Streptomyces is a very important industrial bacterium, which produces two thirds of clinically relevant secondary metabolites. It is considered as a “multicellular” prokaryotic model that includes programmed cell death (PCD) and sporulation in solid cultures. Streptomyces industrial fermentations are mainly produced in liquid cultures (large bioreactors) conditions in which most species do not sporulate, and it was traditionally considered that there was no differentiation. It has been almost unanimously accepted that mycelial morphology in liquid fermentation was correlated with the production of secondary metabolites, albeit the cause-effect relationship was controversial: some authors hold that cellular aggregation, and hence pellet and clump formation is fundamental to obtaining good production of secondary metabolites. On the other hand, other authors state that there is no relationship between morphology and secondary metabolite production. In conclusion, there was no general consensus correlating morphology with secondary metabolite production, and the lack of a consistent developmental model in Streptomycetes hindered the precise description of reliable phenotypes to analyse and optimize industrial fermentations.
New drugs, especially antibiotics, are urgently required in clinic and the most propitious natural source remained in environmental bacteria, especially Streptomyces. The screening for new secondary metabolites was traditionally performed by means of strain isolations, cultures and bioassays [1], a process tremendously productive during the “Golden Age” of antibiotics (1940s–1960s), but which was becoming more and more difficult once the most common antibiotics were already discovered. The main handicap of this screening approach was the high number of false negatives: i.e. strains discarded as bioactive compound producers, because they did not reached the producing phase in the lab. At present, most research groups working in the search for new bioactive compounds are making it by means of metagenomics: large genome sequencing and identification of biosynthetic pathways by sequence homology. However, this genetic approach is totally dependent on a previous knowledge in order to find homologies, and it is not suitable to discover totally new bioactive compounds.
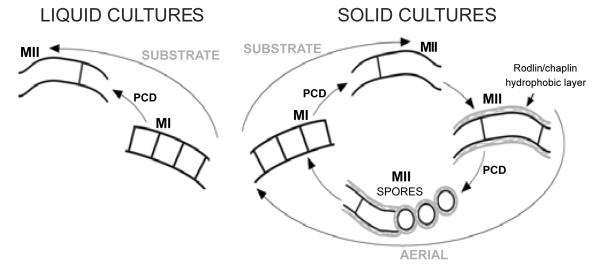
The classical Streptomyces developmental model for confluent solid cultures assumed that differentiation processes took place along the transversal axis of the cultures (bottom-up): completely viable vegetative mycelia (substrate) grew on the surface and inside agar until they underwent a PCD, after which hyphae differentiated to a reproductive (aerial) mycelium characterized by the presence of hydrophobic covers, giving it a characteristic greyish appearance. Substrate and aerial mycelia were multinucleated, but at the end of the cycle, aerial hyphae formed septa and spore chains (Figure 1). In liquid cultures, most Streptomycetes did not sporulate, and it was assumed that there was no differentiation. This developmental model was refined in recent years, describing novel aspects during the pre sporulation phases in solid and liquid cultures [2]. The existence of a previously unidentified compartmentalized mycelium (MI) that initiates the developmental cycle after spore germination was described. The MI suffers a highly ordered cell death (PCD), and the remaining viable segments of this mycelium begin to enlarge as a multinucleated mycelium (MII). In solid cultures, two types of MII have been defined, based on the absence (in early development) or presence (in late development) of the hydrophobic layers characteristic of aerial hyphae. The traditionally denominated substrate mycelium corresponds to MII, lacking hydrophobic layers, and the aerial mycelium, to MII coated with these hydrophobic layers (Figure 1). The only mycelial phases present in liquid cultures were MI and MII without hydrophobic layers (Figure 1). In addition, it was demonstrated that MII is the antibiotic producer mycelium, being the first time than antibiotic production could be associated to differentiation in Streptomyces liquid cultures.
Figure 1. Streptomyces developmental cycle in solid (sporulating) and liquid (non-sporulating) cultures.
MI, first compartmentalized mycelium, vegetative; MII, second multinucleated mycelium, antibiotic producer. The classical nomenclature of substrate and aerial mycelium, and the hydrophobic layers are indicated.
The new Streptomyces developmental cycle opens a new background in which study differentiation, and it will be the key for the analysis and optimization of hyphae differentiation in industrial fermentations [3]. In addition, finding new secondary metabolites is becoming critical in biomedicine, and the only way will be to resume screening from natural strains. The latest discoveries about Streptomyces differentiation will be crucial to design new experimental approaches to optimize the selection process, and reduce the incidence of false negatives.
Acknowledgment
Authors would like to thank to the Erupean Research Council (ERC Starting Grant Strp-differentiation 280304) and to the DGI, Subdirección General de Proyectos de investigación, MICINN (BIO2010-16303) for funding.
References
- 1.Donadio S, Maffioli S, Monciardini P, Sosio M, Jabes D. Antibiotic discovery in the twenty-first century: current trends and future perspectives. J Antibiot (Tokyo) 2010;63:423–430. doi: 10.1038/ja.2010.62. [DOI] [PubMed] [Google Scholar]
- 2.Manteca A, Alvarez R, Salazar N, Yague P, Sanchez J. Mycelium differentiation and antibiotic production in submerged cultures of Streptomyces coelicolor. Appl Environ Microbiol. 2008;74:3877–3886. doi: 10.1128/AEM.02715-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yague P, Manteca A, Simon A, Diaz-Garcia ME, Sanchez J. New method for monitoring programmed cell death and differentiation in submerge Streptomyces cultures. Appl Environ Microbiol. 2010;76:3401–3404. doi: 10.1128/AEM.00120-10. [DOI] [PMC free article] [PubMed] [Google Scholar]