Abstract
Purpose
To ascertain whether single nucleotide polymorphisms (SNPs) in the Vascular Endothelial Growth factor (VEGFA), Complement Factor H (CFH), and LOC387715 genes could predict outcome to anti-VEGF therapy for patients with age related macular degeneration (AMD).
Methods
Patients with “wet” AMD were identified by chart review. Baseline optical coherence tomography (OCT) and visual acuity (VA) data, and at least 6 months of clinical follow up after 3 initial monthly injections of bevacizumab or ranibizumab were required for inclusion. Based on OCT and VA, patients were categorized into two possible clinical outcomes: (a) responders and (b) non-responders. DNA was extracted from saliva and genotyped for candidate SNPs in the VEGFA, LOC387715, and CFH genes. Clinical outcomes were statistically compared to patient genotypes.
Results
101 patients were recruited, and one eye from each patient was included in the analysis. 97% of samples were successfully genotyped for all SNPs. We found a statistically significant association between the LOC387715 A69S TT genotype and outcome based on OCT.
Conclusion
Genetic variation may be associated with outcome in patients receiving anti-VEGF therapy.
Keywords: age related macular degeneration, ARMS2, bevacizumab, complement factor H (CFH), LOC387715, ranibizumab, single nucleotide polymorphisms, vascular endothelial growth factor
Introduction
Exudative (“wet”) age related macular degeneration (AMD) is a leading cause of vision loss in people over 65. An estimated 1.75 million Americans suffer from this severe form of AMD and the number is predicted to reach 2.95 million by 2020.1 Standard therapeutic approach includes the inhibition of vascular endothelial growth factor (VEGFA) with intraocular injections of bevacizumab or ranibizumab.2–4 Although these agents are efficacious, substantial heterogeneity is seen with the implementation of anti-VEGF therapeutics in terms of duration and degree of response.5,6 This heterogeneity and ambiguous duration of treatment demonstrates the need for predictive biomarkers.
Previous work has reported that candidate single nucleotide polymorphisms (SNPs) might serve as prognostic or predictive markers for AMD. A SNP in the LOC387715 gene has previously been correlated with progression from intermediate forms of non-exudative (dry) AMD to wet AMD.7–11 Prior work also demonstrated that SNPS in CFH, LOC387715, and VEGF have been associated with response to anti-VEGF therapy in patients with AMD.9–11 In addition, our group has previously identified SNPs in the VEGFA gene that predicted efficacy and toxicity for bevacizumab in women with metastatic breast cancer.6 In this current study, we sought to determine whether genetic variations could predict outcome as measured by optical coherence tomography (OCT) and visual acuity (VA) in AMD patients receiving anti-VEGF therapy, and to identify a subgroup of patients that may not benefit optimally from this modality of treatment. To our knowledge, studies in which both OCT and VA were investigated in the same study cohort are scarce.
Methods
Patient selection
Patients with wet AMD between 2001-2010 were identified by a retrospective chart review. Inclusion criteria were 1) history of documented exudative AMD with baseline OCT and VA, 2) at least three previous consecutive injections of intravitreal bevacizumab or ranibizumab as initial therapy, and 3) at least 6 monthly follow-ups after the third injection of bevacizumab/ranibizumab. The majority of patients received ranibizumab (supplementary material Table S1). One of four retina specialists determined disease activity for each patient; however, practice patterns were uniform among the four treating physicians. These patients returned monthly for follow up after 3 monthly injections and retreatment was based on the presence or absence of subretinal and intraretinal fluid on OCT. Change in VA did not influence retreatment. Exclusion criteria included: 1) prior vitrectomy surgery or prior therapy (laser, photodynamic therapy, steroid injections), 2) primary pigment epithelial detachment, 3) significant fibrosis, or 4) vitreous hemorrhage.
Phenotype definition and sample acquisition
Patients that met the prespecified inclusion and exclusion criteria were initially categorized into one of two clinical response phenotypes by OCT and VA data from chart review: responder and non-responder (Table 1A and B). Zeiss Stratus Oct™ and Zeiss Cirrus OCT machines (Carl Zeiss Meditech AG, Jena, Germany) performing macular thickness map and line scans were utilized to look for the presence of change. VA was measured using the Snellen eye chart consisting of Sloan letter optotypes at a standard 20 feet. These patients were then invited to provide a saliva specimen for DNA extraction and genotyping. Informed written consent was obtained from all participants. Analysis was not performed on fluorescein angiograms. The study was approved by the Sterling Institutional Review Board and the Indiana University School of Medicine Institutional Review Board, and all patients were consented in person.
Table 1A.
Phenotype based on OCT
| Phenotype | OCT |
|---|---|
| Group 1 (responders) | No fluid after third injection for at least 1 month (no fluid at month 4) |
| Group 2 (non-responders) | Fluid present 1 month after third injection (fluid present at month 4) |
Abbreviation: OCT, optical coherence tomography.
Table 1B.
Phenotype based on VA
| Phenotype | VA |
|---|---|
| Group 1 (responders) | Gained ≥ 3 lines at month 9 |
| Group 2 (non-responders) | Did not gain ≥ 3 lines at month 9 |
Abbreviation: VA, visual acuity.
DNA extraction and genotyping
Candidate SNPs included those in the VEGFA, CFH, and LOC387715 genes (Table 2). The VEGFA SNPs included those that tagged for the common haplotypes or those that were previously shown to have predictive capacity for anti-VEGF therapy in the literature. The CFH and LOC387715 SNP selections were based on those with literature to support an association with AMD at the time this trial was initiated. DNA was extracted from saliva using the Oragene® DNA sample collection kit by DNA Genotek (Ottawa, ON, Canada). Samples were genotyped for candidate SNPs by TaqMan®-based real time-PCR (Applied Biosystems, Foster City, CA, USA). Pre-designed TaqMan SNP genotyping assays were used for LOC387715 A69S, VEGFA −3818G/T, −1498C/T, −2578C/A, −634C/G, −7C/T, and −1154G/A. Custom TaqMan SNP genotyping assays were designed for CFHY402H, VEGFA −2305G/T, and −1210C/A (supplementary material Tables S2 and S3). Samples that were previously sequenced by Sanger sequencing were used as positive controls.
Table 2.
Candidate SNPs
| GENE | Variant | dbSNP ID |
|---|---|---|
| VEGFA | −2578C/A | rs699947 |
| −1154G/A | rs1570360 | |
| −3818G/T | rs833060 | |
| −2305G/T | rs36208049 | |
| −1498C/T | rs833061 | |
| −7C/T | rs25648 | |
| −1210C/A | rs59260042 | |
| −634G/C | rs2010963 | |
| LOC387715 | S69AG/T | rs10490924 |
| CFH | Y402HC/T | rs1061170 |
Abbreviations: SNPs, single nucleotide polymorphisms; VEGFA, vascular endothelial growth factor A; LOC, otherwise known as age-related maculopathy succeptibility gene 2 (ARMS2); CFH, complement factor H; dbSNP, single nucleotide polymorphism database.
Endpoints
Clinical responses (Tables 1A and B) were assessed monthly for 6 months after 3 monthly injections of bevacizumab or ranibizumab. Patient demographics, VA, prior OCT data, as well as prior injection history were collected and utilized to classify each of the enrolled patients into one of the two possible clinical outcomes. For VA, patients with a gain of three lines or greater on the Snellen eye chart after conclusion of the study (9 month time period) were considered “responders” and all others were considered “non-responders”. For OCT, patients that had no sub- or intraretinal fluid present at least one month after the third monthly injection (no fluid at month 4) were considered “responders” and all others (fluid present at month 4) were considered “non-responders”.
Statistical design
There were 101 patients recruited for this study. Analyses for recessive, dominant and allele dose effect were performed for all candidate SNPs. Association between demographics and phenotype was assessed by Fisher’s exact test. Association between genotype and phenotype (responder versus non-responder) was assessed for both VA and OCT. Dominant, recessive, and allele dose effects were assessed using a logistic regression test. P-values < 0.006 were considered statistically significant after correcting for multiple comparisons, which was done using Bonferroni methodology.
Results
Study population, phenotype and demographics
101 patients were recruited based on inclusion and exclusion criteria. Patients were grouped into responders and non-responders based on distinct OCT and VA criteria. Therefore, responders based on OCT were not necessarily responders based on VA and vice versa. Based on OCT data, 79% of the patients were classified as responders and 21% were classified as non-responders. Patient demographics are outlined in Table 3 and demographics by clinical outcome based on OCT and VA are outlined in Table 4 and supplementary Table S4, respectively. VA data was available on only 100 patients. Based on VA, 30% of the patients were classified as responders and 70% were classified as non-responders as outlined in Table 5. There were no significant associations between demographic data points (ie, age, sex, tobacco use, and race) and clinical response.
Table 3.
Patient demographics
| Characteristics | Number of patients |
|---|---|
| Sex | |
| Male | 33 |
| Female | 68 |
| Mean age | 80 |
| Race | |
| White | 101 |
| Other | 0 |
| Tobacco use | |
| None | 84 |
| Past/present use | 17 |
Table 4.
Patient demographics by clinical outcome (OCT)
| Characteristics | Non-responder | Favorable responder phenotype | (P-value) |
|---|---|---|---|
| Total number of patients | 21 | 80 | – |
| Sex | |||
| Male | 4 | 29 | 0.19 |
| Female | 17 | 51 | |
| Mean age | 80 | 80 | 1.00 |
| Race | |||
| White | 21 | 80 | – |
| Other | 0 | 0 | |
| Tobacco use | |||
| None | 20 | 65 | 0.18 |
| Past/present use | 1 | 15 |
Table 5.
Snellen visual acuity (VA) outcomes
| Patient group | Number of patients | Mean initial VA | Mean final VA (at 9 months) |
|---|---|---|---|
| All patients | 100 | 20/80 | 20/60 |
| Responders | 30 | 20/100 | 20/40 |
| (gained ≥ 3 lines) | |||
| Non-responders | 70 | 20/70 | 20/70 |
| (gained < 3 Lines) |
Association by genotype
Ninety nine of 101 patients were successfully genotyped for the VEGFA −2578C/A, −1154G/A, −3818G/T, −1498C/T, −7C/T, and −634G/C SNPs. One hundred of 101 were successfully genotyped for the LOC387715 A69S and VEGFA −2305G/T SNPs. Ninety-seven of 101 were successfully genotyped for CFHY402H. The VEGFA−1210C/A and VEGFA−2305G/T SNPs were excluded, as there were no variants in our population. Genetic analyses for dose, dominant and recessive effects were performed for all other SNPs on all available samples and were evaluated for correlation with outcome (Tables 6, 7, and supplementary Table S5).
Table 6.
Genotype compared to visual acuity (VA)
| Gene/SNP | Dominant analysis
|
Recessive analysis
|
Additive analysis
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| P-value | OR | CI | P-value | OR | CI | P-value | OR | CI | |
| VEGFA-3818G/T | 0.46 | 1.39 | 0.58, 3.31 | 0.11 | 0.32 | 0.08, 1.30 | 0.87 | 0.95 | 0.50, 1.82 |
| VEGFA-2578C/A | 0.80 | 1.14 | 0.42, 3.04 | 0.013 | 0.27 | 0.10, 0.76 | 0.17 | 0.63 | 0.33, 1.22 |
| VEGFA-1498C/T | 0.80 | 1.14 | 0.42, 3.04 | 0.013 | 0.27 | 0.10, 0.76 | 0.17 | 0.63 | 0.33, 1.22 |
| VEGFA-1154A/G | 0.42 | 0.69 | 0.29, 1.68 | – | – | – | 0.10 | 0.59 | 0.31, 1.12 |
| VEGFA-634C/G | 0.80 | 1.12 | 0.47, 2.66 | 0.04 | 0.27 | 0.08, 0.93 | 0.51 | 0.81 | 0.43, 1.53 |
| VEGFA-7C/T | 0.93 | 0.95 | 0.36, 2.54 | 0.09 | 0.33 | 0.09, 1.18 | 0.93 | 0.95 | 0.36, 2.54 |
| CFH Y402H | 0.21 | 0.53 | 0.20, 1.42 | 0.12 | 3.43 | 0.72, 16.30 | 1.00 | 1.00 | 0.53, 1.90 |
| LOC387715 A69S | 0.58 | 1.28 | 0.54, 3.03 | 0.56 | 1.44 | 0.42, 4.91 | 0.50 | 1.23 | 0.67, 2.24 |
Abbreviations: OR, odds ratio; CI, 95% confidence interval; SNP, single nucleotide polymorphism.
Table 7.
Genotype compared to optical coherence tomography (OCT)
| Gene/SNP | Dominant analysis
|
Recessive analysis
|
Additive analysis
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| P-value | OR | CI | P-value | OR | CI | P-value | OR | CI | |
| VEGFA-3818G/T | 1.00 | 1.08 | 0.40, 2.94 | 0.20 | 0.19 | 0.04, 1.96 | 0.71 | 1.06 | 0.78, 1.45 |
| VEGFA-2578C/A | 0.58 | 0.74 | 0.24, 2.27 | 0.76 | 0.65 | 0.16, 2.56 | 0.38 | 0.87 | 0.65, 1.19 |
| VEGFA-1498C/T | 0.58 | 1.56 | 0.40, 6.25 | 0.76 | 1.35 | 0.45, 4.17 | 0.38 | 1.14 | 0.85, 1.56 |
| VEGFA-1154A/G | 0.46 | 3.03 | 0.36, 33.3 | 0.45 | 1.47 | 0.54, 4.17 | 0.70 | 1.06 | 0.79, 1.45 |
| VEGFA-634C/G | 1.00 | 2.78 | 0.32, 25.00 | 0.45 | 0.98 | 0.36, 2.70 | 0.17 | 1.23 | 0.92, 1.67 |
| VEGFA-7C/T | 0.58 | 1.35 | 0.45, 4.17 | 1.00 | – | – | 0.59 | 0.88 | 0.56, 1.41 |
| CFH Y402H | 1.00 | 1.12 | 0.38, 3.45 | 1.00 | 0.95 | 0.24, 4.00 | 0.71 | 1.05 | 0.78, 1.45 |
| LOC387715 A69S | 0.32 | 1.89 | 0.66, 5.26 | 0.0007 | 7.65 | 2.38, 25 | 0.016 | 1.39 | 1.06, 1.85 |
Abbreviations: OR, odds ratio; CI, 95% confidence interval; SNP, single nucleotide polymorphism.
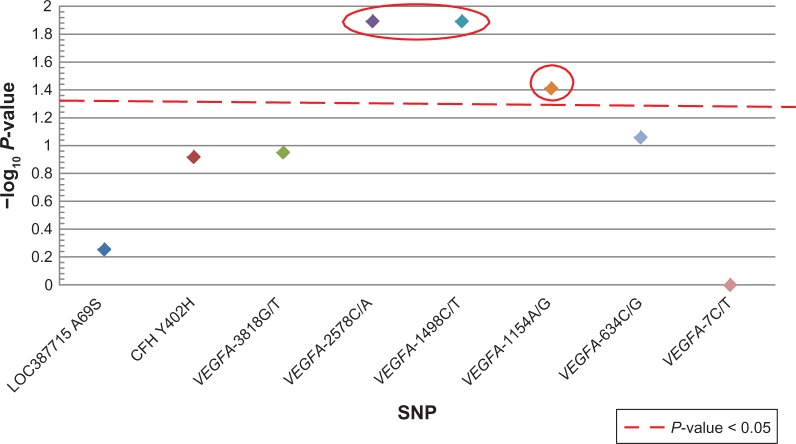
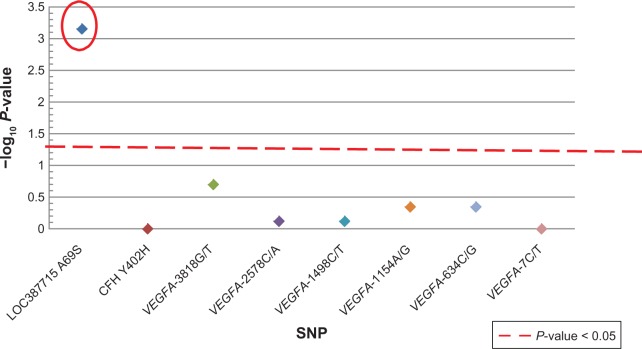
Patients who carried the VEGFA-2578CC, VEGFA -1498TT or the VEGFA-1154GG genotypes were more likely to be non-responders based on VA (Figure 1 and Table 6). However, this association was not statistically significant after correction for multiple comparisons. There were no associations with CFH and LOC387715. Based on OCT, patients who carried the LOC387715 A69S TT genotype were significantly more likely to be classified as a non-responder (9/16) compared to those with the GG and GT genotypes (12/84); (P = 0.00071; odds ratio: 7.69; 95% confidence interval: 2.38–25). There were no significant associations between VEGFA and CFH genotypes with OCT (Figure 2 and Table 7).
Figure 1.
Genotype compared to visual acuity (VA).
Notes: A strong trend between VEGFA -2578CC, VEGFA -1498TT (odds ratio: 3.7, 95% confidence interval: 1.3, 10.2) and VEGFA -1154 GG (odds ratio: 3.7, 95% confidence interval: 1.1, 12.9) and VA. The x-axis indicates the SNP analyzed and the y-axis denotes magnitude of the evidence for association, shown as –log10(P-value). Each colored diamond represents the –log10(P-value) for that specific SNP.
Abbreviations:VEGFA, vascular endothelial growth factor; SNP, single nucleotide polymorphism.
Figure 2.
Genotype compared to OCT.
Notes: A significant association between LOC387715 A69S (odds ratio: 7.69, 95% confidence intervals: 2.38, 25) and OCT response. The colored diamonds represent the correlation between each of the SNPs and OCT.
Abbreviations: SNP, single nucleotide polymorphism; OCT, optical coherrence tomography.
Discussion
There is substantial heterogeneity in the degree and duration of response among AMD patients treated with anti-VEGF therapy. In this retrospective study we examined germline genetic variation in VEGFA, LOC387715 and CFH genes as possible predictive biomarkers for outcome in patients receiving bevacizumab or ranibizumab for the treatment of wet AMD.
Our prior work in metastatic breast cancer had identified several VEGFA SNPs that predicted outcome for systemic bevacizumab.6 While there are clearly some similarities between cancer-mediated angiogenesis and AMD-mediated angiogenesis making cross disease comparisons rational, there are also likely fundamental differences to consider.
In this study we report a strong trend between VEGFA SNPs (-2578CC, -1498TT and -1154) and VA. However, these SNPs lost their significance after correction for multiple comparisons. One of the limitations of this study was sample size and the resultant statistical power. Therefore, it is possible that this study was underpowered to detect associations between genotype and phenotype of smaller effect size. An additional potential limitation of this study was relying upon Snellen VA as opposed to ETDRS vision charts. It is possible that using ETDRS vision charts would have led to different statistical outcomes due to the variable letter size and letters per line on the Snellen chart. However, we used “three lines or greater” on the Snellen eye chart as the best metric available given the retrospective nature of this study.
Other groups have also evaluated these SNPs in the AMD setting. A study by Imai et al10 found a correlation between VEGFA -2578C/A and VA changes in response to anti-VEGF therapy. Similar to our study, VEGFA-2578 C carriers were more likely to be non-responders based on VA. Although our results would support this finding, our data would suggest that the effect is modest. Additionally, this effect was not seen when response was measured by OCT (measuring retinal thickness), which is similar to what we observed. Another study by Boltz et al12 reported significant findings with VEGFA-634 and VEGFA IVS -99 (rs3024997). However, these results were not significant after correction for multivariate analysis.
We also evaluated OCT as a second phenotype. At the time of this trial, OCT was commonly used as a surrogate for the frequency and duration of therapy independent of changes in VA. The goal of using OCT as the endpoint was to predict the duration and frequency of therapy necessary to achieve good vision. This is very different from a biomarker for VA which predicts for vision outcome regardless of the intensity/frequency of therapy and which is impacted by other variables. We identified a statistically significant association between LOC387715 A69S TT carriers and the non-responder phenotype based on OCT. Those patients with two variant alleles had a markedly higher likelihood of not responding to therapy (recessive effect) and each variant allele added to this likelihood (allele-dose effect).
As the exact function of LOC387715 is yet to be elucidated, the role of the non-synonymous LOC387715 A69S SNP in the pathogenesis of AMD remains unknown. Previous studies, however, have clearly demonstrated an association with LOC387715 A69S and poor outcome independent of therapy (ie, a prognostic marker)7,8,13. The results here would further suggest that this genotype does poorly even with the use of standard anti-VEGF therapy; thus making it a powerful predictive biomarker as well. Brantley et al9 demonstrated a correlation between the CFH CC genotype and worse outcome after anti-VEGF injections compared with the alternate genotypes. They did not, however, find a significant association between LOC387715 A69S and treatment outcome. In congruence with our results, a more recent study showed a correlation between the LOC387715 A69S TT genotype and a poor response to ranibizumab injections with no correlation between CFH genotypes and response.14 There are several variables that might explain the differing results among these studies. First, the treatment duration and dosing were not uniform. In our study, response was assessed after 3 monthly injections; whereas Brantley et al used 6-week intervals.9 More importantly, the phenotype was markedly different between the three studies. The prior studies used changes in VA as the endpoint whereas the associations in this study were between genotype and OCT in addition to genotype and VA.
Although we used both ranibizumab and bevacizumab, both CATT (Comparison of AMD Treatment Trial) and IVAN (Inhibit VEGF in Age-related choroidal Neovascularization) suggest that there is no statistically significant difference in visual acuity improvement between bevacizumab and ranibizumab in the treatment of neovascular AMD.15,16
In conclusion, we have demonstrated a genetic association between the LOC387715 A69S TT genotype and a group of patients that may not respond well to bevacizumab or ranibizumab injections. This study would suggest that patients carrying this genotype should be candidates for studies evaluating alternative or novel therapeutic approaches to wet AMD. Prior studies have also suggested that other (non-VEGF directed) interventions such as photodynamic therapy (PDT) may also be influenced by genotype.17–19 Thus, personalized therapy based on genotype may provide a rational direction for selection of therapeutic approach. Alternatively, this patient cohort may simply represent ideal candidates for trials of higher dose anti-VEGF therapy. Further studies to confirm this genotype-phenotype association are underway by our collaborative group.
Supplementary materials
Table S1.
Number of patients receiving bevacizumab, ranibizumab, or both
| Anti-VEGF therapy used | Number of patients |
|---|---|
| Bevacizumab | 17 |
| Ranibizumab | 81 |
| Both | 3 |
Table S2.
Predesigned TaqMan™ assays
| dbSNP ID | Assay ID |
|---|---|
| rs1570360 | C_1647379_10 |
| rs25648 | C_791476_10 |
| rs833060 | C_1647392_20 |
| rs699947 | C_8311602_10 |
| rs833061 | C 1647381 10 |
| rs2010963 | C_8311614_10 |
| rs10490924 | C_29934973_20 |
Abbreviation: dbSNP ID, single nucleotide polymorphism database identification.
Table S3.
Custom made TaqMan™ assays
| dbSNP ID | Primers (forward/reverse) | Reporter sequences |
|---|---|---|
| rs36208049 | GGAGAAGTAGC CAAGGGATCCT/GCCCAGACTCA TAGCTCATCTTC | TCGTCTCAGCTCCCCCA/TCGTCTCAGATCCCCCA |
| rs59260042 | TCGAGCTTCCC CTTCATTGC/GGACAGGCG AGCCTCAG | CCGCAGCCCGCC/CCGCATCCCGCC |
| rs1061170 | TGTTATGGTCCTTAGG AAAATGTTATTTTCCTT/GGCAGGCAACGTCTAT AGATTTACC | CTTTCTTCCATGATTTTG/TTTCTTCCATAATTTTG |
Abbreviation: dbSNP ID, single nucleotide polymorphism database identification.
Table S4.
Patient demographics by clinical outcome (visual acuity)
| Characteristics | Non-responder | Favorable responder phenotype | (P-value) |
|---|---|---|---|
| Total number of patients | 70 | 30 | – |
| Sex | |||
| Male | 26 | 8 | 0.36 |
| Female | 44 | 22 | |
| Mean age | 79 | 81 | 0.44 |
| Race | |||
| White | 70 | 30 | – |
| Other | 0 | 0 | |
| Tobacco use | |||
| None | 57 | 27 | 0.38 |
| Past/present use | 13 | 3 |
Table S5.
Non-responder/responder ratio by genotype
| Gene/SNP | Genotype (never responder/favorable responder)
|
||
|---|---|---|---|
| 1* | 2* | 3* | |
| VEGF-3818G/T | 10/51 | 10/39 | 0/9 |
| VEGF-2578C/A | 6/25 | 11/54 | 3/20 |
| VEGF-1498C/T | 3/20 | 11/54 | 6/25 |
| VEGF-1154A/G | 1/12 | 9/45 | 10/42 |
| VEGF-634C/G | 9/45 | 10/43 | 1/11 |
| VEGF-7C/T | 14/74 | 6/25 | 0/0 |
| CFH Y402H | 6/31 | 11/51 | 3/15 |
| LOC387715 A69S | 7/45 | 5/39 | 9/16 |
Notes:
1, Homozygous wild type; 2, Heterozygous variant; 3, Homozygous variant.
Abbreviation: SNP, single nucleotide polymorphism.
Footnotes
Disclosure
John W Kitchens has been a speaker/consultant for Genentech and Regeneron and a consultant for Thrombogenics, Allergan, Alcon, and Synergetics. He also holds stock in Synergetics. Bryan P Schneider is on the Genentech speaker’s bureau and advisory board (compensated). The authors report no other conflicts of interest in this work.
References
- 1.Friedman DS, O’Colmain BJ, Munoz B, et al. The Eye Diseases Prevalence Research Group. Prevalence of age-related macular degeneration in the United States. Arch Ophthalmol. 2004;122(4):564–572. doi: 10.1001/archopht.122.4.564. [DOI] [PubMed] [Google Scholar]
- 2.Brown DM, Kaiser PK, Michels M, et al. Ranibizumab versus verteporfin for neovascular age-related macular degeneration. N Engl J Med. 2006;355(14):1432–1444. doi: 10.1056/NEJMoa062655. [DOI] [PubMed] [Google Scholar]
- 3.Rosenfeld PJ, Brown DM, Heier JS, et al. Ranibizumab for neovascular age-related macular degeneration. N Engl J Med. 2006 Oct 5;355(14):1419–1431. doi: 10.1056/NEJMoa054481. [DOI] [PubMed] [Google Scholar]
- 4.Avery RL, Pieramici DJ, Rabena MD, Castellarin AA, Nasir MaA, Giust MJ. Intravitreal bevacizumab (Avastin) for neovascular age-related macular degeneration. Ophthalmology. 2006;113(3):363–372. doi: 10.1016/j.ophtha.2005.11.019. [DOI] [PubMed] [Google Scholar]
- 5.Chiang A, Regillo CD. Preferred therapies for neovascular age-related macular degeneration. Curr Opin Ophthalmol. 2011;22(3):199–204. doi: 10.1097/ICU.0b013e32834597d9. [DOI] [PubMed] [Google Scholar]
- 6.Schneider BP, Wang M, Radovich M, et al. Association of vascular endothelial growth factor and vascular endothelial growth factor receptor-2 genetic polymorphisms with outcome in a trial of paclitaxel compared with paclitaxel plus bevacizumab in advanced breast cancer: ECOG 2100. J Clin Oncol. 2008;26(28):4672–4678. doi: 10.1200/JCO.2008.16.1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Seddon JM, Francis PJ, George S, Schultz DW, Rosner B, Klein ML. Association of CFH Y402H and LOC387715 A69S with progression of age-related macular degeneration. JAMA. 2007;297(16):1793–1800. doi: 10.1001/jama.297.16.1793. [DOI] [PubMed] [Google Scholar]
- 8.Rivera A, Fisher SA, Fritsche LG, et al. Hypothetical LOC387715 is a second major susceptibility gene for age-related macular degeneration, contributing independently of complement factor H to disease risk. Hum Mol Genet. 2005;14(21):3227–3236. doi: 10.1093/hmg/ddi353. [DOI] [PubMed] [Google Scholar]
- 9.Brantley MA, Jr, Fang AM, King JM, Tewari A, Kymes SM, Shiels A. Association of complement factor H and LOC387715 genotypes with response of exudative age-related macular degeneration to intravitreal bevacizumab. Ophthalmology. 2007;114(12):2168–2173. doi: 10.1016/j.ophtha.2007.09.008. [DOI] [PubMed] [Google Scholar]
- 10.Imai D, Mori K, Horie-Inoue K, et al. CFH, VEGF, and PEDF genotypes and the response to intravitreous injection of bevacizumab for the treatment of age-related macular degeneration. J Oc Biol DisInfor. 2010;3(2):53–59. doi: 10.1007/s12177-010-9055-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee AY, Raya AK, Kymes SM, Shiels A, Brantley MA. Pharmacogenetics of complement factor H (Y402H) and treatment of exudative age-related macular degeneration with ranibizumab. Bri J Ophthalmol. 2009;93(5):610–613. doi: 10.1136/bjo.2008.150995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boltz A, Ruibeta M, Jonas JB, et al. Role of vascular endothelial growth factor polymorphisms in the treatment success in patients with wet age-related macular degeneration. Ophthalmology. 2012;119(8):1615–1620. doi: 10.1016/j.ophtha.2012.02.001. [DOI] [PubMed] [Google Scholar]
- 13.Shuler RK, Jr, Hauser MA, Caldwell J, et al. Neovascular age-related macular degeneration and its association with LOC387715 and complement factor H polymorphism. Arch Ophthalmol. 2007;125(1):63–67. doi: 10.1001/archopht.125.1.63. [DOI] [PubMed] [Google Scholar]
- 14.Teper SJ, Nowinska A, Pilat J, Palucha A, Wylegala E. Involvement of genetic factors in the response to a variable-dosing ranibizumab treatment regimen for age-related macular degeneration. Mol Vis. 2010;16:2598–2604. [PMC free article] [PubMed] [Google Scholar]
- 15.Chakravarthy U, Harding SP, Rogers CA, et al. Ranibizumab versus bevacizumab to treat neovascular age-related macular degeneration: one-year findings from the IVAN randomized trial. Ophthalmology. 2012;119(7):1399–1411. doi: 10.1016/j.ophtha.2012.04.015. [DOI] [PubMed] [Google Scholar]
- 16.Martin DF, Maguire MG, Fine SL, et al. Ranibizumab and bevacizumab for treatment of neovascular age-related macular degeneration: two-year results. Ophthalmology. 2012;119(7):1388–1398. doi: 10.1016/j.ophtha.2012.03.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sakurada Y, Kubota T, Imasawa M, Mabuchi F, Tanabe N, Iijima H. Association of LOC387715A69S genotype with visual prognosis after photodynamic therapy for polypoidal choroidal vasculopathy. Retina. 2010;30(10):1616–1621. doi: 10.1097/IAE.0b013e3181e587e3. [DOI] [PubMed] [Google Scholar]
- 18.Bessho H, Honda S, Kondo N, Negi A. The association of age-related maculopathy susceptibility 2 polymorphisms with phenotype in typical neovascular age-related macular degeneration and polypoidal choroidal vasculopathy. Mol Vis. 2011;17:977–982. [PMC free article] [PubMed] [Google Scholar]
- 19.Park DH, Kim IT. LOC387715/HTRA1 variants and the response to combined photodynamic therapy with intravitreal bevacizumab for polypoidal choroidal vasculopathy. Retina. 2012;32(2):299–307. doi: 10.1097/IAE.0b013e318225290f. [DOI] [PubMed] [Google Scholar]