Abstract
Microorganisms produce a remarkable selection of bioactive small molecules. The study and exploitation of these secondary metabolites has traditionally been restricted to the cultivable minority of bacteria. Rapid advances in meta-omics challenge this paradigm. Breakthroughs in metagenomic library methodologies, direct sequencing, single cell genomics, and natural product-specific bioinformatic tools now facilitate the retrieval of previously inaccessible biosynthetic gene clusters. Similarly, metaproteomic developments enable the direct study of biosynthetic enzymes from complex microbial communities. Additional methods within and beyond meta-omics are also in development. This review discusses recent reports in these arenas and how they can be utilized to characterize natural product biosynthetic gene clusters and pathways.
Introduction
Microbial natural products and their derivatives account for more than half of currently marketed antibiotics [1,2]. Unfortunately, less than 1% of prokaryotic species are capable of laboratory cultivation using standard techniques, historically limiting the discovery and study of a host of bioactive secondary metabolites [3,4**]. The field of meta-omics now provides culture-independent approaches to study previously elusive microorganisms and harness the potential of associated novel natural products.
Meta-omics utilizes genomic, proteomic, metabolomic, and transcriptomic toolsets to transcend cultivation limitations by studying the collective material of organisms from environmental samples. Sometimes an environmental sample consists of a host organism and associated symbiotic microbiota, jointly referred to as a holobiont [5,6*]. The collective material from holobionts can consequently be coined under the holo- prefix. For the sake of discussing other types of environmental samples in addition to host/microbial consortia, the more general metaprefix will be used herein.
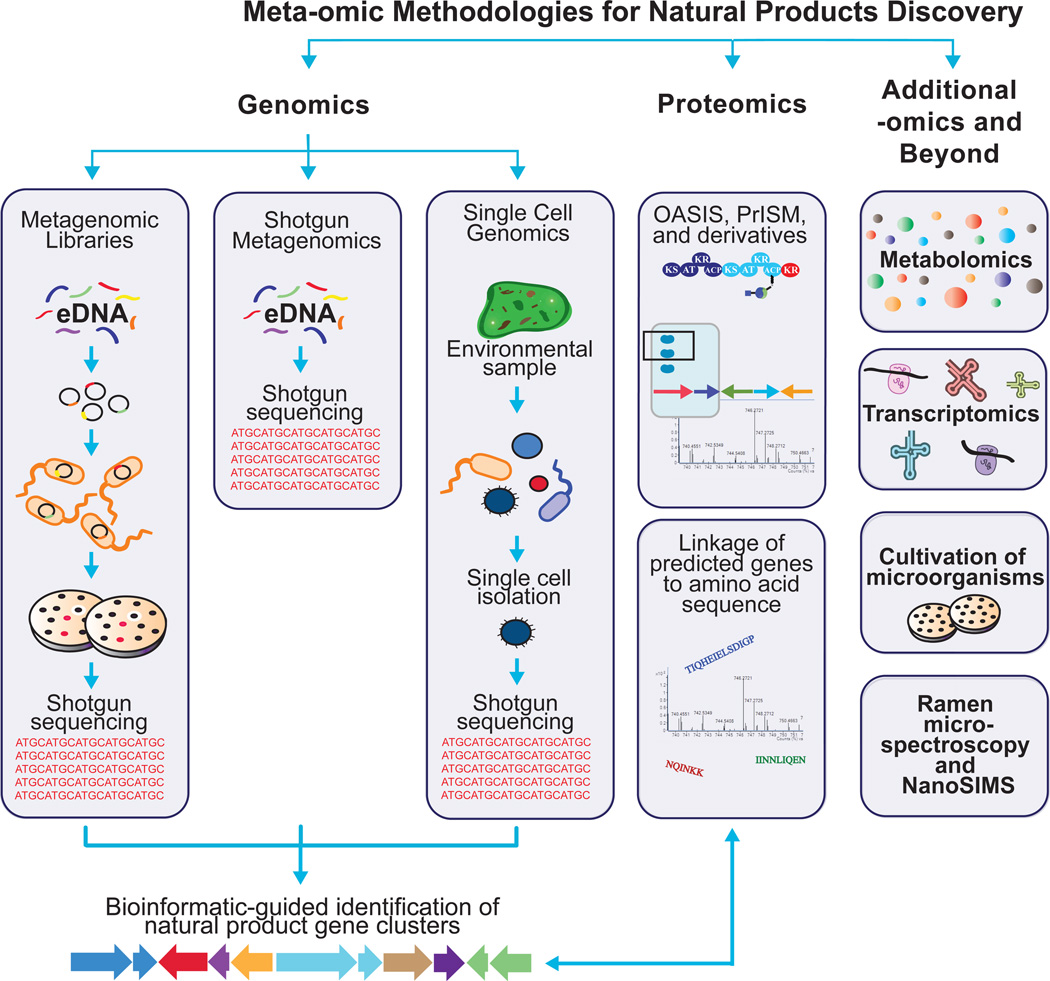
When applied to natural products research, meta-omic technologies can enable the characterization of the biosynthetic pathways of microorganisms that remain incapable of being cultured in the laboratory. Many of these techniques rely on the study of nonribosomal peptide synthetase (NRPS) or polyketide synthase (PKS) enzymes that are frequently contained within the gene clusters of biomedically intriguing natural products. The following review (Figure 1) covers established methodologies and recent advances in natural product meta-omics.
Figure 1.
An overview of meta-omic methodologies for natural products discovery.
Metagenomic approaches: Construction and screening of eDNA libraries
Construction of clone libraries derived from environmental DNA (eDNA) is the most traditional metagenomic approach to sequencing biosynthetic gene clusters. Although the creation and screening of libraries can be time-consuming, it can offer distinct advantages over more innovative methods. Depending on the vector utilized during library construction, clones can contain anywhere from 30–300 kb DNA fragments [7–11]. Most metabolic systems fall into this range, increasing the likelihood of obtaining an intact cluster within a single clone. This is especially appealing since heterologous expression of the full cluster in a culturable host organism could lead to the biosynthesis of the target compound, as demonstrated in recent studies [12,13,14*]. Even if a single clone does not contain an intact cluster, the target pathway can be reconstituted from multiple clones through Red/ET recombineering; this was recently demonstrated by the Müller group for the heterologous expression of the tubulysin biosynthetic gene cluster [15**].
It is important to note that libraries are not the only route to obtaining intact clusters for heterologous studies. Several extensive reviews have focused on alternative methods that enable manipulation of large DNA fragments for natural product biosynthesis in amenable hosts [16–18,19**]. Commonly employed methods can be cloning-dependent, involve DNA recombination, or rely on synthetic gene clusters. Despite these alternatives, metagenomic library construction can still be advantageous. Even if resultant clones do not enable metabolite production, they facilitate DNA sequencing and characterization of target biosynthetic genes.
Library assembly first involves isolation and shearing of total genomic DNA from an environmental sample and insertion of fragmented DNA into a selected vector. It is much more difficult to work with vectors capable of retaining larger fragments. Consequently, natural product pathway studies have traditionally relied on cosmid [7] and lower copy number fosmid [8] vectors capable of accommodating DNA inserts around 40 kb. However, due to the extensive size of many gene clusters, several groups utilize larger vehicles capable of housing up to 300 kb DNA fragments [9], including the bacterial [10] and P1-derived artificial chromosome vectors [11].
Following insertion of DNA fragments into a preferred vector, recombinant DNA is introduced into a host organism. Choice of microorganism is especially important when attempting to confer metabolite production in a heterologous host, since codon bias, differences in key regulatory elements, or the absence of critical precursor substrates or cofactors can inhibit or prevent natural product biosynthesis. Consequently, stark differences in secondary metabolite production have been reported between E. coli and Streptomyces lividans [14*] and even between E. coli and other proteobacteria [13].
After introduction of recombinant DNA into a host organism, resulting clones are screened for DNA fragments of interest. Several recent reviews have thoroughly explored specific library screens used in natural products research [4**,20–22]. In addition, Owen et al. recently developed an E. coli reporter strain to identify clones containing 4’-phosphopantetheinyl transferase (PPTase) genes, which are frequently contained within biosynthetic gene clusters [23*].
Positive clones from a selected screen are subsequently sequenced and mined for biosynthetic genes. Although more labor-intensive, the metagenomic library approach is still effective and commonly employed. This traditional tactic has most recently served in the identification of secondary metabolites derived from soil bacteria [24**], the detection of a siderophore gene cluster [12], and the discovery of a novel natural product family [25**]. The following sections on metagenomics will cover less traversed paths to the discovery of biosynthetic gene clusters.
Metagenomic approaches: Direct eDNA sequencing
Frequently referred to as ‘shotgun metagenomics,’ direct sequencing of eDNA is made possible by the rapid advancement and increasing affordability of next generation sequencing (NGS) technologies. In shotgun metagenomics, the laborious step of library construction can be bypassed in favor of immediately sequencing isolated eDNA. A number of sequencing platforms exist, each with distinct advantages and pitfalls. Roche 454 and Illumina/Solexa are commonly used in shotgun metagenomics [26,27] while newer systems like Pacific Biosciences (PacBio) and Ion Torrent are promising, yet remain relatively underutilized.
Shotgun metagenomics is quickly becoming the method of choice in varied applications. Direct sequencing of metagenomic DNA has been used in the human microbiome project [28], and has helped to identify novel biomass-degrading enzymes from compost [29] and cow rumen [30]. Likewise, an increasing number of natural product pathway studies are beginning to rely on shotgun metagenomics. A series of pioneering studies by the Schmidt group combined metagenomic library construction with direct sequencing to investigate the complex microbiome associated with coral reefs and the marine tunicate Lissoclinum patella [31,32,33**]. Additionally, Roche 454 pyrosequencing was recently used to analyze the microbiome of the marine sponge Arenosclera brasiliensis [34]. A further study conducted by our group utilized Roche 454 shotgun sequencing of the Ecteinascidia turbinata tunicate metagenome to identify the biosynthetic gene cluster of the chemotherapeutic natural product ET-743 [35**].
The major current challenge of the shotgun metagenomic approach is effectively processing the massive output of sequencing data. Pinpointing a target gene cluster amidst the collective genomes of a complex microbial consortium can be daunting. However, current bioinformatic tools (described below) as well as the continued development of new ones form the basis of an effective data mining process. Despite this challenge, direct eDNA sequencing represents a powerful and expanding tool. The popularity of this approach is expected to rise significantly as advancing technology, increasing affordability, and better bioinformatics analysis pipelines improve effectiveness and throughput.
Metagenomic approaches: Single cell genomics
In contrast to traditional metagenomic approaches that work with the collective DNA of multiple organisms, single cell genomics is designed to assess the genomes of individual microbial cells isolated from environmental samples. The technology is dependent on multiple displacement amplification (MDA), which can produce the micrograms of DNA necessary for sequencing applications from the femtograms present in an individual cell [36*].
Single cell genomics begins with the isolation of a microbial cell fraction from an environmental sample and separation of an individual prokaryotic cell through microfluidics, flow cytometry, or micromanipulation [36*]. Screening is typically either combined with cell sorting prior to MDA or conducted subsequently through PCR of amplified isolated genomes. Screening characteristically involves markers such as 16S rRNA gene sequences, rpoB, or recA although other indicators can also be used [36*].
Despite this impressive technology, few studies have applied single cell genomics to natural product pathway discovery and analysis research. The first notable studies involved single cell sorting of microbial cell fractions from the marine sponge Aplysina aerophoba [37,38*]. In both studies, PCR screening of the amplified genomes of single cells led to the identification of putative biosynthetic genes of uncultivable symbionts. A more recent study combined metagenomic library construction with single cell sorting to identify the apratoxin A gene cluster from the marine cyanobacterium Moorea bouillonii [39**],[40].
While single cell methods are subject to amplification bias and the formation of chimeric sequences, there are distinct advantages to isolating and sequencing a single genome over the collective genomes of an environmental sample. Single cell genomics enables biosynthetic gene clusters to be directly linked to any taxonomic information uncovered from the genome, potentially leading to the selection of a suitable host for heterologous expression. Similarly, the identification of metabolic genes could shed light on the conditions or substrates needed to successfully culture the target “unculturable” microorganism in the laboratory. Although not derived from single cell data, several studies have used genomic analysis to guide cultivation efforts. For example, an extensive genomic analysis suggested that members of the SAR11 α- proteobacterial clade were deficient in assimilatory sulphate reduction genes, leading Tripp et. al. to demonstrate that an exogenous reduced sulphur source was required for growth of these microorganisms [41].
Analysis and exploitation of metagenomic data
After acquiring sequencing data, the three aforementioned metagenomic methods require genome assembly, mining for biosynthetic gene clusters, and deep annotation. Mining sequencing data can either be discovery or target-driven [21]. Discovery-driven mining seeks to identify novel gene clusters and ultimately structurally unique natural products. If taking the eDNA library approach, discovery-driven mining can begin with the screening of resultant clones before samples are even submitted for sequencing [4**]. For the remaining metagenomic approaches described above, mining of sequencing data can be performed in combination with genome assembly and annotation. Several recent reviews describe common bioinformatic tools for these applications [42,43]. Fortunately, there is also a selection of applications specific to natural products research (highlighted in Table I). Examples of recently developed tools include NP. searcher [44], PKSIIIpred [45], the antibiotics and Secondary Metabolite Analysis Shell (antiSMASH) [46**], and the Natural Product Domain Seeker (NaPDOS) [47**].
Table 1.
Highlighted natural product-specific tools for bioinformatic-guided analysis of sequencing data
| Analysis Tool | Description | Access | Reference |
|---|---|---|---|
| 2metDB (secondary metabolism database) |
|
http://nrps.igs.umaryland.edu/nrps/2metdb/Welcome.html | [71] |
| antiSMASH |
|
http://antismash.secondarymetabolites.org/ | [46**] |
| BAGEL2 (Bacteriocin Genome Mining Tool 2) |
|
http://bagel2.molgenrug.nl | [72] |
| CLUSEAN (CLUster SEquence ANalyzer) |
|
http://redmine.secondarymetabolites.org/projects/clusean | [73] |
| NaPDoS |
|
http://napdos.ucsd.edu/ | [47**] |
| NP.searcher |
|
http://dna.sherman.lsi.umich.edu | [44] |
| PKSIIIpred |
|
http://type3pks.in/prediction/ | [45] |
| SBSPKS (Structure-Based PKS Analysis) |
|
http://nii.ac.in/sbspks.html | [74] |
| SMURF (Secondary Metabolite Unknown Regions Finder) |
|
http://jcvi.org/smurf/ | [75] |
Conversely, targeted-driven mining is more difficult. This approach seeks to identify the elusive gene clusters of well-studied natural products. For example, although compounds such as bryostatin, aplidine, and ET-743 are already undergoing clinical trials, their biosynthetic pathways remain elusive due to the inability to culture the producing microorganisms in the laboratory [48,49]. One option is to use the aforementioned natural product bioinformatic tools to narrow down candidate gene clusters. Alternatively, target natural products are often thought to be of bacterial origin because their structures resemble compounds produced by thoroughly studied, cultivable bacteria with well-characterized biosynthetic pathways. These pathways and gene clusters can provide a genetically conserved roadmap for mining sequencing data of the target gene cluster. Our group took this approach with the ET-743 gene cluster, using conserved NRPS modules involved in the biosynthesis of three well-studied bacterial-derived natural products (safracin, saframycin MX1, saframycin A) resembling ET-743 to guide the mining of the tunicate-symbiont microbial consortium metagenome. This led to the identification of a contig containing the NRPS genes responsible for the biosynthesis of the tetrahydroisoquinoline core, and ultimately the bulk of the ET-743 biosynthetic gene cluster [35**].
Metaproteomic approaches to the study of biosynthetic gene clusters
Metaproteomic methods for studying biosynthetic gene clusters and pathways are rapidly advancing, spurring a selection of recent reviews that summarize available technologies [50–52]. In particular, the Orthogonal Active Site Identification System (OASIS) and Proteomic Interrogation of Secondary Metabolism (PrISM) have had a substantial impact on the application of proteomic technologies to natural products research. Developed by Meier and coworkers, OASIS utilizes chemical probes that target the active sites of PKS and NRPS enzymes, leading to the enrichment of complex proteomic samples prior to liquid chromatography (LC) tandem mass spectrometry (MS2) [53]. First conceived by the Kelleher group, the PrISM approach similarly takes advantage of the significant size of most PKS and NRPS enzymes and the unique 4’-phosphopantetheine (4’-PPant)-associated ions to profile novel biosynthetic gene clusters [54].
A subsequent study by Meier and coworkers [55**] recently sought to rectify the shortcomings of both approaches, and other 4’-PPant labeling strategies [56,57]. After enrichment of proteomic samples for PKS and NRPS enzymes, selective fragmentation enabled identification of carrier protein peptides. The authors also introduced an optimized pipeline that facilitates targeted identification of carrier protein peptides from low resolution MS2 data. In addition, a machine learning method was presented that permits the identification of target peptides solely from MS2 fragmentation data. Similarly, a recent study by Evans and coworkers demonstrated the effectiveness of the PrISM method through the discovery of koranimine, a natural product associated with a Bacillus species [58]. Although the sensitivity of these approaches will have to be adapted for metaproteomic applications, these collective studies enable harnessing of proteomic technologies to investigate secondary metabolism.
In an additional approach, metaproteomic techniques can be utilized in combination with metagenomics to directly link predicted biosynthetic genes to amino acid sequence and expression in host-symbiont systems. Our group demonstrated the effectiveness of this process by detecting the expression of predicted ET-743 biosynthetic genes in metaproteomic analysis of the tunicate/microbial consortium [35].
Additional omics and beyond
Metabolomics has conventionally focused on identification of biomarkers associated with a specific phenotype. Consequently, it has traditionally been considered mutually exclusive to natural products research despite the shared objective of both fields to identify and characterize metabolites of biological significance [59**,60]. However, metabolomics offers powerful toolsets that could enable natural product chemists to process more complex samples. Therefore, recent efforts by several groups have attempted to bridge the technical gaps between these two important disciplines. Metabolomics technologies have recently been employed to guide the identification of microbial strains containing novel secondary metabolite chemistry [61]. Comparative analysis of metabolite profiles between knockout and wild-type microorganisms also led to the discovery of myxoprincomide from Myxococcus xanthus with high-resolution mass spectrometry [62*] and products associated with the gliotoxin gene cluster of the pathogen Aspergillus fumigatus with 2D NMR spectroscopy (DANS) [63*]. Additional work by Forseth and Schroeder highlighted several specific methodologies used for natural product comparative metabolomics [64]. While the aforementioned studies relied on the ability to culture the associated microorganisms in the laboratory, they provide a foundation for the development of meta-metabolomic natural product approaches. Conversely, transcriptomics involves the investigation of total RNA transcripts of an environmental sample. Although metatranscriptomics methods are becoming popular in the analysis of microbial communities [65,66] and even symbiont-host interactions [67], applications to natural products are still in development.
Outside of the –omics arena, novel methodologies to study elusive gene clusters are continually in development. Several recent reviews discuss efforts to enable the cultivation of “unculturable bacteria,” thereby facilitating access to a host of novel gene clusters [68–70]. Although not yet applied to the natural products of uncultivable bacteria, Ramen microspectroscopy and nano-scale secondary ion mass spectrometry (NanoSIMS) also show promise in the study of individual microorganisms from complex microbial consortia [66].
Conclusions
Meta-omic technologies are rapidly advancing, facilitating the study of previously inaccessible microorganisms and their associated natural product biosynthetic systems. Metagenomic breakthroughs in library construction, direct sequencing of eDNA, and single cell technologies make the acquisition of sequencing data significantly more straightforward, while advancements in bioinformatics facilitate rapid mining for biosynthetic gene clusters. The field of metaproteomics similarly continues to provide further paths to the discovery and analysis of secondary metabolite biosynthesis. Additionally, metabolomic toolsets for natural products discovery in cultivable bacteria are becoming mainstream, providing the groundwork for secondary meta-metabolomic methodology development. In the near-term, it is likely that metagenomic and proteomic approaches will be applied for broad discovery of new natural product pathways. In conjunction with versatile heterologous expression tools, these combined approaches will enable access to broad new chemical diversity resources with new applications in medicine and industry.
Highlights.
Meta-omics permits the study of the biosynthetic gene clusters of uncultivable microbes.
Unculturable microbe genomes can be sequenced via libraries, directly, or by single cell methods.
Natural product-specific bioinformatic tools assist metagenomic sequencing data analysis.
OASIS, PrISM, and derivatives help pave the way for metaproteomic methodologies.
New techniques link metabolomics to natural products research.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
- 1.Newman DJ, Cragg GM. Natural products as sources of new drugs over the last 25 years. J Nat Prod. 2007;70:461–477. doi: 10.1021/np068054v. [DOI] [PubMed] [Google Scholar]
- 2.Lachance H, Wetzel S, Waldmann H. Role of natural products in drug discovery. In: Rankovic Z, Morphy R, editors. Lead Generation Approaches in Drug Discovery. John Wiley & Sons, Inc; 2010. pp. 187–229. [Google Scholar]
- 3.D'Onofrio A, Crawford JM, Stewart EJ, Witt K, Gavrish E, Epstein S, Clardy J, Lewis K. Siderophores from neighboring organisms promote the growth of uncultured bacteria. Chem Biol. 2010;17:254–264. doi: 10.1016/j.chembiol.2010.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Piel J. Approaches to capturing and designing biologically active small molecules produced by uncultured microbes. Annu Rev Microbiol. 2011;65:431–453. doi: 10.1146/annurev-micro-090110-102805. This comprehensive review covers a variety of techniques used to investigate the biosynthetic gene clusters of uncultivable microorganisms and their associated natural products, with an emphasis on metagenomic library assembly and screening options.
- 5.Rosenberg E, Koren O, Reshef L, Efrony R, Zilber-Rosenberg I. The role of microorganisms in coral health, disease and evolution. Nat Rev Microbiol. 2007;5:355–362. doi: 10.1038/nrmicro1635. [DOI] [PubMed] [Google Scholar]
- 6. Rosenberg E, Zilber-Rosenberg I. Symbiosis and development: the hologenome concept. Birth Defects Res C Embryo Today. 2011;93:56–66. doi: 10.1002/bdrc.20196. This intriguing review outlines the hologenome theory of evolution, describing how microbial symbionts and their hosts have developed mutually beneficial relationships. Several classes of holobionts are discussed along with the implications of this important paradigm.
- 7.Collins J, Hohn B. Cosmids: a type of plasmid gene-cloning vector that is packagable in vitro in bacteriophage lambda heads. Proc Natl Acad Sci U S A. 1978;75:4242–4246. doi: 10.1073/pnas.75.9.4242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim UJ, Shizuya H, de Jong PJ, Birren B, Simon MI. Stable propagation of cosmid sized human DNA inserts in an F factor based vector. Nucleic Acids Res. 1992;20:1083–1085. doi: 10.1093/nar/20.5.1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Alduina R, Gallo G. Artificial chromosomes to explore and to exploit biosynthetic capabilities of actinomycetes. J Biomed Biotechnol. 2012;2012:462049. doi: 10.1155/2012/462049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shizuya H, Birren B, Kim UJ, Mancino V, Slepak T, Tachiiri Y, Simon M. Cloning and stable maintenance of 300-kilobase-pair fragments of human DNA in Escherichia coli using an F-factor-based vector. Proc Natl Acad Sci U S A. 1992;89:8794–8797. doi: 10.1073/pnas.89.18.8794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ioannou PA, Amemiya CT, Garnes J, Kroisel PM, Shizuya H, Chen C, Batzer MA, de Jong PJ. A new bacteriophage P1-derived vector for the propagation of large human DNA fragments. Nat Genet. 1994;6:84–89. doi: 10.1038/ng0194-84. [DOI] [PubMed] [Google Scholar]
- 12.Fujita MJ, Kimura N, Sakai A, Ichikawa Y, Hanyu T, Otsuka M. Cloning and heterologous expression of the vibrioferrin biosynthetic gene cluster from a marine metagenomic library. Biosci Biotechnol Biochem. 2011;75:2283–2287. doi: 10.1271/bbb.110379. [DOI] [PubMed] [Google Scholar]
- 13.Craig JW, Chang FY, Kim JH, Obiajulu SC, Brady SF. Expanding small-molecule functional metagenomics through parallel screening of broad-host-range cosmid environmental DNA libraries in diverse proteobacteria. Appl Environ Microbiol. 2010;76:1633–1641. doi: 10.1128/AEM.02169-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. McMahon MD, Guan C, Handelsman J, Thomas MG. Metagenomic analysis of Streptomyces lividans reveals host-dependent functional expression. Appl Environ Microbiol. 2012;78:3622–3629. doi: 10.1128/AEM.00044-12. Several clones from soil metagenomic libraries were found to be bioactive in S. lividans but not E. coli, providing tangible evidence of the importance of host selection in metagenomic library construction.
- 15. Chai Y, Shan S, Weissman KJ, Hu S, Zhang Y, Müller R. Heterologous expression and genetic engineering of the tubulysin biosynthetic gene cluster using Red/ET recombineering and inactivation mutagenesis. Chem Biol. 2012;19:361–371. doi: 10.1016/j.chembiol.2012.01.007. Unable to acquire the intact tubulysin gene cluster within a single clone, the authors utilize Red/ET recombineering to reconstitute the cluster from two cosmids. Subsequent expression of the gene cluster in the native strain and two heterologous hosts provide a foundation for biosynthetic pathway elucidation and possible future manipulation to manufacture novel analogs
- 16.Zotchev SB, Sekurova ON, Katz L. Genome-based bioprospecting of microbes for new therapeutics. Curr Opin Biotechnol. 2012;23:941–947. doi: 10.1016/j.copbio.2012.04.002. [DOI] [PubMed] [Google Scholar]
- 17.Rodriguez E, Menzella HG, Gramajo H. Heterologous production of polyketides in bacteria. Methods Enzymol. 2009;459:339–365. doi: 10.1016/S0076-6879(09)04615-1. [DOI] [PubMed] [Google Scholar]
- 18.Wenzel SC, Müller R. Recent developments towards the heterologous expression of complex bacterial natural product biosynthetic pathways. Curr Opin Biotechnol. 2005;16:594–606. doi: 10.1016/j.copbio.2005.10.001. [DOI] [PubMed] [Google Scholar]
- 19. Zhang H, Boghigian BA, Armando J, Pfeifer BA. Methods and options for the heterologous production of complex natural products. Nat Prod Rep. 2011;28:125–151. doi: 10.1039/c0np00037j. This detailed review covers advancements in heterologous expression of natural product gene clusters, explaining host selection and common molecular toolsets. Specific discussion and examples of successful heterologous expression are provided, organized by type of gene cluster (PKS, NRPS, or mixed) and chosen host organism.
- 20.Taupp M, Mewis K, Hallam SJ. The art and design of functional metagenomic screens. Curr Opin Biotechnol. 2011;22:465–472. doi: 10.1016/j.copbio.2011.02.010. [DOI] [PubMed] [Google Scholar]
- 21.Brady SF, Simmons L, Kim JH, Schmidt EW. Metagenomic approaches to natural products from free-living and symbiotic organisms. Nat Prod Rep. 2009;26:1488–1503. doi: 10.1039/b817078a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ekkers DM, Cretoiu MS, Kielak AM, Elsas JD. The great screen anomaly--a new frontier in product discovery through functional metagenomics. Appl Microbiol Biotechnol. 2012;93:1005–1020. doi: 10.1007/s00253-011-3804-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Owen JG, Robins KJ, Parachin NS, Ackerley DF. A functional screen for recovery of 4'-phosphopantetheinyl transferase and associated natural product biosynthesis genes from metagenome libraries. Environ Microbiol. 2012;14:1198–1209. doi: 10.1111/j.1462-2920.2012.02699.x. The authors present an effective screen to detect 4’-phosphopanteteinyl transferase genes using an E. coli reporter strain, enabling the discovery of novel natural product biosynthetic gene clusters from metagenomic libraries.
- 24. Feng Z, Kallifidas D, Brady SF. Functional analysis of environmental DNA-derived type II polyketide synthases reveals structurally diverse secondary metabolites. Proc Natl Acad Sci U S A. 2011;108:12629–12634. doi: 10.1073/pnas.1103921108. In this study, the authors demonstrate how metagenomic library construction and screening can provide novel drug leads. Soil metagenomic cosmid libraries were screened for type II PKS gene clusters. Subsequent heterologous expression of three systems in Streptomyces albus led to the identification three separate secondary metabolites, including a unique compound with a pentacyclic ring system and a new KB-3346-5 derivative.
- 25. Freeman MF, Gurgui C, Helf MJ, Morinaka BI, Uria AR, Oldham NJ, Sahl HG, Matsunaga S, Piel J. Metagenome Mining Reveals Polytheonamides as Posttranslationally Modified Ribosomal Peptides. Science. 2012;338:387–390. doi: 10.1126/science.1226121. In this investigation, isolation of polytheonamide biosynthetic genes from a sponge metagenomic library provides evidence of a new family of natural products, termed“proteusins” by the authors.
- 26.Petrosino JF, Highlander S, Luna RA, Gibbs RA, Versalovic J. Metagenomic pyrosequencing and microbial identification. Clin Chem. 2009;55:856–866. doi: 10.1373/clinchem.2008.107565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Luo C, Tsementzi D, Kyrpides N, Read T, Konstantinidis KT. Direct comparisons of Illumina vs. Roche 454 sequencing technologies on the same microbial community DNA sample. PLoS ONE. 2012;7:e30087. doi: 10.1371/journal.pone.0030087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Weinstock GM. Genomic approaches to studying the human microbiota. Nature. 2012;489:250–256. doi: 10.1038/nature11553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Allgaier M, Reddy A, Park JI, Ivanova N, D'haeseleer P, Lowry S, Sapra R, Hazen TC, Simmons BA, VanderGheynst JS, et al. Targeted discovery of glycoside hydrolases from a switchgrass-adapted compost community. PLoS ONE. 2010;5:e8812. doi: 10.1371/journal.pone.0008812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hess M, Sczyrba A, Egan R, Kim TW, Chokhawala H, Schroth G, Luo S, Clark DS, Chen F, Zhang T, et al. Metagenomic discovery of biomass-degrading genes and genomes from cow rumen. Science. 2011;331:463–467. doi: 10.1126/science.1200387. [DOI] [PubMed] [Google Scholar]
- 31.Donia MS, Fricke WF, Partensky F, Cox J, Elshahawi SI, White JR, Phillippy AM, Schatz MC, Piel J, Haygood MG, et al. Complex microbiome underlying secondary and primary metabolism in the tunicate-Prochloron symbiosis. Proc Natl Acad Sci U S A. 2011;108:E1423–E1432. doi: 10.1073/pnas.1111712108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Donia MS, Fricke WF, Ravel J, Schmidt EW. Variation in tropical reef symbiont metagenomes defined by secondary metabolism. PLoS ONE. 2011;6:e17897. doi: 10.1371/journal.pone.0017897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kwan JC, Donia MS, Han AW, Hirose E, Haygood MG, Schmidt EW. Genome streamlining and chemical defense in a coral reef symbiosis. Proc Natl Acad Sci U S A. 2012;109:20655–20660. doi: 10.1073/pnas.1213820109. In this report, direct sequencing of the Lissoclinum partella tunicate metagenome links a microbial symbiont to the production of patellazoles and suggests that continued biosynthesis of the natural product preserves symbiosis. This study represents the most recent effort by the Schmidt group (see [31,32]) to employ shotgun metagenomic sequencing to understand the role of bacterial symbionts in the production of secondary metabolites isolated from marine invertebrates.
- 34.Trindade-Silva AE, Rua C, Silva GG, Dutilh BE, Moreira AP, Edwards RA, Hajdu E, Lobo-Hajdu G, Vasconcelos AT, Berlinck RG, et al. Taxonomic and functional microbial signatures of the endemic marine sponge Arenosclera brasiliensis. PLoS ONE. 2012;7:e39905. doi: 10.1371/journal.pone.0039905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Rath CM, Janto B, Earl J, Ahmed A, Hu FZ, Hiller L, Dahlgren M, Kreft R, Yu F, Wolff JJ, et al. Meta-omic characterization of the marine invertebrate microbial consortium that produces the chemotherapeutic natural product ET-743. ACS Chem Biol. 2011;6:1244–1256. doi: 10.1021/cb200244t. In this paper, a unique combination of metagenomic and metaproteomic methodologies elucidates the biosyntetic pathway of the chemotherapeutic natural product ET-743 and links the compound’s production to a microbial symbiont. This study provides a potential framework for the meta-omic characterization of natural product gene clusters.
- 36. Lasken RS. Genomic sequencing of uncultured microorganisms from single cells. Nat Rev Microbiol. 2012;10:631–640. doi: 10.1038/nrmicro2857. This thorough review covers the evolving field of single cell genomics, describing methodologies, technological advancements, and an increasing number of potential applications.
- 37.Siegl A, Hentschel U. PKS and NRPS gene clusters from microbial symbiont cells of marine sponges by whole genome amplification. Environ Microbiol Rep. 2009;2:507–513. doi: 10.1111/j.1758-2229.2009.00057.x. [DOI] [PubMed] [Google Scholar]
- 38. Siegl A, Kamke J, Hochmuth T, Piel J, Richter M, Liang C, Dandekar T, Hentschel U. Single-cell genomics reveals the lifestyle of Poribacteria, a candidate phylum symbiotically associated with marine sponges. ISME J. 2011;5:61–70. doi: 10.1038/ismej.2010.95. Microbial cell fractions from the Mediterranean sponge Aplysina aerophoba were sorted with fluorescence-activated cell sorting. The genome from a single Poribacteria cell was amplified and sequenced. Genomic analysis yielded two PKS genes and additional intriguing artifacts, exemplifying the effectiveness of single cell genomics in the characterization of biosynthetic gene clusters.
- 39. Grindberg RV, Ishoey T, Brinza D, Esquenazi E, Coates RC, Liu WT, Gerwick L, Dorrestein PC, Pevzner P, Lasken R, et al. Single cell genome amplification accelerates identification of the apratoxin biosynthetic pathway from a complex microbial assemblage. PLoS ONE. 2011;6:e18565. doi: 10.1371/journal.pone.0018565. The authors combine metagenomic library construction and single cell genomics to identify the apratoxin A biosynthetic gene cluster from the cyanobacterium Lyngbya bouillonii (recently reclassified as Moorea bouillonii [40]). This innovative study represents one of the first efforts to apply single cell methodologies to natural products research.
- 40.Engene N, Rottacker EC, Kaštovský J, Byrum T, Choi H, Ellisman MH, Komárek J, Gerwick WH. Moorea producens gen. nov., sp. nov. and Moorea bouillonii comb. nov., tropical marine cyanobacteria rich in bioactive secondary metabolites. Int J Syst Evol Microbiol. 2012;62:1171–1178. doi: 10.1099/ijs.0.033761-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tripp HJ, Kitner JB, Schwalbach MS, Dacey JWH, Wilhelm LJ, Giovannoni SJ. SAR11 marine bacteria require exogenous reduced sulphur for growth. Nature. 2008;452:741–744. doi: 10.1038/nature06776. [DOI] [PubMed] [Google Scholar]
- 42.Teeling H, Glöckner FO. Current opportunities and challenges in microbial metagenome analysis--a bioinformatic perspective. Brief Bioinform. 2012;13:728–742. doi: 10.1093/bib/bbs039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Scholz MB, Lo CC, Chain PS. Next generation sequencing and bioinformatic bottlenecks: the current state of metagenomic data analysis. Curr Opin Biotechnol. 2012;23:9–15. doi: 10.1016/j.copbio.2011.11.013. [DOI] [PubMed] [Google Scholar]
- 44.Li MH, Ung PM, Zajkowski J, Garneau-Tsodikova S, Sherman DH. Automated genome mining for natural products. BMC Bioinformatics. 2009;10:185. doi: 10.1186/1471-2105-10-185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mallika V, Sivakumar KC, Jaichand S, Soniya EV. Kernel based machine learning algorithm for the efficient prediction of type III polyketide synthase family of proteins. J Integr Bioinform. 2010;7:143. doi: 10.2390/biecoll-jib-2010-143. [DOI] [PubMed] [Google Scholar]
- 46. Medema MH, Blin K, Cimermancic P, de Jager V, Zakrzewski P, Fischbach MA, Weber T, Takano E, Breitling R. antiSMASH: rapid identification, annotation and analysis of secondary metabolite biosynthesis gene clusters in bacterial and fungal genome sequences. Nucleic Acids Res. 2011;39:W339–W346. doi: 10.1093/nar/gkr466. One of the latest developments in natural product bioinformatics, user-friendly antiSMASH enables the detection and functional annotation of biosynthetic gene clusters directly from sequencing data. In addition to PKS/NRPS identification and analysis tools, antiSMASH provides an extensive comparative database of elucidated gene clusters and the ability to annotate accessory biosynthetic genes.
- 47. Ziemert N, Podell S, Penn K, Badger JH, Allen E, Jensen PR. The natural product domain seeker NaPDoS: a phylogeny based bioinformatic tool to classify secondary metabolite gene diversity. PLoS ONE. 2012;7:e34064. doi: 10.1371/journal.pone.0034064. Representing another recent advancement in the bioinformatic characterization of biosynthetic gene clusters, NaPDoS permits the identification and classification of PKS ketosynthase and NRPS condensation domains from sequencing data. NaPDoS takes advantage of the conservation in these domains in a phylogenetic approach that can guide studies towards gene clusters or microorganisms containing new domain lineages and novel mechanistic biochemistry.
- 48.Gerwick WH, Moore BS. Lessons from the past and charting the future of marine natural products drug discovery and chemical biology. Chem Biol. 2012;19:85–98. doi: 10.1016/j.chembiol.2011.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Piel J. Bacterial symbionts: prospects for the sustainable production of invertebratederived pharmaceuticals. Curr Med Chem. 2006;13:39–50. [PubMed] [Google Scholar]
- 50.Slattery M, Ankisetty S, Corrales J, Marsh-Hunkin KE, Gochfeld DJ, Willett KL, Rimoldi JM. Marine proteomics: a critical assessment of an emerging technology. J Nat Prod. 2012;75:1833–1877. doi: 10.1021/np300366a. [DOI] [PubMed] [Google Scholar]
- 51.Meier JL, Burkart MD. Proteomic analysis of polyketide and nonribosomal peptide biosynthesis. Curr Opin Chem Biol. 2011;15:48–56. doi: 10.1016/j.cbpa.2010.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chiang YM, Chang SL, Oakley BR, Wang CC. Recent advances in awakening silent biosynthetic gene clusters and linking orphan clusters to natural products in microorganisms. Curr Opin Chem Biol. 2011;15:137–143. doi: 10.1016/j.cbpa.2010.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Meier JL, Niessen S, Hoover HS, Foley TL, Cravatt BF, Burkart MD. An orthogonal active site identification system (OASIS) for proteomic profiling of natural product biosynthesis. ACS Chem Biol. 2009;4:948–957. doi: 10.1021/cb9002128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bumpus SB, Evans BS, Thomas PM, Ntai I, Kelleher NL. A proteomics approach to discovering natural products and their biosynthetic pathways. Nat Biotechnol. 2009;27:951–956. doi: 10.1038/nbt.1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Meier JL, Patel AD, Niessen S, Meehan M, Kersten R, Yang JY, Rothmann M, Cravatt BF, Dorrestein PC, Burkart MD, et al. Practical 4 ′ -phosphopantetheine active site discovery from proteomic samples. J Proteome Res. 2011;10:320–329. doi: 10.1021/pr100953b. The authors expand on OASIS and PrISM methodologies in the development of a proteomic method to enrich samples for PSK/NRPS peptides, detect PPant active sites, and identify peptides using lower resolution MS2 data. A machine learning method is also introduced that can enable quick detection of PPant peptides from only MS2 fragmentation data. This work takes an important step toward making proteomic characterization of biosynthetic gene clusters amenable to technology available in most natural product laboratories.
- 56.Meier JL, Mercer AC, Burkart MD. Fluorescent profiling of modular biosynthetic enzymes by complementary metabolic and activity based probes. J Am Chem Soc. 2008;130:5443–5445. doi: 10.1021/ja711263w. [DOI] [PubMed] [Google Scholar]
- 57.Mercer AC, Meier JL, Torpey JW, Burkart MD. In vivo modification of native carrier protein domains. Chembiochem. 2009;10:1091–1100. doi: 10.1002/cbic.200800838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Evans BS, Ntai I, Chen Y, Robinson SJ, Kelleher NL. Proteomics-based discovery of koranimine, a cyclic imine natural product. J Am Chem Soc. 2011;133:7316–7319. doi: 10.1021/ja2015795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Robinette SL, Brushweiler R, Schroeder FC, Edison AS. NMR in metabolomics and natural products research : two sides of the same coin. Acc Chem Res. 2012;45:288–297. doi: 10.1021/ar2001606. This extensive review describes the similarities and differences between conventional metabolomics and natural products research. Authors discuss developing NMR-based methodologies that bridge the void between both disciplines and forecast the challenges that will need to be overcome in future studies.
- 60.Forseth RR, Schroeder FC. NMR-spectroscopic analysis of mixtures: from structure to function. Curr Opin Chem Biol. 2011;15:38–47. doi: 10.1016/j.cbpa.2010.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hou Y, Braun DR, Michel CR, Klassen JL, Adnani N, Wyche TP, Bugni TS. Microbial strain prioritization using metabolomics tools for the discovery of natural products. Anal Chem. 2012;84:4277–4283. doi: 10.1021/ac202623g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Cortina NS, Krug D, Plaza A, Revermann O, Müller R. Myxoprincomide: a natural product from Myxococcus xanthus discovered by comprehensive analysis of the secondary metabolome. Angew Chem Int Ed Engl. 2012;51:811–816. doi: 10.1002/anie.201106305. A recent example of comparative metabolomics reliant on liquid chromatography joined with high-resolution mass spectrometry. Assessing the metabolite profiles of wild-type and knock out strains of Myxococcus xanthus leads to the identification of novel PKS/NRPS secondary metabolite myxoprincomide.
- 63. Forseth RR, Fox EM, Chung D, Howlett BJ, Keller NP, Schroeder FC. Identification of cryptic products of the gliotoxin gene cluster using NMR-based comparative metabolomics and a model for gliotoxin biosynthesis. J Am Chem Soc. 2011;133:9678–9681. doi: 10.1021/ja2029987. This study exemplifies the power of 2D NMR spectroscopy (DANS) in comparative metabolomics. Novel metabolites produced by the gliotoxin gene cluster of Aspergillus fumigatus are discovered by evaluating the metabolite profiles of a wild-type and a transcriptional regulator knockout strain.
- 64.Forseth RR, Schroeder FC. Correlating secondary metabolite production with genetic changes using differential analysis of 2D NMR spectra. Methods Mol Biol. 2012;944:207–219. doi: 10.1007/978-1-62703-122-6_15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Radax R, Rattei T, Lanzen A, Bayer C, Rapp HT, Urich T, Schleper C. Metatranscriptomics of the marine sponge Geodia barretti: tackling phylogeny and function of its microbial community. Environ Microbiol. 2012;14:1308–1324. doi: 10.1111/j.1462-2920.2012.02714.x. [DOI] [PubMed] [Google Scholar]
- 66.Su C, Lei L, Duan Y, Zhang KQ, Yang J, et al. Culture-independent methods for studying environmental microorganisms: methods, application, and perspective. Appl Microbiol Biotechnol. 2012;93:993–1003. doi: 10.1007/s00253-011-3800-7. [DOI] [PubMed] [Google Scholar]
- 67.Stewart FJ, Dmytrenko O, Delong EF, Cavanaugh CM. Metatranscriptomic analysis of sulfur oxidation genes in the endosymbiont of solemya velum. Front Microbiol. 2011;2:134. doi: 10.3389/fmicb.2011.00134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pham VH, Kim J. Cultivation of unculturable soil bacteria. Trends Biotechnol. 2012;30:475–484. doi: 10.1016/j.tibtech.2012.05.007. [DOI] [PubMed] [Google Scholar]
- 69.Stewart EJ. Growing unculturable bacteria. J Bacteriol. 2012;194:4151–4160. doi: 10.1128/JB.00345-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Vartoukian SR, Palmer RM, Wade WG. Strategies for culture of 'unculturable' bacteria. FEMS Microbiol Lett. 2010;309:1–7. doi: 10.1111/j.1574-6968.2010.02000.x. [DOI] [PubMed] [Google Scholar]