Abstract
In plant effector-triggered immunity (ETI), intracellular nucleotide binding-leucine rich repeat (NLR) receptors are activated by specific pathogen effectors. The Arabidopsis TIR (Toll-Interleukin-1 receptor domain)-NLR (denoted TNL) gene pair, RPS4 and RRS1, confers resistance to Pseudomonas syringae pv tomato (Pst) strain DC3000 expressing the Type III-secreted effector, AvrRps4. Nuclear accumulation of AvrRps4, RPS4, and the TNL resistance regulator EDS1 is necessary for ETI. RRS1 possesses a C-terminal “WRKY” transcription factor DNA binding domain suggesting that important RPS4/RRS1 recognition and/or resistance signaling events occur at the nuclear chromatin. In Arabidopsis accession Ws-0, the RPS4Ws/RRS1Ws allelic pair governs resistance to Pst/AvrRps4 accompanied by host programed cell death (pcd). In accession Col-0, RPS4Col/RRS1Col effectively limits Pst/AvrRps4 growth without pcd. Constitutive expression of HA-StrepII tagged RPS4Col (in a 35S:RPS4-HS line) confers temperature-conditioned EDS1-dependent auto-immunity. Here we show that a high (28°C, non-permissive) to moderate (19°C, permissive) temperature shift of 35S:RPS4-HS plants can be used to follow defense-related transcriptional dynamics without a pathogen effector trigger. By comparing responses of 35S:RPS4-HS with 35S:RPS4-HS rrs1-11 and 35S:RPS4-HS eds1-2 mutants, we establish that RPS4Col auto-immunity depends entirely on EDS1 and partially on RRS1Col. Examination of gene expression microarray data over 24 h after temperature shift reveals a mainly quantitative RRS1Col contribution to up- or down-regulation of a small subset of RPS4Col-reprogramed, EDS1-dependent genes. We find significant over-representation of WRKY transcription factor binding W-box cis-elements within the promoters of these genes. Our data show that RRS1Col contributes to temperature-conditioned RPS4Col auto-immunity and are consistent with activated RPS4Col engaging RRS1Col for resistance signaling.
Keywords: resistance gene pair, temperature shift, EDS1 signaling, biotic stress, programed cell death, transcriptional reprograming
INTRODUCTION
A critical layer of plant innate immunity is conferred by intracellular nucleotide binding-leucine rich repeat (NLR) receptors that guard against disease-promoting activities of pathogen effectors during infection (Dodds and Rathjen, 2010). Genes encoding NLR proteins represent the most diverse gene family in plants, probably as a result of pathogen selection pressure (Meyers et al., 2003; Yue et al., 2012). NLR receptors behave as ATP-driven molecular switches which become activated directly by physical association with an effector or indirectly through effector perturbations of a receptor-guarded co-factor (Maekawa et al., 2011; Bernoux et al., 2011a). Receptor activation triggers a robust anti-microbial response which is often accompanied by localized host programed cell death (pcd), although pathogen resistance can be uncoupled from pcd (Maekawa et al., 2011; Heidrich et al., 2012).
The NLR receptor family is broadly divided into two sub-classes based on different N-terminal putative signaling domains containing either Toll-Interleukin-1 receptor (TIR) homology, or a coiled-coil (CC) or other features, referred to, respectively, as TNLs and CNLs (Maekawa et al., 2011; Bernoux et al., 2011a). TNL and CNL receptor types signal in different ways for resistance (Wiermer et al., 2005; Venugopal et al., 2009). However, they all converge on the transcriptional machinery to amplify gene expression programs which operate in basal resistance against virulent (non-recognized) pathogens (Tao et al., 2003; Bartsch et al., 2006). Only a handful of TNL and CNL receptors have been characterized and many questions remain about where and how NLR are activated inside cells and the sequence of downstream signaling events leading to disease resistance. A number of functional NLR representatives from both sub-classes are nucleo-cytoplasmic and there is compelling evidence that NLR nucleo-cytoplasmic partitioning is important for full triggering of an immune response (Heidrich et al., 2012). Moreover, the Arabidopsis TNL protein SNC1 (Zhu et al., 2010b), tobacco TNL receptor N (Padmanabhan et al., 2013) and barley CNL receptor MLA1 (Chang et al., 2013) interact with transcription factors, suggesting a short route to the transcriptional machinery.
All functionally characterized TNL receptors depend on the nucleo-cytoplasmic immune regulator EDS1 (enhanced disease sensitivity1) for triggering resistance and pcd (Wiermer et al., 2005) and associations between several TNLs and EDS1 have been detected in Arabidopsis and tobacco, suggesting that EDS1 is part of an immune receptor signaling complex (Bhattacharjee et al., 2011; Heidrich et al., 2011; Kim et al., 2012). EDS1, in direct association with its signaling partner PAD4 (phytoalexin deficient4), is essential for basal resistance against virulent pathogens, measured as a slowing of pathogen growth without obvious TNL recognition or pcd (Jirage et al., 1999; Feys et al., 2001; Rietz et al., 2011). Based on interactions detected between EDS1 and Pseudomonas syringae Type III-secreted effectors AvrRps4 and HopA1, it was proposed that TNL receptors might guard the EDS1–PAD4 basal resistance machinery against interference by pathogen effectors as well as co-opting EDS1 as an early signaling component for execution of effector-triggered immunity (ETI; Bhattacharjee et al., 2011; Heidrich et al., 2011).
We are studying ETI in Arabidopsis mediated by the TNL receptor gene pair, RPS4 and RRS1, in recognition of AvrRps4 derived from leaf-infecting P. syringae pv pisi (Hinsch and Staskawicz, 1996; Gassmann et al., 1999; Birker et al., 2009; Narusaka et al., 2009). Particular allelic forms of the same RPS4 RRS1 pair also recognize an unrelated YopJ family effector, PopP2, secreted by root-infecting Ralstonia solanacearum bacteria (Deslandes et al., 2003; Narusaka et al., 2009). RPS4 accumulates as a nucleo-cytoplasmic protein associating with endo-membranes (Wirthmueller et al., 2007; Bhattacharjee et al., 2011). Notably, RPS4 nuclear accumulation conferred by a C-terminal NLS is essential for resistance to P. syringae pv tomato (Pst) expressing AvrRps4 (Pst/AvrRps4), although RPS4 nucleo-cytoplasmic partitioning does not rely on the presence of either AvrRps4 or EDS1 (Wirthmueller et al., 2007; Heidrich et al., 2011). RRS1 is an atypical TNL in that it also possesses a C-terminal “WRKY” transcription factor DNA binding domain (Deslandes et al., 2002) known to recognize W-box consensus sequences within the promoters of defense-related genes (Rushton et al., 2010; Chen et al., 2013; Logemann et al., 2013). Analysis of the auto-immune phenotype of an rrs1 (slh1) single amino acid insertion mutation in the WRKY domain abolishing DNA binding in vitro, led to the idea that RRS1 exists as an auto-inhibited form at the chromatin in healthy tissues (Noutoshi et al., 2005). An effector trigger might then cause an RRS1 conformational switch to initiate resistance signaling. Other studies established that RRS1 interacts with R. solanacearum effector PopP2 (Deslandes et al., 2003; Tasset et al., 2010). PopP2 has an auto-acetyltransferase activity and this enzymatic function, coupled with recognition by a resistant RRS1-R allelic form, appear to be necessary for triggering resistance (Tasset et al., 2010). By contrast, AvrRps4 has no known enzyme activity but is proteolytically cleaved inside plant cells to produce an 11 kDa α-helical CC C-terminal fragment which is essential for RPS4/RRS1 recognition (Sohn et al., 2009,2012). While association between AvrRPS4 and EDS1 was reported based on fluorescence resonance energy transfer–fluorescence life-time imaging (FRET–FLIM) and co-immunoprecipitation assays in tobacco and Arabidopsis (Bhattacharjee et al., 2011; Heidrich et al., 2011), another study argued against AvrRps4–EDS1 association based on negative interaction data (Sohn et al., 2012). Clearly, much needs to be resolved about the configurations of receptor pre-activation and signaling complexes and their precise relationship with the transcriptional machinery.
Resistance conditioned by TNL receptors is acutely sensitive to temperature with higher temperatures suppressing activated immune responses (Yang and Hua, 2004; Wang et al., 2009; Kim et al., 2010; Zhu et al., 2010a; Alcazar and Parker, 2011). Previously, we described an HA-StrepII epitope tagged RPS4 over-expression line (35S:RPS4-HS) in Arabidopsis accession Columbia (Col-0) which displays EDS1-dependent auto-immunity and stunting at 22°C, consistent with EDS1 being recruited coincidently or immediately downstream of activated RPS4 (Wirthmueller et al., 2007; Heidrich et al., 2011). Here we establish that auto-immunity in the 35S:RPS4-HS plants grown at 22°C or shifted from a suppressive (28°C) to permissive (19°C) temperature depends fully on EDS1 and partially on RRS1Col. We have used the 28–19°C temperature shift to induce RPS4Col immunity and examine transcriptional reprograming in leaf tissues. This reveals a mainly quantitative contribution of RRS1Col to up- and down-regulation of a discrete set of EDS1-dependent genes. The data suggest that RRS1 acts positively and at an early stage of RPS4 auto-immunity.
MATERIALS AND METHODS
PLANT MATERIALS AND GROWTH CONDITIONS
All mutant and transgenic lines used were in Arabidopsis accessions Columbia (Col-0) or Wassilewskija (Ws-0). Col eds1-2 (Bartsch et al., 2006), rps4-2 (Wirthmueller et al., 2007), rrs1-11 (Birker et al., 2009), Ws eds1-1 (Parker et al., 1996), rps4-21, rrs1-1, and rps4-21/rrs1-1 (Narusaka et al., 2009) mutant lines, 35S:RPS4-HS and 35S:RPS4-HS eds1-2 (Wirthmueller et al., 2007) have been described. The 35S:RPS4-HS rrs1-11 line was generated by crossing 35S:RPS4-HS with rrs1-11. Plants were grown in soil in chambers under a 10/14 h day/night cycle (150–200 μE/m2s) and ∼65% relative humidity at 19, 22, or 28°C.
BACTERIAL STRAINS
Bacterial strains Pst strain DC3000 and Pst DC3000 expressing AvrRps4 (Pst/AvrRps4) were obtained from R. Innes (Indiana University, Bloomington, USA) and grown as described (Hinsch and Staskawicz, 1996). Pst strain DC3000 expressing AvrRps4-HA or the AvrRps4-HA-NLS and AvrRps4-HA-NES variants from a pEDV6 vector, or a non-pathogenic Pseudomonas fluorescens (Pfo) strain for delivery of Type III-secreted effectors (Thomas et al., 2009) expressing AvrRps4-HA in pEDV6, have been described (Heidrich et al., 2011).
BACTERIAL GROWTH ASSAYS
For Pst spray infections, bacteria were adjusted to 1 × 108 cfu/ml in 10 mM MgCl2 containing 0.04 % (v/v) Silwet L-77 (Lehle seeds, USA). In planta bacterial titers were determined 3 h after spray-infection (day 0) and 3 days post-infection (dpi) by shaking leaf disks in 10 mM MgCl2 with 0.01% Silwet L-77 at 28°C for 1 h, as described (Tornero and Dangl, 2001; Garcia et al., 2010). Infected plants were kept in a growth cabinet with a 10/14 h day/night cycle at 23°C. Mean values and standard errors (SEs) were calculated from at least three biological replicates per experiment. In the bacterial growth assays shown in Figure 1A, raw data was log10 transformed and all replicate values from three independent experiments analyzed using a linear model.
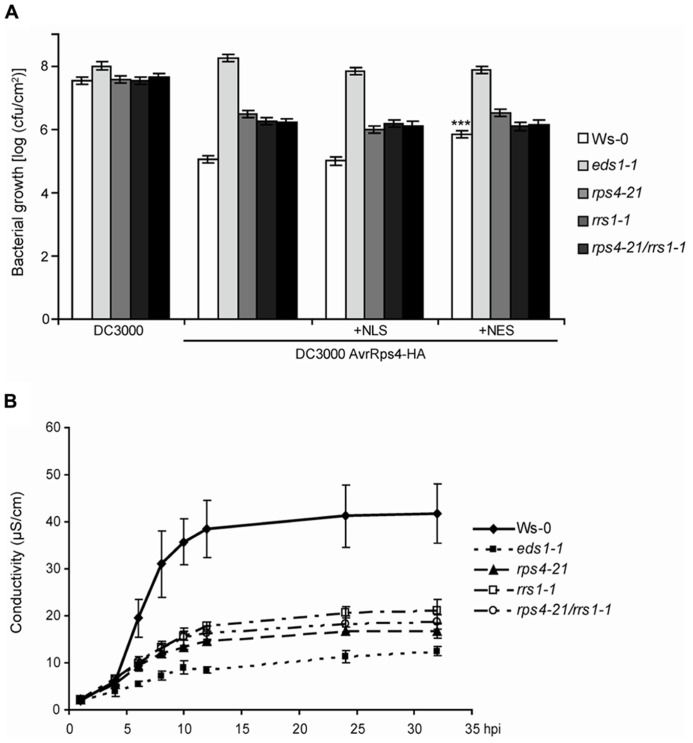
FIGURE 1.
RPS4Ws and RRS1Ws act cooperatively in AvrRps4-triggered bacterial resistance and pcd. (A) Four-week-old plants were spray-inoculated with virulent Pst DC3000 or Pst expressing AvrRps4-HA, AvrRps4-HA-NLS, or AvrRps4-HA-NES variants. Bacterial titers at 3 dpi are shown. All bacterial strains had similar entry rates measured at 3 hpi (data not shown). Replicate values were combined from three independent experiments with similar results and SEs calculated using a linear model. ***Significant difference (p < 0.001). (B) Ion leakage measurements were recorded at the indicated time points in leaf disks of 4-week-old Ws-0, eds1-1, rps4-21, rrs1-1, and rps4-21 rrs1-1 plants after infiltration with Pfo-expressing AvrRps4-HA. Error bars represent standard errors of four samples per genotype. The experiment was performed three times with similar results.
ION LEAKAGE ASSAYS
For conductivity measurements after Pfo infiltration, leaves of 4-week-old-plants were infiltrated with 1.5 × 108 cfu/ml bacteria in 10 mM MgCl2. Leaf disks were collected using a cork borer (6 mm diameter), floated in water for 30 min, and three leaf disks per measurement were subsequently transferred to a microtiter plate containing 3 ml distilled water. Conductivity of the solution was determined with a Horiba Twin B-173 conductivity meter at the indicated time points. Mean values and SE were calculated from four replicate measurements per genotype or bacterial strain. Experiments were repeated at least three times.
PROTEIN IMMUNOBLOTTING
Total protein extracts from Arabidopsis leaves were prepared as previously described (Garcia et al., 2010). Protein concentrations were quantified and separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Proteins were electro-blotted onto nitrocellulose membranes. Equal protein transfer was monitored by staining membranes with Ponceau S (Sigma-Aldrich). Membranes were blocked in a 5%-milk Tris buffer saline-Tween (TBST 20) solution before incubation in a 2% milk-TBST solution containing primary α-HA antibody (3F10; Roche) overnight. The appropriate horseradish peroxidase-conjugated secondary antibody (Santa Cruz Biotechnology) was applied and proteins were detected using enhanced chemiluminiscence reagent (ECL; Pierce Thermo Scientific).
RT-PCR ANALYSIS OF DEFENSE GENE EXPRESSION
Total RNA was extracted from leaf material of 3-week-old plants using TriReagent (Sigma-Aldrich) according to the manufacturer’s protocol. A 1.5 μg of total RNA was incubated with 10 units of RNAse-free DNAse I (Roche) at 37°C for 30 min followed by heat-inactivation of the enzyme at 75°C for 10 min. Reverse transcription was performed with SuperScript II enzyme (Invitrogen) according to the manufacturer’s protocol. The following primer combinations were used for semi-quantitative real-time polymerase chain reaction (RT-PCR). Actin: fw GGCGATGAAGCTCAATCCAAACG, Actin: rev GGTCACGACCAGCAAGATCAAGACG; EDS1: fw TCATACGCAATCCAAATGTTTAC, EDS1: rev AAAAACCTCTCTTGCTCGATCAC; PBS3: fw CAACTTGTTAGAGGAGATCATCACACCC, PBS3: rev CCAGAAGGAGTCATGGATTCTTGTTTA; At5g26920: fw CGGAACAGCCCTAGTTTTCATGGG, At5g26920: rev GAGAAGACGAGAACGGTCCCGTACT; At5g27420: fw CTACTATTATCCGTGTCGGC, At5g27420: rev CGCGTCTAACCCACG.
GENE EXPRESSION MICROARRAY ANALYSIS
Total RNA was prepared from 3.5-week-old plants grown at 28°C and shifted to 19°C for 0, 2, 8, and 24 h, using a QIAGEN Plant RNeasy kit. RNA quality was assessed on a Bioanalyzer (Agilent). Biotinylated cRNA was prepared and hybridized on Affymetrix ATH1-121501 “GeneChip” arrays, as described (Hajheidari et al., 2012). Briefly, biotinylated cRNA was made from 1 μg total RNA using the MessageAmp II-Biotin Enhanced Kit (Ambion). After amplification and fragmentation, 12.5 μg of cRNA were hybridized for 16 h at 45°C. Arrays were subsequently washed and stained in the Affymetrix Fluidics Station 450 using Fluidics Script FS450-004, and scanned with a GeneChip Scanner 3000 7G. For each condition, three Affymetrix ATH1 microarrays were hybridized with independent biological samples. Raw data for gene expression signals was extracted using the Affymetrix GeneChip Operating Software (version 1.4). For further data collection and assessment, R language version 2.15 (bioconductor project) was used. Probe signal values were subjected to GeneChip-robust multiarray average algorithm (GC-RMA; Wu and Irizarry, 2004). Probes which were below the background signal in all samples were not considered for further analysis. The results were analyzed by the following linear model using the lmFit function in the limma package in the R environment: Sgyr = GYgyt + Rr + εgyr, where S is log2 expression value, GY, genotype:time interaction, and random factors; R is biological replicate; ε, residual. The eBayes function in the limma package was used for variance shrinkage in calculating the p-values and the Storey’s q-values were calculated using the q-value function in the q-value package from the p-values (Storey and Tibshirani, 2003). Genes whose expression changes were RRS1-dependent upon temperature shift at any time point (q-values < 0.01 and >2-fold change) were selected (250 genes) for the clustering analysis. Heatmaps were generated by CLUSTER using uncentered Pearson correlation and complete linkage and were visualized by TREEVIEW (Eisen et al., 1998). Promoter sequences of the 250 RRS1-dependent genes were retrieved from the TAIR website1 with fixed 1000 bp sequences upstream of the translational start site. Over representation of the core W-box (TTGACY) was assessed using the promoter bootstrapping (POBO) application2 (Kankainen and Holm, 2004). One thousand pseudo-clusters of 250 genes were generated from the RRS1-dependent genes (Cluster2), all induced/suppressed genes upon temperature shift in Col (q-values < 0.01 and >2-fold change; Cluster 3), and the Arabidopsis genomic background (background). Statistical significance of the t-values generated by POBO was calculated using the linked Graphpad application for a two-tailed comparison: *Comparison of Cluster 2 and background (p < 0.0001); *Comparison of Cluster 2 and Cluster 3 (p < 0.0001); *Comparison of Cluster 3 and background (p < 0.0001). Analysis of gene ontology (GO) terms for the 250 RRS1-dependent genes was performed using Agrico3. Microarray data have been submitted to the Gene Expression Omnibus database (GEO accession no. GSE50019).
RESULTS
ANALYSIS OF RPS4WS AND RRS1WS COOPERATIVITY IN AVRRPS4-TRIGGERED RESISTANCE AND HR
In Arabidopsis accession Ws-0, resistance to Pst strain DC3000 expressing AvrRps4 (Pst/AvrRps4) after bacterial infiltration of leaves relies on genetic cooperation between RPS4Ws and RRS1Ws (Narusaka et al., 2009). We tested whether the RPS4Ws RRS1Ws dual resistance system also operates against spray-inoculated Pst/AvrRps4 which enter leaves through stomata. Suspensions of Pst/AvrRps4 were sprayed onto wild-type Ws-0, Ws eds1-1, the single Ws rps4-21 and rrs1-1 T-DNA insertion mutants or the rps4-21 rrs1-1 double-mutant (Narusaka et al., 2009), and bacterial growth measured in leaves. At 3 h post-inoculation, titers of all bacterial strains were similar (∼5 × 103 cfu/cm2). At 3 days post-inoculation (dpi), the rps4-21 rrs1-1 double-mutant line displayed the same level of intermediate resistance as each rps4-21 and rrs1-1 single mutant, lying between fully resistant Ws-0 and fully susceptible eds1-1 plants (Figure 1A). Therefore, RPS4Ws and RRS1Ws dual resistance to Pst/AvrRps4 also operates after bacterial infection through leaf stomata. Residual EDS1-dependent resistance in rps4-21 rrs1-1 to Pst/AvrRps4 infection (Figure 1A) is conferred by an RPS4- and RRS1-independent mechanism operating in Ws-0 and likely also in accession Col-0 expressing the respective RPS4Col and RRS1Col allelic variants (Birker et al., 2009; Sohn et al., 2012). We showed previously that resistance in Ws-0 and Col-0 to Pst/AvrRps4 could be effectively triggered by an AvrRps4-HA-NLS form targeted to nuclei and that this also required RPS4Col nuclear accumulation (Heidrich et al., 2011). By contrast, enhanced nuclear export of AvrRps4-HA fused to a nuclear export sequence (AvrRps4-HA-NES) triggered low resistance but was able to trigger some pcd. Spray-inoculation of Pst-delivered AvrRps4-HA-NLS or AvrRps4-HA-NES variants (Heidrich et al., 2011) did not alter the partial resistance phenotype of the rps4-21 and rrs1-1 single or rps4-21 rrs1-1 double mutant lines (Figure 1A). Therefore, forced AvrRps4 localization to the nucleus or the cytoplasm does not alleviate the requirement for RPS4Ws or RRS1Ws in limiting bacterial infection or the extent of residual RPS4 and RRS1-independent resistance.
Delivery of AvrRps4 from a non-infectious Pfo strain infiltrated into Ws-0 leaves triggers a strong macroscopic hypersensitive response (HR) which is abolished in Ws eds1-1 mutant plants and reduced in rps4-21 or rrs1-1 mutants (Heidrich et al., 2011; Sohn et al., 2012). Resistance to Pst/AvrRps4 growth in Arabidopsis accession Col-0 is somewhat higher than in Ws-0 and depends on both the RPS4Col and RRS1Col allelic forms (Birker et al., 2009) but is accompanied by an extremely weak HR to Pfo/AvrRps4 (Heidrich et al., 2011; Sohn et al., 2012). Sohn et al. (2012) further showed that Col-0 transformed with a FLAG-tagged RRS1Ws transgene reconstituted a strong HR to infiltrated Pfo/AvrRps4, suggesting that RRS1Ws is a major determinant of AvrRps4-triggered pcd in Ws-0 or is able to boost the existing RPS4Col/RRS1Col low-level pcd response. We performed a quantitative ion leakage assay over 36 h in leaves of Ws-0, the rps4-21 and rrs1-1 single mutants, and rps4-21 rrs1-1 double mutants after leaf infiltration of Pfo/AvrRps4. Ws eds1-1 mutant leaves were infiltrated alongside as a non-responding control. As shown previously (Heidrich et al., 2011), Ws-0 leaves produced a rapid HR reaching a peak at 12–16 h after infiltration, whereas eds1-1 leaves produced base line conductivity of ∼10 μS/cm over the ion leakage time course (Figure 1B). Responses of the single and double rps4-21 rrs1-1 mutants were all intermediate between Ws-0 and eds1-1 (Figure 1B). Therefore, there is genetic cooperativity between RPS4Ws and RRS1Ws in eliciting host pcd and in partially restricting to Pst/AvrRps4 bacterial growth.
RRS1Col CONTRIBUTES TO AUTO-ACTIVATED RPS4Col PLANT STUNTING AND IMMUNITY
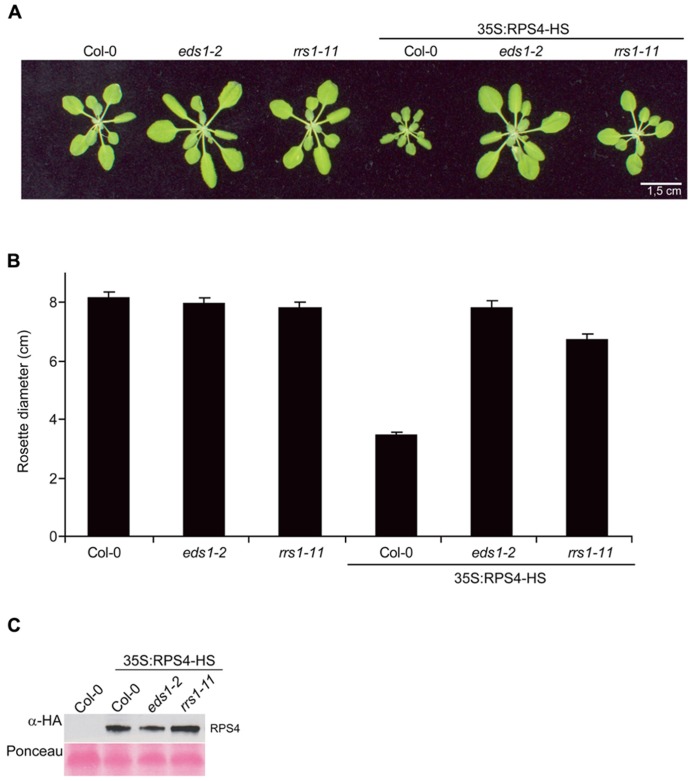
We reported that a Col-0 line constitutively expressing functional HA-StrepII-tagged genomic RPS4Col under control of the CaMV 35S promoter (referred to here as 35S:RPS4-HS) exhibits EDS1-dependent auto-immunity and stunting at 22°C (Wirthmueller et al., 2007; Heidrich et al., 2011). Given the tight functional relationship between the RPS4Ws and RRS1Ws allelic pairs in accession Ws-0, and presumably between RPS4Col and RRS1Col in Col-0 for resistance to Pst/AvrRps4, we investigated whether RRS1Col also has a role in 35S:RPS4-HS-triggered auto-immunity. A Col rrs1 null mutant allele (rrs1-11; Birker et al., 2009) was crossed into the 35S:RPS4-HS background and a line selected that was homozygous for the 35S:RPS4-HS transgene and rrs1-11. The same 35S:RPS4-HS line crossed into a Col eds1-2 null mutant was used as a control with suppressed RPS4 auto-immunity. As anticipated, 35S:RPS4-HS plants were severely stunted after 3–4 weeks growth and 35S:RPS4-HS eds1-2 plants exhibited no growth inhibition at 22°C (Figures 2A,B). Steady-state RPS4-HS protein accumulation in 35S:RPS4-HS eds1-2 was slightly reduced compared to the 35S:RPS4-HS line (Figure 2C). Mutation of RRS1Col caused intermediate 35S:RPS4-HS stunting at 22°C (Figures 2A,B) but did not affect RPS4-HS accumulation (Figure 2C). Therefore, RRS1Col contributes positively to RPS4Col auto-immunity at the level of plant growth inhibition. We concluded that the RRS1Col protein likely plays a role in resistance signaling triggered by an auto-activated RPS4Col receptor, besides its presumed role in AvrRps4 recognition (Birker et al., 2009; Narusaka et al., 2009).
FIGURE 2.
Mutation of RRS1Col partially suppresses 35S:RPS4-HS stunting. (A) Growth at 22°C of representative 3.5-week-old Col-0, eds1-2, and rrs1-11 and the same backgrounds containing the 35S:RPS4-HS transgene. Scale bar, 1.5 cm. (B) Quantification of rosette diameters at 3.5 weeks of lines shown in (A). (C) Immunoblot analysis of total leaf protein extracts separated by SDS-polyacrylamide gel electrophoresis (SDS-PAGE) from the 3.5-week-old 35S:RPS4-HS transgenic leaf tissues in Col-0, eds1-2, and rrs1-11 backgrounds, probed with α-HA antibody. Ponceau S staining shows equal transfer of protein samples to the membrane. Two independent experiments gave similar results.
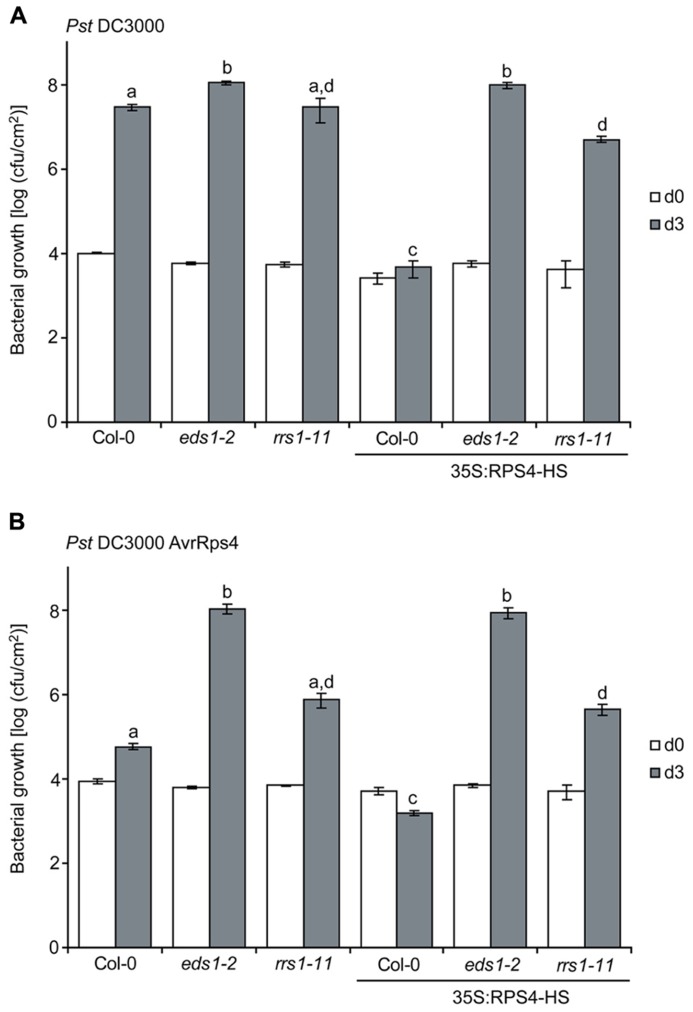
We then tested whether 35S:RPS4-HS plants grown at 22°C display enhanced basal resistance to virulent Pst strain DC3000 and the influence of rrs1-11 compared to eds1-2 on the 35S:RPS4-HS basal resistance phenotype. Col-0 wild-type, eds1-2, and rrs1-11 plants were grown alongside 35S:RPS4-HS, 35S:RPS4-HS eds1-2, and 35S:RPS4-HS rrs1-11 plants for 3.5 weeks at 22°C and then spray-inoculated with Pst DC3000 for bacterial growth assays. The rrs1-11 mutant supported similar Pst DC3000 growth as Col-0 wild type (Figure 3A) and therefore did not exhibit an enhanced disease susceptibility phenotype (which would be indicative of a loss of basal resistance), in contrast to eds1-2 (Figure 3A). The 35S:RPS4-HS plants exhibited strongly enhanced basal resistance to Pst DC3000 which was abolished by eds1-2 and partially suppressed by rrs1-11 (Figure 3A). We concluded that auto-immunity exhibited by 35S:RPS4-HS at 22°C involves RRS1Col for enhancing EDS1-dependent basal resistance responses.
FIGURE 3.
RRS1Col contributes to enhanced basal and AvrRps4-triggered resistance of 35S:RPS4-HS at 22°C. 3.5-week-old plants of the indicated lines grown at 22°C were spray-inoculated with virulent Pst DC3000 (A) or avirulent Pst/AvrRps4 (B) bacteria in the same experiment. Bacterial titers were measured at 3 hpi (d0) indicating bacterial entry rates and at 3 dpi (d3). Standard errors were calculated from three biological samples per genotype. Letters (a,b,c,d) indicate significant differences (p < 0.05) calculated by a Student’s t-test. Experiments were performed independently three times with similar results.
We spray-inoculated the same set of plants with Pst/AvrRps4 and found that the high basal resistance in 35S:RPS4-HS (see Figure 3A) was slightly increased by AvrRps4 and was also fully EDS1-dependent (Figure 3B). The 35S:RPS4-HS rrs1-11 plants displayed intermediate loss of resistance to Pst/AvrRps4 (Figure 3B), suggesting that an RPS4Col RRS1Col-independent mechanism also plays a role in 35S:RPS4-HS immunity to Pst/AvrRps4. The results show that RRS1Col contributes to RPS4Col auto-immunity. In genetically recruiting EDS1 and RRS1Col, while retaining an RRS1Col-independent resistance component (Figure 3B), we reasoned that the 35S:RPS4-HS auto-activated immune system might be useful for measuring RPS4/RRS1-triggered defense pathway transcription dynamics without needing to infect with the pathogen.
A HIGH TO LOW TEMPERATURE SHIFT INDUCES 35S:RPS4-HS AUTO-IMMUNITY
In Arabidopsis, suppression of basal and effector-triggered TNL immunity at high temperature (>25°C) is associated with lowered expression of defense pathway genes, including EDS1, and reduced feed-forward defense amplification (Yang and Hua, 2004; Wang et al., 2009). We therefore investigated whether shifting plants from high temperature (28°C, non-permissive for Arabidopsis TNL resistance) to a lower temperature (19–22°C, permissive for TNL resistance) could be used to turn on RPS4 auto-immunity synchronously in leaf tissues.
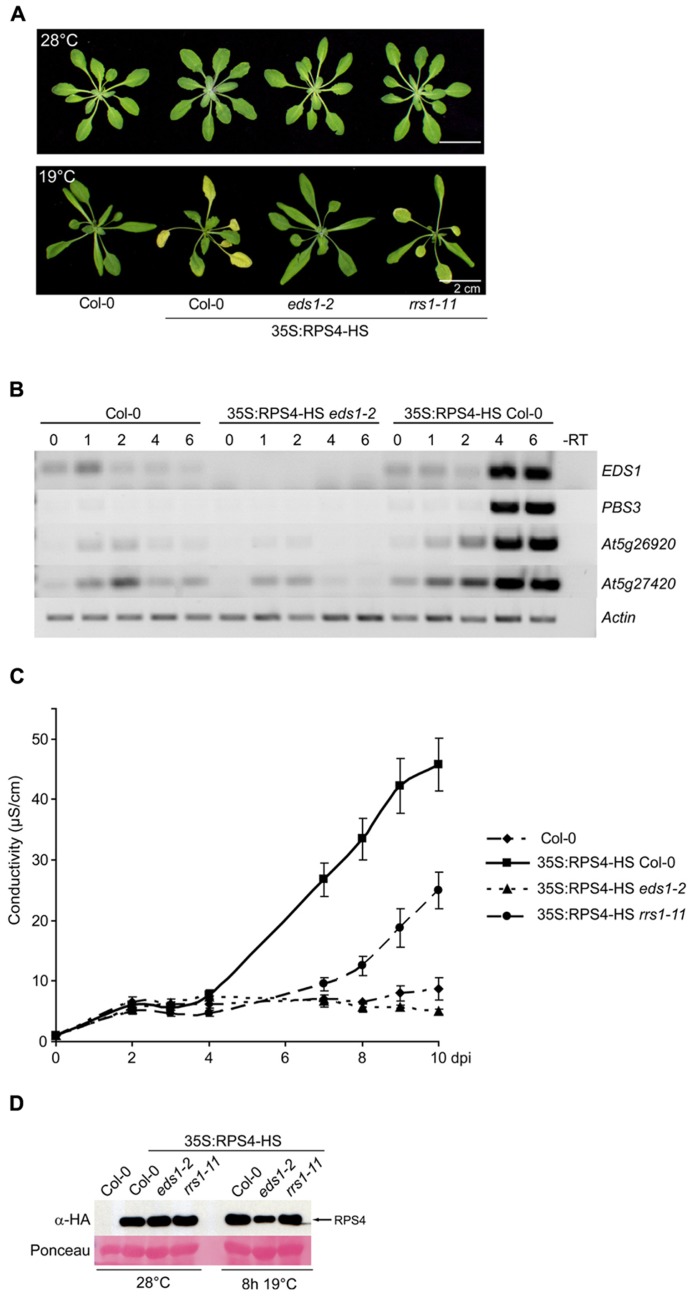
The 35S:RPS4-HS plants grew similarly to wild type Col-0 at 28°C (Figure 4A) and showed no constitutive defense gene expression (Figure 4B). Moving 35S:RPS4-HS plants from 28 to 19°C induced expression of EDS1 itself and several known Pst/AvrRps4-responsive, EDS1-dependent defense-related genes (Bartsch et al., 2006) at 4 and 6 h post-temperature shift (hps; Figure 4B). Col-0 wild type and 35S:RPS4-HS eds1-2 plants subjected to the same temperature change did not show induction of these genes at 4 and 6 hps (Figure 4B). In multiple repeats, the 28 to 19°C temperature shift proved to be an easy and highly reproducible EDS1-requiring defense gene inductive switch for 35S:RPS4-HS plants.
FIGURE 4.
A 28 to 19°C temperature shift induces RPS4-HS auto-immunity. (A) Growth of 3.5-week-old 35S:RPS4-HS plants at 28°C (upper panel) and 6 days after moving to 19°C (lower panel). Scale bars, 2 cm. (B) Semi-quantitative RT-PCR of known Pst/AvrRps4-responsive, EDS1-dependent genes over 0–6 h after temperature shift of Col-0, 35S:RPS4-HS eds1-2, and 35S:RPS4-HS Col-0 plants, as indicated. (C) Ion leakage measurements made over a 10-day period after shift from high to low temperature (dps) in leaf disks of the different 3.5-week-old 35S:RPS4-HS lines and Col-0 wild-type, as indicated. Error bars represent standard errors of four samples per genotype. Three independent experiments gave similar results. (D) Immunoblot analysis of total leaf protein extracts from 3.5-week-old 35S:RPS4-HS lines grown at 28°C and shifted to 19°C for 8 h, separated by SDS-polyacrylamide gel electrophoresis (SDS-PAGE) and probed with α-HA antibody. Ponceau S staining shows equal transfer of protein samples to the membrane.
Macroscopic symptoms of auto-immunity were first seen as leaf chlorosis in 35S:RPS4-HS plants, starting at 3–4 days after the 28 to 19°C temperature shift and showing complete EDS1-dependence (Figure 4A). In conductivity assays for cell death, ion leakage from 35S:RPS4-HS leaf disks started to rise significantly between 4 and 6 days post-shift (dps) but did not increase in 35S:RPS4-HS eds1-2 or wild-type Col-0 (Figure 4C). We tested the 35S:RPS4-HS rrs1-11 line under the same conditions and found that progression of leaf chlorosis (Figure 4A) and ion leakage (Figure 4C) was intermediate between 35S:RPS4-HS and 35S:RPS4-HS eds1-2 plants. Steady-state RPS4-HS protein accumulation was not strongly affected by temperature or the rrs1-11 mutation, but was slightly lower in eds1-2 at 8 h after temperature shift (Figure 4D). Collectively, these data show that RRS1Col contributes to temperature-conditioned 35S:RPS4-HS auto-immunity at the level of leaf chlorosis and pcd.
RRS1Col SUPPORTS TRANSCRIPTIONAL REPROGRAMING OF A DISCRETE SET OF EDS1-DEPENDENT GENES IN TEMPERATURE-SHIFTED 35:RPS4-HS PLANTS
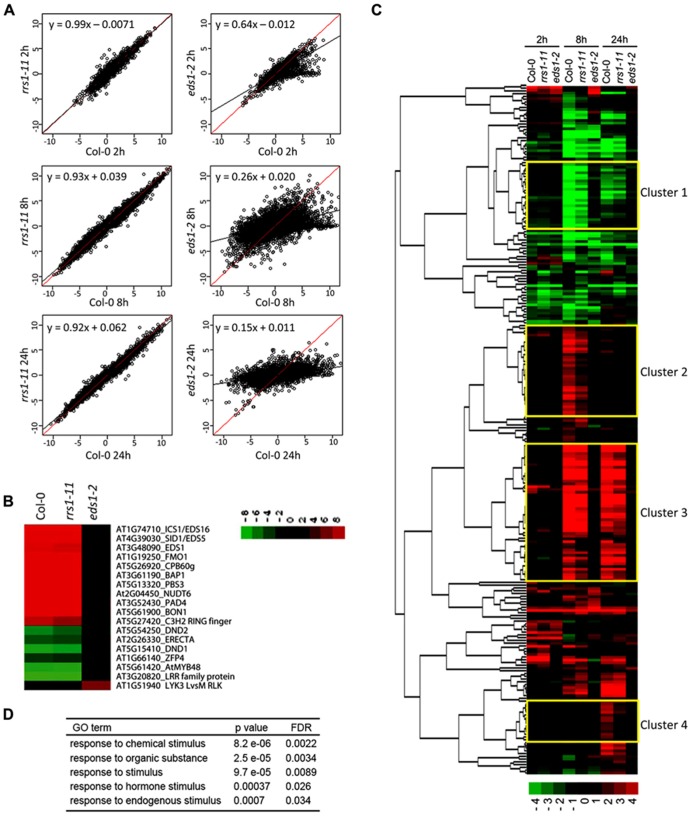
In the above assays, we established that 35S:RPS4-HS 28/19°C-shifted leaf tissues resemble Pst/AvrRps4-infected plants at 22°C with respect to complete EDS1- and partial RRS1Col-dependence in chlorotic and pcd phenotypes. However, the temperature shift will have physiological effects unrelated to immunity (Penfield, 2008; McClung and Davis, 2010). We therefore performed gene expression microarray analysis of 35S:RPS4-HS, 35S:RPS4-HS rrs1-11, and 35S:RPS4-HS eds1-2 leaf mRNAs at 0 h (28°C), 2, 8, and 24 hps to 19°C in order to determine the relative contributions of RRS1Col and EDS1 to temperature-conditioned 35S:RPS4-HS transcriptional reprograming. Profiling of polyA+ RNAs was performed using Affymetrix ATH1 GeneChips (see Materials and Methods). We first selected genes whose expression was significantly up- or down-regulated (q-values < 0.01 and >2-fold change) in 35S:RPS4-HS over all time points compared to non-shifted 35S:RPS4-HS plants at 28°C (t0; 10277 genes in total). Hence, there is extensive reprograming of transcription in 35S:RPS4-HS leaves over 24 hps. We then compared the global gene expression profiles of 35S:RPS4-HS, 35S:RPS4-HS rrs1-11, and 35S:RPS4-HS eds1-2 at 0, 2, 8, and 24 hps by plotting changed transcripts in 35S:RPS4-HS rrs1-11 or 35S:RPS4-HS eds1-2 on a linear regression curve (red) against the regression curve set by 35S:RPS4-HS transcript changes (black; Figure 5A). This analysis shows that expression changes in 35S:RPS4-HS rrs1-11 broadly resemble those of 35S:RPS4-HS over the 24 h time course (Figure 5A). Therefore, loss of RRS1Col function has little effect on RPS4-HS transcriptional reprograming overall. Many gene expression changes in 35S:RPS4-HS at 2 hps (80%) were also similar in 35S:RPS4-HS eds1-2, as seen by the near congruence of the red and black regression curves (Figure 5A). A measurable impact of eds1-2 on expression changes in 35S:RPS4-HS was observed at 8 and 24 hps, with most differences between the two lines established already at 8 hps (Figure 5A). These data show that EDS1 contributes substantially to RPS4-HS-triggered transcriptional reprograming following an early EDS1-independent phase that is likely due to the temperature shift per se and not directly related to RPS4 auto-immunity. We then selected a sample of defense-related genes whose up- or down-regulation was established in a previous gene expression microarray study as EDS1- and PAD4-dependent at 6 h after leaf infiltration with Pst/AvrRps4 bacteria at 22°C (Bartsch et al., 2006; Zhu et al., 2010b). The pattern of AvrRps4-triggered induction or repression of the genes was recapitulated at 8 h post-temperature shift in 35S:RPS4-HS and 35S:RPS4-HS rrs1-11 and was strongly EDS1-dependent, as shown in a heatmap (Figure 5B). This suggests that major defense-related transcriptional changes requiring EDS1 in Pst/AvrRps4-infected tissues are qualitatively similar at 8 hps in the temperature-conditioned RPS4 auto-immune response.
FIGURE 5.
RRS1Col contributes to EDS1-dependent gene expression changes in RPS4Col auto-immunity. Gene expression microarray analysis was performed on leaf RNAs of 3.5-week-old 35S:RPS4-HS transgenic plants in the Col-0, rrs1-11, or eds1-2 backgrounds at 0, 2, 8, and 24 h after temperature shift. (A) Induced or repressed genes (q-values < 0.01 and >2-fold changes) in 35S:RPS4-HS Col-0 upon temperature shift at any time point and the log2 ratios compared to 0 h are plotted. Linear regression lines indicate log2 ratios in 35S:RPS4-HS Col-0 (red) and the 35S:RPS4-HS rrs1-11 or 35S:RPS4-HS eds1-2 mutant lines (black). (B) Log2 gene expression ratios at 8 h after temperature shift compared to 0 h in 35S:RPS4-HS Col-0 plants for previously identified Pst/AvrRps4-triggered EDS1-dependent genes, shown by Heatmap clustering analysis. (C) Heatmap clustering of 250 genes whose expression changes are RRS1-dependent in 35S:RPS4-HS after temperature shift at any time point (q-values < 0.01 and >2-fold change). Expression patterns for the 250 genes in 35S:RPS4-HS Col-0, 35S:RPS4-HS rrs1-11, and 35S:RPS4-HS eds1-2 lines are shown at 2, 8, and 24 h. Highlighted clusters 1–4 are described in the text. (D) GO term analysis of the 250 RRS1-dependent genes.
We investigated whether a subset of the total 10227 genes exhibiting changed expression over the 35S:RPS4-HS temperature shift experiment was affected by rrs1-11 by selecting genes whose up- or down-regulation showed dependence on RRS1Col for at least one time point (q-values < 0.01 and >2-fold change). Altogether, 250 genes fitted this pattern with most showing reduced up-regulation in 35S:RPS4-HS rrs1-11 tissues compared to 35S:RPS4-HS. The 250 genes displayed partial RRS1Col- and strong EDS1-dependence for expression changes, as shown in the heatmap (Figure 5C). Hence, the effect of the rrs1-11 mutation is mainly quantitative in the 35S:RPS4-HS temperature-conditioned system. Analysis of GO terms enriched among the 250 genes shows a high representation of genes responsive to chemical, hormone, and other endogenous stimuli (Figure 5D). In a clustering analysis of the 250 “RRS1Col-dependent” genes (see Materials and Methods), four gene clusters were of interest (Figure 5C). In Cluster 1, genes are grouped that show RRS1Col-dependent repression at 8 and 24 h. Cluster 2 contains genes that are up-regulated at 8 hps and show an RRS1Col contribution to induction. Cluster 3 has genes up-regulated at 8 and 24 hps and showing RRS1Col-dependence at both time points. In Cluster 4, a discrete set of genes displaying RRS1Col-dependence in up-regulation at 24 hps is displayed. Interestingly, distinct sub-clusters of genes with strong RRS1Col-dependence are observed within Clusters 3 and 4 (Figure 5C). We concluded that RRS1Col has a measurable positive effect on expression of a subset of EDS1-dependent genes in 35S:RPS4-HS auto-immunity.
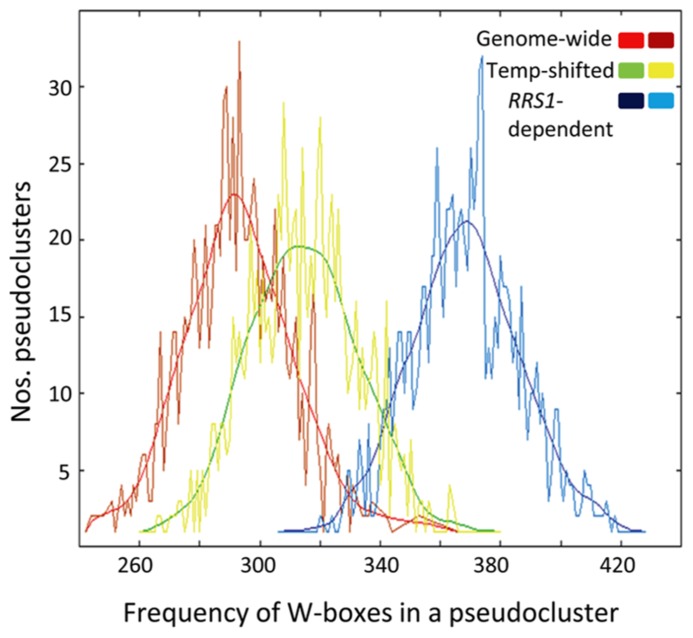
Because RRS1Col encodes a functional TNL receptor with a C-terminal “WRKY” transcription factor DNA-binding domain recognizing W-box elements, we investigated if W-box cis-elements are enriched in the promoters of the 250 RRS1Col-dependent genes. As shown in Figure 6, analysis of the core W-box motif (TTGACY) in promoters of these genes by POBO (Materials and Methods) shows that enrichment of this motif is highly significant (p-value < 0.0001) compared to randomly selected promoters from all Arabidopsis genes. Since the W-box is known to be enriched in promoters of genes that are responsive to biotic stresses (Rushton et al., 2010), we also compared W-box enrichment between promoters of the 250 RRS1Col-dependent genes and promoters from randomly selected 35S:RPS4-HS-regulated genes. The POBO analysis showed that W-boxes remain significantly enriched (p-value < 0.0001) in the promoters of the RRS1Col-dependent genes (Figure 6). These results suggest that RRS1Col acts on a subset of 35S:RPS4-HS reprogramed genes directly or indirectly through the presence of W-box elements in their gene promoters.
FIGURE 6.
W-boxes are highly enriched in promoters of RRS1-dependent genes. POBO analysis of the motif distribution in 1000 bp promoters of RRS1-dependent genes. One thousand pseudo-clusters of the 250 RRS1-dependent genes, genes regulated by the temperature shift (Temp-shifted) and randomly selected genes from 35S:RPS4-HS (Genome-wide) are shown. Jagged lines indicate motif frequencies from which a fitted curve was derived. The W-box (TTGACY) is significantly over-represented in promoters of the RRS1-dependent genes compared to temperature-responsive genes or genes from the genome background with p-values < 0.0001.
DISCUSSION
NLR receptors are usually activated upon specific pathogen effector recognition to trigger a timely and balanced innate immune response. In the absence of a corresponding effector, tight regulation of NLR receptors is enforced by restricting NLR gene expression and ensuring NLR associations with inhibitory co-factors (Heidrich et al., 2012; Staiger et al., 2013). Auto-immunity producing stunting and constitutive activation of resistance and cell death pathways can occur when NLRs are released from inhibition either by NLR over expression or loss-of-function mutations in negative factors (Heidrich et al., 2012; Staiger et al., 2013). An outstanding question is to what extent auto-activated NLR processes mirror those triggered by authentic effector recognition. For TNLs there is compelling evidence that auto-activated receptors connect immediately to a bona fide TNL resistance signaling pathway involving the basal resistance regulator EDS1 (Zhang et al., 2003; Yang and Hua, 2004; Wirthmueller et al., 2007; Huang et al., 2010). Detection of EDS1 in complexes with several NLRs (Bhattacharjee et al., 2011; Heidrich et al., 2011; Kim et al., 2012) is also consistent with EDS1 being an integral and early component of TNL resistance. Thus, effector- and auto-activated TNL signaling steps are likely to be related, although constitutive resistance clearly has deleterious pleiotropic effects on growth and development.
Here we provide evidence that EDS1-dependent auto-immunity in an Arabidopsis RPS4Col over-expression line (35S:RPS4-HS) has a partial requirement for RRS1Col, the genetic partner of RPS4Col in ETI (Birker et al., 2009; Narusaka et al., 2009). This partial dependence on RRS1Col is seen in plants grown at 22°C that exhibit constitutive basal resistance (Figure 3) and after shifting plants from high (28°C) to moderate (19°C) temperature to induce defense-related transcriptional reprograming, chlorosis, and pcd (Figures 4 and 5). Hence, RPS4 auto-immunity does not fully override a requirement for RRS1. Therefore, we reasoned that the dual RPS4–RRS1 resistance system might involve RPS4–RRS1 cooperation beyond initial effector recognition steps to include aspects of downstream resistance signaling. Alternatively, part of the RPS4 auto-activation mechanism involves processes that also occur during effector activation, such as particular NLR conformational transitions (Collier and Moffett, 2009; Lukasik and Takken, 2009). Reduced RPS4Col auto-immunity in rrs1-11 mirrors the intermediate loss of resistance in rrs1-11 mutants to Pst/AvRps4 bacteria (Figures 1 and 3). Therefore, it is possible that in both backgrounds an RPS4/RRS1-independent pathway contributes to the residual resistance (Birker et al., 2009; Sohn et al., 2012). Although the precise nature of effector- and auto-triggered RPS4–RRS1 activation events needs to be resolved, the fact that temperature-induced RPS4 immunity mirrors ETI in displaying complete dependence on EDS1 and partial dependence on RRS1 is significant. The temperature-conditioned RPS4 auto-immune system presents a potentially powerful tool to examine dynamic TNL signaling and transcriptional events in leaf tissues.
Pairing of RPS4 and RRS1 genes and their homologs in a head-to-head tandem arrangement is evolutionarily conserved, underscoring functional significance of the inverted TNL organization (Gassmann et al., 1999; Narusaka et al., 2009). RRS1, a representative of the TNL-A clade, exhibits higher sequence diversity among Arabidopsis accessions than RPS4, as a member of the TNL-B clade (Meyers et al., 2003; Narusaka et al., 2009). This, together with finding that the RRS1 interacts directly with the R. solanacearum effector PopP2 inside nuclei points to RRS1 as a direct effector recognition component, although interaction alone is not sufficient for triggering RRS1 resistance (Deslandes et al., 2003; Tasset et al., 2010). Noutoshi et al. (2005) proposed an attractive model for RRS1 “restraint” and activation based on analysis of an auto-activated slh1 WRKY domain mutation. In the model, RRS1 in non-elicited cells resides at sites on the chromatin as an auto-inhibited NLR. Subsequent studies revealing RRS1–RPS4 genetic cooperativity in resistance to AvrRps4 and PopP2, and an unknown Colletotrichum higginsianum effector (Birker et al., 2009; Narusaka et al., 2009), raised the prospect that effector recognition might be conferred by an auto-inhibited RPS4–RRS1 complex which becomes activated via RPS4–RRS1 conformational changes at the chromatin. Because our data indicate that RRS1 contributes to RPS4 auto-immunity, we propose that signaling events also involve RRS1 with RPS4, as well as EDS1, in what might be a “reconfigured” receptor complex, possibly mediated through TIR–TIR interactions (Mestre and Baulcombe, 2006; Bernoux et al., 2011b). The fact that neither rrs1 nor rps4 null mutant displays constitutive resistance also argues against resistance pathway activation simply being due to release of one or other component from an auto-inhibited complex. An interesting but complicating issue is that EDS1 was found to interact with the AvrRps4 effector in FRET–FLIM and co-immunoprecipitation studies, implying that EDS1 contributes to effector recognition as well as being an integral component of the TNL resistance pathways (Bhattacharjee et al., 2011; Heidrich et al., 2011). Notably, EDS1 interacts with two effectors, AvrRps4 and HopA1, recognized, respectively, by TNLs RPS4/RRS1 and RPS6 (Bhattacharjee et al., 2011; Heidrich et al., 2011). Thus, TNL pre- and post-activation events in these recognition systems might be closely intertwined.
Temperature-induced RPS4 auto-immunity produces an exaggerated transcriptional response compared to ETI probably through an EDS1-dependent transcriptional feed-forward loop (Wang et al., 2009; Zhu et al., 2010a). At 2 h post-temperature shift, analysis of the gene expression microarray data revealed mainly EDS1-independent transcriptional reprograming of 35S:RPS4-HS plants which we attribute to a “temperature” effect (Figure 5A). The small sector (20%) of EDS1-dependent changes at 2 h will be examined in a more detailed expression time series over 1–4 h to identify earliest EDS1 and, potentially, RRS1 effects. At 8 h after temperature shift, transcriptional reprograming was largely EDS1-dependent (Figure 5) and qualitatively similar to ETI for a panel of AvrRps4-triggered EDS1-dependent induced and repressed genes (Figure 5B). A quantitative contribution of RRS1 was detected also at 8 and 24 h after temperature shift in 250 of the EDS1-dependent down and up-regulated genes (Figure 5). An auxiliary role of RRS1 in EDS1-mediated gene expression is reminiscent of the contribution of WRKY18 to NPR1-dependent basal defense responses (Wang et al., 2006) and might reflect a common feature of WRKY-containing transcriptional immune regulators. Notably, several sub-clusters within the RRS1-dependent genes display strong RRS1-dependence in expression at 8 or 24 h (Figure 5C; Clusters 3 and 4). Whether any of these genes are direct targets of RRS1 (or RPS4) is not known but the high representation of W-boxes in their promoter elements (Figure 6) suggests that WRKY-domain protein recruitment might be an important modulator of expression. Current evidence indicates that the dynamics of WRKY transcription factor binding of promoters are complex and likely to involve reconfigurations from repressive to inductive transcription complexes at the chromatin, as well as functional redundancy between WRKY transcription factors (Rushton et al., 2010; Chen et al., 2013; Logemann et al., 2013; Schon et al., 2013).
CONCLUSION
Our data show that RRS1Col positively contributes to RPS4Col auto-immunity induced by a high to moderate temperature shift. The temperature-activated RPS4 over-expression system can help to illuminate the molecular role of RRS1 in this TNL resistance partnership and the hierarchy of defense-related transcriptional reprograming events.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank our colleagues for helpful discussions. This work was supported by The Max-Planck Society, a Deutsche Forschungsgemeinschaft grant in SFB 670 (Jane E. Parker, Katharina Heidrich, Jaqueline Bautor) and a BMBF-funded Plant-KBBE Trilateral “BALANCE” project grant (Jane E. Parker, Servane Blanvillain-Baufumé). Katharina Heidrich was recipient of an IMPRS PhD fellowship.
Footnotes
REFERENCES
- Alcazar R., Parker J. E. (2011). The impact of temperature on balancing immune responsiveness and growth in Arabidopsis. Trends Plant Sci. 16 666–675 10.1016/j.tplants.2011.09.001 [DOI] [PubMed] [Google Scholar]
- Bartsch M., Gobbato E., Bednarek P., Debey S., Schultze J. L., Bautor J., et al. (2006). Salicylic acid-independent ENHANCED DISEASE SUSCEPTIBILITY1 signaling in Arabidopsis immunity and cell death is regulated by the monooxygenase FMO1 and the Nudix hydrolase NUDT7. Plant Cell 18 1038–1051 10.1105/tpc.105.039982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernoux M., Ellis J. G., Dodds P. N. (2011a). New insights in plant immunity signaling activation. Curr. Opin. Plant Biol. 14 512–518 10.1016/j.pbi.2011.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernoux M., Ve T., Williams S., Warren C., Hatters D., Valkov E., et al. (2011b). Structural and functional analysis of a plant resistance protein TIR domain reveals interfaces for self-association, signaling, and autoregulation. Cell Host Microbe 9 200–211 10.1016/j.chom.2011.02.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharjee S., Halane M. K., Kim S. H., Gassmann W. (2011). Pathogen effectors target Arabidopsis EDS1 and alter its interactions with immune regulators. Science 334 1405–1408 10.1126/science.1211592 [DOI] [PubMed] [Google Scholar]
- Birker D., Heidrich K., Takahara H., Narusaka M., Deslandes L., Narusaka Y., et al. (2009). A locus conferring resistance to Colletotrichum higginsianum is shared by four geographically distinct Arabidopsis accessions. Plant J. 60 602–613 10.1111/j.1365-313X.2009.03984.x [DOI] [PubMed] [Google Scholar]
- Chang C., Yu D., Jiao J., Jing S., Schulze-Lefert P., Shen Q. H. (2013). Barley MLA immune receptors directly interfere with antagonistically acting transcription factors to initiate disease resistance signaling. Plant Cell 25 1158–1173 10.1105/tpc.113.109942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Zhang L., Li D., Wang F., Yu D. (2013). WRKY8 transcription factor functions in the TMV-cg defense response by mediating both abscisic acid and ethylene signaling in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 110 E1963–E1971 10.1073/pnas.1221347110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collier S. M., Moffett P. (2009). NB-LRRs work a ``bait and switch'' on pathogens. Trends Plant Sci. 14 521–529 10.1016/j.tplants.2009.08.001 [DOI] [PubMed] [Google Scholar]
- Deslandes L., Olivier J., Peeters N., Feng D. X., Khounlotham M., Boucher C., et al. (2003). Physical interaction between RRS1-R, a protein conferring resistance to bacterial wilt, and PopP2, a type III effector targeted to the plant nucleus. Proc. Natl. Acad. Sci. U.S.A. 100 8024–8029 10.1073/pnas.1230660100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deslandes L., Olivier J., Theulieres F., Hirsch J., Feng D. X., Bittner-Eddy P., et al. (2002). Resistance to Ralstonia solanacearum in Arabidopsis thaliana is conferred by the recessive RRS1-R gene, a member of a novel family of resistance genes. Proc. Natl. Acad. Sci. U.S.A. 99 2404–2409 10.1073/pnas.032485099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodds P. N., Rathjen J. P. (2010). Plant immunity: towards an integrated view of plant-pathogen interactions. Nat. Rev. Genet. 11 539–548 10.1038/nrg2812 [DOI] [PubMed] [Google Scholar]
- Eisen M. B., Spellman P. T., Brown P. O., Botstein D. (1998). Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. U.S.A. 95 14863–14868 10.1073/pnas.95.25.14863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feys B. J., Moisan L. J., Newman M. A., Parker J. E. (2001). Direct interaction between the Arabidopsis disease resistance signaling proteins, EDS1 and PAD4. EMBO J. 20 5400–5411 10.1093/emboj/20.19.5400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia A. V., Blanvillain-Baufume S., Huibers R. P., Wiermer M., Li G., Gobbato E., et al. (2010). Balanced nuclear and cytoplasmic activities of EDS1 are required for a complete plant innate immune response. PLoS Pathog. 6:e1000970. 10.1371/journal.ppat.1000970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gassmann W., Hinsch M. E., Staskawicz B. J. (1999). The Arabidopsis RPS4 bacterial-resistance gene is a member of the TIR-NBS-LRR family of disease-resistance genes. Plant J. 20 265–277 10.1046/j.1365-313X.1999.t01-1-00600.x [DOI] [PubMed] [Google Scholar]
- Hajheidari M., Farrona S., Huettel B., Koncz Z., Koncz C. (2012). CDKF-1 and CDKD protein kinases regulate phosphorylation of serine residues in the C-terminal domain of Arabidopsis RNA Polymerase II. Plant Cell 24 1626–1642 10.1105/tpc.112.096834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidrich K., Blanvillain-Baufume S., Parker J. E. (2012). Molecular and spatial constraints on NB-LRR receptor signaling. Curr. Opin. Plant Biol. 15 385–391 10.1016/j.pbi.2012.03.015 [DOI] [PubMed] [Google Scholar]
- Heidrich K., Wirthmueller L., Tasset C., Pouzet C., Deslandes L., Parker J. E. (2011). Arabidopsis EDS1 connects pathogen effector recognition to cell compartment-specific immune responses. Science 334 1401–1404 10.1126/science.1211641 [DOI] [PubMed] [Google Scholar]
- Hinsch M., Staskawicz B. (1996). Identification of a new Arabidopsis disease resistance locus, RPS4, and cloning of the corresponding avirulence gene, avrRps4, from Pseudomonas syringae pv. pisi. Mol. Plant Microbe Interact. 9 55–61 10.1094/MPMI-9-0055 [DOI] [PubMed] [Google Scholar]
- Huang X., Li J., Bao F., Zhang X., Yang S. (2010). A gain-of-function mutation in the Arabidopsis disease resistance gene RPP4 confers sensitivity to low temperature. Plant Physiol. 154 796–809 10.1104/pp.110.157610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jirage D., Tootle T. L., Reuber T. L., Frost L. N., Feys B. J., Parker J. E., et al. (1999). Arabidopsis thaliana PAD4 encodes a lipase-like gene that is important for salicylic acid signaling. Proc. Natl. Acad. Sci. U.S.A. 96 13583–13588 10.1073/pnas.96.23.13583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kankainen M., Holm L. (2004). POBO, transcription factor binding site verification with bootstrapping. Nucleic Acids Res. 32 W222–W229 10.1093/nar/gkh463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S. H., Gao F., Bhattacharjee S., Adiasor J. A., Nam J. C., Gassmann W. (2010). The Arabidopsis resistance-like gene SNC1 is activated by mutations in SRFR1 and contributes to resistance to the bacterial effector avrRps4. PLoS Pathog. 6:e1001172. 10.1371/journal.ppat.1001172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T. H., Kunz H. H., Bhattacharjee S., Hauser F., Park J., Engineer C., et al. (2012). Natural variation in small molecule-induced TIR-NB-LRR signaling induces root growth arrest via EDS1- and PAD4-complexed R protein VICTR in Arabidopsis. Plant Cell 24 5177–5192 10.1105/tpc.112.107235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logemann E., Birkenbihl R. P., Rawat V., Schneeberger K., Schmelzer E., Somssich I. E. (2013). Functional dissection of the PROPEP2 and PROPEP3 promoters reveals the importance of WRKY factors in mediating microbe-associated molecular pattern-induced expression. New Phytol. 198 1165–1177 10.1111/nph.12233 [DOI] [PubMed] [Google Scholar]
- Lukasik E, Takken F. L. W. (2009). STANDing strong, resistance protein instigators of plant defence. Curr. Opin. Plant Biol. 12 427–436 10.1016/j.pbi.2009.03.001 [DOI] [PubMed] [Google Scholar]
- Maekawa T., Kufer T. A., Schulze-Lefert P. (2011). NLR functions in plant and animal immune systems: so far and yet so close. Nat. Immunol. 12 817–826 10.1038/ni.2083 [DOI] [PubMed] [Google Scholar]
- McClung C. R., Davis S. J. (2010). Ambient thermometers in plants: from physiological outputs towards mechanisms of thermal sensing. Curr. Biol. 20 R1086–R1092 10.1016/j.cub.2010.10.035 [DOI] [PubMed] [Google Scholar]
- Mestre P., Baulcombe D. C. (2006). Elicitor-mediated oligomerization of the tobacco N disease resistance protein. Plant Cell 18 491–501 10.1105/tpc.105.037234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers B. C., Kozik A., Griego A., Kuang H. H., Michelmore R. W. (2003). Genome-wide analysis of NBS-LRR-encoding genes in Arabidopsis. Plant Cell 15 809–834 10.1105/tpc.009308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narusaka M., Shirasu K., Noutoshi Y., Kubo Y., Shiraishi T., Iwabuchi M., et al. (2009). RRS1 and RPS4 provide a dual resistance-gene system against fungal and bacterial pathogens. Plant J. 60 218–226 10.1111/j.1365-313X.2009.03949.x [DOI] [PubMed] [Google Scholar]
- Noutoshi Y., Ito T., Seki M., Nakashita H., Yoshida S., Marco Y., et al. (2005). A single amino acid insertion in the WRKY domain of the Arabidopsis TIR-NBS-LRR-WRKY-type disease resistance protein SLH1 (sensitive to low humidity 1) causes activation of defense responses and hypersensitive cell death. Plant J. 43 873–888 10.1111/j.1365-313X.2005.02500.x [DOI] [PubMed] [Google Scholar]
- Padmanabhan M. S., Ma S., Burch-Smith T. M., Czymmek K., Huijser P., Dinesh-Kumar S. P. (2013). Novel positive regulatory role for the SPL6 transcription factor in N TIR-NB-LRR receptor-mediated plant innate immunity. PLoS Pathog. 9:e1003235. 10.1371/journal.ppat.1003235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker J. E., Holub E. B., Frost L. N., Falk A., Gunn N. D., Daniels M. J. (1996). Characterization of eds1, a mutation in Arabidopsis suppressing resistance to Peronospora parasitica specified by several different RPP genes. Plant Cell 8 2033–2046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penfield S. (2008). Temperature perception and signal transduction in plants. New Phytol. 179 615–628 10.1111/j.1469-8137.2008.02478.x [DOI] [PubMed] [Google Scholar]
- Rietz S., Stamm A., Malonek S., Wagner S., Becker D., Medina-Escobar N., et al. (2011). Different roles of enhanced disease susceptibility1 (EDS1) bound to and dissociated from Phytoalexin Deficient4 (PAD4) in Arabidopsis immunity. New Phytol. 191 107–119 10.1111/j.1469-8137.2011.03675.x [DOI] [PubMed] [Google Scholar]
- Rushton P. J., Somssich I. E., Ringler P., Shen Q. J. (2010). WRKY transcription factors. Trends Plant Sci. 15 247–258 10.1016/j.tplants.2010.02.006 [DOI] [PubMed] [Google Scholar]
- Schon M., Toller A., Diezel C., Roth C., Westphal L., Wiermer M., et al. (2013). Analyses of wrky18 wrky40 plants reveal critical roles of SA/EDS1 signaling and indole-glucosinolate biosynthesis for Golovinomyces orontii resistance and a loss-of resistance towards Pseudomonas syringae pv. tomato avrRPS4. Mol. Plant Microbe Interact. 26 758–767 10.1094/MPMI-11-12-0265-R [DOI] [PubMed] [Google Scholar]
- Sohn K. H., Hughes R. K., Piquerez S. J., Jones J. D. G., Banfield M. J. (2012). Distinct regions of the Pseudomonas syringae coiled-coil effector avrRps4 are required for activation of immunity. Proc. Natl. Acad. Sci. U.S.A. 109 16371–16376 10.1073/pnas.1212332109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohn K. H., Zhang Y, Jones J. D. G. (2009). The Pseudomonas syringae effector protein, avrRPS4, requires in planta processing and the KRVY domain to function. Plant J. 57 1079–1091 10.1111/j.1365-313X.2008.03751.x [DOI] [PubMed] [Google Scholar]
- Staiger D., Korneli C., Lummer M., Navarro L. (2013). Emerging role for RNA-based regulation in plant immunity. New Phytol. 197 394–404 10.1111/nph.12022 [DOI] [PubMed] [Google Scholar]
- Storey J., Tibshirani R. (2003). Statistical methods for identifying differentially expressed genes in DNA microarrays. Methods Mol. Biol. 224 149–157 [DOI] [PubMed] [Google Scholar]
- Tao Y., Xie Z. Y., Chen W. Q., Glazebrook J., Chang H. S., Han B., et al. (2003). Quantitative nature of Arabidopsis responses during compatible and incompatible interactions with the bacterial pathogen Pseudomonas syringae. Plant Cell 15 317–330 10.1105/tpc.007591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasset C., Bernoux M., Jauneau A., Pouzet C., Briere C., Kieffer-Jacquinod S., et al. (2010). Autoacetylation of the Ralstonia solanacearum effector PopP2 targets a lysine residue essential for RRS1-R-mediated immunity in Arabidopsis. PLoS Pathog. 6:e1001202. 10.1371/journal.ppat.1001202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas W. J., Thireault C. A., Kimbrel J. A., Chang J. H. (2009). Recombineering and stable integration of the Pseudomonas syringae pv. syringae 61 hrp/hrc cluster into the genome of the soil bacterium Pseudomonas fluorescens Pf0-1. Plant J. 60 919–928 10.1111/j.1365-313X.2009.03998.x [DOI] [PubMed] [Google Scholar]
- Tornero P., Dangl J. L. (2001). A high-throughput method for quantifying growth of phytopathogenic bacteria in Arabidopsis thaliana. Plant J. 28 475–481 10.1046/j.1365-313X.2001.01136.x [DOI] [PubMed] [Google Scholar]
- Venugopal S. C., Jeong R.-D., Mandal M. K., Zhu S., Chandra-Shekara A. C., Xia Y., et al. (2009). Enhanced disease susceptibility 1 and salicylic acid act redundantly to regulate resistance gene-mediated signaling. PloS Genet. 5:e1000545. 10.1371/journal.pgen.1000545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., Amornsiripanitch N., Dong X. (2006). A genomic approach to identify regulatory nodes in the transcriptional network of systemic acquired resistance in plants. PLoS Pathog. 2:e123. 10.1371/journal.ppat.0020123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Bao Z., Zhu Y., Hua J. (2009). Analysis of temperature modulation of plant defense against biotrophic microbes. Mol. Plant Microbe Interact. 22 498–506 10.1094/MPMI-22-5-0498 [DOI] [PubMed] [Google Scholar]
- Wiermer M., Feys B. J., Parker J. E. (2005). Plant immunity: the EDS1 regulatory node. Curr. Opin. Plant Biol. 8 383–389 10.1016/j.pbi.2005.05.010 [DOI] [PubMed] [Google Scholar]
- Wirthmueller L., Zhang Y., Jones J. D., Parker J. E. (2007). Nuclear accumulation of the Arabidopsis immune receptor RPS4 is necessary for triggering EDS1-dependent defense. Curr. Biol. 17 2023–2029 10.1016/j.cub.2007.10.042 [DOI] [PubMed] [Google Scholar]
- Wu Z. J., Irizarry R. A. (2004). Preprocessing of oligonucleotide array data. Nat. Biotechnol. 22 656–658 10.1038/nbt0604-656b [DOI] [PubMed] [Google Scholar]
- Yang S. H., Hua J. (2004). A haplotype-specific resistance gene regulated by BONZAI1 mediates temperature-dependent growth control in Arabidopsis. Plant Cell 16 1060–1071 10.1105/tpc.020479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue J.-X., Meyers B. C., Chen J.-Q., Tian D., Yang S. (2012). Tracing the origin and evolutionary history of plant nucleotide-binding site-leucine-rich repeat (NBS-LRR) genes. New Phytol. 193 1049–1063 10.1111/j.1469-8137.2011.04006.x [DOI] [PubMed] [Google Scholar]
- Zhang Y. L., Goritschnig S., Dong X. N., Li X. (2003). A gain-of-function mutation in a plant disease resistance gene leads to constitutive activation of downstream signal transduction pathways in suppressor of npr1-1, constitutive 1. Plant Cell 15 2636–2646 10.1105/tpc.015842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y., Qian W., Hua J. (2010a). Temperature modulates plant defense responses through NB-LRR proteins. PLoS Pathog. 6:e1000844. 10.1371/journal.ppat.1000844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z., Xu F., Zhang Y., Cheng Y. T., Wiermer M., Li X. (2010b). Arabidopsis resistance protein SNC1 activates immune responses through association with a transcriptional corepressor. Proc. Natl. Acad. Sci. U.S.A. 107 13960–13965 10.1073/pnas.1002828107 [DOI] [PMC free article] [PubMed] [Google Scholar]