Abstract
Aims: Oxidative stress is thought to be involved in Friedreich's ataxia (FRDA), yet it has not been demonstrated in the target neurons that are first to degenerate. Using the YG8R mouse model of FRDA, microarray and neuritic growth experiments were carried out in the dorsal root ganglion (DRG), the primary site of neurodegeneration in this disease. Results: YG8R hemizygous mice exhibited defects in movement, and DRG neurites had growth defects. Microarray of DRG tissue identified decreased transcripts encoding the antioxidants, including peroxiredoxins, glutaredoxins, and glutathione S-transferase, and these were confirmed by immunoblots and quantitative real-time PCR. Because the decreased gene transcripts are the known targets of the antioxidant transcription factor nuclear factor-E2-related factor-2 (Nrf2), Nrf2 expression was measured; it was significantly decreased at the transcript and protein level in both the DRG and the cerebella of the YG8R hemizygous mouse; further, frataxin expression was significantly correlated with Nrf2 expression. Functionally, in YG8R hemizygous DRG, the total glutathione levels were reduced and explanted cells were more sensitive to the thioredoxin reductase (TxnRD) inhibitor auranofin, a thiol oxidant. In cell models of FRDA, including Schwann and the DRG, frataxin deficiency caused a decreased expression of the Nrf2 protein level in the nucleus, but not a defect in its translocation from the cytosol. Further, frataxin-deficient cells had decreased enzyme activity and expression of TxnRD, which is regulated by Nrf2, and were sensitive the TxnRD inhibitor auranofin. Innovation and Conclusion: These results support a mechanistic hypothesis in which frataxin deficiency decreases Nrf2 expression in vivo, causing the sensitivity to oxidative stress in target tissues the DRG and the cerebella, which contributes to the process of neurodegeneration. Antioxid. Redox Signal. 19, 1481–1493.
Introduction
Friedreich's ataxia (FRDA) is the most common inherited recessive ataxia, affecting ∼1 in 40,000 (16). The most common cause of the disease is the inheritance of two copies of expanded (GAA)n repeats in the first intron of the frataxin gene, resulting in chromatin condensation and reduced expression of frataxin (25, 28, 50). FRDA pathology includes neurodegeneration of the large sensory neurons of the dorsal root ganglion (DRG), and neurodegeneration of the dentate nucleus of the cerebellum, as well as hypertrophic cardiomyopathy (38, 39).
There is substantial support that FRDA is a disease of oxidative stress. FRDA is a virtual phenocopy of ataxia with vitamin E deficiency (AVED), which results from deficiency of the alpha-tocopherol (vitamin E) transporter (9). Vitamin E is an antioxidant that prevents membrane lipid peroxidation (75) and is used to treat patients suffering from FRDA to slow the progression of disease (14). The cells of patients suffering from FRDA have increased sensitivity to oxidative stress, and there is evidence for alteration of the thiol pool (18, 36, 55, 73, 78). There is also evidence for increased markers of oxidative stress in patient tissues (10, 26, 56, 67).
Innovation.
The primary site of neurodegeneration in Friedreich's ataxia (FRDA) is the dorsal root ganglion (DRG), and no microarray study of this crucial target tissue has ever been carried out. In this study, we have performed the first microarray of DRG tissue in an animal model of FRDA, and show that deficiency is a deficiency of multiple thiol antioxidants, and Nrf2 deficiency, and glutathione deficiency in the target tissue. This has potential clinical significance, because drugs that induce Nrf2 are known, and could be protective in FRDA. It also suggests a potential mechanistic basis for the phenotypic overlap between FRDA and ataxia with vitamin E deficiency, since both frataxin (this study) and tocopherols (27) have been suggested to induce Nrf2.
Several model systems with frataxin deficiency exhibit oxidative stress. Bulteau et al. observed a loss of aconitase activity and protein oxidation in Δyfh1 cells (frataxin-depleted yeast cells) (12), and the Δyfh1 cells are sensitive to oxidants (7, 43) under aerobic conditions. Vázquez-Manrique et al. found that frataxin siRNA-knockdown Caenorhabditis elegans are sensitive to oxidative stress (77). Consistently, in Drosophila, overexpression of frataxin increases resistance to oxidative stress (64), and siRNA knockdown of frataxin increases susceptibility to iron toxicity and hydrogen peroxide (5, 6). Mouse conditional knockouts of frataxin in the pancreas and hepatocytes show increased reactive oxygen species (61, 74).
Additionally, the YG8- and YG22-rescue mice (YG8R and YG22R), which express mutant human frataxin, show evidence of increase in oxidative stress of multiple tissues (4). YG8R mice express the expanded mutant alleles of human frataxin, in an animal lacking endogenous mouse frataxin. The mouse was created by breeding mice containing a YAC insert of the human genomic FXN locus bearing a GAA-repeat expansion (3) with heterozygous frataxin knockout mice (21), and then selecting animals with no endogenous mouse frataxin, but with either one (hemizygous) or two (homozygous) copies of the human mutant frataxin locus. YG8R hemizygous mice exhibit mild motor impairment, increased oxidative stress, and demyelination and vacuolization within DRGs, thus providing a suitable model of the human disease (4).
In an effort to understand the pathophysiological mechanism of FRDA in the most relevant target neural tissue, we carried out microarray of the DRGs of the YG8R mouse, which suggested increased oxidative stress in these animals as a result of decreased antioxidant. Defects in the transcripts of multiple thiol-based antioxidant genes were observed, suggesting a frataxin-dependent defect on the antioxidant transcription factor Nrf2, or in the thioredoxin reductase (TxnRD), it regulates or both. We observed a defect in the Nrf2 transcript and protein level in the DRG and the cerebella of YG8R hemizygous mice, providing an explanation for decreased antioxidant gene expression and decreased glutathione level. In multiple cell models, including DRG and Schwann cell and fibroblast and HeLa, we observed a decrease in Nrf2 expression as a consequence of frataxin deficiency, as well as decrease in the thioredoxin reductase 1 (TxnRD1) protein level and enzymatic activity. We suggest that frataxin deficiency leads to Nrf2 deficiency decreasing Nrf2-regulated protective genes, resulting in neurodegeneration, and stimulation of Nrf2 may be a rational therapy in FRDA.
Results
Neurite extension is reduced in YG8R DRG cells
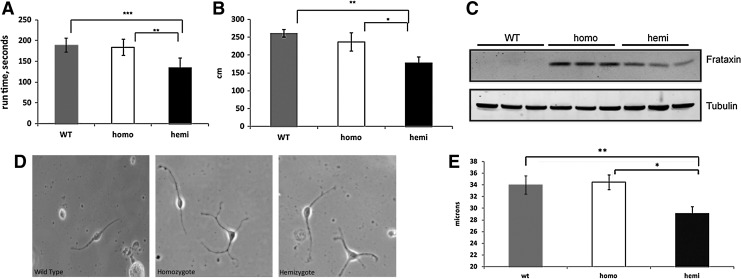
A colony of YG8R mice was established at UC Davis. For our testing, we used both hemizygous mice, containing one allele of the mutant frataxin transgene (and thus the least amount of frataxin) and homozygous mice, containing two alleles of the transgene (and thus higher levels of frataxin), and wild-type (WT) mice. Hemizygous mice in this colony exhibited reduced levels of frataxin expression, movement defects in accelerating rotarod and open-field testing, in agreement with previously published data (Fig. 1A, B), and were deficient in frataxin expression (Fig. 1D) (4).
FIG. 1.
Functional and neuritic defects in UC Davis YG8R mice. (A) Accelerating rotarod performance (4 mice/group); (B) horizontal activity in open-field performance test (4 mice/genotype); (C) frataxin expression in DRG tissue; each lane represents a different mouse. No expression is observed in WT samples, as the antibody is human-specific; (D, E) DRGs from YG8R mice exhibit decreased neurite extension, WT=34.02±1.53 versus homo=34.48±1.29 versus hemi=29.17±1.15 μm, n=3 mice per genotype, 48 neurites counted per mouse. Error bars=SEM. *p<0.05, **p<0.005, ***p<0.0005. Using a 1-tailed Student's t-test. DRG, dorsal root ganglion; WT, wild type.
We also performed neurite extension measurements on DRG cells, the primary site of neurodegeneration in FRDA. Bipolar cells from cultured single-cell suspensions of hemizygous mouse lumbar DRGs extended shorter neurites than from WT and homozygotes (WT=34.02±1.53 vs. homo=34.48±1.29 vs. hemi=29.17±1.15 μm, Fig. 1E), 3 mice per genotype, 48 neurites counted per mouse, demonstrating a growth defect in YG8R DRGs. Thus, the YG8 mice in our facility had behavioral and neuritic defects attributable to frataxin deficiency, and these defects were most severe in hemizygous mice, that is, those with the least frataxin.
YG8R hemizygous DRGs exhibit alterations in thiol-based antioxidant, myelination, and axonal transport transcripts
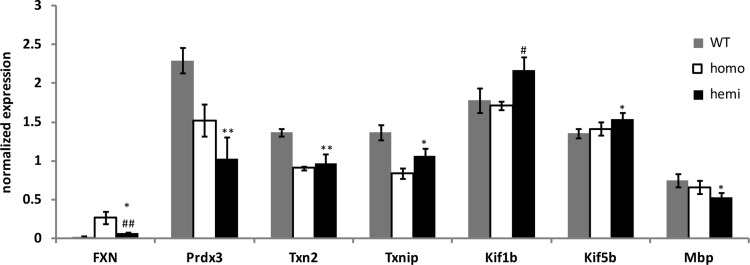
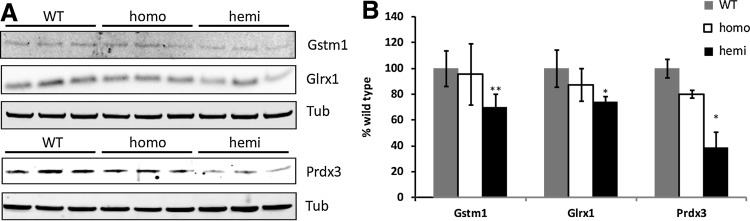
The DRGs from YG8R mice and C57Bl/6 WT controls were dissected and analyzed by microarray analysis. Chip fluorescence analysis using dChip pathway software revealed changes in three functional categories: (i) thiol antioxidants, (ii) myelination, and (iii) axonal transport transcripts (data not shown). Quantitative real time-PCR, using the primers listed in Supplementary Table S1, was used to verify the transcriptional changes in each of these three categories. We found significant decreases in the thiol antioxidant-related genes peroxiredoxin 3 (Prdx3), thioredoxin 2 (Txn2), and thioredoxin-interacting protein (Txnip), a significant decrease in the myelination gene Mbp, and significant increases in the axonal transport genes kinesin family member 1B (Kif1b) and kinesin family member 5B (Kif5b) (Fig. 2). We followed up on the other members of the thiol antioxidant pathway using immunoblot analysis and identified significant decreases in glutathione S-transferase mu 1 (Gstm1), glutaredoxin (Glrx), and Prdx3 (Fig. 3).
FIG. 2.
Quantitative real time-PCR verification of select microarray hits. One nanogram of total RNA was used for amplification, and the values were normalized to GAPDH or Lmnb1 expression. n=3 mice per genotype. Error bars=SEM. *p<0.05 versus WT; **p<0.01 versus WT; #p<0.05 versus homo; ##p<0.01 versus homo using a 1-tailed Student's t-test.
FIG. 3.
Hemizygous YG8R DRGs exhibit reduced levels of thiol antioxidant proteins. (A) Representative immunoblots of Gstm1, Glrx1, and Prx3; (B) densitometry of immunoblots shown in (A). Each lane contains 30 μg protein from a different mouse. Error bars=SEM. *p<0.05 versus WT; **p<0.01 versus WT; using a 1-tailed paired Student's t-test [the bottom blot of (A) is the same blot used in Fig. 1C]. Glrx1, glutaredoxin 1; Gstm1, glutathione S-transferase mu 1.
Nrf2 and Nrf2-controlled targets are decreased in YG8R hemizygous DRGs
Nrf2 is a transcription factor that regulates the expression of cytoprotective genes in response to oxidative stress (15). Nrf2-regulated targets include thiol antioxidant genes such as TxnRD, Gstm1, Txnip, and Glrx1 and glutathione synthesis via glutamate–cysteine ligase, catalytic subunit (Gclc), and glutamate–cysteine ligase, modifier subunit (Gclm) (2, 15, 42, 80). Thus, a frataxin-dependent defect in Nrf2 would explain the observed decreases in thiol antioxidants. Defects in the Nrf2 nuclear translocation (55), and defects in the Nrf2-regulated genes (32), have been reported in Friedreich's fibroblasts, but to this point, no such defects have been demonstrated in neural tissues that degenerate in humans or animal models.
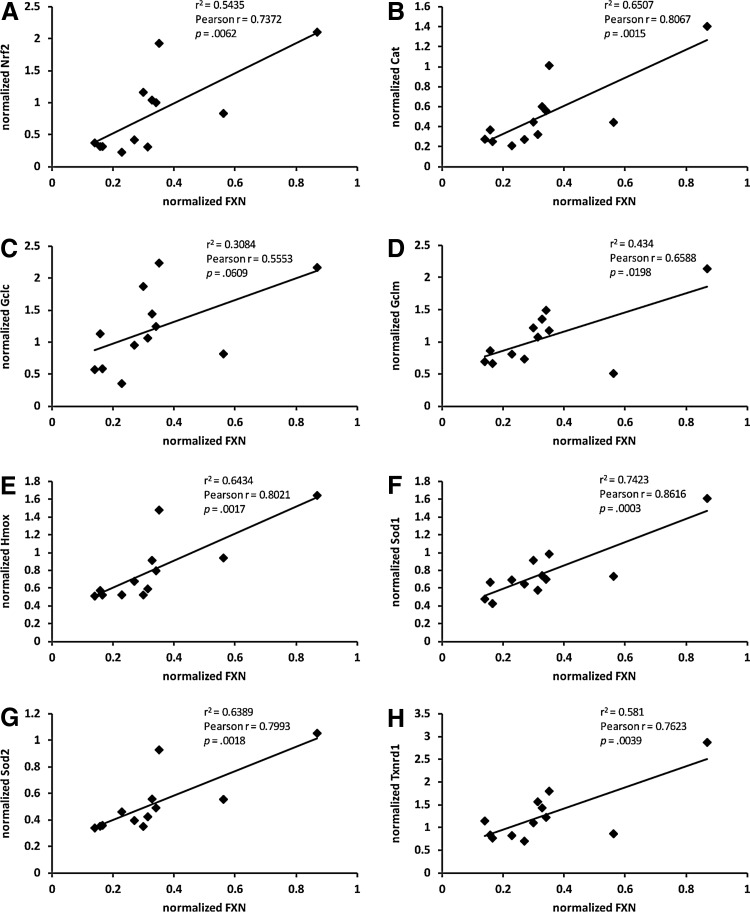
We examined the transcript and protein levels of Nrf2 and several Nrf2-regulated genes and found deficiencies in YG8R hemizygous DRGs. First, the Nrf2 transcript levels were significantly correlated with frataxin expression, and thus mice with decreased frataxin had decreased Nrf2 transcript, p<0.01 (Fig. 4A). Additionally, the transcript levels of the Nrf2-regulated genes heme oxygenase (Hmox), TxnRD1, Gclm, Cat, Cu/Zn superoxide dismutase (Sod1), and Mn superoxide dismutase (Sod2) were also significantly correlated with frataxin expression, p<0.05, while Gclc showed a trend, p=0.061 (Fig. 4B–H). Further, Nrf2 expression was significantly correlated with frataxin expression in the other major neural target tissue, the cerebellum, at the transcript and protein level (Supplementary Fig. S1C).
FIG. 4.
Transcripts encoding Nrf2 and downstream targets are decreased in YG8R DRGs and correlated with frataxin levels. (A–H) Correlation analysis of the frataxin transcript levels with Nrf2 (A), Cat (B) Gclc (C), Gclm (D), Hmox (E), Sod1 (F), Sod2 (G), and TxnRD1 (H). n=12 mice. Statistics were generated using GraphPad Prism software v4.02 for Windows (GraphPad Software). Cat, catalase; Gclc, glutamate–cysteine ligase, catalytic subunit; Gclm, glutamate–cysteine ligase, modifier subunit; Hmox, heme oxygenase; Nrf2, nuclear factor-E2-related factor-2; Sod1, Cu/Zn superoxide dismutase; Sod2, Mn superoxide dismutase; TxnRD1, thioredoxin reductase 1.
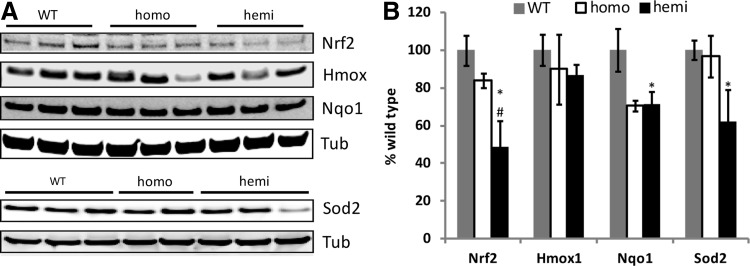
At the protein expression level, immunoblot analysis demonstrated decreased levels of Nrf2, Hmox, NAD(P)H dehydrogenase, quinone 1 (Nqo1), and Sod2 in the hemizygous DRG (Fig. 5). Nrf2 was also significantly decreased at the transcript and protein level in the cerebella of YG8 hemizygous mice (Supplementary Fig. S1A, B), confirming a decrease in two neural target tissues in the animal model (DRG and cerebella). The main chaperone/regulator of Nrf2 expression is Keap1, whose level was not statistically significantly decreased in hemizygotes (Supplementary Fig. S1D, E).
FIG. 5.
Nrf2 and downstream target proteins are decreased in YG8R DRGs. (A) Representative immunoblots of Nrf2, Hmox, Nqo1, and Sod2 in the YG8R DRGs. Each lane represents an individual mouse. (B) Densitometry of blots shown in A, normalized to tubulin. Each lane contains 30 μg protein from a different mouse. Error bars=SEM. *p<0.05 versus WT; #p<0.05 versus homo using a 1-tailed Student's t-test. Nqo1, NAD(P)H dehydrogenase, quinone 1.
YG8R mice exhibit functional alterations in the glutathione status
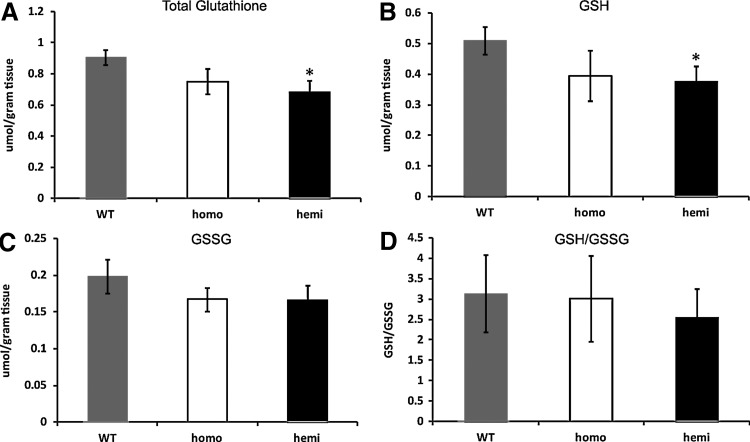
Microarray, qRT-PCR, and immunoblotting demonstrated decreased expression of Nrf2 and Nrf2-regulated genes. Since Nrf2 regulates the genes are involved in glutathione synthesis, an Nrf2 defect would suggest a defect in glutathione synthesis. We measured glutathione levels in the DRGs. The total glutathione (reduced glutathione [GSH]+oxidized GSH [GSSG]) and reduced GSH levels were significantly reduced in hemizygotes, p<0.05, and there was no significant change between GSSG and the GSH/GSSG ratio (Fig. 6). Overall, the GSH levels were similar to previously published data from rat DRGs (19, 76). Thus, there is less GSH to combat oxidative stress in target DRGs in YG8R hemizygous mice.
FIG. 6.
Glutathione levels are altered in YG8R DRGs. Total glutathione, GSH, and GSSG were measured in a pool of 16 DRGs from more than 7 mice per genotype. (A) Total glutathione (GSH+GSSG); (B) GSH; (C) GSSG; (D) GSH/GSSG. Error bars=SEM. *p<0.05 using a one-tailed Student's t-test. GSH, reduced glutathione; GSSG, oxidized glutathione.
Using an anti-glutathione antibody, we investigated protein glutathionylation in the mouse cerebella of mutants and controls, and observed multiple changes as indicated by the arrows (Supplementary Fig. S2). Thus, in target tissues of the cerebella and DRG, there are both decreased glutathione (paragraph above) and altered protein glutathionylation the mouse model of FRDA.
Nrf2 is reduced in frataxin-deficient cells
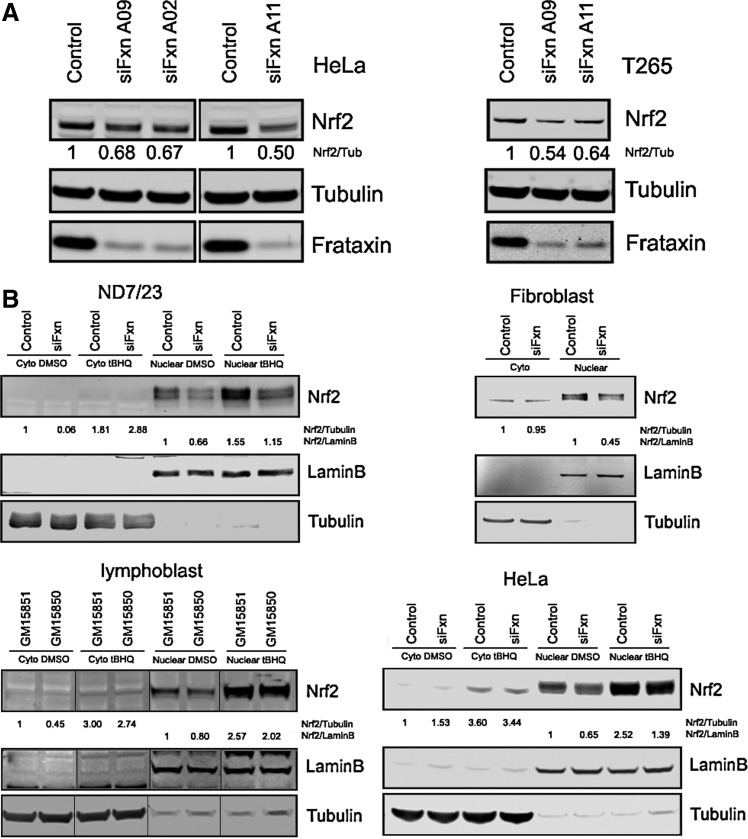
Because DRG tissue is limiting for biochemical studies, and to test the generality of the effect of frataxin deficiency on Nrf2 and TxnRD, we carried out siRNA-mediated knockdown experiments in multiple cell models, including cell models of DRG neurons and the Schwann cells that surround them. We ordered 3 frataxin siRNA oligonucleotides A02, A09, and A11 (Supplementary Table S2). These 3 oligos efficiently knocked down frataxin expression in HeLa cells, and Nrf2 protein decreased from 50% to 32% of the controls (Fig. 7A). Oligo A02 was too toxic to cell proliferation to use in further studies. We transfected frataxin siRNA A09 and A11 into T265 Schwann cells and found that Nrf2 decreased to 46% and 36% of control in the frataxin depleted T265 cells (Fig. 7A).
FIG. 7.
Nrf2 is decreased in frataxin-deficient cells. (A) Nrf2 protein decreased in HeLa and T265 frataxin-depleted cells. (B) No Nrf2 nuclear translocation defect in frataxin-deficient cells. Nuclear extraction and immunopurified anti-Nrf2 antibody demonstrate a substantial nuclear localization of Nrf2 protein, which is inducible by tBHQ, and deficient induction of Nrf2 in the nucleus, but no defect in cytosolic transport to nucleus is evident. Immunoblots contain 30 μg proteins per lane. tBHQ, tert-butylhydroquinone.
Nrf2 nuclear translocation defects were reported in patient's fibroblast cells and Frataxin-depleted SKNAS cells by Paupe et al. (55). Here we knocked down the frataxin gene in the HeLa and fibroblast cells with frataxin siRNA A11 and ND7/23 DRG neuron cells with the rat frataxin siRNA smart pool (Dharmacon), and frataxin expression was clearly inhibited in these cells (Supplementary Fig. S3). We also used FRDA lymphoblasts and controls. These frataxin-deficient cells were incubated with DMSO or 10 nM tert-butylhydroquinone (tBHQ) overnight and fractionated into the cytosol and nuclear extracts, and immunopurified H300 Nrf2 antibody (Santa Cruz), and monoclonal Nrf2 antibody (Epitomics) were used to probe the blots of the extracts. The ND7/23 extracts were also probed with immunopurified Nrf2 (47). Tubulin was used as a cytosol marker, and lamin B was used as a nuclear marker.
We observed a strong Nrf2 signal in the nucleus in all cells, and this signal was induced from 50% to 150% in all cells by tBHQ, a widely used Nrf2 inducer (Fig. 7B). We observed no support for the hypothesis that there is decreased transport of Nrf2 to the nucleus in the frataxin-deficient state in our conditions; this would be observed as increased cytosolic Nrf2 under the tBHQ-induced conditions. Thus, our results in these cell models are consistent with our results in the mouse target tissues the DRG and the cerebella, that is, that frataxin-deficiency causes Nrf2 deficiency at the transcript and protein level.
TxnRD activity and protein are reduced in frataxin-deficient cells
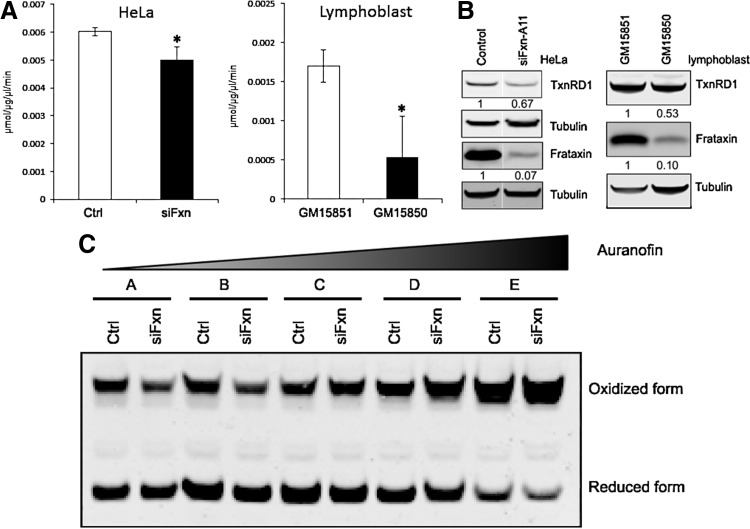
To further characterize Nrf2-dependent defects in frataxin-deficient cells, we examined expression and activity of TxnRD1, an Nrf2-regulated gene (41). TxnRD plays a key role in the thiol antioxidant response, reducing glutathione, peroxiredoxins, and Glrxs in the mitochondria and cytosol (37, 46). We found a significant decrease in the total TxnRD activity (Fig. 8A,) in lysates of frataxin-depleted HeLa cells or lymphoblast cells of patients suffering from FRDA. Immunoblot analysis demonstrated reduced TxnRD1 protein levels in these cells, demonstrating that that some of decreased activity is the result of reduced protein expression (Fig. 8B).
FIG. 8.
TxnRD activity and protein are decreased in frataxin-deficient cells. (A) TxnRD activity analysis in HeLa or lymphoblast cells. HeLa cells were transfected with control or frataxin A11 siRNA; after 72 h, cells were used to TxnRd activity assay. For lymphoblasts, healthy (GM15851) and patient (GM15850) cells were used in this assay. (B) TxnRd immunoblot in HeLa cells and lymphoblasts. The densitometries were normalized with tubulin. (C) Frataxin deficiency shifts the redox state of Prdx2 toward the oxidized isoform. HeLa cells were treated with increasing concentrations of auranofin for 1 h, then treated with an N-ethylmaleimide-containing buffer for 15 min before harvest. A, DMSO (vehicle); B, 10 μM; C, 20 μM; D, 40 μM; E, 80 μM. Immunoblots contain 30 μg protein per lane. Error bars=SEM. *p<0.05 using Student's t-test. Ctrl=control siRNA. siFxn=siRNA oligonucleotide directed against frataxin A11. Prdx2, peroxiredoxin 2.
Multiple thiol antioxidant proteins exist in reduced and oxidized forms, which depend on the cellular redox status. Peroxiredoxins are cysteine-based antioxidants that are reduced by TxnRDs in the cytosol and mitochondria (46). A frataxin-dependent deficiency of TxnRD should have a functional effect on the cellular redox status, and thus frataxin-deficient cells should be less able to reduce oxidized peroxiredoxins. We examined the redox status of Prdx2 in frataxin-depleted HeLa cells and found that increasing concentrations of the TxnRD inhibitor auranofin (30) treatment shifted the protein toward its oxidized isoform. As expected, frataxin deficiency increased this shift (Fig. 8C), especially at higher concentrations of auranofin, indicating that frataxin deficiency further oxidizes this antioxidant molecule in the context of increasing inhibition of TxnRD.
Frataxin-deficient cells are more sensitive to TxnRD inhibitors
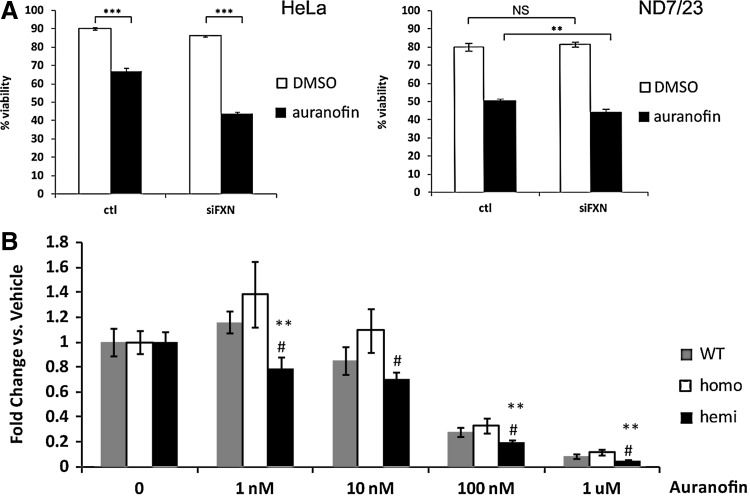
We tested functional alterations in the thiol antioxidants by examining cell sensitivity to compounds affecting the cellular redox poise. Both ND7/23 and HeLa frataxin knockdown cells were significantly more sensitive to the TxnRD inhibitor auranofin (Fig. 9A), as were the DRG cells from the hemizygous mouse model (Fig. 9B). The dose of auranofin required to cause 50% death of DRG cells was ∼40 nM, far lower from the frataxin-depleted cell models (2.5 or 4 μM), suggesting that the DRG environment is inherently sensitive to TxnRD deficiency, which could point to why these cells are selectively vulnerable in the context of whole-organism frataxin deficiency.
FIG. 9.
Frataxin-deficient cells exhibit increased sensitivity to the TxnRD inhibitor auranofin. (A) Left panel: Frataxin-knockdown HeLa cells treated with 4 μM auranofin. Data indicate means of 6 replicates; ***p<0.001. Right panel: Frataxin-knockdown ND7/23 cells treated with 2.5 μM auranofin. Data indicate means of three replicates; **p<0.01. (B) Auranofin-treated YG8R DRG cells. Data indicate means of three replicates per dosage over five experiments, each experiment using one mouse per genotype. Error bars=SEM. **p<0.01 versus WT; #p<0.05 versus homo using a 1-tailed Student's t-test.
Discussion
FRDA is an autosomal recessive disease that causes progressive damage to the nervous system that initiates in the DRG neurons, resulting in a dying back of projections to the cerebellum (29, 31). FRDA is the result of mutations in frataxin, a nuclear-encoded protein expressed predominantly in the mitochondria. In patients suffering from FRDA, the expansion of a GAA trinucleotide repeat located in the first intron of the frataxin gene inhibits its transcription and decreases the level of frataxin protein (17).
Biochemistry studies show that frataxin interacts with IscU/Nfs/Isd11, the core complex for the iron–sulfur cluster (ISC) biogenesis (66, 68), and frataxin deficiency decreases the expression of IscU/Nfs1/Isd11 and aconitase/SDH activities (72, 73). Recent biochemical work suggested that the function of frataxin protein is to activate cysteine desulfurase to provide activated sulfide for the biogenesis of ISCs (11). How the deficiency of frataxin (or ISCs) causes neurodegeneration has been controversial. Major hypotheses have included iron overload (60), bioenergetic failure (59, 63), and oxidative stress (18, 78, 79). Some mutations of ISC components that occur in human patients, that is, Grx5 (81) and ABCB7 (58), cause defects in erythroid differentiation, but not neurodegeneration. These results indicate that ISC deficiency can cause other problems than neurodegeneration. Further, mutations in the ISCU gene were recently found in pedigrees of patients with myopathy syndrome (40, 49), and deficiencies of succinate dehydrogenase and aconitase and iron overload were observed in the patient's skeletal muscle. Although some parts of the phenotype of these diseases initiated by ISC defects overlap with the FRDA phenotype, including tissue iron deposits, and defects in complex II and aconitase, no reports of neurodegeneration were reported in patients with myopathy syndrome. These findings support the view that ISC deficiency does not necessarily cause neurodegeneration.
The idea that oxidative injury occurs in Friedreich's has arisen from multiple sources: patient tissues and cells, animal models, and cell models of the disease. However, how a deficiency in the proven function of frataxin to support ISC biogenesis causes this oxidative injury is not clear. Very recently, Li and Outten have demonstrated that 2Fe-2S ISCs serve as sensors of oxidative stress, and regulate iron in yeast, and this is a possible explanation of the connection between 2Fe-2S clusters and oxidative stress, in which a decreased synthesis of 2Fe-2S clusters may desensitize this signaling pathway (23, 44).
Although many studies of the mechanism have been carried out in cellular, fly, and worm models, we sought to identify molecular changes in what seems to be the most relevant target tissue, that is, the DRG.
In this study, we verified that our colony of YG8R mice recapitulated the behavioral deficits reported by Al-Mahdawi et al. (4), and further demonstrated a functional defect in neuritic growth in the YG8R hemizygous mice. Explanted YG8R hemizygous DRG cells were more sensitive to auranofin (Fig. 9) and diamide (data not shown)—two thiol oxidants. These results indicated an increase in oxidative stress in the YG8R hemizygous DRG cells.
Oxidative stress can be considered an imbalance between the generation of reactive oxygen species and a biological system's ability to detoxify the reactive intermediates or to repair the resulting damage. Oxidative stress is suspected to be important in neurodegenerative diseases, including Lou Gehrig's disease (ALS), Parkinson's disease, Alzheimer's disease, and Huntington's disease (54). In the human body, cells are protected from oxidative stress by multiple enzyme systems.
Glutathione is a major antioxidant, present at millimolar concentrations. In the human body, glutamate and cysteine are synthesized into γ-glutamylcysteine by glutamate cysteine ligase, and this enzyme is a heterodimer composed by two subunits, Gclc and Gclm (52). Glutathione prevents the damage caused by reactive oxygen species such as free radicals and peroxides (57). Microarray data on DRG from the YG8R hemizygous mouse showed that Gclc and Gclm decreased, and further, RT-PCR results showed that the expression of Gclm is correlated with the frataxin transcript levels. Therefore, frataxin decrease causes glutathione synthesis defect in the mouse DRG. We observed a decrease in total GSH (GSH+GSSG) in the YG8R hemizygous DRG, similar to an yeast model (7) that is consistent with the decrease in Gclm. We also found a decrease in reduced GSH, an important peptide in combating oxidative stress in the DRG of hemizygous YG8R mice. Decreased total glutathione and GSH/GSSG ratios were observed in the yeast model and patient lymphoblasts (7, 13, 73), and increased protein glutathionylation was observed in patient motor neurons (71) and patients' lymphoblasts (13). We observed increased protein glutathionylation in the YG8R cerebella (Supplementary Fig. S2), so the glutathione redox status changed in YG8R mouse neural tissues.
The main antioxidant enzymes in human cells are superoxide dismutase (SOD) and catalase (Cat). SOD1 mutation is known to cause neurodegenerative disease ALS (62); SOD functions by converting the superoxide anion into oxygen and hydrogen peroxide (8); and Cat is known to convert hydrogen peroxide to water and oxygen (20). We found the frataxin transcript level to be positively correlated with Sod1, Sod2, and Cat gene transcripts.
In addition to glutathione, Sod, and Cat, there is still a redox system that can protect cells from oxidative stress. The redox system includes thioredoxins and the glutathione system (24). We observed decreases in Glrx1 and GstM1 in YG8R hemizygous DRG, and these two proteins are the components of the glutathione system. We also observed reduction in gene expression of the thioredoxin system, including Txn2, TxnRD1, Txnip, and Prx3. These data indicate that antioxidant protection is disrupted in the YG8R hemizygous DRG, further decreasing the DRG cells' ability to fight oxidative stress.
Several genes whose expression is reduced in YG8R hemizygous DRG (Gclm, Glrx1, GstM1, TxnRD1, Sod, and Cat) are regulated by Nrf2, and we see decreased Nrf2 in the mouse and cellular neural and glial models of frataxin deficiency. The Nrf2 transcript is correlated with the frataxin level, and Nrf2 protein is decreased in the YG8R hemizygous DRG and cerebellum. To further confirm the Nrf2 defect phenotype in YG8R DRG, we checked the expression of Nqo1 and Hmox, two well-known downstream target genes of the Nrf2 transcription factor. As expected, both genes were downregulated in the YG8R hemizygous DRGs.
Because the DRG material is limiting for biochemistry, we studied frataxin-deficient cell lines relevant to neurodegeneration, including the DRG cell line (ND7/23), the Schwann cell line (T265), and the HeLa cell model, and observed a frataxin-dependent decrease in Nrf2. We also studied the nuclear translocation of Nrf2 in several frataxin-deficient cell lines. Our results are consistent with those of Nguyen et al., who showed that Nrf2 is primarily a nuclear protein, and that steady-state levels are controlled by transient shuttling of Keap1 into the nucleus to promote Nrf2 ubiquitinylation (51). Our results are consistent with those of Haugen, Prospero et al. 2011 (32), who observed a defect in the Nrf2-driven genes in fibroblasts of patients suffering from FRDA, but our results are not consistent with those of Paupe et al., who observed a decrease in the cytosolic-to-nuclear transport of Nrf2 (55). We clearly see a tBHQ-dependent induction of Nrf2 in both the cytosol and nucleus of 50%–150%, and frataxin knockdown decreases the levels of Nrf2. If frataxin deficiency caused a decrease in nuclear translocation, we should be observe an increase in cytosolic Nrf2 in frataxin knockdown cells in the condition of induction, and we did not observe any nuclear translocation defects under our conditions in any experiments. We do note that all commercially available preparations of Nrf2 antibodies are of poor quality (except the Epitomics Nrf2 antibody), and so our results were confirmed with immunopurified Nrf2 antibodies, which we purified ourselves, and also received as kind gifts from Professors McMahon and Schmidt. Thus, our results are consistent with those of Nguyen, that is, that Nrf2 is a nuclear protein, and we do not observe any support for the idea of a frataxin-dependent defect in the cytosolic-to-nuclear translocation in frataxin-deficient cells. We do observe a frataxin-dependent defect in Nrf2 expression in the DRG and cerebella of the mouse model, and in five frataxin-deficient cell models, including DRG/ND7/23, Schwann/T265, HeLa, fibroblasts, and patient-derived lymphoblasts.
Cytoskeletal defects have been observed in previous studies of Friedreich's patient fibroblasts. Increased glutathionlyation of actin fibers and impaired microfilament organization have been demonstrated in patient fibroblasts (53, 55). In patient spinal cord samples, increased levels of the dynamic (tyrosinated) form of tubulin and increased levels of phosphorylated neurofilament were observed (71). We observed decreased neurite extension in cultured YG8R DRG cells, whereas microarray and qRT-PCR demonstrated increases in Kif1b and Kif5b, the genes involved in axonal transport of the mitochondria (33). Defects in axonal transport of mitochondria have been observed in a Drosophila model of FRDA (69). One possible explanation is that as a result of decreased Nrf2 and consequent decreases in the mitochondrial oxidative status, that delivery of healthy mitochondria to the growth tip is inhibited, decreasing neuritic growth. Consistent with this idea, we observe increased axonal transport genes Kifs in mutant mice, possibly a response to more oxidatively damaged mitochondria, which may necessitate an increase in axonal transport of fresh, healthier mitochondria.
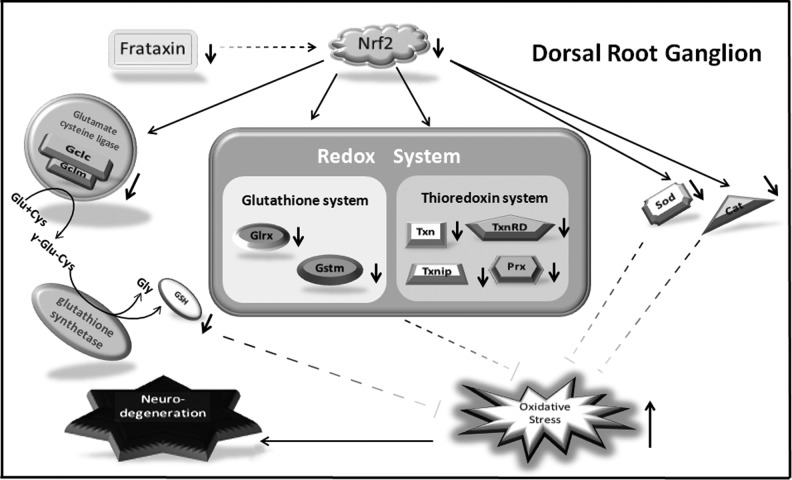
Taken together, these data suggest that frataxin deficiency→Nrf2 deficiency→defect in antioxidant protection→oxidative stress (Fig. 10). The closest clinical phenotype to FRDA is that of AVED (9), an inherited deficiency of the transporter of the antioxidant vitamin E, which has recently been shown to stimulate Nrf2 (27). Nrf2 deficiency is known to cause inflammation and cell death (35).
FIG. 10.
A model for frataxin deficiency with neurodegeneration in the DRG cells.
Most neurodegenerative processes are the result of contributions of multiple cell types (34), and in the nervous system, glial Nrf2 expression is primarily responsible for Nrf2-mediated protection of neurons from oxidative stress (70). Schwann cell demyelination of DRGs occurs in YG8R mice (4), and we observe decreased transcription of the myelin basic protein in DRG tissue. The most precise analytical neuropathological studies available of human FRDA patient autopsies have shown that satellite/Schwann cells become reactive and invade the DRG neurons in the Friedreich's pathophysiological process (39), opening the possibility that an inflammatory reaction between the DRG neurons and Schwann cells may be a part of the disease process; however, this is speculation at this point. We demonstrated previously that frataxin knockdown of Schwann cell lines in particular evoked the induction of inflammatory transcripts and cytokines, which were reduced by pre-exposure to anti-inflammatory drugs (45).
In summary, based on the microarray of target DRGs and functional follow-up, our data support the hypothesis that frataxin deficiency leads to Nrf2 deficiency in DRG tissue, which leads to a defect in the antioxidant status that precedes neurodegeneration. We speculate that this defect may lead to neuroinflammatory changes and death of DRG neurons and those pharmacological inducers of Nrf2 could be of benefit in FRDA.
Materials and Methods
Biochemical reagents were purchased from Sigma, Invitrogen, or Bio-Rad. Microarray chips were purchased from Affymetrix. All animal procedures were approved by the Institutional Animal Care and Use Committee at UC Davis. The median age of mice used for all studies was 6 months. Behavior testing was performed at the UC Davis Mouse Behavioral Analysis Laboratory.
Cell culture
The human Schwann cell lines (T265), human epithelial carcinoma cell line (HeLa), the DRG cell line (ND7/23, rat DRG/mouse neuroblastoma hybrid), and human fibroblast cell line (GM02153) were maintained in DMEM, supplemented with 10% FBS and 2 mM glutamine. Lymphoblasts (GM15850 and GM15851) were grown in the RPMI-1640 medium (Life Technologies) supplemented with 15% fetal bovine serum, 2 mM sodium pyruvate, 2 mM glutamine, and 50 mg/ml uridine. All cells were maintained at 37°C in a humidified atmosphere containing 5% CO2.
DRGs were dissected and placed in ice-cold HBSS during dissection. Single-cell suspensions were made by treating DRGs sequentially with papain and collagenase (Worthington Biochemicals) for 20 min at 37°C. Cells were resuspended and cultured in a C2 medium.
For neurite extension assays, single-cell DRG suspensions (L1–L4) were plated on poly-d-lysine-coated dishes and incubated for 3 days. Bipolar cells were photographed at 40× magnification using a Zeiss Axiovert 200 microscope, and the neurite length measured using NIH ImageJ software with NeuronJ plugin (1, 48).
For cell viability assays, cells were plated at 250 cells per well, allowed to settle overnight, and then treated with the drug or vehicle for 24 h. The cell viability was measured using CellTiter-Glo (Promega).
GSH/GSSG measurements
DRGs (T10-L4) were isolated and snap-frozen in liquid nitrogen. the GSH/GSSG levels were measured using the Bioxytech GSH/GSSG-412 kit (Oxis International). Briefly, 16 DRGs were homogenized in 250 μl of 5% metaphosphoric acid. Of this, 80 μl was mixed with 8 μl M2VP and used for GSSG measurements and the remainder for GSH measurements according to the manufacturer's recommendations.
Microarray and qRT-PCR
Total RNA was extracted from DRG tissue using an RNeasy Micro kit (Qiagen) and quantified by a NanoDrop 2000c Spectrophotometer (Thermo Scientific). Two hundred fifty nanograms RNA was labeled for hybridization to MOE430 2.0 microarray chips (Affymetrix) using MessageAmp III (Ambion). Data were analyzed using dChip (65). Samples were normalized to the median intensity chip and fluorescence values generated using the perfect match/mismatch difference model. Probe sets with a pCall >0%, fluorescence >25 in at least one condition, and p<0.05 were considered significantly altered.
Quantitative PCR was performed using the Superscript III One Step kit (Invitrogen) in a Roche Lightcycler 480 (Roche Diagnostics) or using the Sensimix Capillary One-Step kit (Bioline) in a Roche Lightcycler 1.2. Standard curves were generated for each primer set, and samples fitted to the linear portion of the curve. Primer sequences are listed in Supplementary Table S1.
siRNA and immunoblotting
HeLa, T265, and human fibroblasts were transfected with control siRNA (Allstar-negative siRNA; Qiagen) or frataxin siRNA (Invitrogen; sequence in Supplementary Table S2). ND7/23 cells were transfected with control siRNA smartpool or rat frataxin siRNA smartpool (Dharmacon). In these transfections, 10 nM siRNA was transfected into cells by Lipofectamine RNAiMax (Invitrogen) according to the manufacturer's directions.
Mouse tissues and cell pellets were homogenized with a cell lysis buffer (Cell Signaling) with complete protease inhibitors (Roche) and PSMF. Thirty micrograms of lysates were analyzed on 4%–12% Bis–Tris gels (Invitrogen). Electrophoresis was carried out according to the manufacturer's recommendations. After electrophoresis, the proteins were transferred to nitrocellulose membranes by the iBlot device (Invitrogen), blocked with an Odyssey blocking buffer (LI-COR Biotechnology) for 1 h, and incubated overnight with the following primary antibodies in a blocking buffer: frataxin, Nrf2 (C-20 for mouse and H-300 for human; Santa Cruz Biotech, Santa Cruz, CA); Nrf2 (Epitomics); Nrf2 for ND7/23 cells [a kind gift from Dr. McMahon (47)]; tubulin (Sigma); Gstm1, Mbp, TxnRD, and S100a8 (Abcam); Grlx1 (R&D Systems); Prx3 (AbFrontier); Nqo1 (Epitomics), Hmox and Sod2 (Stressgen/Enzo), and Lamin B1 (Proteintech). Afterward, the membranes were incubated with a corresponding pair of IRDye 680CW and IRDye 800CW-coupled secondary antibodies (LI-COR). Proteins were visualized with the Odyssey infrared imager and software (LI-COR).
Nrf2 antibody immunopurification and nuclear extraction
Recombinant His-Nrf2 protein was expressed from the pET20-Nrf2 plasmid [a kind gift from Prof. Edward E. Schmidt in the BL21 Star cells (Invitrogen) and purified using a B-Per 6xHis protein purification kit (Pierce/Thermo Scientific]. Anti-Nrf2 (H-300; Santa Cruz) was immunopurified using this recombinant fusion protein. Briefly, the purified fusion protein was run on a 4%–12% NuPAGE Bis–Tris gel (Invitrogen) and then electroblotted onto a nitrocellulose membrane using the iBlot. The blot was stained with Ponceau S red, and the His-Nrf2 band was cut out and washed with TBST until the Ponceau S gone. The blot was blocked by the Odyssey blocking buffer for 2 h and washed with TBST twice. Anti-Nrf2 was added on the blot and incubated overnight at 4°C. The blot was washed three times with TBST and eluted with 1 ml 100 mM glycine buffer, pH 2–2.5 (Santa Cruz Biotech), and then 100 μL 1 M Tris, pH 8.0, was added to the elute to adjust the final pH to 7.0. Transfected cells were fractionated with a nuclear extraction kit (Active Motif). Forty micrograms of lysates was analyzed with the immunopurified Nrf2 antibody (H300) or rabbit monoclonal Nrf2 antibody (Epitomics). ND7/23 cells were probed with the purified Nrf2 antibody from Dr. Mcmahon (47). Tubulin was used as the cytosolic marker and lamin B as the nuclear marker.
TxnRD activity
TxnRD activity was measured using the Thioredoxin Reductase Assay Kit (Cayman Chemicals) according to the manufacturer's directions.
Redox isoform method
The redox isoforms of Prdx2 were assayed as previously described (22). Briefly, HeLa cells were treated with auranofin for 1 h, and then trypsinized, washed, and resuspended in a freshly made buffer (100 mM NEM, 40 mM HEPES, pH 7.4, 50 mM NaCl, 1 mM EDTA, 1 mM EGTA, plus protease inhibitors) for 15 min, and CHAPS was added to a final concentration of 1% for 20 min on ice. Thirty micrograms of lysates was combined with an LDS sample buffer (Invitrogen) in the absence of a reducing agent and resolved on a 4%–12% NuPAGE Bis–Tris gel and analyzed by immunoblotting.
Supplementary Material
Abbreviations Used
- AVED
ataxia with vitamin E deficiency
- Cat
catalase
- DRG
dorsal root ganglion
- FRDA
Friedreich's ataxia
- Fxn
frataxin
- Gclc
glutamate–cysteine ligase, catalytic subunit
- Gclm
glutamate–cysteine ligase, modifier subunit
- Glrx1
glutaredoxin 1
- GSH
reduced glutathione
- GSSG
oxidized glutathione
- Gstm1
glutathione S-transferase-mu 1
- Hmox
heme oxygenase
- ISC
iron–sulfur cluster
- Kif1b
kinesin family member 1B
- Kif5b
kinesin family member 5B
- Mbp
myelin basic protein
- Nqo1
NAD(P)H dehydrogenase, quinone 1
- Nrf2
nuclear factor-E2-related factor-2
- Prdx2
peroxiredoxin 2
- Prdx3
peroxiredoxin 3
- siFxn
siRNA oligonucleotide directed against frataxin
- SOD
superoxide dismutase
- Sod1
Cu/Zn superoxide dismutase
- Sod2
Mn superoxide dismutase
- tBHQ
tert-butylhydroquinone
- Txn2
thioredoxin 2
- Txnip
thioredoxin-interacting protein
- TxnRD1
thioredoxin reductase 1
- WT
wild type
Acknowledgments
We thank Khalida Sabeur for performing IVF and rederivation of the YG8R mice, Polly Pearce for the managing the mouse colony, and Peter Takeuchi and Mari Golub for their help with behavioral testing at the UC Davis Mouse Behavioral Analysis core. Purified Nrf2 antibodies and plasmids were kind gifts from Drs. McMahon and Eric Schmidt. This work was supported by NIA 5R01AG16719-12 and USPHS EY12245, AG11967, AG16719, and AG23311 to G.A.C.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Abramoff MD. Magalhaes PJ. Ram SJ. Image Processing with ImageJ. Biophotonics Int. 2004;11:36–42. [Google Scholar]
- 2.Adair-Kirk TL. Atkinson JJ. Griffin GL. Watson MA. Kelley DG. DeMello D. Senior RM. Betsuyaku T. Distal airways in mice exposed to cigarette smoke: Nrf2-regulated genes are increased in Clara cells. Am J Respir Cell Mol Biol. 2008;39:400–411. doi: 10.1165/rcmb.2007-0295OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Al-Mahdawi S. Pinto RM. Ruddle P. Carroll C. Webster Z. Pook M. GAA repeat instability in Friedreich ataxia YAC transgenic mice. Genomics. 2004;84:301–310. doi: 10.1016/j.ygeno.2004.04.003. [DOI] [PubMed] [Google Scholar]
- 4.Al-Mahdawi S. Pinto RM. Varshney D. Lawrence L. Lowrie MB. Hughes S. Webster Z. Blake J. Cooper JM. King R. Pook MA. GAA repeat expansion mutation mouse models of Friedreich ataxia exhibit oxidative stress leading to progressive neuronal and cardiac pathology. Genomics. 2006;88:580–590. doi: 10.1016/j.ygeno.2006.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Anderson PR. Kirby K. Hilliker AJ. Phillips JP. RNAi-mediated suppression of the mitochondrial iron chaperone, frataxin, in Drosophila. Hum Mol Genet. 2005;14:3397–3405. doi: 10.1093/hmg/ddi367. [DOI] [PubMed] [Google Scholar]
- 6.Anderson PR. Kirby K. Orr WC. Hilliker AJ. Phillips JP. Hydrogen peroxide scavenging rescues frataxin deficiency in a Drosophila model of Friedreich's ataxia. Proc Natl Acad Sci U S A. 2008;105:611–616. doi: 10.1073/pnas.0709691105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Auchere F. Santos R. Planamente S. Lesuisse E. Camadro JM. Glutathione-dependent redox status of frataxin-deficient cells in a yeast model of Friedreich's ataxia. Hum Mol Genet. 2008;17:2790–2802. doi: 10.1093/hmg/ddn178. [DOI] [PubMed] [Google Scholar]
- 8.Bannister JV. Bannister WH. Rotilio G. Aspects of the structure, function, and applications of superoxide dismutase. CRC Crit Rev Biochem. 1987;22:111–180. doi: 10.3109/10409238709083738. [DOI] [PubMed] [Google Scholar]
- 9.Benomar A. Yahyaoui M. Meggouh F. Bouhouche A. Boutchich M. Bouslam N. Zaim A. Schmitt M. Belaidi H. Ouazzani R. Chkili T. Koenig M. Clinical comparison between AVED patients with 744 del A mutation and Friedreich ataxia with GAA expansion in 15 Moroccan families. J Neurol Sci. 2002;198:25–29. doi: 10.1016/s0022-510x(02)00057-6. [DOI] [PubMed] [Google Scholar]
- 10.Bradley JL. Blake JC. Chamberlain S. Thomas PK. Cooper JM. Schapira AH. Clinical, biochemical and molecular genetic correlations in Friedreich's ataxia. Hum Mol Genet. 2000;9:275–282. doi: 10.1093/hmg/9.2.275. [DOI] [PubMed] [Google Scholar]
- 11.Bridwell-Rabb J. Winn AM. Barondeau DP. Structure-function analysis of friedreich's ataxia mutants reveals determinants of frataxin binding and activation of the Fe-S assembly complex. Biochemistry. 2011;50:7265–7274. doi: 10.1021/bi200895k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bulteau AL. Dancis A. Gareil M. Montagne JJ. Camadro JM. Lesuisse E. Oxidative stress and protease dysfunction in the yeast model of Friedreich ataxia. Free Radic Biol Med. 2007;42:1561–1570. doi: 10.1016/j.freeradbiomed.2007.02.014. [DOI] [PubMed] [Google Scholar]
- 13.Bulteau AL. Planamente S. Jornea L. Dur A. Lesuisse E. Camadro JM. Auchere F. Changes in mitochondrial glutathione levels and protein thiol oxidation in yfh1 yeast cells and the lymphoblasts of patients with Friedreich's ataxia. Biochim Biophys Acta. 2012;1822:212–225. doi: 10.1016/j.bbadis.2011.11.003. [DOI] [PubMed] [Google Scholar]
- 14.Calabrese V. Lodi R. Tonon C. D'Agata V. Sapienza M. Scapagnini G. Mangiameli A. Pennisi G. Stella AM. Butterfield DA. Oxidative stress, mitochondrial dysfunction and cellular stress response in Friedreich's ataxia. J Neurol Sci. 2005;233:145–162. doi: 10.1016/j.jns.2005.03.012. [DOI] [PubMed] [Google Scholar]
- 15.Calkins MJ. Johnson DA. Townsend JA. Vargas MR. Dowell JA. Williamson TP. Kraft AD. Lee JM. Li J. Johnson JA. The Nrf2/ARE pathway as a potential therapeutic target in neurodegenerative disease. Antioxid Redox Signal. 2009;11:497–508. doi: 10.1089/ars.2008.2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Campuzano V. Montermini L. Lutz Y. Cova L. Hindelang C. Jiralerspong S. Trottier Y. Kish SJ. Faucheux B. Trouillas P. Authier FJ. Durr A. Mandel JL. Vescovi A. Pandolfo M. Koenig M. Frataxin is reduced in Friedreich ataxia patients and is associated with mitochondrial membranes. Hum Mol Genet. 1997;6:1771–1780. doi: 10.1093/hmg/6.11.1771. [DOI] [PubMed] [Google Scholar]
- 17.Campuzano V. Montermini L. Molto MD. Pianese L. Cossee M. Cavalcanti F. Monros E. Rodius F. Duclos F. Monticelli A. Zara F. Canizares J. Koutnikova H. Bidichandani SI. Gellera C. Brice A. Trouillas P. De Michele G. Filla A. De Frutos R. Palau F. Patel PI. Di Donato S. Mandel JL. Cocozza S. Koenig M. Pandolfo M. Friedreich's ataxia: autosomal recessive disease caused by an intronic GAA triplet repeat expansion. Science. 1996;271:1423–1427. doi: 10.1126/science.271.5254.1423. [DOI] [PubMed] [Google Scholar]
- 18.Chantrel-Groussard K. Geromel V. Puccio H. Koenig M. Munnich A. Rotig A. Rustin P. Disabled early recruitment of antioxidant defenses in Friedreich's ataxia. Hum Mol Genet. 2001;10:2061–2067. doi: 10.1093/hmg/10.19.2061. [DOI] [PubMed] [Google Scholar]
- 19.Chao PL. Fan SF. Chou YH. Lin AM. N-acetylcysteine attenuates arsenite-induced oxidative injury in dorsal root ganglion explants. Ann N Y Acad Sci. 2007;1122:276–288. doi: 10.1196/annals.1403.020. [DOI] [PubMed] [Google Scholar]
- 20.Chelikani P. Fita I. Loewen PC. Diversity of structures and properties among catalases. Cell Mol Life Sci. 2004;61:192–208. doi: 10.1007/s00018-003-3206-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cossee M. Puccio H. Gansmuller A. Koutnikova H. Dierich A. LeMeur M. Fischbeck K. Dolle P. Koenig M. Inactivation of the Friedreich ataxia mouse gene leads to early embryonic lethality without iron accumulation. Hum Mol Genet. 2000;9:1219–1226. doi: 10.1093/hmg/9.8.1219. [DOI] [PubMed] [Google Scholar]
- 22.Cox AG. Brown KK. Arner ES. Hampton MB. The thioredoxin reductase inhibitor auranofin triggers apoptosis through a Bax/Bak-dependent process that involves peroxiredoxin 3 oxidation. Biochem Pharmacol. 2008;76:1097–1109. doi: 10.1016/j.bcp.2008.08.021. [DOI] [PubMed] [Google Scholar]
- 23.Dardalhon M. Kumar C. Iraqui I. Vernis L. Kienda G. Banach-Latapy A. He T. Chanet R. Faye G. Outten CE. Huang ME. Redox-sensitive YFP sensors monitor dynamic nuclear and cytosolic glutathione redox changes. Free Radic Biol Med. 2012;52:2254–2265. doi: 10.1016/j.freeradbiomed.2012.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Das KC. White CW. Redox systems of the cell: possible links and implications. Proc Natl Acad Sci U S A. 2002;99:9617–9618. doi: 10.1073/pnas.162369199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Durr A. Cossee M. Agid Y. Campuzano V. Mignard C. Penet C. Mandel JL. Brice A. Koenig M. Clinical and genetic abnormalities in patients with Friedreich's ataxia. N Engl J Med. 1996;335:1169–1175. doi: 10.1056/NEJM199610173351601. [DOI] [PubMed] [Google Scholar]
- 26.Emond M. Lepage G. Vanasse M. Pandolfo M. Increased levels of plasma malondialdehyde in Friedreich ataxia. Neurology. 2000;55:1752–1753. doi: 10.1212/wnl.55.11.1752. [DOI] [PubMed] [Google Scholar]
- 27.Feng Z. Liu Z. Li X. Jia H. Sun L. Tian C. Jia L. Liu J. alpha-Tocopherol is an effective Phase II enzyme inducer: protective effects on acrolein-induced oxidative stress and mitochondrial dysfunction in human retinal pigment epithelial cells. J Nutr Biochem. 2010;21:1222–1231. doi: 10.1016/j.jnutbio.2009.10.010. [DOI] [PubMed] [Google Scholar]
- 28.Filla A. De Michele G. Cavalcanti F. Pianese L. Monticelli A. Campanella G. Cocozza S. The relationship between trinucleotide (GAA) repeat length and clinical features in Friedreich ataxia. Am J Hum Genet. 1996;59:554–560. [PMC free article] [PubMed] [Google Scholar]
- 29.Finocchiaro G. Baio G. Micossi P. Pozza G. di Donato S. Glucose metabolism alterations in Friedreich's ataxia. Neurology. 1988;38:1292–1296. doi: 10.1212/wnl.38.8.1292. [DOI] [PubMed] [Google Scholar]
- 30.Gromer S. Arscott LD. Williams CH., Jr. Schirmer RH. Becker K. Human placenta thioredoxin reductase. Isolation of the selenoenzyme, steady state kinetics, and inhibition by therapeutic gold compounds. J Biol Chem. 1998;273:20096–20101. doi: 10.1074/jbc.273.32.20096. [DOI] [PubMed] [Google Scholar]
- 31.Harding AE. Hewer RL. The heart disease of Friedreich's ataxia: a clinical and electrocardiographic study of 115 patients, with an analysis of serial electrocardiographic changes in 30 cases. Q J Med. 1983;52:489–502. [PubMed] [Google Scholar]
- 32.Haugen AC. Di Prospero NA. Parker JS. Fannin RD. Chou J. Meyer JN. Halweg C. Collins JB. Durr A. Fischbeck K. Van Houten B. Altered gene expression and DNA damage in peripheral blood cells from Friedreich's ataxia patients: cellular model of pathology. PLoS Genet. 2010;6:e1000812. doi: 10.1371/journal.pgen.1000812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hirokawa N. Noda Y. Intracellular transport and kinesin superfamily proteins, KIFs: structure, function, and dynamics. Physiol Rev. 2008;88:1089–1118. doi: 10.1152/physrev.00023.2007. [DOI] [PubMed] [Google Scholar]
- 34.Ilieva H. Polymenidou M. Cleveland DW. Non-cell autonomous toxicity in neurodegenerative disorders: ALS and beyond. J Cell Biol. 2009;187:761–772. doi: 10.1083/jcb.200908164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Innamorato NG. Rojo AI. Garcia-Yague AJ. Yamamoto M. de Ceballos ML. Cuadrado A. The transcription factor Nrf2 is a therapeutic target against brain inflammation. J Immunol. 2008;181:680–689. doi: 10.4049/jimmunol.181.1.680. [DOI] [PubMed] [Google Scholar]
- 36.Jauslin ML. Wirth T. Meier T. Schoumacher F. A cellular model for Friedreich Ataxia reveals small-molecule glutathione peroxidase mimetics as novel treatment strategy. Hum Mol Genet. 2002;11:3055–3063. doi: 10.1093/hmg/11.24.3055. [DOI] [PubMed] [Google Scholar]
- 37.Kalinina EV. Chernov NN. Saprin AN. Involvement of thio-, peroxi-, and glutaredoxins in cellular redox-dependent processes. Biochemistry (Mosc) 2008;73:1493–1510. doi: 10.1134/s0006297908130099. [DOI] [PubMed] [Google Scholar]
- 38.Koeppen AH. Michael SC. Knutson MD. Haile DJ. Qian J. Levi S. Santambrogio P. Garrick MD. Lamarche JB. The dentate nucleus in Friedreich's ataxia: the role of iron-responsive proteins. Acta Neuropathol. 2007;114:163–173. doi: 10.1007/s00401-007-0220-y. [DOI] [PubMed] [Google Scholar]
- 39.Koeppen AH. Morral JA. Davis AN. Qian J. Petrocine SV. Knutson MD. Gibson WM. Cusack MJ. Li D. The dorsal root ganglion in Friedreich's ataxia. Acta Neuropathol. 2009;118:763–776. doi: 10.1007/s00401-009-0589-x. [DOI] [PubMed] [Google Scholar]
- 40.Kollberg G. Tulinius M. Melberg A. Darin N. Andersen O. Holmgren D. Oldfors A. Holme E. Clinical manifestation and a new ISCU mutation in iron-sulphur cluster deficiency myopathy. Brain. 2009;132:2170–2179. doi: 10.1093/brain/awp152. [DOI] [PubMed] [Google Scholar]
- 41.Kong L. Tanito M. Huang Z. Li F. Zhou X. Zaharia A. Yodoi J. McGinnis JF. Cao W. Delay of photoreceptor degeneration in tubby mouse by sulforaphane. J Neurochem. 2007;101:1041–1052. doi: 10.1111/j.1471-4159.2007.04481.x. [DOI] [PubMed] [Google Scholar]
- 42.Kwak MK. Wakabayashi N. Itoh K. Motohashi H. Yamamoto M. Kensler TW. Modulation of gene expression by cancer chemopreventive dithiolethiones through the Keap1-Nrf2 pathway. Identification of novel gene clusters for cell survival. J Biol Chem. 2003;278:8135–8145. doi: 10.1074/jbc.M211898200. [DOI] [PubMed] [Google Scholar]
- 43.Lefevre S. Brossas C. Auchere F. Boggetto N. Camadro JM. Santos R. Apn1 AP-endonuclease is essential for the repair of oxidatively damaged DNA bases in yeast frataxin-deficient cells. Hum Mol Genet. 2012;21:4060–4072. doi: 10.1093/hmg/dds230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li H. Outten CE. Monothiol CGFS Glutaredoxins and BolA-like Proteins: [2Fe-2S] Binding Partners in Iron Homeostasis. Biochemistry. 2012;51:4377–4389. doi: 10.1021/bi300393z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lu C. Schoenfeld R. Shan Y. Tsai HJ. Hammock B. Cortopassi G. Frataxin deficiency induces Schwann cell inflammation and death. Biochim Biophys Acta. 2009;1792:1052–1061. doi: 10.1016/j.bbadis.2009.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Maher P. Redox control of neural function: background, mechanisms, and significance. Antioxid Redox Signal. 2006;8:1941–1970. doi: 10.1089/ars.2006.8.1941. [DOI] [PubMed] [Google Scholar]
- 47.McMahon M. Lamont DJ. Beattie KA. Hayes JD. Keap1 perceives stress via three sensors for the endogenous signaling molecules nitric oxide, zinc, and alkenals. Proc Natl Acad Sci U S A. 2010;107:18838–18843. doi: 10.1073/pnas.1007387107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Meijering E. Jacob M. Sarria JC. Steiner P. Hirling H. Unser M. Design and validation of a tool for neurite tracing and analysis in fluorescence microscopy images. Cytometry A. 2004;58:167–176. doi: 10.1002/cyto.a.20022. [DOI] [PubMed] [Google Scholar]
- 49.Mochel F. Knight MA. Tong WH. Hernandez D. Ayyad K. Taivassalo T. Andersen PM. Singleton A. Rouault TA. Fischbeck KH. Haller RG. Splice mutation in the iron-sulfur cluster scaffold protein ISCU causes myopathy with exercise intolerance. Am J Hum Genet. 2008;82:652–660. doi: 10.1016/j.ajhg.2007.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Montermini L. Richter A. Morgan K. Justice CM. Julien D. Castellotti B. Mercier J. Poirier J. Capozzoli F. Bouchard JP. Lemieux B. Mathieu J. Vanasse M. Seni MH. Graham G. Andermann F. Andermann E. Melancon SB. Keats BJ. Di Donato S. Pandolfo M. Phenotypic variability in Friedreich ataxia: role of the associated GAA triplet repeat expansion. Ann Neurol. 1997;41:675–682. doi: 10.1002/ana.410410518. [DOI] [PubMed] [Google Scholar]
- 51.Nguyen T. Sherratt PJ. Nioi P. Yang CS. Pickett CB. Nrf2 controls constitutive and inducible expression of ARE-driven genes through a dynamic pathway involving nucleocytoplasmic shuttling by Keap1. J Biol Chem. 2005;280:32485–32492. doi: 10.1074/jbc.M503074200. [DOI] [PubMed] [Google Scholar]
- 52.Njalsson R. Norgren S. Physiological and pathological aspects of GSH metabolism. Acta Paediatr. 2005;94:132–137. doi: 10.1111/j.1651-2227.2005.tb01878.x. [DOI] [PubMed] [Google Scholar]
- 53.Pastore A. Tozzi G. Gaeta LM. Bertini E. Serafini V. Di Cesare S. Bonetto V. Casoni F. Carrozzo R. Federici G. Piemonte F. Actin glutathionylation increases in fibroblasts of patients with Friedreich's ataxia: a potential role in the pathogenesis of the disease. J Biol Chem. 2003;278:42588–42595. doi: 10.1074/jbc.M301872200. [DOI] [PubMed] [Google Scholar]
- 54.Patel VP. Chu CT. Nuclear transport, oxidative stress, and neurodegeneration. Int J Clin Exp Pathol. 2011;4:215–229. [PMC free article] [PubMed] [Google Scholar]
- 55.Paupe V. Dassa EP. Goncalves S. Auchere F. Lonn M. Holmgren A. Rustin P. Impaired nuclear Nrf2 translocation undermines the oxidative stress response in Friedreich ataxia. PLoS One. 2009;4:e4253. doi: 10.1371/journal.pone.0004253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Piemonte F. Pastore A. Tozzi G. Tagliacozzi D. Santorelli FM. Carrozzo R. Casali C. Damiano M. Federici G. Bertini E. Glutathione in blood of patients with Friedreich's ataxia. Eur J Clin Invest. 2001;31:1007–1011. doi: 10.1046/j.1365-2362.2001.00922.x. [DOI] [PubMed] [Google Scholar]
- 57.Pompella A. Visvikis A. Paolicchi A. De Tata V. Casini AF. The changing faces of glutathione, a cellular protagonist. Biochem Pharmacol. 2003;66:1499–1503. doi: 10.1016/s0006-2952(03)00504-5. [DOI] [PubMed] [Google Scholar]
- 58.Pondarre C. Campagna DR. Antiochos B. Sikorski L. Mulhern H. Fleming MD. Abcb7, the gene responsible for X-linked sideroblastic anemia with ataxia, is essential for hematopoiesis. Blood. 2007;109:3567–3569. doi: 10.1182/blood-2006-04-015768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Puccio H. Simon D. Cossee M. Criqui-Filipe P. Tiziano F. Melki J. Hindelang C. Matyas R. Rustin P. Koenig M. Mouse models for Friedreich ataxia exhibit cardiomyopathy, sensory nerve defect and Fe-S enzyme deficiency followed by intramitochondrial iron deposits. Nat Genet. 2001;27:181–186. doi: 10.1038/84818. [DOI] [PubMed] [Google Scholar]
- 60.Radisky DC. Babcock MC. Kaplan J. The yeast frataxin homologue mediates mitochondrial iron efflux. Evidence for a mitochondrial iron cycle. J Biol Chem. 1999;274:4497–4499. doi: 10.1074/jbc.274.8.4497. [DOI] [PubMed] [Google Scholar]
- 61.Ristow M. Mulder H. Pomplun D. Schulz TJ. Muller-Schmehl K. Krause A. Fex M. Puccio H. Muller J. Isken F. Spranger J. Muller-Wieland D. Magnuson MA. Mohlig M. Koenig M. Pfeiffer AF. Frataxin deficiency in pancreatic islets causes diabetes due to loss of beta cell mass. J Clin Invest. 2003;112:527–534. doi: 10.1172/JCI18107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rosen DR. Mutations in Cu/Zn superoxide dismutase gene are associated with familial amyotrophic lateral sclerosis. Nature. 1993;364:362. doi: 10.1038/364362c0. [DOI] [PubMed] [Google Scholar]
- 63.Rotig A. de Lonlay P. Chretien D. Foury F. Koenig M. Sidi D. Munnich A. Rustin P. Aconitase and mitochondrial iron-sulphur protein deficiency in Friedreich ataxia. Nat Genet. 1997;17:215–217. doi: 10.1038/ng1097-215. [DOI] [PubMed] [Google Scholar]
- 64.Runko AP. Griswold AJ. Min KT. Overexpression of frataxin in the mitochondria increases resistance to oxidative stress and extends lifespan in Drosophila. FEBS Lett. 2008;582:715–719. doi: 10.1016/j.febslet.2008.01.046. [DOI] [PubMed] [Google Scholar]
- 65.Schadt EE. Li C. Ellis B. Wong WH. Feature extraction and normalization algorithms for high-density oligonucleotide gene expression array data. J Cell Biochem Suppl. 2001;(Suppl 37):120–125. doi: 10.1002/jcb.10073. [DOI] [PubMed] [Google Scholar]
- 66.Schmucker S. Martelli A. Colin F. Page A. Wattenhofer-Donze M. Reutenauer L. Puccio H. Mammalian frataxin: an essential function for cellular viability through an interaction with a preformed ISCU/NFS1/ISD11 iron-sulfur assembly complex. PLoS One. 2011;6:e16199. doi: 10.1371/journal.pone.0016199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Schulz JB. Dehmer T. Schols L. Mende H. Hardt C. Vorgerd M. Burk K. Matson W. Dichgans J. Beal MF. Bogdanov MB. Oxidative stress in patients with Friedreich ataxia. Neurology. 2000;55:1719–1721. doi: 10.1212/wnl.55.11.1719. [DOI] [PubMed] [Google Scholar]
- 68.Shan Y. Napoli E. Cortopassi G. Mitochondrial frataxin interacts with ISD11 of the NFS1/ISCU complex and multiple mitochondrial chaperones. Hum Mol Genet. 2007;16:929–941. doi: 10.1093/hmg/ddm038. [DOI] [PubMed] [Google Scholar]
- 69.Shidara Y. Hollenbeck PJ. Defects in mitochondrial axonal transport and membrane potential without increased reactive oxygen species production in a Drosophila model of Friedreich ataxia. J Neurosci. 2010;30:11369–11378. doi: 10.1523/JNEUROSCI.0529-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Shih AY. Johnson DA. Wong G. Kraft AD. Jiang L. Erb H. Johnson JA. Murphy TH. Coordinate regulation of glutathione biosynthesis and release by Nrf2-expressing glia potently protects neurons from oxidative stress. J Neurosci. 2003;23:3394–3406. doi: 10.1523/JNEUROSCI.23-08-03394.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sparaco M. Gaeta LM. Santorelli FM. Passarelli C. Tozzi G. Bertini E. Simonati A. Scaravilli F. Taroni F. Duyckaerts C. Feleppa M. Piemonte F. Friedreich's ataxia: oxidative stress and cytoskeletal abnormalities. J Neurol Sci. 2009;287:111–118. doi: 10.1016/j.jns.2009.08.052. [DOI] [PubMed] [Google Scholar]
- 72.Stehling O. Elsasser HP. Bruckel B. Muhlenhoff U. Lill R. Iron-sulfur protein maturation in human cells: evidence for a function of frataxin. Hum Mol Genet. 2004;13:3007–3015. doi: 10.1093/hmg/ddh324. [DOI] [PubMed] [Google Scholar]
- 73.Tan G. Napoli E. Taroni F. Cortopassi G. Decreased expression of genes involved in sulfur amino acid metabolism in frataxin-deficient cells. Hum Mol Genet. 2003;12:1699–1711. doi: 10.1093/hmg/ddg187. [DOI] [PubMed] [Google Scholar]
- 74.Thierbach R. Schulz TJ. Isken F. Voigt A. Mietzner B. Drewes G. von Kleist-Retzow JC. Wiesner RJ. Magnuson MA. Puccio H. Pfeiffer AF. Steinberg P. Ristow M. Targeted disruption of hepatic frataxin expression causes impaired mitochondrial function, decreased life span and tumor growth in mice. Hum Mol Genet. 2005;14:3857–3864. doi: 10.1093/hmg/ddi410. [DOI] [PubMed] [Google Scholar]
- 75.Traber MG. Atkinson J. Vitamin E, antioxidant and nothing more. Free Radic Biol Med. 2007;43:4–15. doi: 10.1016/j.freeradbiomed.2007.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Varija D. Kumar KP. Reddy KP. Reddy VK. Prolonged constriction of sciatic nerve affecting oxidative stressors & antioxidant enzymes in rat. Indian J Med Res. 2009;129:587–592. [PubMed] [Google Scholar]
- 77.Vazquez-Manrique RP. Gonzalez-Cabo P. Ros S. Aziz H. Baylis HA. Palau F. Reduction of Caenorhabditis elegans frataxin increases sensitivity to oxidative stress, reduces lifespan, and causes lethality in a mitochondrial complex II mutant. FASEB J. 2006;20:172–174. doi: 10.1096/fj.05-4212fje. [DOI] [PubMed] [Google Scholar]
- 78.Wong A. Yang J. Cavadini P. Gellera C. Lonnerdal B. Taroni F. Cortopassi G. The Friedreich's ataxia mutation confers cellular sensitivity to oxidant stress which is rescued by chelators of iron and calcium and inhibitors of apoptosis. Hum Mol Genet. 1999;8:425–430. doi: 10.1093/hmg/8.3.425. [DOI] [PubMed] [Google Scholar]
- 79.Wong A. Yang J. Danielson S. Gellera C. Taroni F. Cortopassi G. Sensitivity of FRDA lymphoblasts to salts of transition metal ions. Antioxid Redox Signal. 2000;2:461–465. doi: 10.1089/15230860050192233. [DOI] [PubMed] [Google Scholar]
- 80.Wu KC. Cui JY. Klaassen CD. Beneficial role of Nrf2 in regulating NADPH generation and consumption. Toxicol Sci. 2011;123:590–600. doi: 10.1093/toxsci/kfr183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ye H. Jeong SY. Ghosh MC. Kovtunovych G. Silvestri L. Ortillo D. Uchida N. Tisdale J. Camaschella C. Rouault TA. Glutaredoxin 5 deficiency causes sideroblastic anemia by specifically impairing heme biosynthesis and depleting cytosolic iron in human erythroblasts. J Clin Invest. 2010;120:1749–1761. doi: 10.1172/JCI40372. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.