Abstract
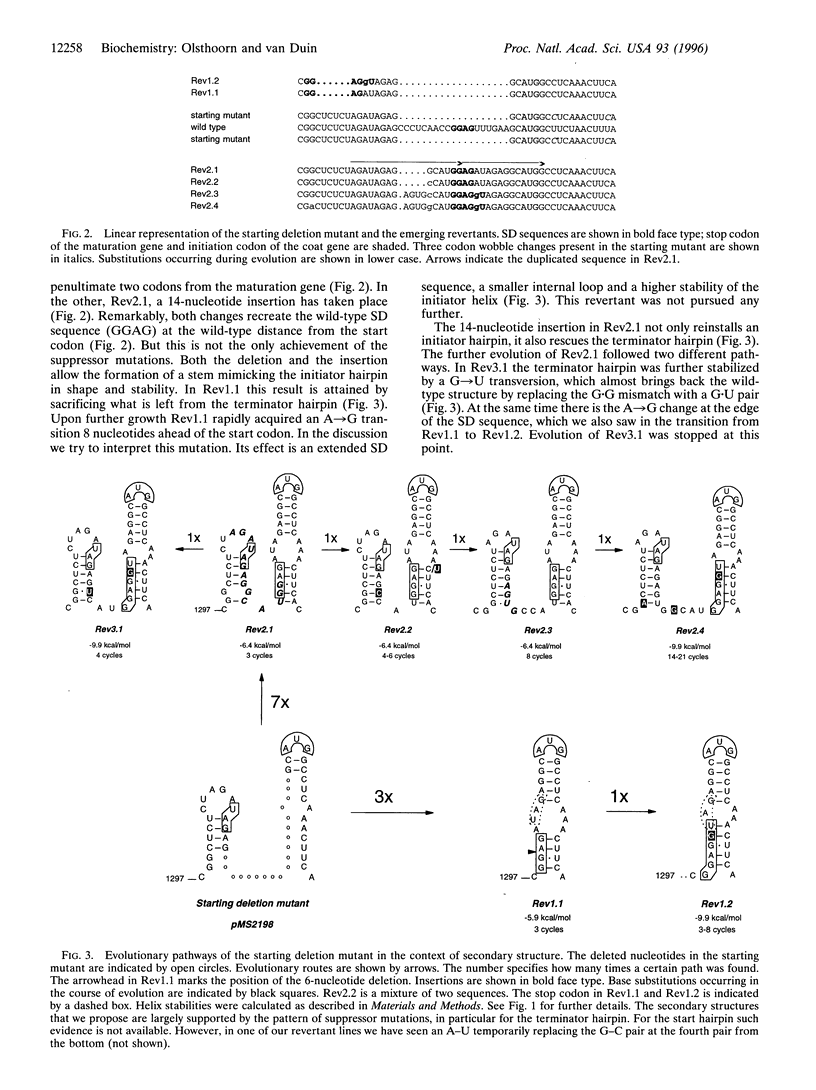
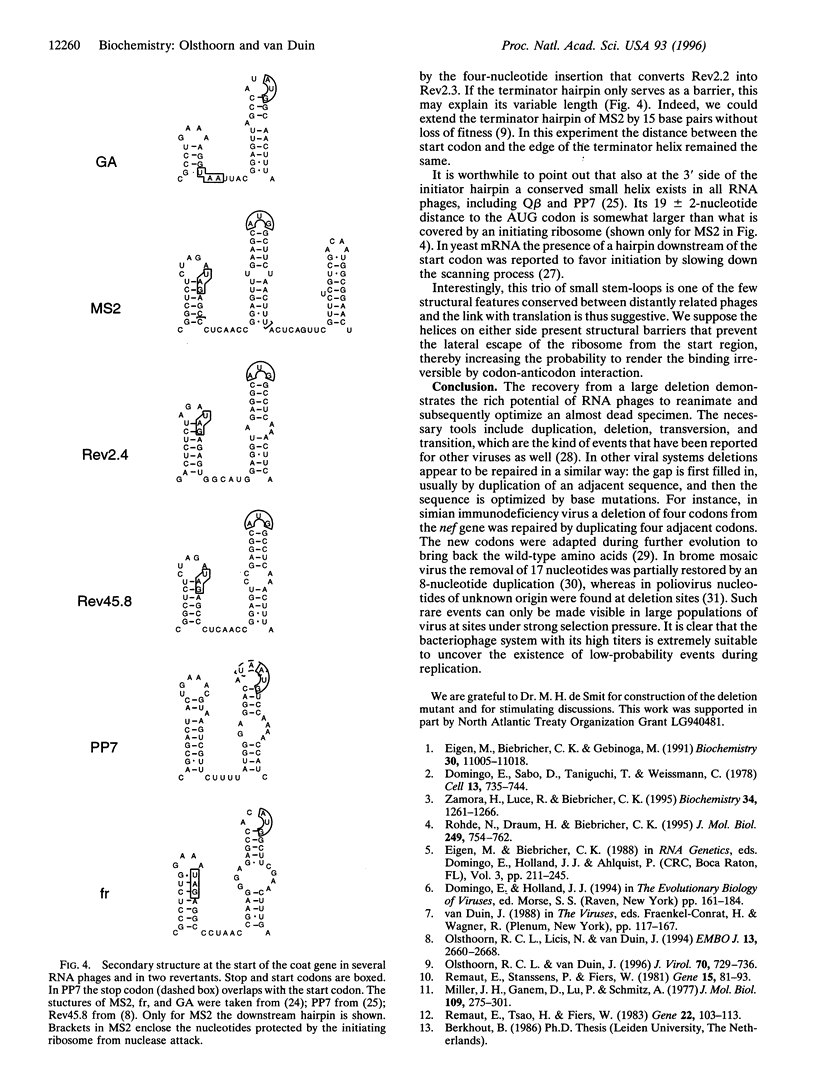
The intercistronic region between the maturation and coat-protein genes of RNA phage MS2 contains important regulatory and structural information. The sequence participates in two adjacent stem-loop structures, one of which, the coat-initiator hairpin, controls coat-gene translation and is thus under strong selection pressure. We have removed 19 out of the 23 nucleotides constituting the intercistronic region, thereby destroying the capacity of the phage to build the two hairpins. The deletion lowered coat-protein yield more than 1000-fold, and the titer of the infectious clone carrying the deletion dropped 10 orders of magnitude as compared with the wild type. Two types of revertants were recovered. One had, in two steps, recruited 18 new nucleotides that served to rebuild the two hairpins and the lost Shine-Dalgarno sequence. The other type had deleted an additional six nucleotides, which allowed the reconstruction of the Shine-Dalgarno sequence and the initiator hairpin, albeit by sacrificing the remnants of the other stem-loop. The results visualize the immense genetic repertoire created by, what appears as, random RNA recombination. It would seem that in this genetic ensemble every possible new RNA combination is represented.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adhin M. R., van Duin J. Scanning model for translational reinitiation in eubacteria. J Mol Biol. 1990 Jun 20;213(4):811–818. doi: 10.1016/S0022-2836(05)80265-7. [DOI] [PubMed] [Google Scholar]
- Batschelet E., Domingo E., Weissmann C. The proportion of revertant and mutant phage in a growing population, as a function of mutation and growth rate. Gene. 1976;1(1):27–32. doi: 10.1016/0378-1119(76)90004-4. [DOI] [PubMed] [Google Scholar]
- Coffin J. M. Structure, replication, and recombination of retrovirus genomes: some unifying hypotheses. J Gen Virol. 1979 Jan;42(1):1–26. doi: 10.1099/0022-1317-42-1-1. [DOI] [PubMed] [Google Scholar]
- Domingo E., Sabo D., Taniguchi T., Weissmann C. Nucleotide sequence heterogeneity of an RNA phage population. Cell. 1978 Apr;13(4):735–744. doi: 10.1016/0092-8674(78)90223-4. [DOI] [PubMed] [Google Scholar]
- Eigen M., Biebricher C. K., Gebinoga M., Gardiner W. C. The hypercycle. Coupling of RNA and protein biosynthesis in the infection cycle of an RNA bacteriophage. Biochemistry. 1991 Nov 19;30(46):11005–11018. doi: 10.1021/bi00110a001. [DOI] [PubMed] [Google Scholar]
- Freier S. M., Kierzek R., Jaeger J. A., Sugimoto N., Caruthers M. H., Neilson T., Turner D. H. Improved free-energy parameters for predictions of RNA duplex stability. Proc Natl Acad Sci U S A. 1986 Dec;83(24):9373–9377. doi: 10.1073/pnas.83.24.9373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gmyl A. P., Pilipenko E. V., Maslova S. V., Belov G. A., Agol V. I. Functional and genetic plasticities of the poliovirus genome: quasi-infectious RNAs modified in the 5'-untranslated region yield a variety of pseudorevertants. J Virol. 1993 Oct;67(10):6309–6316. doi: 10.1128/jvi.67.10.6309-6316.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W. S., Temin H. M. Retroviral recombination and reverse transcription. Science. 1990 Nov 30;250(4985):1227–1233. doi: 10.1126/science.1700865. [DOI] [PubMed] [Google Scholar]
- Kozak M. Circumstances and mechanisms of inhibition of translation by secondary structure in eucaryotic mRNAs. Mol Cell Biol. 1989 Nov;9(11):5134–5142. doi: 10.1128/mcb.9.11.5134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai M. M. RNA recombination in animal and plant viruses. Microbiol Rev. 1992 Mar;56(1):61–79. doi: 10.1128/mr.56.1.61-79.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J. H., Ganem D., Lu P., Schmitz A. Genetic studies of the lac repressor. I. Correlation of mutational sites with specific amino acid residues: construction of a colinear gene-protein map. J Mol Biol. 1977 Jan 15;109(2):275–298. doi: 10.1016/s0022-2836(77)80034-x. [DOI] [PubMed] [Google Scholar]
- Olsthoorn R. C., Garde G., Dayhuff T., Atkins J. F., Van Duin J. Nucleotide sequence of a single-stranded RNA phage from Pseudomonas aeruginosa: kinship to coliphages and conservation of regulatory RNA structures. Virology. 1995 Jan 10;206(1):611–625. doi: 10.1016/s0042-6822(95)80078-6. [DOI] [PubMed] [Google Scholar]
- Olsthoorn R. C., Licis N., van Duin J. Leeway and constraints in the forced evolution of a regulatory RNA helix. EMBO J. 1994 Jun 1;13(11):2660–2668. doi: 10.1002/j.1460-2075.1994.tb06556.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsthoorn R. C., Zoog S., van Duin J. Coevolution of RNA helix stability and Shine-Dalgarno complementarity in a translational start region. Mol Microbiol. 1995 Jan;15(2):333–339. doi: 10.1111/j.1365-2958.1995.tb02247.x. [DOI] [PubMed] [Google Scholar]
- Olsthoorn R. C., van Duin J. Random removal of inserts from an RNA genome: selection against single-stranded RNA. J Virol. 1996 Feb;70(2):729–736. doi: 10.1128/jvi.70.2.729-736.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peliska J. A., Benkovic S. J. Mechanism of DNA strand transfer reactions catalyzed by HIV-1 reverse transcriptase. Science. 1992 Nov 13;258(5085):1112–1118. doi: 10.1126/science.1279806. [DOI] [PubMed] [Google Scholar]
- Remaut E., Stanssens P., Fiers W. Plasmid vectors for high-efficiency expression controlled by the PL promoter of coliphage lambda. Gene. 1981 Oct;15(1):81–93. doi: 10.1016/0378-1119(81)90106-2. [DOI] [PubMed] [Google Scholar]
- Remaut E., Tsao H., Fiers W. Improved plasmid vectors with a thermoinducible expression and temperature-regulated runaway replication. Gene. 1983 Apr;22(1):103–113. doi: 10.1016/0378-1119(83)90069-0. [DOI] [PubMed] [Google Scholar]
- Rohde N., Daum H., Biebricher C. K. The mutant distribution of an RNA species replicated by Q beta replicase. J Mol Biol. 1995 Jun 16;249(4):754–762. doi: 10.1006/jmbi.1995.0334. [DOI] [PubMed] [Google Scholar]
- Skripkin E. A., Adhin M. R., de Smit M. H., van Duin J. Secondary structure of the central region of bacteriophage MS2 RNA. Conservation and biological significance. J Mol Biol. 1990 Jan 20;211(2):447–463. doi: 10.1016/0022-2836(90)90364-R. [DOI] [PubMed] [Google Scholar]
- Smirnyagina E., Hsu Y. H., Chua N., Ahlquist P. Second-site mutations in the brome mosaic virus RNA3 intercistronic region partially suppress a defect in coat protein mRNA transcription. Virology. 1994 Feb;198(2):427–436. doi: 10.1006/viro.1994.1054. [DOI] [PubMed] [Google Scholar]
- Whatmore A. M., Cook N., Hall G. A., Sharpe S., Rud E. W., Cranage M. P. Repair and evolution of nef in vivo modulates simian immunodeficiency virus virulence. J Virol. 1995 Aug;69(8):5117–5123. doi: 10.1128/jvi.69.8.5117-5123.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamora H., Luce R., Biebricher C. K. Design of artificial short-chained RNA species that are replicated by Q beta replicase. Biochemistry. 1995 Jan 31;34(4):1261–1266. doi: 10.1021/bi00004a020. [DOI] [PubMed] [Google Scholar]
- de Smit M. H., van Duin J. Secondary structure of the ribosome binding site determines translational efficiency: a quantitative analysis. Proc Natl Acad Sci U S A. 1990 Oct;87(19):7668–7672. doi: 10.1073/pnas.87.19.7668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Smit M. H., van Duin J. Translational initiation on structured messengers. Another role for the Shine-Dalgarno interaction. J Mol Biol. 1994 Jan 7;235(1):173–184. doi: 10.1016/s0022-2836(05)80024-5. [DOI] [PubMed] [Google Scholar]



