Abstract
We demonstrate that the binding sites for highly conserved transcription factors vary extensively between human and mouse. We mapped the binding of four tissue-specific transcription factors (FOXA2, HNF1A, HNF4A, HNF6) to 4,000 orthologous gene pairs in hepatocytes purified from human and mouse livers. Despite the conserved function of these factors, from 41% to 89% of their binding events appear to be species-specific. When the same protein binds the promoters of orthologous genes, approximately two-thirds of the binding sites do not align.
Elements of transcriptional regulation have central roles in evolution 1-3. In many cases, conserved biological processes are controlled by evolutionarily conserved regulatory programs while evolving phenotypes are associated with cross-species variation in transcription regulation4. However, in the absence of suitable genome-wide data, it is unclear what fraction of all protein-DNA interactions are under either positive or negative selective pressure1. A preliminary effort to compare genome-wide binding sites for two stem cell-specific transcription factors in human and mouse has suggested that large differences exist between mouse and human 5, 6 yet because the data were obtained using different methodologies, there remains the possibility that observed changes are the result of purely technical differences.
To compare systematically the binding of transcriptional regulators to promoter regions across species, we designed carefully matched ChIP-chip experiments 7 in human and mouse. We created custom DNA microarrays that array ten kilobases of sequence surrounding the known transcription start sites of over 4,000 orthologous pairs of mouse and human genes. These genes were selected because their orthology could be unambiguously assigned and oligonucleotides could be designed to represent the putative regulatory regions at high density (Figure 1A, Supplementary Methods). Forty-seven hand-curated, tissue-specific genes were included in the array design as controls.
Figure 1.
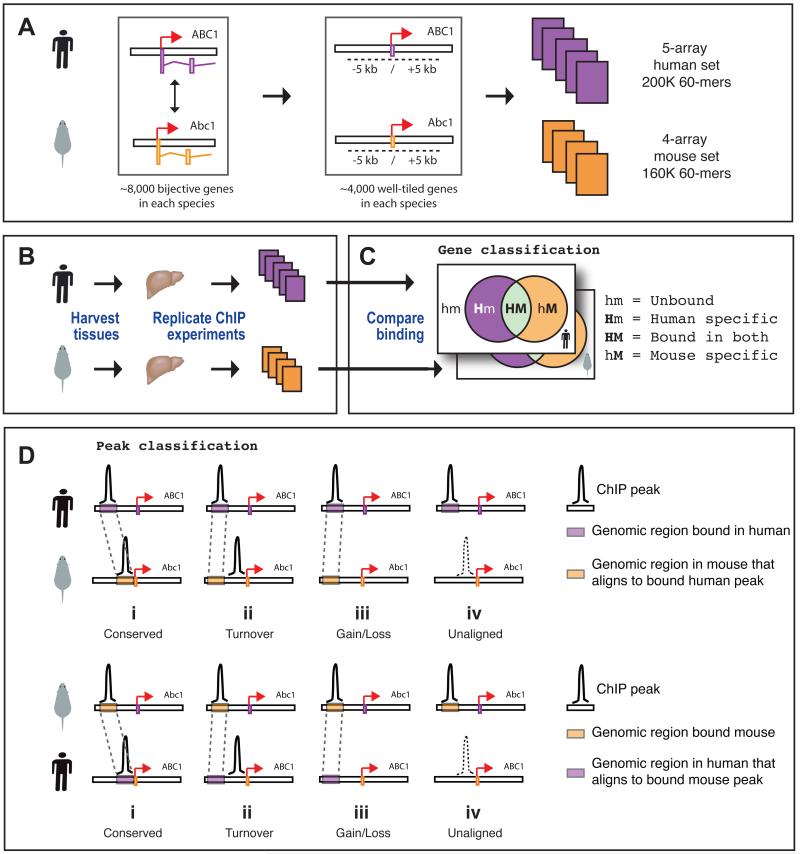
Strategy to analyze transcription factor-DNA interactions in mouse and human. (A) Panel 1: approximately 8,000 high-confidence human (purple) and mouse (orange) gene orthologs were identified. Panel 2: 60-mer oligonucleotides were designed against a ten kilobase region centered around the complete set of transcription start sites in both species (colored boxes on genome track); orthologous genes with incomplete coverage, low oligonucleotide quality, or substantial gaps in one or both species were removed from the final design (Supplementary Methods). Panel 3: a human 5-array set and a mouse 4-array set capturing the transcription start sites for approximately 4,000 genes in each species were created using these oligonucleotides. (B) Mouse and human hepatocytes were isolated from liver samples and used in chromatin immunoprecipitations, which were hybridized against the array sets. (C) Gene-centric analysis classifies orthologous gene pairs by whether they are not bound in either species (hm), bound uniquely in human (Hm), bound in both species (HM), or bound uniquely in mouse (hM). (D) Peak-specific analysis classifies peaks relative to whether corresponding aligned regions exist in the second species and whether these aligned regions are bound. The four possible outcomes are shown in both the human-to-mouse and the mouse-to-human panels: In the first three cases (i, ii, iii) the aligned locus is present in the arrayed region of the ortholog. Case i (conserved): the aligned regions are bound in both species; Case ii (turnover): the orthologous gene is bound, but not at the aligned locus; Case iii (gain/loss): no binding is detected in the arrayed region of the second species, including the aligned sequence; Case iv (unaligned): the aligned sequence is not present within the arrayed region and therefore we cannot definitively classify the binding event, regardless of the presence or absence of a binding event in the other species.
Chromatin immunoprecipitations were performed independently in primary hepatocytes directly isolated from mouse and human liver using antibodies against four tissue-specific transcription factors (FOXA2, HNF1A, HNF4A, HNF6) involved in liver development and regulation (Figure 1B, Table S1) 7. Hepatocytes were chosen as a representative tissue for these experiments because (1) they are functionally and structurally conserved among mammals 8; (2) their gene expression programs are similar across species (Table S1); (3) their gene expression patterns are largely unperturbed by isolation procedures 9; and (4) the transcription factors responsible for hepatocyte development and function are highly conserved 8. We amplified and fluorescently labeled the DNA from these binding experiments, hybridized it to the microarrays, and then scored binding events 10.
Several possible outcomes can be distinguished when comparing a binding event in one species with the data from the second species (Figure 1). First, one can determine if a particular transcription factor binds anywhere within the arrayed region of the human and/or mouse ortholog (gene-centric approach) (Figure 1C). Second, one can determine if the positions of individual binding event are maintained, to the resolution limits of the ChIP assay (peak-centric approach). Since DNA sequences may have undergone rearrangements between human and mouse, we consider whether a binding event detected in one species occurs at the corresponding aligned region in the second species, resulting in the four possible outcomes (Figure 1D).
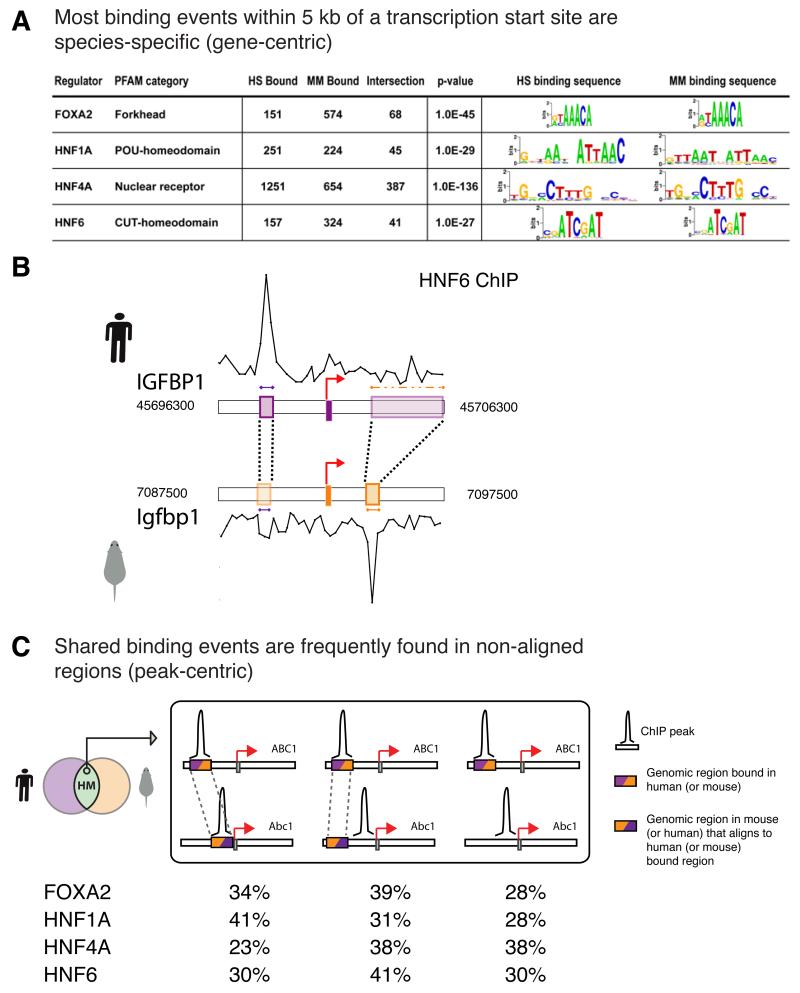
Surprisingly, we found that between 41% and 89% of the orthologous promoters bound by a protein in one species are not bound by the same protein in the second species, depending on the transcription factor (Figure 2A, Figure S1, Table S2). In some of these cases a transcription factor may continue to regulate both orthologs through binding sites that lie beyond the greater than ten kilobases of promoter sequence represented on our arrays. The sets of genes pairs with promoters that are bound in both species by each factor (HM category from Figure 1B) are significantly enriched for an independently determined set of liver-specific genes (Figure S1), consistent with known functional conservation of the transcription factors we profiled. The extent of species-specific binding is much greater than would be expected based on our experimentally determined error rates and does not depend on the computational techniques used to identify bound regions (Figure S2, Table S2).
Figure 2.
(A) Number of genes bound by liver master regulators in each species, p-value (using a hypergeometric distribution) that the cross-species overlap is due to random chance, and the THEME-derived binding motifs in human and mouse. (B) The location of binding events varies between species. Here, ChIP enrichments are shown as traces. The 500 base pair sequence underlying the ChIP peak in each species is colored by species (purple human, orange mouse) and aligned with the corresponding sequence in the second species using dashed lines. For clarity, mouse ChIP enrichments are displayed as a negative y-axis, but orientation of the transcription start site is left to right. IGFPB1 is bound by HNF6 in both species, but the binding events do not align. The human sequence aligned to the mouse HNF6 peak in IGFBP1 contains large insertions overlapping a substantial portion of the human first intron (outlined with a dashed orange box), and is not bound by HNF6. (C) Shared binding events are frequently found in non-aligned regions. From left to right: aligned regions (shown as colored boxes) that are bound in both species (Figure 1D, case i); aligned regions present on both human and mouse arrays but bound only in one species (Figure 1D, case ii); regions bound in both species, but lacking aligned sequences on the orthologous array (Figure 1D, case iv, with a binding peak present). Typically, only about a third of the binding events detected in both species occur in sequences that align to each other (Table S3).
To estimate the maximum variation in binding that could be attributed to environmental and intra-species genetic (as opposed to inter-species evolutionary) sources, we compared HNF6 genomic occupancy in primary human hepatocytes to corresponding HNF6 occupancy in the human cell line HepG2 (Table S2). Despite the fact that HepG2 cells are an immortalized hepatocellular carcinoma that is severely aneuploid and which has been propagated in culture for over two decades 11, we found that 66% of the genes bound in primary human liver were bound in HepG2. In contrast, only 26% of orthologous gene pairs bound by HNF6 in human hepatocytes are also bound in orthologous regions in mouse hepatocytes.
Using the THEME algorithm 12 we determined that the observed changes in binding patterns across species do not arise from changes in the DNA-binding specificity of the transcription factors, and transcription factor binding in each species is highly correlated with the presence of sequences matching the protein’s motif (Table S3, Figure S3). To determine whether binding differences between orthologs arise from sequence differences at potential binding sites, we scanned previously reported mouse-human genome alignments13 for conserved motif sequences. As expected, the frequency of conserved motif sequences near binding peaks is highest for conserved peaks (case i; Figure 1C); the frequency of conserved motif sequences is lower near binding events that are unique to one species, but still above background (cases ii (turnover) and iii (gain/loss)) (Table S3). The conserved sequences that are not bound in our assay may be functional in both species under particular conditions, during alternative developmental stages, or in tissues not analyzed in our study.
Most crucially, the location of binding events varies widely between species in ways that cannot be predicted from human-mouse sequence alignments alone. For instance, the binding site for HNF6 at IGFBP1 shifts over four kilobases from the promoter region in human to the first intron in mouse (Figure 2B). More broadly, of the 41 orthologous pairs of promoters that are bound by HNF1A in both species, there are 47 binding events in human and 51 binding events in mouse. Of these, only 20 occur in sequences that are aligned to each other. The fraction of aligned binding events is even lower for other factors (Figure 2C, Table S3).
Our findings have implications for the use of mouse as a model organism. For example, HNF1A bound strongly to SEL1L in human liver, yet this binding is entirely absent from the corresponding mouse region (Figure S1). Polymorphisms around the SEL1L locus appear to influence the onset of disease in individuals with MODY3 diabetes, which is caused by haploinsufficiency of HNF1A 14. The lack of HNF1A binding in mouse suggests that this susceptibility may be species-specific. In contrast to the variation in cross-species binding sites, the location of binding events within a species is robust to substantial environmental and genetic perturbations. Of genes bound in both human hepatocytes and the human carcinoma cell line HepG2, over 95% had peaks within 100 nucleotides of each other (Table S2).
The in vivo binding of four distinct tissue-specific transcription factors (FOXA2, HNF1A, HNF4A, and HNF6) responsible for liver gene expression has diverged substantially between human and mouse. The most striking feature of this divergence is the high mobility of transcription factor binding sites. Analyzing genomic regions that are bound by the same factors in both species reveals that approximately two-thirds of the binding events are not aligned between the mouse and human genomes. The cross-species variation cannot be explained by changes in the sequence specificity of the transcription factors, nor can it be predicted based solely on the conservation of binding sequences in the two species. Other effects, including the concentration of these transcription factors, other interacting proteins and chromatin modifications are likely to contribute to the observed variations 15 (Supplementary Note). Differences between human and mouse physiology and behavior may also contribute to the observed binding changes, and these physiological and behavioral differences will affect all studies that use mouse as a model for human biology. The striking and unexpected plasticity of transcription factor binding indicates that attempts to map accurately functional genomic elements responsible for gene expression will require direct measurements of transcription factor occupancy in multiple species.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to S. Strom and K. Dorko (U-Pittsburg) for human liver samples (DK92310); T. DiCesare, S. Gupta, R. Kumar, J. Rodriguez, K. Walker for technical assistance; R. Young and G. Gerber for helpful discussions. Supported by funding from NIH (DK68655, DK70813: DTO; DK076284: RDD), Cancer Research UK (DTO), and the Whitaker Foundation (EF).
Footnotes
DATA ACCESSION NUMBER
ArrayExpress E-TABM-108, publicly released.
SUPPORTING WEBSITE
- Analysis files:
-
○Binary binding call files for all genes using two error models with two binding thresholds for each method
-
○Motif discovery input files
-
○
- Downloadable, .pdf file with graphs of binding data for all curated tissue-specific genes
REFERENCES
- 1.Bird CP, Stranger BE, Dermitzakis ET. Functional variation and evolution of non-coding DNA. Curr Opin Genet Dev. 2006;16:559–64. doi: 10.1016/j.gde.2006.10.003. [DOI] [PubMed] [Google Scholar]
- 2.Moses AM, et al. Large-Scale Turnover of Functional Transcription Factor Binding Sites in Drosophila. PLoS Comput Biol. 2006:2. doi: 10.1371/journal.pcbi.0020130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Prabhakar S, Noonan JP, Paabo S, Rubin EM. Accelerated evolution of conserved noncoding sequences in humans. Science. 2006;314:786. doi: 10.1126/science.1130738. [DOI] [PubMed] [Google Scholar]
- 4.King MC, Wilson AC. Evolution at two levels in humans and chimpanzees. Science. 1975;188:107–16. doi: 10.1126/science.1090005. [DOI] [PubMed] [Google Scholar]
- 5.Boyer LA, et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–56. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Loh YH, et al. The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat Genet. 2006;38:431–40. doi: 10.1038/ng1760. [DOI] [PubMed] [Google Scholar]
- 7.Odom DT, et al. Core transcriptional regulatory circuitry in human hepatocytes. Mol Syst Biol. 2006;2:2006–0017. doi: 10.1038/msb4100059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zaret KS. Regulatory phases of early liver development: paradigms of organogenesis. Nat Rev Genet. 2002;3:499–512. doi: 10.1038/nrg837. [DOI] [PubMed] [Google Scholar]
- 9.Richert L, et al. Gene expression in human hepatocytes in suspension after isolation is similar to the liver of origin, is not affected by hepatocyte cold storage and cryopreservation, but is strongly changed after hepatocyte plating. Drug Metab Dispos. 2006;34:870–9. doi: 10.1124/dmd.105.007708. [DOI] [PubMed] [Google Scholar]
- 10.Qi Y, et al. High-resolution computational models of genome binding events. Nature Biotechnology. 2006 doi: 10.1038/nbt1233. [DOI] [PubMed] [Google Scholar]
- 11.Natarajan AT, Darroudi F. Use of human hepatoma cells for in vitro metabolic activation of chemical mutagens/carcinogens. Mutagenesis. 1991;6:399–403. doi: 10.1093/mutage/6.5.399. [DOI] [PubMed] [Google Scholar]
- 12.Macisaac KD, et al. A hypothesis-based approach for identifying the binding specificity of regulatory proteins from chromatin immunoprecipitation data. Bioinformatics. 2006;22:423–9. doi: 10.1093/bioinformatics/bti815. [DOI] [PubMed] [Google Scholar]
- 13.Schwartz S, et al. Human-mouse alignments with BLASTZ. Genome Res. 2003;13:103–7. doi: 10.1101/gr.809403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim SH, et al. Identification of a locus for maturity-onset diabetes of the young on chromosome 8p23. Diabetes. 2004;53:1375–84. doi: 10.2337/diabetes.53.5.1375. [DOI] [PubMed] [Google Scholar]
- 15.Guccione E, et al. Myc-binding-site recognition in the human genome is determined by chromatin context. Nat Cell Biol. 2006;8:764–70. doi: 10.1038/ncb1434. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.