Abstract
TCP family of plant-specific transcription factors regulates plant form through control of cell proliferation and differentiation. This gene family is comprised of two groups, class I and class II. While the role of class II TCP genes in plant development is well known, data about the function of some class I TCP genes is lacking. We studied a group of phylogenetically related class I TCP genes: AtTCP7, AtTCP8, AtTCP22, and AtTCP23. The similar expression pattern in young growing leaves found for this group suggests similarity in gene function. Gene redundancy is characteristic in this group, as also seen in the class II TCP genes. We generated a pentuple mutant tcp8 tcp15 tcp21 tcp22 tcp23 and show that loss of function of these genes results in changes in leaf developmental traits. We also determined that these factors are able to mutually interact in a yeast two-hybrid assay and regulate the expression of KNOX1 genes. To circumvent the issue of genetic redundancy, dominant negative forms with SRDX repressor domain were used. Analysis of transgenic plants expressing AtTCP7-SRDX and AtTCP23-SRDX indicate a role of these factors in the control of cell proliferation.
Keywords: Arabidopsis, TCP, transcription factor, leaf development, SRDX construct
Introduction
Plant architecture and organ form rely on developmental processes which involve the control of cell proliferation, cell growth and cell differentiation (Ingram and Waites, 2006). TCP factors are involved in the coordination of cell proliferation and cell differentiation, having roles in several aspects of plant development, such as regulation of the shoot apical meristem (SAM) (Koyama et al., 2010; Li et al., 2012b), leaf development (Palatnik et al., 2003; Ori et al., 2007), and lateral branching (Doebley et al., 1997; Aguilar-Martínez et al., 2007).
TCPs are a plant-specific family of transcription factors, named after the transcription factors TEOSINTE BRANCHED1 (TB1) in maize, CYCLOIDEA (CYC) in Antirrhinum majus, and PCF1 and PCF2 (for PROLIFERATING CELL NUCLEAR ANTIGEN FACTOR1 and 2) in rice (Navaud et al., 2007; Busch and Zachgo, 2009).
TCP genes code for proteins with a 59-amino acid non-canonical basic helix-loop-helix (bHLH), the TCP domain, involved in DNA binding and dimerization (Cubas et al., 1999; Aggarwal et al., 2010). TCP proteins can be classified into two groups based on differences in the structure of the TCP domain, known as class I (or PCF or TCP-P) and class II (or TCP-C) [reviewed in Uberti Manassero et al. (2013)]. Mainly based on the TCP domain structure, class II TCP proteins can be subdivided into the CIN-like clade, with genes involved in lateral organ development such as CINCINNATA (CIN) in Antirrhinum (Nath et al., 2003), and the CYC/TB1 clade, with genes controlling axillary meristem development, such as TB1 in maize (Doebley et al., 1997), and its orthologs in Arabidopsis (Arabidopsis thaliana) BRANCHED1 (BRC1/TCP18) and BRANCHED2 (BRC2/TCP12) (Aguilar-Martínez et al., 2007; Finlayson, 2007). Both class I and class II include members that can function as transcriptional activators and repressors (Uberti Manassero et al., 2013).
There are 24 predicted TCP proteins in the Arabidopsis genome, 13 class I and 11 class II proteins (Cubas, 2002). Functional analysis has been reported for most of the class II TCP genes. A subset of them, AtTCP2, AtTCP3, AtTCP4, AtTCP10, and AtTCP24, are regulated by miR319a, produced by the JAW locus (Palatnik et al., 2003). In jaw-D, miR319a is overexpressed, which in turns down-regulates AtTCP2, AtTCP3, AtTCP4, AtTCP10, and AtTCP24, resulting in plants which exhibit aberrant cell division, causing a serrated and crinkly leaf margin phenotype (Palatnik et al., 2003). On the contrary, forms of these five mRNAs that are resistant to the action of miR319a induce defects in the formation of a functional shoot meristem and cause fusion of cotyledons (Palatnik et al., 2003; Koyama et al., 2007; Nag et al., 2009). AtTCP3 is proposed to regulate the redundant CUC (for CUP-SHAPED COTYLEDON) genes, CUC1 and CUC2, that function to delimit boundaries between lateral organs (Koyama et al., 2007). miRNA319-targeted TCPs interact with ASYMMETRIC LEAVES 2 (AS2) and regulate the expression of KNOTTED1-like HOMEOBOX (KNOX) genes BREVIPEDICELLUS (BP) and KNAT2 (Li et al., 2012b). In tomato, a compound leaved species, LANCEOLATE (LA), an AtTCP4 homolog, controls compound leaf development. In the mutant Lanceolate, a miR319 resistant version of the TCP gene stimulates precocious cell differentiation, resulting in simple leaved plants (Ori et al., 2007). Mutant plants for BRC1/AtTCP18 exhibit a loss of apical dominance while tcp12 mutants show a milder phenotype (Aguilar-Martínez et al., 2007; Finlayson, 2007). BRC1 interacts with the florigen proteins FLOWERING LOCUS T (FT) and TWIN SISTER OF FT (TSF) to suppress the floral transition in axillary buds (Niwa et al., 2013). In case of AtTCP1, the Arabidopsis homolog of CYC, the mutant tcp1 shows no obvious phenotype (Cubas et al., 2001).
TCP proteins have been shown to mediate hormonal action in some instances. AtTCP1 regulates DWARF4, which produces an enzyme involved in brassinosteroid biosynthesis (Guo et al., 2010). AtTCP3 activates the expression of the auxin signaling repressor IAA3/SHY2 (for INDOLE-3-ACETIC ACID3/SHORT HYPOCOTYL2, Koyama et al., 2010). AtTCP4 regulates the expression of the jasmonate biosynthesis gene LIPOXYGENASE2 (Schommer et al., 2008) while the tomato LANCEOLATE acts to promote gibberellic acid biosynthesis (Yanai et al., 2011).
Much less is known about the function of class I TCP proteins in plant development. Of these, AtTCP21/CHE (for CCA1 HIKING EXPEDITION) is involved in the regulation of circadian clock activity (Pruneda-Paz et al., 2009). Also AtTCP15 and AtTCP11 interact with the core circadian component PRR5 (Giraud et al., 2010). AtTCP15 also regulates endoreduplication by modulating the expression of key cell-cycle genes (Li et al., 2012a). AtTCP16 is involved in pollen development (Takeda et al., 2006). AtTCP8 is found as one interactor factor with PNM1 (PPR protein to the nucleus and mitochondria 1), a potential coordinator of the expression of mitochondrial and nuclear genomes (Hammani et al., 2011). The expression of a repressor form of AtTCP11 (Viola et al., 2011) or AtTCP20 (Hervé et al., 2009) produces plants with pleiotropic developmental alterations. AtTCP14 regulates embryonic growth potential (Tatematsu et al., 2008; Rueda-Romero et al., 2012). AtTCP15, along with its closest homolog AtTCP14, has a role in internode length and leaf shape (Kieffer et al., 2011). Both AtTCP14 and AtTCP15 are found to interact with SPINDLY (SPY) to promote cytokinin responses (Steiner et al., 2012a) and increase the sensibility to this hormone when overexpressed in tomato (Steiner et al., 2012b).
Interplay between class I and class II TCP proteins has also been reported. Class I and class II proteins have distinct but overlapping binding sites: G(T/C)GGNCCC (Kosugi and Ohashi, 2002). Both classes may perform their functions by interacting with similar sets of target genes (Kosugi and Ohashi, 2002; Li et al., 2005). CIN-like TCP proteins regulate the expression of CUC genes through binding of the promoters of miR164, ASYMMETRIC LEAVES1 (AS1), IAA3/SHY2, and the SMALL AUXIN UP RNA (SAUR) gene At1g29460 (Koyama et al., 2010), while AtTCP15, a class I TCP factor, also indirectly regulates CUC genes through binding the promoters of IAA3/SHY2 and the SAUR gene At1g29460 (Uberti-Manassero et al., 2012).
Here we study the developmental role of class I TCP genes AtTCP7 (At5g23280), AtTCP8 (At1g58100), AtTCP22 (At1g72010), and AtTCP23 (At1g35560). AtTCP7, AtTCP8, AtTCP22, and AtTCP23 are expressed mainly in growing leaf tissues. Analysis of the pentuple mutant tcp8 tcp15 tcp21 tcp22 tcp23 reveals the high level of genetic redundancy in the TCP gene family. These genes also interact with each other at the protein level. Plants with SRDX repression domain fusions for AtTCP7 and AtTCP23 show similar pleiotropic developmental alterations.
Results
Genetic redundancy is a characteristic of class I TCP factors
Despite the described role of some TCP factors in plant development [reviewed in Uberti Manassero et al. (2013)] little is known about the function of a subset of class I TCP factors, AtTCP7, AtTCP8, AtTCP22, and AtTCP23. We determined the phylogenetic relationships of class I TCP factors using sequences from Arabidopsis, tomato, and rice (Figure 1 and Supplemental Figure 1). This analysis shows that AtTCP7, AtTCP8, AtTCP14, AtTCP15, AtTCP21, AtTCP22, and AtTCP23 form a distinct clade when compared to the rest of the factors in this subfamily such as AtTCP20, AtTCP9, AtTCP19, AtTCP11, AtTCP6, and AtTCP16. AtTCP7 and AtTCP21 are close homologs and show a 62% of sequence similarity at the protein level. AtTCP14 and AtTCP15 cluster together (Figure 1). Due to high sequence similarity, AtTCP8, AtTCP22, and AtTCP23 do not show clear phylogenetic relationships in this subgroup of class I TCP factors.
Figure 1.
Phylogenetic tree of class I TCP genes. Phylogenetic tree of class I TCP proteins from Arabidopsis, tomato, and rice. Protein sequences were analyzed with the maximum-likelihood method by MEGA 5 software. Bootstrap values after 1000 replicates higher than 50% are shown. The tree was rooted using the translated sequence of the maize teosinte-branched1 (tb1) gene as outgroup. In bold letters are the genes analyzed in this study.
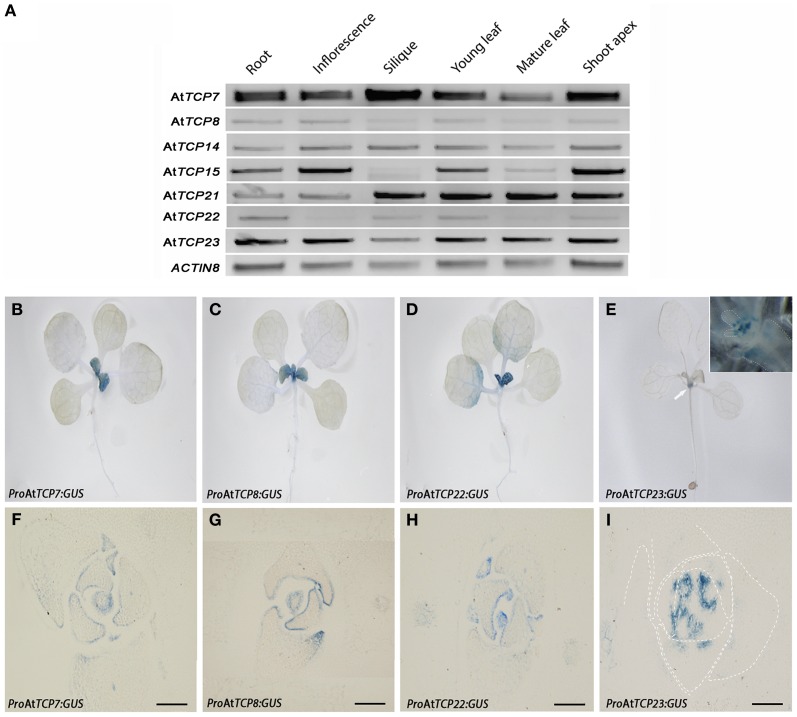
We analyzed the expression pattern of class I TCP genes by RT-PCR in several tissues of Arabidopsis wild type plants (Figure 2A). AtTCP7, AtTCP14, AtTCP21, and AtTCP23 are expressed in all the tissues analyzed. AtTCP15 and AtTCP8 are expressed at low levels in siliques and mature leaves, while AtTCP22 is not detected in inflorescences and mature leaves (Figure 2A). Expression data from Genevestigator (Hruz et al., 2008), the Arabidopsis eFP browser at BAR (Winter et al., 2007), AtGenExpress (Schmid et al., 2005), and GeneCAT (Mutwil et al., 2008) supports our RT-PCR analysis for AtTCP8, AtTCP14, AtTCP15, AtTCP21, and AtTCP23, while for AtTCP7 and AtTCP22 there is no expression data yet available in these resources. Is interesting to note that according to this data AtTCP8 shows a high expression level in dry seeds.
Figure 2.
Expression pattern of class I TCP genes. (A) RT-PCR analysis of the tissue expression of class I TCP genes. ACTIN8 was used as control gene. (B–I) Analysis of the expression pattern using the reporter marker uidA fused to the promoters of the genes in 12 days-old T3 transgenic plants. (B) ProAtTCP7:GUS; (C) ProAtTCP8:GUS; (D) ProAtTCP22:GUS; (E) ProAtTCP23:GUS. Inset, shoot apex, dashed lines indicate leaf primordia or leaf petiole margins. Arrow indicates the localization of the GUS signal. (F–I) Cross-sections of 12 days-old T3 lines. (F) ProAtTCP7:GUS; (G) ProAtTCP8:GUS; (H) ProAtTCP22:GUS; (I) ProAtTCP23:GUS. Dashed lines indicate leaf primordia. Bars = 100 μm.
To determine the spatial expression of class I TCP factors, we fused the complete 5′ gene promoter regions of AtTCP7, AtTCP8, AtTCP22, and AtTCP23 to the coding sequence of the uidA marker gene and analyzed the GUS expression pattern in 12 days-old plants of T3 lines (Figures 2B–I). We found that the localization of GUS is restricted to young emerging leaves in plants ProAtTCP7:GUS (Figure 2B), ProAtTCP8:GUS (Figure 2C) and ProAtTCP22:GUS (Figure 2D). In plants ProAtTCP23:GUS the GUS signal is detected at the base of growing leaves (Figure 2E). Analysis of cross-sections showed that the GUS signal is located at the surface of the lateral organs and mainly in the adaxial side of the young leaves for plants ProAtTCP7:GUS (Figure 2F), ProAtTCP8:GUS (Figure 2G), and ProAtTCP22:GUS (Figure 2H). In plants ProAtTCP23:GUS the signal is found at the base of the growing leaves (Figure 2I). We did not detect signal at later stages of development in structures such as stem internodes, inflorescences, or flowers (data not shown). Thus, the class I TCP factors we studied have a similar expression pattern hinting at a similar function for these genes.
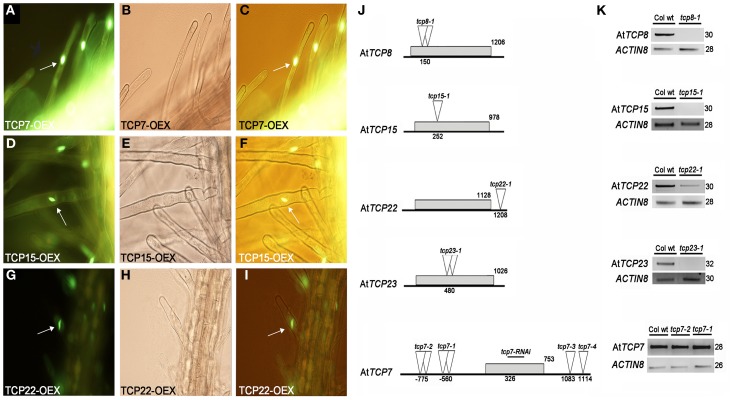
TCP genes are reported to code for a plant-specific family of transcription factors (Cubas et al., 1999). We analyzed the subcellular location of AtTCP7, AtTCP15, and AtTCP22 through fusions of the coding sequences of the corresponding genes to the marker GFP. We found that the fusion proteins are localized in the nucleus in case of AtTCP7 (Figures 3A–C), AtTCP15 (Figures 3D–F), and AtTCP22 (Figures 3G–I), suggesting a role of these TCP factors as transcription factors.
Figure 3.
Subcellular localization of class I TCP factors in root hair cells and characterization of class I TCP alleles. (A–C) AtTCP7, (D–F) AtTCP15, (G–I) AtTCP22. (A,D,G) The GFP signal is located at the nuclei (B,E,H) Bright-field images. (C,F,I) Merged images. Arrows indicate nuclear location. (J) Scheme of the genes and position of the insertions. Gray boxes indicate the coding sequence of the genes. Triangles indicate the insertion lines. Double triangles indicate a tandem insertion. The fragment used for RNAi lines for tcp7-RNAi is indicated. Numbers indicate the position of the insertions or the length of the coding sequences respect to the start codon. (K) RT-PCR analysis of the alleles. ACTIN8 was used as internal control. Number of cycles is indicated.
To determine the function of selected class I TCP genes, we characterized T-DNA insertion lines (Figure 3J). AtTCP7, AtTCP8, AtTCP15, AtTCP22, and AtTCP23 are intron-less genes. For AtTCP8, AtTCP15, and AtTCP23 we used the null alleles tcp8-1, tcp15-1, and tcp23-1, respectively (Figure 3K). For AtTCP22, the allele tcp22-1 showed a reduction in the level of the gene product (Figure 3K). In case of AtTCP7 we used four available insertion lines, however none of them was located in the coding sequence. RT-PCR expression analysis of the alleles tcp7-1 and tcp7-2 indicated that insertions outside the coding region do not lead to a reduction of transcript levels of AtTCP7 compared with the wild type (Figure 3K).
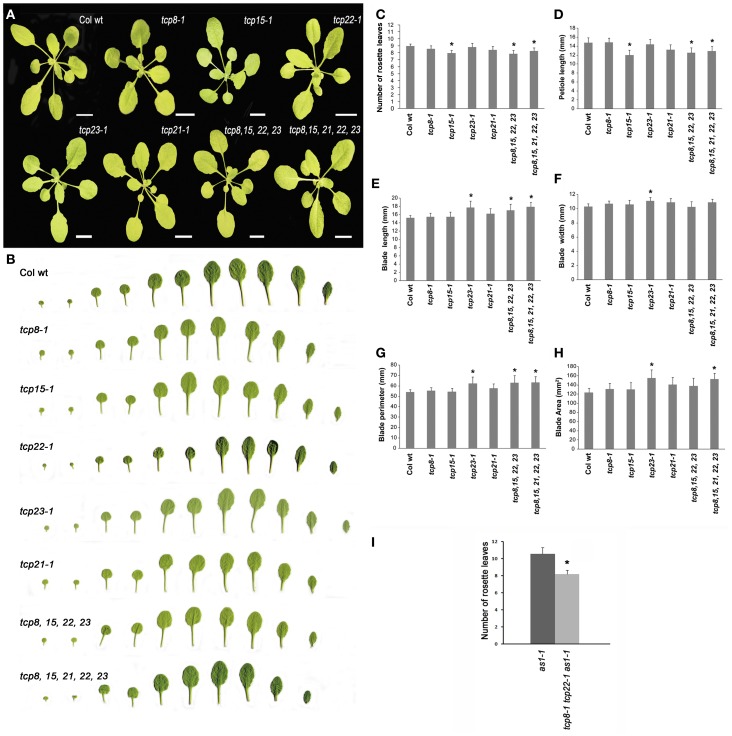
Phenotypically the single mutants are overall similar to the wild type (Figures 4A,B). We determined however that tcp15-1 has reduced number of rosette leaves (Figure 4C). A quantitative analysis of leaf traits revealed that tcp15-1 has shorter petiole length than wild type (Figure 4D). Kieffer et al. (2011) found a slightly shorter fruit pedicel length and shorter inflorescence height in this mutant. We also found that tcp23-1 has increased blade length (Figure 4E), blade width (Figure 4F), blade perimeter (Figure 4G) and blade area (Figure 4H) compared to wild type. However we found that tcp8-1 and tcp21-1 plants are indistinguishable from wild type. For AtTCP7 we made RNAi lines (tcp7-RNAi) but the transgenic plants were similar to the wild type. As the analyzed single mutants only exhibited subtle phenotypic changes compared to the wild type we made multiple mutant combinations in the class I TCP genes reasoning that loss of function at multiple class I TCP genes would reveal more evident changes in plant development. We analyzed the quadruple mutant tcp8-1 tcp15-1 tcp22-1 tcp23-1 and the pentuple mutant tcp8-1 tcp15-1 tcp22-1 tcp23-1 tcp21-1. The overt phenotype of both the quadruple and pentuple mutant was similar to the wild type (Figures 4A,B), although we found that these multiple mutants have fewer rosette leaves (Figure 4C) and bigger leaves than wild type (Figures 4E,G,H).
Figure 4.
Phenotypic characterization of class I TCP gene mutants. (A) Rosettes at bolting stage of Col wt, the single mutants tcp8-1, tcp15-1, tcp22-1, tcp23-1, and tcp21-1, the quadruple mutant tcp8-1 tcp15-1 tcp22-1 tcp23-1 (tcp8, 15, 22, 23) and the pentuple mutant tcp8-1 tcp15-1 tcp22-1 tcp23-1 tcp21-1 (tcp8, 15, 21, 22, 23). Bars are 1 cm. (B) Leaf series of plants at bolting stage of Col wt and the mutants tcp8-1, tcp15-1, tcp22-1, tcp23-1, tcp21-1, tcp8, 15, 22, 23, and tcp8, 15, 22, 23, 21. (C–H) Characterization of leaf traits in Col wt and class I TCP gene mutants. Number of rosette leaves (C), petiole length (D), blade length (E), blade width (F), blade perimeter (G), and blade area (H). (I) Number of rosette leaves in AS1 allele as1-1 and in the triple mutant as1-1 tcp8-1 tcp22-1. n = 13 to 28 in (C–H); n = 30 to 33 in (I). Asterisks (*) indicate statistically significant differences determined by Student's t-test, p < 0.05.
Mutations in AS1 produce leaves with altered leaf shape, with the leaf lamina highly lobed (Byrne et al., 2000). CIN-like members of class II TCP genes also regulate leaf shape (Nath et al., 2003; Palatnik et al., 2003). Indeed, CIN-like TCP genes regulate the expression of AS1 (Koyama et al., 2007). We wanted to determine if class I TCP mutations interact with the leaf shape phenotype of as1. We used the double mutant tcp8 tcp22 to generate the triple mutant as1 tcp8 tcp22. We found that the number of rosette leaves produced in the triple mutant plants is reduced compared with the number of rosette leaves in as1 while the leaf shape resemble the leaves of as1 (Figure 4I), reinforcing the finding of class I TCP genes as regulators of leaf initiation rate.
In summary, this analysis indicates that single mutations in class I TCP genes have mild phenotypes. The quadruple and pentuple mutants have increased defects relating to number of leaves and leaf size while still maintaining normal morphology, indicating that other members in this subgroup of genes may act redundantly with the five genes analyzed.
Class I TCP proteins can regulate the expression of cell-cycle and KNOX1 genes and can interact with other members of the same class
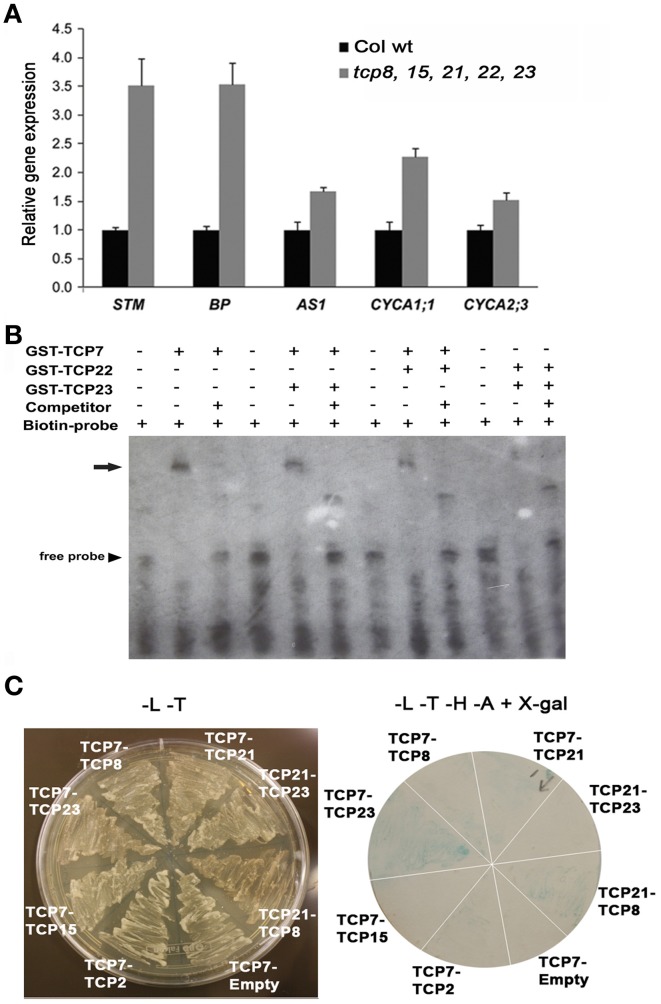
It has been reported that some Arabidopsis class I TCP genes such as AtTCP20 (Li et al., 2005), AtTCP15 (Kieffer et al., 2011; Li et al., 2012a), and AtTCP14 (Kieffer et al., 2011) modulate the expression of cell-cycle genes. Furthermore, it has been shown that class I TCP genes can regulate the expression of genes involved in SAM and leaf development, acting in part redundantly with the class II TCP genes controlling the same set of genes (Uberti-Manassero et al., 2012). We wanted to determine if the expression of genes involved in cell-cycle control and SAM maintenance are affected in the pentuple tcp mutant tcp8-1 tcp15-1 tcp22-1 tcp23-1 tcp21-1. We found that genes involved in the control of SAM maintenance and leaf form such as the KNOX1 genes SHOOT-MERISTEMLESS (STM) and BP and to a lesser extent AS1 are overexpressed in the pentuple tcp mutant (Figure 5A). We also determined that genes regulating the cell-cycle such as CYCA1;1 and CYCA2;3 are up-regulated in the pentuple tcp mutant (Figure 5A). It is known that ectopic expression of KNOX1 genes produces leaf lobing (Chuck et al., 1996). Although in the tcp pentuple mutant we found no dramatic leaf lobing alteration, the mutant plants have bigger leaf blades, and the up-regulation of cell-cycle genes can partially explain this increased leaf size.
Figure 5.
Class I TCP factors regulate KNOX1 genes and are capable of protein–protein interaction. (A) Gene expression analysis by qPCR in Col wt and the pentuple mutant tcp8, 15, 21, 22, 23. Error bars are SD of three biological replicates. (B) EMSA of the K-box region of the STM promoter. Tested were factors AtTCP7, AtTCP22 and AtTCP23 in combinations indicated. Arrow indicates shifted bands. (C) Yeast-two hybrid assay to test for TCP protein–protein interactions. Left panel, co-transformed yeast grown in media without aminoacids Leucine and Tryptophan (-L-T media); right panel, β –Gal filter assay: co-transformed yeast were replica-plated in media without Leucine, Tryptophan, Histidine and Adenine and plus X-gal (-L-T-H-A+X-gal). The factors assayed were class I TCP factors AtTCP7, AtTCP8, AtTCP15, AtTCP21 and AtTCP23 and the class II factors AtTCP2 and AtTCP3. Interactions with empty vectors were used as negative controls.
The up-regulation of KNOX1 genes in the tcp pentuple mutant indicates that class I TCP proteins could act directly or indirectly as regulators of KNOX1 genes. We tested if class I TCP genes directly bind the STM promoter using electrophoretic mobility shift assay (EMSA). We used a probe of 76 bp that belongs to the K-box, a highly conserved region in the STM 5′-flanking sequence involved in the coordination of leaf development (Uchida et al., 2007). We found that AtTCP7 is capable to bind the K-box in vitro (Figure 5B). Mechanistically TCP factors can act as dimers and there is evidence that the DNA binding capacity is increased when they form heterodimers (Kosugi and Ohashi, 2002; Viola et al., 2011). We found that heterodimers AtTCP7-AtTCP23, AtTCP7-AtTCP22, and AtTCP22-AtTCP23 are also able to bind the K-box (Figure 5B).
This interaction between TCP factors can account for the high degree of genetic redundancy found in this family. We wanted to analyze if members of class I TCP genes analyzed in this study are able of protein–protein interaction using a yeast two-hybrid assay (Figure 5C and Table 1). We found that AtTCP7 interacts with AtTCP23, AtTCP21, and to a lesser extent with AtTCP8. We also found interaction between AtTCP8 and AtTCP21 (Figure 5C and Table 1). Class II TCP factors AtTCP2 and AtTCP3 do not show interaction with any of the class I TCP factors tested. Taken together these results indicate that class I TCP factors can have a similar gene function by virtue of their ability to form higher order complexes to regulate similar downstream targets.
Table 1.
Protein–protein interactions between TCP factors using yeast-two hybrid.
| pGBKT7- | |||||||
|---|---|---|---|---|---|---|---|
| TCP7 | TCP8 | TCP15 | TCP21 | TCP23 | TCP2 | Empty | |
| pGADT7-TCP7 | + | − | + | +++ | − | − | |
| pGADT7-TCP8 | − | ++ | − | − | − | ||
| pGADT7-TCP15 | − | − | |||||
| pGADT7-TCP21 | − | − | − | − | |||
| pGADT7-TCP23 | − | − | − | ||||
| pGADT7-TCP2 | − | ||||||
| pGADT7-TCP3 | − | − | − | − | − | ||
| Empty | − | ||||||
Signs are for strong (+++), medium (++), mild (+) and no interaction (−), respectively.
Dominant negative forms of class I TCP genes affect lateral organ growth
The lack of a severe phenotype in the pentuple mutant can be ascribed to genetic redundancy, particularly prominent in the TCP gene family (Schommer et al., 2008; Koyama et al., 2010). One way to circumvent genetic redundancy and to analyze the function of transcription factors is to transform them into dominant negative forms by fusing the coding sequences of the genes to the repressor domain SRDX, producing loss-of-function phenotypes even in the presence of redundant genes (Hiratsu et al., 2003). To further analyze the function of class I TCP genes, we generated transgenic lines with fusions of the coding sequences of AtTCP7 and AtTCP23 to the SRDX repressor domain and compared their phenotypes with the wild type (Figure 6). At the seedling stage lateral organs (cotyledons and first true leaves) of TCP7-SRDX and TCP23-SRDX plants show an upward growth (Figures 6A–C). At the bolting stage compared to the wild type (Figure 6A), TCP7-SRDX and TCP23-SRDX rosette leaves are hyponastically curved and smaller (Figures 6E,F). The basis of the differences in leaf size and form can be due to an altered cell growth pattern. We therefore analyzed the leaf epidermis and found that the leaves of the transgenic lines have groups of small cells on both adaxial (Figures 6G–I) and abaxial (Figures 6J–L) surfaces. Flowers are also affected in the transgenic plants (Figures 6M–R). Compared with the wild type (Figures 6M,N) the flowers of TCP7-SRDX (Figures 6O,P) and TCP23-SRDX (Figures 6Q,R) plants have shorter pedicels and are closed, with irregularly shaped larger sepals, shorter petals and less trichomes. Despite these alterations, the flowers have a normal number of sepals, petals, stamens, and carpels (data not shown). Gene expression analysis through qPCR on SAM gene markers indicated that STM is up-regulated in TCP7-SRDX (Figure 6S) and TCP23-SRDX (Figure 6T) transgenic plants, while the expression of the cell-cycle marker CYCA1;1 is not changed and the expression of CYCA2;3 is down-regulated (Figures 6S,T). In summary, dominant negative forms of AtTCP7 and AtTCP23 produce similar phenotypes with alterations in lateral organ growth, possibly based in differences in cell proliferation. Therefore is likely that AtTCP7 and AtTCP23 both function in regulating cell proliferation.
Figure 6.
Characterization of transgenic plants harboring class I TCP genes AtTCP7 and AtTCP23 fused to the repressor domain SRDX. (A–C) Phenotype of 10 days-old seedlings of TCP7-SRDX and TCP23-SRDX transgenic lines. (A) Col wt. (B) TCP7-SRDX. (C) TCP23-SRDX. (D) Plants at bolting stage of Col wt; (E) of TCP7-SRDX and (F) of TCP23-SRDX. (G–L) Leaf epidermis nail polish impressions in Col wt (G,J), TCP7-SRDX (H,K) and TCP23-SRDX (I,L) plants. (G–I) Adaxial side; (J–L) abaxial side. (M) Inflorescence and (N) isolated flower of a Col wt plant. (O) Inflorescence and (P) flower of a TCP7-SRDX transgenic plant. (Q) Inflorescence and (R) flower in a TCP23-SRDX transgenic plant. (S) qPCR expression analysis of some gene markers comparing Col wt plants with TCP7-SRDX plants and (T) with TCP23-SRDX plants. Error bars are SD of three biological replicates. Scale bars are: (D), 1 cm; (E), (F), 0.5 cm; (G–L), 50 μm.
In conclusion, our results expand our knowledge of a group of members of the TCP gene family which has important roles in plant development. The lack of a severe phenotype in multiple mutants, the phenotype of the leaf epidermis and leaf shape found in dominant negative forms in these genes, their expression in young leaves, the demonstrated protein-protein interaction between selected members, the altered KNOX1 gene expression in the knockout or dominant negative mutations, indicates that they may function in the control of leaf development by regulating similar gene networks.
Discussion
The phylogenetic analysis showed that AtTCP14, AtTCP15, AtTCP23, AtTCP22, AtTCP8, AtTCP7, and AtTCP21 form a subgroup within the class I of TCP factors, AtTCP20, AtTCP9, and AtTCP19 form another clade while AtTCP11, AtTCP6, and AtTCP16 are more distantly related. The phylogenetic relationships of these factors can account for a related function. AtTCP11 and AtTCP16 have a role in pollen development (Takeda et al., 2006; Viola et al., 2011) and AtTCP9 and AtTCP20 act regulating the expression of the jasmonic acid biosynthesis gene LOX2 (Danisman et al., 2012). AtTCP7 and AtTCP21 are close homologs. This raises the question if these two genes share a similar function. AtTCP21 (CHE) has a role in the control of the circadian rhythm, interacting with the core clock component TOC1 (for TIMING OF CAB EXPRESSION1). Giraud et al. (2010) analyzed the interaction of all the TCP factors with known components of the circadian clock. Interestingly they found that class I TCP factors AtTCP15 and AtTCP11 interact with TOC1 and other components of the core circadian clock, however they found no interactions in case of AtTCP21 or AtTCP7. AtTCP3, a member of the class II TCP factors, was also found to interact with TOC1 (Giraud et al., 2010). Moreover, another component of the core clock, CCA1 (for CIRCADIAN CLOCK ASSOCIATED1), binds to the promoter of CHE (Pruneda-Paz et al., 2009), however the promoter of AtTCP7 lacks this CCA-1 binding site. Furthermore, the expression pattern of these genes appears to be different. Using promoter reporter fusions to GUS it was found that AtTCP21 is expressed ubiquitously throughout the plant (Pruneda-Paz et al., 2009) while our data indicate that the expression of AtTCP7 is more restricted to growing new leaves. Taken together is possible that these genes have different functions.
Weak phenotypic alteration or absence of such alteration in single mutants of class II TCP genes is attributed to genetic redundancy (Schommer et al., 2008; Koyama et al., 2010). Mild or no phenotypes are present in loss of function of class I TCP genes such as AtTCP21 (Pruneda-Paz et al., 2009), AtTCP14 (Kieffer et al., 2011), AtTCP15 (Kieffer et al., 2011; Li et al., 2012a; Uberti-Manassero et al., 2012) and in this study for AtTCP8 and AtTCP23. In this case the analysis of multiple mutants is useful to analyze the function of these genes. Other multiple mutants have been developed in the CIN-like clade of TCP genes. Koyama et al. (2010) developed the triple mutant tcp3 tcp4 tcp10, the quadruple mutant tcp3 tcp4 tcp5 tcp10 and the pentuple mutant tcp3 tcp4 tcp5 tcp10 tcp13. These mutants show dose-dependent changes mainly in leaf morphology, however our analysis shows that the pentuple mutant tcp8 tcp15 tcp21 tcp22 tcp23 has no dramatic differences compared with the wild type. This can indicate that analysis of higher order mutants are needed to more understand the function of these genes. For example in the analysis of the gene family of 9 members LONELY GUY (LOG) of cytokinin-activating enzymes, the sexptuple mutants log1 log2 log3 log4 log5 log8 or log1 log2 log3 log4 log5 log7 show no visible phenotype, however the septuple mutant log1 log2 log3 log4 log5 log7 log8 show a extremely severe retardation of shoot and root growth with defects in the maintenance of the apical meristems (Tokunaga et al., 2012). AtTCP7 could not be included in our multiple mutant analysis due to a lack of an available T-DNA knockout insertion line. Further, tcp22-1 is not a null allele and still retains some gene expression. The gene function of these two genes or another class I TCP genes such as AtTCP14 can be sufficient for normal plant growth. Future studies on the role of class I TCP genes would have to focus on multiple mutants such as the pentuple mutant tcp8 tcp14 tcp15 tcp22 tcp23, the sextuple mutant tcp8 tcp14 tcp15 tcp21 tcp22 tcp23 or higher order multiple mutants.
Class II TCP factors and AS2 interact to directly regulate the expression of KNOX1 genes (Li et al., 2012b). Due to the interplay between the two classes of TCP genes in regulating the expression of downstream genes (Danisman et al., 2012; Uberti-Manassero et al., 2012) it is tempting to argue that class I TCP factors can also regulate KNOX1 gene expression. Our data indicates that KNOX1 gene expression is altered in the pentuple mutant tcp8 tcp15 tcp21 tcp22 tcp23. In vitro studies indicate that class I TCP factors can bind the promoter of STM, a KNOX1 gene. Furthermore, yeast-two hybrid experiments show that class I TCP factors are able to interact, as we found for AtTCP7 with AtTCP23, and AtTCP8 with AtTCP21. Heterodimerization improves the action of TCP factors, as reported by Kosugi and Ohashi (2002). Viola et al. (2011) showed interaction between the class I TCP factors AtTCP11 and AtTCP15 and Danisman et al. (2012) reported that AtTCP20 interacts with AtTCP8 and AtTCP22. TCP factors have been recognized as intrinsically disordered proteins (IDPs) and can also form multimers and interact with numerous other proteins (Valsecchi et al., 2013), which can account for the activity of these factors in multiple pathways (Babu et al., 2011; Valsecchi et al., 2013).
There is an interplay between AS1-AS2, TCP, and KNOX1 genes. AS1-AS2 form a complex (Xu et al., 2003) able to bind the promoters of KNOX1 genes BP and KNAT2 (Guo et al., 2008). Class II TCP factors bind the promoter of AS1 (Koyama et al., 2010). Class II TCP factors interact with AS2 (Li et al., 2012b) and are able to bind the promoters of KNOX1 genes BP and KNAT2 (Li et al., 2012b). We have determined that class I TCP factors are able to bind the promoter of KNOX1 gene STM. We have also determined that class I TCP mutants affect the leaf initiation rate. This phenotype is enhanced in an as1 mutant background, that indicates that AS1 could have a role in the control of leaf initiation rate. Leaf initiation rate is influenced by cell division rate (Itoh et al., 1998; Wang et al., 2008) and TCP genes have a role in the control of cell division (Li et al., 2005). The number of rosette leaves is correlated with flowering time in natural accessions (Alonso-Blanco et al., 1998). However Salomé et al. (2011) indicate that flowering time and number of leaves can be genetically uncoupled. Recently it has been reported that the TCP gene BRC1 interacts with flowering genes FT and TSF (Niwa et al., 2013). It can be interesting to determine if class I TCP genes interact with flowering genes and AS1-AS2 factors.
The generation of chimeric repressor mutants with the SRDX domain has been used to analyze the function of a number of genes, such as TCP genes including AtTCP1 (Guo et al., 2010), AtTCP3, AtTCP2, AtTCP4, AtTCP5, AtTCP10, AtTCP13, AtTCP17, AtTCP24 (Koyama et al., 2007), AtTCP11 (Viola et al., 2011), AtTCP14 (Kieffer et al., 2011), AtTCP15 (Kieffer et al., 2011; Li et al., 2012a; Uberti-Manassero et al., 2012), and AtTCP20 (Hervé et al., 2009). In this study we fused the repressor domain SRDX to the coding sequences of AtTCP7 and AtTCP23. Both types of transgenic plants show similar phenotypes, mainly characterized by an upward growth of the leaves and smaller rosette size. The similarity of phenotypes in AtTCP7-SRDX and AtTCP23-SRDX lines indicates that they likely have similar functions. Koyama et al. (2007) found similar phenotypes in SRDX constructs for the TCP genes AtTCP2, AtTCP3, AtTCP4, AtTCP5, AtTCP10, AtTCP13, AtTCP17, and AtTCP24, showing that all of them act redundantly as regulators of the expression of boundary-specific genes.
The phenotype of AtTCP7-SRDX and AtTCP23-SRDX plants is not similar to that seen for SRDX constructs of the class II TCP genes mentioned above (Koyama et al., 2007); instead it resembles to a certain extent the phenotype of plants expressing AtTCP15-SRDX (Uberti-Manassero et al., 2012) and AtTCP11-SRDX (Viola et al., 2011), in particular the upward leaf growth. However the phenotype of the inflorescences differs markedly, while flowers of AtTCP15-SRDX show exposed gynoecia (Kieffer et al., 2011; Uberti-Manassero et al., 2012), the flowers of AtTCP7-SRDX and AtTCP23-SRDX are closed. Is interesting to note that the phenotype of AtTCP15-SRDX described is with the use of the endogenous promoter of AtTCP15 (Kieffer et al., 2011; Uberti-Manassero et al., 2012), while the phenotype of AtTCP7-SRDX and AtTCP23-SRDX plants is with the use of a constitutive promoter. AtTCP15-SRDX plants under a constitutive promoter can produce plants with a very strong phenotype with SAM arrest (Uberti-Manassero et al., 2012), phenotype not found in AtTCP7-SRDX and AtTCP23-SRDX plants.
The role of TCP factors in the control of cell division arrest and leaf development is well documented (Nath et al., 2003; Ori et al., 2007). Other members of the class I TCP factors such as the rice PCF1 and PCF2 (Kosugi and Ohashi, 1997), AtTCP20 (Hervé et al., 2009) and AtTCP14 and AtTCP15 (Kieffer et al., 2011; Li et al., 2012a) have a documented role in the control of cell proliferation. Our analysis indicates that the cell-cycle marker CYCA2;3 is down-regulated in AtTCP7-SRDX and AtTCP23-SRDX plants. Li et al. (2012a) also found that this marker is down-regulated. The leaf phenotype of AtTCP7-SRDX and AtTCP23-SRDX plants, the altered epidermal cell patterning, and altered cell-cycle gene expression suggest that class I TCP genes analyzed in this study can also regulate cell growth and leaf development by acting on cell-cycle genes.
The class I TCP genes analyzed in this study show a similar expression pattern, with the exception of AtTCP23. We used the complete 5′ promoter region for the study of the spatial expression pattern. However the actual expression of AtTCP23 could require regulatory regions in other regions of the gene. It is likely that the encoded proteins retain similar functions as the plants with the constructs AtTCP7-SRDX and AtTCP23-SRDX have similar phenotypes.
Materials and methods
Plant materials and growth conditions
Alleles tcp8-1 (SAIL_656_F11), tcp7-2 (SAIL_180_B09), and tcp23-1 (SAIL_443_F02) were obtained from the Syngenta Arabidopsis Insertion Library (SAIL) (Sessions et al., 2002). Alleles tcp15-1 (SALK_011491C) and tcp22-1 (SALK_027490) were obtained from the Salk Institute Genomic Analysis Laboratory (SIGnAL) (Alonso et al., 2003) through the Arabidopsis Biological Resource Center (ABRC). The mutant allele for AtTCP21 was che-1 (Pruneda-Paz et al., 2009). The allele tcp7-4 (WISCDSLOX240D10) was obtained from the Wisconsin DsLox collection (Woody et al., 2007). tcp7-1 (GK-511D02) belongs to the GABI-Kat collection (Li et al., 2007). The gene allele tcp7-3 was obtained from the INRA-Versailles T-DNA collection (Beschtold et al., 1993). tcp7-3 is in Wassilewskija (Ws) background, all the rest are in Columbia-0 (Col-0) ecotype background. as1-1 was from Uchida et al. (2007).
Plants were grown under long-day conditions (16 h light/8 h dark) at 22°C in a soil mixture (Sunshine Mix1). For the phenotypic analysis plants were grown in chambers at 23°C at 50% humidity and 60–70 μmoles light and long day conditions.
For in vitro culture, Arabidopsis seeds were surface-sterilized in 70% bleach, before transfer on agar plates containing half-strength salts and vitamins, 1.5% sucrose, and 0.8% agar, with or without kanamycin (50 μg/ml). Plants transferred to soil were grown under the same growth chamber conditions as mentioned above.
Phylogenetic analysis
A region of 69 aminoacids spanning the highly conserved TCP box was used to build the phylogenetic trees. We used the sequences of the 24 TCP genes in Arabidopsis and the 25 genes in rice respectively. We followed the notation as in Martín-Trillo and Cubas (2010). For tomato we isolated the sequences from the ITAG 2.3 release of the sequenced tomato genome (http://solgenomics.net/organism/Solanum_lycopersicum/genome; Supplemental Table 1). The sequence alignments were made using Muscle (http://www.ebi.ac.uk/Tools/msa/muscle/). The tree was made using MEGA5 software (Tamura et al., 2011; http://www.megasoftware.net/) using the maximum likelihood method with 1000 bootstrap replicates.
Construction of transgenes and plant transformation
To generate RNAi lines for AtTCP7, a fragment of 195 bp of the coding sequence was amplified using primers M2LRNA2f (5′-TATCCGTGGAGCCACCAATT CTACTTCT-3′) and M2LRNA2r (5′-GCTGGTGTTGTCGTAAAGGTCTCATCTT-3′) and cloned into pCR8/GW/TOPO TA entry vector (Invitrogen) to generate pCR8-TCP7RNAi. As destination vector we used pK7GWIWG2(II) (Karimi et al., 2002).
For the GFP fusions with AtTCP7, AtTCP15, and AtTCP22, we cloned the coding sequences of these genes into the entry vector pCR8/GW/TOPO TA and LR cloned into the plasmid pK2GW7 (Karimi et al., 2002) generating TCP7-OEX, TCP15-OEX, and TCP22-OEX, respectively (for primers used see Supplemental Table 4).
For the SRDX constructs, the coding sequence of AtTCP7 was cloned into the entry vector pCR8/GW/TOPO TA using forward primer M2Lf (5′-ATGTCTATTAACAACAACAACAACAACA-3′) and the reverse primer TCPSRDXrev (5′-TTACGCAAAGCCGAGGCGAAGTTCGAGATCAAGATCGAGACGTGGATCTTCCTCTCTTCGATCCGA-3′) without the stop codon of AtTCP7 and with the EAR peptide sequence LDLDLELRLGFA plus the terminator codon TAA (Hiratsu et al., 2003). The construct was LR cloned into the plasmid pK2GW7 generating TCP7-SRDX. Similarly, for AtTCP23 we used primers P23f (5′-ATGGAGTCCCACAACAACAACCAGAGCA-3′) and TCP23SRDXrev (5′-TTACGCAAAGCCGAGGCGAAGTTCGAGATCAAGATCGAGAGGAGAACCATCTATAGTAGGATTTTG-3′). The PCR fragment was cloned into pCR8/GW/TOPO TA and LR cloned in pK2GW7 to generate TCP23-SRDX.
To generate ProAtTCP7:GUS, ProAtTCP8:GUS, ProAtTCP2:GUS, and ProAtTCP23:GUS constructs, respectively, 1990, 1588, 2180, and 2494 bp of the complete 5′ intergenic region preceding the ORFs was amplified from Col-0 genomic DNA using primers indicated in Supplemental Table 4 and inserted into the binary vector pKGWFS7 (Karimi et al., 2002). Constructs were introduced into Agrobacterium strain GV3101 (pMP90) (Koncz and Schell, 1986) by electroporation. Plant transformation was done using the “floral dip” method (Clough and Bent, 1998).
Subcellular localization analysis
To determine the cellular location of class I TCP factors, the GFP signal of roots in 50% glycerol of transgenic plants TCP7-OEX, TCP15-OEX, and TCP22-OEX was observed under an epifluorescence microscope (Nikon Eclipse E600, Nikon; excitation at 488 nm and emission at 495–575 nm).
Gene expression analysis
Total RNA was extracted with RNeasy Plant Mini Kit (QIAGEN), and treated with on-column DNase (QIAGEN). cDNAs were generated with QuantiTect Reverse Transcription Kit (Qiagen). qPCR was performed on a MyiQ two-color real time PCR detection system with iQ SYBR Green supermix (Bio-Rad). Real-time PCR experiments were repeated using three biological replicates and one technical replicate. For tissue expression analysis of class I TCP genes the roots, young leaves, mature leaves and shoot apices were taken from 2-week old plants, the inflorescences were taken from the tip of the main stem. To analyze expression levels in mutant and transgenic backgrounds, three sets of 5–6 pooled entire 2-week old plants were collected and analyzed. ACTIN8 was used as an endogenous control gene. The number of cycles for the RT-PCR was 28 for AtTCP7, AtTCP8, AtTCP14, AtTCP15, AtTCP21, and AtTCP22, 30 for AtTCP23 and 26 for ACTIN8. Primers used for RT-PCR and qPCR are specified in Supplemental Table 2.
Construction of multiple mutants
To generate multiple tcp mutants, we made first the double mutants tcp8-1 tcp15-1 and tcp22-1 tcp23-1. Double homozygous recessive plants were crossed and from F3 progenies we selected by genotyping the quadruple mutant tcp8-1 tcp15-1 tcp22-1 tcp23-1. This multiple mutant was crossed with tcp21-1 and through selection by genotyping we isolated the pentuple mutant tcp8-1 tcp15-1 tcp22-1 tcp23-1 tcp21-1.
The primers used for the genotyping of the mutants are indicated in Supplemental Table 3. For tcp8-1 we used the combination tcp8-1Lp and tcp8-1Rp for the wild type copy and tcp8-1Lp and LB1SAIL and tcp8-1Rp and LB1SAIL for the insertions. Similarly, for tcp15-1, we used the combinations tcp15-1Lp and tcp15-1Rp for the wild type copy and tcp15-1Lp and LBa1 for the mutation. For tcp22-1, we used primers tcp22-1Lp and tcp22-1Rp for the wild type copy and tcp22-1Rp and LBa1 for the insertion. For tcp23-1, we used the combinations tcp23-1Lp and tcp23-1Rp for the wild type copy and tcp23-1Lp and LB1SAIL and tcp23-1Rp and LB1SAIL for the insertions. For tcp7-1, we used the primers TCP7p1000 and M2LPROr for the wild type copy and TCP7p1000 and NS343_GK-LB and M2LPROr and NS344_GK-RB for the insertions. For tcp7-2, the primers NS256_S18Lp and NS257_S18Rp for the wild type copy and NS256_S18Lp and LB1SAIL and NS257_S18Rp and LB1SAIL for the insertions. For tcp7-3, the primers NS254_S17Lp and NS255_S17Rp for the wild type copy and NS255_S17Rp and NS290_RB4INRA for the mutant copy. In case of tcp7-4, the combinations NS254_S17Lp and NS255_S17Rp for the wild type copy and NS255_S17Rp and NS314_p745Wisc for the mutant copy. For tcp21-1 or che-1, the primers NS550_CHEgF and NS551_CHEgR for the wild type copy and NS550_CHEgF and LBa1 for the insertion. For the design of the primers we used the web tool at http://signal.salk.edu/tdnaprimers.2.html. All the positions of the insertions were determined by sequencing.
GUS histochemical assays
For histochemical GUS activity assays, plants were submerged in 90% acetone for 20 min. on ice, washed twice with 50mM sodium phosphate buffer (pH 7.2) and subsequently incubated in staining solution (0.1 M sodium phosphate buffer (pH 7.2), 0.5 mM Fe(CN)2, 0.5 mM Fe(CN)3, 0.1% Tween-20 and 2 mM 5-bromo-4-chloro-3-indolyl-β-d-glucuronide) at 37°C for 6 h. Plants were cleared and dehydrated through a graded ethanol series. For sections, FAA-fixed and dehydrated material was embedded in Paraplast Plus (McCormick Scientific, St. Louis, MO). The sections were dewaxed by immersion in Histoclear and mounted in Permount (Fisher Scientific).
Yeast two-hybrid assay
For the yeast two-hybrid, the full-length coding sequences of AtTCP7, AtTCP8, AtTCP15, AtTCP21, AtTCP23, AtTCP2, and AtTCP3 were amplified by PCR using Pwo polymerase (Roche) and cloned into either pGBKT7 bait vector (fused with Gal4 DNA-binding domain; BD) or pGADT7 prey vector (fused with GAL4 activation domain; AD, Clontech, http://clontech.com). The constructs were verified by sequencing and co-transformed into yeast strain AH109. To test for interactions, the bait and prey constructs co-expressed in yeast were subjected to β–Galactosidase filter assay according to the manufacturer's instructions (Clontech).
EMSA
We amplified the coding sequences of AtTCP7, AtTCP15, AtTCP22, and AtTCP23, and cloned them into the pCR8/GW/TOPO TA cloning vector (Invitrogen). We generated GST-fusion–protein-expressing constructs through LR reactions between the pCR8 entry clones and the destination vectors pDEST15 (Invitrogen) containing an N-terminal GST tag. All the fusion proteins and empty control plasmids were expressed in Escherichia coli strain BL21 (DE3).
The probe used in EMSA is 76 bp long, from −239 to −315 bp upstream of the starting codon of STM and was prepared by annealing the two biotin-labeled anti-parallel primers Kboxf 5′-AACTATCACTATTTAGAATTTTCAATGTGGAAAAGGAAGCTGATTGTTGAAGCATAAATCCCGGGAGACCACTTTT-3′ and Kboxr 5′-AAAAGTGGTCTCCCGGGATTTATGCTTCAACAATCAGCTTCCTTTTCCACATTGAAAATTCTAAATAGTGATAGTT-3′. The same two unlabeled primers were annealed for competition. For the EMSA, recombinant GST-tagged proteins on pDEST15 were purified on an immobilized glutathione column (Pierce). Binding reactions and detection of biotin-labeled probe was performed according to the manufacturer's instructions (LightShift Chemiluminescent EMSA Kit, Pierce).
Phenotypic analysis
For the number of rosette leaves the plants were at bolting stage with 0.5 cm of the main stem elongated. We did not take account of the cotyledons. The petiole length, blade length, blade width, blade perimeter and blade area were determined using the sixth leaf of plants at bolting stage. The data were analyzed using LeafJ (Maloof et al., 2013) with ImageJ software (http://rsb.info.nih.gov/ij/). For leaf epidermis analysis, dental resin imprints and nail polish copies (Kagan et al., 1992) were taken from the medial leaf lamina zone from rosette leaves of Col wt, TCP7-SRDX, and TCP23-SRDX plants and analyzed by light microscopy in a Nikon Eclipse E600 (Nikon).
Accession numbers
Sequence data from this article can be found in The Arabidopsis Information Resource (TAIR; http://www.arabidopsis.org/) or GenBank/EMBL databases under the following accession numbers: AtTCP7 (At5g23280), AtTCP8 (At1g58100), AtTCP22 (At1g72010), and AtTCP23 (At1g35560).
Author contributions
José A. Aguilar-Martínez and Neelima Sinha designed the research. José A. Aguilar-Martínez performed the research. José A. Aguilar-Martínez and Neelima Sinha analyzed the data and wrote the paper.
Acknowledgments
We thank Clayton Chang and Duy Dang for technical assistance, M. Jones for providing the che-1 mutant seeds, Cristina Nieto Garcia for helpful discussions and members of Sinha lab for helpful discussions and comments on the manuscript. This work was supported by a postdoctoral fellowship from Spanish Ministry of Science (MICINN) to José A. Aguilar-Martínez.
Supplementary material
The Supplementary Material for this article can be found online at: http://www.frontiersin.org/journal/10.3389/fpls.2013.00406/abstract
Sequence alignment of the TCP box of class I TCP factors.
List of TCP genes in the tomato genome.
List of primers used in this study for gene expression analysis.
List of primers used to genotype tcp mutants in Arabidopsis.
List of primers for genetic constructs generated in this study.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
- Aggarwal P., Das Gupta M., Joseph A. P., Chatterjee N., Srinivasan N., Nath U. (2010). Identification of specific DNA binding residues in the TCP family of transcription factors in Arabidopsis. Plant Cell 22, 1174–1189 10.1105/tpc.109.066647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguilar-Martínez J. A., Poza-Carrión C., Cubas P. (2007). Arabidopsis BRANCHED1 acts as an integrator of branching signals within axillary buds. Plant Cell 19, 458–472 10.1105/tpc.106.048934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso J. M., Stepanova A. N., Leisse T. J., Kim C. J., Chen H., Shinn P., et al. (2003). Genome-Wide insertional mutagenesis of Arabidopsis thaliana. Science 301, 653–657 10.1126/science.1086391 [DOI] [PubMed] [Google Scholar]
- Alonso-Blanco C., El-Assal S. E., Coupland G., Koornneef M. (1998). Analysis of natural allelic variation at flowering time loci in the landsberg erecta and cape verde islands ecotypes of Arabidopsis thaliana. Genetics 149, 749–764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babu M. M., van der Lee R., de Groot N. S., Gsponer J. (2011). Intrinsically disordered proteins: regulation and disease. Curr. Opin. Struct. Biol. 21, 432–440 10.1016/j.sbi.2011.03.011 [DOI] [PubMed] [Google Scholar]
- Beschtold N., Ellis J., Pelletier G. (1993). In planta Agrobacterium-mediated gene transfer by filtration of adult Arabidopsis thaliana plants. C. R. Acad. Sci. (Paris) 316, 1194–1199 [Google Scholar]
- Busch A., Zachgo S. (2009). Flower symmetry evolution: towards understanding the abominable mystery of angiosperm radiation. Bioessays 31, 1181–1190 10.1002/bies.200900081 [DOI] [PubMed] [Google Scholar]
- Byrne M. E., Barley R., Curtis M., Arroyo J. M., Dunham M., Hudson A., et al. (2000). Asymmetric leaves1 mediates leaf patterning and stem cell function in Arabidopsis. Nature 408, 967–971 10.1038/35050091 [DOI] [PubMed] [Google Scholar]
- Chuck G., Lincoln C., Hake S. (1996). KNAT1 induces lobed leaves with ectopic meristems when overexpressed in Arabidopsis. Plant Cell 8, 1277–1289 10.2307/3870301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S. J., Bent A. F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743 10.1046/j.1365-313x.1998.00343.x [DOI] [PubMed] [Google Scholar]
- Cubas P. (2002). Role of TCP genes in the evolution of key morphological characters in angiosperms, in Developmental Genetics and Plant Evolution, eds Cronk Q., Hawkins J., Bateman R. M. (London: Taylor and Francis; ), 247–266 10.1201/9781420024982.ch13 [DOI] [Google Scholar]
- Cubas P., Coen E., Zapater J. (2001). Ancient asymmetries in the evolution of flowers. Curr. Biol. 11, 1050–1052 10.1016/S0960-9822(01)00295-0 [DOI] [PubMed] [Google Scholar]
- Cubas P., Lauter N., Doebley J., Coen E. (1999). The TCP domain: a motif found in proteins regulating plant growth and development. Plant J. 18, 215–222 10.1046/j.1365-313X.1999.00444.x [DOI] [PubMed] [Google Scholar]
- Danisman S., Wal F., Dhondt S., Waites R., Folter S., Bimbo A., et al. (2012). Arabidopsis Class I and Class II TCP transcription factors regulate jasmonic acid metabolism and leaf development antagonistically. Plant Physiol. 159, 1511–1523 10.1104/pp.112.200303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doebley J., Stec A., Hubbard L. (1997). The evolution of apical dominance in maize. Nature 386, 485–488 10.1038/386485a0 [DOI] [PubMed] [Google Scholar]
- Finlayson S. A. (2007). Arabidopsis TEOSINTE BRANCHED1-LIKE 1 regulates axillary bud outgrowth and is homologous to monocot teosinte branched1. Plant Cell Physiol. 48, 667–677 10.1093/pcp/pcm044 [DOI] [PubMed] [Google Scholar]
- Giraud E., Ng S., Carrie C., Duncan O., Low J., Lee C. P., et al. (2010). TCP transcription factors link the regulation of genes encoding mitochondrial proteins with the circadian clock in Arabidopsis thaliana. Plant Cell 22, 3921–3934 10.1105/tpc.110.074518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo M., Thomas J., Collins G., Timmermans M. (2008). Direct repression of KNOX loci by the ASYMMETRIC LEAVES1 complex of Arabidopsis. Plant Cell 20, 48–58 10.1105/tpc.107.056127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Z., Fujioka S., Blancaflor E. B., Miao S., Gou X., Li J. (2010). TCP1 modulates brassinosteroid biosynthesis by regulating the expression of the key biosynthetic gene DWARF4 in Arabidopsis thaliana. Plant Cell 22, 1161–1173 10.1105/tpc.109.069203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammani K., Gobert A., Hleibieh K., Choulier L., Small I., Giegé P. (2011). An Arabidopsis dual-localized pentatricopeptide repeat protein interacts with nuclear proteins involved in gene expression regulation. Plant Cell 23, 730–740 10.1105/tpc.110.081638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hervé C., Dabos P., Bardet C., Jauneau A., Auriac M. C., Ramboer A., et al. (2009). In Vivo Interference with AtTCP20 function induces severe plant growth alterations and deregulates the expression of many genes important for development. Plant Physiol. 149, 1462–1477 10.1104/pp.108.126136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiratsu K., Matsui K., Koyama T., Ohme-Takagi M. (2003). Dominant repression of target genes by chimeric repressors that include the EAR motif, a repression domain, in Arabidopsis. Plant J. 34, 733–739 10.1046/j.1365-313X.2003.01759.x [DOI] [PubMed] [Google Scholar]
- Hruz T., Laule O., Szabo G., Wessendorp F., Bleuler S., Oertle L., et al. (2008). Genevestigator V3: a reference expression database for the meta-analysis of transcriptomes. Adv. Bioinformatics 2008, 420747 10.1155/2008/420747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingram G. C., Waites R. (2006). Keeping it together: co-ordinating plant growth. Curr. Opin. Plant Biol. 9, 12–20 10.1016/j.pbi.2005.11.007 [DOI] [PubMed] [Google Scholar]
- Itoh J. I., Hasegawa A., Kitano H., Nagato Y. (1998). A recessive heterochronic mutation, plastochron1, shortens the plastochron and elongates the vegetative phase in rice. Plant Cell 10, 1511–1522 10.2307/3870615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagan M. L., Novoplansky N., Sachs T. (1992). Variable cell lineages form the functional pea epidermis. Ann. Bot. 69, 303–312 [Google Scholar]
- Karimi M., Inze D., Depicker A. (2002). GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 7, 193–195 10.1016/S1360-1385(02)02251-3 [DOI] [PubMed] [Google Scholar]
- Kieffer M., Master V., Waites R., Davies B. (2011). TCP14 and TCP15 affect internode length and leaf shape in Arabidopsis. Plant J. 68, 147–158 10.1111/j.1365-313X.2011.04674.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koncz C., Schell J. (1986). The promoter of TL -DNA gene 5 controls the tissue-specific expression of chimeric genes carried by a novel type of Agrobacterium binary vector. Mol. Gen. Genet. 204, 383–396 10.1007/BF00331014 [DOI] [Google Scholar]
- Kosugi S., Ohashi Y. (1997). PCF1 and PCF2 specifically bind to cis elements in the rice proliferating cell nuclear antigen gene. Plant Cell 9, 1607–1619 10.2307/3870447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosugi S., Ohashi Y. (2002). DNA binding and dimerization specificity and potential targets for the TCP protein family. Plant J. 30, 337–348 10.1046/j.1365-313X.2002.01294.x [DOI] [PubMed] [Google Scholar]
- Koyama T., Furutani M., Tasaka M., Ohme-Takagi M. (2007). TCP transcription factors control the morphology of shoot lateral organs via negative regulation of the expression of boundary-specific genes in Arabidopsis. Plant Cell 19, 473–484 10.1105/tpc.106.044792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyama T., Mitsuda N., Seki M., Shinozaki K., Ohme-Takagi M. (2010). TCP transcription factors regulate the activities of ASYMMETRIC LEAVES1 and miR164, as well as the auxin response, during differentiation of leaves in Arabidopsis. Plant Cell 22, 3574–3588 10.1105/tpc.110.075598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C., Potuschak T., Colón-Carmona A., Gutiérrez R. A., Doerner P. (2005). Arabidopsis TCP20 links regulation of growth and cell division control pathways. Proc. Natl. Acad. Sci. U.S.A. 102, 2978–12983 10.1073/pnas.0504039102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Rosso M. G., Viehoever P., Weisshaar B. (2007). GABI-Kat SimpleSearch: an Arabidopsis thaliana T-DNA mutant database with detailed information for confirmed insertions. Nucleic Acids Res. 35, D874–D878 10.1093/nar/gkl753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Li B., Dong A. (2012a). The Arabidopsis transcription factor AtTCP15 regulates endoreduplication by modulating expression of key cell-cycle genes. Mol. Plant 5, 270–280 10.1093/mp/ssr086 [DOI] [PubMed] [Google Scholar]
- Li Z., Li B., Shen W. H., Huang H., Dong A. (2012b). TCP transcription factors interact with AS2 in the repression of class-I KNOX genes in Arabidopsis thaliana. Plant J. 71, 99–107 10.1111/j.1365-313X.2012.04973.x [DOI] [PubMed] [Google Scholar]
- Maloof J. N., Nozue K., Mumbach M. R., Palmer C. M. (2013). LeafJ: an imagej plugin for semi-automated leaf shape measurement. J. Vis. Exp. 71, e50028 10.3791/50028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martín-Trillo M., Cubas P. (2010). TCP genes: a family snapshot ten years later. Trends Plant Sci. 15, 31–39 10.1016/j.tplants.2009.11.003 [DOI] [PubMed] [Google Scholar]
- Mutwil M., Obro J., Willats W. G., Persson S. (2008). GeneCAT: novel webtools that combine BLAST and co-expression analyses. Nucleic Acids Res 36, W320–W326 10.1093/nar/gkn292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nag A., King S., Jack T. (2009). miR319a targeting of TCP4 is critical for petal growth and development in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 106, 22534–22539 10.1073/pnas.0908718106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nath U., Crawford B. C. W., Carpenter R., Coen E. (2003). Genetic control of surface curvature. Science 299, 1404–1407 10.1126/science.1079354 [DOI] [PubMed] [Google Scholar]
- Navaud O., Dabos P., Carnus E., Tremousaygue D., Hervé C. (2007). TCP transcription factors predate the emergence of land plants. J. Mol. Evol. 65, 23–33 10.1007/s00239-006-0174-z [DOI] [PubMed] [Google Scholar]
- Niwa M., Daimon Y., Kurotani K., Higo A., Pruneda-Paz J. L., Breton G., et al. (2013). BRANCHED1 interacts with FLOWERING LOCUS T to repress the floral transition of the axillary meristems in Arabidopsis. Plant Cell 25, 1228–1242 10.1105/tpc.112.109090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ori N., Cohen A. R., Etzioni A., Brand A., Yanai O., Shleizer S., et al. (2007). Regulation of LANCEOLATE by miR319 is required for compound-leaf development in tomato. Nat. Genet. 39, 787–791 10.1038/ng2036 [DOI] [PubMed] [Google Scholar]
- Palatnik J. F., Allen E., Wu X., Schommer C., Schwab R., Carrington J. C., et al. (2003). Control of leaf morphogenesis by microRNAs. Nature 425, 257–263 10.1038/nature01958 [DOI] [PubMed] [Google Scholar]
- Pruneda-Paz J. L., Breton G., Para A., Kay S. A. (2009). A functional genomics approach reveals CHE as a component of the Arabidopsis circadian clock. Science 323, 1481–1485 10.1126/science.1167206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rueda-Romero P., Barrero-Sicilia C., Gómez-Cadenas A., Carbonero P., Oñate-Sánchez L. (2012). Arabidopsis thaliana DOF6 negatively affects germination in non-after-ripened seeds and interacts with TCP14. J. Exp. Bot. 63, 1937–1949 10.1093/jxb/err388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salomé P. A., Bomblies K., Laitinen R. A., Yant L., Mott R., Weigel D. (2011). Genetic architecture of flowering-time variation in Arabidopsis thaliana. Genetics 188, 421–433 10.1534/genetics.111.126607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid M., Davison T. S., Henz S. R., Pape U. J., Demar M., Vingron M., et al. (2005). A gene expression map of Arabidopsis development. Nat. Genet. 37, 501–506 10.1038/ng1543 [DOI] [PubMed] [Google Scholar]
- Schommer C., Palatnik J. F., Aggarwal P., Chételat A., Cubas P., Farmer E. E., et al. (2008). Control of jasmonate biosynthesis and senescence by miR319 targets. PLoS Biol. 6:e230 10.1371/journal.pbio.0060230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sessions A., Burke E., Presting G., Aux G., McElver J., Patton D., et al. (2002). A high-throughput Arabidopsis reverse genetics system. Plant Cell 14, 2985–2994 10.1105/tpc.004630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner E., Efroni I., Gopalraj M., Saathoff K., Tseng T.-S., Kieffer M., et al. (2012a). The Arabidopsis O-linked N-acetylglucosamine transferase SPINDLY interacts with Class I TCPs to facilitate cytokinin responses in leaves and flowers. Plant Cell 24, 96–108 10.1105/tpc.111.093518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner E., Yanai O., Efroni I., Ori N., Eshed Y., Weiss D. (2012b). Class I TCPs modulate cytokinin-induced branching and meristematic activity in tomato. Plant Signal. Behav. 7, 807–810 10.4161/psb.20606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda T., Amano K., Ohto M.-A., Nakamura K., Sato S., Kato T., et al. (2006). RNA interference of the Arabidopsis putative transcription factor TCP16 gene results in abortion of early pollen development. Plant Mol. Biol. 61, 165–177 10.1007/s11103-006-6265-9 [DOI] [PubMed] [Google Scholar]
- Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. (2011). MEGA5: molecular evolutionary genetics analysis using likelihood, distance, and parsimony methods. Mol. Biol. Evol. 28, 2731–2739 10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatematsu K., Nakabayashi K., Kamiya Y., Nambara E. (2008). Transcription factor AtTCP14 regulates embryonic growth potential during seed germination in Arabidopsis thaliana. Plant J. 53, 42–52 10.1111/j.1365-313X.2007.03308.x [DOI] [PubMed] [Google Scholar]
- Tokunaga H., Kojima M., Kuroha T., Ishida T., Sugimoto K., Kiba T., et al. (2012). Arabidopsis lonely guy (LOG) multiple mutants reveal a central role of the LOG-dependent pathway in cytokinin activation. Plant J. 69, 355–365 10.1111/j.1365-313X.2011.04795.x [DOI] [PubMed] [Google Scholar]
- Uberti-Manassero N. G., Lucero L. E., Viola I. L., Vegetti A. C., Gonzalez D. H. (2012). The class I protein AtTCP15 modulates plant development through a pathway that overlaps with the one affected by CIN-like TCP proteins. J. Exp. Bot. 63, 809–823 10.1093/jxb/err305 [DOI] [PubMed] [Google Scholar]
- Uberti Manassero N. G., Viola I. L., Welchen E., Gonzalez D. H. (2013). TCP transcription factors: architectures of plant form. Biomol. Concepts 4, 111–127 10.1515/bmc-2012-0051 [DOI] [PubMed] [Google Scholar]
- Uchida N., Townsley B., Chung K., Sinha N. (2007). Regulation of SHOOT MERISEMLESS genes via an upstream-conserved noncoding sequence coordinates leaf development. Proc. Natl. Acad. Sci. U.S.A. 104, 15953–15958 10.1073/pnas.0707577104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valsecchi I., Guittard-Crilat E., Maldiney R., Habricot Y., Lignon S., Lebrun R., et al. (2013). The intrinsically disordered C-terminal region of Arabidopsis thaliana TCP8 transcription factor acts both as a transactivation and self-assembly domain. Mol. BioSyst. 9, 2282–2295 10.1039/C3MB70128J [DOI] [PubMed] [Google Scholar]
- Viola I., Uberti-Manassero N., Ripoll R., Gonzalez D. (2011). The Arabidopsis class I TCP transcription factor AtTCP11 is a developmental regulator with distinct DNA-binding properties due to the presence of a threonine residue at position 15 of the TCP domain. Biochem. J. 435, 143–155 10.1042/BJ20101019 [DOI] [PubMed] [Google Scholar]
- Wang J.-W., Schwab R., Czech B., Mica E., Weigel D. (2008). Dual effects of miR156-targeted SPL genes andCYP78A5/KLUH on plastochron length and organ size in Arabidopsis thaliana. Plant Cell 20, 1231–1243 10.1105/tpc.108.058180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter D., Vinegar B., Nahal H., Ammar R., Wilson G. V., Provart N. J. (2007). An ‘Electronic Fluorescent Pictograph’ browser for exploring and analyzing large-scale biological data sets. PLoS ONE 2:e718 10.1371/journal.pone.0000718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woody S., Austin-Phillips S., Amasino R., Krysan P. (2007). The WiscDsLox T-DNA collection: an Arabidopsis community resource generated by using an improved high-throughput T-DNA sequencing pipeline. J. Plant Res. 120, 157–165 10.1007/s10265-006-0048-x [DOI] [PubMed] [Google Scholar]
- Xu L., Xu Y., Dong A., Sun Y., Pi L., Xu Y., et al. (2003). Novel as1 and as2 defects in leaf adaxial-abaxial polarity reveal the requirement for ASYMMETRIC LEAVES1 and 2 and ERECTA functions in specifying leaf adaxial identity. Development 130, 4097–4107 10.1242/dev.00622 [DOI] [PubMed] [Google Scholar]
- Yanai O., Shani E., Russ D., Ori N. (2011). Gibberellin partly mediates LANCEOLATE activity in tomato. Plant J. 68, 571–582 10.1111/j.1365-313X.2011.04716.x [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Sequence alignment of the TCP box of class I TCP factors.
List of TCP genes in the tomato genome.
List of primers used in this study for gene expression analysis.
List of primers used to genotype tcp mutants in Arabidopsis.
List of primers for genetic constructs generated in this study.