Summary
Previous evidence has shown that Parkinson disease (PD) has a heritable component, but only a small proportion of the total genetic contribution to PD has been identified. Genetic heterogeneity complicates the verification of proposed PD genes and the identification of new PD susceptibility genes. Our approach to overcome the problem of heterogeneity is to study a population isolate, the mid-western Amish communities of Indiana and Ohio. We performed genome-wide association and linkage analyses on 798 individuals (31 with PD), who are part of a 4,998 member pedigree. Through these analyses, we identified a region on chromosome 5q31.3 that shows evidence of association (p-value < 1 × 10−4) and linkage (multipoint HLOD = 3.77). We also found further evidence of linkage on chromosomes 6 and 10 (multipoint HLOD 4.02 and 4.35 respectively). These data suggest that locus heterogeneity, even within the Amish, may be more extensive than previously appreciated.
Keywords: Parkinson Disease, founder population, genome wide association study, genome wide linkage study
Introduction
Parkinson disease (PD) is the second most common neurodegenerative disorder in adults and was originally defined as a motor disease based on the four hallmark characteristics: resting tremor, bradykinesia, rigidity, and postural instability (Gelb et al. 1999). However, it has since been realized that individuals with PD are affected by many other symptoms. Non-motor symptoms of PD include dementia, pain, mood disorders, sleep difficulties, and autonomic dysfunction. Not all symptoms are present for each individual diagnosed with PD, but the number and severity of symptoms tend to accumulate over time. The average age at diagnosis of PD is 70.5 years and most frequently occurs after the age of 60 (Van Den Eeden et al. 2003).
The clinical expression and physiological systems involved in Parkinson disease are also complex, suggesting variability in the etiology. Development of PD in an individual has been attributed to environmental factors, genetic factors, and interactions between the two, although we still understand only a small part of all the components in play (Lai et al. 2002; Klein and Westenberger 2012; Marras and Goldman 2011; Hamza et al. 2011). Many environmental factors have been implicated in PD, but only a few are consistent across studies (pesticides as a risk factor, smoking and caffeine as protective factors) (Lai et al. 2002; Hancock et al. 2007; Hancock et al. 2008). In addition, an abundance of genetic risk factors have been identified, and many have been replicated and confirmed by subsequent studies. Loci have been identified that act under autosomal dominant (e.g. SNCA, LRRK2) and autosomal recessive (e.g. PRKN, DJ1, PINK1) models, and that function as strong risk factors for development of the disease and other associations with common polymorphisms (e.g. variants in GBA, MAPT) (Klein and Westenberger 2012; Lill et al. 2012; International Parkinson Disease Genomics Consortium et al. 2011; IPDGC and WTCCC2 2011). At least 13 loci have been confirmed as PD-related loci and they have been identified through linkage, candidate gene, genome-wide association, and exome sequencing studies. These results are encouraging and exciting; however, there is still a large portion of the genetic component of PD that remains to be uncovered. In light of this, we have undertaken a study in an isolated founder population, from mid-western Amish communities in Ohio and Indiana, and conducted genome-wide association and linkage analyses to detect additional PD-related loci.
The Amish population is a promising population to study the genetic risk of PD for several reasons. The first PD-related loci were identified in genetically isolated founder populations (Polymeropoulos et al. 1997). We hypothesize that the genetics of PD may be less heterogeneous in the Amish due to the shared ancestry and minimal gene flow. Also, the Amish share a very homogenous lifestyle, including minimal caffeine and tobacco use, due to cultural and religious beliefs. This homogenous background may allow us to more easily detect genetic effects by reducing confounding effects of environmental factors. We have analyzed 798 individuals, who can all be connected into one 4,998-member pedigree. In this dataset we see a higher average kinship coefficient among pairs of affected individuals (.0177) compared to pairs of unaffected individuals (.0149). We performed a Wilcoxon rank-sum (Mann Whitney) test and found this difference to be statistically significant (p < 1 × 10−4), strongly suggesting that PD is heritable in the Amish and likely a valuable population to uncover future knowledge of the underlying genetics of PD.
Methods
Subjects
Methods for ascertainment were reviewed and approved by the individual Institutional Review Boards of the respective institutions. Informed consent was obtained from participants recruited from Amish communities with which we have had established working relationship for over 10 years. These communities are in Elkhart, LaGrange, Adams, and surrounding Indiana counties, and Holmes and surrounding Ohio counties. To date we have enrolled over 2,200 Amish individuals with 32 of these diagnosed with PD.
Clinical Evaluation
The spectrum of clinical symptoms for this pedigree was previously described (Cummings et al. 2011; Lee et al. 2008). A standardized interview for PD was conducted by a board-certified genetic counselor or genetic study research associate with participating individuals or a knowledgeable family informant. Individuals were screened for a history of encephalitis, dopamine-blocking medication exposure within one year before diagnosis, symptoms of normal pressure hydrocephalus (dementia, gait difficulty, and urinary incontinence), or a clinical course with unusual features suggestive of atypical or secondary parkinsonism. Individuals with a positive symptom history of PD and apparently unaffected individuals (mostly siblings) were personally examined by a board-certified neurologist with subspecialty training in movement disorders, and many have been examined more than once to ensure the diagnoses are accurate over time. At these secondary interviews, participants were evaluated for a history of exposure to substances known or suspected to cause parkinsonism, including heavy metals and pesticides. Participants were classified as affected, unaffected or unknown, using published diagnosis criteria based on clinical history and neurologic examination (Gelb et al. 1999). Affected individuals had at least two cardinal signs of PD (resting tremor, bradykinesia, or rigidity) and no atypical features of parkinsonism. Individuals with unknown status had only one sign of PD, a history of atypical clinical features, or both. Unaffected individuals had no signs of PD. Age at onset was self-reported and defined as the age at which onset of the first symptom suggestive of PD was noted by the affected individual. Levodopa responsiveness was determined based on physician and patient observations. Individuals with uncertain symptom benefit or who never received levodopa therapy were classified as having an unknown response.
The severity of extrapyramidal signs and symptoms were evaluated by Hoehn-Yahr staging (Hoehn and Yahr 1998) and the Unified Parkinson Disease Rating Scale (UPDRS-motor subscale) UPDRS-III (Fahn et al. 1987). When available, reports of brain imaging studies were reviewed to confirm the absence of hydrocephalus or vascular parkinsonism. Dementia was assessed by the memory-orientation-concentration test (Short-Blessed Test (SBT)) (Katzman et al. 1983). Diagnosis of progressive supranuclear palsy was determined from the NINDS-PSP International Workgroup clinical criteria (Litvan et al. 1996).
Genotyping
Genotyping and quality control (QC) in this dataset were performed as described previously (Cummings et al. 2012). Briefly, genome-wide genotyping was performed on 830 DNA samples using the Affymetrix 6.0 GeneChip ® Human Mapping 1 million array set (Affymetrix ®, Inc Santa Clara, CA). DNA for this project was allocated by the respective DNA banks at both the Hussman Institute of Human Genomics (HIHG) at the University of Miami and the Center for Human Genetics Research (CHGR) at Vanderbilt University. Genomic DNA was quantitated via the ND-8000 spectrophotometer and DNA quality was evaluated via gel electrophoresis. The genomic DNA (250ng/5ul) samples were processed according to standard Affymetrix procedures for the processing of the Affymetrix 6.0 GeneChip array. The arrays were then scanned using the GeneChip Scanner 3000 7G operated by the Affymetrix® GeneChip® Command Console® (AGCC) software. The data were processed for genotype calling using the Affymetrix® Power Tools (APT) software using the Birdseed calling algorithm version 2.0 Affymetrix®, Inc Santa Clara, CA.
Strict QC procedures were applied to both samples and SNPs to ensure the accuracy of our data prior to analyses. Sample QC included visualization of each DNA sample via agarose to ensure high quality samples prior to inclusion on the array and CEPH samples and duplicate samples plated across multiple arrays to check reproducibility across the arrays. Samples with call rates <95% were re-examined to ensure quality of genotypes; if the call rate for a sample was still <95% after this examination, we attempted to rerun the array with a new DNA sample. Nine samples failed at this point and were dropped due to low genotyping efficiency. The Anabaptist Genealogy DataBase (AGDB) (Agarwala et al. 2003) was used to determine the relationships between individuals; three samples were excluded because they did not connect with the rest of the samples and the relationship of these individuals could not be accounted for in analyses. Samples with genetic sex (based on X chromosome heterozygosity rates) contrary to reported sex were excluded from analyses (16 samples). Sex mismatches were not correlated with genotyping efficiency or sample quality (DNA source, date of collection, or degradation). Three samples appeared aberrantly connected in the pedigree based on genotype data and were also excluded.
SNP QC comprised checking call rates and minor allele frequencies (MAF). 76,816 SNPs with call rates < 98% were dropped, along with 206,970 SNPs with MAF ≤ 0.05. Due to the relatedness in this dataset we did not test SNPs for Hardy-Weinberg equilibrium. After this extensive QC, 798 samples and 622,812 SNPs were available for analysis. All samples are part of one 4,998-member pedigree with many consanguineous loops. The AGDB provided the pedigree information using an “all common paths” database query with all genotyped individuals.
Statistical Analysis
Association analysis was conducted using the Modified Quasi-Likelihood Score (MQLS) (Thornton and McPeek 2007) test for all SNPs. The MQLS test is analogous to a chi-square test for case-control data, but adjusts for correlations among individuals based on pairwise kinship coefficients estimated from the pedigree structure. The MQLS test can be run on all samples without dividing the pedigree, which offers a great advantage by incorporating all pedigree data into a single analysis. Allele frequencies adjusted for relationships are also calculated by this program. Association analysis included all individuals in the 4,998-member pedigree, including the 798 genotyped individuals (31 affected, 123 unaffected, 647 unknown individuals). To test the validity of the MQLS test in our pedigree, we performed simulation studies using this same pedigree structure and null data to assess the type 1 error rate using MQLS for association. The genomic inflation factor for this analysis is 1.04. Type 1 error rates were not inflated (submitted data, Cummings et al.).
Linkage analyses were run using Merlin for autosomal chromosomes and MINX (Merlin in X) for the X chromosome (Abecasis et al. 2002). Due to the large size and substantial consanguinity of the pedigree, it was essential to cut the pedigree into smaller sub-pedigrees that were computationally feasible to analyze. To do this we used PedCut (Liu et al. 2008) to find an optimal set of sub-pedigrees with a bit size limit of 24 and a maximal number of subjects of interest in each pedigree. This procedure resulted in 10 sub-pedigrees (261 individuals, 85 genotyped) for analysis with an average of 8.5 genotyped individuals (3 genotyped affected) per sub-pedigree (Supplementary Table 1). Parametric two-point heterogeneity logarithm of the odds of linkage (HLOD) scores were computed using affecteds-only autosomal dominant and recessive models. Disease allele frequency was estimated at 1% under the dominant model and 20% under the recessive model. Under the dominant model, penetrances of 0 for no disease alleles and 0.0001 for one or two disease alleles were used. For the recessive model, penetrances of 0 for zero or one disease allele and 0.0001 for two disease alleles were used.
The number of SNPs and samples in our dataset prevented us from running the entire dataset in multipoint linkage analysis due to the complexity of the pedigree and extensive computational time. Regions showing evidence for linkage, i.e. containing at least one two-point HLOD ≥ 3.0, or with evidence for linkage (two-point HLOD ≥ 2.0) and association (MQLS p < 1 × 10−4) were followed up with parametric multipoint linkage analysis (also using Merlin). For the multipoint analyses, a seven megabase region surrounding each significant SNP(s) was used. If a clear LOD score peak was not observed in the results (e.g. the maximum LOD score was at one end, or the peak did not drop enough to define a 1-LOD-down support interval), we widened the region until we could define the complete peak. SNPs were pruned for linkage disequilibrium (LD) in each region so all pair-wise r2 values were < 0.16 between SNPs (Boyles et al. 2005). The LD from the HapMap CEPH samples (parents only) was used for pruning. Initially, we chose to use this population for pruning because we were concerned that the consanguinity of the pedigree would cause us to overestimate LD between the SNPs and, subsequently, over-prune the data. Because the HapMap CEPH samples may not be an exact representation of LD in our Amish population, we further tested pruning using the data from this Amish dataset, but no significant difference was seen (data not shown). Because linkage analyses can be affected when breaking larger pedigrees into a series of smaller ones, we performed simulation studies assuming no linkage (e.g. null distribution) and using the same large pedigree structure and the same pedigree splitting method. We determined empirical cut-offs for significance in our linkage studies to maintain a nominal type I error rate. We found only 2.5% of the multipoint linkage scans generated a maximum HLOD > 3.0 (submitted data, Cummings et al.). Possible subhaplotypes and co-inheritance were determined manually and using Merlin. HaploPainter (Thiele et al. 2005) was used to draw sub-pedigrees with associated haplotypes.
Computations were performed using either the CHGR computational cluster or the Advanced Computing Center for Research and Education (ACCRE) cluster at Vanderbilt University. All map positions are listed in megabases (Mb) and refer to NCBI36/hg18, March 2006 positions.
Results
Association analysis
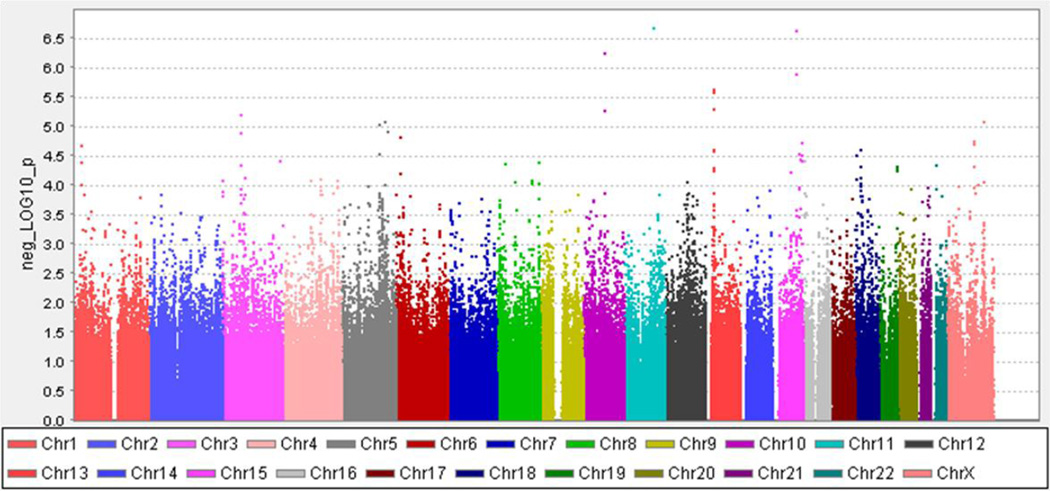
622,812 SNPs were analyzed. No SNPs met a genome-wide significance cutoff of p < 5 × 10−8 (Figure 1). However, 70 SNPs representing 35 regions were identified at p < 1 × 10−4 across 16 chromosomes. 16 of the 35 regions had at least two SNPs significant at this level. The three most significant SNPs are located on chromosomes 10q22.1 (COL13A1), 11q21 (CCDC82), and 15q25.1 (TMC3) (Table 1). All three SNPs are intronic.
Figure 1.
MQLS association results
Table 1.
MQLS top results
| Chromosome | SNP | Map (Mb) |
MQLS p-value |
Gene | Minor Allele |
MAF Affecteds |
MAF Unaffecteds |
MAF Overall |
|---|---|---|---|---|---|---|---|---|
| 11q21 | rs7118648 | 96.11 | 2.16 × 10−7 | CCDC82 | G | 0.21 | 0.03 | 0.03 |
| 15q25.1 | rs3935740 | 81.63 | 2.31 × 10−7 | TMC3 | T | 0.29 | 0.08 | 0.08 |
| 10q22.1 | rs17497526 | 71.58 | 5.50 × 10−7 | COL13A1 | G | 0.42 | 0.15 | 0.15 |
MAF: minor allele frequency; All MAF reported are adjusted for relationships using MQLS
Linkage analysis
Five SNPs had a two-point HLOD > 3.0, all under a recessive model (Table 2). All SNPs had an alpha = 1. The highest HLOD (3.67) was seen on chromosome 5q23.2 in an intron of the MEGF10 gene, with a minor allele frequency (MAF) adjusted for relatedness of 0.08 in unaffecteds and 0.27 in affecteds. Two other SNPs in this gene had an HLOD ≥ 2.5. Multipoint linkage analysis resulted in a peak HLOD of 1.81.
Table 2.
Top linkage (two-point) results
| Chromosome | SNP | Map (Mb) |
HLOD (DOM) |
HLOD (REC) |
Gene | Minor Allele |
MAF Affecteds |
MAF Unaffecteds |
MAF Overall |
|---|---|---|---|---|---|---|---|---|---|
| 5q23.2 | rs17165041 | 126.70 | 2.51 | 3.67 | MEGF10 | G | 0.28 | 0.08 | 0.08 |
| 9q21.33 | rs1439054 | 86.46 | 2.44 | 3.01 | - | G | 0.26 | 0.10 | 0.09 |
| 10p14 | rs2186063 | 10.53 | 2.29 | 3.48 | - | T | 0.46 | 0.18 | 0.18 |
| 10p12.31 | rs1926693 | 22.47 | 2.11 | 3.14 | - | G | 0.16 | 0.06 | 0.06 |
| 19q13.12 | rs16970293 | 40.59 | 2.80 | 3.57 | LOC100128682 | C | 0.15 | 0.04 | 0.04 |
DOM: dominant model; REC: recessive model; MAF: minor allele frequency;
All MAF reported are adjusted for relationships using MQLS
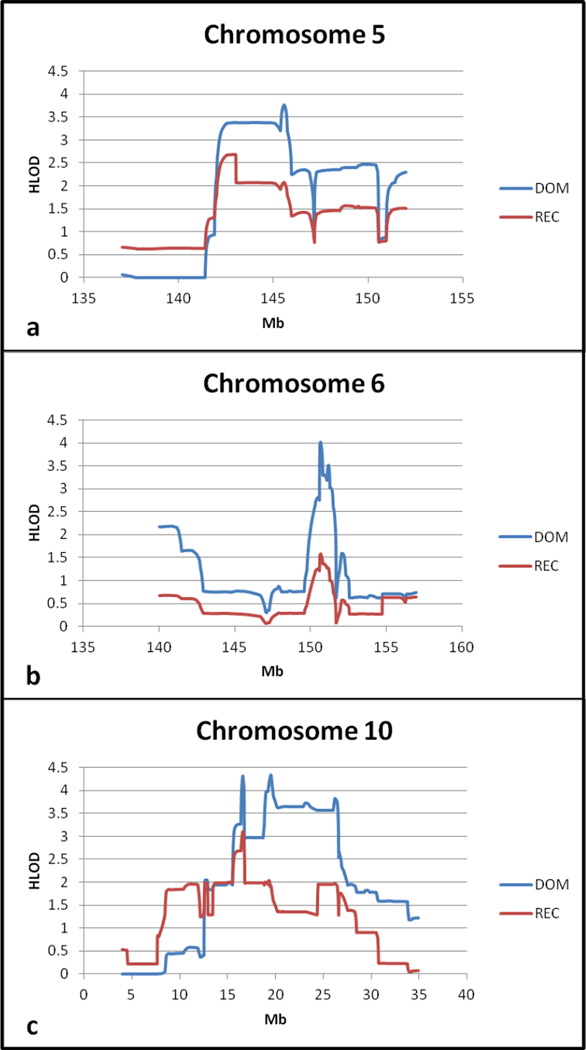
The next highest two-point HLOD was for rs16970293 on chromosome 19q13.12 in a non-coding gene (LOC100128682). Multipoint linkage analysis in this region resulted in a peak HLOD of 1.95 (recessive model). Other significant SNPs were located on chromosomes 9q21.33, 10p14, and 10p12.31, all in intergenic regions. The multipoint HLOD for chromosome 9 was less than 1.0, suggesting further investigation was unlikely to uncover new knowledge about PD. However, multipoint analysis of the regions on chromosome 10 showed a strong linkage peak (dominant HLOD 4.35) (Figure 2c, Table 4).
Figure 2.
Multipoint HLOD scores for significant regions
Table 4.
Most significant multipoint linkage results (HLOD > 3)
| Chromosome | Map (Mb) | Peak HLOD | Model | Alpha | 1-LOD down SI (Mb) |
|---|---|---|---|---|---|
| 5q32 | 145.56 | 3.77 | DOM | 0.69 | (142.08, 146.89) |
| 6q25.1 | 150.70 | 4.02 | DOM | 0.69 | (150.59, 151.35) |
| 10p12.33 | 19.50 | 4.35 | DOM | 1.00 | (16.40, 26.55) |
SI: support interval; DOM: dominant model
Previously we identified linkage peaks in this population on chromosomes 6, 19, 21, and 22 (Cummings et al. 2011). In the current dataset with additional information, the two-point HLODs for these regions were between 1.5 and 1.99, and they did not meet our criteria for follow-up. However, because of the previous implication of these regions, we ran multipoint analyses. We did not see further evidence for linkage on chromosomes 19 and 22, but we did observe increased HLOD scores on chromosomes 6 (dominant HLOD 4.02) (Figure 2b, Table 4) and 21 (dominant HLOD 2.83).
Overlap
To integrate the knowledge gained from performing linkage and association analyses, we looked at the overlap between the two. Six SNPs in five regions showed evidence of linkage (two-point HLOD ≥ 2.0) and association (MQLS p < 1 × 10−4) (Table 3). Alpha values for the HLOD scores were equal to one. Two regions are in genes (FHIT, MEGF10), the others are intergenic. Multipoint linkage analyses were run for each of these regions. The region on chromosome 5q23.2 was analyzed as described above. Regions at 3p14.2, 4q34.2, and 18q11.2 showed no further evidence for linkage based on multipoint analysis. However, chromosome 5q31.3 showed greater evidence for linkage with an HLOD of 3.77 under a dominant model (Figure 2a, Table 4).
Table 3.
Overlapping linkage (two-point) and association results
| Chromosome | SNP | Map (Mb) |
HLOD (DOM) |
HLOD (REC) |
MQLS p-value |
Gene | Minor Allele |
MAF Affecteds |
MAF Unaffecteds |
MAF Overall |
|---|---|---|---|---|---|---|---|---|---|---|
| 3p14.2 | rs41355450 | 60.09 | 2.19 | 2.62 | 4.56 × 10−5 | FHIT | C | 0.33 | 0.12 | 0.12 |
| 4q34.2 | rs6848215 | 177.72 | 1.55 | 2.35 | 8.16 × 10−5 | - | A | 0.77 | 0.47 | 0.46 |
| 5q23.2 | rs17165041 | 126.70 | 2.51 | 3.67 | 2.97 × 10−5 | MEGF10 | G | 0.28 | 0.08 | 0.08 |
| 5q23.2 | rs17673147 | 126.70 | 2.04 | 2.71 | 9.48 × 10−6 | MEGF10 | A | 0.23 | 0.06 | 0.07 |
| 5q31.3 | rs17403174 | 142.93 | 1.66 | 2.10 | 9.59 × 10−5 | - | C | 0.26 | 0.09 | 0.09 |
| 18q11.2 | rs299238 | 18.56 | 1.55 | 2.28 | 4.67 × 10−5 | - | T | 0.75 | 0.45 | 0.45 |
DOM: dominant model; REC: recessive model; MAF: minor allele frequency;
All MAF reported are adjusted for relationships using MQLS
Discussion
The region on chromosome 5q31.3 has been linked to PD in several previous studies, although this study is the first to implicate this region in the Amish population. It was initially identified in an analysis of 174 families by Scott et al. (2001), and this is the seventh study that has implicated this region since that time (Martinez et al. 2004; Hicks et al. 2002; Li et al. 2002; Krygowska-Wajs et al. 2005; Rosenberger et al. 2007; Pankratz et al. 2002). Foroud et al combined samples from two of these studies into one dataset for a more powered linkage analysis and analyzed 20 microsatellite markers across 79 cM in this region and found no additional evidence of linkage (2006). However, these two studies each identified the 5q peak after removing families with other PD-linked loci (parkin, chromosome 2) (Pankratz et al. 2002; Martinez et al. 2004), but the combined study appears to have used all families and did not condition for either of these two loci. Association studies published to-date have not identified a marker in this region (Foltynie et al. 2005; Maraganore et al. 2005). In light of previous analyses of this region, we propose that a more in-depth study of this region in this Amish population and in other familial datasets may prove enlightening. There are many genes in this region. Of particular interest is synphilin-1 in this region (Marx et al. 2003; Myhre et al. 2008; Maraganore et al. 2003), which encodes a protein that interacts with alpha-synuclein, the main component of Lewy bodies. No study has conclusively identified an allele significantly associated with PD in this gene. However, most studies have analyzed synphilin-1 only for idiopathic PD; it is possible it could play a role in familial PD.
The most significant linkage region we identified was on chromosome 10p12.31. There are several candidate genes in this region: RAB18, which has been linked to Warburg Micro syndrome, a developmental disorder with brain abnormalities (Bem et al. 2011); SLC39A12, a zinc transporter; and ITGA8, an integrin receptor expressed in the brain. ITGA8 was recently associated with PD in a large meta-analysis (Lill et al. 2012).
This region on chromosome 10 was also identified in an earlier linkage analysis we performed using microsatellite markers in a subset of the current dataset (Lee et al. 2008). It did not show up in the initial two-point genome-wide linkage scan we performed here, but this may be due to variances in coverage. The current study is the most in-depth of the studies we have performed for PD in this population.
The region on chromosome 6 was only identified in multipoint linkage in this study because it was previously implicated in the Amish (Cummings et al. 2011). This suggests that there may be other linked loci in this population that we missed because the two-point HLOD scores were not high enough to meet our criteria for follow-up. Ideally, we would analyze all markers across the genome that we have genotyped using multipoint linkage analysis. At the current point in time, however, this is not feasible due to time and computational requirements.
Of the 10 sub-pedigrees we analyzed in multipoint linkage analysis, between three and five showed evidence for linkage (LOD ≥ 0.5) in each of the four significant regions (Table 5, Supplementary Figure 1). While there was some overlap, the set of sub-pedigrees linked to each region varied, and two sub-pedigrees did not link to any of the regions (sub-pedigrees one and six). This may be the result of the relatively small size (and hence power) of the subpedigrees, or a greater level of heterogeneity for PD loci in the Amish than previously suspected. Drawings of sub-pedigrees 4 and 9 with possible haplotypes at the peak HLOD scores on chromosomes 5 and 6, respectively, are shown in supplementary data (Supplementary Figures 2, 3).
Table 5.
Peak LOD scores by sub-pedigree for the most significant multipoint linkage regions
| Chromosome | Sub-ped 1 |
Sub-ped 2 |
Sub-ped 3 |
Sub-ped 4 |
Sub-ped 5 |
Sub-ped 6 |
Sub-ped 7 |
Sub-ped 8 |
Sub-ped 9 |
Sub-ped 10 |
|---|---|---|---|---|---|---|---|---|---|---|
| 5 | 0.2904 | −1.4653 | 0.0437 | 2.1626 | 1.5572 | −0.9193 | −0.1998 | 0 | 1.1993 | 0.9418 |
| 6 | 0.2862 | 0.6731 | −0.2512 | −0.9409 | −0.1427 | −0.9279 | 1.078 | 0.8798 | 3.0017 | 0.9414 |
| 10 | 0.2904 | −0.0658 | 1.45 | 2.1581 | −0.143 | 0.4807 | 1.1975 | 0 | 1.3069 | −0.4649 |
Sub-ped: sub-pedigrees
Conclusions
Simulation studies suggest the HLOD scores we have observed on chromosomes 5, 6, and 10 are unlikely to be false positives. In each case, there is additional evidence that a true locus could exist in that region. Although we were somewhat surprised to find significant evidence of linkage to multiple loci, we are encouraged that the strong linkage signals represent true loci. We suggest that this population is a prime candidate for follow-up in these regions due to the homogenous environment and a more homogenous genetic background than the general population. These benefits and the large size of the family studied may allow us to more easily detect and decode a true effect in these regions. We suggest an in-depth study of the Amish population particularly to investigate the importance of chromosome 5q31 as a risk locus, as this is a more genetically and environmentally homogenous study population and may lead to more conclusive decisions in this region.
Supplementary Material
Sub-pedigree 4 is shown with possible haplotype combinations for the peak region of co-inheritance on chromosome 5. The six SNPs in this haplotype are at the location of the maximum HLOD score. Genders have been randomized to protect privacy.
Sub-pedigree 9 is shown with possible haplotype combinations for the peak region of co-inheritance on chromosome 6. The four SNPs in this haplotype are at the location of the maximum HLOD score. Genders have been randomized to protect privacy.
Acknowledgements
We would like to thank the participants of this study who have so graciously allowed us to visit with them and have participated in studies with us for over 10 years. We would like to acknowledge additional work for this study that was performed using the Vanderbilt Center for Human Genetics Research Core facilities: the Genetic Studies Ascertainment Core, the DNA Resources Core, and the Computation Genomics Core. This study was supported by the National Institutes of Health grants AG019085 (to JLH and MAP-V) and AG019726 (to WKS), and a grant from the Michael J. Fox Foundation (to JLH). Some of the samples used in this study were collected while WKS, JRG, and MAP-V were faculty members at Duke University. The authors would like to thank L. L. McFarland, C. Knebusch, and the late C. E. Jackson for their contributions to the overall Amish projects.
Reference List
- International Parkinson's Disease Genomics Consortium (IPDGC) & Wellcome Trust Case Control Consortium 2 (WTCCC2) A two-stage meta-analysis identifies several new loci for Parkinson's disease. PLoS Genet. 2011;7:e1002142. doi: 10.1371/journal.pgen.1002142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abecasis GR, Cherny SS, Cookson WO, Cardon LR. Merlin--rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet. 2002;30:97–101. doi: 10.1038/ng786. [DOI] [PubMed] [Google Scholar]
- Agarwala R, Biesecker LG, Schaffer AA. Anabaptist genealogy database. Am J Med Genet C Semin Med Genet. 2003;121:32–37. doi: 10.1002/ajmg.c.20004. [DOI] [PubMed] [Google Scholar]
- Bem D, Yoshimura S, Nunes-Bastos R, Bond FC, Kurian MA, Rahman F, Handley MT, Hadzhiev Y, Masood I, Straatman-Iwanowska AA, Cullinane AR, McNeill A, Pasha SS, Kirby GA, Foster K, Ahmed Z, Morton JE, Williams D, Graham JM, Dobyns WB, Burglen L, Ainsworth JR, Gissen P, Müller F, Maher ER, Barr FA, Aligianis IA. Loss-of-function mutations in RAB18 cause Warburg micro syndrome. Am J Hum Genet. 2011;88:499–507. doi: 10.1016/j.ajhg.2011.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyles AL, Scott WK, Martin ER, Schmidt S, Li YJ, Ashley-Koch A, Bass MP, Schmidt M, Pericak-Vance MA, Speer MC, Hauser ER. Linkage disequilibrium inflates type I error rates in multipoint linkage analysis when parental genotypes are missing. Hum Hered. 2005;59:220–227. doi: 10.1159/000087122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummings AC, Jiang L, Velez Edwards DR, McCauley JL, Laux R, McFarland LL, Fuzzell D, Knebusch C, Caywood L, Reinhart-Mercer L, Nations L, Gilbert JR, Konidari I, Tramontana M, Cuccaro ML, Scott WK, Pericak-Vance MA, Haines JL. Genome-wide association and linkage study in the amish detects a novel candidate late-onset Alzheimer disease gene. Ann Hum Genet. 2012;76:342–351. doi: 10.1111/j.1469-1809.2012.00721.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummings AC, Lee SL, McCauley JL, Jiang L, Crunk A, McFarland LL, Gallins PJ, Fuzzell D, Knebusch C, Jackson CE, Scott WK, Pericak-Vance MA, Haines JL. A genome-wide linkage screen in the Amish with Parkinson disease points to chromosome 6. Ann Hum Genet. 2011;75:351–358. doi: 10.1111/j.1469-1809.2011.00643.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahn S, Elton R. members of the UPDRS Development Committee. Recent Developments in Parkinson's Disease. In: Fahn S, Marsden CD, Jenner P, Teychenne P, editors. Recent Developments in Parkinson's Disease. New York: Raven Press; 1987. pp. 153–163. [Google Scholar]
- Foltynie T, Hicks A, Sawcer S, Jonasdottir A, Setakis E, Maranian M, Yeo T, Lewis S, Brayne C, Stefansson K, Compston A, Gulcher J, Barker RA. A genome wide linkage disequilibrium screen in Parkinson's disease. J Neurol. 2005;252:597–602. doi: 10.1007/s00415-005-0686-2. [DOI] [PubMed] [Google Scholar]
- Foroud T, Pankratz N, Martinez M. Chromosome 5 and Parkinson disease. Eur J Hum Genet. 2006;14:1106–1110. doi: 10.1038/sj.ejhg.5201666. [DOI] [PubMed] [Google Scholar]
- Gelb DJ, Oliver E, Gilman S. Diagnostic criteria for Parkinson disease. Arch Neurol. 1999;56:33–39. doi: 10.1001/archneur.56.1.33. [DOI] [PubMed] [Google Scholar]
- Hamza TH, Chen H, Hill-Burns EM, Rhodes SL, Montimurro J, Kay DM, Tenesa A, Kusel VI, Sheehan P, Eaaswarkhanth M, Yearout D, Samii A, Roberts JW, Agarwal P, Bordelon Y, Park Y, Wang L, Gao J, Vance JM, Kendler KS, Bacanu SA, Scott WK, Ritz B, Nutt J, Factor SA, Zabetian CP, Payami H. Genome-wide gene-environment study identifies glutamate receptor gene GRIN2A as a Parkinson's disease modifier gene via interaction with coffee. PLoS Genet. 2011;7:e1002237. doi: 10.1371/journal.pgen.1002237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock DB, Martin ER, Mayhew GM, Stajich JM, Jewett R, Stacy MA, Scott BL, Vance JM, Scott WK. Pesticide exposure and risk of Parkinson's disease: a family-based case-control study. BMC Neurol. 2008;8:6. doi: 10.1186/1471-2377-8-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock DB, Martin ER, Stajich JM, Jewett R, Stacy MA, Scott BL, Vance JM, Scott WK. Smoking, caffeine, and nonsteroidal anti-inflammatory drugs in families with Parkinson disease. Arch Neurol. 2007;64:576–580. doi: 10.1001/archneur.64.4.576. [DOI] [PubMed] [Google Scholar]
- Hicks AA, Petursson H, Jonsson T, Stefansson H, Johannsdottir HS, Sainz J, Frigge ML, Kong A, Gulcher JR, Stefansson K, Sveinbjornsdottir S. A susceptibility gene for late-onset idiopathic Parkinson's disease. Ann Neurol. 2002;52:549–555. doi: 10.1002/ana.10324. [DOI] [PubMed] [Google Scholar]
- Hoehn MM, Yahr MD. Parkinsonism: onset, progression, and mortality. Neurology. 1998;50:318. doi: 10.1212/wnl.50.2.318. 1967. [DOI] [PubMed] [Google Scholar]
- International Parkinson Disease Genomics Consortium. Nalls MA, Plagnol V, Hernandez DG, Sharma M, Sheerin UM, Saad M, Simon-Sanchez J, Schulte C, Lesage S, Sveinbjornsdottir S, Stefansson K, Martinez M, Hardy J, Heutink P, Brice A, Gasser T, Singleton AB, Wood NW. Imputation of sequence variants for identification of genetic risks for Parkinson's disease: a meta-analysis of genome-wide association studies. Lancet. 2011;377:641–649. doi: 10.1016/S0140-6736(10)62345-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katzman R, Brown T, Fuld P, Peck A, Schechter R, Schimmel H. Validation of a short Orientation-Memory-Concentration Test of cognitive impairment. Am J Psychiatry. 1983;140:734–739. doi: 10.1176/ajp.140.6.734. [DOI] [PubMed] [Google Scholar]
- Klein C, Westenberger A. Genetics of Parkinson's disease. Cold Spring Harb Perspect Med. 2012;2:a008888. doi: 10.1101/cshperspect.a008888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krygowska-Wajs A, Kachergus JM, Hulihan MM, Farrer MJ, Searcy JA, Booij J, Berendse HW, Wolters ECh, Wszolek ZK. Clinical and genetic evaluation of 8 Polish families with levodopa-responsive parkinsonism. J Neural Transm. 2005;112:1487–1502. doi: 10.1007/s00702-005-0290-8. [DOI] [PubMed] [Google Scholar]
- Lai BC, Marion SA, Teschke K, Tsui JK. Occupational and environmental risk factors for Parkinson's disease. Parkinsonism Relat Disord. 2002;8:297–309. doi: 10.1016/s1353-8020(01)00054-2. [DOI] [PubMed] [Google Scholar]
- Lee SL, Murdock DG, McCauley JL, Bradford Y, Crunk A, McFarland L, Jiang L, Wang T, Schnetz-Boutaud N, Haines JL. A genome-wide scan in an Amish pedigree with parkinsonism. Ann Hum Genet. 2008;72:621–629. doi: 10.1111/j.1469-1809.2008.00452.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YJ, Scott WK, Hedges DJ, Zhang F, Gaskell PC, Nance MA, Watts RL, Hubble JP, Koller WC, Pahwa R, Stern MB, Hiner BC, Jankovic J, Allen FA, Jr, Goetz CG, Mastaglia F, Stajich JM, Gibson RA, Middleton LT, Saunders AM, Scott BL, Small GW, Nicodemus KK, Reed AD, Schmechel DE, Welsh-Bohmer KA, Conneally PM, Roses AD, Gilbert JR, Vance JM, Haines JL, Pericak-Vance MA. Age-of-onset in two common neurodegenerative diseases is genetically controlled. Am J Hum Genet. 2002;70:985–993. doi: 10.1086/339815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lill CM, Roehr JT, McQueen MB, Kavvoura FK, Bagade S, Schjeide BM, Schjeide LM, Meissner E, Zauft U, Allen NC, Liu T, Schilling M, Anderson KJ, Beecham G, Berg D, Biernacka JM, Brice A, DeStefano AL, Do CB, Eriksson N, Factor SA, Farrer MJ, Foroud T, Gasser T, Hamza T, Hardy JA, Heutink P, Hill-Burns EM, Klein C, Latourelle JC, Maraganore DM, Martin ER, Martinez M, Myers RH, Nalls MA, Pankratz N, Payami H, Satake W, Scott WK, Sharma M, Singleton AB, Stefansson K, Toda T, Tung JY, Vance J, Wood NW, Zabetian CP, 23andMe Genetic Epidemiology of Parkinson's Disease Consortium; International Parkinson's Disease Genomics Consortium; Parkinson's Disease GWAS Consortium; Wellcome Trust Case Control Consortium 2, Young P, Tanzi RE, Khoury MJ, Zipp F, Lehrach H, Ioannidis JP, Bertram L. Comprehensive research synopsis and systematic meta-analyses in Parkinson's disease genetics: The PDGene database. PLoS Genet. 2012;8:e1002548. doi: 10.1371/journal.pgen.1002548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litvan I, Agid Y, Calne D. Clinical research criteria for the diagnosis of progressive supranuclear gaze palsy. Neurology. 1996;47:1–9. doi: 10.1212/wnl.47.1.1. [DOI] [PubMed] [Google Scholar]
- Liu F, Kirichenko A, Axenovich TI, van Duijn CM, Aulchenko YS. An approach for cutting large and complex pedigrees for linkage analysis. Eur J Hum Genet. 2008;16:854–860. doi: 10.1038/ejhg.2008.24. [DOI] [PubMed] [Google Scholar]
- Maraganore DM, de Andrade M, Lesnick TG, Strain KJ, Farrer MJ, Rocca WA, Pant PV, Frazer KA, Cox DR, Ballinger DG. High-resolution whole-genome association study of Parkinson disease. Am J Hum Genet. 2005;77:685–693. doi: 10.1086/496902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maraganore DM, Farrer MJ, Lesnick TG, de Andrade M, Bower JH, Hernandez D, Hardy JA, Rocca WA. Case-control study of the alpha-synuclein interacting protein gene and Parkinson's disease. Mov Disord. 2003;18:1233–1239. doi: 10.1002/mds.10547. [DOI] [PubMed] [Google Scholar]
- Marras C, Goldman SM. Genetics meets environment: evaluating gene-environment interactions in neurologic diseases. Semin Neurol. 2011;31:553–561. doi: 10.1055/s-0031-1299793. [DOI] [PubMed] [Google Scholar]
- Martinez M, Brice A, Vaughan JR, Zimprich A, Breteler MM, Meco G, Filla A, Farrer MJ, Bétard C, Hardy J, De Michele G, Bonifati V, Oostra B, Gasser T, Wood NW, Dürr A French Parkinson's Disease Genetics Study Group; European Consortium on Genetic Susceptibility in Parkinson's Disease. Genome-wide scan linkage analysis for Parkinson's disease: the European genetic study of Parkinson's disease. J Med Genet. 2004;41:900–907. doi: 10.1136/jmg.2004.022632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marx FP, Holzmann C, Strauss KM, Li L, Eberhardt O, Gerhardt E, Cookson MR, Hernandez D, Farrer MJ, Kachergus J, Engelender S, Ross CA, Berger K, Schols L, Schulz JB, Riess O, Kruger R. Identification and functional characterization of a novel R621C mutation in the synphilin-1 gene in Parkinson's disease. Hum Mol Genet. 2003;12:1223–1231. doi: 10.1093/hmg/ddg134. [DOI] [PubMed] [Google Scholar]
- Myhre R, Klungland H, Farrer MJ, Aasly JO. Genetic association study of synphilin-1 in idiopathic Parkinson's disease. BMC Med Genet. 2008;9:19. doi: 10.1186/1471-2350-9-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pankratz N, Nichols WC, Uniacke SK, Halter C, Rudolph A, Shults C, Conneally PM, Foroud T Parkinson Study Group. Genome screen to identify susceptibility genes for Parkinson disease in a sample without parkin mutations. Am J Hum Genet. 2002;71:124–135. doi: 10.1086/341282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polymeropoulos MH, Lavedan C, Leroy E, Ide SE, Dehejia A, Dutra A, Pike B, Root H, Rubenstein J, Boyer R, Stenroos ES, Chandrasekharappa S, Athanassiadou A, Papapetropoulos T, Johnson WG, Lazzarini AM, Duvoisin RC, Di Iorio G, Golbe LI, Nussbaum RL. Mutation in the alpha-synuclein gene identifed in families with Parkinson's disease. Science. 1997;276:2045–2047. doi: 10.1126/science.276.5321.2045. [DOI] [PubMed] [Google Scholar]
- Rosenberger A, Sharma M, Muller-Myhsok B, Gasser T, Bickeboller H. Meta analysis of whole-genome linkage scans with data uncertainty: an application to Parkinson's disease. BMC Genet. 2007;8:44. doi: 10.1186/1471-2156-8-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott WK, Nance MA, Watts RL, Hubble JP, Koller WC, Lyons K, Pahwa R, Stern MB, Colcher A, Hiner BC, Jankovic J, Ondo WG, Allen FH, Jr, Goetz CG, Small GW, Masterman D, Mastaglia F, Laing NG, Stajich JM, Slotterbeck B, Booze MW, Ribble RC, Rampersaud E, West SG, Gibson RA, Middleton LT, Roses AD, Haines JL, Scott BL, Vance JM, Pericak-Vance MA. Complete genomic screen in Parkinson disease: evidence for multiple genes. JAMA. 2001;286:2239–2244. doi: 10.1001/jama.286.18.2239. [DOI] [PubMed] [Google Scholar]
- Thiele H, Nurnberg P. HaploPainter: a tool for drawing pedigrees with complex haplotypes. Bioinformatics. 2005;21:1730–1732. doi: 10.1093/bioinformatics/bth488. [DOI] [PubMed] [Google Scholar]
- Thornton T, McPeek MS. Case-control association testing with related individuals: a more powerful quasi-likelihood score test. Am J Hum Genet. 2007;81:321–337. doi: 10.1086/519497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Den Eeden SK, Tanner CM, Bernstein AL, Fross RD, Leimpeter A, Bloch DA, Nelson LM. Incidence of Parkinson's disease: variation by age, gender, and race/ethnicity. Am J Epidemiol. 2003;157:1015–1022. doi: 10.1093/aje/kwg068. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Sub-pedigree 4 is shown with possible haplotype combinations for the peak region of co-inheritance on chromosome 5. The six SNPs in this haplotype are at the location of the maximum HLOD score. Genders have been randomized to protect privacy.
Sub-pedigree 9 is shown with possible haplotype combinations for the peak region of co-inheritance on chromosome 6. The four SNPs in this haplotype are at the location of the maximum HLOD score. Genders have been randomized to protect privacy.