Abstract
Bacteriophages are traditionally used for the development of phage display technology. Recently, their nanosized dimensions and ease with which genetic modifications can be made to their structure and function have put them in the spotlight towards their use in a variety of biosensors. In particular, the expression of any protein or peptide on the extraluminal surface of bacteriophages is possible by genetically engineering the genome. In addition, the relatively short replication time of bacteriophages offers researchers the ability to generate mass quantities of any given bacteriophage-based biosensor. Coupled with the emergence of various biomarkers in the clinic as a means to determine pathophysiological states, the development of current and novel technologies for their detection and quantification is imperative. In this review, we categorize bacteriophages by their morphology into M13-based filamentous bacteriophages and T4- or T7-based icosahedral bacteriophages, and examine how such advantages are utilized across a variety of biosensors. In essence, we take a comprehensive approach towards recent trends in bacteriophage-based biosensor applications and discuss their outlook with regards to the field of biotechnology.
Keywords: biosensing, M13 bacteriophage, T4 bacteriophage, bacterial detection, Escherichia coli, SPR sensor
Introduction
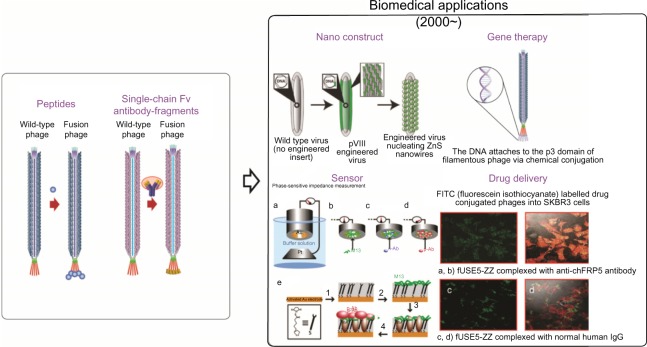
Since it was first reported in 1985 in Science,1 phage display technology has played a key role towards the development of antigen-specific peptides. Specifically, bacteriophages are used to choose select peptides with high specificity and affinity towards a desired antigen from a pool consisting of a large number of potential peptides. Their subsequent use for the recognition of specific target molecules and biomarkers has yielded a variety of economical, rapid, and efficient applications in fields such as vaccine development, enzyme inhibition, inflammation, and cancer research. In particular, the readily available information on the genomic makeup of various bacteriophages has enabled researchers to easily engineer foreign entities (ie, proteins, peptides, and so on) on the extraluminal surface, which in turn has enabled their widespread use for phage display applications. While bacteriophages have continued to receive interest for these reasons, the last 10 years in particular have seen an emergence of novel bacteriophage-based biosensor applications in the form of nanotemplates or nanoprobes (Figure 1).2,3
Figure 1.
Traditional and recent applications of bacteriophages.
Notes: Bacteriophages, conventionally used in phage display technology for the selection of peptides or antibody fragments, have emerged in the development of a variety of biomedical applications such as the fabrication of nanowire-based secondary battery cells, gene therapy constructs, sensors, and drug delivery vehicles.14,32,66–69 The nano construct is adapted from Proc Natl Acad Sci U S A. Mao C, Flynn CE, Hayhurst A, et al. Viral assembly of oriented quantum dot nanowires. 100(12):6946–6951, Copyright (2003) with permission from PMC.68 Sensor is adapted from Anal Chem. Yang LC, Tam PY, Murray BJ, et al. Virus electrodes for universal biodetection. 78:3265–3270, Copyright (2006) with permission from the American Chemical Society.69 Drug delivery is adapted from Antimicrob Agents Chemother. Yacoby I, Bar H, Benhar I. Targeted drug-carrying bacteriophages as antibacterial nanomedicines. 51(6):2156–2163, Copyright (2007) with permission from BioMed Central.14
Abbreviations: Fv, variable fragment; DNA, deoxyribonucleic acid; PMC, PubMed Central.
The most widely used bacteriophages can be categorized into filamentous (M13, fd) and icosahedral (T7, T4) phages, and are largely composed of two segments: a capsid and tail fiber region (Figure 2). Here, the former contains the genomic material (for example, deoxyribonucleic acid [DNA] or ribonucleic acid), and the latter binds to receptors on the surface of its host (for example, bacteria) to inject its genome for subsequent host infection, and ultimately, the replication and proliferation of bacteriophages.4–7 Provided that there is adequate culture of the host, an economical, rapid, and reproducible means to mass produce genetically-engineered bacteriophages is possible,8 thereby providing an excellent alternative to conventional artificially-synthesized nanoconstructs (ie, nanoparticles, nanorods, and nanotubes). Additionally, their morphological variations (dimensions ranging from 40 nm to 2 μm with linear or polygonal capsid structures), the ease with which genetic modifications can be made to their structure and functionality, and their biocompatibility confer further advantages over traditional nanoconstructs.9 In essence, bacteriophages have transcended their mere use in phage display technology to play an expanded role in the development of various applications such as biosensing, cellular imaging, vaccine development, and drug and gene delivery.10–15
Figure 2.
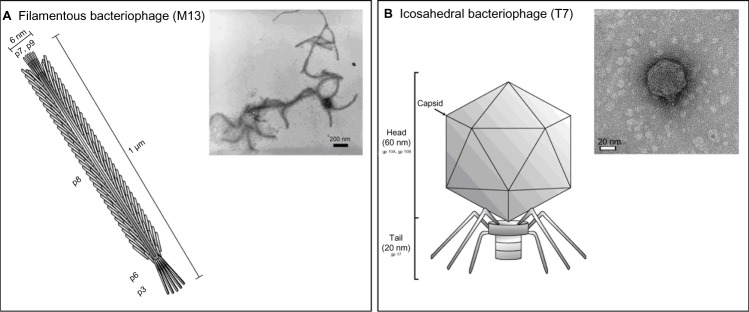
Categorization of bacteriophages.
Notes: (A) Schematic representation of M13, a representative filamentous bacteriophage (inset: transmission electron microscopy image of multiple M13 bacteriophages). (B) Schematic representation of T7, a representative icosahedral bacteriophage (inset: transmission electron microscopy image of a single T7 bacteriophage). Reprinted with permission from Mao C, Liu A, Cao B. Virus-based chemical and biological sensing. Angew Chem Int Ed Engl. 2009;48(37):6790–6810. Copyright 2009 John Wiley and Sons.
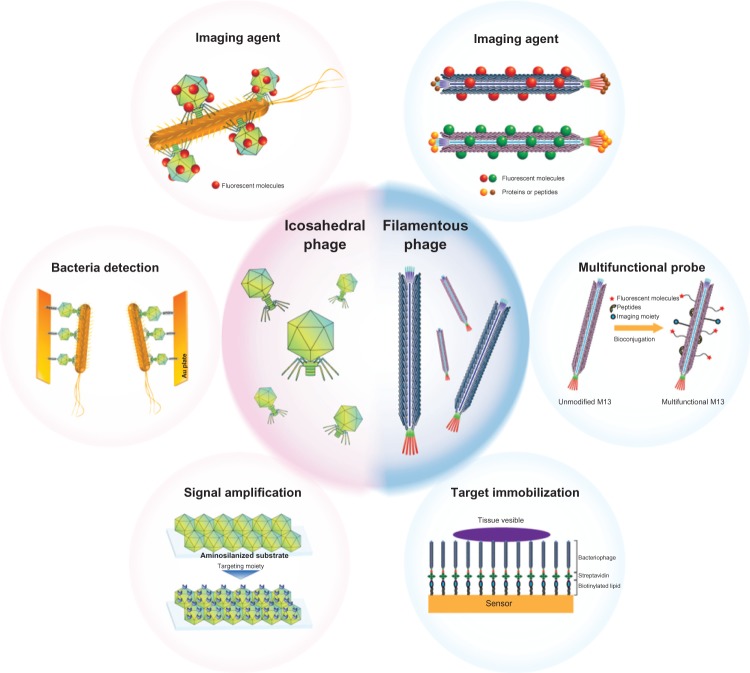
Herein, we categorize bacteriophages by their morphology into M13-based filamentous phages and T4- or T7-based icosahedral phages. We then examine recent technological trends, as well as bacteriophage development as biosensors, and discuss their prospective use in the field of biotechnology (Figure 3).
Figure 3.
Current applications of bacteriophages in biotechnology.
Notes: Icosahedral bacteriophages (ie, T4 and T7) are often used as imaging agents, targeting moieties for the detection of bacteria, and substrates for the amplification of signal. Filamentous bacteriophages (ie, M13) are often used as imaging agents, multifunctional probes, and components in a variety of sensors.
Filamentous bacteriophages as biomedical nanoprobes
M13, a representative type of filamentous bacteriophage, resembles a long thread of string with its 6 nm thick and 1 μm long dimensions (Figure 2A). Approximately 3,000 p8 subunits, a type of major coat protein, form a cylinder to encapsulate the M13 single-stranded DNA. Furthermore, each phage is capped by five p3 and p6 subunits on one end, and by five p7 and p9 subunits at the other end.17–21 Among the sequences for the major and minor coat proteins, those for p3, p8, and p9 are particularly amenable for genetic modification for the consequent expression of recombinant coat protein subunits on the surface of the filamentous phage. Specifically, up to three different types of peptides can be expressed on the surface via corresponding modifications to the p3, p8, and p9 sequences. Such flexibility imparts multiple functions to the bacteriophage, and not surprisingly, has paved the way for the use of filamentous bacteriophage for a variety of multifunctional nanoprobes.22–29
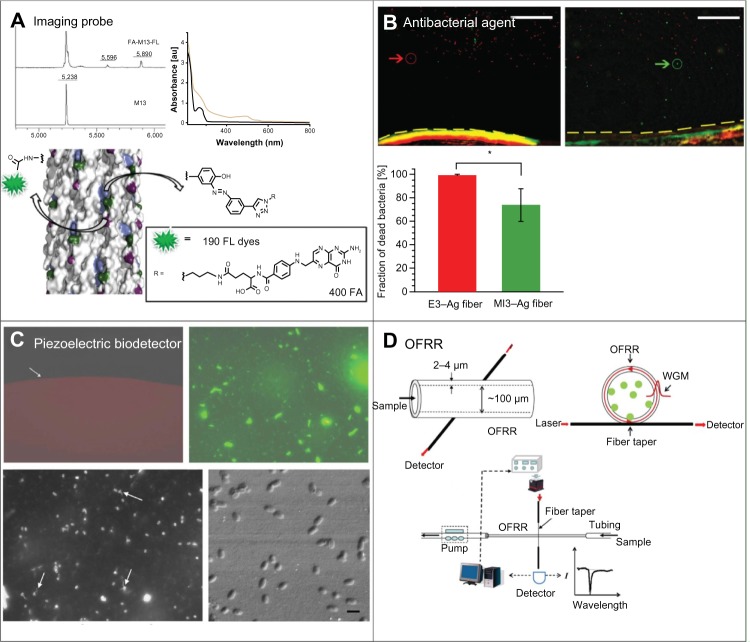
The most common form of filamentous bacteriophage-based biosensor is engineered by conjugating targeting moieties and fluorescent molecules onto the capsid surface, ultimately for the detection of target analyte and for the transduction of signal via fluorescence.30,31 For example, Li et al9 constructed a bacteriophage capable of targeting a HeLa contaminant KB cell line and emitting fluorescence upon target binding (Figure 4A). Specifically, the p8 domain is modified for the surface expression of folic acid, while lysine groups and N-termini on the bacteriophage surface are conjugated to carboxylic acid and glutamic acid groups on fluorescent molecules. Approximately 190 fluorescent molecules were determined to be conjugated onto each p8 subunit, effectively increasing detection sensitivity. In addition to the use of filamentous bacteriophages as imaging modalities, others have used them as theranostic (ie, a combination of therapeutics and diagnostics) agents capable of the delivery of drugs.32 For instance, Ghosh et al16 modified the p3, p8, and p9 domains for the detection and visualization of a prostate cancer cell line. Specifically, secreted protein, acidic and rich in cysteine-binding peptides are expressed on p3 subunits, doxorubicin onto p8 subunits, and Alexa Fluor® 488 (Life Technologies, Carlsbad, CA, USA) onto p9 subunits for the detection of prostate cancer cells, their treatment, and their visualization, respectively.
Figure 4.
Examples of filamentous bacteriophage-based biomedical applications.
Notes: (A) Bacteriophages engineered to display both folic acid and fluorescent molecules on their surface are used to detect and image a HeLa contaminant KB cell line. Upper left: MALDI–TOF MS of wild-type M13 and dual-modified M13. Upper right: UV–Vis absorbance spectra of wild-type M13 (black) and dual-modified M13 (orange) after purification. Lower: surface structure of M13 conjugated with fluorescent dyes via amine and tyrosine residues. Reprinted from Bioconjug Chem. Li K, Chen Y, Li S, et al. Chemical modification of M13 bacteriophage and its application in cancer cell imaging. 21:1369–1377, Copyright (2010) with permission from the American Chemical Society.9 (B) Bacteriophages are coated with Ag and used for the selective killing of bacterial cells (red: e3 bacteriophage; green: M13 bacteriophage) Upper left: E3 bacteriophage–Ag fiber. Upper right: M13 bacteriophage–Ag fiber. Lower: live–dead cell counts. Scale bar =100 μm. Reprinted from Adv Funct Mater. Mao JY, Belcher AM, Van Vliet KJ. Genetically engineered phage fibers and coatings for antibacterial applications. 20(2):209–214, Copyright (2010) with permission from WILEY-VCH Verlag GmbH and Co. KGaA.36 (C) E2 bacteriophages are used for the detection of Salmonella typhimurium via a modified QCM sensor. Upper left: a clean resonator (control) is operated in real-time. Upper right: image of resonator following phage deposition. Lower left: a fluorescence image of S. typhimurium attached to the surface of a resonator. Lower right: a SEM image of S. typhimurium bound to the bacteriophage immobilized on the sensor surface. Reprinted from Biosens Bioelectron. Olsen EV, Sorokulova IB, Petrenko VA, Chen IH, Barbaree JM, Vodyanoy VJ. Affinity-selected filamentous bacteriophage as a probe for acoustic wave biodetectors of S. typhimurium. 21(8):1434–1442, Copyright (2006) with permission from Elsevier.71 (D) R5C2 bacteriophages are modified to express biotin and used to improve conventional OFRR sensor capabilities for the detection of streptavidin. Upper left: a microsized glass capillary with a thin wall (<4 μm) makes up the OFRR sensor. Upper right: formation of a ring resonator via the capillary cross section. Lower: experimental scheme for the detection of biomolecules. Reprinted from Biosens Bioelectron. Zhu H, white IM, Suter JD, Fan X. Phage-based label-free biomolecule detection in an opto-fluidic ring resonator. 24(3):461–466, Copyright (2008) with permission from Elsevier.47
Abbreviations: Ag, silver; OFFR, optofluidic resonator; WGM, whole-genome mapping; MALDI–TOF MS, matrix-assisted laser desorption/ionization–time-of-flight mass spectrometer; UV–Vis, ultraviolet–visible spectroscopy; SEM, scanning electron microscopy.
Another common type of genetically-engineered filamentous bacteriophage-based sensor involves the conjugation of nanoparticles to the capsid, in which the unique properties of each type of nanoparticle is exploited in an application-specific manner.33–35 In one instance, Lee et al10 engineered a bacteriophage with a peptide library against a specific antigen on the p3 subunit and gold (Au) nanoparticles on its p8 subunit. By measuring the ultraviolet absorbance from the engineered bacteriophage, the authors demonstrated the capacity to detect protein down to 25 fmoles. In another example, Mao et al36 expressed a peptide on the p8 subunits of E3 and M13 bacteriophages, capable of electrostatically binding to silver particles for antibacterial applications (Figure 4B). By fabricating a silver-coated phage for the selective killing of bacterial cells, the authors demonstrate 99% and 74% bacterial cell death via the use of E3 and M13 bacteriophages, respectively.
Another type of filamentous bacteriophage-based biosensor uses the piezoelectric effect of quartz crystal microbalance (QCM) sensors. Generally coated with a layer of metal such as Au, QCM sensors enable the real-time analysis of biomolecular interactions down to the nano-gram scale. Conventional QCM methods, however, have disadvantages such as limited sensitivity due to the internal friction generated from the direct binding of analyte to the transducer. To overcome such issues, modified QCM biosensors have been developed based on the immobilization of bacteriophages onto the surface of QCM. For instance, Huang et al37 expressed a peptide with an affinity for Salmonella typhimurium on the surface of E2 bacteriophages, and immobilize the modified bacteriophage onto a QCM sensor (Figure 4C). The authors demonstrated a detection limit of 102 cells mL−1 within an assay time of 3 minutes. Similarly, Olsen et al38 Expressed the VTPPTQHQ peptide, specific for Salmonella typhimurium, onto the 7b1 filamentous phage, and immobilized the bacteriophage onto a QCM sensor for subsequent resonance measurements.
Traditional methods used for the detection of bacteria such as the enumeration of colonies in vitro, the use of polymerase chain reaction, or enzyme-linked immune sorbent assays, are generally time-consuming and require a large number of samples. The detection of pathogens in the field, however, requires the real-time detection of the target analyte in-situ. Accordingly, the use of bacteriophage-based magnetoelastic (ME) resonators is emerging.39–41 ME resonators – composed of a material that lengthens and shortens under an external magnetic field – emit a characteristic resonance frequency upon exposure to the external magnetic field and release an oscillatory signal, which is ultimately detected via a pickup coil. Bacteriophage nanoprobes used in bacteriophage-based ME biosensors are genetically modified to bind to a target pathogen and are immobilized onto the resonator. Upon capture of the target pathogen, the ME biosensor is placed under increasing load, and the resulting decrease in resonance frequency is measured as a means to analyze the number of pathogens.37,42 For example, Li et al42 immobilized E2 phage, genetically modified to express a peptide against S. typhimurium, onto the surface of ME resonators, and they used the modified ME biosensor to demonstrate the real-time ability to track in situ S. typhimurium levels on the surface of a tomato. The binding of the modified bacteriophage onto S. typhimurium is confirmed via scanning electron microscopy and a detection sensitivity of 50 colony-forming units/mL−1 is obtained. In another example, Shen et al43 immobilized the JRB7 phage, modified to express a library against Bacillus anthracis spores onto a ME resonator, and used the system to take real-time measurements of in vitro biomarker concentrations from the solution.
The use of fluorescent tags in biosensors is quite often a time-consuming process that alters the physical and chemical properties of the target analyte. To overcome such obstacles, optical sensors, such as the use of surface plasmon resonance (SPR), provide real-time measurements on biomolecular interactions without the need for fluorescent tags. SPR sensors, however, are relatively large in size, complicated in their structure, and require an external microfluidic channel, as well as an optical detection system.44 Optofluidic ring resonators compensate for such weaknesses by combining microfluidic channels with microsized ring resonators to reduce the overall size of the sensor (<4 μm), thereby offering an alternative fluorescent label-free method for the real-time detection of the target analyte.45 Zhu et al46 engineered an OFRR biosensor that utilizes R5C2 filamentous phages – genetically modified to express biotin – as receptors for the detection of streptavidin, a protein tetramer with a high affinity for biotin (Figure 4D). By demonstrating a detection sensitivity down to 100 pM and a dissociation constant of 25 pM, the authors present a cheap and highly-sensitive platform for the detection of biomolecules.
Icosahedral bacteriophages as biomedical nanoprobes
While filamentous bacteriophages can be engineered to express peptides or proteins on their surface, their secretion and consequent interaction with the bacteria cell membrane needs to be taken into consideration. While such nonlytic filamentous bacteriophages secrete newly synthesized phages by merging with the host cellular membrane, icosahedral bacteriophages lyse the host cell following infection of the host and replication of the viral genome. Unlike nonlytic filamentous bacteriophages that secrete newly synthesized phages by merging with the host cellular membrane, icosahedral bacteriophages lyse the host cell following infection of the host and replication of the viral genome.47 This lytic cycle of icosahedral bacteriophages ensures that the expression of foreign proteins or peptides on the capsid surface is not hindered by the need to consider their interaction with the bacterial cell membrane. In other words, without the need to consider the effect of such foreign constructs on bacteriophage uptake and its distribution from the host, researchers have a much wider spectrum of constructs to choose from when engineering a bacteriophage.48–50 In addition, Krumpe et al51 demonstrated a lower rate of genetic mutation or sequence bias when expressing a peptide library on the capsid of T7 icosahedral bacteriophages than on that of M13 filamentous bacteriophages. Furthermore, icosahedral bacteriophages do not exhibit issues of aggregation, as observed with filamentous bacteriophages (Figure 1A), making them appropriate for use in applications that require monodispersed nanoprobes. Finally, icosahedral bacteriophages have complex structural symmetry denoted by their capsid (icosahedral symmetry) and tail fibers (helical symmetry). This inherent anisotropy is in contrast to traditional isotropic nanoconstructs (quantum dots, nanoparticles, nanotubes, liposomes, and so on),52 making it possible to engineer a true Janus nanoconstruct (ie, a different peptide or protein displayed on the capsid from the tail fiber) with multiple functions incorporated into one construct.53 Such icosahedral bacteriophage attributes will most likely generate growing interest among researchers to yield a variety of unique nanosensors in the future.
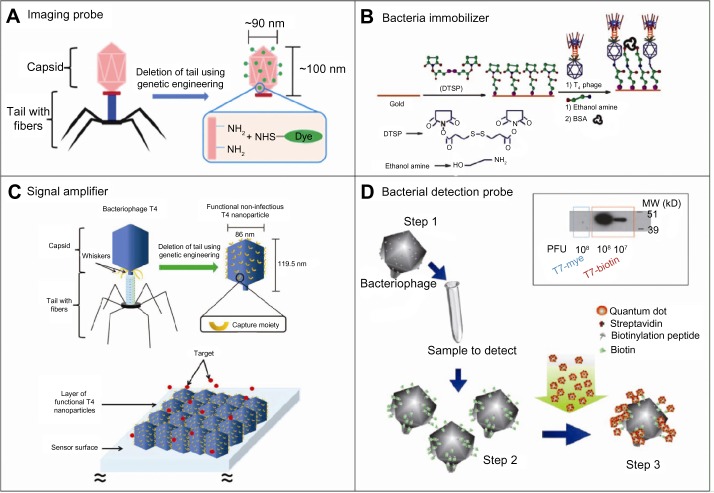
An examination of recent literature detailing the use of icosahedral bacteriophages as nanosensors shows that the most common form of icosahedral bacteriophage-based biosensors utilizes the spherical profile of the capsid as an alternative to conventional nanoparticles, onto which various peptides or protein subunits are expressed.54–56 For example, Robertson et al48 engineered a T4-based nanoprobe with applications in cellular imaging and flow cytometry by conjugating fluorescent dyes to the capsid. By inducing the expressing of NH2 groups on the capsid for subsequent NH2–N-hydroxysuccinimide (NHS) conjugation with Alexa Fluor® 546 (Life Technologies), the authors demonstrate cellular uptake of their proof-of-concept bacteriophage for a variety of imaging applications (Figure 5A). In another example, Barrett et al57 conjugated or expressed a variety of structures – streptavidin, biotin, thiol groups, amine groups, and so on – onto the capsid of a T7 icosahedral bacteriophage. Others, such as Merril et al,58 expressed protein on the capsid of a T4 bacteriophage for the targeting of specific biomolecules, nucleic acids, or protein biomarkers. In yet another example, Ren and Lai59 expressed a peptide on the capsid of a T4 bacteriophage for the detection of a cancer antigen. There have also been slight modifications to such applications, as seen with Liu et al,60 who engineered a T7-based bacteriophage for imaging applications by replacing the DNA within the bacteriophage with a fluorescent dye.
Figure 5.
Examples of icosahedral bacteriophage-based biomedical applications.
Notes: (A) The tail fibers of T4 bacteriophages are genetically removed to generate nanoparticles composed of T4 capsids, which are subsequently conjugated with fluorescent molecules for use as a nanoprobe. Reprinted from Bioconjugate chemistry. Robertson KL, Soto CM, Archer MJ, Odoemene O, Liu JL. Engineered T4 viral nanoparticles for cellular imaging and flow cytometry. 22(4):595–604, Copyright (2011) with permission from American Chemical Society.48 (B) The head of a T4 bacteriophage is conjugated to a gold surface via DTSP–amine, leaving the tail available for the detection of bacteria. Reprinted from Analyst. Arya SK, Singh A, Naidoo R, Wu P, McDermott MT, Evoy S. Chemically immobilized T4-bacteriophage for specific Escherichia coli detection using surface plasmon resonance. 136(3):486–492, Copyright (2011) with permission from the Royal Society of Chemistry.62 (C) T4 bacteriophages, which have been conjugated with a targeting moiety on its head, are dispersed onto an aminosilanized substrate for the detection of bacteria. Reprinted from Proc Natl Acad Sci U S A. Archer MJ, Liu JL. Bacteriophage t4 nanoparticles as materials in sensor applications: variables that influence their organization and assembly on surfaces. 9(8):6298–6311, Copyright (2009) with permission from MDPI.65 (D) T7 bacteriophages, which have been conjugated with quantum dots on its head, are used for the detection of bacteria. Reprinted from Proc Natl Acad Sci U S A. Edgar R, McKinstry M, Hwang J, et al. High-sensitivity bacterial detection using biotin-tagged phage and quantum-dot nanocomplexes. 103(13):4841–4845, Copyright (2006) with permission from PMC.3,64
Abbreviations: DTSP, Dithiobis N-succinimidyl propionate; BSA, bovine serum albumin; PFU, plaque-forming unit; MDPI, Multidisciplinary Digital Publishing Institute; PMC, PubMed Central.
Another commonly utilized form of icosahedral bacteriophages used as a biosensor is the immobilization of bacteriophage onto a platform for the subsequent detection of bacteria via its tail fiber region.61 For example, Arya et al62 assembled T4 bacteriophages onto an Au platform and demonstrated the ability to detect bacteria. Specifically, the Au platform is functionalized with dithiobis (succinimidyl propionate) for the covalent immobilization of the T4 bacteriophage via amine groups that were genetically introduced on the T4 capsid. Once the bacteriophages are bound to the Au platform, the tail fibers – orientated outwards – detect bacteria, all of which leads to a change in SPR signal (Figure 5B).62 In another example, Boss and Lieberman63 developed a T4-based bacteriophage biosensor for the detection of bacteria by utilizing magnetic nanoparticles. Here, amine groups on magnetic nanoparticles form covalent bonds with alkylamine compounds genetically introduced on the T4 capsids, and the resulting modified bacteriophages are used with conventional sensors, such as SPR or QCM, as a means to improve their detection capacity. A slightly different approach was taken by Archer and Liu,3 who removed the tail fibers of T4 bacteriophages to form capsid-only constructs.64 After conjugating targeting moieties onto the capsids, the authors optimized the pH to reduce modified T4 aggregation upon their immobilization onto an amino-silanized sensor platform. Here, the authors demonstrated severe aggregation at pH 5.6 that decreases in degree as the pH shifts towards 8.8, at which point the modified bacteriophages are equally-distributed onto the sensor platform (Figure 5C). Ultimately, the authors validated the use of their modified bacteriophage with SPR or QCM systems as a means to improve conventional biosensor capabilities.
Recently, examples of multifunctional nanoprobes – engineered by conjugating nanoparticles onto the bacteriophage capsid – have emerged. For example, Edgar et al65 conjugated quantum dots onto a T7 bacteriophage capsid for the detection of bacteria with a sensitivity of down to 10 cells/mL−1. Here, the modified bacteriophages are generated by expressing biotinylated peptide on the capsid for subsequent binding to streptavidin-coated quantum dots (Figure 5D). Similarly, Caberoy et al66 engineered a modified T7 bacteriophage for the detection of Tubby-like protein 1. Specifically, the authors expressed both 3C protease cleavage sites and biotinylation tags onto the capsid; upon selection of bacteriophages bound to streptavidin, the authors cleaved the 3C site to generate a biotinylated complementary DNA library.
Future opportunities for bacteriophages in biosensor applications
Viruses have been used in a variety of chemical and biological sensors, and new applications continue to emerge. Bacteriophages, in particular, are one of the most successful viruses yet to have been applied towards biosensors, and can be grouped into the following three categories regardless of their filamentous or icosahedral classification: 1) specific peptides or proteins discovered through phage display technology are genetically-introduced onto the extraluminal surface of the bacteriophage for the detection of target analyte. In general, biosensors based on the use of nonlytic bacteriophages (ie, M13) form the majority of this class. 2) Specific bacterial cell markers are detected upon bacterial cell rupture via the use of a lytic phage, which infects the host, replicates its genome, and induces host cell lysis. 3) Bacteriophages act a nanoscaffolding material for the transportation of functional molecules or their immobilization in a variety of sensing devices.
The translation of bacteriophage-based biosensors from in vitro to in vivo settings must be preceded by toxicity examinations to eliminate any safety concerns. Furthermore, the sensitivity of the general public to genetically engineered bacteriophages must be taken into account, and this is arguably one of the largest hurdles in the field. However, by coupling the aforementioned advantages of bacteriophages with the continued development of micro-and nanobased measurements and analytical devices, we predict a concurrent increase in safety awareness and subsequent improvements. Finally, it is important to note that the use of bacteriophages is not limited to the aforementioned categories. Bacteriophages with varying functions that express desired peptides or proteins on their surface can be engineered through the simple manipulation of the bacteriophage genome, and ultimately be used for the rapid and economical mass production of bacteriophage-based biosensors. Here, an appropriate understanding of the bacteriophage that covers a wide area of topics such as the structure and function of individual components and its life cycle must accompany such novel applications. In summary, we envision that the combination of such bacteriophages with analytical nanofabricated devices will emerge in novel applications as a powerful nanoscale tool for the detection of a wide spectrum of targets, ranging from biological agents (ie, proteins, bacteria, spores, viruses, toxins, and so on) to chemical agents or even explosives.
Acknowledgments
The authors would like to gratefully acknowledge the financial support provided by the Korea Institute of Science and Technology (KIST Institutional Project #2E23720 and Dream Project # 2V02840).
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Smith GP. Filamentous fusion phage: novel expression vectors that display cloned antigens on the virion surface. Science. 1985;228(4705):1315–1317. doi: 10.1126/science.4001944. [DOI] [PubMed] [Google Scholar]
- 2.Deutscher SL. Phage display in molecular imaging and diagnosis of cancer. Chem Rev. 2010;110(5):3196–3211. doi: 10.1021/cr900317f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Archer MJ, Liu JL. Bacteriophage t4 nanoparticles as materials in sensor applications: variables that influence their organization and assembly on surfaces. Sensors (Basel) 2009;9(8):6298–6311. doi: 10.3390/s90806298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Petrenko VA. Landscape phage as a molecular recognition interface for detection devices. Microelectronics J. 2008;39(2):202–207. doi: 10.1016/j.mejo.2006.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Singh A, Arutyunov D, Szymanski CM, Evoy S. Bacteriophage based probes for pathogen detection. Analyst. 2012;137(15):3405–3421. doi: 10.1039/c2an35371g. [DOI] [PubMed] [Google Scholar]
- 6.Georgieva Y, Konthur Z. Design and screening of M13 phage display cDNA libraries. Molecules. 2011;16(2):1667–1681. doi: 10.3390/molecules16021667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carnazza S, Guglielmino S. Phage Display as a Tool for Synthetic Biology: Nanotechnology Science and Technology Series. New York, NY: Nova Science Pub Inc; 2010. [Google Scholar]
- 8.Schofield DA, inventor, Guild Associates, Inc, assignee Biological detection system and method. Nov 11, 2010. United States patent US 20100285460.
- 9.Li K, Chen Y, Li S, et al. Chemical modification of M13 bacteriophage and its application in cancer cell imaging. Bioconjug Chem. 2010;21(7):1369–1377. doi: 10.1021/bc900405q. [DOI] [PubMed] [Google Scholar]
- 10.Lee JH, Domaille DW, Cha JN. Amplified protein detection and identification through DNA-conjugated M13 bacteriophage. ACS Nano. 2012;6(6):5621–5626. doi: 10.1021/nn301565e. [DOI] [PubMed] [Google Scholar]
- 11.Cho W, Fowler JD, Furst EM. Targeted binding of the M13 bacteriophage to thiamethoxam organic crystals. Langmuir. 2012;28(14):6013–6020. doi: 10.1021/la300522g. [DOI] [PubMed] [Google Scholar]
- 12.Wang T, D’Souza GG, Bedi D, et al. Enhanced binding and killing of target tumor cells by drug-loaded liposomes modified with tumor-specific phage fusion coat protein. Nanomedicine (Lond) 2010;5(4):563–574. doi: 10.2217/nnm.10.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jayanna PK, Bedi D, Deinnocentes P, Bird RC, Petrenko VA. Landscape phage ligands for PC3 prostate carcinoma cells. Protein Eng Des Sel. 2010;23(6):423–430. doi: 10.1093/protein/gzq011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yacoby I, Bar H, Benhar I. Targeted drug-carrying bacteriophages as antibacterial nanomedicines. Antimicrob Agents Chemother. 2007;51(6):2156–2163. doi: 10.1128/AAC.00163-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Palaniappan KK, Ramirez RM, Bajaj VS, Wemmer DE, Pines A, Francis MB. Molecular imaging of cancer cells using a bacteriophage-based 129Xe NMR biosensor. Angew Chem Int Ed Engl. 2013;52(18):4849–4853. doi: 10.1002/anie.201300170. [DOI] [PubMed] [Google Scholar]
- 16.Ghosh D, Kohli AG, Moser F, Endy D, Belcher AM. Refactored M13 bacteriophage as a platform for tumor cell imaging and drug delivery. ACS Synth Biol. 2012;1(12):576–582. doi: 10.1021/sb300052u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Naik RR, Jones SE, Murray CJ, McAuliffe C, Vaia RA, Stone MO. Peptide templates for nanoparticle synthesis derived from polymerase chain reaction-driven phage display. Adv Funct Mater. 2004;14(1):25–30. [Google Scholar]
- 18.Merzlyak A, Lee SW. Phage as templates for hybrid materials and mediators for nanomaterial synthesis. Curr Opin Chem Biol. 2006;10(3):246–252. doi: 10.1016/j.cbpa.2006.04.008. [DOI] [PubMed] [Google Scholar]
- 19.Staquicini FI, Sidman RL, Arap W, Pasqualini R. Phage display technology for stem cell delivery and systemic therapy. Adv Drug Deliv Rev. 2010;62(12):1213–1216. doi: 10.1016/j.addr.2010.09.014. [DOI] [PubMed] [Google Scholar]
- 20.Clementi N, Mancini N, Solforosi L, Castelli M, Clementi M, Burioni R. Phage display-based strategies for cloning and optimization of monoclonal antibodies directed against human pathogens. Int J Mol Sci. 2012;13(7):8273–8292. doi: 10.3390/ijms13078273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Clackson T, Lowman HB, editors. Phage Display: A Practical Approach. New York, NY: Oxford University Press; 2004. [Google Scholar]
- 22.Niu Z, Bruckman MA, Harp B, Mello CM, Wang Q. Bacteriophage M13 as a scaffold for preparing conductive polymeric composite fibers. Nano Res. 2008;1(3):235–241. [Google Scholar]
- 23.Mulchandani A, Myung NV. Conducting polymer nanowires-based label-free biosensors. Curr Opin Biotechnol. 2011;22(4):502–508. doi: 10.1016/j.copbio.2011.05.508. [DOI] [PubMed] [Google Scholar]
- 24.Lu TK, Collins JJ. Engineered bacteriophage targeting gene networks as adjuvants for antibiotic therapy. Proc Natl Acad Sci U S A. 2009;106(12):4629–4634. doi: 10.1073/pnas.0800442106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hilderbrand SA, Kelly KA, Niedre M, Weissleder R. Near infrared fluorescence-based bacteriophage particles for ratiometric pH imaging. Bioconjug Chem. 2008;19(8):1635–1639. doi: 10.1021/bc800188p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sawada R, Peterson CY, Gonzalez AM, et al. A phage-targeting strategy for the design of spatiotemporal drug delivery from grafted matrices. Fibrogenesis Tissue Repair. 2011;4:7. doi: 10.1186/1755-1536-4-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ma Y, Nolte RJ, Cornelissen JJ. Virus-based nanocarriers for drug delivery. Adv Drug Deliv Rev. 2012;64(9):811–825. doi: 10.1016/j.addr.2012.01.005. [DOI] [PubMed] [Google Scholar]
- 28.Steinmetz NF. Viral nanoparticles in drug delivery and imaging. Mol Pharm. 2013;10(1):1–2. doi: 10.1021/mp300658j. [DOI] [PubMed] [Google Scholar]
- 29.Nanduri V, Balasubramanian S, Sista S, Vodyanoy VJ, Simonian AL. Highly sensitive phage-based biosensor for the detection of beta-galactosidase. Anal Chim Acta. 2007;589(2):166–172. doi: 10.1016/j.aca.2007.02.071. [DOI] [PubMed] [Google Scholar]
- 30.Suthiwangcharoen N, Li T, Li K, Thompson P, You S, Wang Q. M13 bacteriophage-polymer nanoassemblies as drug delivery vehicles. Nano Res. 2011;4(5):483–493. [Google Scholar]
- 31.Chen I, Choi YA, Ting AY. Phage display evolution of a peptide substrate for yeast biotin ligase and application to two-color quantum dot labeling of cell surface proteins. J Am Chem Soc. 2007;129(20):6619–6625. doi: 10.1021/ja071013g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Larocca D, Baird A, Kassner P, inventors, Selective Genetics, Inc, assignee Methods using genetic package display for selecting internalizing ligands for gene delivery. Sep 17, 2002. United States patent US 6541527.
- 33.Kim HJ, Ahn KC, González-Techera A, González-Sapienza GG, Gee SJ, Hammock BD. Magnetic bead-based phage anti-immunocomplex assay (PHAIA) for the detection of the urinary biomarker 3-phenoxybenzoic acid to assess human exposure to pyrethroid insecticides. Anal Biochem. 2009;386(1):45–52. doi: 10.1016/j.ab.2008.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ghosh D, Lee Y, Thomas S, et al. M13-templated magnetic nanoparticles for targeted in vivo imaging of prostate cancer. Nat Nanotechnol. 2012;7(10):677–682. doi: 10.1038/nnano.2012.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Avery KN, Schaak JE, Schaak RE. M13 bacteriophage as a biological scaffold for magnetically-recoverable metal nanowire catalysts: combining specific and nonspecific interactions to design multifunctional nanocomposites. Chem Mater. 2009;21(11):2176–2178. [Google Scholar]
- 36.Mao JY, Belcher AM, Van Vliet KJ. Genetically engineered phage fibers and coatings for antibacterial applications. Adv Funct Mater. 2010;20(2):209–214. [Google Scholar]
- 37.Huang S, Yang H, Lakshmanan RS, et al. Sequential detection of Salmonella typhimurium and Bacillus anthracis spores using magnetoelastic biosensors. Biosens Bioelectron. 2009;24(6):1730–1736. doi: 10.1016/j.bios.2008.09.006. [DOI] [PubMed] [Google Scholar]
- 38.Olsen EV, Sykora JC, Sorokulova IB, et al. Phage fusion proteins as bioselective receptors for piezoelectric sensors. ECS Transactions. 2007;2(19):9–25. [Google Scholar]
- 39.Li S, Johnson M, Wan J, Petrenko V, Chin BA. Microfabricated magnetoelastic biosensors for the detection of Bacillus anthracis spores. ECS Transactions. 2008;16(11):177–185. [Google Scholar]
- 40.Johnson ML, Wan J, Huang S, et al. A wireless biosensor using microfabricated phage-interfaced magnetoelastic particles. Sens Actuators A Phys. 2008;144(1):38–47. [Google Scholar]
- 41.Park MK, Park JW, Wikle HC, III, Chin BA. Evaluation of phage-based magnetoelastic biosensors for direct detection of Salmonella typhimurium on spinach leaves. Sens Actuators B Chem. 2013;176:1134–1140. [Google Scholar]
- 42.Li S, Li Y, Chen H, et al. Direct detection of Salmonella typhimurium on fresh produce using phage-based magnetoelastic biosensors. Biosens Bioelectron. 2010;26(4):1313–1319. doi: 10.1016/j.bios.2010.07.029. [DOI] [PubMed] [Google Scholar]
- 43.Shen W, Lakshmanan RS, Mathison LC, Petrenko VA, Chin BA. Phage coated magnetoelastic micro-biosensors for real-time detection of Bacillus anthracis spores. Sens Actuators B Chem. 2009;137(2):501–506. [Google Scholar]
- 44.Scherma G, Kish L, inventors, United State of America, as Represented by the Secretary of the Navy, assignee Bacteria identification by phage induced impedance fluctuation analysis. Oct 4, 2012. United States patent US 20120252003.
- 45.Zhu H, White IM, Sutter JD, Zourob M, Fan X. Miniaturized opto-fluidic ring resonator for sensitive label-free viral detection. Proc SPIE. 2008;6896(15):1–6. doi: 10.1039/b716834a. [DOI] [PubMed] [Google Scholar]
- 46.Zhu H, White IM, Suter JD, Fan X. Phage-based label-free biomolecule detection in an opto-fluidic ring resonator. Biosens Bioelectron. 2008;24(3):461–466. doi: 10.1016/j.bios.2008.04.028. [DOI] [PubMed] [Google Scholar]
- 47.Rosenberg A, Griffin K, Studier W, et al. T7 select phage display system: a powerful new protein display system based on the bacteriophage T7. Innovations. 1996;6:1–6. [Google Scholar]
- 48.Robertson KL, Soto CM, Archer MJ, Odoemene O, Liu JL. Engineered T4 viral nanoparticles for cellular imaging and flow cytometry. Bioconjug Chem. 2011;22(4):595–604. doi: 10.1021/bc100365j. [DOI] [PubMed] [Google Scholar]
- 49.Nanduri V, Sorokulova IB, Samoylov AM, Simonian AL, Petrenko VA, Vodyanoy V. Phage as a molecular recognition element in biosensors immobilized by physical adsorption. Biosens Bioelectron. 2007;22(6):986–992. doi: 10.1016/j.bios.2006.03.025. [DOI] [PubMed] [Google Scholar]
- 50.Shabani A, Marquette CA, Mandeville R, Lawrence MF. Carbon microarrays for the direct impedimetric detection of Bacillus anthracis using Gamma phages as probes. Analyst. 2013;138(5):1434–1440. doi: 10.1039/c3an36830k. [DOI] [PubMed] [Google Scholar]
- 51.Krumpe LR, Atkinson AJ, Smythers GW, et al. T7 lytic phage-displayed peptide libraries exhibit less sequence bias than M13 filamentous phage-displayed peptide libraries. Proteomics. 2006;6(15):4210–4222. doi: 10.1002/pmic.200500606. [DOI] [PubMed] [Google Scholar]
- 52.Kim BY, Rutka JT, Chan WC. Nanomedicine. N Engl J Med. 2010;363(25):2434–2443. doi: 10.1056/NEJMra0912273. [DOI] [PubMed] [Google Scholar]
- 53.Jiang S, Chen Q, Tripathy M, Luijten E, Schweizer KS, Granick S. Janus particle synthesis and assembly. Adv Mater Weinheim. 2010;22(10):1060–1071. doi: 10.1002/adma.200904094. [DOI] [PubMed] [Google Scholar]
- 54.Hashemi H, Pouyanfard S, Bandehpour M, et al. Immunization with M2e-displaying T7 bacteriophage nanoparticles protects against influenza A virus challenge. PLoS ONE. 2012;7(9):e45765. doi: 10.1371/journal.pone.0045765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Oślizło A, Miernikiewicz P, Piotrowicz A, et al. Purification of phage display-modified bacteriophage T4 by affinity chromatography. BMC Biotechnol. 2011;11:59. doi: 10.1186/1472-6750-11-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Malys N, Chang DY, Baumann RG, Xie D, Black LW. A bipartite bacteriophage T4 SOC and HOC randomized peptide display library: detection and analysis of phage T4 terminase (gp17) and late sigma factor (gp55) interaction. J Mol Biol. 2002;319(2):289–304. doi: 10.1016/S0022-2836(02)00298-X. [DOI] [PubMed] [Google Scholar]
- 57.Barrett RW, Cwirla SE, Dower WJ, Gallop M, Woiwode T, inventors, Xenoport, Inc, assignee Compounds displayed on icosahedral phage and methods of using same. Jun 12, 2007. United States patent US 7229757.
- 58.Merril C, inventor, Merril C., assignee High sensitivity phage display biomolecule detection. United States Patent US. 2002 Aug 15;20020110806 [Google Scholar]
- 59.Ren ZJ, Lai D, inventors, Ren ZJ, Lai D., assignee Novel recombinant T4 phage particle containing HIV, H. pylori on cancer antigens and uses thereof. Dec 4, 2008. United States patent US 20080299082.
- 60.Liu CM, Jin Q, Sutton A, Chen L. A novel fluorescent probe: europium complex hybridized T7 phage. Bioconjug Chem. 2005;16(5):1054–1057. doi: 10.1021/bc050108+. [DOI] [PubMed] [Google Scholar]
- 61.Gervais L, Gel M, Allain B, et al. Immobilization of biotinylated bacteriophages on biosensor surfaces. Sens Actuators B Chem. 2007;125(2):615–621. [Google Scholar]
- 62.Arya SK, Singh A, Naidoo R, Wu P, McDermott MT, Evoy S. Chemically immobilized T4-bacteriophage for specific Escherichia coli detection using surface plasmon resonance. Analyst. 2011;136(3):486–492. doi: 10.1039/c0an00697a. [DOI] [PubMed] [Google Scholar]
- 63.Boss PA, Lieberman SH, inventors, The United States of America as Represented by the Secretary of the Navy, assignee Technique for orienting and binding phage for bacteria detection. Dec 15, 2009. United States patent US 7632637.
- 64.Liu JL, Robertson KL, Soto CM, inventors, The Government of the United States of America, as represented by the Secretary of the Navy, assignee Bacteriophage T4 nanoparticles for eukaryotic delivery. Aug 23, 2012. United States patent US 20120214867.
- 65.Edgar R, McKinstry M, Hwang J, et al. High-sensitivity bacterial detection using biotin-tagged phage and quantum-dot nanocomplexes. Proc Natl Acad Sci U S A. 2006;103(13):4841–4845. doi: 10.1073/pnas.0601211103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Caberoy NB, Zhou Y, Jiang X, Alvarado G, Li W. Efficient identification of tubby-binding proteins by an improved system of T7 phage display. J Mol Recognit. 2010;23(1):74–83. doi: 10.1002/jmr.983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Clackson T, Hoogenboom HR, Griffiths AD, Winter G. Making antibody fragments using phage display libraries. Nature. 1991;352(6336):624–628. doi: 10.1038/352624a0. [DOI] [PubMed] [Google Scholar]
- 68.Mao C, Flynn CE, Hayhurst A, et al. Viral assembly of oriented quantum dot nanowires. Proc Natl Acad Sci U S A. 2003;100(12):6946–6951. doi: 10.1073/pnas.0832310100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yang LM, Tam PY, Murray BJ, et al. Virus electrodes for universal biodetection. Anal Chem. 2006;78(10):3265–3270. doi: 10.1021/ac052287u. [DOI] [PubMed] [Google Scholar]
- 70.Mao C, Liu A, Cao B. Virus-based chemical and biological sensing. Angew Chem Int Ed Engl. 2009;48(37):6790–6810. doi: 10.1002/anie.200900231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Olsen EV, Sorokulova IB, Petrenko VA, Chen IH, Barbaree JM, Vodyanoy VJ. Affinity-selected filamentous bacteriophage as a probe for acoustic wave biodetectors of Salmonella typhimurium. Biosens Bioelectron. 2006;21(8):1434–1442. doi: 10.1016/j.bios.2005.06.004. [DOI] [PubMed] [Google Scholar]