Abstract
SirT1 is a class III histone deacetylase that has been implicated in metabolic and reactive oxygen species control. In the vasculature it has been shown to decrease endothelial superoxide production, prevent endothelial dysfunction and atherosclerosis. However, the mechanisms that mediate SirT1 antioxidant functions remain to be characterized. The transcription factor FoxO3a and the transcriptional coactivator peroxisome proliferator activated receptor γ-coactivator 1α (PGC-1α) have been shown to induce the expression of antioxidant genes and to be deacetylated by SirT1. Aims: Here we investigated SirT1 regulation of antioxidant genes and the roles played by FoxO3a and PGC-1α in this regulation. Results: We found that SirT1 regulates the expression of several antioxidant genes in bovine aortic endothelial cells, including Mn superoxide dismutase (MnSOD), catalase, peroxiredoxins 3 and 5 (Prx3, Prx5), thioredoxin 2 (Trx2), thioredoxin reductase 2 (TR2), and uncoupling protein 2 (UCP-2) and can be localized in the regulatory regions of these genes. We also found that knockdown of either FoxO3a or PGC-1α prevented the induction of antioxidant genes by SirT1 over-expression. Furthermore, SirT1 increased the formation of a FoxO3a/PGC-1α complex as determined by co-immunoprecipitation (IP) assays, concomitantly reducing H2O2-dependent FoxO3a and PGC-1α acetylation. Data showing that FoxO3a knockdown increases PGC-1α acetylation levels and vice versa, suggest that SirT1 activity on FoxO3a and PGC-1α may be dependent of the formation of a FoxO3a/PGC-1α complex. Innovation: A unifying mechanism for SirT1 activities is suggested. Conclusion: We show that SirT1 regulation of antioxidant genes in vascular endothelial cells depends on the formation of a FoxO3a/PGC-1α complex. Antioxid. Redox Signal. 19, 1507–1521.
Introduction
All the risk factors considered for atherosclerosis have been related to increased endothelial reactive oxygen species (ROS) levels (51). Excessive production of ROS in the vascular endothelium contributes to the appearance of endothelial dysfunction and progression of atherosclerosis (14, 18). Endothelial dysfunction is characterized by the decrease in nitric oxide (NO) bioavailability and subsequent impair of endothelium-dependent vasorelaxation and mitochondrial biogenesis (7, 39). ROS are also necessary for normal endothelial cell function and have been shown to play an important role in angiogenesis (17, 48). In fact, abnormally high endothelial ROS levels have also been associated with dysfunctional angiogenic processes like those associated with tumors (47) and diabetic retinopathy (4). Under normal physiological conditions, the main source of ROS in the cell is the mitochondrial electric transport chain (22). In pathological states, increased mitochondrial ROS results in the activation of NADPH oxidases (NOX), by still unclear mechanisms (12). NOX activation dramatically increases cellular ROS production and is responsible for oxidative stress-dependent tissue damage (24).
Innovation.
The notion that oxidative stress is closely linked to the aging process has been challenged by studies that highlight the major role played by proteins that control genomic stability, such as SirT1. Here we show that SirT1 plays a central role in oxidative stress control and provides a central link between genomic stability and reactive oxygen species suggesting a unifying mechanism for the aging process that is also central in the regulation of vascular homeostasis.
ROS levels are controlled by a complex system of defenses that limit the accumulation of oxidants, including superoxide dismutases, catalase, peroxiredoxins, thioredoxins and thioredoxin reductases, and the enzymes involved in the control of the levels and redox state of glutathione (27). Given the pivotal role played by the mitochondria in ROS control, this organelle is particularly well provisioned with antioxidant proteins, including uncoupling protein 2 (UCP-2), that prevents mitochondrial ROS generation, detoxifying enzymes like Mn superoxide dismutase (MnSOD), glutathione peroxidase 4 (Gpx4), peroxiredoxin 5 (Prx5), thioredoxin 2 (Trx2) and thioredoxin reductase 2 (TR2), and the mitochondrial isoforms of peroxiredoxin 3 (Prx3) and glutation peroxidase 1 (GPx1), among others (27).
In the last years, several reports have highlighted the role of SirT1 as a key regulator of endothelial homeostasis. SirT1 is a class III histone deacetylase. It is highly sensitive to cellular redox and nutritional status and it is known to control genomic stability and cellular metabolism. It deacetylates both histone and nonhistone proteins with diverse cellular and tissue functions (10). In the vasculature SirT1 has been shown to regulate the angiogenesis process (35), and to prevent endothelial dysfunction and atherosclerosis (44). In this context, SirT1 has been shown to reduce endothelial superoxide production, to inhibit Nuclear Factor-κB (NF-κB) signaling, and to reduce expression of adhesion molecules. Both NF-kB activity and expression of adhesion molecules are hallmarks of endothelial oxidative stress, suggesting that SirT1 protective activity might be related to induction of antioxidants (45). However, its role in oxidative control remains poorly characterized.
Previous studies have shown that SirT1 deacetylates the transcription factor FoxO3a, and this regulation increased FoxO3a ability to induce resistance to oxidative stress (8). MnSOD and catalase are well-characterized targets of the FoxO family of transcription factors, including FoxO1a, FoxO3a and FoxO4. It has been later evidenced that SirT1 activity is associated with increased levels of MnSOD in cardiomyocytes (46) and catalase both in the heart (1), where the induction was shown to be FoxO1a-dependent, and renal tubular cells (15), where it was proposed to be linked to FoxO3a activation. Furthermore, FoxO factors have also been shown to play a key role in endothelial homeostasis regulating angiogenesis (36). In endothelial cells, FoxO3a positively regulates a set of major antioxidant genes through its interaction with the transcriptional co-activator peroxisome proliferator activated receptor γ-coactivator 1α (PGC-1α) (30). In turn, PGC-1α had been previously shown to protect endothelial cells from ROS-induced endothelial dysfunction and cell death (49). Importantly, PGC-1α, a master metabolic regulator, is another SirT1 target (25). SirT1 was shown to deacetylate PGC-1α in response to caloric restriction and enhance its activity on fatty acid oxidation and gluconeogenesis genes (13, 38). Of note, PGC-1α expression is positively regulated by SirT1 (2).
Therefore, we decided to characterize SirT1 role in the regulation of antioxidant systems in endothelial cells. We found that SirT1 positively regulates the expression of a set of antioxidant genes and it is recruited to the promoter regions of those genes. Importantly, SirT1 requires both PGC-1α and FoxO3a to regulate antioxidant gene expression. We observed that SirT1 deacetylates PGC-1α and FoxO3a in response to oxidative stress and enhances the formation and stability of the PGC-1α/FoxO3a transcriptional complex.
Results
SirT1 regulates the expression of oxidative stress protection genes
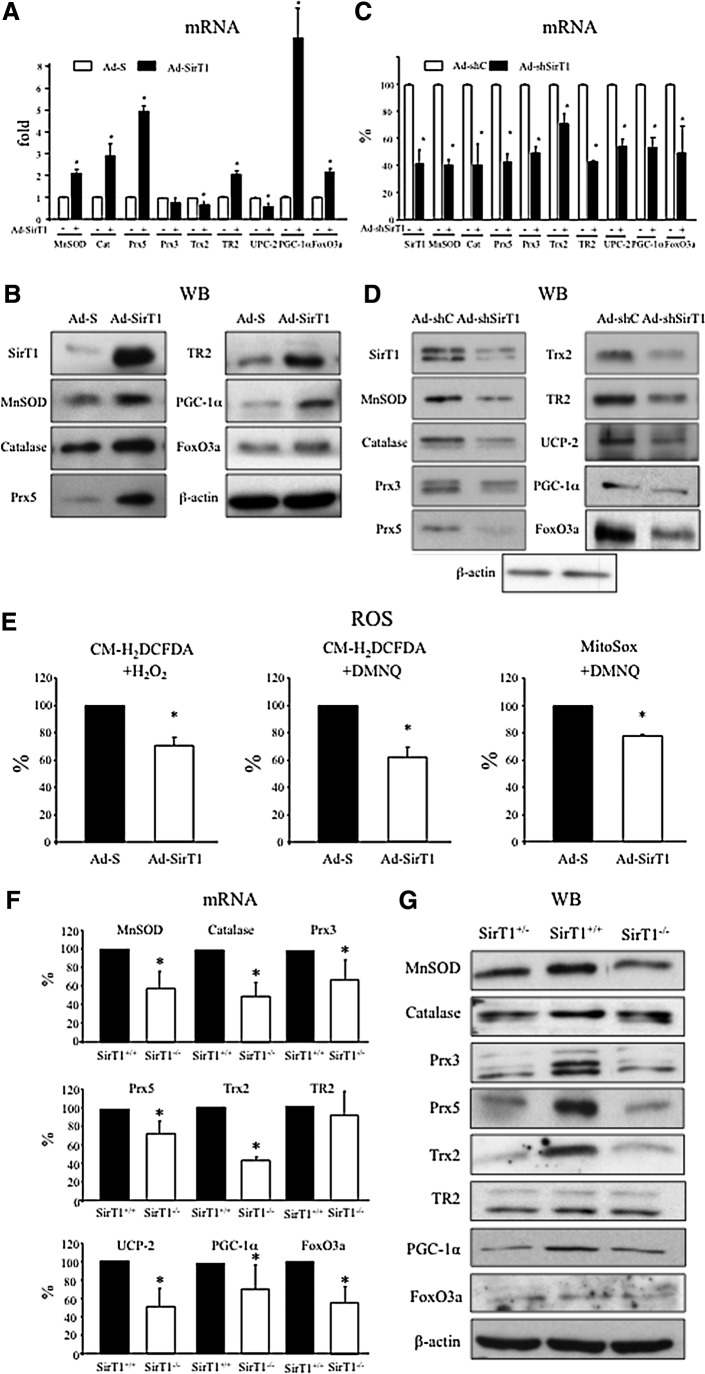
To characterize the role of SirT1 in the regulation of antioxidant genes, we first used a recombinant adenovirus (Ad-SirT1) to over-express SirT1 in bovine aortic endothelial cells (BAEC) and analyzed by qRT-PCR and Western Blot the levels of a group of antioxidant genes regulated by both PGC-1α and FoxO3a, namely MnSOD, catalase, Prx3, Prx5, Trx2, TR2, and UCP-2. Full-size Western Blot membranes, including molecular weight markers for all proteins included in the study are shown in Supplementary Fig. S1.
We found that SirT1 over-expression increased the mRNA and protein levels of MnSOD, catalase, Prx5, and TR2. Importantly, SirT1 also induced the expression of PGC-1α and FoxO3a. However, SirT1 over-expression failed to increase the expression levels of Prx3 and it actually decreased Trx2 and UCP-2 mRNA levels (Fig. 1A, B and Supplementary Fig. S2). This result suggested that SirT1 was in fact a regulator of, at least, some antioxidant genes in endothelial cells, and induction of PGC-1α and FoxO3a mRNA expression could play a role in this regulation. We also tested whether SirT1 over-expression modulated the levels of another important antioxidant gene transcriptional regulator, Nrf2, but SirT over-expression was not found to significantly change Nrf2 levels (Supplementary Fig. S3).
FIG. 1.
SirT1 regulates the expression of oxidative stress protection genes. (A–D) Analysis of oxidative stress protection genes in bovine aortic endothelial cells (BAEC) that were infected o/n with recombinant adenovirus as indicated and harvested 24 h postinfection. (A, C) qRT-PCR mRNA expression analysis. (B, D) Protein analysis by Western Blot. (E) SirT1 dependent changes in ROS levels were monitored in control and SirT1-overexpressing BAEC, treated with 50 μM H2O2 for 4 h or 20 μM DMNQ for 2 h, as indicated. Cells were labeled with CM-H2DCFDA and analyzed by flow cytometry, (left and middle panels), or labeled with MitoSOX Red and after fixation, analyzed by confocal microscopy. (F-G) Analysis of oxidative stress protection genes in serum deprived SirT1+/+ and SirT1−/− MEFs. BAEC (F) qRT-PCR. (G) Western Blot. Data are from ≥3 independent experiments. Data are means+SD (*) p≤0.05 versus control. The blots presented correspond to a representative experiment.
Next, we knocked-down SirT1 in BAEC using a recombinant adenovirus that drives the expression of a SirT1 specific shRNA (shSirT1). We observed that Ad-shSirT1 decreased the levels of not only MnSOD, catalase, Prx5, TR2, PGC-1α and FoxO3a, but also of Prx3, Trx2 and UCP-2, suggesting that Prx3, Trx2 and UCP-2 are also positively regulated by SirT1 (Fig. 1C, D and Supplementary Fig. S2). These results indicate that endogenous SirT1 is required to maintain the basal expression levels of a whole set of oxidative stress protection genes in endothelial cells and supports the idea that its regulation of PGC-1α and FoxO3a expression levels is likely to be relevant in the process.
Cell viability was not significantly affected by SirT1 over-expression or knock-down, indicating that antioxidant gene regulation is not likely to be a side-effect of altered cell viability (Supplementary Fig. S4).
To evaluate if SirT1 regulation of antioxidant genes actually increased the antioxidant capacity of the cells, we treated BAEC with a moderate concentration of H2O2 (50 μM), and evaluated the effect of SirT1 in total ROS levels using the fluorescent probe chloromethyl-2′,7′-dichlorofluorescein diacetate (CM-H2DCFDA). We found that SirT1 reduced the DCF signal in H2O2-treated cells, suggesting that it actually increased the cellular detoxification capacity (Fig. 1E, left panel). Next, we tested the capacity of SirT1 to control ROS accumulation in the presence of an internal source of ROS. BAEC were treated with 2,3-dimethoxy-1-naphthoquinone (DMNQ), an inhibitor of the mitochondrial electron transport chain that drives the formation of mitochondrial O2− in a dose-dependent manner, and we found that SirT1 reduced DCF accumulation (Fig. 1E, middle panel). To elucidate the impact of SirT1 in mitochondrial ROS, we tested SirT1 activity in DMNQ treated cells labeled with MitoSOX, a relatively specific probe for mitochondrial O2− (it has been noted that it can also react with other oxidants) (55). We observed that SirT1 also significantly reduced mitochondrial O2− accumulation, supporting the idea that SirT1 increases the mitochondrial detoxification capacity (Fig. 1E, right panel).
Next, we determined the expression levels of the antioxidant genes under study in SirT1+/+, and SirT1−/− MEFs (11). Analysis by qRT-PCR and Western Blot showed that the expression levels of the oxidative stress protection genes studied as well as PGC-1α and FoxO3a are lower in SirT1−/− MEFs than in SirT1+/+ MEFs, supporting the notion that SirT1 is a general regulator of antioxidant genes and is required to maintain the basal expression levels of the antioxidant system (Fig. 1F–G and Supplementary Fig. S2). Interestingly, it was noted that SirT1+/- MEFs also showed an important reduction in the levels of antioxidant proteins, further stressing the relevance of SirT1 on their regulation (Fig. 1G).
Furthermore, when we tested the effect of SirT1 absence in total ROS levels using the fluorescent probe CM-H2DCFDA, we found that SirT1−/− MEFs had higher levels of ROS than SirT1+/+ MEFs (Supplementary Fig. S5). Importantly, it was also noted that SirT1 expression is increased in oxidative stress conditions (Supplementary Fig. S6). The specificity of the antibody used against SirT1, was tested using whole cell extracts from SirT1+/+ and SirT1−/− MEFs by Western Blot (Supplementary Fig. S7).
SirT1 is recruited to promoter regions of antioxidant genes
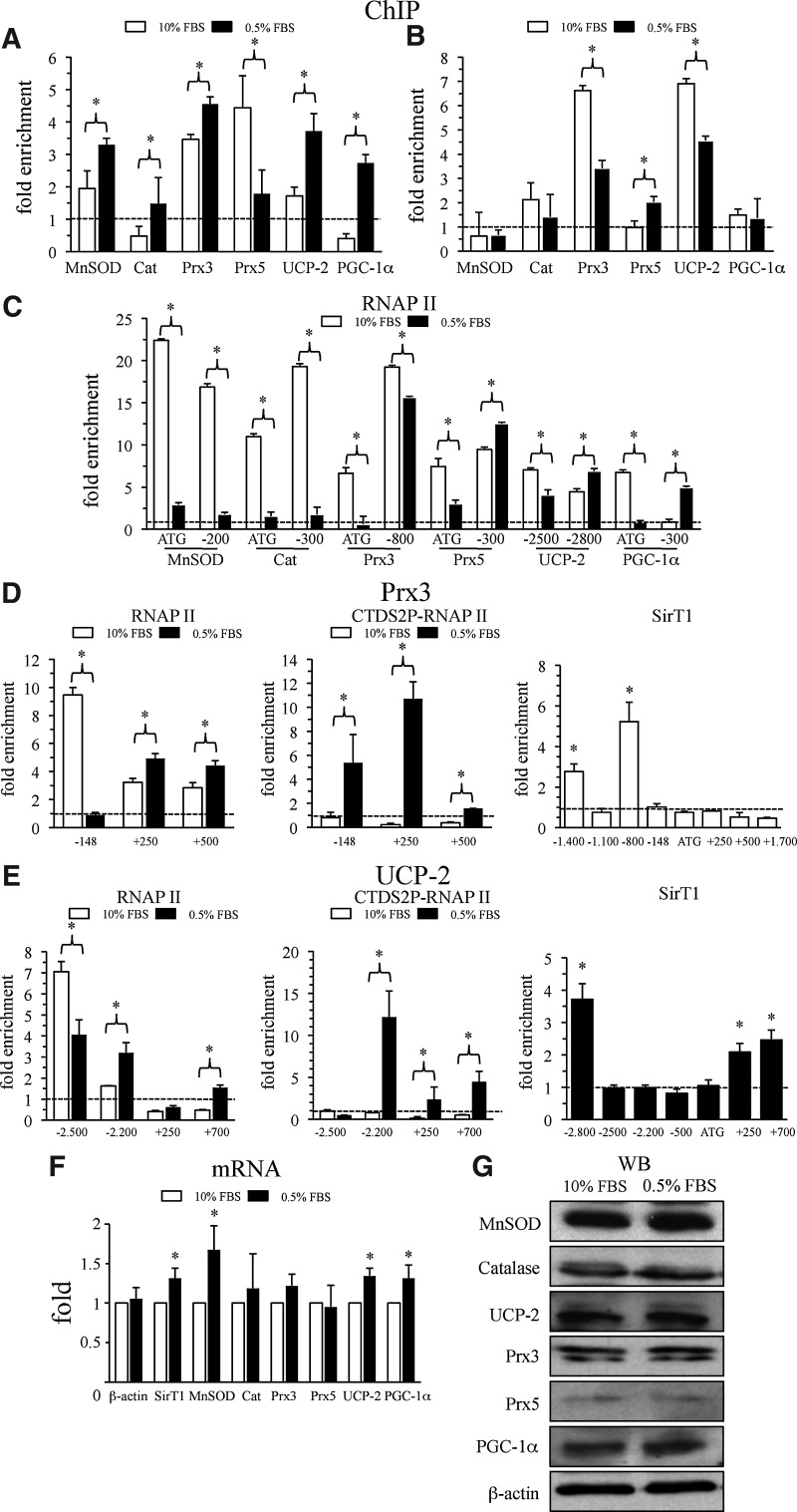
To investigate the mechanisms involved in SirT1 regulation, we tested SirT1 association with promoter regions of the target genes using chromatin immunoprecipitation (ChIP). 10% fetal bovine serum (FBS) and 0.5% FBS conditions were used as a simple strategy to modulate SirT1 activity (8). We found that SirT1 did associate with the promoter regions of sod2 (MnSOD), catalase, prx3, prx5, ucp-2, pgc-1α, and foxO3a (trx2, tr2 were not analyzed). Serum starvation generally increased SirT1 recruitment with the remarkable exception of the Prx5 gene, where it had the opposite effect (Fig. 2A and Supplementary Fig. S8). These data suggested that SirT1 is likely to be directly involved in the regulation of mitochondrial antioxidant genes, including PGC-1α itself.
FIG. 2.
SirT1 is recruited to promoter regions of ROS detoxification genes upon serum starvation. (A–E) Chromatin immunoprecipitation assays of promoter regions corresponding to the indicated ROS detoxification genes. Cross-linked chromatin Promoter occupancy was analyzed by qPCR using specific primers. β-actin CDS was used to control for non specific enrichment. (A) SirT1 binding sites. (B) H4K16 acetylation levels at SirT1 sites. (C) Pol II recruitment. The position of the analyzed fragments is indicated, using as reference (+1), the position of the ATG. (D, E) Analysis of prx3 and ucp-2 genes. Left panels, analysis of Pol II at the transcriptional start region and along the gene. Central panel, analysis of “elongating” Pol II levels (Pol II phosphorylated in the Ser2 positions of the CTD repeats of the Pol II largest subunit) at the transcriptional start region and downstream of the ATG. Right panels, SirT1 recruitment along the genes, upstream and downstream of the ATG. The position of the analyzed fragments is indicated, using as reference (+1), the position of the ATG. (F) mRNA and (G) protein levels of SirT1 target genes in confluent BAEC cells in 10% fetal bovine serum (FBS) or serum deprived (0.5% FBS) o/n were analyzed by qRT-PCR and western blot, respectively. Data are from ≥3 independent experiments. Data are means+SD (*) p≤0.05 versus control. The blots presented correspond to a representative experiment.
Next, we decided to evaluate if SirT1 recruitment was associated with a reduction in histone acetylation. We carried out ChIP assays using antibodies against the acetylated form of histone H4 on lysine 16 (H4K16Ac), a position highly sensitive to SirT1 activity (50). We found that changes in acetylation inversely correlated with changes in SirT1 recruitment (more SirT1/less acetylation). This result suggests that SirT1 deacetylates H4K16 positions upon recruitment to antioxidant genes (Fig. 2B).
Histone deacetylation is normally associated with gene silencing and reduced recruitment of RNA polymerase II (RNAP II). We monitored RNAP II recruitment by ChIP and found that in positions proximal to the transcription start site (ATG primers for all genes except for ucp-2/-2500 since in this case the ATG is in exon 3), serum deprivation resulted in a dramatic decrease in RNAP II recruitment, suggesting that SirT1-dependent histone deacetylation negatively impacts on RNAP II recruitment (Fig. 2C).
Since serum deprivation did not decrease antioxidant gene expression and actually increased it in some cases (Fig. 2F–G and Supplementary Fig. S2), we first analyzed if the pattern of RNAP II recruitment was the same or different at the up stream SirT1 binding sites. We found that the upstream sites where SirT1 binding had been identified showed two distinct patterns. At sod-2, catalase and prx3, RNAP II binding was also reduced in starvation conditions, but only very moderately at the prx3 promoter. On the other hand, RNAP II recruitment was actually increased by serum deprivation at the prx5, ucp-2 and pgc-1α promoters, suggesting that SirT1 recruitment could, at least in some cases, be associated to increased RNAP II recruitment at alternative start sites, despite of reduced acetylation on the site itself (Fig. 2C).
To understand how these two recruitment patterns resulted in a common regulation pattern, we selected two representative genes, prx3 and ucp-2, for further analysis, where SirT1 binding, RNAP II recruitment, and levels of elongating RNAP II were monitored along the gene (Fig. 2D–E). As already indicated, in positions proximal to the transcription start site, serum deprivation resulted in a reduction in RNAP II recruitment, but downstream, it resulted in an increase in RNAP II levels. Reduced recruitment at the start site and increased levels downstream could be indicative of increased transcription rates. To test this idea we monitored the levels of elongating RNAP II by ChIP, using antibodies against the phosphorylated form of the C-terminal domain of RNAP II (CTD2P). We found dramatic increases in “elongating RNAP II” in serum starvation conditions in both prx3 and ucp-2 genes, suggesting that reduced RNAP II recruitment at the start site was compensated by increased promoter clearance. SirT1 binding could only be detected at promoter sites in the prx3 gene, but it was also detected within the transcriptional unit in the ucp-2 gene. These results might explain how SirT1 recruitment can be associated with a sustained or increased gene expression despite of a general reduction in RNA II recruitment induced by serum deprivation, as observed (Fig. 2F–G, Supplementary Table S1).
SirT1 requires PGC-1α and FoxO3a to induce the expression of oxidative stress genes
SirT1 controls the expression of a set of antioxidant genes that have been previously described as direct targets of the transcriptional regulators PGC-1α and FoxO3a (30, 49). Importantly, these two factors are regulated by SirT1 both at transcriptional (see above) and post-transcriptional levels (8, 38). These results prompted us to investigate to what extent SirT1 activity on antioxidant genes was dependent on these two regulators.
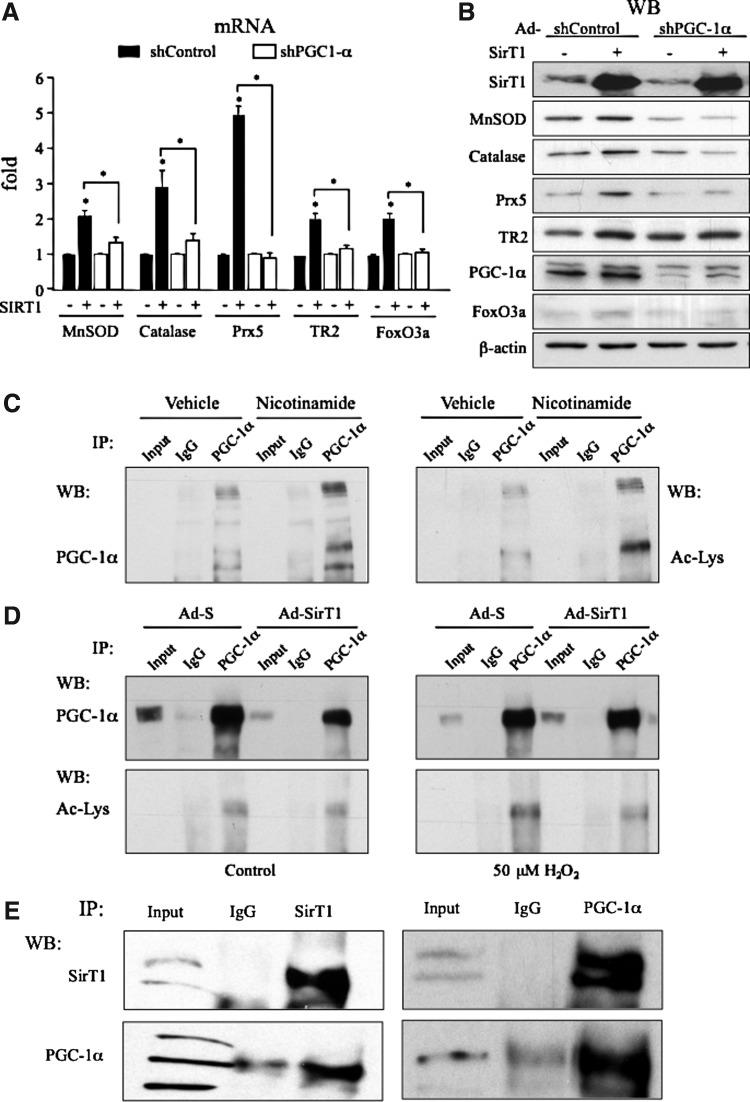
To evaluate if PGC-1α was required for SirT1 activity, we tested the effect of knocking down PGC-1α (Ad-shPGC-1α) when SirT1 was over-expressed in BAEC. Analysis of antioxidant gene expression by qRT-PCR and Western Blot showed that SirT1 induction of MnSOD, catalase, Prx5, TR2, and FoxO3a was clearly reduced in PGC-1α knocked-down cells (Fig. 3A, B and Supplementary Fig. S2). These results indicate that PGC-1α is required for SirT1 regulation of a set of genes involved in mitochondrial ROS detoxification.
FIG. 3.
SirT1 requires PGC-1α to induce the expression of oxidative stress protection genes. (A, B) BAEC were coinfected o/n with Ad-SirT1 and Ad-shPGC-1α, or the respective control adenoviruses. 12 h postinfection, cells were deprived of serum o/n and harvested. Expression of the oxidative stress protection genes was analyzed by qRT-PCR (A) and Western Blot (B). (C, D). BAEC whole cell extracts were subjected to immunoprecipitation (IP) with a specific PGC-1α antibody or normal IgG, as a control, and analyzed by western blot as indicated. Input indicates a 1/10 of whole cell extract not subjected to immunoprecipitation. (C) BAEC were infected o/n with Ad-PGC-1α and 8 h postinfection were treated with 10 mM nicotinamide for 15 h. (D) BAEC were coinfected o/n with Ad-SirT1 and Ad-PGC-1α, or the respective control adenoviruses. 24 h postinfection cells were deprived of serum o/n, and then treated with 50 μM H2O2 or vehicle for 4 h. (E) Confluent BAEC were grown in galactose media and in the absence of serum for 48 h. Whole cell extracts were subjected to immunoprecipitation (IP) with specific PGC-1α or SirT1 antibody or normal IgG, as a control, and analyzed by western blot as indicated. Data are from ≥3 independent experiments. Data are means+SD (*) p≤0.05 versus control. The blots presented correspond to a representative experiment.
We have already shown that SirT1 directly regulates PGC-1α expression. To evaluate if the regulation by SirT1 of PGC-1α activity could also play a role, we first tested SirT1 ability to deacetylate PGC-1α in BAEC. First, we used BAEC that over-expressed PGC-1α and treated them with the pan-sirtuin inhibitor nicotinamide. Nicotinamide increased acetylated PGC-1α, suggesting that PGC-1α acetylation is regulated by sirtuins in endothelial cells. Nicotinamide treatment also resulted in an increase in high molecular weight forms of Ac-PGC-1α and likely corresponded to polyubiquitinated PGC-1α (3, 31). Therefore, this result suggests that sirtuins could also play a role in PGC-1α protein stabilization in endothelial cells (Fig. 3C).
It has been described that in response to oxidative stress PGC-1α acetylation levels are dramatically increased (3). To directly evaluate the effect of SirT1 on PGC-1α acetylation, we tested the effect of SirT1 over-expression in oxidative stress conditions. We found that, SirT1 significantly reduced the levels of PGC-1α acetylation and SirT1 knock-down increased it, suggesting that SirT1 regulates PGC-1α acetylation in response to oxidative stress (Fig. 3D and Supplementary Fig. S9). Importantly, SirT1 over-expression also reduced PGC-1α ubiquitination in oxidative stress conditions (Supplementary Fig. S10).
We then used co-immunoprecipitation (co-IP) assays to test SirT1/PGC-1α interaction. We cultured BAEC in media in which glucose was substituted with galactose, for 48 h, to boost mitochondrial metabolism and induce PGC-1α levels. In these conditions, we were able to identify SirT1 in PGC-1α immunoprecipitates and vice versa, confirming that SirT1 and PGC-1α interact in endothelial cells (Fig. 3E). To test whether SirT1/PGC-1α interaction was redox sensitive, we pretreated the cells with the antioxidant EUK-89; however, no significant differences were observed (Supplementary Fig. S11).
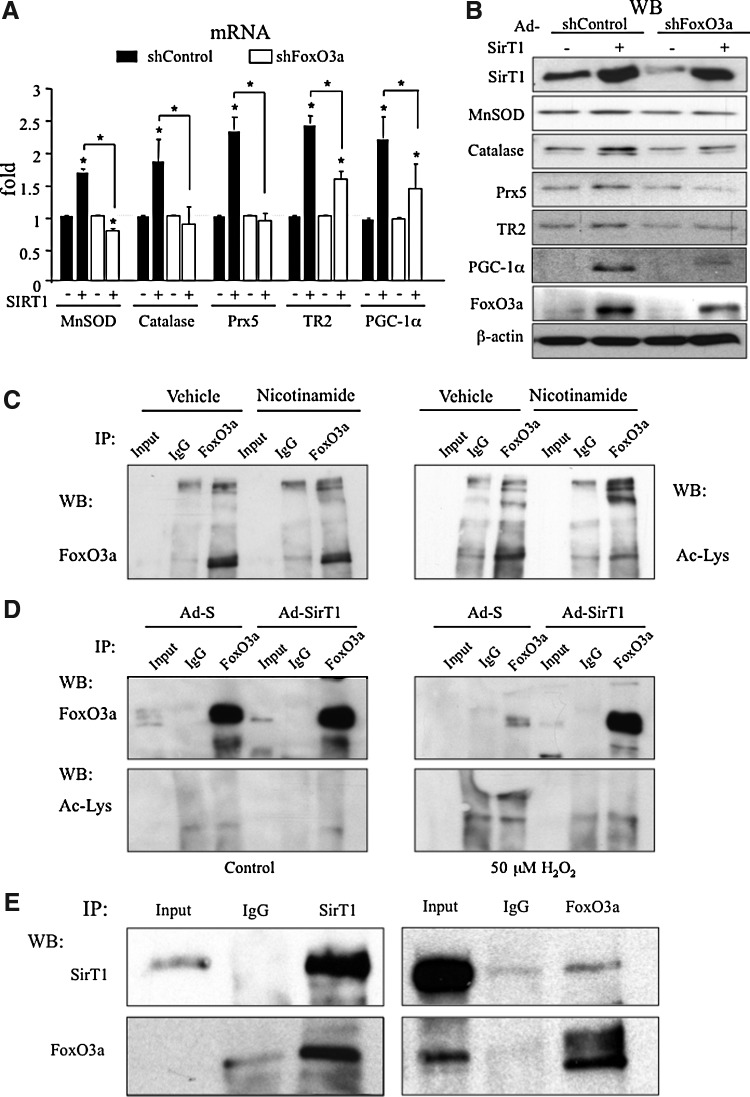
To evaluate if FoxO3a was required for SirT1 activity, we tested the effect of knocking down FoxO3a (Ad-shFoxO3a) when SirT1 was over-expressed in BAEC. Gene expression analysis showed that SirT1 ability to induce the expression of MnSOD, catalase, Prx5, TR2 and PGC-1α was clearly reduced in FoxO3a knocked-down cells (Figure 4A, B and Supplementary Fig. S2). These results indicate that FoxO3a is required for SirT1 regulation of antioxidant genes.
FIG. 4.
FoxO3a is required for SirT1-induction of antioxidant genes. (A, B) BAEC were first infected o/n with Ad-SirT1 or a control adenovirus, 36 h later cells were infected o/n with Ad-shFoxO3a or a control adenovirus and finally serum deprived for 8 h. Expression levels of ROS detoxification genes were determined by qRT-PCR (A) and Western Blot (B). (*) p≤0.05 versus control. (C, D) BAEC whole cell extracts were subjected to immunoprecipitation (IP) with a specific FoxO3a (C) or anti-HA (D) antibody or normal rabbit IgG, as a control, and analyzed by western blot as indicated. Input indicates a 1/10 of whole cell extract not subjected to immunoprecipitation. (C) BAEC were treated with 10 mM nicotinamide for 15 h (D) BAEC were coinfected o/n with Ad-SirT1 and Ad-FoxO3a-WT. 24 h postinfection the cells were serum deprived o/n, and then treated for 4 h with 50 μM H2O2 or vehicle (E) Confluent BAEC were grown in galactose media in the absence of serum for 48 h. Whole cell extracts were subjected to immunoprecipitation (IP) with a specific FoxO3a or SirT1 antibody or normal IgG, as a control, and analyzed by western blot as indicated. Data are from ≥3 independent experiments. Data are means+SD (*) p≤0.05 versus control. The blots presented correspond to a representative experiment.
We have already shown that SirT1 regulates FoxO3a expression. To determine if regulation by SirT1 of FoxO3a activity could also play a role on SirT1 antioxidant activity, we tested whether SirT1 regulates FoxO3a acetylation in endothelial cells. Nicotinamide treatment did not increase acetylated FoxO3a levels, but resulted in an increase in high molecular weight Ac-FoxO3a that could correspond to polyubiquitinated FoxO3a (9). Therefore, this result suggests that sirtuins could also play a role in FoxO3a protein stabilization in endothelial cells (Fig. 4C). In fact, SirT1 was found to reduce FoxO3a ubiquitination in oxidative stress conditions (Supplementary Fig. S10).
To test the effect of SirT1 on the acetylation status of FoxO3a, we tested the effect of SirT1 over-expression. As already described (8), FoxO3a is acetylated in response to oxidative stress and SirT1 over-expression significantly reduced its acetylation (Fig. 4D). Therefore, these results confirm that SirT1 regulates FoxO3a acetylation in endothelial cells (Fig. 4D).
Finally, we tested Sirt1 and FoxO3a interaction in endothelial cells with co-IP assays. We cultured BAEC in media in which glucose was substituted with galactose, for 48 h, a condition that induces FoxO3a levels. In these conditions, we were able to identify FoxO3a in SirT1 IPs, together with PGC-1α, and vice versa, confirming that SirT1 and FoxO3a interact in endothelial cells (Fig. 4E and Supplementary Fig. S12). To test whether SirT1/FoxO3a interaction was redox sensitive, we pretreated the cells with the antioxidant EUK-89; however, no significant differences were observed (Supplementary Fig. S11).
SirT1 regulates the formation of a PGC-1α/FoxO3a complex
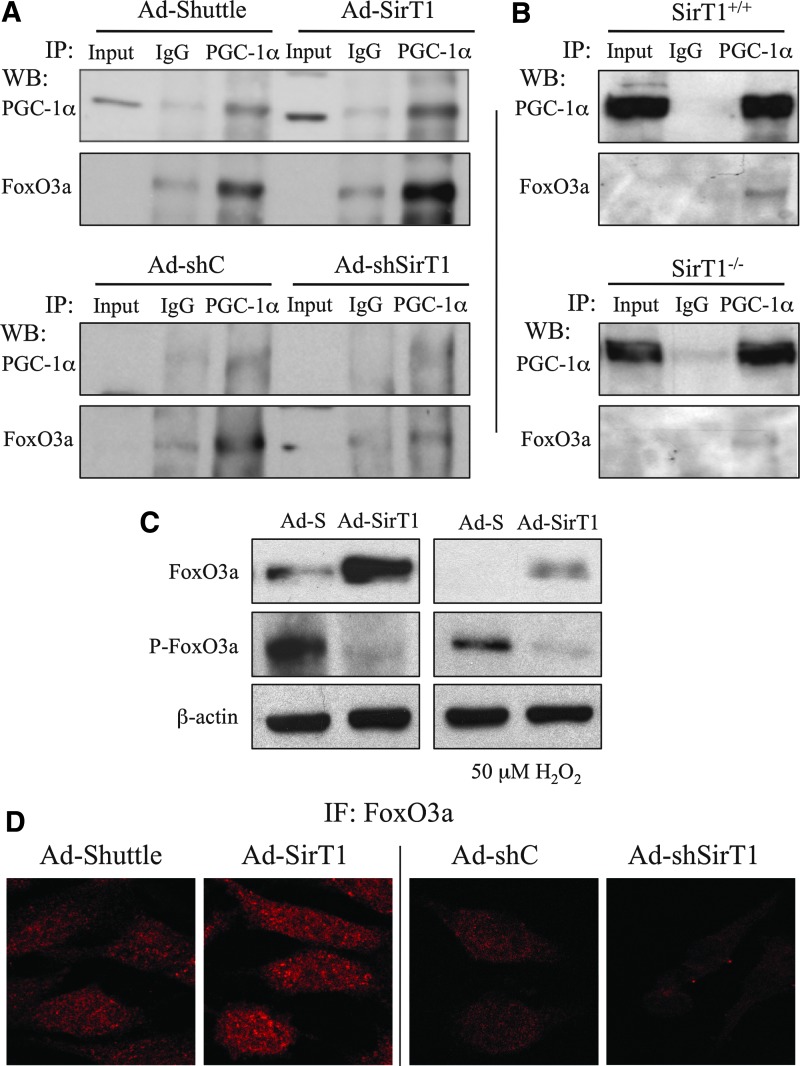
We have previously shown that PGC-1α and FoxO3a form a complex to regulate antioxidant genes in endothelial cells (30). Taking into account that SirT1 regulation of oxidative stress genes requires both PGC-1α and FoxO3a, we decided to test if SirT1 regulates the formation of the PGC-1α/FoxO3a complex in response to oxidative stress in BAEC. First, we tested the effect of SirT1 over-expression (Ad-SirT1) and found that SirT1 increased the levels of FoxO3a associated with PGC-1α in these conditions (Fig. 5A upper panel and Supplementary Fig. S2). Conversely, knock-down of SirT1 (Ad-shSirT1) had the opposite effect (Fig. 5A lower panel and Supplementary Fig. S2) suggesting that SirT1 positively regulates the formation of a PGC-1α/FoxO3a complex.
FIG. 5.
SirT1 regulates the formation of a PGC-1α/FoxO3a complex. (A) BAEC were coinfected o/n with Ad-PGC-1α and the indicated adenoviruses. 12 h postinfection, cells were deprived of serum o/n, and then treated for 4 h with 50 μM H2O2. Whole cell extracts were subjected to immunoprecipitation (IP) with a specific PGC-1α antibody or normal IgG, as a control, and analyzed by Western Blot as indicated. Input indicates a 1/10 of whole cell extract not subjected to immunoprecipitation. (B) SIRT1+/+ and SIRT1−/− MEFs were infected o/n with Ad-PGC1α and 12 h postinfection cells were deprived of serum o/n. Cell extracts were immunoprecipitated with an specific anti-PGC1α antibody (PGC-1α) or rabbit IgG as control (IgG) and analyzed by western blot. (C) BAEC were coinfected o/n with Ad-FoxO3a-WT and Ad-SirT1 or the corresponding controls, 12 h postinfection cells were deprived of serum o/n, and then treated for 4 h with 50 μM H2O2. Protein levels were analyzed by Western Blot (D) BAEC were infected o/n with Ad-SirT1, Ad-shSirT1 or the corresponding control adenovirus, 12 h postinfection, cells were deprived of serum o/n, and then fixed and analyzed by immunofluorescence with FoxO3a specific antibodies. The blots presented correspond to a representative experiment from ≥3 independent experiments.
To further investigate the role of SirT1 in the regulation of the PGC-1α/FOXO3a complex formation, we used SIRT1+/+ and SIRT1−/− MEFs that over-expressed PGC-1α (Ad-PGC-1α), and monitored the amount of FoxO3a that co-IP with PGC-1α. We observed that the amount of FoxO3a found in PGC-1α IPs was lower in SirT1−/− MEFs than in SirT1+/+ MEFs, supporting the previous observation that SirT1 is a positive regulator of FoxO3a/PGC-1α complex formation (Fig. 5B and Supplementary Fig. S2).
SirT1 regulates FoxO3a levels and phosphorylation
Phosphorylation is an important mechanism for the regulation of FoxOs subcellular localization and activity (9). It has been proposed that phosphorylation and acetylation are interdependent mechanisms that interact to regulate FoxO proteins in response to different stimuli (37). Therefore, we decided to test if SirT1 could play a role on FoxO3a phosphorylation. To that end we over-expressed SirT1 in BAEC (Ad-SirT1) and tested by Western Blot the phosphorylation levels of FoxO3a. We found that SirT1 over-expression resulted in a dramatic decrease in FoxO3a phosphorylation, and a significant increase in total FoxO3a levels (Fig. 5C). Similarly, immunofluorescence (IF) assays showed that SirT1 over-expression increased nuclear FoxO3a levels and shSirT1 had the opposite effect (Fig. 5D and Supplementary Fig. S13).
PGC-1α regulates FoxO3a level of acetylation and vice versa
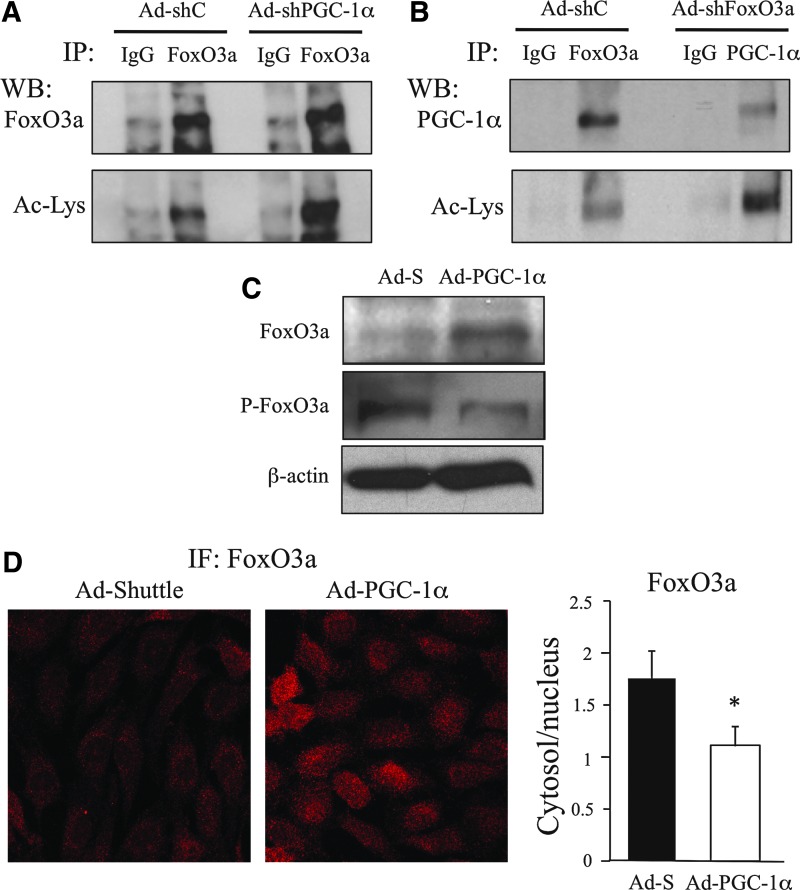
Two alternative mechanisms could explain SirT1 activity on the PGC-1α/FoxO3a complex, SirT1 could act either on PGC-1α and FoxO3a individually or on a preformed complex. To determine whether the acetylation status of either PGC-1α or FoxO3a could be influenced by the formation of a PGC-1α/FoxO3a complex, we tested FoxO3a acetylation levels in PGC-1α knocked-down BAEC and PGC-1α acetylation levels in FoxO3a knocked-down BAEC. We found that suppression of PGC-1α expression resulted in a dramatic increase in FoxO3a acetylation (Fig. 6A and Supplementary Fig. S2). Similarly, shRNA-mediated knockdown of FoxO3a induced an increase in acetylated PGC-1α (Fig. 6B and Supplementary Fig. S2). This result might suggest that SirT1 is likely to be active on the PGC-1α/FoxO3a complex, since in the absence of one of these factors, the other might not be deacetylated.
FIG. 6.
FoxO3a and PGC-1α influence each other's level of acetylation. (A, B) BAEC were infected o/n with Ad-shPGC-1α (A) Ad-shFoxO3a (B) or the corresponding control adenoviruses, 12 h postinfection, cells were deprived of serum o/n and harvested. Whole cell extracts were subjected to immunoprecipitation (IP) with FoxO3a (A) or PGC-1α (B) specific antibodies, or normal IgG, as a control. Acetylation levels were analyzed by western blot. (C, D) BAEC were infected o/n with Ad-PGC-1α or the control Ad-S and 48 h postinfection the cells were harvested. (C) FoxO3a phosphorylation was analyzed in WCE by western blot. (D) Immunofluorescence analysis of FoxO3a. Left panel confocal microscopy images. Right panel, FoxO3a cytoplasm/nuclear ratio. Data are from ≥3 independent experiments. Data are means+SD (*) p≤0.05 versus control.
We have shown that FoxO3a deacetylation by SirT1 reduces FoxO3a Akt-dependent phosphorylation. Now we tested whether PGC-1α over-expression had the same effect. We found that over-expression of PGC-1α resulted in a significant reduction in Akt-dependent FoxO3a phosphorylation, that was accompanied by an increase in total FoxO3a levels (Fig. 6C). To determine if this reduced phosphorylation resulted in a change of FoxO3a cytosolic/nuclear ratios, the effect of PGC-1α over-expression on FoxO3a localization was analyzed by IF. We found that PGC-1α significantly increased the nuclear localization of FoxO3a (Fig. 6D). This result further suggests that formation of a PGC-1α/FoxO3a complex plays role on the activity of their modifying enzymes, in particular SirT1.
PGC-1α over-expression cannot compensate for SirT1 deficiency in the regulation of antioxidant genes
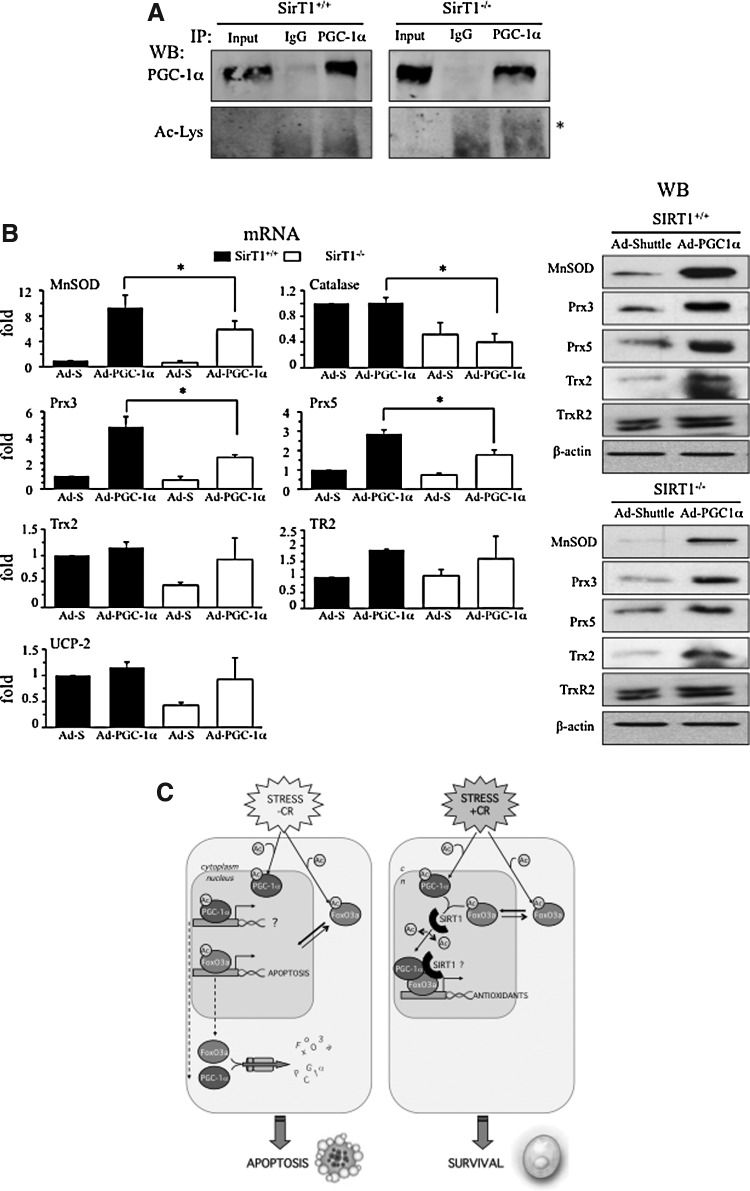
To evaluate to what extent SirT1 activity on antioxidant genes is dependent on its activity on the PGC-1α/FoxO3a complex, we tested whether PGC-1α over-expression could compensate for SirT1 loss in SirT1−/− MEF. We first tested the level of acetylation of PGC-1α in SirT1+/+ and SirT1−/− MEFs infected with Ad-PGC-1α, and observed that as expected PGC-1α acetylation was higher in SirT1−/− MEF (Fig. 7A). However, when we compared the effect of PGC-1α over-expression in SirT1+/+ and SirT1−/− MEFs, the fold induction of antioxidant genes by PGC-1α was not significantly different in SirT1+/+ than in SirT1−/− MEFs, although both the basal and induced levels were lower in SirT1−/− MEFs in all genes tested (Fig. 7B and Supplementary Fig. S2). These results suggest that a part of SirT1 loss of activity on antioxidant genes cannot be compensated by artificially increasing the formation of a FoxO3a/PGC-1α complex and could involve H4K16 deacetylation of the regulated promoters upon SirT1 recruitment.
FIG. 7.
PGC-1α activity is reduced in the absence of SirT1. SIRT1+/+ and SIRT1−/− MEFs were infected o/n with Ad-PGC1α or a control adenovirus (Ad-Shuttle), 12 h postinfection cells were deprived of serum o/n and harvested. (A) Cell extracts were immunoprecipitated with a specific anti-PGC1α antibody (PGC-1α) or IgG as control (IgG) and PGC-1α acetylation was analyzed by western blot. (B) qRT-PCR and Western Blot analysis of oxidative stress protection proteins. Control samples were assigned the value of 1. Data are from ≥3 independent experiments. Data are means+SD (*) p≤0.05 versus control. (C) Schematic diagram of the proposed model. In stress conditions, if SirT1 is not active, both PGC-1α and FoxO3a are acetylated and targeted for degradation. In the presence of SirT1, deacetylation of PGC-1α and FoxO3a facilitates the formation of an active transcriptional complex that induces antioxidant genes and facilitates survival in stress conditions.
Discussion
The identification of the key transcriptional factors involved in the regulation of oxidative stress defense mechanisms is of great importance in the context of the vascular endothelium, where high ROS levels play a determinant role in endothelial dysfunction and therefore, in the appearance and progression of atherosclerosis as well as in the control of the angiogenesis process. In this study, we show that SirT1 positively regulates the expression of a group of oxidative stress protection genes in vascular endothelial cells and provide evidence that at least for some of these genes this effect is likely to be mediated by its regulation of the PGC-1α/FoxO3a transcriptional complex (Fig. 7C).
SirT1 had previously been involved in the regulation of two ROS detoxification enzymes, MnSOD, and catalase (1, 15, 46). Here we show that SirT1 is a regulator of a set of antioxidant genes in primary endothelial cells and in mouse embryonic fibroblasts (MEFs), suggesting that it is a general non cell-type specific activity. Furthermore, it shows that SirT1 prevents the accumulation of ROS produced as a result of the mitochondrial activity, and highlights the tight coupling between the systems that control metabolism and ROS homeostasis.
It has been previously reported that SirT1 is a negative regulator of UCP-2 expression (5) in pancreatic cells. This discrepancy might be related to the specific role played by UCP-2 in the pancreas and to the observed UCP-2 down-regulation observed when SirT1 was over-expressed in BAEC.
In muscle cells, SirT1 is recruited to the PGC-1α promoter and positively regulates its expression (2). Here we show that SirT1 can induce the expression not only of PGC-1α but also of FoxO3a in endothelial cells. Notably, although FoxOs post-transcriptional regulation has been extensively studied, very little is known about their transcriptional regulation. Our results suggests that SirT1 activity on a group of antioxidant genes can at least in part be dependent on the induction of these two key regulators of antioxidant gene expression.
We have investigated the mechanisms that mediate SirT1 activity and we have found that SirT1 associates with the regulatory regions of antioxidant genes. SirT1 binding was associated with reduced H4K16Ac, a hallmark of SirT1 activity. It is generally considered that reduced acetylation is associated with gene silencing (29). However, an important number of studies carried out mainly in yeast have shown that histone deacetylation and, in particular, H4K16 deacetylation can play a positive role in gene expression regulation (41). The mechanisms of action described so far, involve the deacetylation of histones within the coding regions of active genes to ensure proper transcription initiation, by repressing aberrant initiation within the coding region (19), and promoter specific effects, still poorly characterized (42). Our results show that SirT1 deacetylation of H4K16 is accompanied by a general reduction in RNAP II levels in the promoter proximal regions but this does not result in decreased gene expression. Importantly, we found that SirT1 recruitment was associated with increased levels of elongating RNAP II suggesting that SirT1 activity might be contributing to repressing aberrant initiation and or increased recruitment of elongation factors.
Although the mechanisms involved in the transcriptional activation by histone deacetylases are still unclear, there is some evidence that the lysine specificity of the deacetylase may be a critical determinant. In particular, H4K16 appears to be an important target for deacetylases playing an activating role (53). It appears that unacetylated H4K16 in the context of acetylation at other sites is important for transcription, but unacetylated H4K16 in the context of other unacetylated residues is important for heterochromatin formation. So far, the only characterized example in mammals in which H4K16Ac is linked to gene silencing is rDNA regulation (54). A recent study has shed some light on the mechanism through which acetylated H4K16Ac plays a dual role in gene silencing: it recruits Sir2-4 and repels Sir3 (32). Therefore, acetylation would serve to recruit SirT1 to the target gene and this recruitment may result in enhanced or decreased transcription depending on whether or not additional silencing/activating factors are subsequently recruited. However, experimental evidence for this type of regulation in mammals is missing.
Our results suggest that SirT1 is likely to act at least, in part, by directly binding to promoter of target genes, probably recruited through its interaction with transcription factors. Although SirT1 acts predominantly as a corepressor, there is evidence that SirT1 can also serve as a coactivator. For example, SirT1 recycles Tat to its unacetylated form and acts as a transcriptional coactivator during Tat transactivation (33).
Our previous work had shown that a FoxO3a/PGC-1α complex regulates several antioxidant genes (30). We have now found that PGC-1α and FoxO3a knock-down drastically reduced SirT1 induction of the antioxidant genes studied; an indication that both PGC-1α and FoxO3a are necessary partners of SirT1. The mechanism involved could be related at least, in part, to the observed induction of both FoxO3a and PGC-1α, but could also depend on SirT1 deacetylation of these factors. We found that in endothelial cells SirT1 over-expression reduces both FoxO3a and PGC-1α acetylation in response to oxidative stress. It has been previously noted that FoxO4 is acetylated by p300 and deacetylated by SirT1 in response to stress (21), FoxO3a deacetylation by SirT1 in response to oxidative stress has also been described (8). It has also been shown that PGC-1α is acetylated and targeted for degradation in oxidative stress conditions, and SirT1 was proposed to prevent this effect (3).
Our results also suggest that while acetylation of PGC-1α and FoxO3a in response to stress promotes their ubiquitination and degradation, deacetylation by SirT1 promotes protein stabilization. Nicotinamide experiments showed a nicotinamide-induced increase in the acetylated high molecular weight material present in both PGC-1α and FoxO3a IPs that likely corresponds to polyubiquitinated forms of these proteins (31, 34). Furthermore, oxidative stress increased the acetylation and decreased the levels of both PGC-1α and FoxO3a, while SirT1 over-expression reversed this effect. This result suggests that SirT1 can regulate PGC-1α and FoxO3a cellular levels not only at the transcriptional level as previously shown, but also by modulating its protein stability/degradation.
Our results on FoxO3a seem to contrast with previous observations on FoxO1, since it was originally proposed that FoxO1 acetylation increased its protein levels by reducing its degradation by the proteasome (20). However, recent observations showed that increased acetylation of FoxO1 leads to increased phosphorylation (37), a well characterized trigger for ubiquitination and proteasomal degradation (9). Importantly, we also showed that SirT1 protects FoxO3a from Akt phosphorylation both in basal an oxidative stress conditions.
To further investigate how SirT1 deacetylation of both FoxO3a and PGC-1α could play a role in SirT1 induction of antioxidant genes, we tested the effect of SirT1 on the formation of the FoxO3a/PGC-1α complex and found that SirT1 increased the formation of the PGC-1α/FoxO3a complex. We envisioned that SirT1 could work through two alternative mechanisms. SirT1 could deacetylate FoxO3a and PGC-1α independently and the deacetylated proteins could have an enhanced affinity for each other. Alternatively, SirT1 could be part of a ternary complex recruited to the target promoters. Since our SirT1 ChIP data supported this second model, we challenged this hypothesis testing whether FoxO3a and PGC-1α could modulate each other's level of acetylation and found that PGC-1α decreases FoxO3a acetylation, while FoxO3a reduces PGC-1α acetylation, supporting the notion that SirT1 is part of a ternary complex together with PGC-1α and FoxO3a.
The model that emerges is that oxidative stress would induce the acetylation of both PGC-1α and FoxO3a, and this acetylation would result in the accelerated ubiquitination and degradation of both factors, unless SirT1 is active. SirT1 would interact with FoxO3a and PGC-1α, deacetylate both and form with them a ternary complex that would be recruited to antioxidant genes, enhancing the cell survival in stress conditions. Therefore, the key determinant in the ROS response would be to what extent SirT1 is activated (16).
Nrf2 is another important transcriptional regulator of antioxidant gene expression. Importantly, it has been recently shown that Nrf2 pays also a role in metabolic control (52). These observations make it a putative relevant partner in the regulatory circuits controlled by SirT1, PGC-1α, and FoxO factors. However, this possibility remains to be investigated.
Taken together, the findings of the present study uncover a novel role for SirT1 as a critical regulator of endothelial oxidative stress protection systems and ROS homeostasis. Our results also highlight a potential therapeutic role of SirT1 regulation in vascular homeostasis. Therefore, the current availability of highly specific chemical SirT1 activators is of great value and seems now to make it a real possibility.
Material and Methods
Cell culture
BAEC were isolated and cultured as previously described (23, 26). BAEC were used from passages 3–6. On day 13.5 of gestation, embryos from C57BL6 SirT1+/− x C57BL6 SirT1+/− crosses were taken to isolate MEF that were prepared and cultured as described (40). MEF were used from passages 1–4. The SirT1+/− mice used were part of a colony established within the CNIC animal facility. The animal experimental protocols were approved by the Institutional Animal Care and Use Committee of the CNIC All protocols used conform to the Declaration of Helsinki and to the Guide for the Care and Use of Laboratory Animals published by the US National Institute of Health (NIH publication No. 85-23).
Adenoviral vectors and infections
Recombinant adenoviruses Ad-PGC-1α, Ad-SirT1, Ad-FoxO3a, and Ad-shFoxO3a have been previously described (30, 38, 43, 49). Silencing Ad-shPGC1α and Ad-shSirt1 were constructed using the pSilencer™ Adeno 1.0 Cytomegalovirus Promoter System from Ambion (Carlsbad, CA). The oligonucleotides used are listed in the Supp. Inf. Cells were infected with the adenoviral vectors at a multiplicity of infection of 10–25 for 12–16 h. 24 or 48 h postinfection cells were washed 3× with PBS and harvested.
Western blot
Whole-cell lysates were obtained as described (28). 20–25 μg of whole-cell protein extracts were resolved on 8%–15% SDS-PAGE gels, transferred onto PVDF Hybond-P membranes (Amersham Biosciences, Fairfield, CT) and incubated with specific antibodies against PGC-1α (Cayman Chemical), FoxO3a (Cell Signalling, Danvers, MA and Santa Cruz, Santa Cruz, CA), P-FoxO3a (Thr32) (Millipore, Mendota Heights, MN), SirT1 (Santa Cruz), Prx5 (Santa Cruz), Trx2, TR2, and Prx3 (LabFrontier, Daehyun-dong Seodaemun-gu Seoul, Korea), Catalase (Chemicon International, Temecula, CA), MnSOD (Stressgen, Victoria, British Columbia, Canada), UCP-2 (Santa Cruz and Biolegend, San Diego, CA), Acetylated-Lysine (Cell Signaling, Danvers, MA), β-actin (Sigma, Dublin, OH) and HA (Hybridoma 12CA5).). β-actin was used as a loading control.
Co-immunoprecipitations
Protein-protein interactions were analyzed by co-immunoprecipitation experiments. Whole-cell extracts were prepared and subjected to immunoprecipitation with specific antibodies against PGC-1α (Santa Cruz H-300, Santa Cruz, CA), HA-tagged FoxO3a (mouse monoclonal 12CA5) and SirT1 (Santa Cruz) or a control normal rabbit or mouse IgG as previously described (28). The immunoprecipitates were assessed for the presence of PGC-1α (Cayman Chemical, Ann Arbor, MI), FoxO3a (Santa Cruz), and SirT1 (Santa Cruz) and for the levels of Lys acetylation (Cell Signaling) by Western Blot with specific antibodies.
Chromatin immunoprecipitation
ChIP experiments were carried out essentially as previously described (49) with some modifications (Supp. Info.).
Determination of relative mRNA levels
RNA was isolated using TRIzol® Reagent (Invitrogen, Carlsbad, CA). 1 μg of total RNA was reverse transcribed by extension of random primers with M-MLV (Promega, Madison, WI). Relative expression levels were determined by real-time PCR (qRT-PCR) in a 7900 Sequence Detection System (Applied Biosystems, Carlsbad, CA). Primers were as previously described (7). The amount of PCR product was normalized to 18S rRNA levels.
ROS analysis
Cells were incubated with 2 μM 5-chloromethyl-2′7′-dichlorodihydrofluorescein diacetate acetyl ester (CM-H2DCFDA; Molecular Probes, Carlsbad, CA) for 30 min at 37°C. Cells were then trypsinized and resuspended in PBS. Fluorescence intensity was measured by flow cytometry.
Mitochondrial superoxide was measured by labeling cells with MitoSOX Red (Molecular Probes, Carlsbad, CA, USA, essentially as described (6). Briefly, BAEC were grown in coverslips, incubated with 3 μM MitoSOX Red for 10 min, fixed with paraformaldehyde and analyzed by fluorescence microscopy (Leica TCS SP5, Buffalo Grove, IL).
Immunofluorescence
BAEC were grown on coverslips, fixed with 3.7% formaldehyde, permeabilized with 0.1% Triton, and then incubated consecutively with primary antibody (α-FoxO3a) and secondary antibody (αIgG rabbit ALEXA-488 conjugate). Cells were counterstained with TO-PRO-3, mounted and examined by confocal microscopy (Zeiss LSM 700, Obercochen, Germany), as previously described (6).
Statistical analysis
The Kolmogorov-Smirnov test was used to test for normality of the sample's values distribution. Sample values, are shown as mean±standard deviation (SD). Comparison of continuous variables from two or more independent groups was evaluated by analysis of variance with Bonferroni correction for multiple comparisons. Statistical analyses were performed using SPSS for Windows, release 17.0 (SPSS, Inc., Chicago, IL). Levene's test for equality of variances and t-test for equality of means were used. All p-values refer to two-tailed tests of significance; p, 0.05 was considered significant.
Supplementary Material
Abbreviations Used
- Ac
acetylated
- Ad
recombinant adenovirus
- ApoE−/−
apolipoprotein E
- BAEC
bovine aortic endothelial cells
- ChIP
chromatin immunoprecipitation
- CM-H2DCFDA
chloromethyl-2′,7′-dichlorofluorescein diacetate
- co-IP
co-immunoprecipitation
- DMNQ
2,3-dimethoxy-1-naphthoquinone
- FBS
fetal bovine serum
- Gpx1
glutathione peroxidase 1
- Gpx4
glutathione peroxidase 4
- H4
histone 4
- IF
immunofluorescence
- K16
lysine 16
- MEFs
mouse embryonic fibroblasts
- MnSOD
Mn superoxide dismutase
- NF-κB
nuclear factor-κB
- NO
nitric oxide
- NOX
NADPH oxidases
- PGC-1α
peroxisome proliferator activated receptor γ-coactivator 1α
- Prx3
peroxiredoxin 3
- Prx5
peroxiredoxin 5
- RNAP II
RNA polymerase II
- ROS
reactive oxygen species
- sh
small hairpin
- TR2
thioredoxin reductase 2
- Trx2
thioredoxin 2
- UCP-2
uncoupling protein 2
Acknowledgments
This work was supported by the Spanish “Ministerio de Economía y Competitividad” (grants SAF2009-07599 to M.M, SAF 2009-07520 to S.L. and CSD 2007-00020 to M.M. and S.L.) and the “Comunidad Autónoma de Madrid” (grant S2010/BMD-2361 to M.M.). We thank Dr. M. Serrano (CNIO) for SirT1+/- mice and L. Durán for technical support.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Alcendor RR. Gao S. Zhai P. Zablocki D. Holle E. Yu X. Tian B. Wagner T. Vatner SF. Sadoshima J. Sirt1 regulates aging and resistance to oxidative stress in the heart. Circ Res. 2007;100:1512–1521. doi: 10.1161/01.RES.0000267723.65696.4a. [DOI] [PubMed] [Google Scholar]
- 2.Amat R. Planavila A. Chen SL. Iglesias R. Giralt M. Villarroya F. SIRT1 controls the transcription of the peroxisome proliferator-activated receptor-gamma Co-activator-1alpha (PGC-1alpha) gene in skeletal muscle through the PGC-1alpha autoregulatory loop and interaction with MyoD. J Biol Chem. 2009;284:21872–21880. doi: 10.1074/jbc.M109.022749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anderson RM. Barger JL. Edwards MG. Braun KH. O'Connor CE. Prolla TA. Weindruch R. Dynamic regulation of PGC-1alpha localization and turnover implicates mitochondrial adaptation in calorie restriction and the stress response. Aging Cell. 2008;7:101–111. doi: 10.1111/j.1474-9726.2007.00357.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arden GB. Sivaprasad S. Hypoxia and oxidative stress in the causation of diabetic retinopathy. Curr Diabetes Rev. 2011;7:291–304. doi: 10.2174/157339911797415620. [DOI] [PubMed] [Google Scholar]
- 5.Bordone L. Motta MC. Picard F. Robinson A. Jhala US. Apfeld J. McDonagh T. Lemieux M. McBurney M. Szilvasi A. Easlon EJ. Lin SJ. Guarente L. Sirt1 regulates insulin secretion by repressing UCP2 in pancreatic beta cells. PLoS Biol. 2006;4:e31. doi: 10.1371/journal.pbio.0040031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Borniquel S. Garcia-Quintans N. Valle I. Olmos Y. Wild B. Martinez-Granero F. Soria E. Lamas S. Monsalve M. Inactivation of Foxo3a and subsequent downregulation of PGC-1 alpha mediate nitric oxide-induced endothelial cell migration. Mol Cell Biol. 2010;30:4035–4044. doi: 10.1128/MCB.00175-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Borniquel S. Valle I. Cadenas S. Lamas S. Monsalve M. Nitric oxide regulates mitochondrial oxidative stress protection via the transcriptional coactivator PGC-1alpha. FASEB J. 2006;20:1889–1891. doi: 10.1096/fj.05-5189fje. [DOI] [PubMed] [Google Scholar]
- 8.Brunet A. Sweeney LB. Sturgill JF. Chua KF. Greer PL. Lin Y. Tran H. Ross SE. Mostoslavsky R. Cohen HY. Hu LS. Cheng HL. Jedrychowski MP. Gygi SP. Sinclair DA. Alt FW. Greenberg ME. Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science. 2004;303:2011–2015. doi: 10.1126/science.1094637. [DOI] [PubMed] [Google Scholar]
- 9.Calnan DR. Brunet A. The FoxO code. Oncogene. 2008;27:2276–2288. doi: 10.1038/onc.2008.21. [DOI] [PubMed] [Google Scholar]
- 10.Canto C. Auwerx J. PGC-1alpha, SIRT1 and AMPK, an energy sensing network that controls energy expenditure. Curr Opin Lipidol. 2009;20:98–105. doi: 10.1097/MOL.0b013e328328d0a4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cheng HL. Mostoslavsky R. Saito S. Manis JP. Gu Y. Patel P. Bronson R. Appella E. Alt FW. Chua KF. Developmental defects and p53 hyperacetylation in Sir2 homolog (SIRT1)-deficient mice. Proc Natl Acad Sci U S A. 2003;100:10794–10799. doi: 10.1073/pnas.1934713100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dikalov S. Cross talk between mitochondria and NADPH oxidases. Free Radic Biol Med. 2011;51:1289–1301. doi: 10.1016/j.freeradbiomed.2011.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gerhart-Hines Z. Rodgers JT. Bare O. Lerin C. Kim SH. Mostoslavsky R. Alt FW. Wu Z. Puigserver P. Metabolic control of muscle mitochondrial function and fatty acid oxidation through SIRT1/PGC-1alpha. EMBO J. 2007;26:1913–1923. doi: 10.1038/sj.emboj.7601633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gori T. Munzel T. Oxidative stress and endothelial dysfunction: therapeutic implications. Ann Med. 2011;43:259–272. doi: 10.3109/07853890.2010.543920. [DOI] [PubMed] [Google Scholar]
- 15.Hasegawa K. Wakino S. Yoshioka K. Tatematsu S. Hara Y. Minakuchi H. Washida N. Tokuyama H. Hayashi K. Itoh H. Sirt1 protects against oxidative stress-induced renal tubular cell apoptosis by the bidirectional regulation of catalase expression. Biochem Biophys Res Commun. 2008;372:51–56. doi: 10.1016/j.bbrc.2008.04.176. [DOI] [PubMed] [Google Scholar]
- 16.Herranz D. Serrano M. SIRT1: recent lessons from mouse models. Nat Rev Cancer. 2010;10:819–823. doi: 10.1038/nrc2962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang SS. Zheng RL. Biphasic regulation of angiogenesis by reactive oxygen species. Pharmazie. 2006;61:223–229. [PubMed] [Google Scholar]
- 18.Kawano N. Emoto M. Mori K. Yamazaki Y. Urata H. Tsuchikura S. Motoyama K. Morioka T. Fukumoto S. Shoji T. Koyama H. Okuno Y. Nishizawa Y. Inaba M. Association of endothelial and vascular smooth muscle dysfunction with cardiovascular risk factors, vascular complications, and subclinical carotid atherosclerosis in type 2 diabetic patients. J Atheroscler Thromb. 2012;19:276–284. doi: 10.5551/jat.10629. [DOI] [PubMed] [Google Scholar]
- 19.Kim T. Buratowski S. Dimethylation of H3K4 by Set1 recruits the Set3 histone deacetylase complex to 5′ transcribed regions. Cell. 2009;137:259–272. doi: 10.1016/j.cell.2009.02.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kitamura YI. Kitamura T. Kruse JP. Raum JC. Stein R. Gu W. Accili D. FoxO1 protects against pancreatic beta cell failure through NeuroD and MafA induction. Cell Metab. 2005;2:153–163. doi: 10.1016/j.cmet.2005.08.004. [DOI] [PubMed] [Google Scholar]
- 21.Kobayashi Y. Furukawa-Hibi Y. Chen C. Horio Y. Isobe K. Ikeda K. Motoyama N. SIRT1 is critical regulator of FOXO-mediated transcription in response to oxidative stress. Int J Mol Med. 2005;16:237–243. [PubMed] [Google Scholar]
- 22.Kowaltowski AJ. de Souza-Pinto NC. Castilho RF. Vercesi AE. Mitochondria and reactive oxygen species. Free Radic Biol Med. 2009;47:333–343. doi: 10.1016/j.freeradbiomed.2009.05.004. [DOI] [PubMed] [Google Scholar]
- 23.Lamas S. Michel T. Brenner BM. Marsden PA. Nitric oxide synthesis in endothelial cells: evidence for a pathway inducible by TNF-alpha. Am J Physiol. 1991;261:C634–C641. doi: 10.1152/ajpcell.1991.261.4.C634. [DOI] [PubMed] [Google Scholar]
- 24.Lassegue B. Griendling KK. NADPH oxidases: functions and pathologies in the vasculature. Arterioscler Thromb Vasc Biol. 2010;30:653–661. doi: 10.1161/ATVBAHA.108.181610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lin J. Handschin C. Spiegelman BM. Metabolic control through the PGC-1 family of transcription coactivators. Cell Metab. 2005;1:361–370. doi: 10.1016/j.cmet.2005.05.004. [DOI] [PubMed] [Google Scholar]
- 26.Lopez-Ongil S. Saura M. Rodriguez-Puyol D. Rodriguez-Puyol M. Lamas S. Regulation of endothelial NO synthase expression by cyclosporin A in bovine aortic endothelial cells. Am J Physiol. 1996;271:H1072–H1078. doi: 10.1152/ajpheart.1996.271.3.H1072. [DOI] [PubMed] [Google Scholar]
- 27.Monsalve M. Borniquel S. Valle I. Lamas S. Mitochondrial dysfunction in human pathologies. Front Biosci. 2007;12:1131–1153. doi: 10.2741/2132. [DOI] [PubMed] [Google Scholar]
- 28.Monsalve M. Wu Z. Adelmant G. Puigserver P. Fan M. Spiegelman BM. Direct coupling of transcription and mRNA processing through the thermogenic coactivator PGC-1. Mol Cell. 2000;6:307–316. doi: 10.1016/s1097-2765(00)00031-9. [DOI] [PubMed] [Google Scholar]
- 29.Oberdoerffer P. Michan S. McVay M. Mostoslavsky R. Vann J. Park SK. Hartlerode A. Stegmuller J. Hafner A. Loerch P. Wright SM. Mills KD. Bonni A. Yankner BA. Scully R. Prolla TA. Alt FW. Sinclair DA. SIRT1 redistribution on chromatin promotes genomic stability but alters gene expression during aging. Cell. 2008;135:907–918. doi: 10.1016/j.cell.2008.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Olmos Y. Valle I. Borniquel S. Tierrez A. Soria E. Lamas S. Monsalve M. Mutual dependence of Foxo3a and PGC-1alpha in the induction of oxidative stress genes. J Biol Chem. 2009;284:14476–14484. doi: 10.1074/jbc.M807397200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Olson BL. Hock MB. Ekholm-Reed S. Wohlschlegel JA. Dev KK. Kralli A. Reed SI. SCFCdc4 acts antagonistically to the PGC-1alpha transcriptional coactivator by targeting it for ubiquitin-mediated proteolysis. Genes Dev. 2008;22:252–264. doi: 10.1101/gad.1624208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Oppikofer M. Kueng S. Martino F. Soeroes S. Hancock SM. Chin JW. Fischle W. Gasser SM. A dual role of H4K16 acetylation in the establishment of yeast silent chromatin. EMBO J. 2011;30:2610–2621. doi: 10.1038/emboj.2011.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pagans S. Pedal A. North BJ. Kaehlcke K. Marshall BL. Dorr A. Hetzer-Egger C. Henklein P. Frye R. McBurney MW. Hruby H. Jung M. Verdin E. Ott M. SIRT1 regulates HIV transcription via Tat deacetylation. PLoS Biol. 2005;3:e41. doi: 10.1371/journal.pbio.0030041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Plas DR. Thompson CB. Akt activation promotes degradation of tuberin and FOXO3a via the proteasome. J Biol Chem. 2003;278:12361–12366. doi: 10.1074/jbc.M213069200. [DOI] [PubMed] [Google Scholar]
- 35.Potente M. Ghaeni L. Baldessari D. Mostoslavsky R. Rossig L. Dequiedt F. Haendeler J. Mione M. Dejana E. Alt FW. Zeiher AM. Dimmeler S. SIRT1 controls endothelial angiogenic functions during vascular growth. Genes Dev. 2007;21:2644–2658. doi: 10.1101/gad.435107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Potente M. Urbich C. Sasaki K. Hofmann WK. Heeschen C. Aicher A. Kollipara R. DePinho RA. Zeiher AM. Dimmeler S. Involvement of Foxo transcription factors in angiogenesis and postnatal neovascularization. J Clin Invest. 2005;115:2382–2392. doi: 10.1172/JCI23126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Qiang L. Banks AS. Accili D. Uncoupling of acetylation from phosphorylation regulates FoxO1 function independent of its subcellular localization. J Biol Chem. 2010;285:27396–27401. doi: 10.1074/jbc.M110.140228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rodgers JT. Lerin C. Haas W. Gygi SP. Spiegelman BM. Puigserver P. Nutrient control of glucose homeostasis through a complex of PGC-1alpha and SIRT1. Nature. 2005;434:113–118. doi: 10.1038/nature03354. [DOI] [PubMed] [Google Scholar]
- 39.Schulz E. Jansen T. Wenzel P. Daiber A. Munzel T. Nitric oxide, tetrahydrobiopterin, oxidative stress, and endothelial dysfunction in hypertension. Antioxid Redox Signal. 2008;10:1115–1126. doi: 10.1089/ars.2007.1989. [DOI] [PubMed] [Google Scholar]
- 40.Serrano M. Lin AW. McCurrach ME. Beach D. Lowe SW. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell. 1997;88:593–602. doi: 10.1016/s0092-8674(00)81902-9. [DOI] [PubMed] [Google Scholar]
- 41.Shahbazian MD. Grunstein M. Functions of site-specific histone acetylation and deacetylation. Annu Rev Biochem. 2007;76:75–100. doi: 10.1146/annurev.biochem.76.052705.162114. [DOI] [PubMed] [Google Scholar]
- 42.Sharma VM. Tomar RS. Dempsey AE. Reese JC. Histone deacetylases RPD3 and HOS2 regulate the transcriptional activation of DNA damage-inducible genes. Mol Cell Biol. 2007;27:3199–3210. doi: 10.1128/MCB.02311-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Skurk C. Maatz H. Kim HS. Yang J. Abid MR. Aird WC. Walsh K. The Akt-regulated forkhead transcription factor FOXO3a controls endothelial cell viability through modulation of the caspase-8 inhibitor FLIP. J Biol Chem. 2004;279:1513–1525. doi: 10.1074/jbc.M304736200. [DOI] [PubMed] [Google Scholar]
- 44.Stein S. Matter CM. Protective roles of SIRT1 in atherosclerosis. Cell Cycle. 2011;10:640–647. doi: 10.4161/cc.10.4.14863. [DOI] [PubMed] [Google Scholar]
- 45.Stein S. Schafer N. Breitenstein A. Besler C. Winnik S. Lohmann C. Heinrich K. Brokopp CE. Handschin C. Landmesser U. Tanner FC. Luscher TF. Matter CM. SIRT1 reduces endothelial activation without affecting vascular function in ApoE−/− mice. Aging (Albany NY) 2010;2:353–360. doi: 10.18632/aging.100162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tanno M. Kuno A. Yano T. Miura T. Hisahara S. Ishikawa S. Shimamoto K. Horio Y. Induction of manganese superoxide dismutase by nuclear translocation and activation of SIRT1 promotes cell survival in chronic heart failure. J Biol Chem. 2010;285:8375–8382. doi: 10.1074/jbc.M109.090266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tertil M. Jozkowicz A. Dulak J. Oxidative stress in tumor angiogenesis- therapeutic targets. Curr Pharm Des. 2010;16:3877–3894. doi: 10.2174/138161210794454969. [DOI] [PubMed] [Google Scholar]
- 48.Ushio-Fukai M. Nakamura Y. Reactive oxygen species and angiogenesis: NADPH oxidase as target for cancer therapy. Cancer Lett. 2008;266:37–52. doi: 10.1016/j.canlet.2008.02.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Valle I. Alvarez-Barrientos A. Arza E. Lamas S. Monsalve M. PGC-1alpha regulates the mitochondrial antioxidant defense system in vascular endothelial cells. Cardiovasc Res. 2005;66:562–573. doi: 10.1016/j.cardiores.2005.01.026. [DOI] [PubMed] [Google Scholar]
- 50.Vaquero A. Sternglanz R. Reinberg D. NAD+-dependent deacetylation of H4 lysine 16 by class III HDACs. Oncogene. 2007;26:5505–5520. doi: 10.1038/sj.onc.1210617. [DOI] [PubMed] [Google Scholar]
- 51.Victor VM. Apostolova N. Herance R. Hernandez-Mijares A. Rocha M. Oxidative stress and mitochondrial dysfunction in atherosclerosis: mitochondria-targeted antioxidants as potential therapy. Curr Med Chem. 2009;16:4654–4667. doi: 10.2174/092986709789878265. [DOI] [PubMed] [Google Scholar]
- 52.Vomhof-Dekrey EE. Picklo MJ., Sr. The Nrf2-antioxidant response element pathway: a target for regulating energy metabolism. J Nutr Biochem. 2012;23:1201–1206. doi: 10.1016/j.jnutbio.2012.03.005. [DOI] [PubMed] [Google Scholar]
- 53.Wang A. Kurdistani SK. Grunstein M. Requirement of Hos2 histone deacetylase for gene activity in yeast. Science. 2002;298:1412–1414. doi: 10.1126/science.1077790. [DOI] [PubMed] [Google Scholar]
- 54.Zhou Y. Grummt I. The PHD finger/bromodomain of NoRC interacts with acetylated histone H4K16 and is sufficient for rDNA silencing. Curr Biol. 2005;15:1434–1438. doi: 10.1016/j.cub.2005.06.057. [DOI] [PubMed] [Google Scholar]
- 55.Zielonka J. Kalyanaraman B. Hydroethidine- and MitoSOX-derived red fluorescence is not a reliable indicator of intracellular superoxide formation: another inconvenient truth. Free Radic Biol Med. 2010;48:983–1001. doi: 10.1016/j.freeradbiomed.2010.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.