Abstract
Identifying risk factors for Alzheimer’s disease, such as carrying the APOE-4 allele, and understanding their contributions to disease pathophysiology or clinical presentation is critical for establishing and improving diagnostic and therapeutic strategies. A first-degree family history of Alzheimer’s disease represents a composite risk factor, which reflects the influence of known and unknown susceptibility genes and perhaps non-genetic risks. There is emerging evidence that investigating family history risk associated effects may contribute to advances in Alzheimer’s disease research and ultimately clinical practice.
Keywords: Alzheimer’s disease, APOE Genotype, Family history, Genetic risk, Risk factors, Neuroimaging
Introduction
Alzheimer’s disease is the most important cause of dementia and affects one in 10 individuals over the age of 65. By 2050, there will be an estimated one million new Alzheimer’s disease patients per year in the United States alone, and it remains unclear how the expected increase in healthcare costs will be covered (Hebert et al. 2001; Mount and Downton 2006). These data illustrate that improving early detection and treatment of Alzheimer’s disease may be essential for the future wellbeing of society. The development of new pharmacological strategies, such as targeting Alzheimer’s disease neuropathological hallmarks – amyloid plaques and neurofibrillary tangles – has not yet resulted in a clinically effective therapy (Wisniewski and Boutajangout 2010; Holmes et al. 2008). The main reason for this is limited knowledge about the underlying pathophysiology; subsequently, interventions may fail or come too late in the course of the disease to benefit patients.
Pathological studies show changes in neuronal integrity that appear years, maybe decades before the onset of cognitive symptoms (Braak and Braak 1997; Ohm et al. 1995). Recent neuroimaging and biochemical research has revealed various brain structural and functional characteristics that could be useful for future diagnostic procedures. However, Alzheimer’s disease has a complex polygenic background and studying the neurobiological effects associated with the presence of a single genetic risk factor will often result in variable research results. Unknown gene-gene and gene-environment interactions are likely to modulate such a factor’s influence on brain structure and function, and they may also determine the clinical relevance of these changes for an individual person.
The e4 allele of the apolipoprotein E gene (APOE-4) on Chromosome 19 is a perfect example for this scientific challenge. As the best-established genetic risk factor for late-onset sporadic Alzheimer’s disease it is the primary target of numerous studies investigating the disease’s underlying molecular neuropathology, macroscopic brain anatomy and brain function characteristics, cognition and behavior changes, clinical progression and treatment response. Whereas some of the conflicting data might be attributable to the risk allele’s antagonistically pleiotropic effects across different stages of the life span (Tuminello and Han 2011), others likely illustrate the influence of unknown variables on APOE-4 associated effects. The APOE gene is clearly remarkable, however, for Alzheimer’s disease there may be unidentified susceptibility genes of equal or even larger effect size when compared to APOE (Daw et al. 2000).
Research on Alzheimer’s disease genetics is rapidly growing. There are fully penetrant forms of the disease due to mutations in the amyloid precursor protein (APP) or presenelin (PSEN1, PSEN2) genes. In these inherited autosomal dominant forms of the disease, the development of the disease as well as the approximate age at onset can be predicted (Murrell et al. 2006). Investigating these patients and presymptomatic mutation carriers can provide insights into Alzheimer’s disease pathophysiology and the relationship of the underlying genetic mutations to other susceptibility genes, such as APOE-4 (Ringman et al. 2011).
Recent genome wide association studies (GWAS) aim to detect new susceptibility genes for the common sporadic, late-onset variant of Alzheimer’s disease. In addition to APOE-4, GWAS studies have confirmed new susceptibility loci in the CLU, PICALM and CR1 gene regions (Lambert et al. 2009; Harold et al. 2009). Seshadri and colleagues (2010) recently identified two new loci near BIN1 and EXOC3L2/BLOC1S3/MARK4, also confirming CLU and PICALM. The authors noted, however, that these new loci did not improve Alzheimer’s disease risk prediction. Although they may not be clinically useful, they could implicate new biological pathways important for future research (Seshadri et al. 2010). The identified genes encode such proteins as clusterin (CLU) or the complement component (3b/4b) receptor 1 (CR1), and it remains unclear how these mutations exactly contribute to Alzheimer’s disease pathology. Epigenetic mechanisms add to the increasing complexity of processes involved in Alzheimer’s disease development and clinical expression. These mechanisms regulate the transcriptional activity of genes, and epigenetics also provide a means by which environmental factors can influence gene expression (Mastroeni et al. 2011). Epigenetic regulation of gene expression includes histone modifications, DNA methylation or RNA-related mechanisms, and these mechanisms may contribute to the aging process itself and the development of dementia (for review see Mastroeni et al. 2011). Furthermore, the development of Alzheimer’s disease could be associated with specific risk factor clusters composed of genetic and environmental variables rather than single factors, and pathogenic effects may vary depending on age, gender or ethnicity.
Alzheimer’s disease has a high heritability (Gatz et al. 2006). There could be many genes contributing to the familial clustering of the disease, and at the individual level, APOE-4 may or may not be a part of this pattern. A first-degree family history of Alzheimer’s disease is associated with a greater risk for developing the disease (van Duijn et al. 1991; Fratiglioni et al. 1993). It remains an interesting question whether or under which circumstances a family history associated risk exists in addition to or interacts with APOE-4 genetic risk (Jarvik and Wijsman 1994; Cupples et al. 2004; van Duijn et al. 1994). Recent neurobiological and neurocognitive research data provide evidence that modeling family history risk in disease prediction should be accompanied by recognizing this risk factor in all investigations aimed at expanding our knowledge on Alzheimer’s disease neuropathology and clinical presentation.
The family history risk factor could be conceptualized as a composite factor (Fig. 1) reflecting the influence of known and unknown susceptibility genes. Whether there is an observed family history of the disease also depends on the age-dependent penetrance of a susceptibility gene. An individual may have a genetic risk that is never expressed and thus would not be observed as a family history risk. Furthermore, non-genetic risks (Borenstein et al. 2006) could be reflected in the family history risk factor as well, as these traits are likely to be passed on through generations, and could even have direct effects on gene expression (Robinson et al. 2008). In this review we will focus on the possible relationship between family history risk and the APOE-4 allele, highlighting neuroimaging, other biological, and neurocognitive research data. These studies focus on cognitively healthy people, since disease-associated processes could prevent detection of the individual effects associated with or modulated by family history risk.
Fig. 1.
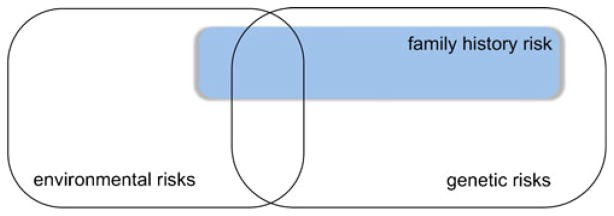
Alzheimer’s disease and the ‘family history’ risk factor. Alzheimer’s disease has a high heritability, but the influence of environmental variables on disease development and clinical course is also substantial (Gatz et al. 2006). A first-degree family history of Alzheimer’s disease can be conceptualized as a composite risk factor, reflecting the influence of known and yet unknown genetic risks. Furthermore, specific environmental risk factors (such as low socioeconomic status; Borenstein et al. 2006) may contribute to the familial clustering of the disease as well, since these risks may be passed on to the next generation. Note: The individual size of a rectangle in the figure does not reflect an exact value
Functional MRI
Many functional magnetic resonance imaging (fMRI) studies in people at risk for Alzheimer’s disease focus on the APOE-4 allele, utilizing cognitive tasks believed to be most susceptible to early changes in a possible future cognitive decline’s preclinical stage. In a study by Bookheimer and colleagues (2000) healthy participants differing in APOE-4 carrier status were asked to learn and recall unrelated word pairs during fMRI scanning. When compared with subjects not carrying the risk allele, the authors revealed greater brain activity during memory tasks among APOE-4 carriers in several brain regions. During recall, the APOE-4 carriers showed about twice as much fMRI signal increase in the hippocampus when compared with non-carriers. The hyper-activity could be a compensatory mechanism used to aid task performance (Bookheimer et al. 2000). Ringman and colleagues (2011) investigated cognitively unimpaired people with APOE-4 genetic risk and participants with fully penetrant familial Alzheimer’s disease mutations. The authors found that APOE-4 associated affects on brain activity may be at least in part independent of Alzheimer’s disease risk, which would not be in line with an exclusively compensatory mechanism (Ringman et al. 2011). Although a number of studies revealed greater brain activity during cognitive tasks in APOE-4 allele carriers when compared with non-carriers (Bondi et al. 2005; Wishart et al. 2006; Fleisher et al. 2005) others did not find this association. Trivedi and colleagues (2006) showed reduced hippocampal and medial temporal lobe activity among APOE-4 carriers in a novel versus familiar item encoding task. The authors highlight that less activity in this area would be in line with the reduced glucose metabolism known to be associated with Alzheimer’s disease pathology. Lind and colleagues (2006) detected an APOE-4 allele dose dependent activity decrease in the parietal cortex during a semantic categorization task.
There are possible explanations for these contrasting findings, such as the utilization of different cognitive tasks, or the participants’ varying age across studies. In contrast to middle-aged and older participants, younger APOE4-carriers have shown a memory task-associated blood oxygen level dependent (BOLD) signal increase relative to non-carriers in fMRI studies (Filbey et al. 2006; Filippini et al. 2009). Recently Filippini and colleagues (2011) showed that age interacts with APOE-4 genetic risk. The authors demonstrated a decrease of APOE-4 associated hyperactivity with a more advanced age using a memory encoding task. Trivedi and colleagues (2008) found that an age-related decline in brain activation in the ventral temporal lobes and the hippocampus during novel picture encoding was not strongly modulated by APOE-4 genotype or a positive family history of the disease. However, APOE-4 carriers who also had a positive family history of Alzheimer’s disease showed an age-related increase in fMRI activation in the right hippocampus during the encoding task (Trivedi et al. 2008). These data suggest that family history and APOE-4 may exert unique effects on brain activity. In line with this finding, Bassett and colleagues (2006) demonstrated an APOE-4 independent neural activity increase in the frontal and temporal lobes during memory encoding in healthy individuals with a family history of Alzheimer’s disease.
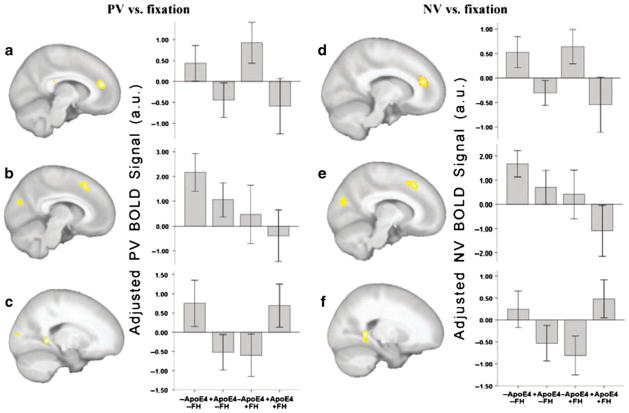
The family history risk factor could have influenced the results of previous fMRI studies investigating healthy people at APOE-4 genetic risk. About half of the subjects in the Bookheimer and colleagues’ (2000) study but all of the participants recruited by Trivedi and colleagues (2006) had a family history of Alzheimer’s disease. The number of fMRI studies in healthy people, in which APOE genotype and family history have been modeled as separate factors, is very limited. Johnson and colleagues (2006) found a greater response to novel items in the mesial temporal lobe and fusiform gyrus bilaterally among middle-aged participants without a family history of Alzheimer’s disease. Although authors did not find a family history by APOE genotype interaction at the predefined statistical threshold, direct comparison of APOE-4 carriers differing in family history status revealed significantly greater fMRI signal change in the right ventral temporal lobe as well as the hippocampus bilaterally associated with the absence of a family history among APOE-4 allele carriers. Xu and colleagues (2009) used an episodic recognition task utilizing previously learned (PV) and novel (NV) faces. The authors found a stronger fMRI signal among subjects not having a family history of Alzheimer’s disease in the dorsal cuneus and medial frontal cortices. An interaction effect between the factors family history and APOE genotype was found in the fusiform gyrus bilaterally (Xu et al. 2009) (Fig. 2).
Fig. 2.
Functional MRI (from Xu et al. 2009, The influence of parental history of Alzheimer’s disease and apolipoprotein E e4 on the BOLD signal during recognition memory, Brain, 2009, 132 (2):383–91, by permission of Oxford University Press). In a 2×2 ANCOVA analysis, the APOE4, FH and their interaction effects showed similar clusters with both PV and NV responses [P<0.05 (corrected for cluster size)]. a and d A larger response was observed in the -APOE4 group compared to the +APOE4 in the left anterior cingulate cortex to PV (a) or NV faces (d). b and e A larger response was observed in the -FH group compared to the +FH group in the left medial superior frontal gyrus (signal shown in the plot) and left cuneus. There is a clear declining trend in PV or NV response amplitude with the accumulation of Alzheimer’s disease risk factors (+FH and +APOE4). c and f The interaction between FH and APOE4 showed significance in the bilateral fusiform–parahippocampal gyrus with both PV (c) and NV faces (f)
Structural MRI and DTI
Recent advances in MRI technology and image analysis now enable researchers to detect risk condition-associated changes in radiological brain anatomy. Although not all studies could find an APOE-4 related effect among healthy people (Cherbuin et al. 2008), there is evidence that subtle APOE-4 allele associated changes in brain morphology exist, at least in brain regions most susceptible to Alzheimer’s disease pathology, such as the entorhinal cortex and the hippocampus. Mueller and colleagues (2008) reported a smaller dentate gyrus and CA3 hippocampus subfield associated with APOE-4 genetic risk. Cortical thickness measures revealed a thinner entorhinal cortex and subiculum in cognitively healthy APOE-4 carriers prior to volumetric changes (Burggren et al. 2008). There is still a debate about whether or not APOE-4 associated structural brain characteristics represent a genetically determined neuroanatomical feature, or whether they may also reflect pathological processes. Reduced entorhinal cortical thickness among children and young adults carrying the APOE-4 allele suggest that it is at least in part a neuroanatomical feature, a static risk factor, so that less thinning would be required to reach a critical threshold in neural integrity (Shaw et al. 2007). Longitudinal studies demonstrate higher rates of whole brain atrophy (Chen et al. 2007), hippocampal atrophy (Jak et al. 2007), and greater hippocampal and entorhinal cortical thinning (Donix et al. 2010b) among APOE-4 allele carriers when compared with non-carriers. It is possible that the APOE-4 allele dose influences atrophy rates (Chen et al. 2007), but most investigators have been unable to recruit a sufficient number of cognitively healthy elderly homozygous APOE-4 allele carriers.
In comparison to fMRI studies, investigations of APOE-4 associated brain structure changes are in general less variable. The intuitive relationship of ‘risk’ and ‘atrophy’ in neurodegeneration research may leave little room for contrasting data. Espeseth and colleagues (2008) found a thicker cortex in APOE-4 carriers when compared non non-carriers in several frontal and temporal regions, although the rate of cortical thickness decline over time was still accelerated in people at genetic risk (Espeseth et al. 2008). It is possible that non-intuitive findings are not reported and confounding variables are rarely modeled. However, the few studies available suggest a substantial APOE-4 independent contribution of a first-degree family history for Alzheimer’s disease on brain anatomy.
Among siblings discordant for Alzheimer’s disease, the heritability for cerebral atrophy and white matter lesions is high and cannot be explained by APOE status alone (Lunetta et al. 2007). Honea and colleagues (2010) revealed decreased gray matter volume in the precuneus and frontal cortices among cognitively healthy people with a maternal family history of Alzheimer’s disease compared with subjects having a paternal or no family history risk. In a subsequent longitudinal investigation the authors showed decreased whole-brain gray matter as well as precuneus and hippocampal atrophy among subjects with a maternal family history (Honea et al. 2011). In both studies, the authors controlled for APOE-4 carrier status, gender, and age. The existence of APOE-4 independent family history effects on brain structure are in line with a recent study demonstrating independent and additive contributions of APOE-4 genotype and family history risk on hippocampal subfield and entorhinal cortical thickness (Donix et al. 2010a). The family history risk explained a greater proportion of the unique variance in cortical thickness than the APOE-4 carrier status (Donix et al. 2010a) (Fig. 3).
Fig. 3.
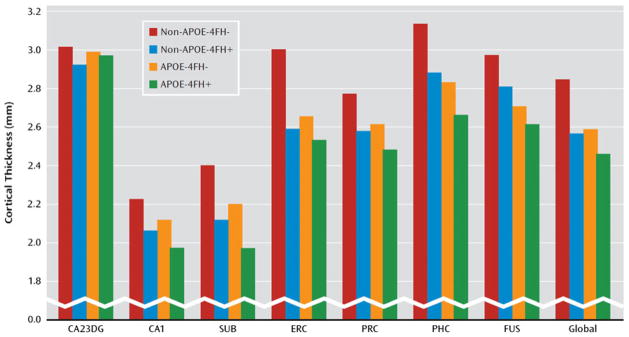
Structural MRI (from Donix et al. 2010a. Reprinted with permission from the American Journal of Psychiatry, (Copyright ©2010). American Psychiatric Association). The figure displays cortical thickness values for the possible APOE and family history risk factor combinations among cognitively healthy subjects. CA23DG cornu ammonis fields 2, 3 and dentate gyrus; CA1 cornu ammonis field 1; SUB subiculum; ERC entorhinal cortex; PRC perirhinal cortex; PHC parahippocampal cortex; FUS fusiform cortex; Global average cortical thickness across all medial temporal lobe subregions; FH family history; “+” = positive; “−” = negative
The number of diffusion tensor imaging (DTI) studies investigating healthy people at APOE-4 genetic risk is on the rise. Heise and colleagues (2011) found a general reduction of fractional anisotropy and an increase in mean diffusivity values among APOE-4 carriers compared with non-carriers. The authors did not find an interaction between genotype and age for these measures, which could suggest that differences in white matter structure do not undergo significant differential changes with age (Heise et al. 2011). Brown and colleagues (2011) reported an accelerated age-related loss of mean local interconnectivity and regional interconnectivity decreases in the precuneus, the medial orbitofrontal cortex, and the lateral parietal cortex among elderly healthy APOE-4 carriers when compared with non-carriers. An APOE-4 associated age-related reduction in small worldness, which reflects the balance between local interconnectivity and global integration, was also driven primarily by the loss of interconnectivity in specific brain regions, whereas global integration was relatively spared (Brown et al. 2011). These two DTI studies illustrate how specific DTI measures may be differentially susceptible to possible APOE-4 associated white matter changes in aging. Other studies could also detect white matter changes associated with APOE-4 genetic risk (Persson et al. 2006; Nierenberg et al. 2005; Ryan et al. 2011; Honea et al. 2009). Persson and colleagues (2006) showed a decline in fractional anisotropy in the posterior corpus callosum and the medial temporal lobe in healthy APOE-4 carriers.
Recent DTI studies also focus on the effects of the family history risk factor on white matter integrity. Whereas two studies investigated high-risk individuals, having both family history and APOE-4 risk factors (Gold et al. 2010; Smith et al. 2010), Bendlin and colleagues (2010) investigated these risks separately. The authors demonstrated an association of the family history risk factor with lower fractional anisotropy in several regions including the hippocampus. There was an additive effect of APOE-4 and family history risks, but no main effect of APOE-4 genotype, which may indicate that unknown risk factors contained in family history are associated with changes in white matter integrity (Bendlin et al. 2010).
PET
Using fluorodeoxyglucose (FDG)-PET, Reiman and colleagues (2001) demonstrated an abnormally low cerebral metabolic rate for glucose (CMRglc) among cognitively normal middle-aged and elderly APOE-4 carriers in the same brain regions as patients with probable Alzheimer’s disease. The authors showed that this effect was modulated by the APOE-4 allele dose (Reiman et al. 2005), and detectable even in young adults (Reiman et al. 2004). Although this association is well established, CMRglc reductions can be modified by additional variables such as having a maternal family history of Alzheimer’s disease (Mosconi et al. 2007), and an advanced maternal age at birth (Mosconi et al. 2011). Mosconi and colleagues (2007) showed posterior cingulate/precuneus, parietotemporal, frontal and medial temporal CMRglc reductions among people with a maternal family history of Alzheimer’s disease when compared with participants having a paternal or no family history. The effect remained significant after accounting for age, gender, and APOE genotype (Mosconi et al. 2007). In a longitudinal investigation Mosconi and colleagues (2009) additionally demonstrated greater CMRglc declines in these regions among participants with a maternal family history of Alzheimer’s disease. There is evidence for a ‘FDG-endophenotype’, which may be useful in Alzheimer’s disease prediction research (Mosconi et al. 2007; Mosconi et al. 2009; During et al. 2011).
Amyloid-beta and tau labeling PET ligands allow in vivo detection of neuropathological features in people at risk for Alzheimer’s disease. Healthy subjects carrying the APOE-4 allele show increased Pittsburgh Compound-B (PIB) mean cortical binding potential in an allele dose dependant pattern in several frontal, temporal, posterior cingulate/precuneus, parietal, and basal ganglia brain regions (Reiman et al. 2009). With the amyloid plaque and tau labeling PET ligand 2-(1-{6-[(2-[F-18]fluoroethyl)(methyl)amino]-2-naphthyl}ethylidene)malononitrile (FDDNP), Small and colleagues (2009) showed an increased PET signal in the frontal cortex of APOE-4 carriers without dementia. In a recent PIB-PET study, Mosconi and colleagues (2010b) found a family history effect, after controlling for age, gender, education, and APOE status. Subjects with a maternal family history showed higher PIB retention in various brain regions compared with subjects having a paternal or no family history of Alzheimer’s disease. A paternal family history was associated with increased PIB retention only in the posterior cingulate and frontal cortex, with an intermediate level between participants with a maternal and no family history (Mosconi et al. 2010b) (Fig. 4).
Fig. 4.
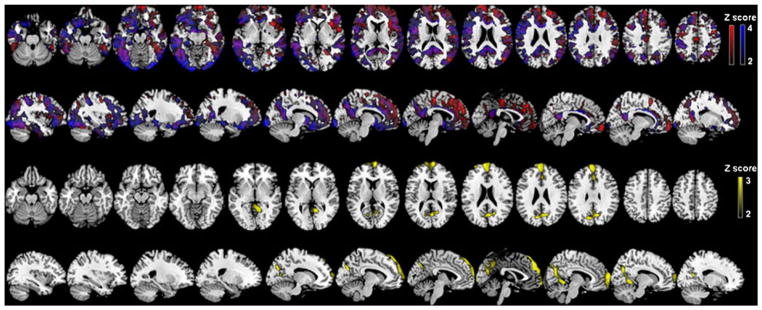
PIB-PET (from Mosconi et al. 2010b. Reprinted with permission from the National Academy of Sciences). Statistical parametric maps showing higher Pittsburgh Compound B (PIB) retention in cognitively normal subjects with a maternal family history of Alzheimer’s disease than in subjects with a paternal or no family history (upper two rows), and in cognitively normal subjects with a paternal family history of Alzheimer’s disease than in subjects with no family (lower two rows). Areas with higher PIB retention are represented on color-coded scales, reflecting Z scores. All results remained significant after controlling for age, gender, education, and APOE status
Cerebrospinal Fluid Biomarkers
Sunderland and colleagues (2004) revealed significantly lower cerebrospinal fluid (CSF) beta-amyloid(1-42) but not tau levels among elderly cognitively healthy APOE-4 allele carriers compared with subjects not carrying the risk allele. Glodzik-Sobanska and colleagues (2009) showed an age by APOE-4 allele interaction in hyperphosphorylated tau (P-tau 231) levels, and a total tau concentration increase with age. Another study demonstrated age effects on CSF beta-amyloid(1-42) and phosphorylated tau (P-tau 181) levels, and APOE-4 effects on beta-amyloid(1-42) levels in cognitively normal middle aged and elderly participants (Popp et al. 2010). Whereas the study by Sunderland and colleagues (2004) reported the healthy group’s enrichment with people having a family history of Alzheimer’s disease, Mosconi and colleagues (2010a) investigated a family history effect directly. The authors demonstrated higher F-isoprostanes levels (which is a marker for oxidative stress) and reduced beta-amyloid(42/40) CSF levels in healthy subjects with a maternal family history of Alzheimer’s disease when compared with subjects having a paternal or no family history risk (Mosconi et al. 2010a). The latter groups themselves did not differ in these measures. There were also no group differences for P-tau 231 and total tau. The results remained significant after controlling for age, gender, education, and APOE genotype (Mosconi et al. 2010a).
Physiological Measures
The APOE proteins’ major role in neural lipid metabolism, as well as in repair and plasticity processes suggest multiple mechanisms of how the APOE-4 allele may contribute to neurodegeneration (Saunders 2000; Teter 2004; Mahley et al. 2006). Caselli and colleagues (2011) found an interaction of APOE-4 allele dose and the impact of cerbrovascular risk factors on brain function. There is evidence for an association between the APOE-4 allele and large as well as small vessel vascular injury. APOE-4 carriers have a higher risk for carotid atherosclerosis and myocardial infarction (Anand et al. 2009; Elosua et al. 2004). In a neuropathological study, Yip and colleagues (2005) found more small-vessel arteriolosclerosis, microinfarcts of the deep nuclei, as well as higher neuritic senile plaque density, and amyloid angiopathy in APOE-4 carriers with autopsy-proven Alzheimer’s disease. The relationship between APOE-4 and vascular factors in predicting cognitive and functional decline could be complex. Mielke and colleagues (2011) found a 3-way interaction among stroke, APOE-4 genotype and time in predicting decline in the Mini-Mental State Exam (Folstein et al. 1975).
Using the amyloid-labeling PET ligand PIB, Langbaum and colleagues (2011) did not find an APOE-4 effect in the association of high blood pressure with higher brain amyloid burden in healthy late middle-aged persons. The authors suggest that the family history of Alzheimer’s disease status could have influenced the results. Van Exel and colleagues (2009) showed that hypertension, indices of vascular disease, and the expression of an innate pro-inflammatory cytokine profile in middle age are early risk factors of Alzheimer’s disease in old age. These findings were not modulated by lifestyle factors, such as stress level, fat intake, current smoking or physical activity. They were also independent of the participants’ APOE genotype but associated with a parental family history of Alzheimer’s disease (van Exel et al. 2009).
Neurocognitive Data
The majority of studies investigating healthy people at risk for Alzheimer’s disease aim at identifying changes in cognitive abilities associated with a specific risk condition, such as carrying the APOE-4 allele. In a meta-analysis Wisdom and colleagues (2011) demonstrated poorer cognitive performance in episodic memory, executive functioning, perceptual speed, and global cognitive measures among APOE-4 carriers when compared with non-carriers. Older age exacerbated the effects. These data are in line with a previous meta-analysis, although the authors did not find the age effect (Small et al. 2004). Longitudinal studies investigating APOE-4 associated changes in cognition are relatively rare. Caselli and colleagues (2007) found higher rates of cognitive decline in several domains among middle-aged homozygous APOE-4 carriers when compared with heterozygous carriers or non-carriers prior to the development of mild cognitive impairment or Alzheimer’s disease. In a high-functioning elderly sample, APOE-4 carriers were twice as likely to have declined on a global cognitive score in a seven-year follow-up investigation when compared with non-carriers (Bretsky et al. 2003). Not all studies can confirm an association of APOE-4 genetic risk and greater cognitive decline over time (Bunce et al. 2004). This could be due to variations in the participants’ age, or the inclusion of subjects with preclinical dementia (Bunce et al. 2004). It is also intuitive to assume that having a family history of Alzheimer’s disease may exert APOE-4 independent effects on cognition. There are only few studies available that investigate a family history effect on cognition in healthy people while accounting for APOE-4 risk. Furthermore, these studies focus on single tests, such as a word list learning task for which a family history effect could be detected (La Rue et al. 2008), or global scales (Mini-Mental State Exam) in which the subjects’ performance did not seem to be modulated by this risk factor (Hayden et al. 2009). In a recent longitudinal study subjects with a parental family history of Alzheimer’s disease had lower baseline scores in processing speed, executive functioning, memory encoding, and delayed memory when compared with participants without this risk factor (Donix et al. 2011). APOE-4 carrier status but not family history risk had a longitudinal effect on memory performance (Donix et al. 2011).
Discussion
A positive first-degree family history of Alzheimer’s disease increases the risk to develop the disease (van Duijn et al. 1991; Fratiglioni et al. 1993; Cupples et al. 2004). Family history and APOE-4 genetic risks highly co-occur (Zintl et al. 2009) and it remains controversial whether both risk factors contribute additively to Alzheimer’s disease development, whether they interact and overlap. This points to some intrinsic limitations of the family history approach. The pattern of risk variables embodied in family history risk is presumably heterogeneous across different subjects and study populations, and the interaction potential of these variables with other given factors, such as the APOE-4 allele, may vary on the individual level. Relatives with dementias other than Alzheimer’s disease could be misclassified based on clinical criteria, and healthy relatives could develop Alzheimer’s disease in the future. Moreover, in the presence of both risk factors, it cannot be determined clinically whether the familial clustering of the disease is mainly driven by the APOE-4 allele. However, it has been demonstrated across various neurobiological and clinical studies that family history associated effects are often dissociable from APOE-4 related effects.
Data from fMRI studies suggest that a different family history status may even influence the direction of neural activity changes during cognitive tasks. The studies by Johnson and colleagues (2006) and Xu and colleagues (2009) are great examples for how interesting fMRI findings could have been overlooked if one would chose not to model the individual effects of APOE-4 and family history risks. Studies of APOE-4 associated effects as they relate to fMRI findings have yielded variable results, especially when compared with structural MRI findings. Such variability likely reflects several challenges and limitations. The biological basis of BOLD signal dynamics may reflect the interplay of neural activity, metabolism, blood volume, blood flow and subsequent oxygenation changes (Bandettini and Ungerleider 2001). Logothetis and colleagues (2001) showed that a spatially localized increase in the BOLD signal directly and monotonically reflects an increase in neural activity, specifically local field potential rather than the neuronal spiking activity. However, it is possible that the BOLD signal and local field potential dissociate. Caution is necessary when interpreting BOLD data, and making direct inferences about underlying neural activity (Ekstrom 2010). BOLD signal increase during cognitive tasks in people at risk for Alzheimer’s disease may be interpreted as ‘compensatory’, reflecting the recruitment of additional neural resources to aid task performance. Decreased BOLD signal could be interpreted as an indication of regional neuronal loss in the same risk population, however, BOLD signal decrease could also reflect better brain efficiency. Correlations with cognitive performance scores from tasks performed during fMRI scanning can help guide investigators to meaningful interpretations of the BOLD signal. It is obvious that fMRI itself cannot be used to determine whether, and under which circumstances one of these hypotheses is more likely to be true than others. Many questions about Alzheimer’s disease risk factors and their association with BOLD signal changes and directionality may remain unanswered. However, there is evidence that APOE-4 and family history risk can influence the BOLD response. In general, age, APOE-4, and family history risk need consideration when interpreting neuronal function in the context of BOLD signal direction.
Structural MRI investigations show clear evidence of independent and/or additive effects of family history and APOE-4 risks on regional cortical thickness patterns (Donix et al. 2010a) or white matter integrity (Bendlin et al. 2010). This might be helpful to more precisely determine the unique APOE-4 associated effects on brain morphology. Neurocognitive profiles suggest family history effects in various cognitive domains, whereas APOE-4 effects may preferentially occur in memory tasks (Donix et al. 2011). This would be in line with the hypothesis that the presumable diversity of risk factors embodied in family history risk may involve factors less closely related to Alzheimer’s disease pathology.
An interesting finding from structural MRI, PET and CSF data is the possible significance of having a maternal rather than paternal family history of Alzheimer’s disease (Honea et al. 2010; Mosconi et al. 2007; Mosconi et al. 2010a). It indicates that modeling family history risk does not only allow to better isolate and describe APOE-4 related phenomena; it may also enhance our understanding of sporadic Alzheimer’s disease genetic transmission. Mosconi and colleagues (2007) suggest the possibility of a mitochondrial DNA inheritance pattern, which would be in line with the mechanisms contributing to reduced brain (glucose) metabolism and changes in the oxidative microenvironment (Mosconi et al. 2010a; Lin and Beal 2006).
In summary, the reviewed data indicate that APOE-4 carrier status modulates brain structure and function. The underlying molecular mechanisms often remain unknown because of the limited knowledge about the physiological functions of APOE proteins, which play an important role in neural lipid metabolism. In contrast to the e2 and e3 isoforms, the APOE-4 allele has poorer functionality, is conformationally unstable, increases amyloid production and tau phosphorylation and may even have direct neurotoxic effects (Mahley et al. 2006). The APOE-4 allele is associated with reduced neural repair and plasticity (Teter 2004) and may be a general risk factor for neurodegenerative diseases (Blazquez et al. 2006; van Duijn et al. 1994; Chapman et al. 2001). The mechanisms through which a family history of Alzheimer’s disease becomes a risk factor may be even more complex. On the one hand, family history risk may reflect the presence of genetic risk factors ranging from established susceptibility genes with only partially known roles (e.g., APOE) to yet unknown genetic variables. On the other hand, family history risk could also reflect non-genetic risks, such as low socioeconomic status (Borenstein et al. 2006) that may be passed on through generations, as well as shared environmental risks that family members may be exposed to (e.g., mold in family dwellings, exposure to toxins, etc.). Future research might further detail familial risks (e.g., first degree vs. second degree relatives, potential influence of environmental factors) and the underlying risk mechanisms.
For researchers it may be important to recognize family history effects in studies aimed at investigating APOE-4 associated changes irrespective of the research modality. Until we can better separate the single variables that likely contribute to family history risk, the composite risk factor may be a practical approach to control for yet unknown risk variables in Alzheimer’s disease research. Many studies investigating healthy people at risk for Alzheimer’s disease focus on single risk factors, such as the APOE-4 allele. This could result in both overestimation and masking of APOE-4 related effects. For clinicians, these data could be useful to avoid oversimplifying assumptions about APOE-4 genetic risk. APOE is a remarkable susceptibility gene, but the extensive literature on APOE could reduce the awareness for other important risk conditions and their influence on brain function including other genetic factors as well as modifiable environmental risks. Studies show that, for example, vascular risks may contribute to the family history risk factor. Strategies aimed at enhancing vascular health could be important for the prevention of Alzheimer’s disease (Luchsinger 2008). This should encourage clinicians to proactively educate and advise their patients.
Dubois and colleagues (2007) proposed revised research criteria that require significant episodic memory impairment and the presence of supporting biomarkers to diagnose Alzheimer’s disease. This specifically acknowledges the potential of today’s biomedical research techniques to help establish a diagnosis as early as possible. Multimodal neuroimaging could help to determine changes in brain morphology and function that may occur as a result of risk factors, aging, and neuropathology. These data, in concert with CSF and neurocognitive variables, could be helpful to establish prediction models for future cognitive decline. From a clinical perspective it is important to know how and how much dementia risk conditions contribute to brain structure and function changes. This could determine the subjective weight we may attribute to risk factors in clinical evaluations, which ultimately influences clinical decisions. If we examine the APOE genotype status, we do not disclose this information to our patients. Because of the substantial amount of available APOE research data we believe in the impact an APOE-4 allele may have, although the risk allele is not sufficient to cause the disease or to fully determine its clinical course. The family history risk factor may be useful to account for yet unknown genetic and perhaps non-genetic risks for Alzheimer’s disease. Across various fields of investigation recent data show remarkable effects associated with this risk factor. For family history risk there is no disclosure we could chose to avoid, but this should not prevent us from recognizing it’s value in Alzheimer’s disease research and clinical practice.
Acknowledgments
Funding Supported by NIH grant P01-AG025831, AG13308, P50 AG 16570, MH/AG58156, MH52453; AG10123; M01-RR00865, General Clinical Research Centers Program; the Fran and Ray Stark Foundation Fund for Alzheimer’s Disease Research; the Sence Foundation; and the McMahan Foundation.
Footnotes
Financial Disclosures The University of California, Los Angeles, owns a U.S. patent (6,274,119) entitled “Methods for Labeling b-Amyloid Plaques and Neurofibrillary Tangles,” and Dr. Small is among the inventors, has received royalties, and may receive royalties on future sales. Dr. Small reports having served as a consultant and/or having received lecture fees from Dakim, Forest, Lilly, and Novartis. Dr. Small also reports having received stock options from Dakim. Drs. Bookheimer and Donix report no financial relationships with commercial interests.
Contributor Information
Markus Donix, Email: markus.donix@uniklinikum-dresden.de, Department of Psychiatry and Psychotherapy, Universitätsklinikum Carl Gustav Carus, Technische Universität Dresden, 01307 Dresden, Germany. DZNE, German Center for Neurodegenerative Diseases, Dresden, Germany.
Gary W. Small, David Geffen School of Medicine at UCLA, Department of Psychiatry and Biobehavioral Sciences, Semel Institute, Los Angeles, CA 90095, USA. UCLA Longevity Center, Los Angeles, CA 90095, USA
Susan Y. Bookheimer, David Geffen School of Medicine at UCLA, Center for Cognitive Neurosciences, Semel Institute, Los Angeles, CA 90095, USA. David Geffen School of Medicine at UCLA, Department of Psychiatry and Biobehavioral Sciences, Semel Institute, Los Angeles, CA 90095, USA. Department of Psychology, David Geffen School of Medicine at UCLA, Los Angeles, CA 90095, USA
References
- Anand SS, Xie C, Pare G, Montpetit A, Rangarajan S, McQueen MJ, et al. Genetic variants associated with myocardial infarction risk factors in over 8000 individuals from five ethnic groups: The INTERHEART Genetics Study. Circulation Cardiovascular Genetics. 2009;2(1):16–25. doi: 10.1161/CIRCGENETICS.108.813709. [DOI] [PubMed] [Google Scholar]
- Bandettini PA, Ungerleider LG. From neuron to BOLD: new connections. Nature Neuroscience. 2001;4(9):864–866. doi: 10.1038/nn0901-864. [DOI] [PubMed] [Google Scholar]
- Bassett SS, Yousem DM, Cristinzio C, Kusevic I, Yassa MA, Caffo BS, et al. Familial risk for Alzheimer’s disease alters fMRI activation patterns. Brain. 2006;129(Pt 5):1229–1239. doi: 10.1093/brain/awl089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendlin BB, Ries ML, Canu E, Sodhi A, Lazar M, Alexander AL, et al. White matter is altered with parental family history of Alzheimer’s disease. Alzheimer’s & Dementia. 2010;6(5):394–403. doi: 10.1016/j.jalz.2009.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blazquez L, Otaegui D, Saenz A, Paisan-Ruiz C, Emparanza JI, Ruiz-Martinez J, et al. Apolipoprotein E epsilon4 allele in familial and sporadic Parkinson’s disease. Neuroscience Letters. 2006;406(3):235–239. doi: 10.1016/j.neulet.2006.07.037. [DOI] [PubMed] [Google Scholar]
- Bondi MW, Houston WS, Eyler LT, Brown GG. fMRI evidence of compensatory mechanisms in older adults at genetic risk for Alzheimer disease. Neurology. 2005;64(3):501–508. doi: 10.1212/01.WNL.0000150885.00929.7E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bookheimer SY, Strojwas MH, Cohen MS, Saunders AM, Pericak-Vance MA, Mazziotta JC, et al. Patterns of brain activation in people at risk for Alzheimer’s disease. The New England Journal of Medicine. 2000;343(7):450–456. doi: 10.1056/NEJM200008173430701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borenstein AR, Copenhaver CI, Mortimer JA. Early-life risk factors for Alzheimer disease. Alzheimer Disease and Associated Disorders. 2006;20(1):63–72. doi: 10.1097/01.wad.0000201854.62116.d7. [DOI] [PubMed] [Google Scholar]
- Braak H, Braak E. Staging of Alzheimer-related cortical destruction. International Psychogeriatrics. 1997;9(Suppl 1):257–261. discussion 269–272. [PubMed] [Google Scholar]
- Bretsky P, Guralnik JM, Launer L, Albert M, Seeman TE. The role of APOE-epsilon4 in longitudinal cognitive decline: MacArthur Studies of Successful Aging. Neurology. 2003;60(7):1077–1081. doi: 10.1212/01.wnl.0000055875.26908.24. [DOI] [PubMed] [Google Scholar]
- Brown JA, Terashima KH, Burggren AC, Ercoli LM, Miller KJ, Small GW, et al. Brain network local interconnectivity loss in aging APOE-4 allele carriers. Proceedings of the National Academy of Sciences USA. 2011 doi: 10.1073/pnas.1109038108. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunce D, Fratiglioni L, Small BJ, Winblad B, Backman L. APOE and cognitive decline in preclinical Alzheimer disease and non-demented aging. Neurology. 2004;63(5):816–821. doi: 10.1212/01.wnl.0000137041.86153.42. [DOI] [PubMed] [Google Scholar]
- Burggren AC, Zeineh MM, Ekstrom AD, Braskie MN, Thompson PM, Small GW, et al. Reduced cortical thickness in hippocampal subregions among cognitively normal apolipoprotein E e4 carriers. NeuroImage. 2008;41(4):1177–1183. doi: 10.1016/j.neuroimage.2008.03.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caselli RJ, Reiman EM, Locke DE, Hutton ML, Hentz JG, Hoffman-Snyder C, et al. Cognitive domain decline in healthy apolipoprotein E epsilon4 homozygotes before the diagnosis of mild cognitive impairment. Archives of Neurology. 2007;64(9):1306–1311. doi: 10.1001/archneur.64.9.1306. [DOI] [PubMed] [Google Scholar]
- Caselli RJ, Dueck AC, Locke DE, Sabbagh MN, Ahern GL, Rapcsak SZ, et al. Cerebrovascular risk factors and preclinical memory decline in healthy APOE epsilon4 homozygotes. Neurology. 2011;76(12):1078–1084. doi: 10.1212/WNL.0b013e318211c3ae. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman J, Korczyn AD, Karussis DM, Michaelson DM. The effects of APOE genotype on age at onset and progression of neurodegenerative diseases. Neurology. 2001;57(8):1482–1485. doi: 10.1212/wnl.57.8.1482. [DOI] [PubMed] [Google Scholar]
- Chen K, Reiman EM, Alexander GE, Caselli RJ, Gerkin R, Bandy D, et al. Correlations between apolipoprotein E epsilon4 gene dose and whole brain atrophy rates. The American Journal of Psychiatry. 2007;164(6):916–921. doi: 10.1176/ajp.2007.164.6.916. [DOI] [PubMed] [Google Scholar]
- Cherbuin N, Anstey KJ, Sachdev PS, Maller JJ, Meslin C, Mack HA, et al. Total and regional gray matter volume is not related to APOE*E4 status in a community sample of middle-aged individuals. The Journals of Gerontology Series A, Biological Sciences and Medical Sciences. 2008;63(5):501–504. doi: 10.1093/gerona/63.5.501. [DOI] [PubMed] [Google Scholar]
- Cupples LA, Farrer LA, Sadovnick AD, Relkin N, Whitehouse P, Green RC. Estimating risk curves for first-degree relatives of patients with Alzheimer’s disease: the REVEAL study. Genetics in Medicine. 2004;6(4):192–196. doi: 10.1097/01.gim.0000132679.92238.58. [DOI] [PubMed] [Google Scholar]
- Daw EW, Payami H, Nemens EJ, Nochlin D, Bird TD, Schellenberg GD, et al. The number of trait loci in late-onset Alzheimer disease. American Journal of Human Genetics. 2000;66(1):196–204. doi: 10.1086/302710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donix M, Burggren A, Suthana N, Siddarth P, Ekstrom A, Krupa A, et al. Family history of Alzheimer’s disease and hippocampal structure in healthy people. The American Journal of Psychiatry. 2010a;167(11):1399–1406. doi: 10.1176/appi.ajp.2010.09111575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donix M, Burggren AC, Suthana NA, Siddarth P, Ekstrom AD, Krupa AK, et al. Longitudinal changes in medial temporal cortical thickness in normal subjects with the APOE-4 polymorphism. NeuroImage. 2010b;53:37–43. doi: 10.1016/j.neuroimage.2010.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donix M, Ercoli LM, Siddarth P, Brown JA, Martin-Harris L, Burggren AC, et al. Influence of Alzheimer disease family history and genetic risk on cognitive performance in healthy middle-aged and older people. American Journal of Geriatric Psychiatry. 2011 doi: 10.1097/JGP.0b013e3182107e6a. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubois B, Feldman HH, Jacova C, Dekosky ST, Barberger-Gateau P, Cummings J, et al. Research criteria for the diagnosis of Alzheimer’s disease: revising the NINCDS-ADRDA criteria. Lancet Neurology. 2007;6(8):734–746. doi: 10.1016/S1474-4422(07)70178-3. [DOI] [PubMed] [Google Scholar]
- During EH, Osorio RS, Elahi FM, Mosconi L, de Leon MJ. The concept of FDG-PET endophenotype in Alzheimer’s disease. Neurological Science. 2011;32(4):559–569. doi: 10.1007/s10072-011-0633-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekstrom A. How and when the fMRI BOLD signal relates to underlying neural activity: the danger in dissociation. Brain Research Reviews. 2010;62(2):233–244. doi: 10.1016/j.brainresrev.2009.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elosua R, Ordovas JM, Cupples LA, Fox CS, Polak JF, Wolf PA, et al. Association of APOE genotype with carotid atherosclerosis in men and women: the Framingham Heart Study. Journal of Lipid Research. 2004;45(10):1868–1875. doi: 10.1194/jlr.M400114-JLR200. [DOI] [PubMed] [Google Scholar]
- Espeseth T, Westlye LT, Fjell AM, Walhovd KB, Rootwelt H, Reinvang I. Accelerated age-related cortical thinning in healthy carriers of apolipoprotein E epsilon 4. Neurobiology of Aging. 2008;29(3):329–340. doi: 10.1016/j.neurobiolaging.2006.10.030. [DOI] [PubMed] [Google Scholar]
- Filbey FM, Slack KJ, Sunderland TP, Cohen RM. Functional magnetic resonance imaging and magnetoencephalography differences associated with APOEepsilon4 in young healthy adults. Neuroreport. 2006;17(15):1585–1590. doi: 10.1097/01.wnr.0000234745.27571.d1. [DOI] [PubMed] [Google Scholar]
- Filippini N, MacIntosh BJ, Hough MG, Goodwin GM, Frisoni GB, Smith SM, et al. Distinct patterns of brain activity in young carriers of the APOE-epsilon4 allele. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(17):7209–7214. doi: 10.1073/pnas.0811879106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filippini N, Ebmeier KP, MacIntosh BJ, Trachtenberg AJ, Frisoni GB, Wilcock GK, et al. Differential effects of the APOE genotype on brain function across the lifespan. Neuro-Image. 2011;54(1):602–610. doi: 10.1016/j.neuroimage.2010.08.009. [DOI] [PubMed] [Google Scholar]
- Fleisher AS, Houston WS, Eyler LT, Frye S, Jenkins C, Thal LJ, et al. Identification of Alzheimer disease risk by functional magnetic resonance imaging. Archives of Neurology. 2005;62(12):1881–1888. doi: 10.1001/archneur.62.12.1881. [DOI] [PubMed] [Google Scholar]
- Folstein MF, Folstein SE, McHugh PR. Mini-Mental-State: a practical method for grading the cognitive state of patients for the clinician. Journal of Psychiatric Research. 1975;12:189–198. doi: 10.1016/0022-3956(75)90026-6. [DOI] [PubMed] [Google Scholar]
- Fratiglioni L, Ahlbom A, Viitanen M, Winblad B. Risk factors for late-onset Alzheimer’s disease: a population-based, case-control study. Annals of Neurology. 1993;33(3):258–266. doi: 10.1002/ana.410330306. [DOI] [PubMed] [Google Scholar]
- Gatz M, Reynolds CA, Fratiglioni L, Johansson B, Mortimer JA, Berg S, et al. Role of genes and environments for explaining Alzheimer disease. Archives of General Psychiatry. 2006;63(2):168–174. doi: 10.1001/archpsyc.63.2.168. [DOI] [PubMed] [Google Scholar]
- Glodzik-Sobanska L, Pirraglia E, Brys M, de Santi S, Mosconi L, Rich KE, et al. The effects of normal aging and ApoE genotype on the levels of CSF biomarkers for Alzheimer’s disease. Neurobiology of Aging. 2009;30(5):672–681. doi: 10.1016/j.neurobiolaging.2007.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gold BT, Powell DK, Andersen AH, Smith CD. Alterations in multiple measures of white matter integrity in normal women at high risk for Alzheimer’s disease. NeuroImage. 2010;52(4):1487–1494. doi: 10.1016/j.neuroimage.2010.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harold D, Abraham R, Hollingworth P, Sims R, Gerrish A, Hamshere ML, et al. Genome-wide association study identifies variants at CLU and PICALM associated with Alzheimer’s disease. Nature Genetics. 2009;41(10):1088–1093. doi: 10.1038/ng.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayden KM, Zandi PP, West NA, Tschanz JT, Norton MC, Corcoran C, et al. Effects of family history and apolipoprotein E epsilon4 status on cognitive decline in the absence of Alzheimer dementia: the Cache County Study. Archives of Neurology. 2009;66(11):1378–1383. doi: 10.1001/archneurol.2009.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert LE, Beckett LA, Scherr PA, Evans DA. Annual incidence of Alzheimer disease in the United States projected to the years 2000 through 2050. Alzheimer Disease and Associated Disorders. 2001;15(4):169–173. doi: 10.1097/00002093-200110000-00002. [DOI] [PubMed] [Google Scholar]
- Heise V, Filippini N, Ebmeier KP, Mackay CE. The APOE varepsilon4 allele modulates brain white matter integrity in healthy adults. Molecular Psychiatry. 2011;16(9):908–916. doi: 10.1038/mp.2010.90. [DOI] [PubMed] [Google Scholar]
- Holmes C, Boche D, Wilkinson D, Yadegarfar G, Hopkins V, Bayer A, et al. Long-term effects of Abeta42 immunisation in Alzheimer’s disease: follow-up of a randomised, placebo-controlled phase I trial. Lancet. 2008;372(9634):216–223. doi: 10.1016/S0140-6736(08)61075-2. [DOI] [PubMed] [Google Scholar]
- Honea RA, Vidoni E, Harsha A, Burns JM. Impact of APOE on the healthy aging brain: a voxel-based MRI and DTI study. Journal of Alzheimer’s Disease. 2009;18(3):553–564. doi: 10.3233/JAD-2009-1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honea RA, Swerdlow RH, Vidoni ED, Goodwin J, Burns JM. Reduced gray matter volume in normal adults with a maternal family history of Alzheimer disease. Neurology. 2010;74(2):113–120. doi: 10.1212/WNL.0b013e3181c918cb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honea RA, Swerdlow RH, Vidoni ED, Burns JM. Progressive regional atrophy in normal adults with a maternal history of Alzheimer disease. Neurology. 2011;76(9):822–829. doi: 10.1212/WNL.0b013e31820e7b74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jak AJ, Houston WS, Nagel BJ, Corey-Bloom J, Bondi MW. Differential cross-sectional and longitudinal impact of APOE genotype on hippocampal volumes in nondemented older adults. Dementia and Geriatric Cognitive Disorders. 2007;23(6):382–389. doi: 10.1159/000101340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvik GP, Wijsman EM. Alzheimer’s disease and the family effect. Nature Genetics. 1994;8(2):115. doi: 10.1038/ng1094-115a. [DOI] [PubMed] [Google Scholar]
- Johnson SC, Schmitz TW, Trivedi MA, Ries ML, Torgerson BM, Carlsson CM, et al. The influence of Alzheimer disease family history and apolipoprotein E epsilon4 on mesial temporal lobe activation. Journal of Neuroscience. 2006;26(22):6069–6076. doi: 10.1523/JNEUROSCI.0959-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert JC, Heath S, Even G, Campion D, Sleegers K, Hiltunen M, et al. Genome-wide association study identifies variants at CLU and CR1 associated with Alzheimer’s disease. Nature Genetics. 2009;41(10):1094–1099. doi: 10.1038/ng.439. [DOI] [PubMed] [Google Scholar]
- Langbaum JB, Chen K, Launer LJ, Fleisher AS, Lee W, Liu X, et al. Blood pressure is associated with higher brain amyloid burden and lower glucose metabolism in healthy late middle-age persons. Neurobiology of Aging. 2011 doi: 10.1016/j.neurobiolaging.2011.06.020. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Rue A, Hermann B, Jones JE, Johnson S, Asthana S, Sager MA. Effect of parental family history of Alzheimer’s disease on serial position profiles. Alzheimer’s & Dementia. 2008;4(4):285–290. doi: 10.1016/j.jalz.2008.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin MT, Beal MF. Mitochondrial dysfunction and oxidative stress in neurodegenerative diseases. Nature. 2006;443(7113):787–795. doi: 10.1038/nature05292. [DOI] [PubMed] [Google Scholar]
- Lind J, Persson J, Ingvar M, Larsson A, Cruts M, Van Broeckhoven C, et al. Reduced functional brain activity response in cognitively intact apolipoprotein E epsilon4 carriers. Brain. 2006;129(Pt 5):1240–1248. doi: 10.1093/brain/awl054. [DOI] [PubMed] [Google Scholar]
- Logothetis NK, Pauls J, Augath M, Trinath T, Oeltermann A. Neurophysiological investigation of the basis of the fMRI signal. Nature. 2001;412(6843):150–157. doi: 10.1038/35084005. [DOI] [PubMed] [Google Scholar]
- Luchsinger JA. Adiposity, hyperinsulinemia, diabetes and Alzheimer’s disease: an epidemiological perspective. European Journal of Pharmacology. 2008;585(1):119–129. doi: 10.1016/j.ejphar.2008.02.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunetta KL, Erlich PM, Cuenco KT, Cupples LA, Green RC, Farrer LA, et al. Heritability of magnetic resonance imaging (MRI) traits in Alzheimer disease cases and their siblings in the MIRAGE study. Alzheimer Disease and Associated Disorders. 2007;21(2):85–91. doi: 10.1097/WAD.0b013e3180653bf7. [DOI] [PubMed] [Google Scholar]
- Mahley RW, Weisgraber KH, Huang Y. Apolipoprotein E4: a causative factor and therapeutic target in neuropathology, including Alzheimer’s disease. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(15):5644– 5651. doi: 10.1073/pnas.0600549103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastroeni D, Grover A, Delvaux E, Whiteside C, Coleman PD, Rogers J. Epigenetic mechanisms in Alzheimer’s disease. Neurobiology of Aging. 2011;32(7):1161–1180. doi: 10.1016/j.neurobiolaging.2010.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mielke MM, Leoutsakos JM, Tschanz JT, Green RC, Tripodis Y, Corcoran CD, et al. Interaction between vascular factors and the APOE epsilon4 allele in predicting rate of progression in Alzheimer’s disease. Journal of Alzheimer’s Disease. 2011;26(1):127–134. doi: 10.3233/JAD-2011-110086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosconi L, Brys M, Switalski R, Mistur R, Glodzik L, Pirraglia E, et al. Maternal family history of Alzheimer’s disease predisposes to reduced brain glucose metabolism. Proceedings of the National Academy of Sciences of the United States of America. 2007;104(48):19067–19072. doi: 10.1073/pnas.0705036104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosconi L, Mistur R, Switalski R, Brys M, Glodzik L, Rich K, et al. Declining brain glucose metabolism in normal individuals with a maternal history of Alzheimer disease. Neurology. 2009;72(6):513–520. doi: 10.1212/01.wnl.0000333247.51383.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosconi L, Glodzik L, Mistur R, McHugh P, Rich KE, Javier E, et al. Oxidative stress and amyloid-beta pathology in normal individuals with a maternal history of Alzheimer’s. Biological Psychiatry. 2010;68(10):913–921. doi: 10.1016/j.biopsych.2010.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosconi L, Rinne JO, Tsui WH, Berti V, Li Y, Wang H, et al. Increased fibrillar amyloid-{beta} burden in normal individuals with a family history of late-onset Alzheimer’s. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(13):5949–5954. doi: 10.1073/pnas.0914141107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosconi L, Tsui W, Murray J, McHugh P, Li Y, Williams S, et al. Maternal age affects brain metabolism in adult children of mothers affected by Alzheimer’s disease. Neurobiology of Aging. 2011 doi: 10.1016/j.neurobiolaging.2011.03.003. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mount C, Downton C. Alzheimer disease: progress or profit? Nature Medicine. 2006;12(7):780–784. doi: 10.1038/nm0706-780. [DOI] [PubMed] [Google Scholar]
- Mueller SG, Schuff N, Raptentsetsang S, Elman J, Weiner MW. Selective effect of Apo e4 on CA3 and dentate in normal aging and Alzheimer’s disease using high resolution MRI at 4T. NeuroImage. 2008;42(1):42–48. doi: 10.1016/j.neuroimage.2008.04.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murrell J, Ghetti B, Cochran E, Macias-Islas MA, Medina L, Varpetian A, et al. The A431E mutation in PSEN1 causing familial Alzheimer’s disease originating in Jalisco State, Mexico: an additional fifteen families. Neurogenetics. 2006;7(4):277–279. doi: 10.1007/s10048-006-0053-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nierenberg J, Pomara N, Hoptman MJ, Sidtis JJ, Ardekani BA, Lim KO. Abnormal white matter integrity in healthy apolipoprotein E epsilon4 carriers. Neuroreport. 2005;16(12):1369–1372. doi: 10.1097/01.wnr.0000174058.49521.16. [DOI] [PubMed] [Google Scholar]
- Ohm TG, Muller H, Braak H, Bohl J. Close-meshed prevalence rates of different stages as a tool to uncover the rate of Alzheimer’s disease-related neurofibrillary changes. Neuroscience. 1995;64(1):209–217. doi: 10.1016/0306-4522(95)90397-p. [DOI] [PubMed] [Google Scholar]
- Persson J, Lind J, Larsson A, Ingvar M, Cruts M, Van Broeckhoven C, et al. Altered brain white matter integrity in healthy carriers of the APOE epsilon4 allele: a risk for AD? Neurology. 2006;66(7):1029–1033. doi: 10.1212/01.wnl.0000204180.25361.48. [DOI] [PubMed] [Google Scholar]
- Popp J, Lewczuk P, Frommann I, Kolsch H, Kornhuber J, Maier W, et al. Cerebrospinal fluid markers for Alzheimer’s disease over the lifespan: effects of age and the APOEepsilon4 genotype. Journal of Alzheimer’s Disease. 2010;22(2):459–468. doi: 10.3233/JAD-2010-100561. [DOI] [PubMed] [Google Scholar]
- Reiman EM, Caselli RJ, Chen K, Alexander GE, Bandy D, Frost J. Declining brain activity in cognitively normal apolipoprotein E epsilon 4 heterozygotes: a foundation for using positron emission tomography to efficiently test treatments to prevent Alzheimer’s disease. Proceedings of the National Academy of Sciences of the United States of America. 2001;98(6):3334–3339. doi: 10.1073/pnas.061509598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiman EM, Chen K, Alexander GE, Caselli RJ, Bandy D, Osborne D, et al. Functional brain abnormalities in young adults at genetic risk for late-onset Alzheimer’s dementia. Proceedings of the National Academy of Sciences of the United States of America. 2004;101(1):284–289. doi: 10.1073/pnas.2635903100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiman EM, Chen K, Alexander GE, Caselli RJ, Bandy D, Osborne D, et al. Correlations between apolipoprotein E epsilon4 gene dose and brain-imaging measurements of regional hypometabolism. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(23):8299–8302. doi: 10.1073/pnas.0500579102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiman EM, Chen K, Liu X, Bandy D, Yu M, Lee W, et al. Fibrillar amyloid-beta burden in cognitively normal people at 3 levels of genetic risk for Alzheimer’s disease. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(16):6820–6825. doi: 10.1073/pnas.0900345106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringman JM, Medina LD, Braskie M, Rodriguez-Agudelo Y, Geschwind DH, Macias-Islas MA, et al. Effects of risk genes on BOLD activation in presymptomatic carriers of familial Alzheimer’s disease mutations during a novelty encoding task. Cerebral Cortex. 2011;21(4):877–883. doi: 10.1093/cercor/bhq158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson GE, Fernald RD, Clayton DF. Genes and social behavior. Science. 2008;322(5903):896–900. doi: 10.1126/science.1159277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan L, Walther K, Bendlin BB, Lue LF, Walker DG, Glisky EL. Age-related differences in white matter integrity and cognitive function are related to APOE status. NeuroImage. 2011;54(2):1565–1577. doi: 10.1016/j.neuroimage.2010.08.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders AM. Apolipoprotein E and Alzheimer disease: an update on genetic and functional analyses. Journal of Neuropathology and Experimental Neurology. 2000;59(9):751–758. doi: 10.1093/jnen/59.9.751. [DOI] [PubMed] [Google Scholar]
- Seshadri S, Fitzpatrick AL, Ikram MA, DeStefano AL, Gudnason V, Boada M, et al. Genome-wide analysis of genetic loci associated with Alzheimer disease. JAMA: The Journal of the American Medical Association. 2010;303(18):1832–1840. doi: 10.1001/jama.2010.574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw P, Lerch JP, Pruessner JC, Taylor KN, Rose AB, Greenstein D, et al. Cortical morphology in children and adolescents with different apolipoprotein E gene polymorphisms: an observational study. Lancet Neurology. 2007;6(6):494–500. doi: 10.1016/S1474-4422(07)70106-0. [DOI] [PubMed] [Google Scholar]
- Small BJ, Rosnick CB, Fratiglioni L, Backman L. Apolipoprotein E and cognitive performance: a meta-analysis. Psychology and Aging. 2004;19(4):592–600. doi: 10.1037/0882-7974.19.4.592. [DOI] [PubMed] [Google Scholar]
- Small GW, Siddarth P, Burggren AC, Kepe V, Ercoli LM, Miller KJ, et al. Influence of cognitive status, age, and APOE-4 genetic risk on brain FDDNP positron-emission tomography imaging in persons without dementia. Archives of General Psychiatry. 2009;66(1):81–87. doi: 10.1001/archgenpsychiatry.2008.516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CD, Chebrolu H, Andersen AH, Powell DA, Lovell MA, Xiong S, et al. White matter diffusion alterations in normal women at risk of Alzheimer’s disease. Neurobiology of Aging. 2010;31(7):1122–1131. doi: 10.1016/j.neurobiolaging.2008.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunderland T, Mirza N, Putnam KT, Linker G, Bhupali D, Durham R, et al. Cerebrospinal fluid beta-amyloid1-42 and tau in control subjects at risk for Alzheimer’s disease: the effect of APOE epsilon4 allele. Biological Psychiatry. 2004;56(9):670–676. doi: 10.1016/j.biopsych.2004.07.021. [DOI] [PubMed] [Google Scholar]
- Teter B. ApoE-dependent plasticity in Alzheimer’s disease. Journal of Molecular Neuroscience. 2004;23(3):167–179. doi: 10.1385/JMN:23:3:167. [DOI] [PubMed] [Google Scholar]
- Trivedi MA, Schmitz TW, Ries ML, Torgerson BM, Sager MA, Hermann BP, et al. Reduced hippocampal activation during episodic encoding in middle-aged individuals at genetic risk of Alzheimer’s disease: a cross-sectional study. BMC Medicine. 2006;4:1–14. doi: 10.1186/1741-7015-4-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trivedi MA, Schmitz TW, Ries ML, Hess TM, Fitzgerald ME, Atwood CS, et al. fMRI activation during episodic encoding and metacognitive appraisal across the lifespan: risk factors for Alzheimer’s disease. Neuropsychologia. 2008;46(6):1667–1678. doi: 10.1016/j.neuropsychologia.2007.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuminello ER, Han SD. The apolipoprotein e antagonistic pleiotropy hypothesis: review and recommendations. Int J Alzheimers Dis. 2011;2011:726197. doi: 10.4061/2011/726197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Duijn CM, Clayton D, Chandra V, Fratiglioni L, Graves AB, Heyman A, et al. Familial aggregation of Alzheimer’s disease and related disorders: a collaborative re-analysis of case-control studies. International Journal of Epidemiology. 1991;20(Suppl 2):S13–S20. doi: 10.1093/ije/20.supplement_2.s13. [DOI] [PubMed] [Google Scholar]
- van Duijn CM, de Knijff P, Cruts M, Wehnert A, Havekes LM, Hofman A, et al. Apolipoprotein E4 allele in a population-based study of early-onset Alzheimer’s disease. Nature Genetics. 1994;7(1):74–78. doi: 10.1038/ng0594-74. [DOI] [PubMed] [Google Scholar]
- van Exel E, Eikelenboom P, Comijs H, Frolich M, Smit JH, Stek ML, et al. Vascular factors and markers of inflammation in offspring with a parental history of late-onset Alzheimer disease. Archives of General Psychiatry. 2009;66(11):1263–1270. doi: 10.1001/archgenpsychiatry.2009.146. [DOI] [PubMed] [Google Scholar]
- Wisdom NM, Callahan JL, Hawkins KA. The effects of apolipoprotein E on non-impaired cognitive functioning: a meta-analysis. Neurobiology of Aging. 2011;32(1):63–74. doi: 10.1016/j.neurobiolaging.2009.02.003. [DOI] [PubMed] [Google Scholar]
- Wishart HA, Saykin AJ, Rabin LA, Santulli RB, Flashman LA, Guerin SJ, et al. Increased brain activation during working memory in cognitively intact adults with the APOE epsilon4 allele. The American Journal of Psychiatry. 2006;163(9):1603–1610. doi: 10.1176/ajp.2006.163.9.1603. [DOI] [PubMed] [Google Scholar]
- Wisniewski T, Boutajangout A. Vaccination as a therapeutic approach to Alzheimer’s disease. The Mount Sinai Journal of Medicine. 2010;77(1):17–31. doi: 10.1002/msj.20156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu G, McLaren DG, Ries ML, Fitzgerald ME, Bendlin BB, Rowley HA, et al. The influence of parental history of Alzheimer’s disease and apolipoprotein E epsilon4 on the BOLD signal during recognition memory. Brain. 2009;132(Pt 2):383–391. doi: 10.1093/brain/awn254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yip AG, McKee AC, Green RC, Wells J, Young H, Cupples LA, et al. APOE, vascular pathology, and the AD brain. Neurology. 2005;65(2):259–265. doi: 10.1212/01.wnl.0000168863.49053.4d. [DOI] [PubMed] [Google Scholar]
- Zintl M, Schmitz G, Hajak G, Klunemann HH. ApoE genotype and family history in patients with dementia and cognitively intact spousal controls. American Journal of Alzheimer’s Disease and Other Dementias. 2009;24(4):349–352. doi: 10.1177/1533317509333040. [DOI] [PMC free article] [PubMed] [Google Scholar]