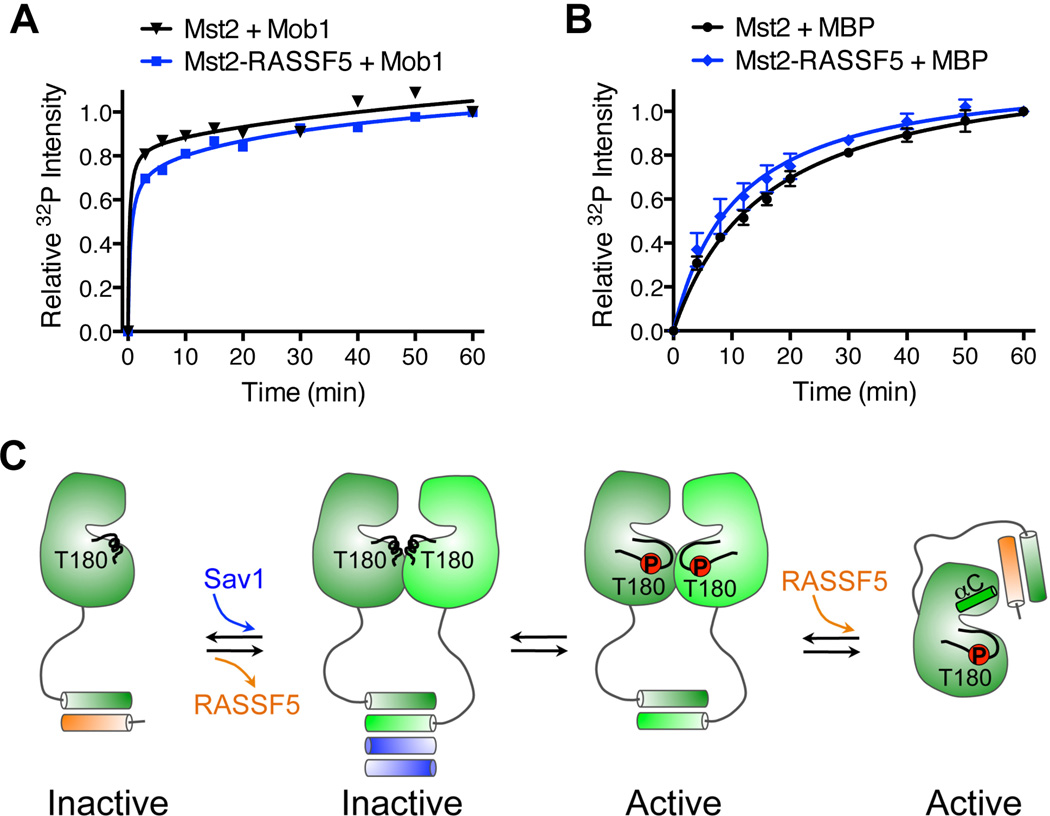
If RASSF5 simply prevents Mst2 autophosphorylation and activation, then binding of RASSF5 to the already fully activated Mst2 kinase should have no effect on the kinase activity. Indeed, in the presence of RASSF5, the activated Mst2 phosphorylated either Mob1 or myelin basic protein (MBP) similarly as Mst2 alone did (Figures 7A and 7B). Therefore, RASSF5 does not inhibit the already activated Mst2 kinase.
Figure 7. RASSF5 Does Not Inhibit the Kinase Activity of Mst2 that Has Already Undergone Autoactivation.
(A) The kinetic profiles of Mob1 phosphorylation by the full-length Mst2 and the Mst2-RASSF5 complex containing already activated Mst2, as monitored by 32P incorporation.
(B) The kinetic profiles of myelin basic protein (MBP) phosphorylation by activated full-length Mst2, and the Mst2-RASSF5 complex, as monitored by 32P incorporation. Data are representative of four independent experiments.
(C) Model for the dual modes of Mst2 regulation by RASSF5.
Collectively, our results support the following model for Mst1/2 regulation by RASSFs (Figure 7C). When the Hippo pathway is off, RASSFs form heterodimers with Mst1/2 through heterotypic SARAH interactions, thus blocking the homodimerization and trans-autophosphorylation of Mst1/2. In this context, RASSFs act as inhibitors of apoptosis. When the Hippo pathway is on, Mst1/2 undergo homodimerization and autoactivation, possibly through Sav1-dependent displacement of RASSFs. RASSFs can then bind to already activated Mst1/2. This binding does not inhibit the kinase activity of Mst1/2, but may instead promote the function of Mst1/2 through unknown mechanisms. In this capacity, RASSFs act as activators of apoptosis and tumor suppressors. Thus, RASSFs are double-edged swords in the Hippo pathway. Depending on the timing of their binding to Mst1/2 relative to Mst1/2 autophosphorylation, RASSFs can either function as inhibitors or activators of Mst1/2 and the Hippo pathway.
CONCLUSIONS
The Hippo pathway is a major regulatory pathway of cell proliferation and survival. Dysregulation of the Hippo pathway is intimately linked to cancer. Our study provides key structural and mechanistic insights into the activation of Mst1/2, the upstream kinases in this pathway. In particular, we have discovered important roles of RASSFs in regulating Mst1/2 in vitro. Binding of RASSFs to unphosphorylated, inactive Mst1/2 blocks their autoactivation. Binding of RASSFs to the already activated Mst1/2 does not inhibit their kinase activities, but may simulate Mst1/2 function through other mechanisms. Our discovery provides a potential explanation for the perplexing dual functions of RASSFs in vivo. By virtue of their ability to interact with both Mst1/2 and Ras proteins, RASSFs are at the intersection of the Hippo and Ras pathways. Temporal regulation of the binding between Mst1/2 and RASSFs affords an opportunity for the Hippo pathway to respond to and integrate diverse cellular signals.
EXPERIMENTAL PROCEDURES
Plasmids, Protein Expression, and Purification
The kinase domain of human Mst2 (Mst2KD, residues 16–313) was cloned into a modified pET28 vector (EMD Millipore) that included a tobacco etch virus (TEV) cleavage site at the N terminus. The full-length Mst2 (Mst2FL, residues 9–491) and linker-deletion Mst2 (Mst2ΔL, residues 9–313 and 428–491) were cloned into a modified pET29 vector (EMD Millipore) that included an N-terminal His6-tag without protease cleavage site. The C-terminal domain of Mst2 (Mst2CTD, residues 314–491) and the SARAH domain of human RASSF5 (RASSF5SARAH, residues 366–413) were cloned into a modified pGEX-6P vector (GE Healthcare) that included a TEV cleavage site at their N-termini. The Mst2 mutants were generated with the QuikChange mutagenesis kit (Stratagene). All constructs were verified by DNA sequencing.
When expressed alone in bacteria, the yield of Mst2 was extremely low. To obtain large quantities of soluble Mst2 proteins, the pET28-Mst2KD, pET29-Mst2FL or pET29-Mst2ΔL plasmid was co-transformed into BL21(DE3) with pGEX-6p–Mst2CTD to produce N-terminal His6-tagged Mst2 proteins. Mst2KD was purified with Ni2+-NTA agarose resin (Qiagen) and cleaved with TEV to remove the His6 tag. His6-Mst2FL and His6-Mst2ΔL were purified with Ni2+-NTA agarose resin. These proteins were further purified by a resource-Q anion exchange column and followed by a Superdex 200 gel filtration column (GE Healthcare). Purified Mst2 proteins were concentrated to 20 mg/ml in a buffer containing 20 mM Tris (pH 7.7), 100 mM NaCl, 2 mM MgCl2, and 5 mM TCEP.
The Mst2ΔL/D146N-RASSF5SARAH complex was obtained by co-expression of pET29-Mst2ΔL/D146N and pGEX-6p–RASSF5SARAH in the E. coli strain BL21(DE3). Mst2ΔL/D146N-RASSF5SARAH was purified with Glutathione Sepharose 4B beads (GE Healthcare) and cleaved with TEV to remove the GST moiety. Mst2ΔL/D146N-RASSF5SARAH was further purified by a Superdex 200 gel filtration column and then concentrated to 10 mg/ml in a buffer containing 20 mM Tris (pH 7.7), 100 mM NaCl, 2 mM MgCl2 and 5 mM TCEP. The seleno-methionine labeled Mst2ΔL/D146N-RASSF5SARAH was produced using the methionine biosynthesis inhibition method (Van Duyne et al., 1993).
Crystallization, Data Collection, and Structure Determination
Mst2KD/D146N was crystallized at 4°C using the hanging-drop vapor-diffusion method with a reservoir solution containing 0.2 M sodium citrate, 15% (w/v) PEG 3350, and 0.1 M HEPES (pH 7.7). The crystals were cryo-protected with the reservoir solution supplemented with 25% (v/v) glycerol and then flash-cooled in liquid nitrogen. Crystals diffracted to a minimum Bragg spacing (dmin) of 2.42 Å and exhibited the symmetry of space group C2 with cell dimensions of a = 227.2 Å, b = 69.8 Å, c = 148.1 Å and β = 108.6° and contained six Mst2KD/D146N molecules per asymmetric unit.
Mst2ΔL/D146N-RASSF5SARAH was crystallized at 20°C using the hanging-drop vapor-diffusion method with a reservoir solution containing 0.1 M Bis-Tris propane (pH 8.5), 200 mM Na2SO4, and 20% (w/v) PEG 3350. The crystals were cryo-protected with the reservoir solution supplemented with 25% glycerol and then flash-cooled in liquid nitrogen. Native crystals diffracted to a minimum Bragg spacing (dmin) of 3.05 Å and exhibited the symmetry of space group C2 with cell dimensions of a = 123.4 Å, b = 237.1 Å, c = 95.9 Å and β = 100.7° and contained four Mst2-RASSF5 heterodimers per asymmetric unit. The selenomethionine-derivatized Mst2ΔL/D146N-RASSF5SARAH crystals were obtained in the same way and had similar cell parameters. The selenomethionine-derivatized crystals only diffracted to a minimum Bragg spacing of 3.55 Å. All crystals showed significant anisotropic diffraction. SAD data on the selenomethionine Mst2ΔL/D146N-RASSF5SARAH crystals were collected at 3.55 Å with weak anomalous signal present to about 5 Å.
All diffraction data were collected at beamline 19-ID (SBC-CAT) at the Advanced Photon Source (Argonne National Laboratory, Argonne, Illinois, USA) and processed with HKL3000 (Otwinowski and Minor, 1997). Phases for Mst2KD/D146N were obtained by molecular replacement with Phaser using the crystal structure of human Mst1 kinase domain (PDB code: 3COM) as the search model (McCoy et al., 2007). Iterative model building and refinement were performed with COOT and Phenix, respectively (Adams et al., 2010; Emsley and Cowtan, 2004). The final model for Mst2KD/D146N (Rwork = 19.3%, Rfree = 23.1%) contained 1,646 residues in six monomers, 279 water molecules, and 4 glycerols. Two residues, L33 and D164, are outliers in the Ramachandran plot as defined in the program MolProbity (Davis et al., 2007). Both are located in surface loops with poor electron density.
Phases for Mst2ΔL/D146N-RASSF5SARAH were first obtained by molecular replacement with Phaser using Mst2KD/D146N structure as the search model. Phenix was used to build and refine a partial model only containing the Mst2 kinase domain. The resulting electron density map clearly showed several helices from the SARAH domains, yet the primary sequence for these helices was impossible to assign due to poor side chain density, and the density for the RASSF5SARAH helices was weak and disconnected. The MR-SAD module of Phaser was used to calculate initial phases to 3.6 Å from the MR solution and the known 36 Se sites in the Mst2ΔL/D146N kinase domains from a selenium-SAD experiment, and to locate the remaining 16 Se sites in the SARAH domains of Mst2ΔL/D146N. Iterative cycles of solvent flattening and four-fold averaging in Parrot, automatic model building in Buccaneer and phase calculation and refinement in the MR-SAD module of Phaser resulted in a model with 1,483 residues (89%) built and assigned (Winn et al., 2011). The side-chain assignment of RASSF5SARAH was further confirmed by packing interactions with the Mst2ΔL/D146N SARAH domain. Completion of the model and final refinement was performed using the native data to 3.05 Å. The final model for Mst2ΔL/D146N-RASSF5SARAH (Rwork = 19.9%, Rfree = 24.4%) contained 1,551 residues in four monomers, 4 molecules of Mg2+-AMP-PNP, 3 sodium ions, and 9 sulfate ions. Three residues, E31, T174, and E311, are outliers in the Ramachandran plot as defined in the program MolProbity and are located in surface loops with weak electron density. Data collection and structure refinement statistics are summarized in Table 1.
Analytical Ultracentrifugation
All sedimentation velocity analytical ultracentrifugation experiments were performed in an An-50Ti rotor at 20°C using a Beckman-Coulter Optima XL-I analytical ultracentrifuge. 400 µl of protein samples in the gel filtration buffer (20 mM Tris, pH 7.7, 100 mM NaCl, 2 mM MgCl2, 1 mM TCEP) at three different concentrations in the range of 2 – 100 µM were placed in one sector of centrifugation cells that housed dual-sector charcoal-filled Epon centerpieces. The same volumes of the gel filtration buffer were placed in the other sector of centerpieces as reference. The sealed centrifugation cells and the rotor were incubated at the experimental temperature under vacuum for at least two hours prior to centrifugation. Samples were centrifuged at 50,000 rpm until the boundary region reached the bottom of cells. Concentration profiles of the proteins were monitored using the on-board absorption optical system tuned to 280 nm and, where appropriate, the Rayleigh interferometer. The partial specific volume (0.7421 ml/g) based on the amino acid sequence, the solution density (1.00309 g/ml), and the viscosity (0.010177 P) of the buffer were calculated using SEDNTERP (Laue, 1992). Data were directly fitted and optimized to the monomer-dimer self-association model with the Marquardt-Levenberg methods using the Lamm equation with explicit reaction kinetics in SEDPHAT (http://www.analyticalultracentrifugation.com) (Brautigam, 2011; Schuck, 2003). A global analysis was performed for datasets obtained at different protein concentrations to calculate the dissociation constant for each protein sample.
NMR Spectroscopy
All NMR spectra were acquired at 30°C on a Varian INOVA 600 MHz four-channel spectrometer equipped with pulsed-field gradient triple resonance probe using H2O/D2O 95:5 (v/v) as the solvent. Samples typically contained 0.1 mM 15N-labeled protein in the NMR buffer consisting of 20 mM sodium phosphate (pH 6.8), 100 mM KCl, 2 mM MgCl2, and 1 mM DTT. The data were processed and analyzed with NMRPipe and NMRView (Delaglio et al., 1995; Johnson and Blevins, 1994).
In Vitro Kinase Assays and Autophosphorylation
For in vitro kinase assays, 0.025-0.4 µM Mst2 kinase or its mutants either alone or in complex with RASSF5 was incubated in the kinase buffer containing 50 mM Tris (pH 7.5), 150 mM NaCl, 10 mM MgCl2 , 1 mM ATP, 1 mM DTT, and 0.1 µCi/µl γ-32P-ATP with 40 µM substrate (Mob1, or Mst2KD/D146N, or MBP) for 30 min at room temperature. The reaction was terminated by adding 5X SDS sample buffer and followed by boiling for 5 min in 100°C water bath. The mixture was separated by SDS-PAGE and analyzed by autoradiography.
For autophosphorylation experiments, the wild-type Mst2 protein purified from E. coli was treated with Lambda (λ) protein phosphatase (New England Biolabs) at 30°C for 40 min to remove the phosphorylation at T180. The reaction was stopped by adding phosphatase inhibitor Na3VO4 to a final concentration of 3 mM. The treated Mst2 was then incubated at room temperature in the kinase buffer containing 50 mM Tris (pH 7.5), 150 mM NaCl, 10 mM MgCl2, 1 mM ATP, 1 mM DTT, and 1 mM Na3VO4. At the indicated time intervals, aliquots were removed and mixed with 5X SDS sample buffer. The autophosphorylation product was detected by Western blot using a specific antibody against phospho-T180 of Mst2 (Cell Signaling) and quantified with the Odyssey LI-COR imaging system.
Cell Culture and Transfection
HEK293 cells were grown in Dulbecco’s modified Eagle’s medium (DMEM; Invitrogen) supplemented with 10% fetal bovine serum (FBS). Plasmid transfections were performed with the Effectene reagent (Qiagen) according to manufacturer’s instructions. HA-YAP, Flag-Mst2, Myc-Lats1 and Myc-Lats2 have been described previously (Dong et al., 2007). Flag-Mst2I193A and Flag-Mst2F231A were made using the QuikChange mutagenesis kit (Stratagene). All clones were verified by DNA sequencing. Immunoprecipitation was performed as previously described (Yu et al., 2010). The Western blots were developed using the Amersham ECL chemiluminescence system (GE Healthcare).
In Vitro Binding Assays
To assay Mst2 homodimerization, same amounts of plasmids encoding Myc- or HA-tagged Mst2 WT or point mutants were translated in reticulocyte lysate in the presence of 35S-methionine. Affi-Prep protein A beads (Bio-Rad) covalently coupled to α-HA monoclonal antibodies (Roche) were incubated with 35S-labeled Mst2, and washed three times with TBS containing 0.05% Tween. The bound proteins were separated by SDS-PAGE and analyzed with a phosphor imager (Fujifilm).
To assay the binding between human Mst2 and RASSF5 proteins, the full-length WT or point mutants of Mst2 were translated in reticulocyte lysate in the presence of 35S-methionine. Purified GST-RASSF5SARAH was bound to glutathione-agarose beads (GE Healthcare), incubated with 35S-labeled Mst2 proteins, and washed three times with TBS containing 0.05% Tween. The proteins retained on the beads were analyzed by SDS-PAGE followed by autoradiography. Glutathione-agarose beads bound to GST were used as controls.
Supplementary Material
ACKNOWLEDGMENTS
We thank Hongtao Yu for discussion and critical reading of the manuscript. We also thank Boning Gao for providing the RASSF5 plasmid. Results shown in this study are derived from work performed at Argonne National Laboratory, Structural Biology Center at the Advanced Photon Source. Argonne is operated by University of Chicago Argonne, LLC, for the U.S. Department of Energy, Office of Biological and Environmental Research under contract DE-AC02-06CH11357. This work was supported in part by grants from the National Institutes of Health (GM085004 to X.L. and EY015708 to D.P.). J. Y. is a recipient of Young Investigator Award from the Children’s Tumor Foundation. D. P. is an investigator of the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
ACCESSION NUMBERS
The atomic coordinates and structure factors for human Mst2 kinase domain and Mst2-RASSF5 have been deposited in the Protein Data Bank with the PDB ID codes 4LG4 and 4LGD, respectively.
The authors declare no conflicts of interest.
REFERENCES
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoyama Y, Avruch J, Zhang XF. Nore1 inhibits tumor cell growth independent of Ras or the MST1/2 kinases. Oncogene. 2004;23:3426–3433. doi: 10.1038/sj.onc.1207486. [DOI] [PubMed] [Google Scholar]
- Avruch J, Xavier R, Bardeesy N, Zhang XF, Praskova M, Zhou D, Xia F. Rassf family of tumor suppressor polypeptides. J Biol Chem. 2009;284:11001–11005. doi: 10.1074/jbc.R800073200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avruch J, Zhou D, Fitamant J, Bardeesy N, Mou F, Barrufet LR. Protein kinases of the Hippo pathway: Regulation and substrates. Semin Cell Dev Biol. 2012;23:770–784. doi: 10.1016/j.semcdb.2012.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badouel C, Garg A, McNeill H. Herding Hippos: regulating growth in flies and man. Curr Opin Cell Biol. 2009;21:837–843. doi: 10.1016/j.ceb.2009.09.010. [DOI] [PubMed] [Google Scholar]
- Brautigam CA. Using Lamm-Equation modeling of sedimentation velocity data to determine the kinetic and thermodynamic properties of macromolecular interactions. Methods. 2011;54:4–15. doi: 10.1016/j.ymeth.2010.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper WN, Hesson LB, Matallanas D, Dallol A, von Kriegsheim A, Ward R, Kolch W, Latif F. RASSF2 associates with and stabilizes the proapoptotic kinase MST2. Oncogene. 2009;28:2988–2998. doi: 10.1038/onc.2009.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creasy CL, Ambrose DM, Chernoff J. The Ste20-like protein kinase, Mst1, dimerizes and contains an inhibitory domain. J Biol Chem. 1996;271:21049–21053. doi: 10.1074/jbc.271.35.21049. [DOI] [PubMed] [Google Scholar]
- Davis IW, Leaver-Fay A, Chen VB, Block JN, Kapral GJ, Wang X, Murray LW, Arendall WB, 3rd, Snoeyink J, Richardson JS, et al. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35:W375–W383. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- Dong J, Feldmann G, Huang J, Wu S, Zhang N, Comerford SA, Gayyed MF, Anders RA, Maitra A, Pan D. Elucidation of a universal size-control mechanism in Drosophila and mammals. Cell. 2007;130:1120–1133. doi: 10.1016/j.cell.2007.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar BA. From cell structure to transcription: Hippo forges a new path. Cell. 2006;124:267–273. doi: 10.1016/j.cell.2006.01.005. [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Guo C, Tommasi S, Liu L, Yee JK, Dammann R, Pfeifer GP. RASSF1A is part of a complex similar to the Drosophila Hippo/Salvador/Lats tumor-suppressor network. Curr Biol. 2007;17:700–705. doi: 10.1016/j.cub.2007.02.055. [DOI] [PubMed] [Google Scholar]
- Halder G, Johnson RL. Hippo signaling: growth control and beyond. Development. 2011;138:9–22. doi: 10.1242/dev.045500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao Y, Chun A, Cheung K, Rashidi B, Yang X. Tumor suppressor LATS1 is a negative regulator of oncogene YAP. J Biol Chem. 2008;283:5496–5509. doi: 10.1074/jbc.M709037200. [DOI] [PubMed] [Google Scholar]
- Harvey K, Tapon N. The Salvador-Warts-Hippo pathway - an emerging tumour-suppressor network. Nat Rev Cancer. 2007;7:182–191. doi: 10.1038/nrc2070. [DOI] [PubMed] [Google Scholar]
- Harvey KF, Zhang X, Thomas DM. The Hippo pathway and human cancer. Nat Rev Cancer. 2013;13:246–257. doi: 10.1038/nrc3458. [DOI] [PubMed] [Google Scholar]
- Hergovich A. Mammalian Hippo signalling: a kinase network regulated by protein-protein interactions. Biochem Soc Trans. 2012;40:124–128. doi: 10.1042/BST20110619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong W, Guan KL. The YAP and TAZ transcription co-activators: Key downstream effectors of the mammalian Hippo pathway. Semin Cell Dev Biol. 2012;23:785–793. doi: 10.1016/j.semcdb.2012.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Wu S, Barrera J, Matthews K, Pan D. The Hippo signaling pathway coordinately regulates cell proliferation and apoptosis by inactivating Yorkie, the Drosophila Homolog of YAP. Cell. 2005;122:421–434. doi: 10.1016/j.cell.2005.06.007. [DOI] [PubMed] [Google Scholar]
- Hwang E, Ryu KS, Paakkonen K, Guntert P, Cheong HK, Lim DS, Lee JO, Jeon YH, Cheong C. Structural insight into dimeric interaction of the SARAH domains from Mst1 and RASSF family proteins in the apoptosis pathway. Proc Natl Acad Sci U S A. 2007;104:9236–9241. doi: 10.1073/pnas.0610716104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda M, Kawata A, Nishikawa M, Tateishi Y, Yamaguchi M, Nakagawa K, Hirabayashi S, Bao Y, Hidaka S, Hirata Y, et al. Hippo pathway-dependent and -independent roles of RASSF6. Sci Signal. 2009;2:ra59. doi: 10.1126/scisignal.2000300. [DOI] [PubMed] [Google Scholar]
- Jin Y, Dong L, Lu Y, Wu W, Hao Q, Zhou Z, Jiang J, Zhao Y, Zhang L. Dimerization and cytoplasmic localization regulate Hippo kinase signaling activity in organ size control. J Biol Chem. 2012;287:5784–5796. doi: 10.1074/jbc.M111.310334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson BA, Blevins RA. NMRView: A computer program for visualization and analysis of NMR data. J Biomol NMR. 1994;4:603–614. doi: 10.1007/BF00404272. [DOI] [PubMed] [Google Scholar]
- Khokhlatchev A, Rabizadeh S, Xavier R, Nedwidek M, Chen T, Zhang XF, Seed B, Avruch J. Identification of a novel Ras-regulated proapoptotic pathway. Curr Biol. 2002;12:253–265. doi: 10.1016/s0960-9822(02)00683-8. [DOI] [PubMed] [Google Scholar]
- Laue TM, Shah B, Ridgeway R, Pelletier SL. Computer-aided interpretation of analytical sedimentation data for proteins. Cambridge, UK: The Royal Society of Chemistry; 1992. [Google Scholar]
- Luo X. Snapshots of a hybrid transcription factor in the Hippo pathway. Protein Cell. 2010;1:811–819. doi: 10.1007/s13238-010-0105-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matallanas D, Romano D, Yee K, Meissl K, Kucerova L, Piazzolla D, Baccarini M, Vass JK, Kolch W, O'Neill E. RASSF1A elicits apoptosis through an MST2 pathway directing proapoptotic transcription by the p73 tumor suppressor protein. Mol Cell. 2007;27:962–975. doi: 10.1016/j.molcel.2007.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. Journal of applied crystallography. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh HJ, Lee KK, Song SJ, Jin MS, Song MS, Lee JH, Im CR, Lee JO, Yonehara S, Lim DS. Role of the tumor suppressor RASSF1A in Mst1-mediated apoptosis. Cancer Res. 2006;66:2562–2569. doi: 10.1158/0008-5472.CAN-05-2951. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Processing X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Pan D. The hippo signaling pathway in development and cancer. Dev Cell. 2010;19:491–505. doi: 10.1016/j.devcel.2010.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pervushin K. Impact of transverse relaxation optimized spectroscopy (TROSY) on NMR as a technique in structural biology. Quarterly reviews of biophysics. 2000;33:161–197. doi: 10.1017/s0033583500003619. [DOI] [PubMed] [Google Scholar]
- Pirruccello M, Sondermann H, Pelton JG, Pellicena P, Hoelz A, Chernoff J, Wemmer DE, Kuriyan J. A dimeric kinase assembly underlying autophosphorylation in the p21 activated kinases. J Mol Biol. 2006;361:312–326. doi: 10.1016/j.jmb.2006.06.017. [DOI] [PubMed] [Google Scholar]
- Polesello C, Huelsmann S, Brown NH, Tapon N. The Drosophila RASSF homolog antagonizes the hippo pathway. Curr Biol. 2006;16:2459–2465. doi: 10.1016/j.cub.2006.10.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Praskova M, Khoklatchev A, Ortiz-Vega S, Avruch J. Regulation of the MST1 kinase by autophosphorylation, by the growth inhibitory proteins, RASSF1 and NORE1, and by Ras. Biochem J. 2004;381:453–462. doi: 10.1042/BJ20040025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter AM, Pfeifer GP, Dammann RH. The RASSF proteins in cancer; from epigenetic silencing to functional characterization. Biochim Biophys Acta. 2009;1796:114–128. doi: 10.1016/j.bbcan.2009.03.004. [DOI] [PubMed] [Google Scholar]
- Schuck P. On the analysis of protein self-association by sedimentation velocity analytical ultracentrifugation. Analytical biochemistry. 2003;320:104–124. doi: 10.1016/s0003-2697(03)00289-6. [DOI] [PubMed] [Google Scholar]
- Song H, Oh S, Oh HJ, Lim DS. Role of the tumor suppressor RASSF2 in regulation of MST1 kinase activity. Biochem Biophys Res Commun. 2010;391:969–973. doi: 10.1016/j.bbrc.2009.11.175. [DOI] [PubMed] [Google Scholar]
- Staley BK, Irvine KD. Hippo signaling in Drosophila: recent advances and insights. Dev Dyn. 2012;241:3–15. doi: 10.1002/dvdy.22723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudol M, Shields DC, Farooq A. Structures of YAP protein domains reveal promising targets for development of new cancer drugs. Semin Cell Dev Biol. 2012;23:827–833. doi: 10.1016/j.semcdb.2012.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tommasi S, Dammann R, Zhang Z, Wang Y, Liu L, Tsark WM, Wilczynski SP, Li J, You M, Pfeifer GP. Tumor susceptibility of Rassf1a knockout mice. Cancer Res. 2005;65:92–98. [PubMed] [Google Scholar]
- Van Duyne GD, Standaert RF, Karplus PA, Schreiber SL, Clardy J. Atomic structures of the human immunophilin FKBP-12 complexes with FK506 and rapamycin. J Mol Biol. 1993;229:105–124. doi: 10.1006/jmbi.1993.1012. [DOI] [PubMed] [Google Scholar]
- Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AG, McCoy A, et al. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Zheng Y, Dong J, Klusza S, Deng WM, Pan D. Kibra functions as a tumor suppressor protein that regulates Hippo signaling in conjunction with Merlin and Expanded. Dev Cell. 2010;18:288–299. doi: 10.1016/j.devcel.2009.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Li L, Lei Q, Guan KL. The Hippo-YAP pathway in organ size control and tumorigenesis: an updated version. Genes Dev. 2010a;24:862–874. doi: 10.1101/gad.1909210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Li L, Tumaneng K, Wang CY, Guan KL. A coordinated phosphorylation by Lats and CK1 regulates YAP stability through SCF(beta-TRCP) Genes Dev. 2010b;24:72–85. doi: 10.1101/gad.1843810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Wei X, Li W, Udan RS, Yang Q, Kim J, Xie J, Ikenoue T, Yu J, Li L, et al. Inactivation of YAP oncoprotein by the Hippo pathway is involved in cell contact inhibition and tissue growth control. Genes Dev. 2007;21:2747–2761. doi: 10.1101/gad.1602907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Ye X, Yu J, Li L, Li W, Li S, Yu J, Lin JD, Wang CY, Chinnaiyan AM, et al. TEAD mediates YAP-dependent gene induction and growth control. Genes Dev. 2008;22:1962–1971. doi: 10.1101/gad.1664408. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.