Abstract
Mutations in the FOXP2 gene cause speech and language impairments, accompanied by structural and functional abnormalities in brain regions underlying speech-related sensory-motor processing, including the striatum and cerebellum. The sequence and expression patterns of FOXP2 are highly conserved among higher vertebrates. In the zebra finch brain, FoxP2 is expressed in Area X, a striatal nucleus required for vocal learning, and reduced FoxP2 expression impairs dendritic development and vocal learning. The FoxP2 gene encodes a transcription factor that controls the expression of many downstream genes. However, how FOXP2 gene expression is regulated is not clearly understood. miRNAs regulate gene expression post-transcriptionally by targeting the 3′-untranslated regions (UTRs) of mRNAs, leading to translational suppression or mRNA degradation. In this study, we identified miR-9 and miR-140-5p as potential regulators of the FoxP2 gene. We show that both miR-9 and miR-140-5p target specific sequences in the FoxP2 3′-UTR and downregulate FoxP2 protein and mRNA expression in vitro. We also show that the expression of miR-9 and miR-140-5p in Area X of the zebra finch brain is regulated during song development in juvenile zebra finches. We further show that in adult zebra finches the expression of miR-9 and miR-140-5p in Area X is regulated as a function of the social context of song behavior in males singing undirected songs. Our findings reveal a post-transcriptional mechanism that regulates FoxP2 expression and suggest that social vocal behavior can influence the basal ganglia circuit controlling vocal learning via a miRNA-FoxP2 gene regulatory network.
Introduction
Mutations in the human FOXP2 gene cause speech and language impairments characterized by core deficits in coordinating and sequencing complex orofacial movements underlying speech production (Vargha-Khadem et al., 1998; Lai et al., 2001; MacDermot et al., 2005; Feuk et al., 2006; Zeesman et al., 2006). The FOXP2 gene encodes a transcription factor that regulates the expression of many downstream genes, which in turn have important roles in nervous system development and function (Shu et al., 2001; Spiteri et al., 2007; Vernes et al., 2007, 2011; Konopka et al., 2009). One current hypothesis suggests that an adequate amount of functional FOXP2 is required for the proper development of the corticostriatal and/or cortical-cerebellar circuits responsible for learning and execution of motor sequencing underlying speech (Fisher and Scharff, 2009).
Similarly to speech and language development in humans, zebra finches learn to sing by imitating songs of an adult tutor (usually the father) early in life (Immelmann, 1969; Eales, 1985; Doupe and Kuhl, 1999; Tchernichovski et al., 2001) and they devote a set of interconnected brain nuclei to vocal learning and production (Nottebohm et al., 1976; Bottjer et al., 1989). The sequences and expression patterns of the FOXP2 gene are highly conserved between humans and zebra finches (Lai et al., 2001, 2003; Haesler et al., 2004; Teramitsu et al., 2004). In the zebra finch brain, FoxP2 is expressed in Area X, a striatal nucleus required for vocal learning (Scharff and Nottebohm, 1991; Haesler et al., 2004; Teramitsu et al., 2004; Doupe et al., 2005), and reducing FoxP2 expression in Area X impairs dendritic development and vocal learning in juvenile zebra finches (Haesler et al., 2007; Schulz et al., 2010). Adult male zebra finches sing two types of songs depending on the social context: a “directed” courtship song toward a female zebra finch, or an “undirected” song when singing alone. These two types of songs trigger distinct neuronal firing activities and gene expression patterns in Area X (Jarvis et al., 1998; Hessler and Doupe, 1999; Teramitsu and White, 2006; Kojima and Doupe, 2011). FoxP2 expression in Area X is downregulated in males singing undirected songs, but not in males singing female-directed songs (Teramitsu and White, 2006).
MicroRNAs (miRNAs) are small nonprotein-coding RNA molecules that regulate gene expression post-transcriptionally by targeting specific sequences in the 3′-UTRs of messenger RNAs (mRNAs), leading to translational suppression or mRNA degradation (Lee et al., 1993; Reinhart et al., 2000; Bartel, 2004). In this study, we present evidence that miR-9 and miR-140-5p, two miRNAs known to be expressed in the striatum of mammals (Landgraf et al., 2007; Pena et al., 2009), target FoxP2 3′-UTR and downregulate FoxP2 protein and mRNA expression. We also show that the expression of miR-9 and miR-140-5p in Area X of the zebra finch brain is regulated during vocal development. We further show that in adult zebra finches, the expression of miR-9 and miR-140-5p in Area X is regulated by song behavior in a social context-dependent manner.
Materials and Methods
Nomenclature.
Following the convention for the nomenclature of the forkhead family of genes (Kaestner et al., 2000), we refer to proteins as FOXP2 in humans, Foxp2 in rodents, and FoxP2 in other species, and the corresponding genes and mRNAs in italics.
Animals.
Animal usage was approved by the Louisiana State University Health Sciences Center Institutional Animal Care and Use Committee. For developmental studies, juveniles with documented birth date were obtained from our breeding colony.
Cloning of the zebra finch FoxP2 3′-UTR.
Total RNA was extracted from embryonic zebra finch brain and mRNA was purified as described previously (Li et al., 2007). mRNA was reversely transcribed (SuperScript Choice System; Invitrogen) into cDNA using an oligo-dT primer [5′-atat-GCGGCCGCNotI-AG-T18-(A, C, G)-3]. Based on the homology to the human FOXP2 3′-UTR sequence obtained from the NCBI database (variant II, accession No. NM148898), we extracted the zebra finch FoxP2 3′-UTR sequence (3846 bp) from the zebra finch genome (reference assembly 3.2.4), and designed PCR primer. We amplified two overlapping cDNA fragments, and ligated the two fragments to obtain a full-length FoxP2 3′-UTR. The following primers were used: fragment 1 forward primer: 5′-TTCAGCGATCGCAAAGGGACTTGAGAAACCTCAAAATG-3′ (a Sgf1 restriction site is underlined) and fragment 1 reverse primer: 5′-GCTTTCAGAAAGCCT TTTGGGTAAGTGGGAAAACCC-3′; fragment 2 forward primer: 5′-TTCCCCCAACCAAGGGACCTCATAACCTG-3′ and fragment 2 reverse primer: 5′-ATTGCGGCCGCTTATAAATGGATCAAAAAGAATTATATTTTTTAT-3′ (a NotI restriction site is underlined). The overlapping sequence between these two fragments contained an NdeI restriction site. PCR conditions were as follows: 1 min at 95°C and 35 cycles of 30 s at 95°C, 30 s at 55°C, and 2 min at 72°C, followed by 10 min at 72°C. PfuUltra High-Fidelity DNA Polymerase (Agilent) was used and the PCR products were gel purified. Fragment 1 was inserted into the TOPO PCR cloning vector (Invitrogen) to create the plasmid TOPO-FoxP2 3′-UTR-1. Fragment 2 was digested by restriction enzymes NdeI and NotI and inserted into the NdeI and NotI sites of the TOPO-FoxP2 3′-UTR-1 to generate the TOPO-FoxP2 3′-UTR 1 + 2 construct. Finally, the complete 3846 nt zebra finch FoxP2 3′-UTR sequence was excised from the TOPO-FoxP2 3′-UTR 1 + 2 plasmid (NotI and SgfI) and inserted into the psiCHECK-2 vector (Promega) to obtain the psiCHECK-2-FoxP2 3′-UTR plasmid construct. At each stage of cloning, all plasmids were sequence validated.
Identification of miRNA binding sites.
The miRNA target prediction software TargetScan was used to identify miRNA binding sites in the FoxP2 3′-UTR. One hundred and ninety-three zebra finch miRNAs, which were recently annotated by profiling miRNA expression in zebra finch brain and other tissues (Luo et al., 2012), were included in the binding site search. Sequence elements within the FoxP2 3′-UTR that matched perfectly to the “seed sequences” of miR-9 and miR-140-5p were accepted as putative binding sites for these miRNAs. Since miRNA binding sites are often conserved in vertebrates, we performed similar sequence analysis of the 3′-UTRs of the FOXP2 genes in chicken, mouse, chimpanzee, and human.
Mutagenesis of the miR-9 and miR-140-5p binding sites.
Mutagenesis was performed using the QuikChange II XL Site-Directed Mutagenesis Kit (Agilent) following the manufacturer's instructions. To modify the two miR-9 binding sites in the FoxP2 3′-UTR, we made three mutant constructs containing mutations in binding site 1, binding site 2, and binding sites 1 + 2 (psiCHECK-FoxP2-3′-UTR-miR-9mb1, psiCHECK-FoxP2-3′-UTR-miR-9mb2, and psiCHECK-FoxP2-3′-UTR-miR-9mb1 + 2). Specific PCR primers were designed using the QuikChangePrimer Design tool and synthesized by BioBasic. For each miR-9 binding site, we mutated 3 nt in two rounds of PCR-based mutagenesis. For example, we mutated 2 nt in the first round and used the mutated plasmid as a template for the second round of PCR to mutate a third nucleotide. The miR-140-5p binding site was mutated similarly in two rounds of PCR mutagenesis. The final mutated plasmids were sequence validated.
Cloning of the zebra finch FoxP2 coding region for protein and mRNA expression.
For expression assays, we cloned the full-length zebra finch FoxP2 protein-coding region and the 3′-UTR into the expression vector pcDNA3. We first divided the entire clone into three segments with lengths of 1.6, 2.4, and 2.1 kb. Each segment was individually PCR amplified using the following primers: segment 1 forward primer: 5′-AAGCTAGCAGAAAAAGACTTTCCCACGCACAGC-3′ (NheI site underlined) and segment 1 reverse primer: 5′-ATTGGATCCTCTGAATGTCTTCTTCGAATGGCTCC-3′ (BstbI and BamHI sites underlined); segment 2 forward primer: 5′-GGAGCCATTCGAAGAAGACATTCAGAC-3′ (BstbI site underlined) and segment 2 reverse primer: 5′- ATTGGATCCACCATCACATGGAACTGGTACATATGT-3′ (BamHI and NdeI sites underlined); and segment 3 forward primer: 5′-GGCTGCTAGAGCTTAACATATGTAC-3′ (NdeI site underlined) and segment 3 reverse primer: 5′-ATTGGATCCTTATAAATGGATCAAAAAGAATTATATTTTTTAT-3′ (BamHI site underlined). Primers in the coding regions were designed based on previously published sequences (Haesler et al., 2004; Teramitsu et al., 2004), and primers in the 3′-UTR region were designed based on our own results. cDNAs made from zebra finch embryonic brains were used as template for segments 1 and 2, and the psiCHECK-2-FoxP2 3′-UTR plasmid was used as template for segment 3. Notably, the overlapping region between segments 1 and 2 contains a BstbI restriction site, and the overlapping region between segments 2 and 3 contains an NdeI restriction site. Thus, BstbI and NdeI sites were used as junction sites to ligate segments 1, 2, and 3. The final product was inserted into the NheI and BamHI sites of the pcDNA3.1 expression vector and sequence verified. To make expression plasmid constructs containing mutated FoxP2 3′-UTR, we replaced the wild-type 3′-UTR with 3′-UTRs carrying mutated miR-9 or miR-140-5p binding sites as described above.
Dual luciferase assays.
SH-SY5Y cells were grown in Dulbecco's Modified Eagle Medium/F12 supplemented with 10% fetal bovine serum. Plasmid constructs carrying wild-type or mutant FoxP2 3′-UTRs in the psiCHECK vector (100 ng) were cotransfected with miR-9 or miR-140-5p mimics or a control miRNA duplex (100 nm final concentration) into SH-SY5Y cells using Lipofectamine 2000 (Invitrogen). Cells were harvested 48 h later and luciferase activity was assayed using the Dual-Luciferase Reporter 1000 System (Promega) following the manufacturer's protocol. Renilla luciferase (R-Luc) activity was normalized to the internal control firefly luciferase (f-Luc) activity to control transfection efficiency. Each transfection was performed in triplicate (three wells), and the luciferase activities in the three wells were averaged; this averaged result was used as the value for that transfection. The transfection was independently performed three times; thus, for the experiment, n = 3. The following miRNA duplexes used in the luciferase reporter assays and in FoxP2 protein and mRNA assays (see below) were purchased from GenePharma: 5′-UUCUCCGAACGUGUCACGUTT-3′, 5′-ACGUGACACGUUCGGAGAATT-3′ (negative control miRNA); 5′-UCUUUGGUUAUCUAGCUGUAUGA-3′, 5′-AUACAGCUAGAUAACAAAGAUU-3′ (miR-9); 5′-CAGUGGUUUUACCCUAUGGUAG-3′, 5′-ACCAUAGGGUAAAACCACUGUU-3′ (miR-40–5p); and 5′-UAAGGCACGCGGUGAAUGCC-3′, 5′-CAUUCACCGCGUGCCUUAUU-3′ (miR-124).
Western blot.
Plasmid constructs containing the FoxP2 protein-coding region with wild-type or mutant 3′-UTRs (2 μg) and miR-9 or miR-140-5p or a control miRNA duplex (100 nm final concentration) were cotransfected into SH-SY5Y cells using Lipofectamine 2000 (Invitrogen). Cells were harvested 72 h later and proteins were isolated for Western blot. Protein samples (40 μg per lane) were separated by SDS-PAGE (12%) and then transferred to PVDF membranes (Bio-Rad). The membranes were probed with a goat anti-Foxp2 antibody (Santa Cruz Biotechnology, sc-21069) and signals were detected using an ECL detection system (GE Healthcare). As an internal control, β-actin was probed with a mouse anti-β-actin antibody (Santa Cruz Biotechnology, sc-47778). Transfection and Western blots were performed three times for each miRNA. Quantification was performed using the NIH ImageJ software.
Area X tissue dissection and RNA isolation.
Frozen zebra finch brains were cut into 100 μm sagittal sections using a cryostat. Each section (attached to a glass slide) was semifixed (70% ice-cold ETOH/PBS, 30”) and examined under a dissecting microscope. Area X, a round structure ∼1 mm in diameter, visibly stands out from the surrounding areas. We used a syringe needle to carve and pick out Area X tissue and quickly transfer it into lysis buffer for total RNA isolation. In general, Area X of adult male zebra finches can be easily visualized. The youngest juveniles included were 20 d old, an age at which we can confidently distinguish Area X from surrounding tissues. As needed, cresyl violet-staining of serial sagittal sections was performed to help define the medial and lateral edges of Area X. If there was any doubt, a section was not used. For each section, an area adjacent to Area X was also collected as a control. Total RNA was isolated from dissected tissues using TRIZOL reagent (Invitrogen). The quality of RNA samples was determined on an Agilent Bioanalyzer 2000 and RNA was quantified using a Nanodrop spectrophotometer.
Quantitative real-time PCR.
To quantify miRNA, reverse transcription and quantitative PCR (qPCR) were performed using the TaqMan@ microRNA assay Kit following the manufacturer's protocol (Applied Biosystems). Briefly, reverse transcription was performed in a 15 μl reaction mix containing 10 ng of total RNA, 3 μl of miRNA primer mix, 1 mm dNTP, 50 U reverse transcriptase, and 3.8 u. RNase inhibitor. Reactions were incubated at 16°C for 30 min, 42°C for 30 min followed by 85°C for 5 min. The PCR was performed in a 10 μl reaction volume containing 0.5 μl of miRNA primer and TaqMan probe mix, 1.33 μl of RT product (diluted fivefold), and 10 μl of TaqMan Universal PCR Master Mix. Cycling conditions were as follows: 10 min at 95°C followed by 40 cycles of 15 s at 95°C and 1 min at 60°C. U6 small RNA was used as an internal control following the manufacture's recommendation. We confirmed that U6 expression levels did not change during development or as a function of singing behavior. Specificities of U6 small RNA and miRNA primers have been extensively tested and established by the manufacturer. Sequences of U6 small RNA, miR-9, and miR-140-5p are 100% conserved among zebra finch, human, and mouse. The Catalog Numbers of reagents are as follows: miR-9: #4427975, 000583; miR-140-5p: #4427975, 001187; and U6: #4427975, 001973. To measure FoxP2 and ZENK mRNA levels, reverse transcription was performed using 50 ng of total RNA with the iScript Reverse Transcription Supermix kit (Bio-Rad). qPCR was performed using the iQ SYBR Green Supermix (Bio-Rad) with the following primers: FoxP2 forward primer: 5′-GACTCATTTCCATCCCACC-3′ and reverse primer: 5′-GGTCTAGCCCTCCATGTTTA-3′ (NCBI sequence ID: NM_001048263); ZENK forward primer: 5′-CGCACAGCGTATTCTTC-3′ and reverse primer: 5′-TAGCCACTTGGGTTTGA-3′ (NCBI sequence ID: EF052676.1); and GAPDH forward primer: 5′-ATGGCATCCAAGGAGTAA-3′ and reverse primer: 5′-GGAGACAAGGGAACAGAAC-3′ (NCBI sequence ID: NM_001198610). For all samples, reverse transcription and qPCR were performed two times and qPCR was performed in triplicate. GAPDH was used as a reference gene after determining that it did not change expression during development or as a function of song behavior. The specificity of the PCR products of FoxP2, ZENK, and GAPDH were validated by three methods: (1) dissociation curve analysis showed a single peak for each PCR product; (2) gel electrophoresis showed that each PCR resulted in a single band of the correct size: 153 nt for FoxP2, 124 nt for ZENK, and 145 nt for GAPDH; and (3) PCR products recovered from the gel were sequenced, and all exhibited correct sequences (data not shown). Relative gene expression levels between experimental groups were determined using the comparative Ct (2−ΔΔCt) method after normalizing to reference genes (Livak and Schmittgen, 2001).
In situ hybridization.
Perfused brains were cut into 12 μm sagittal sections and sections were treated with proteinase K for 20 min, fixed in 4% paraformaldehyde (PFA) for 10 min, followed by acetylation. Brain sections were hybridized with digoxigenin-labeled locked nucleic acid (LNA) modified miRNA detection probes (Exiqon) in 40 μl of hybridization buffer (50% formamide, 4×SSC, 5× Denhardt's, 0.25 μg/μl tRNA, 0.5 μg/μl salmon sperm DNA, 20 μg/μl blocking reagent, 2.5 mm chaps and 0.1% Tween-20) at 38−45°C overnight. Slides were then washed two times in a wash solution (50% formamide, 1×SSC, and 0.1% Tween 20) 30 min each, at 35°C followed by washing in 0.2×SSC for 15 min at RT. After incubating in a 3% hydrogen peroxide solution for 30 min at RT, sections were rinsed three times (PBS, 0.1% Tween 20), 1 min each. Sections were then incubated with an antibody blocking solution [0.1 g of block reagent (Roche), 5% serum, and 0.1%Tween 20 in TBS] for 1 h at room temperature (RT), and then incubated with the anti-digoxigenin antibody in blocking solution (1:500 dilution, Roche) for 1 h at RT, followed by washing twice, 15 min each in TNT buffer (10 mm Tris-HCl, 150 mm NaCl and 0.1% Tween-20). The sections were incubated in TSA Plus Cy3 System (PerkinElmer) working solution for 10 min, and then washed three times, 5 min each, with TNT buffer and were mounted (Vector Laboratories) for microscopic analysis. miRNA detection probes used were as follows: a negative control probe (5′-GTGTAACACGTCTATACGCCCA-3′, Exiqon, 99004-01); a miR-9 detection probe (5′-TCATACAGCTAGATAACCAAAGA-3′, Exiqon, 88078-05); a customer designed miR-9 detection probe with four nucleotides mutated (5′-TCATAGAGCTACATAACCATACA-3′, underlined are mutated nucleotides); and a double-digoxigenin labeled miR-140-5p detection probe (5′-CTACCATAGGGTAAAACCACT-3′, Exiqon, 21309-15).
Song behavior.
Adult male zebra finches (120–150 d old) were used. Following methods used previously by other groups (Jarvis et al., 1998; Teramitsu and White, 2006; Kojima and Doupe, 2011), song behavior of birds was observed individually, one at a time, for a 2 h time period in the morning (between 8:00 A.M. and 10:00 A.M. right after lights were switched on). The nonsinging control birds either did not sing naturally or sang <10 bouts during the observation time. If a control bird started to sing, an observer sitting nearby waved a hand at them to interrupt singing. Prior work has shown that distracting birds from singing does not significantly alter gene expression in Area X compared with birds that do not sing naturally (Hilliard et al., 2012). To quantify song production, an observer sitting nearby counted the number of song bouts produced by each bird and a bird that sang 100–150 song bouts per hour during a 2 h observation time was accepted into the singing groups. For the undirected song group, male birds were observed individually during the 2 h observation period in the absence of any special treatment; they sang alone and sang spontaneously. Since no other birds were around, songs of this group were 100% undirected songs. For the directed song group, female-directed singing was encouraged by presenting several females to a male in turn (10–15 min each) in a nearby cage over the 2 h period. Birds that sang >90% songs toward females accompanied with typical male courtship behaviors including body posture and orientation were accepted into the directed-song group. After behavioral observation, birds were killed, and brains were collected and frozen immediately for subsequent processing.
Results
Characterization of the FoxP2 3′-UTR and identification of miRNA binding sites
Northern blot analysis detects several FoxP2 transcripts, including a major transcript of 6.5 kb, in the zebra finch brain and other tissues (Haesler et al., 2004). However, the zebra finch FoxP2 protein is ∼710 aa long (Haesler et al., 2004; Teramitsu et al., 2004), raising the possibility that the FoxP2 gene may have a long 3′-UTR, and, therefore, it might be regulated post-transcriptionally by miRNAs. To clone and characterize the full-length sequence of the zebra finch FoxP2 3′-UTR, we used an approach combining bioinformatic sequence analysis with PCR-based cDNA amplification. By searching the National Center for Biotechnology Information (NCBI) database, we obtained a partial sequence of 327 nt, immediately downstream from the zebra finch FoxP2 stop codon (Haesler et al., 2004; Teramitsu et al., 2004). Based on this sequence, we designed a FoxP2 3′-UTR 5′ PCR primer. Other PCR primers were designed based on genomic sequences downstream from the protein coding region, taking advantage of the fact that segments of the zebra finch sequence are highly homologous to the sequence of the last exon (exon 17) of the human FOXP2 gene. We performed PCR amplification using cDNA templates made from embryonic zebra finch brains and assembled a cDNA fragment of 3846 nt (see Materials and Methods). This cDNA fragment mapped to the zebra finch FoxP2 locus, from the first nucleotide immediately downstream of the stop codon and continuing downstream without interruption, and contained a polyadenylation signal at the 3′-end. The 5′-most portion of this fragment aligned perfectly with the first 327 nt of the FoxP2 3′-UTR reported previously (Haesler et al., 2004). This 3846 nt cDNA fragment shared ∼96% sequence identity with the 3′-UTR portion of exon 17 of the human FOXP2 gene. We also searched for, but did not find, sequences of exon-intron junctions in the 3846 nt sequence segment. Thus, this 3846 nt sequence likely represents the zebra finch FoxP2 3′-UTR. We used this sequence for subsequent miRNA binding site analysis.
Binding of an miRNA to its target genes requires complementary base pairing between specific sequence elements in a 3′-UTR and nucleotides 2–8 of the 5′ region of an miRNA, commonly referred to as the seed sequence (Lewis et al., 2003; John et al., 2004; Brennecke et al., 2005; Krek et al., 2005; Bartel, 2009). We searched the FoxP2 3′-UTR for sequences complementary to seed sequences of miRNAs using the commonly used target prediction software package Targetscan (Friedman et al., 2009). One hundred and ninety-three zebra finch miRNAs, which were recently annotated by profiling miRNA expression in zebra finch brain and other tissues (Luo et al., 2012), were included in the binding site search. Sequence analysis revealed numerous miRNA binding sites in the 3846 nt FoxP2 3′-UTR, including two binding sites for miR-9 and one binding site for miR-140-5p, respectively, located 826, 1472, and 51 nt downstream from the stop codon of the protein coding region (Fig. 1A). Both miR-9 and miR-140-5p are known to be expressed in the striatum of mammals, a region where FOXP2 is expressed (Lai et al., 2003; Landgraf et al., 2007; Pena et al., 2009). Because conserved base pairing between miRNA seed sequences and their binding sites are often used to eliminate false-positive predictions (Krek et al., 2005; Lewis et al., 2005) and the sequence of the FOXP2 gene is highly conserved through evolution, we also searched for binding sites for miR-9 and miR-140-5p in the 3′-UTRs of the FOXP2 genes of chickens, mice, chimpanzees, and humans. As expected, the two miR-9 binding sites and the miR-140-5p binding site and their surrounding sequences were conserved in the FOXP2 3′-UTRs at orthologous positions in these vertebrate species (Fig. 1B,C). Together, this molecular cloning and sequence analysis results suggest that FoxP2 is a likely target of miR-9 and miR-140-5p. The conservation of miR-9 and miR-140-5p binding sites in the FOXP2 3′-UTRs among higher vertebrates provides further evidence that the regulatory relationships between FOXP2 and miR-9 and miR-140-5p are likely to be authentic and biologically functional.
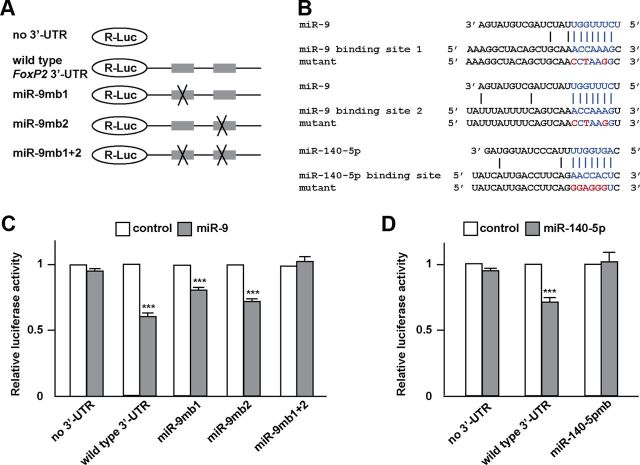
Figure 1.

The zebra finch FoxP2 3′-UTR contains two miR-9 binding sites and one miR-140-5p binding site. A, The two miR-9 binding sites are 826 and 1472 nt downstream from the stop codon and the miR-140-5p binding site is 51 nt downstream from the stop codon. B, C, The sequences of the miR-9 binding sites and the miR-140-5p binding site are conserved in higher vertebrates. The seed sequences of miR-9 and miR-140-5p and their corresponding binding sites are highlighted in blue.
miR-9 and miR-140-5p target specific sequences in the FoxP2 3′-UTR
We used luciferase reporter assays to experimentally determine whether miR-9 and miR-140-5p target their predicted binding sites in the FoxP2 3′-UTR. We inserted the 3846 nt zebra finch FoxP2 3′-UTR sequence downstream of the luciferase gene in a dual luciferase reporter plasmid vector, which allowed us to assess the effects of miR-9 or miR-140-5p on the FoxP2 3′-UTR based on luciferase activity in transfected cells (Fig. 2A). SH-SY5Y cells were used because these cells do not endogenously express FOXP2 (data not shown). We found that miR-9 had no effect on a control construct that did not contain the FoxP2 3′-UTR. In contrast, when co-transfected with the construct containing the FoxP2 3′-UTR (Luc-FoxP2 3′-UTR), miR-9 repressed luciferase activity by 40% compared with the negative control miRNA (t(4) = 18.33, p < 0.001, Student's t test) (Fig. 2C). To determine whether the downregulatory effect of miR-9 was sequence-specific, we made three mutant luciferase reporter constructs (see Materials and Methods), carrying mutations in miR-9 binding site 1 (miR-9mb1), binding site 2 (miR-9mb2), and binding sites 1 plus 2 (miR-9mb1 + 2) (Fig. 2B), and tested these mutant constructs in SH-SY5Y cells. The targeting effects of miR-9 were partially relieved when either binding site 1 or binding site 2 was mutated (t(4) = 9.38, p < 0.001 for binding site 1 mutant; t(4) = 16.5, p < 0.001 for binding site 2 mutant, Student's t test). The targeting effect was completely abolished when both binding sites were mutated (p = 0.445) (Fig. 2C). In a similar experiment, we found that when co-transfected with a construct containing a wild-type FoxP2 3′-UTR, miR-140-5p repressed luciferase activity by 27% compared with the control miRNA (t(4) = 10.79, p < 0.001, Student's t test). This downregulatory effect was completely abolished when the miR-140-5p binding site was mutated (p = 0.65) (Fig. 2D).
Figure 2.
miR-9 and miR-140-5p target specific sequences in the FoxP2 3′-UTR. A, Luciferase reporter constructs containing no FoxP2 3′-UTR, a wild-type FoxP2 3′-UTR, and FoxP2 3′-UTRs with miR-9 binding site 1, binding site 2, or binding sites 1 + 2 mutated. B, Mutated binding sites of miR-9 and miR-140-5p (mutated nucleotides in red). C, miR-9 significantly repressed luciferase activity when co-transfected with a luciferase reporter construct containing a wild-type FoxP2 3′-UTR compared with a control miRNA duplex (***p < 0.001). Repression was partially abolished when miR-9 binding site 1 (miR-9mb1) or binding site 2 (miR-9mb2) was mutated (***p < 0.001 for both mutants), and repression was completely abolished when both binding sites (miR-9mb1 + 2) were mutated (p = 0.445). D, miR-140-5p repressed luciferase activity when co-transfected with a luciferase reporter construct containing a wild-type FoxP2 3′-UTR compared with a control miRNA duplex (***p < 0.001), and repression was abolished when the miR-140-5p binding site (miR-140-5pmb) was mutated (p = 0.65). Transfection was performed three times, thus n = 3 for each experimental group. To control for transfection efficiency, R-Luc activity was normalized to the internal control f-Luc activity. Data are mean ± SEM. Statistical analysis was performed with Student's t test.
miR-9 and miR-140-5p directly regulate FoxP2 protein and mRNA expression
We tested whether miR-9 and miR-140-5p directly regulate zebra finch FoxP2 protein and mRNA expression in cell culture. We cloned the full-length zebra finch FoxP2 protein coding region (isoform I) by PCR amplification using cDNA templates made from embryonic zebra finch brains and primers designed based on previously published sequences (Haesler et al., 2004; Teramitsu et al., 2004). We inserted the protein coding region sequence together with the 3846 nt wild-type 3′-UTR into the pcDNA3 expression vector to obtain a plasmid construct pcDNA3-FoxP2-3′-UTRwt, containing the full-length cDNA. When this plasmid was transfected into SH-SY5Y cells, a FoxP2 protein of ∼80 kDa was detected using an anti-human FOXP2 antibody by Western blot analysis. Using the same antibody, a protein of the same size was also detected in tissue lysates obtained from Area X of a 45-d-old male zebra finch (Fig. 3A), consistent with previous findings by others (Haesler et al., 2004; Miller et al., 2008). We co-transfected the plasmid pcDNA3-FoxP2-3′-UTRwt with miR-9 into SH-SY5Y cells, and found that miR-9 significantly downregulated zebra finch FoxP2 protein expression compared with a control miRNA duplex (t(4) = 35.69, p < 0.01, Student's t test) (Fig. 3B,D). We also made a plasmid construct, pcDNA3-FoxP2-miR-9mb1 + 2, in which both miR-9 binding sites in the FoxP2 3′-UTR were mutated. When this plasmid was co-transfected with miR-9 into SH-SY5Y cells, the downregulatory effect of miR-9 on FoxP2 protein expression was completely abolished (p = 0.944) (Fig. 3B,D). In a similar experiment, miR-140-5p downregulated FoxP2 protein expression when co-transfected with a construct containing a wild-type 3′-UTR (t(4) = 7.73, p < 0.01, Student's t test), and the downregulatory effect was completely abolished when the miR-140-5p binding site was mutated (p = 0.56) (Fig. 3C,E). Additionally, in separate experiments run in parallel, we tested miR-124, a widely expressed brain-specific miRNA that does not have a binding site in the FoxP2 3′-UTR. We found that miR-124 had no effect on FoxP2 protein expression compared with the control miRNA whether the expression construct contained a wild-type or a mutant 3′-UTR (p = 0.774 for wild-type 3′-UTR; p = 0.776 for miR-9 binding site mutant; p = 0.61 for miR-140-5p binding site mutant, Student's t test) (Fig. 3B–E).
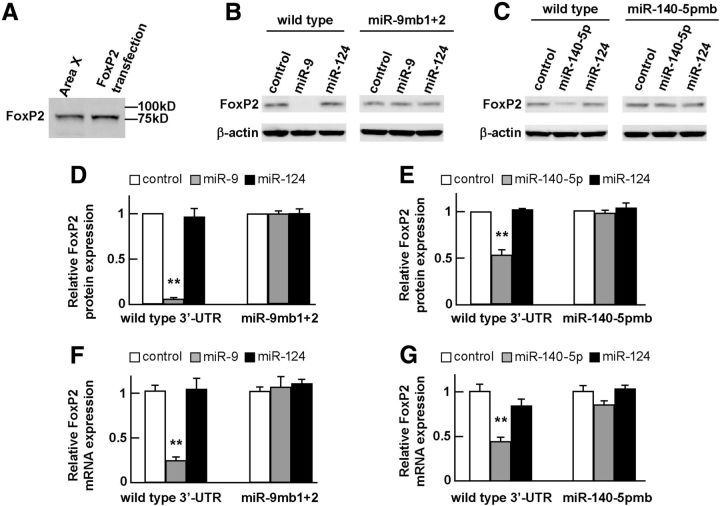
Figure 3.
miR-9 and miR-140-5p downregulate FoxP2 protein and mRNA expression. A, Western blot detection of an ∼80 kd FoxP2 protein from Area X tissue of a 45-d-old male zebra finch and from SH-SY5Y cells transfected with the plasmid pcDNA3-FoxP2-3′-UTRwt, which contained the FoxP2 protein coding region and a wild-type 3′-UTR. B, C, Representative Western blot images showing that miR-9 and miR-140-5p downregulated FoxP2 protein expression in co-transfection assays. β-actin was used as a loading control. D, Quantification of Western blot experiments shows that miR-9, but not miR-124, downregulated FoxP2 protein compared with a control miRNA duplex (**p < 0.01) and that downregulation was abolished when the two miR-9 binding sites in the FoxP2 3′-UTR were mutated (miR-9mb1 + 2) (p = 0.944). E, In similar transfection experiments, miR-140-5p downregulated FoxP2 protein (**p < 0.01), and downregulation was abolished when the miR-140-5p binding site was mutated (miR-140-5pmb) (p = 0.56). F, G, miR-9 and miR-140-5p also downregulated FoxP2 mRNA expression as measured by RT-qPCR (**p < 0.01 for miR-9; **p < 0.01 for miR-140-5p), and downregulation was abolished when the binding sites of miR-9 or miR-140-5p were mutated (p = 0.769 for miR-9; p = 0.085 for miR-140-5p). For RT-qPCR, β-actin was used as an internal control. Transfection was performed three times, thus n = 3 for each experimental group. Data are mean ± SEM; statistical analysis was performed with Student's t test.
We also performed real-time qPCR (RT-qPCR) to measure the effects of miR-9 and miR-140-5p on FoxP2 mRNA expression in SH-SY5Y cells. We found that miR-9 downregulated FoxP2 mRNA expression when it was co-transfected with the plasmid construct containing a wild-type FoxP2 3′-UTR (pcDNA3-FoxP2-3′-UTRwt) compared with a control miRNA (t(4) = 10.39, p < 0.01, Student's t test) (Fig. 3F). The ability of miR-9 to downregulate FoxP2 mRNA expression was abolished when both miR-9 binding sites in the FoxP2 3′-UTR were mutated (p = 0.769) (Fig. 3F). Similarly, we found that miR-140-5p downregulated FoxP2 mRNA expression when it was co-transfected with the plasmid construct containing a wild-type FoxP2 3′-UTR (pcDNA3-FoxP2-3′-UTRwt) compared with a control miRNA (t(4) = 6.70, p < 0.01, Student's t test), and the downregulation effect was abolished when the miR-140-5p binding site in the FoxP2 3′-UTR was mutated (p = 0.085) (Fig. 3G). Again, in these co-transfection assays, miR-124 had no effect on FoxP2 mRNA expression regardless of whether the expression construct contained a wild-type or a mutant 3′-UTR (p = 0.825 for wild-type 3′-UTR; p = 0.365 for miR-9 binding site mutant; p = 0.9225 for miR-140-5p binding site mutant, Student's t test) (Fig. 3F,G).
The expression of miR-9 and miR-140-5p in Area X of the zebra finch brain is regulated during vocal development
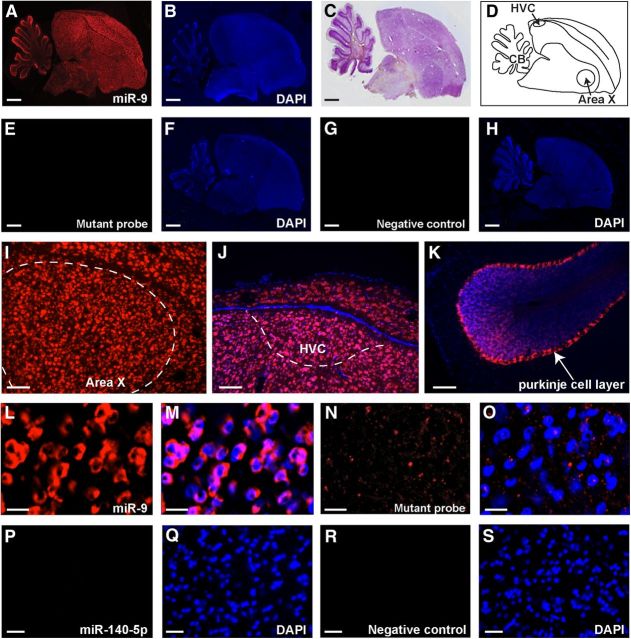
In the zebra finch brain, Area X is a song nucleus located in the dorsal–medial striatum. It is part of the cortico-striatal-thalamo-cortical loop that processes song related sensory-motor learning in juvenile zebra finches (Sohrabji et al., 1990; Scharff and Nottebohm, 1991; Doupe et al., 2005). To establish functional roles of miR-9 and miR-140-5p in the zebra finch brain, we performed in situ hybridization on brain sections of adult male zebra finches (120 d of age or older) using LNA-modified miRNA detection probes. We found that miR-9 was abundantly expressed in most brain regions, including the cortical regions, striatum, dorsal thalamus, and the cerebellum (Fig. 4A–O). Notably, the expression pattern of miR-9 overlapped, but was distinct from that of FoxP2. In the zebra finch brain, FoxP2 is prominently expressed in the striatum, dorsal thalamus, and the Purkinje cell layer of the cerebellum (Haesler et al., 2004; Teramitsu et al., 2004). In addition to these regions, miR-9 is also expressed in regions where low or little FoxP2 is expressed. Within the song control circuit, miR-9 is expressed in major song system nuclei including Area X, HVC (used as a proper name), the vocal motor nucleus robustus arcopallialis (RA), and the lateral magnocellular nucleus of the anterior nidopallium (LMAN). In Area X, a large majority of cells exhibited miR-9 expression (Fig. 4L,M). Despite the fact that we used a LNA-modified probe labeled with digoxygenin at both 5′ and 3′-ends, we could not reliably detect miR-140-5p signal in Area X or the cerebellum, probably due to its low expression.
Figure 4.
In situ hybridization showing the expression of miR-9 in Area X of adult male zebra finches. A, A sagittal brain section hybridized with an miR-9 detection probe (Cy3). B, The same brain section as in A stained with DAPI. C, Cresyl violet staining of an adjacent brain section. D, A schematic diagram indicating the positions of Area X and HVC in a sagittal section of a zebra finch brain. E, G, A mutated miR-9 detection probe and a negative control probe detected no miR-9 signal. F, H, DAPI staining of the same brain sections as in E and G. I–K, miR-9 signal in Area X, HVC, and cerebellum Purkinje cell layer. L, A higher-magnification image showing miR-9 signal in Area X cells. M, The same section as in L showing DAPI-stained nuclei. Note the cytoplasmic localization of the miR-9 signal. N, A higher-magnification image showing that no signal was detected using a mutated miR-9 probe. O, The same section as in N stained with DAPI. P, Area X region hybridized with an miR-140-5p detection probe. Q, DAPI staining of the same section as in P. R, Area X hybridized with a negative control probe. S, DAPI staining of the same section as in R. In all images, Cy3 fluorescence signals are in red and DAPI signals are in blue. Scale bars: A–H, 1 mm; I–K, 100 μm; L–O, 5 μm; P–S, 10 μm.
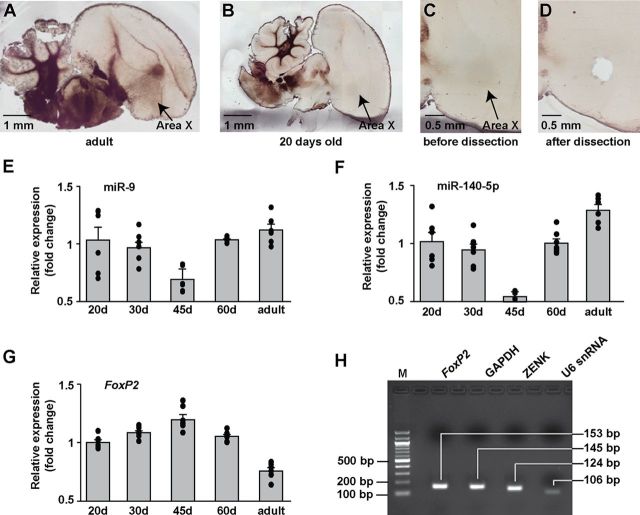
Male juvenile zebra finches begin to sing at ∼30 d of age; they imitate their tutor's song and use auditory feedback to guide their vocal motor outputs. By 90 d, birds have reached sexual maturity, and their songs become stabilized and resemble their tutor's song (Immelmann, 1969; Eales, 1985; Tchernichovski et al., 2001). During this time, a cascade of molecular, cellular, and physiological changes occur in the song system (for review, see Doupe et al., 2004; Mooney, 2009; Kirn, 2010; White, 2010). We examined the expression levels of miR-9 and miR-140-5p in Area X of male zebra finches at 20, 30, 45, 60, and 100 d of age, as these ages represent milestone stages during vocal development. Because singing behavior may activate gene expression, to minimize potential compounding effects, all birds were collected in the morning after room lights were switched on, but before onset of singing. We dissected out Area X tissues (Fig. 5A–D), isolated RNA, and performed RT-qPCR to quantitatively measure the expression levels of miR-9 and miR-140-5p in Area X.
Figure 5.
The expression of miR-9 and miR-140-5p in Area X is regulated during vocal development. A, B, Sagittal brain sections of an adult and a 20-d-old juvenile male zebra finch showing the song nucleus Area X. C, D, The same juvenile brain section as in B before and after dissecting out Area X. E–G, The relative expression levels of miR-9, miR-140-5p, and FoxP2 in Area X in 20-, 30-, 45-, and 60-d-old and adult male zebra finches as measured by RT-qPCR. The relative expression levels in animals 30 d and older were normalized to the 20 d group. U6 small RNA was used as a reference for miR-9 and miR-140-5p, and GAPDH was used as a reference for FoxP2. Neither U6 nor GAPDH changed expression during development. n = 6 for the 20 d, 45 d, and adult groups, and n = 7 for the 30 d and 60 d groups. Data are mean ±SEM; p < 0.001 for miR-9, miR-140-5p, and FoxP2 (one-way ANOVA). H, Gel electrophoresis shows that all qPCRs resulted in single bands with correct sizes of 153 nt for FoxP2, 124 nt for ZENK, 145 nt for GAPDH, and 106 nt for U6.
We found that the expression levels of miR-9 in Area X followed a U-shaped pattern during vocal developmental. At 20 d of age, when Area X is well formed but before juveniles begin to sing, miR-9 was already prominently expressed in Area X; at 45 d of age, when birds are actively learning to sing, miR-9 was at its lowest. As birds practice their song, miR-9 expression increased and reached its highest level when birds reach adulthood and their songs become crystallized (F(4,27) = 7.35, p < 0.001, one-way ANOVA) (Fig. 5E). The expression levels of miR-140-5p, parallel to that of miR-9, decreased from 20 to 45 d of age, and then increased, reaching its highest level in adults (F(4,27) = 26.79, p < 0.001, one-way ANOVA) (Fig. 5F). We also examined FoxP2 mRNA expression in Area X of the same age groups. Consistent with previous findings by others (Haesler et al., 2004), FoxP2 expression increased from 20 to 45 d of age and then decreased as birds matured into adulthood (F(4,27) = 36.1, p < 0.001, one-way ANOVA) (Fig. 5G). We further applied Tukey's honest significant difference (Tukey's HSD) method for multiple comparisons among different age groups. The expression level of miR-9 at 45 d was significantly different from those for other ages (p < 0.01 for 45 d vs 20 d; p < 0.05 for 45 d vs 30 d; p < 0.01 for 45 d vs 60 d; and p < 0.001 for 45 d vs adults, Tukey's HSD). The expression levels of miR-140-5p for the 45 d and the adult groups were significantly different from those for other age groups (p < 0.001 for comparisons between the 45 d and all other age groups; p < 0.01 for adults vs 20 d; p < 0.001 for adults vs 30 d; p < 0.01 for adults vs 60 d, Tukey's HSD). The expression levels of FoxP2 for the 45 d and the adult groups were significantly different from those for other age groups (p < 0.001 for 45 d vs 20 d; p < 0.05 for 45 d vs 30 d; p < 0.01 for 45 d vs 60 d; p < 0.001 for 45 d vs adults; p < 0.001 for comparisons between the adult and all other age groups, Tukey's HSD). The changes in the expression of miR-9 and miR-140-5p in the opposite direction of that of FoxP2 during vocal development support the roles of miR-9 and miR-140-5p in regulating FoxP2. We assessed the specificity of the RT-qPCR products by gel electrophoresis and found that all PCRs resulted in single bands with correct sizes (153 nt for FoxP2, 124 nt for ZENK, 145 nt for GAPDH, and 106 nt for U6) (Fig. 5H). We also sequenced the PCR amplified DNA fragments corresponding to FoxP2, ZENK, GAPDH, and U6, and obtained correct sequences (data not shown).
The expression of miR-9 and miR-140-5p in Area X is regulated as a function of the social context of singing behavior
The higher expression of miR-9 and miR-140-5p in Area X of adult male zebra finches compared with juveniles implicates these miRNAs in the maintenance of the adult song circuit and in song performance. Teramitsu and White have shown that FoxP2 is downregulated in Area X in adult males singing undirected songs (2006). The molecular mechanism of this social context-dependent FoxP2 downregulation is not known. Because miRNAs generally downregulate gene expression, we reasoned that if FoxP2 downregulation is mediated by miR-9 and miR-140-5p, these miRNAs should be upregulated as a function of the same song behavior. To test whether this is the case, we examined the expression of miR-9 and miR-140-5p in Area X of adult male zebra finches singing directed or undirected songs.
Following behavioral paradigms previously used by other groups (Jarvis et al., 1998; Teramitsu and White, 2006; Kojima and Doupe, 2011), we collected three groups of adult male zebra finches: (1) nonsinging controls; (2) birds that sang female-directed songs for 2 h; and (3) birds that sang undirected songs for 2 h. Birds in Group 2 sang >90% female-directed songs, and birds in Group 3 sang 100% undirected songs. Because gene expression levels often correlate with the amount of songs produced (Jarvis and Nottebohm, 1997; Li et al., 2000), to avoid confounding effects introduced by variations in the amount of singing, birds were only accepted into the singing groups if they sang 100–150 bouts per hour during the 2 h observation period. After behavioral observation, brains were collected and Area X tissues were dissected out. We isolated total RNAs and performed RT-qPCR to measure the expression levels of miR-9 and miR-140-5p. We found that miR-9 expression increased significantly in Area X of undirected singers compared with nonsinging controls (t(15) = −4.40, p < 0.001, Student's t test). In contrast, miR-9 expression remained constant in female-directed singers (p = 0.82) (Fig. 6A). In the same set of birds, the expression of miR-140-5p increased significantly in Area X of undirected singers compared with nonsinging controls (t(15) = −7.82, p < 0.001, Student's t test), but did not change in female-directed singers (p = 0.40) (Fig. 6B). In the same birds, consistent with previous findings (Teramitsu and White, 2006), FoxP2 mRNA expression in Area X decreased significantly in undirected singers compared with nonsinging controls (t(15) = 5.49, p < 0.001, Student's t test), but did not change in female-directed singers (p = 0.325) (Fig. 6C).
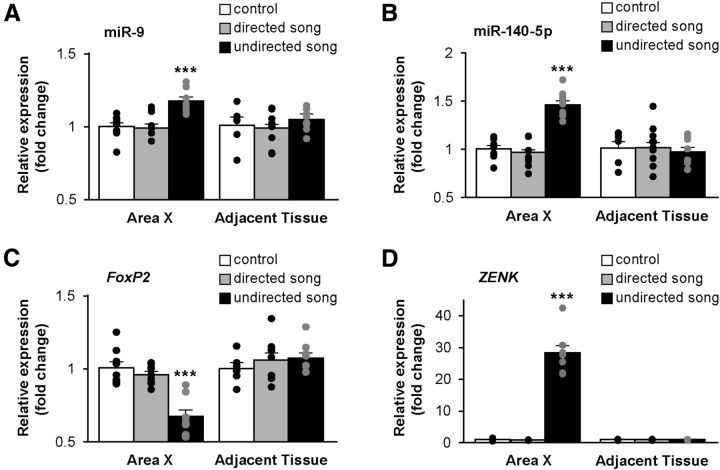
Figure 6.
miR-9 and miR-140-5p are upregulated in Area X of adult male zebra finches singing undirected songs. A, B, Singing undirected but not singing directed songs upregulated the expression of miR-9 (***p < 0.001 for the undirected song group; p = 0.82 for the directed song group) and miR-140-5p (***p < 0.001 for the undirected song group; p = 0.40 for the directed song group) in Area X of adult male zebra finches. C, FoxP2 mRNA expression in Area X was downregulated in adult males singing undirected songs, but was unchanged in birds singing directed songs (***p < 0.001 for the undirected songs group; p = 0.325 for the directed song group). D, The expression of the immediate early gene ZENK in Area X was upregulated in males singing undirected songs (***p < 0.001 for the undirected song group; p = 0.325 for the directed song group). Note there was no change in the expression of miR-9, miR-140-5p, FoxP2, and ZENK in striatal tissues immediately adjacent to Area X in the same set of birds (A–D). The relative expression levels of miR-9, miR-140-5p, FoxP2, and ZENK in all samples were measured by RT-qPCR. U6 small RNA was used as an internal control for miR-9 and miR-140-5p, and GAPDH was used as an internal control for FoxP2 and ZENK. Neither U6 nor GAPDH changed expression by song behavior. n = 8 for the undirected song group; n = 9 for both the nonsinging and the directed song groups. Data are mean ± SEM. Statistical analysis was performed using Student's t test.
The immediate early gene ZENK is known to be highly activated in Area X of adult males upon singing undirected songs (Jarvis et al., 1998). We also tested ZENK mRNA expression in the same set of samples. Consistent with previous findings, ZENK expression was upregulated in males that sang undirected songs compared with nonsinging controls (t(15) = −12.64, p < 0.001, Student's t test), but was not upregulated in birds that sang female-directed songs (p = 0.1396) (Fig. 6D). To further assess whether singing-induced changes in miRNA expression were specific to Area X, we tested whether miR-9 and miR-140-5p were regulated in striatal tissues immediately adjacent to Area X in the same set of birds. We found that the expression of miR-9 and miR-140-5p did not change whether the birds sang or not. Similarly, neither the expression of FoxP2 nor the expression of ZENK changed in striatal tissues adjacent to Area X upon singing either directed or undirected songs (Fig. 6A–D).
Discussion
Our results highlight the importance of sequence motifs in the FoxP2 3′-UTR and the roles of miRNAs in regulating FoxP2 expression. The binding sites of both miR-9 and miR-140-5p in the FoxP2 3′-UTR are conserved in higher vertebrates, including mice, chimpanzees, and humans, suggesting that the regulatory relationships between miR-9 and miR-140-5p and FoxP2 are evolutionarily conserved. The ∼4-kb-long FoxP2 3′-UTR provides numerous opportunities for this gene to be regulated by multiple miRNAs. Recently, Clovis et al. (2012) reported that miR-9 and miR-132 regulate foxp2 expression in mouse embryonic brain. In a separate study, we also found that miR-9 and miR-140-5p regulated the human FOXP2 gene (Li, unpublished data). In humans, a heterozygous missense mutation in the FOXP2 protein coding region (e.g., R553H) results in a FOXP2 protein with compromised DNA binding and transactivation activities. The reduced functional dosage of the FOXP2 protein presumably is the etiological basis underlying speech and language impairments (Lai et al., 2001; Vernes et al., 2006). Our results and the results of Clovis et al. (2012) demonstrating that FOXP2 expression can be quantitatively regulated by multiple miRNAs raise the possibility that sequence variations in the FOXP2 3′-UTR, particularly in miRNA binding sites, contribute to dysregulation and/or functional variations in FOXP2 expression.
The amino acid sequences of the FOXP2 protein are highly conserved; the human FOXP2 protein differs from primate, mouse, and zebra finch versions by two, three, and eight amino acids, respectively (Lai et al., 2001; Zhang et al., 2002; Haesler et al., 2004; Teramitsu et al., 2004). The two amino acid residues that are unique to the human FOXP2 protein are thought to have been evolutionarily selected, and are thought to contribute critically to the unique speech and language capacity of humans (Enard et al., 2002, 2009; Enard, 2011). The nucleotide sequences in the FOXP2 3′-UTRs in humans and zebra finches share 96% sequence identity, but are less conserved compared with the amino acid sequences. In addition to evolutionarily conserved miRNAs (e.g., miR-9 and miR-140-5p), each animal species has evolved its own unique set of miRNAs. Some of these may regulate the FOXP2 gene in a species-specific manner. Thus, subtle sequence variations in the FOXP2 3′-UTRs and differences in miRNA repertoires in combination may influence species specific vocal capabilities.
miR-9 is one of the most abundantly expressed brain-specific vertebrate miRNAs (Lagos-Quintana et al., 2001; Landgraf et al., 2007; Luo et al., 2012), and it is prominently expressed in Area X and other regions in the zebra finch brain (Fig. 4). In contrast, we could not reliably detect miR-140-5p signal in Area X by in situ hybridization, possibly due to its low expression. When RT-qPCR was used to measure miR-9 and miR-140-5p expression levels in adult Area X, the Ct values (defined as the number of amplification cycles needed for reaching a detectable threshold) for miR-9 and miR-140-5p were 22.11 ± 0.02 and 28.78 ± 0.04 (n = 9), respectively, indicating that the expression level of miR-140-5p was much lower than that of miR-9. Similarly, when RNA-sequencing (RNA-seq) was used to measure miRNA expression levels in Area X, miR-9 and miR-140-5p were represented by 75,139 and 357 reads respectively (X. C. Li, unpublished results). The expression pattern of miR-9 overlaps but differs from the expression pattern of FoxP2. For example, miR-9 is abundantly expressed in HVC, where little FoxP2 mRNA is detected (Haesler et al., 2004; Teramitsu et al., 2004), suggesting that functions of miR-9 are not restricted to Area X or to regulating FoxP2. Rather, miR-9 (and possibly miR-140-5p) may have broad functions in the zebra finch brain by regulating target genes in addition to FoxP2.
The expression levels of miR-9 and miR-140-5p in Area X are opposite from that of FoxP2 (Haesler et al., 2004; Teramitsu et al., 2004) (Fig. 5G): lower at 45 d and higher in adults, consistent with their roles in downregulating FoxP2 in Area X. Experimentally reducing FoxP2 expression in Area X impairs neuronal dendritic development and song learning in juvenile zebra finches (Haesler et al., 2007; Schulz et al., 2010). It is likely that miR-9 and miR-140-5p, via regulating FoxP2 and possibly other genes, play a role in modulating dendritic structure and vocal learning. Interestingly, neither the expression of miR-9 and miR-140-5p nor the expression of FoxP2 in Area X is linear during vocal development. It appears that 45 d of age is a turning point, possibly reflecting two overlapping but temporally distinct phases during the structural and functional development in Area X. We note that the fact that the expression of miR-9 and miR-140–5p is in the opposite direction of the expression of FoxP2 is also consistent with the possibility that miR-9 and miR-140-5p are negatively regulated by FoxP2. Recent studies have indicated that miRNAs are among the downstream genes regulated by FOXP2 (Vernes et al., 2011). Further mapping of the promoter sequences of miR-9 and miR-140-5p should reveal molecular mechanisms controlling the expression of these two miRNAs.
The decrease in FoxP2 expression when juveniles mature into adulthood occurs relatively slowly (over weeks or months), whereas the downregulation of FoxP2 in adult males singing undirected songs occurs relatively rapidly (within hours). These different temporal patterns argue that FoxP2 might be regulated by distinct molecular mechanisms in juveniles and adults, and that the downregulation of FoxP2 in undirected singers may be mediated via mRNA degradation rather than transcriptional repression. The upregulation of miR-9 and miR-140-5p in the same birds is consistent with their roles in downregulating FoxP2 posttranscriptionally. Activity-dependent upregulation or downregulation of gene expression in the song system has been observed when birds sing or hear songs (Mello et al., 1995; Jarvis and Nottebohm, 1997; Kimpo and Doupe, 1997; Li et al., 2000; Hilliard et al., 2012). While gene activation is often attributed to Ca2+ influx triggered by neuronal firing (Finkbeiner and Greenberg, 1998; Tao et al., 1998), the mechanisms of gene downregulation are not clear. Song-induced miRNA-mediated mRNA degradation provides a means to rapidly adjust gene expression levels in response to behavioral or environmental changes.
Compared with a directed song, syllables in an undirected song are more variable, and they are accompanied by increased neural activities and more variable neuronal firing patterns in Area X (Jarvis et al., 1998; Hessler and Doupe, 1999; Teramitsu and White, 2006; Kojima and Doupe, 2011). Directed singing is thought to represent a stereotyped “performance” of a learned song, whereas undirected singing reflects “off-duty” practice of a song with higher motor variability, which allows exploratory vocal space (Jarvis et al., 1998; Kojima and Doupe, 2011). The downregulation of FoxP2 in Area X during undirected singing is postulated to relieve repression of downstream genes related to exploratory vocal behavior (Teramitsu and White, 2006). Expanding upon previous findings, our observations reveal a miRNA-FoxP2 gene regulation network that is dynamically regulated each time birds sing a socially engaged song. miR-9 has been implicated in vertebrate embryonic brain development in several recent studies (Shibata et al., 2008; Gao, 2010; Clovis et al., 2012). Broadening the roles for miR-9 and miR-140-5p in adult brains, our results suggest that the functional state of a mature neural circuit for a social vocal behavior is constantly influenced by its ongoing behavioral output via miRNA-mediated gene regulation. We note that FoxP2 expression in Area X is higher in juveniles when the song circuit is more plastic than in adults. It is possible that as the song circuit matures, accompanied by diminished plasticity, the roles of FoxP2 transition from promoting circuit plasticity to maintaining circuit stability. The miRNA-FoxP2 regulatory network in adult Area X may be part of the homeostatic maintenance of the dynamic, yet relatively stable, mature circuit. This process, while replenishing cellular components, may allow moderate circuit plasticity. Further work will be needed to address why miR-9, miR-140-5p, and FoxP2 are regulated as a function of undirected singing, but not directed singing, and how the miRNA-FoxP2 regulatory network contributes to variability in song and neuronal firing patterns.
In summary, our results reveal that two miRNAs, miR-9 and miR-140-5p, regulate FoxP2 expression posttranscriptionally by targeting its 3′-UTR. We also provide evidence, for the first time, that the expression of these two miRNAs is regulated during vocal development and is regulated as a function of the social context of singing behavior in a basal ganglia nucleus required for vocal learning in zebra finches. In mammals, both miR-9 and miR-140-5p are known to be expressed in the striatum (Landgraf et al., 2007; Pena et al., 2009), a region important for speech and language in humans and where FOXP2 is expressed (Lai et al., 2003). As the binding sites for both miR-9 and miR-140-5p in the FOXP2 3′-UTR are evolutionarily conserved, these miRNAs are likely to play a role, via regulating FOXP2, in the development and function of speech and language in humans.
Footnotes
This work was supported by NIH Grant MH081254 and a Young Investigator Award by the Brain Behavior Research Foundation to X.C.L.
The authors declare no competing financial interests.
References
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bottjer SW, Halsema KA, Brown SA, Miesner EA. Axonal connections of a forebrain nucleus involved with vocal learning in zebra finches. J Comp Neurol. 1989;279:312–326. doi: 10.1002/cne.902790211. [DOI] [PubMed] [Google Scholar]
- Brennecke J, Stark A, Russell RB, Cohen SM. Principles of microRNA-target recognition. PLoS Biol. 2005;3:e85. doi: 10.1371/journal.pbio.0030085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clovis YM, Enard W, Marinaro F, Huttner WB, De Pietri Tonelli D. Convergent repression of Foxp2 3′UTR by miR-9 and miR-132 in embryonic mouse neocortex: implications for radial migration of neurons. Development. 2012;139:3332–3342. doi: 10.1242/dev.078063. [DOI] [PubMed] [Google Scholar]
- Doupe AJ, Kuhl PK. Birdsong and human speech: common themes and mechanisms. Annu Rev Neurosci. 1999;22:567–631. doi: 10.1146/annurev.neuro.22.1.567. [DOI] [PubMed] [Google Scholar]
- Doupe AJ, Solis MM, Kimpo R, Boettiger CA. Cellular, circuit, and synaptic mechanisms in song learning. Ann N Y Acad Sci. 2004;1016:495–523. doi: 10.1196/annals.1298.035. [DOI] [PubMed] [Google Scholar]
- Doupe AJ, Perkel DJ, Reiner A, Stern EA. Birdbrains could teach basal ganglia research a new song. Trends Neurosci. 2005;28:353–363. doi: 10.1016/j.tins.2005.05.005. [DOI] [PubMed] [Google Scholar]
- Eales LA. Song learning in zebra finches: some effects of song model availability on what is learnt and when. Anim Behav. 1985;33:1293–1300. doi: 10.1016/S0003-3472(85)80189-5. [DOI] [Google Scholar]
- Enard W. FOXP2 and the role of cortico-basal ganglia circuits in speech and language evolution. Curr Opin Neurobiol. 2011;21:415–424. doi: 10.1016/j.conb.2011.04.008. [DOI] [PubMed] [Google Scholar]
- Enard W, Przeworski M, Fisher SE, Lai CS, Wiebe V, Kitano T, Monaco AP, Pääbo S. Molecular evolution of FOXP2, a gene involved in speech and language. Nature. 2002;418:869–872. doi: 10.1038/nature01025. [DOI] [PubMed] [Google Scholar]
- Enard W, Gehre S, Hammerschmidt K, Hölter SM, Blass T, Somel M, Brückner MK, Schreiweis C, Winter C, Sohr R, Becker L, Wiebe V, Nickel B, Giger T, Müller U, Groszer M, Adler T, Aguilar A, Bolle I, Calzada-Wack J, et al. A humanized version of Foxp2 affects cortico-basal ganglia circuits in mice. Cell. 2009;137:961–971. doi: 10.1016/j.cell.2009.03.041. [DOI] [PubMed] [Google Scholar]
- Feuk L, Kalervo A, Lipsanen-Nyman M, Skaug J, Nakabayashi K, Finucane B, Hartung D, Innes M, Kerem B, Nowaczyk MJ, Rivlin J, Roberts W, Senman L, Summers A, Szatmari P, Wong V, Vincent JB, Zeesman S, Osborne LR, Cardy JO, et al. Absence of a paternally inherited FOXP2 gene in developmental verbal dyspraxia. Am J Hum Genet. 2006;79:965–972. doi: 10.1086/508902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkbeiner S, Greenberg ME. Ca2+ channel-regulated neuronal gene expression. J Neurobiol. 1998;37:171–189. doi: 10.1002/(SICI)1097-4695(199810)37:1<171::AID-NEU13>3.0.CO%3B2-H. [DOI] [PubMed] [Google Scholar]
- Fisher SE, Scharff C. FOXP2 as a molecular window into speech and language. Trends Genet. 2009;25:166–177. doi: 10.1016/j.tig.2009.03.002. [DOI] [PubMed] [Google Scholar]
- Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao FB. Context-dependent functions of specific microRNAs in neuronal development. Neural Dev. 2010;5:25. doi: 10.1186/1749-8104-5-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haesler S, Wada K, Nshdejan A, Morrisey EE, Lints T, Jarvis ED, Scharff C. FoxP2 expression in avian vocal learners and nonlearners. J Neurosci. 2004;24:3164–3175. doi: 10.1523/JNEUROSCI.4369-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haesler S, Rochefort C, Georgi B, Licznerski P, Osten P, Scharff C. Incomplete and inaccurate vocal imitation after knockdown of FoxP2 in songbird basal ganglia nucleus Area X. PLoS Biol. 2007;5:e321. doi: 10.1371/journal.pbio.0050321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hessler NA, Doupe AJ. Social context modulates singing-related neural activity in the songbird forebrain. Nat Neurosci. 1999;2:209–211. doi: 10.1038/6306. [DOI] [PubMed] [Google Scholar]
- Hilliard AT, Miller JE, Fraley ER, Horvath S, White SA. Molecular microcircuitry underlies functional specification in a basal ganglia circuit dedicated to vocal learning. Neuron. 2012;73:537–552. doi: 10.1016/j.neuron.2012.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Immelmann K. Song development in the zebra finches and other estrilid finches. In: Hinde RA, editor. Bird vocalizations. Cambride, UK: Cambridge UP; 1969. pp. 64–74. [Google Scholar]
- Jarvis ED, Nottebohm F. Motor-driven gene expression. Proc Natl Acad Sci U S A. 1997;94:4097–4102. doi: 10.1073/pnas.94.8.4097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis ED, Scharff C, Grossman MR, Ramos JA, Nottebohm F. For whom the bird sings: context-dependent gene expression. Neuron. 1998;21:775–788. doi: 10.1016/S0896-6273(00)80594-2. [DOI] [PubMed] [Google Scholar]
- John B, Enright AJ, Aravin A, Tuschl T, Sander C, Marks DS. Human MicroRNA targets. PLoS Biol. 2004;2:e363. doi: 10.1371/journal.pbio.0020363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaestner KH, Knochel W, Martinez DE. Unified nomenclature for the winged helix/forkhead transcription factors. Genes Dev. 2000;14:142–146. [PubMed] [Google Scholar]
- Kimpo RR, Doupe AJ. FOS is induced by singing in distinct neuronal populations in a motor network. Neuron. 1997;18:315–325. doi: 10.1016/S0896-6273(00)80271-8. [DOI] [PubMed] [Google Scholar]
- Kirn JR. The relationship of neurogenesis and growth of brain regions to song learning. Brain Lang. 2010;115:29–44. doi: 10.1016/j.bandl.2009.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima S, Doupe AJ. Social performance reveals unexpected vocal competency in young songbirds. Proc Natl Acad Sci U S A. 2011;108:1687–1692. doi: 10.1073/pnas.1010502108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konopka G, Bomar JM, Winden K, Coppola G, Jonsson ZO, Gao F, Peng S, Preuss TM, Wohlschlegel JA, Geschwind DH. Human-specific transcriptional regulation of CNS development genes by FOXP2. Nature. 2009;462:213–217. doi: 10.1038/nature08549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krek A, Grün D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, MacMenamin P, da Piedade I, Gunsalus KC, Stoffel M, Rajewsky N. Combinatorial microRNA target predictions. Nat Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294:853–858. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- Lai CS, Fisher SE, Hurst JA, Vargha-Khadem F, Monaco AP. A forkhead-domain gene is mutated in a severe speech and language disorder. Nature. 2001;413:519–523. doi: 10.1038/35097076. [DOI] [PubMed] [Google Scholar]
- Lai CS, Gerrelli D, Monaco AP, Fisher SE, Copp AJ. FOXP2 expression during brain development coincides with adult sites of pathology in a severe speech and language disorder. Brain. 2003;126:2455–2462. doi: 10.1093/brain/awg247. [DOI] [PubMed] [Google Scholar]
- Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, Pfeffer S, Rice A, Kamphorst AO, Landthaler M, Lin C, Socci ND, Hermida L, Fulci V, Chiaretti S, Foà R, Schliwka J, Fuchs U, Novosel A, Müller RU, et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-Y. [DOI] [PubMed] [Google Scholar]
- Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/S0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Li X, Wang XJ, Tannenhauser J, Podell S, Mukherjee P, Hertel M, Biane J, Masuda S, Nottebohm F, Gaasterland T. Genomic resources for songbird research and their use in characterizing gene expression during brain development. Proc Natl Acad Sci U S A. 2007;104:6834–6839. doi: 10.1073/pnas.0701619104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li XC, Jarvis ED, Alvarez-Borda B, Lim DA, Nottebohm F. A relationship between behavior, neurotrophin expression, and new neuron survival. Proc Natl Acad Sci U S A. 2000;97:8584–8589. doi: 10.1073/pnas.140222497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Luo GZ, Hafner M, Shi Z, Brown M, Feng GH, Tuschl T, Wang XJ, Li X. Genome-wide annotation and analysis of zebra finch microRNA repertoire reveal sex-biased expression. BMC Genomics. 2012;13:727. doi: 10.1186/1471-2164-13-727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDermot KD, Bonora E, Sykes N, Coupe AM, Lai CS, Vernes SC, Vargha-Khadem F, McKenzie F, Smith RL, Monaco AP, Fisher SE. Identification of FOXP2 truncation as a novel cause of developmental speech and language deficits. Am J Hum Genet. 2005;76:1074–1080. doi: 10.1086/430841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mello C, Nottebohm F, Clayton D. Repeated exposure to one song leads to a rapid and persistent decline in an immediate early gene's response to that song in zebra finch telencephalon. J Neurosci. 1995;15:6919–6925. doi: 10.1523/JNEUROSCI.15-10-06919.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JE, Spiteri E, Condro MC, Dosumu-Johnson RT, Geschwind DH, White SA. Birdsong decreases protein levels of FoxP2, a molecule required for human speech. J Neurophysiol. 2008;100:2015–2025. doi: 10.1152/jn.90415.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mooney R. Neurobiology of song learning. Curr Opin Neurobiol. 2009;19:654–660. doi: 10.1016/j.conb.2009.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nottebohm F, Stokes TM, Leonard CM. Central control of song in the canary, Serinus canarius. J Comp Neurol. 1976;165:457–486. doi: 10.1002/cne.901650405. [DOI] [PubMed] [Google Scholar]
- Pena JT, Sohn-Lee C, Rouhanifard SH, Ludwig J, Hafner M, Mihailovic A, Lim C, Holoch D, Berninger P, Zavolan M, Tuschl T. miRNA in situ hybridization in formaldehyde and EDC-fixed tissues. Nat Methods. 2009;6:139–141. doi: 10.1038/nmeth.1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, Horvitz HR, Ruvkun G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- Scharff C, Nottebohm F. A comparative study of the behavioral deficits following lesions of various parts of the zebra finch song system: implications for vocal learning. J Neurosci. 1991;11:2896–2913. doi: 10.1523/JNEUROSCI.11-09-02896.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz SB, Haesler S, Scharff C, Rochefort C. Knockdown of FoxP2 alters spine density in Area X of the zebra finch. Genes Brain Behav. 2010;9:732–740. doi: 10.1111/j.1601-183X.2010.00607.x. [DOI] [PubMed] [Google Scholar]
- Shibata M, Kurokawa D, Nakao H, Ohmura T, Aizawa S. MicroRNA-9 modulates Cajal-Retzius cell differentiation by suppressing Foxg1 expression in mouse medial pallium. J Neurosci. 2008;28:10415–10421. doi: 10.1523/JNEUROSCI.3219-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu W, Yang H, Zhang L, Lu MM, Morrisey EE. Characterization of a new subfamily of winged-helix/forkhead (Fox) genes that are expressed in the lung and act as transcriptional repressors. J Biol Chem. 2001;276:27488–27497. doi: 10.1074/jbc.M100636200. [DOI] [PubMed] [Google Scholar]
- Sohrabji F, Nordeen EJ, Nordeen KW. Selective impairment of song learning following lesions of a forebrain nucleus in the juvenile zebra finch. Behav Neural Biol. 1990;53:51–63. doi: 10.1016/0163-1047(90)90797-A. [DOI] [PubMed] [Google Scholar]
- Spiteri E, Konopka G, Coppola G, Bomar J, Oldham M, Ou J, Vernes SC, Fisher SE, Ren B, Geschwind DH. Identification of the transcriptional targets of FOXP2, a gene linked to speech and language, in developing human brain. Am J Hum Genet. 2007;81:1144–1157. doi: 10.1086/522237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao X, Finkbeiner S, Arnold DB, Shaywitz AJ, Greenberg ME. Ca2+ influx regulates BDNF transcription by a CREB family transcription factor-dependent mechanism. Neuron. 1998;20:709–726. doi: 10.1016/S0896-6273(00)81010-7. [DOI] [PubMed] [Google Scholar]
- Tchernichovski O, Mitra PP, Lints T, Nottebohm F. Dynamics of the vocal imitation process: how a zebra finch learns its song. Science. 2001;291:2564–2569. doi: 10.1126/science.1058522. [DOI] [PubMed] [Google Scholar]
- Teramitsu I, White SA. FoxP2 regulation during undirected singing in adult songbirds. J Neurosci. 2006;26:7390–7394. doi: 10.1523/JNEUROSCI.1662-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teramitsu I, Kudo LC, London SE, Geschwind DH, White SA. Parallel FoxP1 and FoxP2 expression in songbird and human brain predicts functional interaction. J Neurosci. 2004;24:3152–3163. doi: 10.1523/JNEUROSCI.5589-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vargha-Khadem F, Watkins KE, Price CJ, Ashburner J, Alcock KJ, Connelly A, Frackowiak RS, Friston KJ, Pembrey ME, Mishkin M, Gadian DG, Passingham RE. Neural basis of an inherited speech and language disorder. Proc Natl Acad Sci U S A. 1998;95:12695–12700. doi: 10.1073/pnas.95.21.12695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernes SC, Nicod J, Elahi FM, Coventry JA, Kenny N, Coupe AM, Bird LE, Davies KE, Fisher SE. Functional genetic analysis of mutations implicated in a human speech and language disorder. Hum Mol Genet. 2006;15:3154–3167. doi: 10.1093/hmg/ddl392. [DOI] [PubMed] [Google Scholar]
- Vernes SC, Spiteri E, Nicod J, Groszer M, Taylor JM, Davies KE, Geschwind DH, Fisher SE. High-throughput analysis of promoter occupancy reveals direct neural targets of FOXP2, a gene mutated in speech and language disorders. Am J Hum Genet. 2007;81:1232–1250. doi: 10.1086/522238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernes SC, Oliver PL, Spiteri E, Lockstone HE, Puliyadi R, Taylor JM, Ho J, Mombereau C, Brewer A, Lowy E, Nicod J, Groszer M, Baban D, Sahgal N, Cazier JB, Ragoussis J, Davies KE, Geschwind DH, Fisher SE. Foxp2 regulates gene networks implicated in neurite outgrowth in the developing brain. PLoS Genet. 2011;7:e1002145. doi: 10.1371/journal.pgen.1002145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White SA. Genes and vocal learning. Brain Lang. 2010;115:21–28. doi: 10.1016/j.bandl.2009.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeesman S, Nowaczyk MJ, Teshima I, Roberts W, Cardy JO, Brian J, Senman L, Feuk L, Osborne LR, Scherer SW. Speech and language impairment and oromotor dyspraxia due to deletion of 7q31 that involves FOXP2. Am J Med Genet A. 2006;140:509–514. doi: 10.1002/ajmg.a.31110. [DOI] [PubMed] [Google Scholar]
- Zhang J, Webb DM, Podlaha O. Accelerated protein evolution and origins of human-specific features: Foxp2 as an example. Genetics. 2002;162:1825–1835. doi: 10.1093/genetics/162.4.1825. [DOI] [PMC free article] [PubMed] [Google Scholar]