Abstract
Pseudomonas extremaustralis is a versatile Antarctic bacterium, able to grow under microaerobic and anaerobic conditions and is related to several non-pathogenic Pseudomonads. Here we report on the role of the global anaerobic regulator Anr, in the early steps of P. extremaustralis biofilm development. We found that the anr mutant was reduced in its ability to attach, to form aggregates and to display twitching motility but presented higher swimming motility than the wild type. In addition, microscopy revealed that the wild type biofilm contained more biomass and was thicker, but were less rough than that of the anr mutant. In silico analysis of the P. extremaustralis genome for Anr-like binding sites led to the identification of two biofilm-related genes as potential targets of this regulator. When measured using Quantitative Real Time PCR, we found that the anr mutant expressed lower levels of pilG, which encodes a component of Type IV pili and has been previously implicated in cellular adhesion. Levels of morA, involved in signal transduction and flagella development, were also lower in the mutant. Our data suggest that under low oxygen conditions, such as those encountered in biofilms, Anr differentially regulates aggregation and motility thus affecting the first stages of biofilm formation.
Introduction
Biofilms, bacterial assemblies enclosed in a matrix, are found throughout many environmental biological niches. Cells forming these communities have advantages over their planktonic counterparts with respect to protection against both physical and chemical stresses [1]. Bacteria in biofilms are more resistant to antimicrobial agents and immune system surveillance [2], [3]. In addition, these structures provide protection against protozoa predation and environmental stresses such as cold and contaminants [4], [5].
The steps in biofilm formation have been studied extensively in Pseudomonas aeruginosa [6]. Several cellular functions such as motility, adhesion, metabolic switching, exopolysaccharides production and DNA and protein secretion are important during biofilm development. The expression of genes encoding these cellular functions is modulated during biofilm development [7], [8]. The role of global regulator proteins on biofilm formation has been studied in different species, for example, HhA and RpoS in Escherichia coli, CcpA in Bacillus subtilis and Streptococcus mutans and RpoN in P. aeruginosa [9], [10], [11], [12].
Biofilms are heterogeneous structures and microbial metabolism can vary dramatically depending on where cells are found within the biofilm [13]. Oxygen, in particular, can be in limiting supply within the biofilm and oxygen gradients have detected within biofilms. The mechanisms whereby cells sense and respond to oxygen are complex and not fully understood [14]. In Pseudomonas, the redox global regulator Anr controls anaerobic metabolism by activation and repression of targets genes. Denitrification, arginine and pyruvate fermentation, redox state maintenance, fimbria and cytochrome biosynthesis, secretion type III system, oxidative stress resistance and quorum sensing cascades are some of the functions that are known to be modulated by Anr [15], [16], [17], [18], [19], [20], [21], [22]. The role of global regulator Anr in biofilm development in non-pathogenic Pseudomonas species has not been studied yet.
Pseudomonas extremaustralis, a highly stress resistant bacterium isolated from Antarctic temporary water pond, is able to synthesize high amounts of poly(3-hydroxybutyrate) (PHB) [23], [24]. PHB production is important for the transition between biofilm and planktonic lifestyles under cold conditions [5].Biofilms increase diesel degradation in P. extremaustralis [25]. Under microaerobic or anaerobic conditions, P. extremaustralis reduces nitrate but it is unable to perform complete denitrification because it lacks nitrite reductase genes [26], [27].
In this work we analyzed the role of Anr in the early steps of biofilm formation by P. extremaustralis.
Materials and Methods
Bacterial Strains and Growth Conditions
Pseudomonas extremaustralis DSM 25547 and an anr mutant containing a 250-bp deletion and a kanamycin cassette insertion were used throughout this study [26], [27]. A complemented strain was constructed as described before, by inserting the entire wild type anr sequence in the mutant by using a mini-Tn5 delivery system [21]. All cultures were performed using 0.5 NE2 medium supplemented with 0.2% glucose, 0.3%, casaminoacids, 0.08% KNO3, 1 mM MgSO4 and 0.1% micronutrients [28]. Microaerobic cultures were performed in 100-ml hermetically sealed bottles containing 50 ml of culture medium. Bottles were incubated at 30°C and low shaking (75 rpm) to avoid cellular aggregation.
Biofilm Experiments
Biofilms were grown in glass bottom Petri dishes (Mat Tek, 3 mm Petri-dish, 14 mm microwell and 1.0 mm coverglass) with 3 ml of 0.5 NE2 supplemented medium as described above with slow shaking (50 rpm). The medium was inoculated with overnight cultures to give an initial OD600 of 0.025. Culture medium was replaced every 24 h and the OD600 and the CFU/ml of planktonic cells were determined. For further microscopy visualization of the biofilm, 3 ml of low melting point agarose (1%) was added as the culture medium was withdrawn in order to maintain biofilm structure. The experiment was performed by triplicate.
Attachment Experiments
Initial attachment was studied by using 96-well polystyrene microtiter plates (Gibco), as described by O’Toole and Kolter [29]. Briefly, 200 µl of each culture were added to the microplate wells and incubated at 30°C without agitation for 3 h. Non-attached cells were collected and OD600 was measured (absorbance of planktonic cells: APL). Biofilm attached cells were stained with 200 µl 0.1% crystal violet. After 20 min, the unbound crystal violet solution was removed and plates were gently rinsed with water. Subsequently, the crystal violet was extracted from the bound cell with 200 µl 96% ethanol for 20 min and transferred to flat bottom microtiter plates in order to measure the absorbance at 550 nm (absorbance of crystal violet: ACV) in a Tasoh Corp MPR A4i microplate reader. The attachment index was defined as ACV/APL.
Autoaggregation Assays
Autoaggregation and settling assays were performed as described Sherlock et al. [30] with modifications. Briefly, overnight microaerobic cultures were diluted with fresh media and incubated until 0.8 OD600. A 1 ml aliquot was incubated at room temperature without agitation for 3 h and stained with 4′,6-diamidino-2-phenylindole (DAPI) for fluoresce microscopy or with a 4% aqueous solution of uranyl acetate for electronic microscopy. After that, 200 µl from the top 5 mm of the culture was taken (non- settled) while the rest of the culture was vigorously vortexed and the OD600 of both samples was determined. Aggregation % was determined as follows: (OD vortexed-OD non-settled)/OD vortexed × 100.
Microscopy
For DAPI visualization an Olympus BX40 microscope with a UV lamp was used. For transmission electron microscopy (TEM) samples were allowed to adhere to carbon-coated 200 mesh grids and were stained with uranyl acetate. TEM was performed with a Philips EM 201 microscope. Twitching motility was visualized under a Leica DFC300X microscope using contrast phase mode using 400× magnification.
Motility Experiment
Swimming motility was evaluated using a plate assay [31]. 5 µl of an overnight culture was used to inoculate swimming medium plates containing 10 g/l bacto-tryptone, 5 g/l NaCl, 0.3% wt/vol agarose, 0.3% casaminoacids and 0.2% glucose. Swimming distance was measured after 24 h. Twitching motility was visualized using the slide culture method [32]. Briefly, microaerobic cultures were used to point inoculate onto the surface of a LB agar (1%) slice placed on a microscope slide. The inoculum was covered with a coverslip and incubated at 30°C for 15 h in a humid environment. The samples were visualized by contrast phase microscopy.
Quantitative Real Time PCR Experiments
Total RNA was extracted from 24 h old microaerophilic cultures of P. extremaustralis and the anr mutant using the RNeasy Mini kit (Qiagen). After treatment with DNaseI, cDNA was obtained using random hexamers (Promega) and AMV retrotranscriptase following the manufacturer’s instructions. At least three independent cultures were analyzed for each strain. RT-qPCR was performed using a LightCycler (DNA Engine M.J. Research) and Real Time PCR mix (Biocientist, no Rox). Three genes were analyzed using the following primers: pilG 5′TCCCGGTGATCATGCTGTCCTCC3′ and 5′TGTTCTACTGCCGCGAACCCA3′; morA 5′GGTTGCGGGACAACCCCATCG3′ and 5′ GGTGGTGTTACGCGGGCAGTC3′ and 16S rRNA gene 5′AGCTTGCTCCTTGATTCAGC3′ and 5′AAGGGCCATGATGACTTGAC3′ employed as reference for normalization of expression levels of target genes in each strain. The thermocycler conditions were as follows: denaturation at 95°C for 5 min; 40 cycles at 95°C for 25 s, 52.3°C for 15 s, and 72°C for 15 s; with fluorescence acquisition at 80°C in single mode. Relative changes in the expression of individual genes in both strains were obtained through the relative standard curve method [33].
Biofilm Visualization
P. extremaustralis wild type and the anr mutant strain were transformed with plasmids encoding GFP in order to analyze biofilm structure [34]. Biofilms were visualized using an Olympus BX61 microscope equipped with a 620 low-power objective with WG and FITC fluorescence filter cubes (Olympus). Samples were illuminated using a Lambda LS Xenon arc lamp (Sutter Instruments). Images were acquired using a Cooke SensiCam with a Sony Interline chip. The image capture size was 512 x 512 pixels and the Z section step size was 1 µm. Image acquisition, nearest neighbor deconvolution and 2D image production were performed using the SlideBook Software package version 3.0.10.15 (Intelligent Imaging). At least six stacks for each sample were analyzed with COMSTAT2 software [35] available on line using the automatic threshold option.
Bioinformatic Analysis
The complete genome sequence of P. extremaustralis has been deposited at DDBJ/EMBL/GenBank under the accession no. AHIP01000001.1- AHIP01000135.1 [27].The Anr regulon of P. extremaustralis was predicted using the Virtual Footprint tool available in PRODORIC software [36]. Putative target genes were considered only when the Anr-box was located within 300 bp upstream from the start ATG codon, based on previous data with experimental support [37], and in an intergenic genomic zone. Putative σ70 dependent promoters were identified using the Softberry Bprom algorithm (http://linux1.softberry.com/berry.phtml). Sequence logos was performed using 5 Anr-boxes belonging to morA and pilG genes and also those found in previous works to be influenced by Anr in P.extremaustralis anr strain in previous work [26], [27].
Statistics Tests
The significance of each treatment was evaluated by the Student’s t test with P<0.05 considered as significant. In attachment experiments the treatments between strains and time were evaluated by ANOVA test.
Results
Biofilm Formation is Influenced by Anr
The wild type formed well-defined microcolonies by 24 h (data not shown) and had the highest value for total biomass and average thickness after 72 h, though the roughness coefficient was lowest at that time point (Table 1). At 96 h a decrease in total biomass and thickness was observed probably due to a dispersion phenomenon. In contrast, biofilms of the anr mutant strain had significantly lower total biomass as well as average and maximum thickness at all the assayed times (Table 1). The R value was higher, however, for the mutant strain biofilms than for the wild type biofilms, suggesting a more disorganized structure (Table 1). Due to the defects observed in biofilm formation in the mutant strain and the importance of Anr in anaerobic metabolism, we investigated the culturability of planktonic cells. There was not statistical significance in the numbers of culturable planktonic cells for the time points except at 96 h, when the anr strain numbers (2.10±5.10 CFU ml−1) exceeded those of the wild type (6.10±2.10 CFU ml−1) (P<0.05).
Table 1. Biofilm parameters of wild type and anr mutant of P. extremaustralis. Values represent the mean ± SD of three independent experiments.
| Parameters | strain | 24 h | 48 h | 72 h | 96 h |
| Biomass (Bio-volume, µm3. µm−2) | Wild type | 17.2±4.4 | 13.9±3.2 | 23.2±4.6 | 14.2±2.5 |
| anr strain | 2.2±0.3* | 6.1±2.6* | 0.53±0.5* | 1.2±0.8* | |
| Average thickness (µm) | Wild type | 22.5±7.2 | 22.3±6.9 | 34.4±5.5 | 18.3±3.4 |
| anr strain | 2.5±0.8* | 14.7±5.9 | 1.7±1.5* | 2.6±2.0* | |
| Maximum thickness (µm) | Wild type | 100.0±26.3 | 92.0±13.0 | 81.6±11.6 | 68.4±20.0 |
| anr strain | 60.5±10.2* | 63.7±20.0 | 65.2±12.7* | 70.0±11.6 | |
| Roughness coefficient | Wild type | 1.4±0.1 | 1.3±0.3 | 0.9±0.2 | 1.1±0.2 |
| anr strain | 1.6±0.4 | 1.6±0.2 | 1.7±0.4* | 1.6±0.3* |
The asterisk (*) denotes significant differences among strains (P<0.05) using the Student’s t test.
Anr Enhances Attachment, Autoaggregation and Twitching Motility and Decrease Swimming Motility
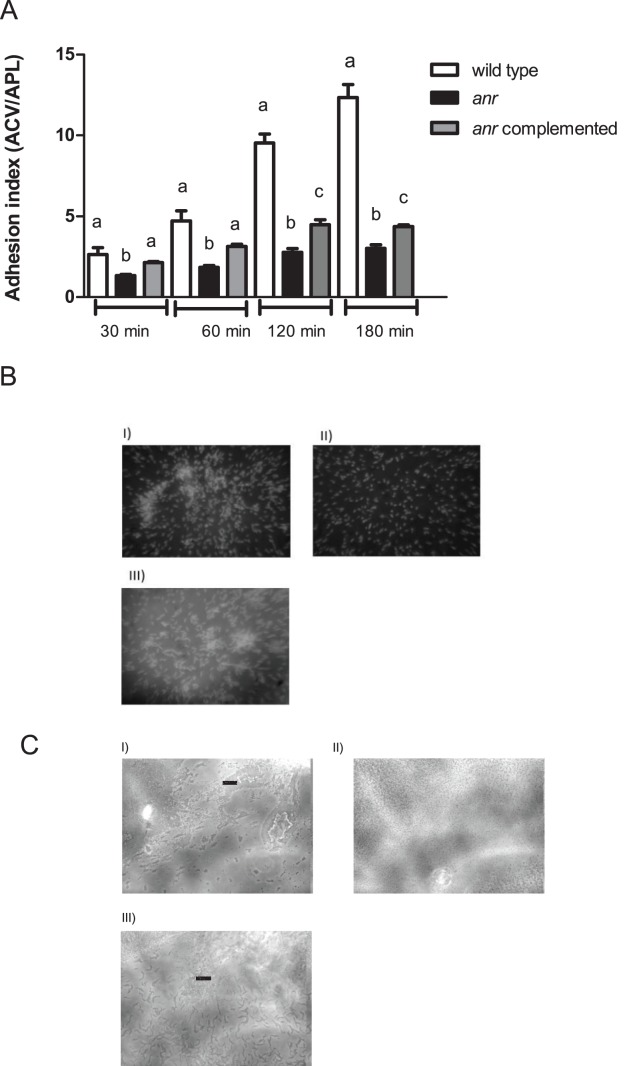
Biofilm development includes different stages, among them; adhesion to surface and cellular aggregation constitute important earlier steps. The wild type strain incremented significantly the adhesion index throughout the experiment (P<0.05) while the anr mutant strain showed a slight increment of this parameter with only significantly differences in the first assayed times. The values of the adherence index were significantly higher in the wild type strain in all times in comparison with the anr mutant strain (Fig. 1A). The complemented strain presented a partial restoration of this phenotype (Fig. 1A).
Figure 1. Anr absences decrease attachment and aggregation in P. extremaustralis.
(A) Attachment to polystyrene plates in 0.5 NE2 medium supplemented with glucose, KNO3 and casaminoacids. Values represent media ± SD of 5 independent experiments with 12 wells per strain. Different letters showed significant differences among strains (P<0.05) using ANOVA (B). Autoaggregation experiment. The cells were incubated during 2 h without agitation and a culture sample was stained with DAPI. I) wild type strain. II) anr mutant strain. III) anr complemented strain. All observations were performed at 1000X. (C). Slide culture assay to investigate twitching motility. Cells were incubated for 15 h. I) wild type strain. II) anr mutant strain. III) anr complemented strain. Arrows showed rafts in the edge of the culture. All observations were performed using contrast phase microscopy at 400× magnification.
Additionally, settling capability, which is a common measure of cell to cell adhesion, was significantly higher in the wild type than the anr mutant, with values of 48±13% of aggregation for the wild type and 17±5% for the anr (P<0.05). This aggregation defect was partially restored by complementation (33±6%). Aggregation by the complemented was significantly different than the anr strain but not significantly different than the wild type (P<0.05 and P>0.05, respectively). Microscopic observation of DAPI stained cells showed that the wild type and the complemented strain aggregated in clumps while mostly single cells were observed in the anr mutant culture (Fig. 1B). Another important feature in biofilm formation is twitching motility. We assayed the twitching capability in cells belonging to microaerobic cultures. The examination of the wild type slide culture revealed the presence of motile rafts of cells at the leading edge of the moving zone while in the mutant strain; the cells were dispersed without forming rafts (Fig. 1C). The complemented strain presented a pattern similar to that observed in the wild type strain (Fig. 1C).
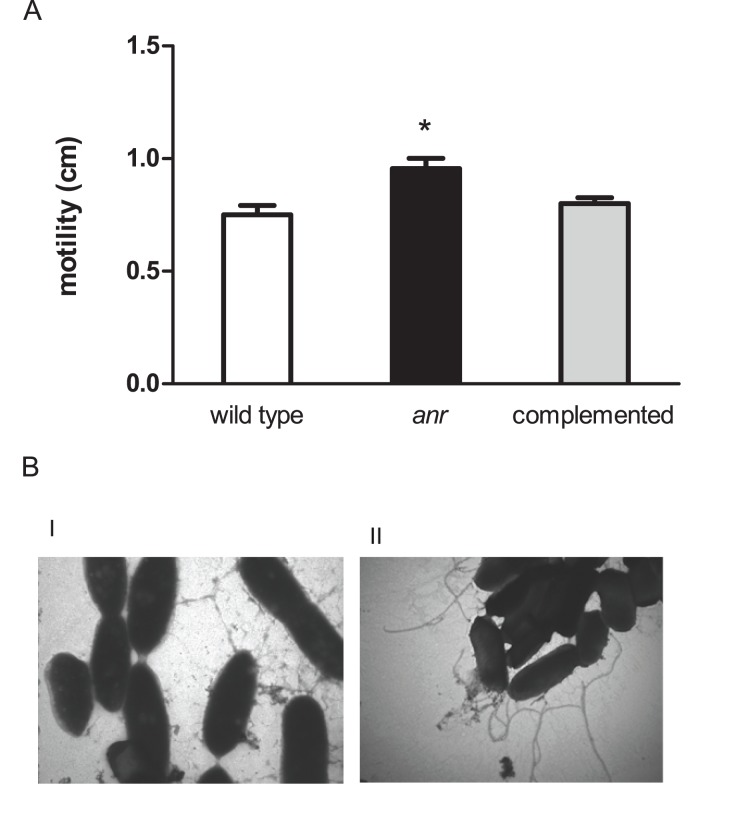
The mutant cells were also visibly more motile than the wild type when viewed under the microscope (data not shown). Given this observation and the importance of motility in the early stages of biofilm development, swimming motility was also evaluated in motility agar. As expected, the wild type and the complemented strain were significantly less motile than the anr mutant strain (Fig. 2A). Consistent with these results, flagellar-like structures were only visible in TEMs of the anr mutant and not in those of the wild type (Fig. 2B).
Figure 2. Swimming motility is increased in anr mutant strain.
(A) Swimming motility. The asterisk (*) denotes significant differences (P<0.05) using the Student’s t test. (B) Transmission electron microscopy. I) wild type strain. II) anr mutant strain. Observations were performed at 46000X magnification.
Anr Controls the Expression of Genes Involved in Biofilm Development
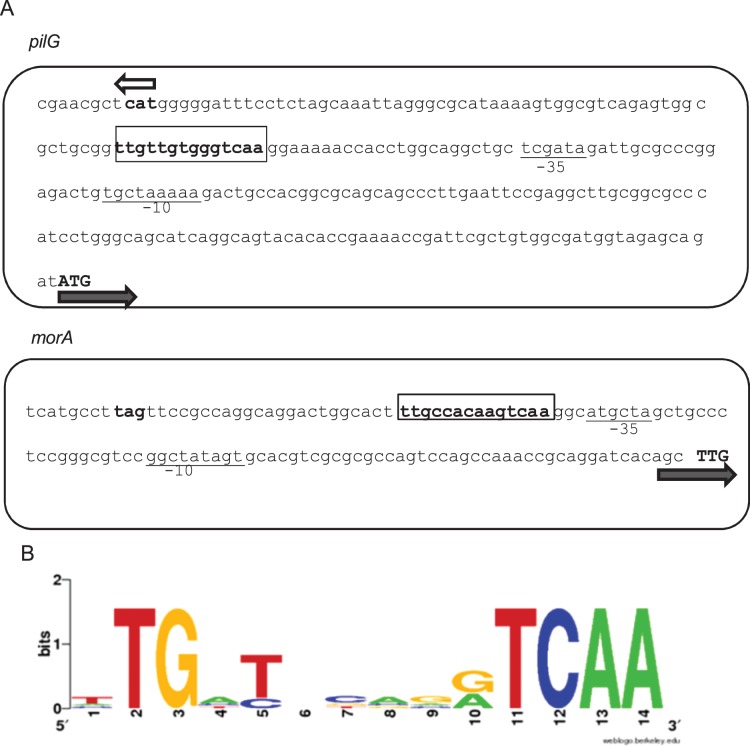
The P. extremaustralis genome sequence was analyzed to determine putative targets genes for Anr regulation. Two target genes involved in different biofilm functions were detected in which the Anr-box was located in an intergenic zone, pilG and morA (Fig. 3). pilG, is relevant to Type IV pili assembly in other Pseudomonads whereas morA encodes a repressor of flagella development [38], [39]. The logos performed with the Anr-boxes located in the putative promoter zone of pilG and morA and also the Anr-boxes of phaBAC, cioA and cooN of P. extremaustralis showed a similar P.aeruginosa Anr-box consensus sequence [37]. We performed Quantitative Real Time PCR experiments to determine if expression of pilG and morA was altered in the anr mutant strain. The expression of both genes in the mutant strain was lower than the wild type (Fig. 4A). The lower pilG expression is in line with the defects on twitching, adhesion and aggregation in the anr mutant strain. The morA altered expression was consistent with the higher swimming motility observed in absence of anr.
Figure 3. Organization of the P. extremaustralis intergenic pilG and morA region showing putative Anr boxes.
(A) The sequences −35 and −10 of a probable σ70 promoter are underlined. The pilG and morA start codons and the start or the stop codon of the neighbors genes are shown by boldface type and arrows indicate the direction of the transcription. Anr boxes are boxed. (B) Sequence logo of 5 Anr boxes located in genes influenced by Anr in P. extremaustralis were used to generate the Anr position weight matrix sequence.
Figure 4. Effect of anr mutation on expression of pilG and morA genes in microaerobic cultures.
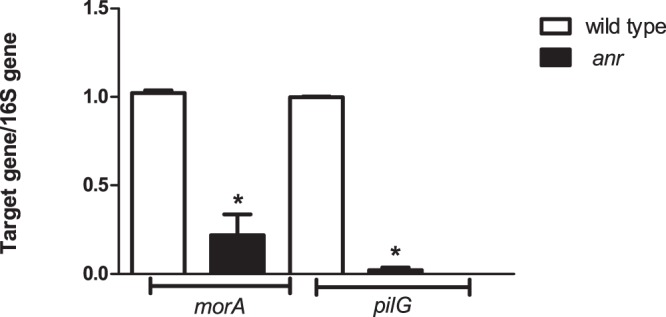
(A) qPCR Real Time experiments were carried out in microaerobic cultures. Values were expressed as the ratio between the raw level of expression of each target gene and the 16S rRNA gene, and represent the mean ± SD of three independent experiments. The asterisk (*) denotes significant differences (P<0.05) using the Student’s t test.
Discussion
Biofilms development constitutes a complex process influenced by a variety of factors including oxygen availability [13]. In Pseudomonas species Anr controls several components of the response to low oxygen availability. In P. aeruginosa, the best studied member of the genus, Anr transcriptional levels were found to be up-regulated in confluent biofilms and it has been demonstrated that Anr controls quorum sensing by regulating the expression of the small regulatory RNA PhrS [14], [22].Recently, Jackson et al. [40] observed that Anr was crucial for P. aeruginosa virulence in a mouse model and that biofilm formation was defective in an anr mutant, but the mechanism behind this deficit was not described. The importance of global regulators that affect biofilm formation has also demonstrated with arcA in Escherichia coli [41]. These reports suggest a relationship between oxygen availability and physiological responses mediated by quorum sensing and biofilm development.
In this work, microscopic analysis showed that an anr mutant of P. extremaustralis had impaired biofilm development. This was not simply due to a growth deficit, since the number of culturable planktonic anr cells was the same or greater than the number of wild type cells. The reduction in biofilm formation can be attributed to two specific phenotypic defects that are known to be important in the early steps of biofilm formation, namely, the anr mutant strain showed lack of cellular aggregation and was highly motile. Newell et al. [42] have shown that biofilm initiation is result of a combined reduction of motility and an increase in adhesion. Thus, both flagella and Type IV pili are important to biofilm development in glucose supplemented cultures [43]. Pili serve to assist in attachment of cells to surfaces and twitching motility whereas flagella have a dual function since are important for attachment but are also involved in dispersion [44]. In the present work, we demonstrated that Anr is involved in aggregation and motility in P. extremaustralis, since an anr mutant was deficient in the attachment to polystyrene plates, autoaggregation, and twitching motility while presented a higher swimming motility which is dependent of flagella.
Several genes encode functions that contribute to biofilm development, including those encoding regulators or components of surface structures such as pili and flagella including, for example pilG, pilA, fliC [44], [45]. Our in silico analysis revealed that several P. extremaustralis, genes relevant to these structures had putative Anr boxes including pilG and morA. Quantitative real time PCR confirmed that both these genes were down regulated in the anr mutant. The reduction of pilG expression levels in the P. extremaustralis anr mutant likely impaired pili biosynthesis and resulted in the aggregation and twitching defects we observed. Something similar has been observed in P. aeruginosa Crc mutants where the mutations in this regulatory protein lead to pili defects and concomitant defects in biofilm formation [46].
Additionally, we found that transcripts of morA, predicted to encode a protein similar to those involved in signal transduction, were less abundant in the P. extremaustralis anr mutant. Studies of morA mutants of P. putida showed increased motility and an inability to develop biofilms [39] which is similar to our observations of the P. extremaustralis anr mutant in this work. Interestingly, in P. aeruginosa morA mutant strains neither motility nor biofilm formation are affected, suggesting a different mechanism in this species [39].
Taken together, the results presented here demonstrate that Anr is involved in regulating the autoaggregation, adhesion and twitching and swimming motility of P. extremaustralis and that the loss of Anr impairs biofilm development. Although different signals also regulate aggregation and motility [47], [48], [49] the importance of Anr in biofilm development indicate oxygen availability as a signal that regulates biofilm development in P. extremaustralis.
Funding Statement
This work was supported by grants from University of Buenos Aires-UBACyT 2011–2014 Proyecto N° 20020100100854, Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET)-PIP (2011) 0338, and Agencia Nacional de Promoción Científica y Técnológica (ANPCyT)-Programación 2007, PICT 0639. N.I.L. is a career investigator from CONICET. P.M.T. received a post-graduate student fellowship from CONICET and a Wood-Wheland fellowship for short term research stays from the International Union of Biochemistry and Molecular Biology. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Decho AW (2000) Microbial biofilms in intertidal systems: an overview. Cont Shelf Res 20: 1257–1273. [Google Scholar]
- 2. Breidenstein EB, de la Fuente-Nunez C, Hancock RE (2011) Pseudomonas aeruginosa: all roads lead to resistance. Trends Microbiol 19: 419–426. [DOI] [PubMed] [Google Scholar]
- 3. Romling U, Balsalobre C (2012) Biofilm infections, their resilience to therapy and innovative treatment strategies. J Intern Med 272: 541–561. [DOI] [PubMed] [Google Scholar]
- 4. Romeo T (2006) When the party is over: a signal for dispersal of Pseudomonas aeruginosa biofilms. J Bacteriol 188: 7325–7327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Tribelli PM, López NI (2011) Poly(3-hydroxybutyrate) influences biofilm formation and motility in the Antarctic novel species Pseudomonas extremaustralis under cold conditions. Extremophiles 15: 541–547. [DOI] [PubMed] [Google Scholar]
- 6. Worlitzsch D, Tarran T, Ulrich M, Schwab U, Cekici A, et al. (2002) Effects of reduced mucus oxygen concentration in airway Pseudomonas infections of cystic fibrosis patients. J Clin Invest 109: 317–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Whiteley M, Gita Bangera M, Bumgarner R, Parsek M, Teitzel G, et al. (2001) Gene expression in Pseudomonas aeruginosa biofilms. Nature 413: 860–864. [DOI] [PubMed] [Google Scholar]
- 8. Williamson K, Richards L, Perez-Osorio A, Pitts B, McInnerney K, et al. (2012) Heterogeneity in Pseudomonas aeruginosa biofilms includes expression of ribosome hibernation factors in the antibiotic-tolerant subpopulation and hypoxia-induced stress response in the metabolically active population. J Bacteriol 194: 2062–2073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wen ZT, Burne RA (2002) Functional genomics approach to identifying genes required for biofilm development by Streptococcus mutans . Appl Environ Microbiol 68: 1196–1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Thompson LS, Webb JS, Rice SA, Kjelleberg S (2003) The alternative sigma factor RpoN regulates the quorum sensing gene rhlI in Pseudomonas aeruginosa . FEMS Microbiol Lett 220: 187–195. [DOI] [PubMed] [Google Scholar]
- 11. Stanley NR, Britton RA, Grossman AD, Lazazzera BA (2003) Identification of catabolite repression as a physiological regulator of biofilm formation by Bacillus subtilis by use of DNA microarrays. J Bacteriol 185: 1951–1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hong SH, Wang X, Wood TK (2010) Controlling biofilm formation, prophage excision and cell death by rewiring global regulator H-NS of Escherichia coli . Microb Biotechnol 3: 344–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Folsom JP, Richards L, Pitts B, Roe F, Ehrlich GD, et al. (2012) Physiology of Pseudomonas aeruginosa in biofilms as revealed by transcriptome analysis. BMC Microbiol 10: 294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Waite RD, Paccanaro A, Papakonstantinopoulou A, Hurst J, Saqi M, et al. (2006) Clustering of Pseudomonas aeruginosa transcriptomes from planktonic cultures, developing and mature biofilms reveals distinct expression profiles. BMC Gen 7: 162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Galimand M, Gamper M, Zimmermann A, Haas D (1991) Positive FNR-like control of anaerobic arginine degradation and nitrate respiration in Pseudomonas aeruginosa. . J Bacteriol 173: 1598–1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Schreiber K, Krieger R, Benkert B, Eschbach M, Arai H, et al. (2007) The anaerobic regulatory network required for Pseudomonas aeruginosa nitrate respiration. J Bacteriol 189: 4310–4314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ugidos A, Morales G, Rial E, Williams HD, Rojo F (2008) The coordinate regulation of multiple terminal oxidases by the Pseudomonas putida ANR global regulator. Environ Microbiol 10: 1690–1702. [DOI] [PubMed] [Google Scholar]
- 18. Vallet-Gely I, Sharp J, Simon Dove L (2007) Local and global regulators linking anaerobiosis to cupA fimbrial gene expression in Pseudomonas aeruginosa . J Bacteriol 189: 8667–8676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. O’Callaghan JF, Reen J, Adams C, O’Gara F (2011) Low oxygen induces the type III secretion system in Pseudomonas aeruginosa via modulation of the small RNAs rsmZ and rsmY . Microbiology 157: 3417–3428. [DOI] [PubMed] [Google Scholar]
- 20. O’Callaghan JJ, Reen F, Adams C, Casey PG, Gahan CG, et al. (2012) A novel host responsive sensor mediates virulence and type III secretion during Pseudomonas aeruginosa-host cell interactions. Microbiology 158: 1057–1070. [DOI] [PubMed] [Google Scholar]
- 21. Tribelli PM, Nikel PI, Oppezzo OJ, Lopez NI (2013) Anr, the anaerobic global regulator, modulates the redox state and oxidative stress resistance in Pseudomonas extremaustralis . Microbiology (UK) 159: 259–268. [DOI] [PubMed] [Google Scholar]
- 22. Sonnleitner E, Gonzalez N, Sorger-Domenigg T, Heeb S, Richter A, et al. (2011) The small RNA PhrS stimulates synthesis of the Pseudomonas aeruginosa quinolone signal. Mol Microbiol 80: 868–885. [DOI] [PubMed] [Google Scholar]
- 23. Ayub ND, Pettinari MJ, Ruiz J, Lopez NI (2004) A polyhydroxybutyrate producing Pseudomonas sp. isolated from Antarctic environments with high stress resistance. Curr Microbiol 49: 170–174. [DOI] [PubMed] [Google Scholar]
- 24. López N, Pettinari MJ, Stackebrandt E, Tribelli PM, Pötter M, et al. (2009) Pseudomonas extremaustralis sp. nov. A poly(3-hydroxybutyrate) producer isolated from an Antarctic environment. Curr Microbiol 59: 514–519. [DOI] [PubMed] [Google Scholar]
- 25. Tribelli PM, Di Martino C, López NI, Raiger Iustman LJ (2012) Biofilm lifestyle enhances diesel bioremediation in the Antarctic polyhydroxyalkanoate producer Pseudomonas extremaustralis . Biodegradation 23: 645–651. [DOI] [PubMed] [Google Scholar]
- 26. Tribelli PM, Méndez B, López NI (2010) The oxygen sensitive global regulator, Anr, is involved in biosynthesis of poly(3-hydroxybutyrate) in Pseudomonas extremaustralis . J Mol Microbiol Biotechnol 19: 180–188. [DOI] [PubMed] [Google Scholar]
- 27. Tribelli PM, Raiger Iustman LJ, Catone MV, Di Martino C, Revale S, et al. (2012) Genome sequence of the polyhydroxybutyrate producer Pseudomonas extremaustralis a highly stress resistant Antarctic bacterium. J Bacteriol 194: 2381–2382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Huisman GW, Wonink E, Meima R, Kazemier B, Terpstra P, et al. (1992) Metabolism of poly(3-hydroxyalkanoates) (PHAs) by Pseudomonas oleovorans. Identification and sequences of genes and function of the encoded proteins in the synthesis and degradation of PHA. J Biol Chem 266: 2191–2198. [PubMed] [Google Scholar]
- 29. O’Toole G, Kolter R (1998) Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol Microbiol 30: 295–304. [DOI] [PubMed] [Google Scholar]
- 30. Sherlock O, Munk Vejborg R, Klemm P (2005) The TibA adhesin/invasin from enterotoxigenic Escherichia coli is self recognizing and induces bacterial aggregation and biofilm formation. Infection and Immunity 73: 1954–1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Tremblay J, Déziel E (2008) Improving the reproducibility of Pseudomonas aeruginosa swarming motility assays. J Basic Microb 48: 509–515. [DOI] [PubMed] [Google Scholar]
- 32. Darzins A (1993) The pilG gene product, required for Pseudomonas aeruginosa Pilus production and twitching motility, is homologous to the enteric, single-domain response regulator CheY. J Bacteriol 175: 5934–5944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Larionov A, Krause A, Miller W (2005) A standard curve based method for relative real time PCR data processing. BMC Bioinformatics 6: 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Cheng H, Walker GC (1998) Succinoglycan is required for initiation and elongation of infection threads during nodulation of alfalfa by Rhizobium meliloti . J Bacteriol 180: 5183–5191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Heydorn A, Nielsen AT, Hentzer M, Sternberg C, Givskov M, et al. (2000) Quantification of biofilm structures by the novel computer program COMSTAT. Microbiology 146: 2395–2407. [DOI] [PubMed] [Google Scholar]
- 36.Münch R, Hiller K, Barg H, Heldt D, Linz S, et al.. (2003) PRODORIC: prokaryotic database of gene regulation. Nucleic Acids Res. 31, 266–269. [DOI] [PMC free article] [PubMed]
- 37. Trunk K, Benkert B, Quäck N, Münch R, Scheer M, et al. (2010) Anaerobic adaptation in Pseudomonas aeruginosa: definition of the Anr and Dnr regulons. Environ Microbiol 12: 1719–1733. [DOI] [PubMed] [Google Scholar]
- 38. Bertrand JJ, West JT, Engel JN (2010) Genetic analysis of the regulation of type IV pilus function by the Chp chemosensory system of Pseudomonas aeruginosa . J Bacteriol 192: 994–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Choy W, Zhou L, Kiu-Choong Syn C, Zhang L, Swarup S (2004) MorA defines a new class of regulators affecting flagellar development and biofilm formation in diverse Pseudomonas species. J Bacteriol 186: 7221–7228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jackson A, Gross MJ, Daniels E, Hampton T, Hammond J, et al. (2013) Anr, and its activation by PlcH activity, in Pseudomonas aeruginosa host colonization and virulence. J Bacteriol doi:10.1128/JB.02169-12. [DOI] [PMC free article] [PubMed]
- 41. Junker L, Peters JE, Hay AG (2006) Global analysis of candidate genes important for fitness in a competitive biofilm using DNA-array-based transposon mapping. Microbiology 152: 2233–2245. [DOI] [PubMed] [Google Scholar]
- 42. Newell PD, Yoshioka S, Hvorecny KL, Monds RD, O’Toole GA (2011) Systematic analysis of diguanylate cyclases that promote biofilm formation by Pseudomonas fluorescens Pf01. J Bacteriol 193: 4685–4698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Klausen M, Heydorn A, Ragas P, Lambertsen L, Aaes-Jørgensen A (2003) Biofilm formation by Pseudomonas aeruginosa wild type, flagella and type IV pili mutants. Mol Microbiol 48: 1511–1524. [DOI] [PubMed] [Google Scholar]
- 44. Shrout JD, Tolker-Nielsen T, Givskov M, Parsek MR (2011) The contribution of cell-cell signaling and motility to bacterial biofilm formation. MRS Bull 36: 367–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Yoon SS, Hennigan R, Hilliard JM, Ochsner A, Parvatiyar K, et al. (2002) Pseudomonas aeruginosa anaerobic respiration in biofilms: relationships to cystic fibrosis pathogenesis. Dev Cell 3: 593–603. [DOI] [PubMed] [Google Scholar]
- 46. O’Toole G, Gibbs KA, Hager PW, Phibbs PV, Kolter R (2000) The global carbon metabolism regulator Crc is a component of a signal transduction pathway required for biofilm development by Pseudomonas aeruginosa. . J Bacteriol 182: 425–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Borlee BR, Goldman AD, Murakami K, Samudrala R, Wozniak DJ, et al. (2010) Pseudomonas aeruginosa uses a cyclic-di-GMP-regulated adhesin to reinforce the biofilm extracellular matrix. Mol Microbiol 75: 827–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Klebensberger J, Birkenmaier A, Geffers R, Kjelleberg S, Philipp B (2009) SiaA and SiaD are essential for inducing autoaggregation as a specific response to detergent stress in Pseudomonas aeruginosa . Environ Microbiol 11: 3073–3086. [DOI] [PubMed] [Google Scholar]
- 49.Guttenplan SB, Kearns DB (2013) Regulation of flagellar motility during biofilm formation. FEMS Microbiol Rev DOI: 10.1111/1574-6976.12018. [DOI] [PMC free article] [PubMed]