Abstract
SLOB (SLOWPOKE-binding protein) modulates the Drosophila SLOWPOKE calcium-activated potassium channel. We have shown previously that SLOB deletion or RNAi knockdown decreases excitability of neurosecretory pars intercerebralis (PI) neurons in the adult Drosophila brain. In contrast, we found that SLOB deletion/knockdown enhances neurotransmitter release from motor neurons at the fly larval neuromuscular junction, suggesting an increase in excitability. Because two prominent SLOB isoforms, SLOB57 and SLOB71, modulate SLOWPOKE channels in opposite directions in vitro, we investigated whether divergent expression patterns of these two isoforms might underlie the differential modulation of excitability in PI and motor neurons. By performing detailed in vitro and in vivo analysis, we found strikingly different modes of regulatory control by the slob57 and slob71 promoters. The slob71, but not slob57, promoter contains binding sites for the Hunchback and Mirror transcriptional repressors. Furthermore, several core promoter elements that are absent in the slob57 promoter coordinately drive robust expression of a luciferase vector by the slob71 promoter in vitro. In addition, we visualized the expression patterns of the slob57 and slob71 promoters in vivo and found clear spatiotemporal differences in promoter activity. SLOB57 is expressed prominently in adult PI neurons, whereas larval motor neurons exclusively express SLOB71. In contrast, at the larval neuromuscular junction, SLOB57 expression appears to be restricted mainly to a subset of glial cells. Our results illustrate how the use of alternative transcriptional start sites within an ion channel modulator locus coupled with functionally relevant alternative splicing can be used to fine-tune neuronal excitability in a cell-specific manner.
Introduction
Neuronal excitability can be modulated profoundly by changes in the properties of ion channels. Many ion channels bind directly to signaling and scaffolding proteins that control channel expression, trafficking, and function (Levitan, 2006; Wu et al., 2010; Vacher and Trimmer, 2011; Jepson et al., 2012). We have identified and characterized a protein that we named SLOB (Schopperle et al., 1998; Zhou et al., 1999), which binds to the Drosophila large conductance calcium-activated potassium channel encoded by the slowpoke locus (Atkinson et al., 1991; Adelman et al., 1992). slowpoke mutations result in elongation of the action potential in Drosophila flight muscle (Elkins et al., 1986; Elkins and Ganetzky, 1988), suggesting that SLOWPOKE channels play a prominent role in action potential repolarization.
At least four slob transcripts can be expressed from the single slob gene as a result of multiple transcriptional start sites and alternative splicing (Jaramillo et al., 2004, 2006), and we have named the SLOB protein isoforms that are produced from these transcripts according to their molecular weights in kilodaltons. When SLOWPOKE is expressed in HEK293 cells, coexpression with SLOB57 shifts the voltage dependence of SLOWPOKE channel activation markedly in the depolarizing direction (Zeng et al., 2005). In contrast, SLOB71 coexpression shifts the voltage dependence more modestly and in the opposite direction (Zeng et al., 2005).
To investigate the role of SLOB modulation of SLOWPOKE channels in vivo, we knocked out or knocked down SLOB expression by P-element mutagenesis or expression of RNAi and recorded in the brains of living flies from pars intercerebralis (PI) neurons (Shahidullah et al., 2009). Knock-out/knockdown of SLOB shifts the voltage dependence of outward current in PI neurons in the hyperpolarizing direction and also produces a decrease in the duration of the PI neuron action potentials, both consistent with an increase in SLOWPOKE channel activity and a corresponding decrease in neuronal excitability (Shahidullah et al., 2009). Subsequently, we examined the effect of SLOB knock-out/knockdown on synaptic transmission in vivo at the fly larval neuromuscular junction (NMJ) and found changes in spontaneous and evoked neurotransmitter release from the presynaptic motor neuron terminals consistent with an increase in neuronal excitability (Ma et al., 2011). One possible explanation for this apparent discrepancy between PI neurons and motor neurons is that the modulation of excitability by SLOB is different in neuronal cell bodies and nerve terminals. Another possibility is that there is either developmental or cell-type-specific regulation of the expression of different SLOB isoforms. To distinguish between these possibilities, we recorded action potential waveforms and voltage-dependent outward currents in motor neuron cell bodies and used promoter analysis in vitro and in vivo to determine the expression patterns of SLOB57 and SLOB71. Our results are consistent with the idea that preferential activity of SLOB57 in PI neurons and of SLOB71 in motor neurons can account for the differential modulation of excitability by SLOB in these two populations of neurons. These findings demonstrate that differential isoform expression from a single locus can be used to bidirectionally alter ion channel modulation and electrical activity within specific cell types.
Materials and Methods
Transcription start site determination.
Transcription start sites (TSSs) were mapped using RNA ligase-mediated 5′ rapid amplification of cDNA ends (RLM-RACE) (Ambion/Invitrogen). Total RNA was extracted from a minimum of 30 yw fly heads using the UltraSpec RNA isolation system following the recommendations of the manufacturer (Biotecx Laboratories). RLM-RACE was performed with four reverse primers targeted to the following areas: slob71 exon 1, slob71 5′ UTR, slob57 5′ UTR, and the 168 bp alternatively spliced exon (exon 3 of slob57 or slob71). RLM-RACE products were run on agarose gels, and individual products were extracted. Products were then TOPO cloned (Invitrogen) and sequenced to identify TSSs.
Generation of luciferase constructs.
Genomic DNA was extracted from yw fly heads. A total of 30 fly heads were homogenized in a buffer containing 7 m urea, 2% SDS, 50 mm Tris, pH 7.5, 10 mm EDTA, pH 8.0, and 0.35 m NaCl. DNA was extracted using phenol chloroform extraction followed by ethanol precipitation and resuspension in 30 μl of water. Luciferase reporter constructs were created by sequential cloning of promoter regions from genomic DNA. An ∼2 kb region (−1966 to +81) surrounding the TSS for slob71 was amplified from genomic DNA from yw fly heads by PCR (Advantage cDNA PCR Kit and Polymerase; Clontech) using primers engineered to contain KpnI and BglII restriction enzyme flanking sites (primers available on request). The promoter construct containing the 2.2 kb region (−1877 to +333) surrounding the TSS for slob57 was similarly created. Smaller constructs were generated using the same techniques. Fragments of the slob71 promoter were subcloned into the minimal promoter (minP) pGL4.23 vector (Promega) between the KpnI and BglII sites.
Sequences within the downstream promoter element (DPE) and motif ten element (MTE) were mutated in the slob71 −1500 to +81 luciferase construct using Quikchange site-directed mutagenesis (Agilent Technologies). Hunchback (HB) and Mirror (MIRR) target sequences were also mutated with Quikchange. Core sequences within the HB recognition consensus sequence were mutated from ggcacAAAAaaca to ggcacCCCCaaca, and core sequences within the MIRR recognition consensus site were mutated from caaaaAACA to caaaaCCAC in slob71 promoters. All constructs were confirmed by sequencing at the Kimmel Cancer Center genomics core facility (Thomas Jefferson University, Philadelphia, PA).
Fly genetics.
For the construction of slob57–Gal4 or slob71–Gal4 promoter insertion transgenic flies, promoter region cDNAs were cloned into the commercially available pUAST vector. For slob57, we used base pairs −1016 to +333 relative to the TSS; for slob71, base pairs −1500 to +81 relative to the TSS were used. Multiple transgenic lines were generated by Rainbow Transgenic Flies. Individual lines were mapped and crossed to flies homozygous for either UAS–cd8::gfp or UAS–gfp–nls insertions (Bloomington Drosophila Stock Center). The slob null allele generated via P-element mediated imprecise excision, as described previously (Shahidullah et al., 2009); it has no detectable slob transcripts corresponding to any slob isoform and is thus a true null allele. Imaging and electrophysiological experiments were performed on adult and/or larval Drosophila of either sex.
Cell culture and transfection.
Drosophila S2 cells were cultured at 25°C in six-well plates with Schneider's Drosophila Media (Invitrogen) supplemented with 10% heat-inactivated fetal bovine serum (Atlanta Biologicals) and penicillin/streptomycin. S2 cells were transfected with the luciferase reporter constructs (2 μg/well) along with a vector expressing β-galactosidase (β-gal; pCMV–LacZ vector from Clontech; 1 μg/well) in duplicate using Lipofectamine (Invitrogen). Luciferase assays were performed according to the instructions of the manufacturer for the luciferase assay kit (Promega). In brief, S2 cells were lysed by freeze/thawing in reporter lysis buffer 48 h after transfection, luciferase reagent was added to the lysate in 96-well plates, and luminescence was immediately measured using a Victor3 luminometer (PerkinElmer Life and Analytical Sciences). Using a β-gal assay kit (Promega), activity of the β-gal enzyme was measured in a spectrophotometer after addition of substrate and incubation at 37°C for 30 min. Luciferase activity was normalized to β-gal activity to control for transfection efficiency. Fold changes were calculated by dividing normalized luciferase activity measures for the slob57 and slob71 promoter constructs by the normalized luciferase activity measured in S2 cells transfected with the pGL4.10 or pGL4.23 empty vectors.
Confocal microscopy of adult and larval Drosophila brains and larval NMJs.
Confocal images were obtained on an Olympus Fluoview confocal microscope. Adult and larval brains were dissected and immunostained as described previously (Wu and Luo, 2006). Briefly, brains were removed, fixed in 4% paraformaldehyde in PBT (1× PBS with 0.3% Triton X-100), and blocked in 5% normal goat serum before incubation with primary antibody. Primary antibody concentrations were as follows: mouse nc82 and anti-REPO (Developmental Studies Hybridoma Bank) at 1:100; and rabbit anti-GFP (Invitrogen) at 1:500. Fluorescently conjugated secondary antibodies (Invitrogen) were used as follows: goat anti-mouse Cy3 at 1:400; and goat anti-rabbit Cy5 at 1:400. Primary and secondary antibodies were applied overnight at 4°C.
Larval neuromuscular preparations were dissected in low calcium solution of the following composition (in mm): 0.2 CaCl2, 70 NaCl, 5 KCl, 20 MgCl2, 10 NaHCO3, 115 sucrose, 5 trehalose, and 5 HEPES, at pH 7.3. Larvae were fixed in 4% paraformaldehyde in PBS for 20 min. Blocking solution and antibodies were applied as above in PBS with 0.15% Triton X-100. Cy3-conjugated HRP (1:200; Jackson ImmunoResearch) was used to label neurons.
Electrophysiological recording from larval motor neurons.
All experiments were performed on wandering late third-instar larvae. Larvae were pinned dorsal side up and bathed in Ca2-free solution that contained the following (in mm): 70 NaCl, 5 KCl, 4 MgCl2, 10 NaHCO3, 5 trehalose, 115 sucrose, and 5 HEPES, pH 7.2. The preparation was visualized using an upright Olympus BX511WI microscope. To access motor neurons in the ventral ganglion, Protease 14 (2 mg/ml extracellular solution; Sigma-Aldrich) was focally applied to the ganglionic sheath by applying positive pressure to a recording electrode with the tip broken to a diameter of ∼10 μm. Treatment of the sheath was performed with constant laminar perfusion, and the debris was removed by applying negative pressure to the electrode (Worrell and Levine, 2008).
Motor neurons were identified though the expression of GFP under the control of the RRA–GAL4 driver (Fujioka et al., 2003), which labels CCa motor neurons. The extracellular recording solution contained the following (in mm): 101 NaCl, 3 KCl, 1.8 CaCl2, 2 MgCl2, 1.25 Na2HPO4, 5 glucose, and 20.7 NaHCO3, pH 7.2. Thin-walled borosilicate electrodes were pulled to a resistance of 4–6 MΩ when filled with intracellular solution containing the following (in mm): 102 K-gluconate, 17 NaCl, 2 CaCl2, 5 EGTA, 0.5 MgCl2, and 10 HEPES, pH 7.2. Standard techniques were used to record macroscopic currents in the whole-cell voltage-clamp mode and voltage in current-clamp mode, with an Axopatch 200A amplifier (Molecular Devices). Data were digitized with a Digidata 1322A interface (Molecular Devices) and stored on a personal computer hard drive for additional analysis with pClamp9 software (Molecular Devices).
Results
SLOB knock-out influences the electrical excitability of larval motor neurons
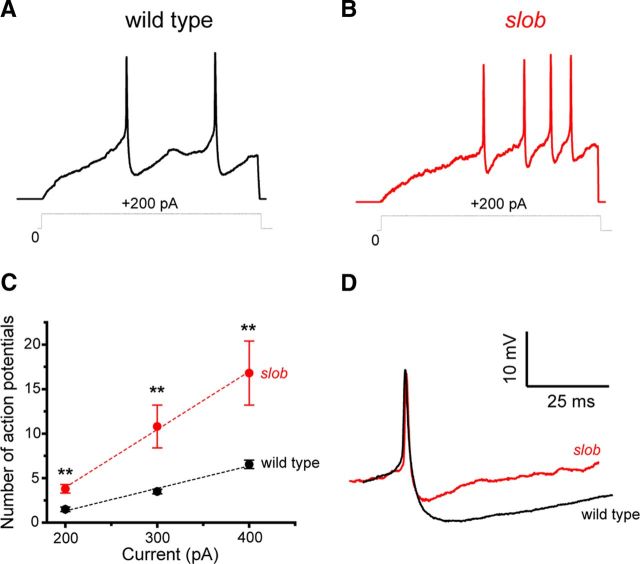
To determine the effects of SLOB on motor neuron excitability, we recorded action potential waveforms under current clamp and voltage-dependent outward currents under voltage clamp in larval motor neuron cell bodies. As shown in Figure 1A, action potentials can be evoked in these neurons by the injection of depolarizing current. Application of tetrodotoxin and zero-calcium solutions demonstrated that these actions potentials are mediated by voltage-gated sodium channels (data not shown). In slob null mutant larvae (Shahidullah et al., 2009), more action potentials are evoked by the same depolarizing stimulus (Fig. 1B) at all stimulus levels examined (Fig. 1C). It is worthy of note that the duration of these fast action potentials is not affected by the knock-out of SLOB (Fig. 1D). Rather, the prominent spike afterhyperpolarization (AHP) that follows the action potential is significantly reduced in slob null larvae [wild-type peak AHP, 8.54 mV ± 0.63 mV (mean ± SEM); slob, 2.78 ± 0.52 mV; p < 0.0005, Mann–Whitney U test], allowing for the higher firing frequency observed in Figure 1B. The AHP is greatly reduced in zero-calcium saline (data not shown), consistent with the notion that it is mediated predominantly by SLOWPOKE channels. This may be contrasted with our previous studies in adult PI neurons, in which SLOB disruption decreases the duration of the much slower action potentials characteristic of these neurosecretory cells but has little effect on the AHP (Shahidullah et al., 2009).
Figure 1.
A, B, Whole-cell patch recording demonstrates the modulation of the frequency and waveform of action potentials by endogenous SLOB in motor neurons in vivo. Action potentials were evoked in motor neuron cell bodies under current-clamp mode. The starting voltage was adjusted to −80 mV by the injection of an appropriate amount of holding current, and action potentials were evoked by the subsequent injection of 200 pA of depolarizing current of 500 ms duration in wild-type (A) and slob null (B) flies. C, The number of action potentials evoked by different depolarizing current injections. Error bars represent SEM. Wild type, n = 10; slob, n = 5. **p < 0.005, Mann–Whitney U test. D, Superimposition of the first spikes evoked by a 200 pA current injection in motor neurons from wild-type and slob null flies.
To further examine the modulation of membrane excitability by SLOB, we recorded voltage-dependent currents under voltage clamp. As shown in Figure 2, A and B, SLOB knock-out appears to have little effect on fast inward currents, although we did not study these systematically. Conversely, voltage-dependent outward currents are smaller in the absence of SLOB, and SLOB knock-out shifts the conductance–voltage (G–V) relationship in the depolarizing direction (at −10 mV; normalized conductance, wild-type, 0.55 ± 0.03; slob, 0.35 ± 0.04; p < 0.005, Mann–Whitney U test; Fig. 2C). Again, this may be contrasted with our previous finding that the G–V relationship in PI neurons is shifted in the hyperpolarizing direction in the absence of SLOB expression (Shahidullah et al., 2009). These results demonstrate that there is little difference in the modulation of excitability by SLOB in neuronal cell bodies versus nerve terminals. Instead, the differential effect of SLOB in PI neurons and motor neurons may be attributable to distinct expression patterns of SLOB isoforms.
Figure 2.
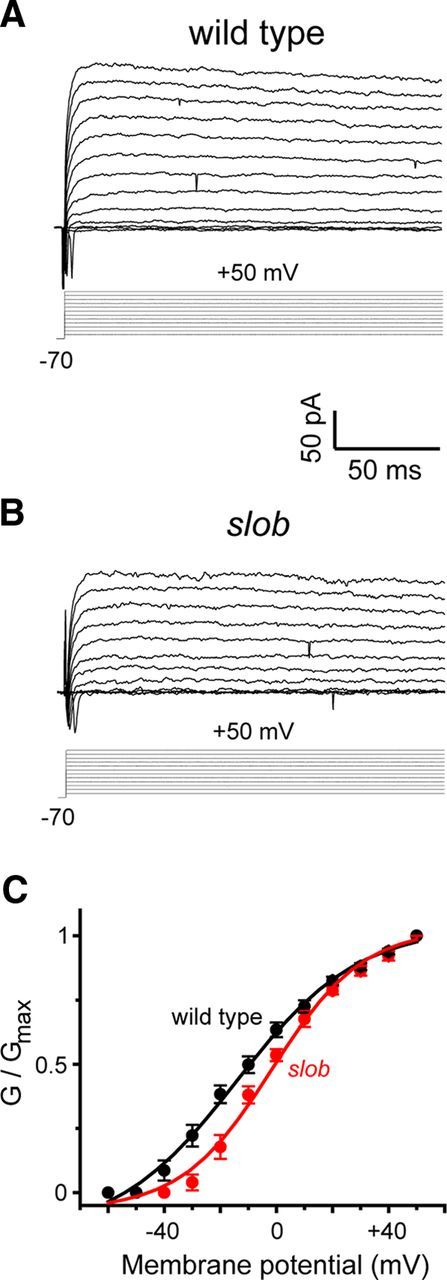
A, B, Whole-cell patch recording demonstrates the modulation of outward currents by endogenous SLOB in motor neurons in vivo. Using voltage-clamp mode, large outward currents were evoked by 150 ms depolarizing voltage steps from a holding potential of −70 mV to potentials ranging from −60 to +50 mV in 10 mV increments, in motor neurons from wild-type (A) and slob null (B) flies. C, G–V relationships for the peak outward currents in the above genotypes. Error bars represent SEM. Wild type, n = 6; slob, n = 7.
Separate promoters control the expression of slob57 and slob71 transcripts
To obtain more detailed information about the regulation of expression of individual SLOB isoforms, experiments to characterize slob promoters were undertaken. To map the TSSs for slob57 and slob71 transcripts, RLM-RACE experiments were conducted with RNA isolated from yw fly heads and primers targeted at different regions of the transcripts. Primers were designed to hybridize to the slob57/51 5′ UTR, slob71/65 5′ UTR, slob71/65 exon 1, and the 168 bp exon that is alternatively spliced in slob57/51 (resulting in slob57 and slob51) and slob71/65 (resulting in slob71 and slob65) (Jaramillo et al., 2006). Two TSSs were identified by RLM-RACE: one for slob57/51 and one for slob71/65. For the sake of simplicity, we will refer to slob57/51 as slob57 and slob71/65 as slob71. Our experiments identified the slob71 TSS sequence as TCA+1 ATC. The A+1 initiator (Inr) is 21 nt upstream of the first 5′ UTR identified in the y1;cn1bw1sp1 background by FlyBase (McQuilton et al., 2012). A separate TSS was determined for slob57 (TCA+1 TTA). The TSS for slob57 is also slightly different from that reported in FlyBase. The A+1 Inr is contained within what FlyBase identifies as the first 5′ UTR, 43 nt downstream of the TSS for slob57 identified in the y1;cn1bw1sp1 background. The slob71 transcript contains long intronic sequences upstream of exon 1; thus, the TSS is ∼15 kb upstream of the slob57 TSS.
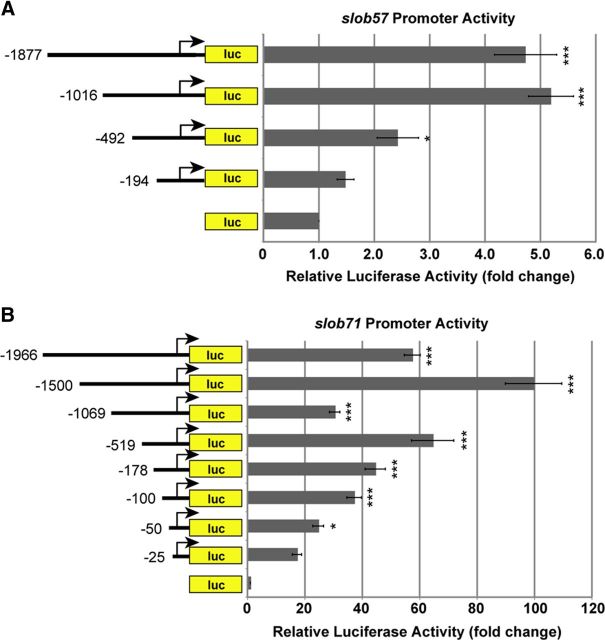
To test whether slob57 and slob71 promoters displayed differences in transcriptional activation, slob57 and slob71 promoter activities were measured using luciferase assays in the Drosophila S2 cell line. S2 cells were transfected with slob57 or slob71 promoter–luciferase constructs containing sequentially smaller regions of each promoter, and luciferase activity was measured as an indicator of promoter activity (Fig. 3). Analysis of sequentially smaller regions of the slob57 promoter revealed that a region 1016 nt upstream and 333 nt downstream of the TSS maximally and significantly increased promoter activity by approximately fivefold compared with empty luciferase vector (p < 0.001; Fig. 3A). Although the slob57 −492 to +333 promoter also significantly increased promoter activity compared with control, activity was approximately half that of the slob57 −1016 to +333 construct, suggesting that sequences important for activation of transcription were missing in this smaller construct. Luciferase activity was not enhanced by a larger promoter construct (slob57 −1877 to +333); thus, the slob57 −1016 to +333 promoter was sufficient for slob57 promoter activity in this model system.
Figure 3.
slob57 and slob71 promoters exhibit different transcriptional activity. Promoter regions upstream of the identified TSSs for slob57 and slob71 were cloned into the pGL4.10[luc2] vector to drive the luciferase (luc) reporter gene. Drosophila S2 cells were transfected with various slob promoter–luc constructs and the pCMV–LacZ vector as an internal control. The relative luciferase activity is the luciferase activity normalized to β-gal activity and is reported as the fold change compared with the empty luc control vector. A, Summary of luciferase activity experiments with slob57 promoters. B, Summary of luciferase activity experiments with slob71 promoters. Relative luciferase activity driven by slob71 promoters is higher than that of slob57. *p < 0.05, ***p < 0.001, relative to the empty luc vector, one-way ANOVA with Bonferroni's post hoc test. n ≥ 3 independent experiments. Error bars represent SEM.
The ability of putative promoter regions surrounding the slob71 TSS to activate transcription was next measured using luciferase activity assays. Insertion of slob71 promoter regions upstream of the luciferase coding sequence resulted in very high levels of luciferase activity, with almost all constructs significantly increasing luciferase activity compared with control and maximal activation occurring from the slob71 −1500 to +81 promoter (Fig. 3B). This construct displayed luciferase activity ∼100-fold greater than control. Interestingly, the pattern of slob71 promoter activity did not mirror promoter length among larger constructs. Maximal activation was achieved from slob71 −1500 to +81, whereas promoter constructs ∼500 nt longer or shorter displayed reduced activity. Promoter activity then increased in the slob71 −519 to +81 construct and declined as promoter constructs became smaller thereafter. Two questions arose from the slob57 and slob71 promoter analyses. First, what sequences are responsible for the high promoter activity of slob71 that is lacking in slob57? Second, what determines the pattern of activation displayed by the slob71 promoter constructs?
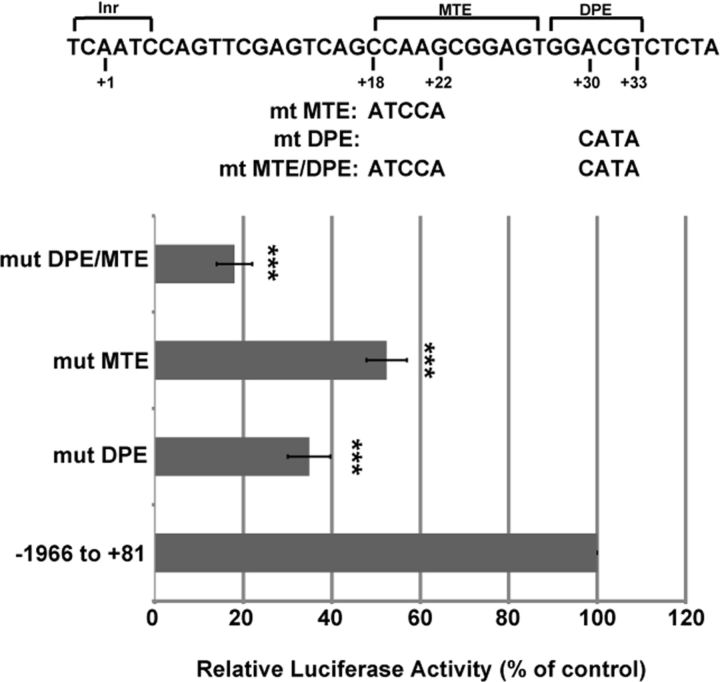
Sequences surrounding the slob57 TSS and slob71 TSS were examined to determine whether differences between core promoter sequences could account for the large difference in promoter activity. Core promoter elements are known to be important regulators of transcription and include the Inr, TATA box, DPE, and MTE (Kutach and Kadonaga, 2000; Lim et al., 2004). In addition to the Inr, two additional core promoter elements (DPE and MTE) were identified in the slob71 promoter, whereas none were found in the slob57 promoter. The DPE is a 6 nt sequence located precisely from +28 to +33 relative to the Inr (Burke and Kadonaga, 1997; Kutach and Kadonaga, 2000), and the MTE is a 10 nt sequence located from +18 to +27 relative to the Inr (Lim et al., 2004). Core nucleotides within each of the DPE and MTE have been identified as critical to promoter activation (Lim et al., 2004). To determine whether the DPE and MTE regulate transcription from the slob71 promoter, core nucleotides of the DPE only, MTE only, or the DPE and MTE together were mutated in the slob71 −1966 to +81 promoter–luciferase construct (Fig. 4). S2 cells were transfected with mutant promoter–luciferase constructs, and promoter activity was measured using luciferase assays. Mutation of +30 to +33 within the DPE of slob71 −1966 to +81 significantly decreased promoter activity to 35 ± 5% of the intact control promoter. Similarly, mutation of +18 to +22 within the MTE of slob71 −1966 to +81 resulted in a significant decrease in promoter activity compared with control (52 ± 5% of control). Mutation of core nucleotides within both the DPE and MTE significantly reduced promoter activity even further to 18 ± 4% of control. Therefore, both the DPE and MTE are important transcriptional activators present in the core promoter of slob71 and contribute greatly to the high levels of promoter activity exhibited by slob71. However, the slob71 −1966 to +81 promoter containing mutations in the DPE and MTE still exhibited ∼17-fold greater activity compared with control empty vector, whereas maximal promoter activity of intact slob57 −1016 to +333 promoter was only approximately fivefold greater than empty vector, suggesting that additional transcriptional mechanisms are present in the slob71 promoter.
Figure 4.
The DPE and MTE are important regulators of transcription from the slob71 promoter. Core nucleotides within the MTE, DPE, or both MTE and DPE were mutated in the slob71 −1966 to +81 promoter. Luciferase activity was measured in Drosophila S2 cells transfected with mutant or intact control constructs. The relative luciferase activity is the luciferase activity normalized to β-gal activity and is reported as the percentage of activity exhibited by the intact control construct. ***p < 0.001 compared with −1966 to +81, one-way ANOVA with Bonferroni's post hoc test. n = 4 independent experiments. Error bars represent SEM.
We next sought to characterize the pattern of transcriptional activity displayed by the slob71 promoter constructs. Based on the increased activity of slob71 −1500 to +81 compared with −1966 to +81, we hypothesized that the −1966 to −1500 fragment region contained repressor elements that resulted in increased activity when eliminated in the −1500 to +81 construct. We also predicted that the −1500 to −1069 fragment contained activator elements and the −1069 to −519 contained repressor elements. To test these predictions, fragments of each promoter construct were inserted upstream of a minP capable of activating a low level of basal luciferase transcription. The effect of isolated promoter regions on minP activity was then measured using luciferase assays. Compared with the luciferase activity exhibited by the control minP luciferase vector, the −1966 to −1500 fragment construct significantly decreased transcriptional activity, supporting the hypothesis that this region contains elements capable of repressing transcription (Fig. 5A). However, there was no difference in luciferase activity from the construct containing the −1500 to −1069 fragment upstream of the minP compared with the minP construct alone. Furthermore, rather than repressing transcription, the −1069 to −519 fragment enhanced transcription of luciferase from the minP. It is possible that these fragments may act differently in isolation and that elements in surrounding regions of the slob71 promoter interact with sequences within these fragments to regulate transcription.
Figure 5.
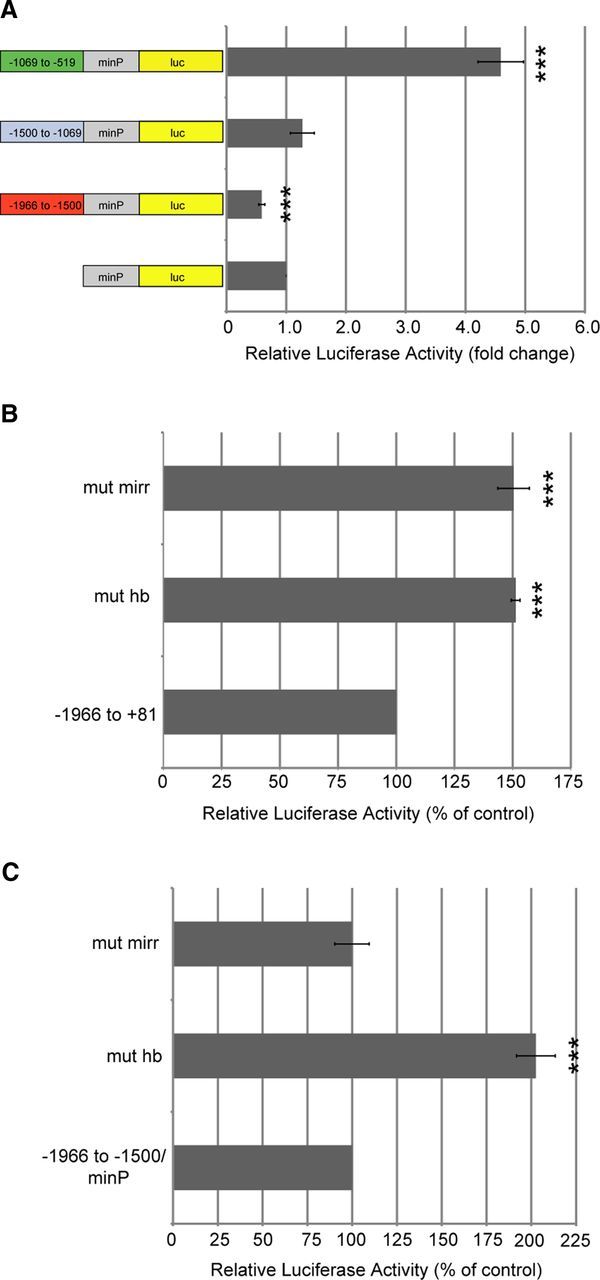
Promoter elements in specific domains of slob71 affect transcriptional activity. A, Promoter fragments of slob71 were inserted upstream of a minP in the pGL4.23[luc2/minP] vector, which drives a low level of basal luciferase expression. Luciferase activity was measured in Drosophila S2 cells transfected with minP–luc or slob71 promoter fragment–minP–luc constructs. The relative luciferase activity is the luciferase activity normalized to β-gal activity and is reported as the fold change compared with the empty minP–luc vector. B, C, Core nucleotides within the HB and MIRR recognition sites were mutated in slob71 promoters. Luciferase activity was measured in Drosophila S2 cells transfected with mutant or intact control constructs. The relative luciferase activity is the luciferase activity normalized to β-gal activity and is reported as the percentage of activity exhibited by the intact control construct. Mutating MIRR or HB sites in the slob71 −1966 to +81 promoter increases relative luciferase activity (B). Mutation of the HB site in the slob71 −1966 to −1500 promoter fragment upstream of the minP increases relative luciferase activity, whereas mutation of the MIRR site has no effect (C). ***p < 0.001 compared with minP–luc or intact control construct, one-way ANOVA with Bonferroni's post hoc test. n = 4 independent experiments. Error bars represent SEM.
Because the slob71 −1966 to −1500 fragment repressed transcription from the minP, this region was further analyzed with MatInspector promoter prediction software (Cartharius et al., 2005) for potential cis-elements capable of transcriptional repression. Two putative repressor elements were identified within this region: an HB recognition consensus sequence and an MIRR recognition consensus sequence. Compared with control slob71 −1966 to +81 with sequences intact, mutation of the core nucleotides within the HB recognition sequence (ggcacAAAAaaca to ggcacCCCCaaca) in the slob71 −1966 to +81 promoter significantly increased transcriptional activity (151 ± 2% of control), as measured by luciferase activity in transfected S2 cells (Fig. 5B). Likewise, mutation of core nucleotides within the MIRR recognition sequence (caaaaAACA to caaaaCCAC) in slob71 −1966 to +81 resulted in increased transcription (150 ± 7% of control). To investigate whether regulation of transcription is modulated by intact HB or MIRR sites interacting with core promoter elements present in slob71 or whether these sites are involved in more general repression, the sites were mutated in the −1966 to −1500 fragment upstream of the general minP. Interestingly, there was a differential effect on transcription activity on mutation of HB or MIRR recognition sites in this promoter construct (Fig. 5C). Mutant HB constructs exhibited significantly enhanced minP activity (202 ± 11% of control), suggesting that intact HB sites function to repress transcription from the minP. In contrast, mutation of the MIRR recognition site had no effect on minP activity (100 ± 10% of control), suggesting that intact MIRR sites more specifically repress transcription from the slob71 promoter. These results demonstrate that, in the context of S2 cells, the slob57 and slob71 promoters display markedly different promoter activities. Some of the differences are presumably attributable to the presence of the DPE and MTE in the slob71 promoter, but it is also possible that S2 cells do not express transcription factors important for high-level expression of the slob57 promoter.
The slob57 and slob71 promoters drive cell-specific expression patterns in the adult and larval nervous systems
To test whether the promoters that control slob57 and slob71 transcription drive expression of functionally opposing SLOB isoforms in distinct cell types in vivo, we generated multiple independent lines of transgenic Drosophila containing either slob57–Gal4 or slob71–Gal4 promoter insertions. We visualized slob57–Gal4 and slob71–Gal4 promoter activity by crossing each strain to a transgenic line containing a membrane-tagged GFP (cd8::gfp) under control of the UAS promoter (Brand and Perrimon, 1993). To control for position effects, we required that at least three independent lines drove highly similar expression patterns. Using this criterion, we then compared the representative expression patterns of slob57–Gal4 and slob71–Gal4 in the adult and larval nervous systems (Figs. 6, 7).
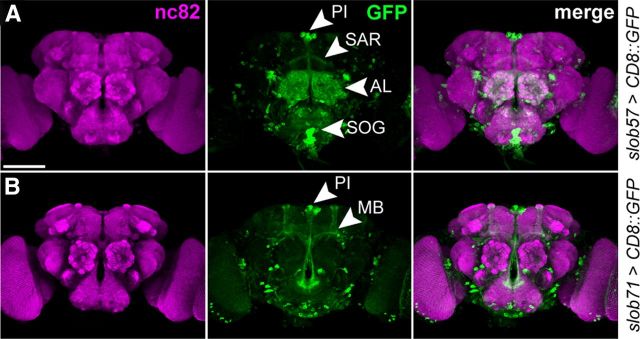
Figure 6.
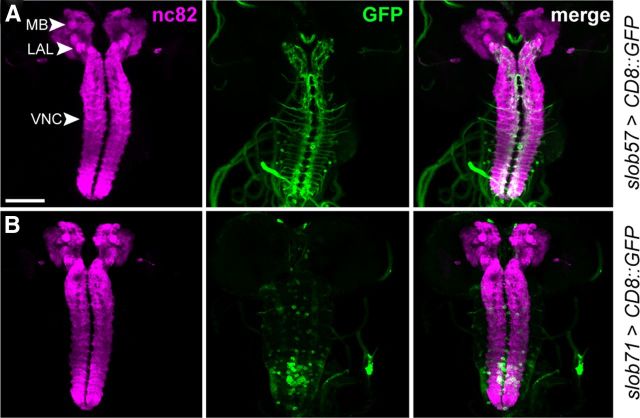
Activity of the slob57 and slob71 promoters in the adult CNS. A, Representative confocal z-stack of CD8::GFP driven by slob57–Gal4 in the adult Drosophila brain. Synaptic neuropil regions are labeled with the presynaptic marker nc82 (anti-Bruchpilot). B, Representative confocal z-stack of CD8::GFP driven by slob71–Gal4. SAR, Superior arch; AL, antennal lobe; SOG, suboesophageal ganglion; MB, mushroom body α, β, and γ lobes. Scale bars, 100 μm.
Figure 7.
Expression of slob57 and slob71 promoters in the third-instar larval nervous system. Similarly to Figure 6, the larval neuropil was labeled with nc82. A, Representative confocal z-stack of CD8::GFP driven by slob57–Gal4 in the larval brain. B, Representative confocal z-stack of CD8::GFP driven by slob71–Gal4. Note the absence of significant CD8::GFP expression in the central brain for both slob57–Gal4 and slob71–Gal4. MB, Mushroom body γ lobes; LAL, larval precursor of the antennal lobe. Scale bar, 100 μm.
In the major neuronal centers of the adult brain, we observed distinct but partially overlapping domains of expression when comparing the activities of the two slob promoters (Fig. 6). slob57–Gal4 was active in the antennal lobes, suboesophageal ganglion, and components of the central complex, including the fan-shaped body and the superior arch (Fig. 6A). slob71–Gal4 was similarly expressed in the central complex but showed no expression in the antennal lobes and suboesophageal neuropil (Fig. 6B). In additional contrast to slob57–Gal4, the slob71 promoter is also clearly active in mushroom body neurons (Fig. 6B).
One shared domain of expression between the two slob promoters is the PI (Fig. 6A,B), with both slob57–Gal4 and slob71–Gal4 driving strong GFP expression in many PI neuronal cell bodies. Thus, two opposing isoforms of SLOB are coexpressed in the PI and presumably many other Drosophila neurons, whereas other neuronal subtypes express only a single SLOB variant.
Intriguingly, when we compared expression of the same slob57–Gal4 and slob71–Gal4 promoter insertions in the larval nervous system, we found an almost complete absence of GFP expression in the central brain when CD8::GFP was driven with either slob promoter (Fig. 7). This result was replicated for multiple independent lines (data not shown) and suggests that slob expression in the CNS is subject to significant developmental regulation.
slob57 and slob71 expression in larval motor neurons and glia
Both slob57–Gal4 and slob71–Gal4 drove GFP expression in the larval ventral nerve cord (VNC) but with distinct patterns of expression (Fig. 7). slob57-positive cell bodies were relatively small and often located in the lateral areas of the VNC, whereas slob71-positive cell bodies appeared larger and were more commonly located in the central region of the VNC. Because the VNC contains cell bodies that project axons to the larval NMJ and loss of slob has been shown previously to enhance synaptic transmission at the NMJ (Ma et al., 2011), we examined the expression of both promoter reporters at the third-instar larval NMJ (Fig. 8).
Figure 8.
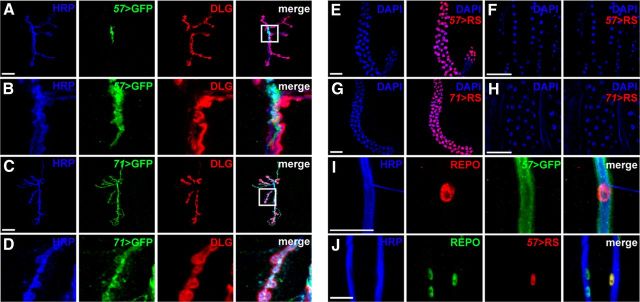
slob57 and slob71 promoter activity in larval glia and motor neurons. A, B, Expression of CD8::GFP (GFP) driven by slob57 (57) at the third-instar larval NMJ. Presynaptic and postsynaptic domains are labeled using antibodies against HRP and DLG, respectively (A). Representative confocal z-stacks are shown. Magnified image of CD8::GFP localization is shown in B. Note the non-overlap between CD8::GFP, HRP, and DLG. C, D, Representative confocal z-stack showing CD8::GFP under control of slob71–Gal4 (71) at the NMJ. Magnified image in D illustrates colocalization of CD8::GFP and the motor neuron marker HRP in synaptic boutons. E, F, Expression of the nuclear marker Red-Stinger (RS) under control of slob57–Gal4 in either larval salivary gland (E) or muscle cell nuclei (F). G, H, Red-Stinger driven by slob71–Gal4 in either larval salivary gland (G) or muscle cell nuclei (H). I, CD8::GFP driven by slob57–Gal4 surrounds axonal bundles and is closely associated with REPO-positive glial nuclei. J, Colocalization of REPO and Red-Stinger driven by slob57–Gal4 in a subset of glial nuclei that surround motor neuron axonal bundles. Scale bars: A, C, I, J, 20 μm; E–H, 100 μm.
We used fluorescently conjugated HRP to label presynaptic motor neurons and antibodies against Discs Large (DLG) to label the postsynaptic subsynaptic reticulum and examined the localization of CD8::GFP driven by slob57–Gal4 and slob71–Gal4 promoter insertions relative to these two synaptic markers (Fig. 8A–D). Interestingly, we found that CD8::GFP driven by slob57–Gal4 was closely associated with a small portion of the HRP signal close to the central axonal branching point but did not directly colocalize with this neuronal marker (Fig. 8A,B). In contrast, CD8::GFP driven by slob71–Gal4 completely colocalized with HRP (Fig. 8C,D), indicating that motor neurons primarily express SLOB71 but not SLOB57.
SLO channel isoforms are expressed both presynaptically and postsynaptically (Atkinson et al., 1998; Ghezzi et al., 2004), and it is unclear whether the modulation of SLO by SLOB occurs in one or both of the synaptic domains. Although our initial results suggested a predominantly presynaptic expression pattern of SLOB71, diffuse postsynaptic expression CD8::GFP using either the slob57–Gal4 or slob71–Gal4 promoter could not be ruled out under these experimental conditions. To further examine whether slob promoters were active postsynaptically at the NMJ, we used the slob57–Gal4 and slob71–Gal4 drivers to activate expression of a highly fluorescent nuclear marker, Red-Stinger (Barolo et al., 2004). As an internal control, we examined expression of Red-Stinger under the control of both drivers in the larval salivary gland, which is clearly labeled when using CD8::GFP to examine the expression of slob57–Gal4 and slob71–Gal4. Correspondingly, robust nuclear localization of Red-Stinger in salivary gland cells was also observed under the control of slob57–Gal4 and slob71–Gal4 (Fig. 8E,G). In contrast, at the same fluorescent settings, we observed no detectable expression of Red-Stinger using either slob promoter in muscle cell nuclei (Fig. 8F,H), indicating that neither SLOB57 nor SLOB71 are strongly expressed postsynaptically at the NMJ and that the regulation of SLO by SLOB at the NMJ is primarily presynaptic.
Finally, we further investigated the intriguing difference in expression patterns of slob57–Gal4 and slob71–Gal4. Although the colocalization of CD8::GFP driven by slob71–Gal4 with HRP indicates expression of SLOB71 in motor neurons, slob57–Gal4 activity is clearly apparent at the synapse but in a non-motor neuron cell type. Because glial cells are often adjacent to motor neurons at the larval NMJ (Brink et al., 2012), we hypothesized that slob57 may be active in a subset of glia that innervate the NMJ. Indeed, we found that CD8::GFP driven by slob57–Gal4 surrounded HRP-positive axonal bundles descending from the larval VNC, which were also closely associated with nuclei positive for the glial marker REPO (Fig. 8I). Furthermore, Red-Stinger driven by slob57–Gal4 labeled a subset of glial nuclei that surround motor neuron axons (Fig. 8J). Together, these data demonstrate that alternative SLOB isoforms are expressed in distinct cell types in the presynaptic cellular complement of the larval NMJ, with SLOB57 localized predominantly to glial cells and SLOB71 expressed in motor neurons.
Discussion
Modulation of neuronal excitability via changes in the properties of membrane ion channels is a ubiquitous and fundamental feature of neuronal function. The pore-forming subunits of many ion channels are often intimately associated with auxiliary subunits and other binding partners, the interaction with which may profoundly influence such channel properties as voltage-dependent gating. The SLOWPOKE large conductance calcium- and voltage-dependent potassium channel is among those channels that are subject to complex modulation by a variety of molecular mechanisms. As a model for understanding the molecular details and physiological consequences of ion channel regulation, we have focused our attention on the modulation of the Drosophila SLOWPOKE channel by its binding partner SLOB.
We previously studied the role of SLOB in vivo by knocking out or knocking down its expression by P-element mutagenesis or targeted expression of SLOB–RNAi, respectively, followed by electrophysiological analysis. We found that PI neurons in adult flies exhibit a decrease in membrane excitability when SLOB is knocked out/knocked down (Shahidullah et al., 2009), consistent with the effects of one SLOB isoform, SLOB57, on SLOWPOKE channels in a heterologous expression system (Zeng et al., 2005). However, to our surprise, glutamatergic synaptic transmission at the fly larval NMJ was enhanced by SLOB knock-out/knockdown (Ma et al., 2011), and we show here that motor neuron cell body excitability is also higher in the absence of SLOB. Because the actions of modulatory proteins are rarely studied in multiple neuronal types, there is limited information regarding such cell-type-specific ion channel modulation. To determine whether differential expression of SLOB isoforms might be responsible for these observations, we embarked on a characterization of the expression patterns of two prominent isoforms in larval and adult Drosophila.
We first characterized the TSSs and promoter sequences that regulate the expression of slob57 and slob71. Two separate TSSs were identified for slob57 and slob71, which are slightly different from those listed in FlyBase for the reference Drosophila genome y1;cn1bw1sp1 (Adams et al., 2000; McQuilton et al., 2012). Interestingly, slob71 promoter activity was much higher than that of slob57, and this effect was primarily dependent on DPE and MTE core promoter elements in slob71 that are absent in slob57. However, a slob71 promoter lacking functional DPE and MTE core promoter elements still exhibited greater promoter activity compared with the slob57 promoter; thus, additional elements that facilitate transcription in the slob71 promoter or repress transcription in slob57 promoter may account for this discrepancy. All combinations of promoter configurations for core promoter elements (Inr, TATA box, DPE, and MTE) alone or in tandem are found in Drosophila (Kutach and Kadonaga, 2000; Lim et al., 2004). The DPE is conserved from Drosophila to humans and is frequently found in Drosophila promoters lacking a TATA box element within the core promoter (Burke and Kadonaga, 1997). Furthermore, the DPE is as widely used as the TATA box in Drosophila promoters (Kutach and Kadonaga, 2000). More recently, the MTE has been identified as a core promoter element and may function alone or in tandem with the DPE (Lim et al., 2004). When both the MTE and DPE are present, one element may compensate for the other. In some genes, the DPE is the stronger activator of transcription, and for others the MTE is more important. In our experiments, mutation of the DPE decreased promoter activity slightly more than mutation of the MTE, suggesting that the DPE plays a larger role in transcriptional activation from the slob71 promoter. However, mutation of either core element significantly decreased promoter activity, and mutation of both the MTE and DPE severely inhibited promoter activity to <20% of the control promoter.
By analyzing fragments of the slob71 promoter, we focused on the domain from −1966 to −1500 as a probable region for repressor elements and identified HB and MIRR sites within this region. The zinc finger transcription factor HB is an important regulator of anterior/posterior development in Drosophila (Tran et al., 2010), and the homeobox transcription factor MIRR also functions in pattern formation during development (Ikmi et al., 2008). HB acts as a repressor for most of its targets in the Drosophila CNS and suppresses promoter activity in murine models as well (Yamazaki et al., 2006; Tran et al., 2010). MIRR has also been shown to directly repress gene expression in Drosophila (Andreu et al., 2012). Our results are consistent with repression of the slob71 promoter by HB and MIRR, because mutation of HB and MIRR recognition sites upregulated promoter activity. Interestingly, repression through the MIRR recognition consensus sequence appears to require specific sequences within the promoter of slob71, because mutation of the MIRR site within a fragment of the slob71 promoter had no effect on promoter activity from the minP. In contrast, repression through the HB recognition site functioned in both the full-length slob71 promoter construct and the fragment construct upstream of the minP, suggesting that repression through the HB site does not require specific core promoter elements within the slob71 promoter. These findings parallel recent work demonstrating that the Lim-homeodomain transcription factor ISLET regulates potassium currents in ventral larval motor neurons via negative control of the Shaker potassium channel (Wolfram et al., 2012). Our results suggest that the HB and MIRR transcription factors may similarly negatively regulate a distinct potassium channel, SLO, albeit indirectly through the inhibition of SLOB71 expression.
The structural differences observed between the slob71 and slob57 promoters are mirrored by distinct spatial patterns of expression in the larval and adult nervous systems of Drosophila. slob71 promoter sequences drive expression in the adult mushroom body and larval motor neurons, which do not exhibit labeling when using slob57 promoter sequences to drive reporter expression. Conversely, the slob57 promoter shows activity in the adult antennal lobes and larval glial cells, which do not exhibit slob71 activity. In Drosophila, many genes that are subject to developmental regulation have multiple transcriptional start sites that drive divergent spatiotemporal patterns of gene expression (Batut et al., 2013). Alternative splicing provides an additional regulatory mechanism to generate functionally distinct protein isoforms from the same locus and has been shown previously to regulate the operational properties of several mammalian channel-interacting proteins (Boland et al., 2003; Van Hoorick et al., 2003; Lewis et al., 2009). In some cases, alternate splice forms can have markedly different effects on channel properties. For example, differentially spliced isoforms of the cyclic nucleotide-gated channel (HCN) interacting protein TRIP8b can either inhibit or potentiate HCN currents by altering the surface trafficking of the channel (Lewis et al., 2009).
The SLOB57 and SLOB71 alternative isoforms also display opposing influences on SLOWPOKE channel function, with SLOB57 inhibiting SLOWPOKE currents and SLOB71 potentiating (Zeng et al., 2005). Spatiotemporal control of the expression of these two isoforms thus provides a mechanism to tune SLOWPOKE function in a cell-specific manner. Consistent with this hypothesis, in larval motor neurons, SLOB acts to potentiate SLOWPOKE currents (Ma et al., 2011), and our data indicate that this effect is mediated by restricted expression of SLOB71 rather than SLOB57, which in turn is controlled by differential activity of the two slob isoform promoters in concert with alternative splicing of slob mRNA. Of note, transcripts encoding SLOWPOKE also undergo tissue-specific, developmentally regulated alternative splicing (Yu et al., 2006). Thus, it is likely that interactions between differentially spliced isoforms of both SLOWPOKE and its regulator SLOB, as well as other spatially and temporally regulated processes such as RNA editing, which modifies the amino-acid sequence of SLOWPOKE channels (Hoopengardner et al., 2003; Jepson et al., 2011), determines the net level of cell-specific SLOWPOKE activity.
Interestingly, there is nonetheless a restricted degree of overlap between the slob71 and slob57 promoters. One prominent cell type exhibiting such overlap is the adult PI, which is strongly labeled using reporters of both promoters. Previous studies have indicated that SLOB expression in the PI acts to inhibit SLOWPOKE channel activity, consistent with the known activity of the SLOB57 isoform (Shahidullah et al., 2009). However, our results suggest that both isoforms are expressed within the PI. It is possible that distinct cells within the PI express different SLOB isoforms. Alternatively, both isoforms may be simultaneously expressed in the same neurons, and the ratio of SLOB57/SLOB71 determines the net effect on SLOWPOKE channel function. Additional experiments will be required to examine in detail the expression of these two SLOB isoforms within the PI and whether particular SLOB isoforms act in a dominant or additive manner.
In conclusion, we have uncovered a regulatory mechanism by which neuronal excitability is differentially tuned via cell-type-specific expression of functionally distinct protein isoforms derived from the same genetic locus and resolved previous results demonstrating apparent opposing activities of SLOB proteins in different cellular contexts. Our findings emphasize both the complexity and the flexibility of ion channel modulation by intimately associated protein binding partners.
Footnotes
This study was funded by National Institute of Neurological Disorders and Stroke Grant NS17910 (I.B.L.) and National Institute of General Medical Sciences Grant R01GM088221 (K.K.) from the National Institutes of Health. We are grateful to Lauren Manderfield for mapping SLOB isoform transcriptional start sites and Karen Monono for confirming the activity of several of the promoter constructs.
The authors declare no competing financial interests.
References
- Adams MD, Celniker SE, Holt RA, Evans CA, Gocayne JD, Amanatides PG, Scherer SE, Li PW, Hoskins RA, Galle RF, George RA, Lewis SE, Richards S, Ashburner M, Henderson SN, Sutton GG, Wortman JR, Yandell MD, Zhang Q, Chen LX, et al. The genome sequence of Drosophila melanogaster. Science. 2000;287:2185–2195. doi: 10.1126/science.287.5461.2185. [DOI] [PubMed] [Google Scholar]
- Adelman JP, Shen KZ, Kavanaugh MP, Warren RA, Wu YN, Lagrutta A, Bond CT, North RA. Calcium-activated potassium channels expressed from cloned complementary DNAs. Neuron. 1992;9:209–216. doi: 10.1016/0896-6273(92)90160-F. [DOI] [PubMed] [Google Scholar]
- Andreu MJ, González-Pérez E, Ajuria L, Samper N, González-Crespo S, Campuzano S, Jiménez G. Mirror represses pipe expression in follicle cells to initiate dorsoventral axis formation in Drosophila. Development. 2012;139:1110–1114. doi: 10.1242/dev.076562. [DOI] [PubMed] [Google Scholar]
- Atkinson NS, Robertson GA, Ganetzky B. A component of calcium-activated potassium channels encoded by the Drosophila slo locus. Science. 1991;253:551–555. doi: 10.1126/science.1857984. [DOI] [PubMed] [Google Scholar]
- Atkinson NS, Brenner R, Bohm RA, Yu JY, Wilbur JL. Behavioral and electrophysiological analysis of Ca-activated K-channel transgenes in Drosophila. Ann NY Acad Sci. 1998;860:296–305. doi: 10.1111/j.1749-6632.1998.tb09057.x. [DOI] [PubMed] [Google Scholar]
- Barolo S, Castro B, Posakony JW. New Drosophila transgenic reporters: insulated P-element vectors expressing fast-maturing RFP. Biotechniques. 2004;36:436–440. 442. doi: 10.2144/04363ST03. [DOI] [PubMed] [Google Scholar]
- Batut P, Dobin A, Plessy C, Carninci P, Gingeras TR. High-fidelity promoter profiling reveals widespread alternative promoter usage and transposon-driven developmental gene expression. Genome Res. 2013;23:169–180. doi: 10.1101/gr.139618.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boland LM, Jiang M, Lee SY, Fahrenkrug SC, Harnett MT, O'Grady SM. Functional properties of a brain-specific NH2-terminally spliced modulator of Kv4 channels. Am J Physiol. 2003;285:C161–C170. doi: 10.1152/ajpcell.00416.2002. [DOI] [PubMed] [Google Scholar]
- Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- Brink DL, Gilbert M, Xie X, Petley-Ragan L, Auld VJ. Glial processes at the Drosophila larval neuromuscular junction match synaptic growth. PLoS One. 2012;7:e37876. doi: 10.1371/journal.pone.0037876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke TW, Kadonaga JT. The downstream core promoter element, DPE, is conserved from Drosophila to humans and is recognized by TAFII60 of Drosophila. Genes Dev. 1997;11:3020–3031. doi: 10.1101/gad.11.22.3020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartharius K, Frech K, Grote K, Klocke B, Haltmeier M, Klingenhoff A, Frisch M, Bayerlein M, Werner T. MatInspector and beyond: promoter analysis based on transcription factor binding sites. Bioinformatics. 2005;21:2933–2942. doi: 10.1093/bioinformatics/bti473. [DOI] [PubMed] [Google Scholar]
- Elkins T, Ganetzky B. The roles of potassium currents in Drosophila flight muscles. J Neurosci. 1988;8:428–434. doi: 10.1523/JNEUROSCI.08-02-00428.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elkins T, Ganetzky B, Wu CF. A Drosophila mutation that eliminates a calcium-dependent potassium current. Proc Natl Acad Sci U S A. 1986;83:8415–8419. doi: 10.1073/pnas.83.21.8415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujioka M, Lear BC, Landgraf M, Yusibova GL, Zhou J, Riley KM, Patel NH, Jaynes JB. Even-skipped, acting as a repressor, regulates axonal projections in Drosophila. Development. 2003;130:5385–5400. doi: 10.1242/dev.00770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghezzi A, Al-Hasan YM, Larios LE, Bohm RA, Atkinson NS. slo K(+) channel gene regulation mediates rapid drug tolerance. Proc Natl Acad Sci U S A. 2004;101:17276–17281. doi: 10.1073/pnas.0405584101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoopengardner B, Bhalla T, Staber C, Reenan R. Nervous system targets of RNA editing identified by comparative genomics. Science. 2003;301:832–836. doi: 10.1126/science.1086763. [DOI] [PubMed] [Google Scholar]
- Ikmi A, Netter S, Coen D. Prepatterning the Drosophila notum: the three genes of the iroquois complex play intrinsically distinct roles. Dev Biol. 2008;317:634–648. doi: 10.1016/j.ydbio.2007.12.034. [DOI] [PubMed] [Google Scholar]
- Jaramillo AM, Zheng X, Zhou Y, Amado DA, Sheldon A, Sehgal A, Levitan IB. Pattern of distribution and cycling of SLOB, Slowpoke channel binding protein, in Drosophila. BMC Neurosci. 2004;5:3. doi: 10.1186/1471-2202-5-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaramillo AM, Zeng H, Fei H, Zhou Y, Levitan IB. Expression and function of variants of slob, slowpoke channel binding protein, in Drosophila. J Neurophysiol. 2006;95:1957–1965. doi: 10.1152/jn.00427.2005. [DOI] [PubMed] [Google Scholar]
- Jepson JE, Savva YA, Yokose C, Sugden AU, Sahin A, Reenan RA. Engineered alterations in RNA editing modulate complex behavior in Drosophila: regulatory diversity of adenosine deaminase acting on RNA (ADAR) targets. J Biol Chem. 2011;286:8325–8337. doi: 10.1074/jbc.M110.186817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jepson JE, Shahidullah M, Lamaze A, Peterson D, Pan H, Koh K. dyschronic, a Drosophila homolog of a deaf-blindness gene, regulates circadian output and Slowpoke channels. PLoS Genet. 2012;8:e1002671. doi: 10.1371/journal.pgen.1002671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutach AK, Kadonaga JT. The downstream promoter element DPE appears to be as widely used as the TATA box in Drosophila core promoters. Mol Cell Biol. 2000;20:4754–4764. doi: 10.1128/MCB.20.13.4754-4764.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levitan IB. Signaling protein complexes associated with neuronal ion channels. Nat Neurosci. 2006;9:305–310. doi: 10.1038/nn1647. [DOI] [PubMed] [Google Scholar]
- Lewis AS, Schwartz E, Chan CS, Noam Y, Shin M, Wadman WJ, Surmeier DJ, Baram TZ, Macdonald RL, Chetkovich DM. Alternatively spliced isoforms of TRIP8b differentially control h channel trafficking and function. J Neurosci. 2009;29:6250–6265. doi: 10.1523/JNEUROSCI.0856-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim CY, Santoso B, Boulay T, Dong E, Ohler U, Kadonaga JT. The MTE, a new core promoter element for transcription by RNA polymerase II. Genes Dev. 2004;18:1606–1617. doi: 10.1101/gad.1193404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H, Zhang J, Levitan IB. Slob, a Slowpoke channel-binding protein, modulates synaptic transmission. J Gen Physiol. 2011;137:225–238. doi: 10.1085/jgp.201010439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McQuilton P, St Pierre SE, Thurmond J, FlyBase Consortium. FlyBase 101—the basics of navigating FlyBase. Nucleic Acids Res. 2012;40:D706–D714. doi: 10.1093/nar/gkr1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schopperle WM, Holmqvist MH, Zhou Y, Wang J, Wang Z, Griffith LC, Keselman I, Kusinitz F, Dagan D, Levitan IB. Slob, a novel protein that interacts with the Slowpoke calcium-dependent potassium channel. Neuron. 1998;20:565–573. doi: 10.1016/S0896-6273(00)80995-2. [DOI] [PubMed] [Google Scholar]
- Shahidullah M, Reddy S, Fei H, Levitan IB. In vivo role of a potassium channel-binding protein in regulating neuronal excitability and behavior. J Neurosci. 2009;29:13328–13337. doi: 10.1523/JNEUROSCI.3024-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran KD, Miller MR, Doe CQ. Recombineering Hunchback identifies two conserved domains required to maintain neuroblast competence and specify early-born neuronal identity. Development. 2010;137:1421–1430. doi: 10.1242/dev.048678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vacher H, Trimmer JS. Diverse roles for auxiliary subunits in phosphorylation-dependent regulation of mammalian brain voltage-gated potassium channels. Pflugers Arch. 2011;462:631–643. doi: 10.1007/s00424-011-1004-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Hoorick D, Raes A, Keysers W, Mayeur E, Snyders DJ. Differential modulation of Kv4 kinetics by KCHIP1 splice variants. Mol Cell Neurosci. 2003;24:357–366. doi: 10.1016/S1044-7431(03)00174-X. [DOI] [PubMed] [Google Scholar]
- Wolfram V, Southall TD, Brand AH, Baines RA. The LIM-homeodomain protein islet dictates motor neuron electrical properties by regulating K(+) channel expression. Neuron. 2012;75:663–674. doi: 10.1016/j.neuron.2012.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worrell JW, Levine RB. Characterization of voltage-dependent Ca2+ currents in identified Drosophila motoneurons in situ. J Neurophysiol. 2008;100:868–878. doi: 10.1152/jn.90464.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu JS, Luo L. A protocol for dissecting Drosophila melanogaster brains for live imaging or immunostaining. Nat Protoc. 2006;1:2110–2115. doi: 10.1038/nprot.2006.336. [DOI] [PubMed] [Google Scholar]
- Wu MN, Joiner WJ, Dean T, Yue Z, Smith CJ, Chen D, Hoshi T, Sehgal A, Koh K. SLEEPLESS, a Ly-6/neurotoxin family member, regulates the levels, localization and activity of Shaker. Nat Neurosci. 2010;13:69–75. doi: 10.1038/nn.2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki K, Crowe DL, Shuler CF. Hunchback sequence binding protein suppresses mouse TGF-beta3 promoter in vitro. Biochem Biophys Res Commun. 2006;346:802–809. doi: 10.1016/j.bbrc.2006.05.178. [DOI] [PubMed] [Google Scholar]
- Yu JY, Upadhyaya AB, Atkinson NS. Tissue-specific alternative splicing of BK channel transcripts in Drosophila. Genes Brain Behav. 2006;5:329–339. doi: 10.1111/j.1601-183X.2005.00164.x. [DOI] [PubMed] [Google Scholar]
- Zeng H, Weiger TM, Fei H, Jaramillo AM, Levitan IB. The amino terminus of Slob, Slowpoke channel binding protein, critically influences its modulation of the channel. J Gen Physiol. 2005;125:631–640. doi: 10.1085/jgp.200509252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Schopperle WM, Murrey H, Jaramillo A, Dagan D, Griffith LC, Levitan IB. A dynamically regulated 14-3-3, Slob, and Slowpoke potassium channel complex in Drosophila presynaptic nerve terminals. Neuron. 1999;22:809–818. doi: 10.1016/S0896-6273(00)80739-4. [DOI] [PubMed] [Google Scholar]