Abstract
Purpose
In head and neck and bladder cancer tumor hypoxia is associated with a poor prognosis, hypoxia modification improves outcome and hypoxic status predicts benefit from treatment. Yet, there is no universal measure of clinical hypoxia. The aim of this study was to investigate whether a 26-gene hypoxia signature predicted benefit from hypoxia-modifying treatment in both cancer types.
Patients and Methods
Samples were available from 157 T2-T4 laryngeal cancer and 185 T1-T4a bladder cancer patients enrolled on the ARCON and BCON phase III randomized trials of radiotherapy alone or with carbogen and nicotinamide (CON) respectively. Customized TaqMan Low Density Arrays (TLDAs) were used to assess expression of the 26-gene signature using quantitative real-time PCR. The median expression of the 26 genes was used to derive a hypoxia score (HS). Patients were categorized as TLDA-HS low (≤median) or TLDA-HS high (>median). The primary outcome measures were regional control (RC; ARCON) and overall survival (OS; BCON).
Results
Laryngeal tumors categorized as TLDA-HS high showed greater benefit from ARCON than TLDA-HS low tumors. Five-year RC was 81% (radiotherapy alone) versus 100% (CON) for TLDA-HS high (P=0.009). For TLDA-HS low five-year RC was 91% (radiotherapy alone) versus 90% (CON) (P=0.90). TLDA-HS did not predict benefit from CON in bladder cancer.
Conclusion
The 26-gene hypoxia signature predicts benefit from hypoxia-modifying treatment in laryngeal cancer. These findings will be evaluated in a prospective clinical trial.
Keywords: Head and neck cancer, bladder cancer, hypoxia, gene signature, radiotherapy
INTRODUCTION
Tumor hypoxia confers resistance to radiotherapy and predicts for a poor treatment outcome (1, 2). A meta-analysis of clinical trials showed that there is level 1a evidence in favor of adding hypoxic modification to radiotherapy for head and neck squamous cell carcinoma (HNSCC) (3). Despite this, hypoxia-modifying therapies have not been adopted as an international standard of care. A recent randomized phase III trial compared accelerated radiotherapy (AR) alone or with the hypoxia-modifying agents carbogen and nicotinamide (ARCON) in laryngeal carcinoma. The ARCON trial showed a significant improvement in five-year regional control with CON (4). Evidence suggests that hypoxic tumors benefit most from hypoxia-modifying therapy (4-8). Similar results have been shown in bladder cancer. The BCON phase III trial showed that addition of CON to radiotherapy significantly improved overall survival in bladder carcinoma (9, 10).
Several methods are used to measure tumor hypoxia. Oxygen electrode (2, 11), imaging approaches (7, 12) and hypoxia-specific markers such as pimonidazole (4, 6, 13) require prospective assessment, limiting the ability to validate findings. Measurement of endogenous markers of hypoxia such as carbonic anhydrase-9 (CA9) (13-16) and hypoxia-inducible factor-1α (HIF-1α) (14, 17, 18) have shown some success. However, approaches using multiplex markers such as gene signatures (8, 19-21) potentially better reflect the complex cellular response to hypoxia and account for the intra-tumor heterogeneity of hypoxia. Previous studies showed the potential of a 15-gene hypoxia signature to predict benefit from hypoxia-modifying therapy in HNSCC (8). A 121-gene hypoxia meta-signature was derived from three independent head and neck cancer datasets. The full and a reduced 26-gene signature (Table 1) had prognostic significance in head and neck, breast and lung cancer (20). The 26-gene signature was taken forward for validation using a TaqMan Low Density Array (TLDA) approach. Technical validation of the signature showed the approach was a reliable and reproducible measure of tumor hypoxia associated with low intra-tumor heterogeneity; the multiplex nature of the biomarker allows buffering of artifact effects which often contribute to variability of single measures (21).
Table 1.
26-gene hypoxia signature
| Gene | Function |
|---|---|
|
| |
| ALDOA | Glucose metabolism |
| ANGPTL4 | Lipid and glucose metabolism |
| ANLN | Cytokenesis |
| BNC1 | Keratinocyte proliferation |
| C20orf20 | Cellular proliferation |
| CA9 | pH regulation |
| CDKN3 | Cellular proliferation |
| COL4A6 | Extracellular-matrix metabolism |
| DCBLD1 | Unknown |
| ENO1 | Glucose metabolism |
| FAM83B | Unknown |
| FOSL1 | Cellular proliferation |
| GNAI1 | Signal transduction |
| HIG2 | Stress response |
| KCTD11 | Apoptosis |
| KRT17 | Keratin production |
| LDHA | Glucose metabolism |
| MPRS17 | Mitochondrial translation |
| P4HA1 | Extracellular-matrix metabolism |
| PGAM1 | Glucose metabolism |
| PGK1 | Glucose metabolism |
| SDC1 | Cellular proliferation |
| SLC16A1 | Glucose metabolism |
| SLC2A1 | Glucose metabolism |
| TPI1 | Glucose metabolism |
| VEGFA | Angiogenesis |
Abbreviations: ALDOA, aldolase A; ANGPTL4, angiopoietin-like 4; ANLN, anillin; BNC1, basonuclin 1; C20orf20, chromosome 20 open reading frame 20; CA9, carbonic anhydrase 9; CDKN3, cyclin-dependent kinase inhibitor 3; COL4A6, collagen, type IV, alpha 6; DCBLD1, discoidin, CUB and LCCL domain containing 1; ENO1, enolase 1; FAM83B, family with sequence similarity 83, member B; FOSL1, FOS-like antigen 1; GNAI1, guanine nucleotide binding protein; HIG2, hypoxia-inducible gene 2; KCTD11, potassium channel tetramerisation domain containing 11; KRT17, keratin 17; LDHA, lactate dehydrogenase A; P4HA1, prolyl 4-hydroxylase; PGAM1, phosphoglycerate mutase 1; PGK1, phosphoglycerate kinase 1; SDC1, syndecan 1; SLC16A1, solute carrier family 16 member 1 (monocarboxylic acid transporter 1); SLC2A1, solute carrier family 2 (facilitated glucose transporter), member 1; TPI1, triosephosphate isomerase 1; VEGFA, vascular endothelial growth factor A.
The primary aim of this study was to investigate the ability of the 26-gene hypoxia signature to predict benefit from hypoxia modification using laryngeal and bladder cancer samples from the ARCON and BCON phase III clinical trials respectively. As a secondary objective, the prognostic ability of the gene signature was also investigated in patients receiving non experimental treatment. Owing to its prognostic ability in multiple cancer types (20), similar gene expression in both cancer types was assumed and subsequently examined.
MATERIALS AND METHODS
Patients, sample size determination and tissue samples
A retrospective study was carried out using formalin-fixed, paraffin-embedded (FFPE) pre-treatment samples obtained from two prospective phase III clinical trials of radiotherapy alone or with CON. The target sample size was 150 patients. Analysis of 150 ARCON patients was required to detect a difference in hazard ratios for hypoxic versus oxygenated tumors with P=0.002 and 90% power. 150 BCON patients were required to detect a difference in hazard rate with P=0.01 and 80% power.
First, tumor biopsies were obtained from 157 patients with T2-T4 laryngeal cancer who participated in the ARCON trial and randomized between April 2001 and February 2008. Samples from six centers in The Netherlands were obtained for 229 of the 345 patients enrolled in ARCON (Supplementary Fig. S1). Second, tumor resections were obtained from 185 patients with high grade T1-T4a transitional cell bladder cancer who participated in the BCON trial and randomized between November 2000 and April 2006. Samples from 11 UK hospitals were obtained for 251 of the 333 patients enrolled in BCON (Supplementary Fig. S2). The study was approved by the Greater Manchester Research Ethics Committee (LREC 09/H1011/40 and LREC 09/H1013/24 respectively). REMARK guidelines for reporting tumor marker studies were followed.
ARCON patients allocated to the accelerated radiotherapy (AR) arm received 68 Gy in 2 Gy fractions to the primary tumor over 36-38 days. Patients allocated to the ARCON arm received 64 Gy in 2 Gy fractions, carbogen (98% O2 + 2% CO2, four minutes before and during daily fractions) and oral nicotinamide (60 mg/kg, 1-1½ hours before each fraction). BCON patients received 55 Gy in 20 fractions in four weeks or 64 Gy in 32 fractions in 6½ weeks daily, five times per week. Carbogen was given five minutes before and during radiotherapy. Nicotinamide (40-60 mg/kg) was given 1½-2 hours before each fraction.
Histopathology
One 4 μm haemotoxylin and eosin (H&E)-stained section from each FFPE block was analyzed. Clinical staging followed TNM AJCC/UICC classifications and grading was according to the World Health Organization guidelines. As recognized prognostic features, tumor necrosis (10, 22) and concurrent carcinoma in situ (pTis) (23) were recorded (BCON). Samples with <10% tumor material were excluded from analysis.
RNA extraction and cDNA synthesis
RNA was extracted from FFPE samples (three 20 μm sections) using the RecoverAll Total Nucleic Acid Isolation Kit (Life Technologies, Warrington, UK), which included DNase I treatment. Reverse transcription of total RNA was performed using the Omniscript Reverse Transcription Kit (Qiagen, West Sussex, UK), oligo-dT primers (Qiagen) and random nonomers (Sigma-Aldrich, Dorset, UK). RNA yield from laryngeal biopsies was low due to small size, so preamplification was used to boost message. Preamplification of cDNA involved pooled TaqMan assays, Preamplification Master Mix (Life Technologies) and a PCR thermal cycler (Veriti 9902, Life Technologies).
Quantitative PCR and generation of hypoxia score
Customized 384-well microfluidic TLDA cards were produced by Life Technologies. Reactions were carried out using TaqMan Gene Expression Master Mix (Life Technologies). Real-time quantitative (qPCR) was performed using the ABI Prism 7900HT qPCR system as per manufacturer’s protocol. Manual Cq values were determined retrospectively using ABI Prism Sequence Detection System (SDS) software (Life Technologies). Relative quantification of gene expression was calculated using the 2−ΔCq method (24). Hypoxia scores (HS) were calculated as the normalized median expression of the 26 hypoxia genes. The geometric mean of three reference genes (GNB2L1, B2M, RPL11) was used for normalization. The three best performing genes (best correlations and lowest median Cq values) (Supplementary Fig. S3 and S4) were selected from five previously validated reference genes (21). A customized program was designed and used to automatically calculate TLDA-HS from SDS files [R package version 2.1.5 (25)]. The group median was used to stratify patients as TLDA-HS low (≤median) or high (>median). This cut-off had the most discriminative prognostic power in multiple cancer types in previous analyses (20). Minus reverse transcriptase and no template controls were analyzed using intron-spanning TaqMan assays (CA9, GNB2L1, RPL24; Life Technologies). All negative controls had Cq values >40 cycles. Scientific analysts were blinded to clinical outcome data.
Previous validation of the 26-gene hypoxia signature using fresh frozen HNSCC biopsies revealed one gene (CDC4A) to have non-linear PCR reaction efficiency, which was removed from the signature (21). This gene was subsequently replaced by KRT17, the next in rank (20). The TLDA-HS (25-gene versus 26-gene) for 33 biopsies (from nine tumors) was significantly paired (Wilcoxon P<0.0001) and there was no significant difference in intra-tumor variation in TLDA-HS. Median coefficient in variance was 24.8% (range 7.1-52.5%; 25 genes) versus 23.1% (range 10.1-49.8%; 26 genes) (paired Wilcoxon P=0.07; Supplementary Fig. S5).
Endpoints and statistics
Primary endpoints for analyses were those which reached significance in the clinical trials: regional control (RC; ARCON) and overall survival (OS; BCON). For ARCON patients, RC was defined as freedom from first regional recurrence. Secondary end points were local control (LC), disease-free survival (DFS), and OS. LC was defined as freedom from first recurrence at the site of primary tumor, DFS was defined as the time to local, regional or distant recurrence, and OS was defined as time to death from any cause. For BCON patients, OS was defined as death from any cause. The secondary end point of relapse-free survival (RFS) was also reported. RFS was taken as time to tumor recurrence in bladder (muscle invasive lesions only), locoregional failure, or death from any cause. Time was set to zero for those with persistent muscle invasive tumor after radiotherapy or if no cystoscopic assessment was ever performed. All endpoints were taken from randomization to event; patients were censored at the last time seen or at five years whichever was earlier. Median follow-up times for the cohorts studied were 59 (RT only) and 60 (CON) months and 61 (AR) and 61 (ARCON) months for BCON and ARCON patients, respectively.
Statistical analyses were performed using SPSS version 19 for Mac (Chicago, IL, USA). Survival estimates were performed using the Kaplan-Meier method and differences compared using the log rank test. Hazard ratios (HR) and 95% confidence intervals (CI) were obtained using Cox’s proportional hazard model. For ARCON patients, the number of events for these outcome parameters was low; hence multivariate analyses were not possible. For BCON patients, in multivariate analyses prognostic features were entered as covariates. The chi-square test was used to compare proportions across the levels of categorical factors and Yates’ correction was used for 2×2 tables; the Mann-Whitney U test was used to compare median values for continuous variables between two groups. Spearman’s rank correlation coefficients were used to assess statistical relationships. P values were two-sided and significance set to P ≤0.05.
RESULTS
Hypoxia gene expression in laryngeal cancer
Tumor sections from 229 patients were received from Nijmegen. The median RNA yield was 52.2 ng/μl (range 1.9-339.0 ng/μl). As a power calculation indicated 150 samples needed to be analyzed, 162 patient samples with the highest RNA quantity were selected. The samples were preamplified and TLDA-HS generated for 157 patients (five patient samples failed to generate a TLDA-HS; 96.9% success rate). Hypoxia gene expression for eight matched non-preamplified and preamplified samples correlated well (Spearman’s rank ρ=0.88, p<0.0001; Supplementary Fig. S6).
In the subset of 157 patients, 77 (49.0%) received AR and 80 (51.0%) ARCON (Table 2). There were similar clinical characteristics for patients in the subset and the main trial (Supplementary Table S1). In the AR arm, radiotherapy was given as planned to most patients (76; 98.7%). In the ARCON arm, 59 patients (73.8%) received all doses of radiotherapy and CON. All analyses were performed on an “intention to treat” basis. There were no statistically significant differences in clinicopathologic features between treatment arms. Supplementary Fig. S7 shows the distribution of TLDA-HS according to randomization arm. The median TLDA-HS for the 157 patients was 0.034 (range 1 × 10−5 to 0.19) with no statistically significant differences in clinicopathologic features between TLDA-HS high and low groups (Supplementary Table S2). TLDA-HS did not correlate with percent tumor in the samples analyzed (rho= −0.105, p=0.217; Supplementary Fig. S7).
Table 2.
ARCON patient clinicopathologic features by randomization arm
| Variable |
AR N=77 |
ARCON N=80 |
P |
|---|---|---|---|
| Gender | 0.06 | ||
| Male | 55 (71.4%) | 68 (85%) | |
| Female | 22 (28.6%) | 12 (15%) | |
| Age (years) | 59 (46-82) | 60 (44-84) | 0.69 |
| T stage | 0.64 | ||
| T2 | 24 (31.1%) | 28 (35.0%) | |
| T3 | 37 (48.1%) | 40 (50.0%) | |
| T4 | 16 (20.8%) | 12 (15.0%) | |
| N stage | 0.33 | ||
| N0 | 41 (53.2%) | 54 (67.5%) | |
| N1 | 14 (18.2%) | 9 (11.3%) | |
| N2a | 3 (3.9%) | 4 (5.0%) | |
| N2b | 5 (6.5%) | 2 (2.5%) | |
| N2c | 14 (18.1%) | 11 (13.7%) | |
| Subsite | 0.25 | ||
| Glottis | 23 (29.9%) | 32 (40%) | |
| Supraglottis | 54 (70.1%) | 48 (60%) | |
| Hb (g/L) | 9.1 (5.8-11.0) | 9.0 (6.1-10.5) | 0.64 |
| No data | 1 (1.3%) | 2 (2.5%) | |
| TLDA-HS | 0.035 (1 × 10−5 to .085) | 0.031 (9 × 10−5 to .19) | 0.72 |
Data represented as n (%) or median (range)
Abbreviations: AR, accelerated radiotherapy; ARCON, AR with carbogen and nicotinamide; Hb, hemoglobin; TLDA-HS, TaqMan Low Density Array-Hypoxia Score. NB. Percent tumor was >30% for all cases.
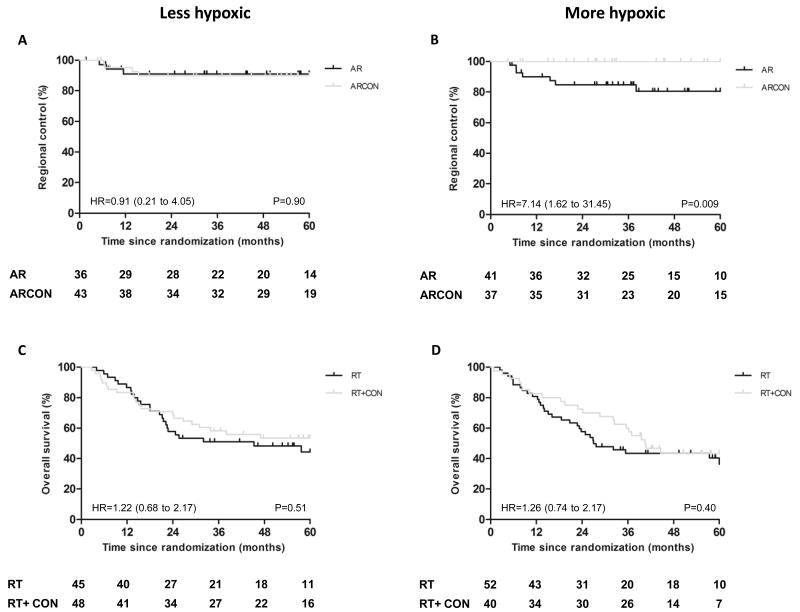
Patients with TLDA-HS high (“more hypoxic” tumors) receiving AR had a significantly lower five-year RC of 81% (7/7 events) compared to 100% (0 events) for those who received ARCON (log rank P=0.009). Patients with TLDA-HS low did not benefit from the addition of hypoxia-modifying therapy to radiotherapy. Respective five-year RC rates for AR and ARCON were 91% (3/7 events) and 90% (4/7 events) (log rank P=0.90) (Fig 1A & B). Patients with less hypoxic tumors (RC 90%) did less well with ARCON than those with more hypoxic tumors (RC 100%; p=0.056). A test for heterogeneity in treatment effect by TLDA-HS strata was not possible due to the low event rate; however survival analysis revealed a distinct difference in treatment efficacy according to each stratum so an interaction may be assumed. TLDA-HS did not predict benefit from hypoxia modification for five-year LC, DFS or OS.
Figure 1.
TLDA-HS predicts benefit from ARCON for laryngeal cancer. Kaplan-Meier plots for regional control of patients with A) less hypoxic (TLDA-HS ≤median) and B) more hypoxic (TLDA-HS >median) laryngeal tumors, and overall survival of patients with C) less hypoxic and D) more hypoxic bladder tumors. Hazard ratios (HR) (95% CI), log rank P values and numbers at risk at each yearly interval are shown.
For prognostication in AR patients, there was no significant difference in five-year RC between TLDA-HS high (80.5%, 7/10 events) and TLDA-HS low (91.0%, 3/10 events) (n=77; log rank P=0.29) (Fig 2A). Of the other clinicopathologic features only N stage was prognostic; increasing N stage reduced five-year RC (log rank P<0.001). TLDA-HS high was not an adverse prognostic factor for five-year LC or DFS. There was a trend towards poorer OS in patients with TLDA-HS high (40.7%, 23/34 events) versus TLDA-HS low (67.5%, 11/34 events) (log rank P=0.06) (Fig. 2B). There were no significant differences in clinicopathologic features between TLDA-HS high and low groups.
Figure 2.
Kaplan-Meier plots for A) regional control and B) overall survival of patients with laryngeal cancer, and C) overall survival and D) relapse-free survival of patients with bladder cancer. Patients stratified according to the median TLDA-HS. Hazard ratios (HR) (95% CI), log rank P values and numbers at risk at each yearly interval are shown.
Hypoxia gene expression in bladder cancer
RNA was extracted from tumor sections from 251 patient blocks available. The median RNA yield was 90.3 ng/μl (range 1.1-502.5 ng/μl). Samples from 41 patients were excluded for: low RNA yield (<33 ng/ul; 39 samples) or reclassification as T4b (two samples). In total, 185 TLDA-HS were generated (25 samples failed; 88.1% success rate). Higher RNA yields meant preamplification before TLDA was not necessary.
In the subset of 185 patients, 97 (52.4%) patients received radiotherapy (RT) and 88 (47.6%) patients RT+CON (Table 3). There were similar clinical characteristics for patients in the subset and the main trial (Supplementary Table S1). In the RT arm, radiotherapy was given as planned to all patients (97; 100%). In the RT+CON arm, 60 patients (68.2%) received all doses of radiotherapy and CON. There were no differences in clinicopathologic features between treatment arms, except a higher proportion of concurrent pTis cases in the RT+CON arm (chi squared P=0.002). Supplementary Fig. S7 shows the distribution of TLDA-HS according to randomization arm. The median TLDA-HS for the 185 patients was 0.021 (range 3 × 10−4 to 0.15). There were no statistically significant differences in clinicopathologic features in TLDA-HS high and low groups (Supplementary Table S3). TLDA-HS did not correlate with percent tumor in the samples analyzed (rho= 0.003, p=0.97; Supplementary Fig. S7).
Table 3.
BCON patient clinicopathologic features by randomization arm
| Variable |
RT n=97 |
RT+CON n=88 |
P |
|---|---|---|---|
| Gender | 0.48 | ||
| Male | 73 (75.3%) | 71 (80.7%) | |
| Female | 24 (24.7%) | 17 (19.3%) | |
| Age (years) | 75.0 (51.5-89.7) | 75.1 (51.5-90.5) | 0.83 |
| T stage | 0.63 | ||
| T1 | 4 (4.1%) | 4 (4.5%) | |
| T2 | 76 (78.4%) | 70 (79.6%) | |
| T3 | 16 (16.5%) | 11 (12.5%) | |
| T4a | 1 (1.0%) | 3 (3.4%) | |
| Necrosis | 0.19 | ||
| Absent | 48 (49.5%) | 53 (60.2%) | |
| Present | 49 (50.5%) | 35 (39.8%) | |
| Concurrent pTis | 0.002 | ||
| Absent | 30 (30.9%) | 10 (11.4%) | |
| Present | 67 (69.1%) | 78 (88.6%) | |
| Hb (g/L) | 13.7 (9.3-17.0) | 14.1 (9.8-17.2) | 0.46 |
| No data | 1 | 0 | |
| TLDA-HS | 0.022 (4 × 10−4 to .148) | 0.019 (3 × 10−4 to .062) | 0.37 |
| Percent tumor | 80% (15-95%) | 80% (30-100%) | 0.34 |
Data are represented as n (%) or median (range).
Abbreviations: RT, radiotherapy, CON, carbogen and nicotinamide; pTis, carcinoma in situ; Hb, hemoglobin; TLDA-HS, TaqMan Low Density Array-Hypoxia Score.
Prediction of treatment benefit from CON showed no significant improvement in five-year OS in patients with TLDA-HS high (36.3%, 31/53 events RT versus 43.4%, 22/53 events RT+CON, log rank P=0.40). Five-year OS was 44.3% (24/46 events) (RT) and 53.4% (22/46 events) (RT+CON) (log rank P=0.51) for TLDA-HS low (Fig. 1C & D). TLDA-HS did not predict benefit from hypoxia modification for five-year RFS.
For prognostication in RT patients, there was no significant difference in five-year OS between TLDA-HS high (35.4%, 29/55 events) and TLDA-HS low (44.0%, 26/55 events; P=0.33; Fig. 2C). Adverse prognostic features of OS were necrosis (log rank P=0.008) and concurrent pTis (log rank P=0.03), which were also independent predictors of poor OS (necrosis P=0.002 and pTis P=0.004). Patients with TLDA-HS high showed a trend towards a poorer RFS (39.0%, 24/41 events, TLDA-HS high versus 52.1%, 17/41 events, TLDA-HS low, log rank P=0.06) (Fig. 2D). Necrosis was the only adverse prognostic feature using this endpoint (log rank P=0.002, multivariate P=0.001). There were no significant differences in clinicopathologic features between TLDA-HS high and low groups.
Exploration of a new cut-off for TLDA-HS using all BCON patients (n=185) showed the lowest quartile (5-year OS 59.5%) had a better prognosis compared to the upper three quartiles (39.0%; log rank P=0.02; Supplementary Fig. S8). TLDA-HS was an independent predictor of 5-year OS (P=0.007 versus pTis P=0.009 and T stage P=0.01) and RFS (P=0.01 versus pTis P=0.009 and T stage P=0.02). Independent verification of this new cut-off using another cohort is necessary to draw conclusions.
Comparison of hypoxia gene expression in laryngeal and bladder cancers
Comparison of laryngeal and bladder cancer data showed similar expression in the two tissues types (Spearman’s rank ρ=0.73, P<0.0001), but greater differential expression in laryngeal samples (Fig. 3). The median fold increase of the 26 genes (between highest and lowest five samples) was 57.2 (range 5.7-8.3 × 103; laryngeal) and 43.3 (range 6.2-1.3 × 103; bladder). For TLDA-HS fold increase was 6.5-fold higher in laryngeal than in bladder cancer (429.6 in laryngeal versus 66.5 in bladder; Mann-Whitney U P<0.0001).
Figure 3.
Gene by gene comparison of the 26-gene hypoxia signature in laryngeal and bladder cancer. A) Median expression of each gene is plotted for 157 ARCON laryngeal and 185 BCON bladder cancer patients. B) Heat map showing gene expression per sample. C) Fold increase in gene expression between lowest five and highest five (▲) ARCON and (●) BCON patient samples.
DISCUSSION
Clinical trials of hypoxia-modifying treatments have not allocated treatment according to hypoxic status despite large inter-tumor variability in hypoxia (13, 26, 27). In this study, classification of patients as “more” or “less” hypoxic based upon expression of a 26-gene hypoxia signature showed that hypoxic laryngeal tumors benefited from hypoxia-modifying CON treatment. Patients had a 19% improvement in five-year RC rate versus those who received radiotherapy alone. Although nodal bulk at diagnosis was an important predictor of poor prognosis and a multivariate analysis could not be performed, there was no difference in N stage distribution for patients with TLDA-HS high versus low scores (p=0.90) suggesting that the apparent benefit of hypoxia modification was not due to patients in the hypoxia group having smaller bulk nodal disease at diagnosis.
In a recent study of 323 HNSCC patients, classification of patients as more hypoxic based upon expression of a 15-gene hypoxia signature independently predicted benefit from the hypoxic radiosensitizer nimorazole (8). Four genes (P4HA1, SLC2A1, KCTD11, ALDOA) overlap with the 26-gene HNSCC hypoxia signature.
Due to the prominent role the larynx plays in swallowing, respiration, communication and protection of the lower airway, quality-of-life considerations are paramount. The larynx preservation rate of 87% for patients receiving ARCON is comparable with patients receiving radiotherapy and concurrent cisplatin (28). In addition, AR and ARCON produce equal levels of toxicity (4). ARCON patients with more hypoxic tumors as measured by TLDA-HS even had a five-year RC of 100%. A previous translational study using the hypoxia marker pimonidazole in 79 ARCON patients also demonstrated this trend (4) indicating the need for a companion biomarker for treatment selection. Lack of translation of treatment benefit from RC to OS or DFS in ARCON patients reflects effective salvage therapy by laryngectomy and neck dissection and patient co-morbidities with consequent increased mortality. The lack of prediction for LC is consistent with the findings from the ARCON trial, which suggested that the lack of benefit from CON for LC might be due to the extra 4 Gy given in the radiotherapy alone arm (4).
Despite similar hypoxia gene expression profiles in laryngeal and bladder cancer, the 26-gene signature did not predict benefit from CON in bladder cancer patients. Differential gene expression is lower in bladder cancer and this reduced inter-tumor variation may explain the loss in predictive ability. It is known that a significant proportion of the transcriptional response to hypoxia is cell-type dependent (29, 30) The rest comprises a conserved hypoxia response independent of hypoxic output (20, 29-31). Chi et al. (2006) examined core hypoxia signatures in primary cells (30), Lendahl et al. (2009) identified a 30-gene core hypoxia signature in cancer cell lines (31) and Buffa et al. (2010) derived a 51-gene common hypoxia signature by creating co-expression networks in three head and neck and five breast cancer datasets (20). Comparison of these core hypoxia targets with our 26-gene HNSCC signature shows an overlap of only three genes (HIG2, P4HA1,VEGFA) with the Lendahl signature and 15 genes (CA9, MPRS17, SLC16A1, C20orf20, ENO1, ALDOA, TPI1, LDHA, SLC2A1, PGK1, P4HA1, VEGFA, PGAM1, ANLN, CDKN3) overlap with the Buffa signature. The highly conserved core hypoxia genes are primarily metabolic genes (e.g. PGK1, LDHA, ENO1, ALDOA). Future work using a conserved core signature or an independently derived bladder-specific signature is under investigation. It is likely that the different cell-specific response to hypoxia is at least in part due to variations in transactivation by HIF, the master transcriptional regulator of the hypoxia response, and epigenetic factors. Supplementary Fig. S9 demonstrates how the TLDA can minimize intra-tumor variation in hypoxia of bladder tumors as seen in HNSCC (27). Hence, modification of the signature for use in this patient population has great potential. Another possible explanation for the lack of predictive value of the signature in bladder cancer is that the patterns of failure are different from laryngeal cancer patients (mainly locoregional versus <10% locoregional for bladder). Although hypoxia modification improves locoregional control and survival, the head and neck cancer meta-analysis showed the benefit for overall survival was less than for locoregional control (3).
This study strongly supports the growing body of evidence advocating pre-treatment measurement of tumor hypoxia for selection of patients most likely to benefit from hypoxia modification in HNSCC. Measurement of the TLDA-HS is a simple procedure; most hospital laboratories are equipped and can perform routine qPCR analyses. There is a high success rate in generating data – 96.9% with preamplification. Multi-gene tests are already available in outpatient settings for breast (32-36) and colon (37) cancer. It was estimated that 12,360 people were diagnosed with and 3,650 people died of laryngeal cancer and 73,510 were diagnosed with and 14,880 people died of bladder cancer in the USA in 2012 (38). Use of this multi-gene test could potentially assist in providing treatment options for patients with hypoxic tumors.
Supplementary Material
STATEMENT OF TRANSLATIONAL RELEVANCE.
The ARCON trial showed that hypoxia-modification improved regional control in patients with laryngeal cancer by 7%. Similarly, the BCON trial showed that hypoxia modification improved overall survival in patients with high risk bladder cancer by 13%. However, growing evidence shows that only hypoxic tumors benefit. Expression of a hypoxia 26-gene signature was examined for its ability to reflect tumor hypoxia and predict benefit from hypoxia-modifying treatment in both cancer types. In laryngeal cancer dichotomization of patients using the gene signature showed those with high expression (>median) exhibited a significant improvement in regional control when treated with radiotherapy plus hypoxia-modification versus radiotherapy alone. This trend was not observed in patients with low expression (≤median) or in patients with bladder cancer. These observations open prospects for future use of the hypoxia gene signature as a predictive assay for selection of patients for hypoxia modification therapies.
ACKNOWLEDGMENTS
The authors thank ARCON and BCON trial investigators, the University of Manchester Clinical Immunology and Molecular Medicine Group for use of Good Clinical Practice facilities, and Ric Swindell for statistical support.
ROLE OF THE FUNDING SUPPORT The work was supported by the Medical Research Council of the UK (grant G0801525), Cancer Research UK (grant C480/A12328), EU Metoxia, and the NIHR Biomedical Research Centre, Oxford. Study sponsors had no involvement in study design, collection, analysis and interpretation of data, or writing or submission of the manuscript.
Financial support: AE, NM, JJI, GNJB and CMLW were funded by the Medical Research Council of the UK (grant G0801525). CMLW was also funded by Cancer Research UK (grant C1094/A11365) and the Experimental Cancer Medicine Centre. JT and CJM were funded by Cancer Research UK (grant C480/A12328). FMB and ALH were funded by Cancer Research UK, EU Metoxia and the NIHR Biomedical Research Centre, Oxford.
Footnotes
Authors’ disclosures of potential conflicts of interest CMLW, ALH, CJM AND FMB are registered as inventors of a patent covering the use of the hypoxia gene signature. The patent reference number is WO 011/076895 A1 and is fully accessible at www.patentlens.net including the list of genes comprising the signature. The patent is registered in the name of Cancer Research Technology. None of the authors have gained financially from the patent and no commercial funding has been provided in the generation of data for this paper.
REFERENCES
- 1.Brizel DM, Dodge RK, Clough RW, Dewhirst MW. Oxygenation of head and neck cancer: changes during radiotherapy and impact on treatment outcome. Radiother Oncol. 1999;53(2):113–7. doi: 10.1016/s0167-8140(99)00102-4. [DOI] [PubMed] [Google Scholar]
- 2.Nordsmark M, Bentzen SM, Rudat V, Brizel D, Lartigau E, Stadler P, et al. Prognostic value of tumor oxygenation in 397 head and neck tumors after primary radiation therapy. An international multi-center study. Radiother Oncol. 2005;77(1):18–24. doi: 10.1016/j.radonc.2005.06.038. [DOI] [PubMed] [Google Scholar]
- 3.Overgaard J. Hypoxic modification of radiotherapy in squamous cell carcinoma of the head and neck--a systematic review and meta-analysis. Radiother Oncol. 2011;100(1):22–32. doi: 10.1016/j.radonc.2011.03.004. [DOI] [PubMed] [Google Scholar]
- 4.Janssens GO, Rademakers SE, Terhaard CH, Doornaert PA, Bijl HP, van den Ende P, et al. Accelerated radiotherapy with carbogen and nicotinamide for laryngeal cancer: results of a phase III randomized trial. J Clin Oncol. 2012;30(15):1777–83. doi: 10.1200/JCO.2011.35.9315. [DOI] [PubMed] [Google Scholar]
- 5.Overgaard J, Eriksen JG, Nordsmark M, Alsner J, Horsman MR. Plasma osteopontin, hypoxia, and response to the hypoxia sensitiser nimorazole in radiotherapy of head and neck cancer: results from the DAHANCA 5 randomised double-blind placebo-controlled trial. Lancet Oncol. 2005;6(10):757–64. doi: 10.1016/S1470-2045(05)70292-8. [DOI] [PubMed] [Google Scholar]
- 6.Kaanders JH, Wijffels KI, Marres HA, Ljungkvist AS, Pop LA, van den Hoogen FJ, et al. Pimonidazole binding and tumor vascularity predict for treatment outcome in head and neck cancer. Cancer Res. 2002;62(23):7066–74. [PubMed] [Google Scholar]
- 7.Rischin D, Hicks RJ, Fisher R, Binns D, Corry J, Porceddu S, et al. Prognostic significance of [18F]-misonidazole positron emission tomography-detected tumor hypoxia in patients with advanced head and neck cancer randomly assigned to chemoradiation with or without tirapazamine: a substudy of Trans-Tasman Radiation Oncology Group Study 98.02. J Clin Oncol. 2006;24(13):2098–104. doi: 10.1200/JCO.2005.05.2878. [DOI] [PubMed] [Google Scholar]
- 8.Toustrup K, Sorensen BS, Nordsmark M, Busk M, Wiuf C, Alsner J, et al. Development of a hypoxia gene expression classifier with predictive impact for hypoxic modification of radiotherapy in head and neck cancer. Cancer Res. 2011;71(17):5923–31. doi: 10.1158/0008-5472.CAN-11-1182. [DOI] [PubMed] [Google Scholar]
- 9.Hoskin PJ, Rojas AM, Bentzen SM, Saunders MI. Radiotherapy with concurrent carbogen and nicotinamide in bladder carcinoma. J Clin Oncol. 2010;28(33):4912–8. doi: 10.1200/JCO.2010.28.4950. [DOI] [PubMed] [Google Scholar]
- 10.Eustace A, Irlam JJ, Taylor J, Denley H, Agrawal S, Choudhury A, Ryder WD, Ord JJ, Harris AL, Rojas AM, Hoskin PJ, West CML. Necrosis predicts benefit from hypoxia-modifying therapy in high risk bladder cancer. Submitted for publication. [DOI] [PMC free article] [PubMed]
- 11.Nordsmark M, Overgaard M, Overgaard J. Pretreatment oxygenation predicts radiation response in advanced squamous cell carcinoma of the head and neck. Radiother Oncol. 1996;41(1):31–9. doi: 10.1016/s0167-8140(96)91811-3. [DOI] [PubMed] [Google Scholar]
- 12.Mortensen LS, Johansen J, Kallehauge J, Primdahl H, Busk M, Lassen P, et al. FAZA PET/CT hypoxia imaging in patients with squamous cell carcinoma of the head and neck treated with radiotherapy: Results from the DAHANCA 24 trial. Radiother Oncol. 2012;105(1):14–20. doi: 10.1016/j.radonc.2012.09.015. [DOI] [PubMed] [Google Scholar]
- 13.Hoogsteen IJ, Lok J, Marres HA, Takes RP, Rijken PF, van der Kogel AJ, et al. Hypoxia in larynx carcinomas assessed by pimonidazole binding and the value of CA-IX and vascularity as surrogate markers of hypoxia. Eur J Cancer. 2009;45(16):2906–14. doi: 10.1016/j.ejca.2009.07.012. [DOI] [PubMed] [Google Scholar]
- 14.Schrijvers ML, van der Laan BF, de Bock GH, Pattje WJ, Mastik MF, Menkema L, et al. Overexpression of intrinsic hypoxia markers HIF1alpha and CA-IX predict for local recurrence in stage T1-T2 glottic laryngeal carcinoma treated with radiotherapy. Int J Radiat Oncol Biol Phys. 2008;72(1):161–9. doi: 10.1016/j.ijrobp.2008.05.025. [DOI] [PubMed] [Google Scholar]
- 15.Koukourakis MI, Giatromanolaki A, Sivridis E, Simopoulos K, Pastorek J, Wykoff CC, et al. Hypoxia-regulated carbonic anhydrase-9 (CA9) relates to poor vascularization and resistance of squamous cell head and neck cancer to chemoradiotherapy. Clin Cancer Res. 2001;7(11):3399–403. [PubMed] [Google Scholar]
- 16.Eriksen JG, Overgaard J. Lack of prognostic and predictive value of CA IX in radiotherapy of squamous cell carcinoma of the head and neck with known modifiable hypoxia: an evaluation of the DAHANCA 5 study. Radiother Oncol. 2007;83(3):383–8. doi: 10.1016/j.radonc.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 17.Aebersold DM, Burri P, Beer KT, Laissue J, Djonov V, Greiner RH, et al. Expression of hypoxia-inducible factor-1alpha: a novel predictive and prognostic parameter in the radiotherapy of oropharyngeal cancer. Cancer Res. 2001;61(7):2911–6. [PubMed] [Google Scholar]
- 18.Palit V, Phillips RM, Puri R, Shah T, Bibby MC. Expression of HIF-1alpha and Glut-1 in human bladder cancer. Oncol Rep. 2005;14(4):909–13. doi: 10.3892/or.14.4.909. [DOI] [PubMed] [Google Scholar]
- 19.Winter SC, Buffa FM, Silva P, Miller C, Valentine HR, Turley H, et al. Relation of a hypoxia metagene derived from head and neck cancer to prognosis of multiple cancers. Cancer Res. 2007;67(7):3441–9. doi: 10.1158/0008-5472.CAN-06-3322. [DOI] [PubMed] [Google Scholar]
- 20.Buffa FM, Harris AL, West CM, Miller CJ. Large meta-analysis of multiple cancers reveals a common, compact and highly prognostic hypoxia metagene. Br J Cancer. 2010;102(2):428–35. doi: 10.1038/sj.bjc.6605450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Betts GN, Eustace A, Patiar S, Valentine HR, Irlam J, Ramachandran A, et al. Prospective technical validation and assessment of intra-tumour heterogeneity of a low density array hypoxia gene profile in head and neck squamous cell carcinoma. Eur J Cancer. 2013;49(1):156–65. doi: 10.1016/j.ejca.2012.07.028. [DOI] [PubMed] [Google Scholar]
- 22.Ord JJ, Agrawal S, Thamboo TP, Roberts I, Campo L, Turley H, et al. An investigation into the prognostic significance of necrosis and hypoxia in high grade and invasive bladder cancer. J Urol. 2007;178(2):677–82. doi: 10.1016/j.juro.2007.03.112. [DOI] [PubMed] [Google Scholar]
- 23.Youssef RF, Lotan Y. Predictors of outcome of non-muscle-invasive and muscle-invasive bladder cancer. ScientificWorldJournal. 2011;11:369–81. doi: 10.1100/tsw.2011.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25(4):402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 25.Dunn PK. Tweedie: Tweedie exponential family models. R package version 2.1.5 2007. [Google Scholar]
- 26.Hoskin PJ, Sibtain A, Daley FM, Wilson GD. GLUT1 and CAIX as intrinsic markers of hypoxia in bladder cancer: relationship with vascularity and proliferation as predictors of outcome of ARCON. Br J Cancer. 2003;89(7):1290–7. doi: 10.1038/sj.bjc.6601260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Betts GN, Eustace A, Patiar S, Valentine HR, Irlam J, Ramachandran A, et al. Prospective technical validation and assessment of intra-tumour heterogeneity of a low density array hypoxia gene profile in head and neck squamous cell carcinoma. Eur J Cancer. 2012 doi: 10.1016/j.ejca.2012.07.028. [DOI] [PubMed] [Google Scholar]
- 28.Forastiere AA, Goepfert H, Maor M, Pajak TF, Weber R, Morrison W, et al. Concurrent chemotherapy and radiotherapy for organ preservation in advanced laryngeal cancer. N Engl J Med. 2003;349(22):2091–8. doi: 10.1056/NEJMoa031317. [DOI] [PubMed] [Google Scholar]
- 29.Benita Y, Kikuchi H, Smith AD, Zhang MQ, Chung DC, Xavier RJ. An integrative genomics approach identifies Hypoxia Inducible Factor-1 (HIF-1)-target genes that form the core response to hypoxia. Nucleic Acids Res. 2009;37(14):4587–602. doi: 10.1093/nar/gkp425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chi JT, Wang Z, Nuyten DS, Rodriguez EH, Schaner ME, Salim A, et al. Gene expression programs in response to hypoxia: cell type specificity and prognostic significance in human cancers. PLoS Med. 2006;3(3):e47. doi: 10.1371/journal.pmed.0030047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lendahl U, Lee KL, Yang H, Poellinger L. Generating specificity and diversity in the transcriptional response to hypoxia. Nat Rev Genet. 2009;10(12):821–32. doi: 10.1038/nrg2665. [DOI] [PubMed] [Google Scholar]
- 32.van ’t Veer LJ, Dai H, van de Vijver MJ, He YD, Hart AA, Mao M, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415(6871):530–6. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- 33.Albain KS, Barlow WE, Shak S, Hortobagyi GN, Livingston RB, Yeh IT, et al. Prognostic and predictive value of the 21-gene recurrence score assay in postmenopausal women with node-positive, oestrogen-receptor-positive breast cancer on chemotherapy: a retrospective analysis of a randomised trial. Lancet Oncol. 2010;11(1):55–65. doi: 10.1016/S1470-2045(09)70314-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Paik S, Tang G, Shak S, Kim C, Baker J, Kim W, et al. Gene expression and benefit of chemotherapy in women with node-negative, estrogen receptor-positive breast cancer. J Clin Oncol. 2006;24(23):3726–34. doi: 10.1200/JCO.2005.04.7985. [DOI] [PubMed] [Google Scholar]
- 35.van de Vijver MJ, He YD, van’t Veer LJ, Dai H, Hart AA, Voskuil DW, et al. A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med. 2002;347(25):1999–2009. doi: 10.1056/NEJMoa021967. [DOI] [PubMed] [Google Scholar]
- 36.Ma XJ, Dahiya S, Richardson E, Erlander M, Sgroi DC. Gene expression profiling of the tumor microenvironment during breast cancer progression. Breast Cancer Res. 2009;11(1):R7. doi: 10.1186/bcr2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Webber EM, Lin JS, Evelyn PW. Oncotype DX tumor gene expression profiling in stage II colon cancer. Application: prognostic, risk prediction. PLoS Curr. 2010;2 doi: 10.1371/currents.RRN1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Neyman N, Altekruse SF, Kosary CL, Ruhl J, Tatalovich Z, Cho H, et al. SEER Cancer Statistics Review, 1975-2009 (Vintage 2009 Populations) National Cancer Institute; Bethesda, MD: http://seer.cancer.gov/csr/1975_2009_pops09/, based on November 2011 SEER data submission, posted to the SEER web site, 2012. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.