Abstract
Since sodium, Na, is a non-essential element for the plant growth, the molecular mechanism of Na+ transport system in plants has remained elusive for the last two decades. The accumulation of Na+ in soil through irrigation for sustainable agricultural crop production, particularly in arid land, and by changes in environmental and climate conditions leads to the buildup of toxic level of salts in the soil. Since the latter half of the twentieth century, extensive molecular research has identified several classes of Na+ transporters that play major roles in the alleviation of ionic stress by excluding toxic Na+ from the cytosol or preventing Na+ transport to the photosynthetic organs, and also in osmotic stress by modulating intra/extracellular osmotic balance. In this review, we summarize the current knowledge of three major Na+ transporters, namely NHX, SOS1, and HKT transporters, including recently revealed characteristics of these transporters.
Keywords: salt tolerance, Na+ transporter, plant, NHX transporters, SOS1 transporters, HKT transporters
INTRODUCTION
Water availability and its quality greatly affect the growth of most of plant species, and keeping a sufficient amount of water in the soil is of paramount importance for agricultural crop production. Constant irrigation often leads to the accumulation of undesirable salts due to the use of poor quality water, particularly in arid lands, and it has been estimated that about 20% of irrigated land is affected by soil salinity (Yeo, 1999). Salinity stress to most plant species mainly accounts for the increase in cytoplasmic osmotic stress and ion (mainly Na+)-specific toxicity (Blumwald, 2000; Blumwald et al., 2000 Munns and Tester, 2008). Overaccumulation of Na+ in the cytosol causes inhibitions of protein synthesis, many enzymatic reactions (Hall and Flowers, 1973; Flowers and Läuchli, 1983; Murguía et al., 1995), and photosynthetic processes (Tsugane et al., 1999; Tsunekawa et al., 2009). Exclusion of Na+ from photosynthetic organs is therefore crucial for the maintenance of an adequate metabolism and efficient carbon fixation.
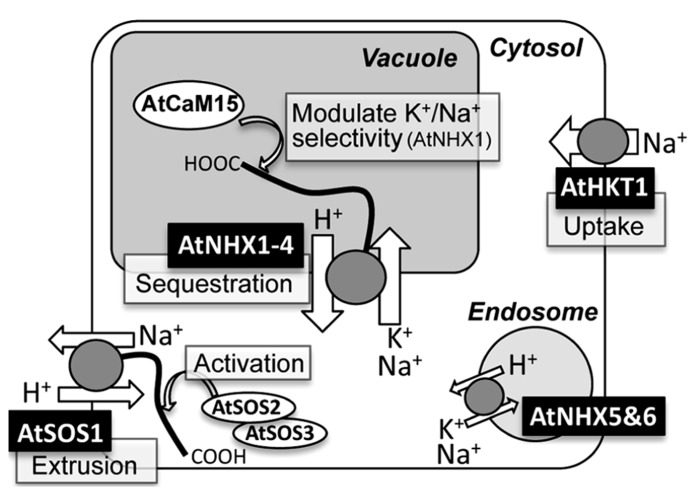
To date, several classes of Na+-transporters have been shown to play central roles in Na+ homeostasis during salinity stress (Apse and Blumwald, 2007; Hauser and Horie, 2010; Uozumi and Schroeder, 2010). Among them, the NHX-, SOS1-, and the Class I HKT-type transporter have drawn particular attention because of their capability of transporting Na+ across cellular membranes (Figure 1). Many studies have demonstrated significant involvements of these Na+ transporters in Na+ sequestration in plant vacuoles, Na+ extrusion from cells, and Na+ circulation for the alleviation of sodium stress under saline conditions. Moreover, some of these Na+ transporters have been shown to improve salinity tolerance of crop plants when their encoding gene is overexpressed (Yamaguchi and Blumwald, 2005; Agarwal et al., 2013).
FIGURE 1.
Schematic representation of subcellular localizations, functions, and regulations of NHX-, SOS1-, and HKT1-type Na+ transporters in Arabidopsis.
In this review, we will provide brief summaries on the properties and physiological roles of three major Na+ transporters that are involved in the plant Na+ tolerance, namely NHX, SOS1, and HKT transporters, from well-established to recently revealed characteristics of these transporters.
NHX TRANSPORTERS
One of the strategies that plant cells employ for the alleviation of excess cytosolic Na+ is to compartmentalize Na+ into the vacuoles, and NHX-type intracellular Na+/H+ exchangers have appeared to be the best-characterized proteins involved in this process so far (Blumwald et al., 2000). Most of NHX proteins studied to date mediate electroneutral Na+/H+ and K+/H+ antiport across the membrane by utilizing the H+ gradient as a driving force and are considered to be responsible for Na+ sequestration into intracellular compartments under salinity stress (Bassil et al., 2012).
Plant genomes often contain multiple intracellular NHX isoforms (Bassil et al., 2012). Intracellular NHX transporters are classified into the IC-NHE/NHX family (Pardo et al., 2006), which is a part of the large cation/proton antiporter 1 (CPA1) family (Säier et al., 2006). These are further subdivided into vacuolar (Class I) or endosomal (Class II) NHXs, according to their sequence similarity and subcellular localizations of representative members (Rodríguez-Rosales et al., 2009; Bassil et al., 2012; Figure 1). Most of the plant species sequenced to date contain both types of NHXs (Rodríguez-Rosales et al., 2009; Bassil et al., 2012), and functional redundancies of vacuolar or endosomal NHXs had been recently reported in Arabidopsis (Bassil et al., 2011a,b; Barragan et al., 2012).
Currently, two distinct topology models have been proposed for AtNHX1. The first model was proposed based on the protease protection analysis of isolated yeast vacuoles expressing full-length AtNHX1 with epitope tags inserted into each hydrophilic loop (Yamaguchi et al., 2003). This model comprised nine transmembrane segments with a C-terminal hydrophilic tail facing toward the vacuolar lumen. Another topology model was proposed on the basis of in vitro translation experiments using truncated AtNHX1 genes, which suggested that several transmembrane segments of AtNHX1 retain similar topogenic properties as human NHE1 (Sato and Sakaguchi, 2005).
The cation selectivity of AtNHX1 appears to be regulated by its C-terminal tail through the binding to a calmodulin-like protein AtCaM15, which interacts with the C-terminus of AtNHX1 in a Ca2+ and pH dependent manner from inside of the vacuole (Yamaguchi et al., 2005; Figure 1). Interaction with AtCaM15 decreases the Na+ transport activity of AtNHX1, while keeping K+ transport activity almost unchanged. Given that the plant vacuole usually keeps a high Ca2+ concentration and an acidic luminal pH, AtNHX1 would favor a K+/H+ antiport mode under normal physiological conditions due to the binding with AtCaM15. However, under salinity stress, which often causes vacuolar alkalization (Okazaki et al., 1996; Gruwel et al., 2001), disassociation of AtCaM15 would result in increased Na+/H+ antiport activity of AtNHX1 and promote compartmentation of Na+ into the vacuoles (Yamaguchi et al., 2005).
Overexpression of genes encoding vacuolar NHXs has conferred salt tolerance to a range of plant species with a concomitant increase in tissue Na+ (Apse et al., 1999; Zhang and Blumwald, 2001; Zhang et al., 2001; Agarwal et al., 2013). These observations, together with the fact that the Arabidopsis atnhx1 mutant exhibited Na+ sensitivity and significantly less vacuolar Na+/H+ antiport activity (Apse et al., 2003), strongly supported the role of vacuolar NHXs in Na+ compartmentation under salinity stress. However, there has been a case where the overexpression of vacuolar NHX led to accumulations of K+ but not Na+ (Leidi et al., 2010). Involvement of endosomal NHXs in salt tolerance has also been demonstrated in tomato and Arabidopsis. LeNHX2 knockdown tomato and atnhx5/atnhx6 double knockout Arabidopsis plants exhibited salt sensitivities (Rodríguez-Rosales et al., 2008; Bassil et al., 2011a). The introduction of the AtNHX5 gene increased salt tolerance and Na+-accumulation in Torenia (Shi et al., 2008). Tomato plants overexpressing LeNHX2 also showed increased salt tolerance. However, an increase of tissue K+ instead of Na+ was observed in this case, suggesting that increased salt tolerance was achieved by improving cellular K+-homeostasis (Huertas et al., 2013).
Besides significant roles in salt tolerance, involvement in pH regulation has been demonstrated for vacuolar NHXs (Fukada-Tanaka et al., 2000; Yamaguchi et al., 2001). Furthermore, vacuolar NHXs appeared to be involved in organ developments. The Arabidopsis atnhx1/atnhx2 double knockout plant exhibited severe defects on the plant growth, development of reproductive organ, and leaf cell expansion under normal conditions (Bassil et al., 2011b; Barragan et al., 2012). These developmental defects seem to be attributed to the role of AtNHXs in K+- rather than Na+-homeostasis, since the nhx1-1/nhx2-1 plant accumulated a lower level of K+ and exhibited less tonoplast K+/H+ antiport activity than the wild type plant. Severe growth defects have also been observed in endosomal NHX knockout/knockdown plants. The LeNHX2 knockdown tomato plant showed severely retarded growth as compared to the wild type (Rodríguez-Rosales et al., 2008). Similarly, Arabidopsis atnhx5/atnhx6 double knockout lines exhibited smaller cell size, slower development of flower organs, and root growth (Bassil et al., 2011a). These results may indicate significant contributions of endosomal NHXs on growth and development, possibly via maintaining appropriate pH and/or cation balance in plant cells. Roles in endosomal trafficking have also been documented (Sottosanto et al., 2004; Bassil et al., 2011a), which have been recently summarized in excellent review articles (Bassil et al., 2012; Qiu, 2012) and therefore will not be discussed further.
SOS1 TRANSPORTERS
Exclusion of Na+ from the cytosol across the plasma membrane is also an important mechanism for the alleviation of cellular Na+ toxicity in plants (Blumwald, 2000). To date, the SOS1 (salt overly sensitive 1 = AtNHX7 in Arabidopsis) transporters are the best-characterized class of transporters attributed with this function (Figure 1). The SOS1 gene has been identified in several plants including Arabidopsis (Wu et al., 1996), rice (Martínez-Atienza et al., 2007), wheat (Xu et al., 2008), tomato (Olías et al., 2009), and Thellungiella salsuginea (Vera-Estrella et al., 2005). The SOS1 gene encodes a large membrane protein (127 kDa, in the case of Arabidopsis AtSOS1) that is categorized into the CPA1 family along with NHXs, although it is more closely related to bacterial NhaP antiporters (Wu et al., 1996; Shi et al., 2000; Brett et al., 2005). AtSOS1 protein is localized in the plasma membrane, and appears to mediate electroneutral Na+/H+ antiport (Shi et al., 2002; Qiu et al., 2002,2003). Hydropathy prediction has indicated that the AtSOS1 protein consists of 12 putative transmembrane segments and a long C-terminal hydrophilic tail (approximately 700 aa; Shi et al., 2000). Although detailed structural properties of AtSOS1 remain elusive, a recent biochemical analysis revealed that AtSOS1 forms a homodimer (Núñez-Ramírez et al., 2012). In addition, Arabidopsis has another SOS1-like gene, termed AtNHX8, which seems to encode a C-terminal truncated isoform of AtSOS1.Despite its high similarity to AtSOS1 (~88% for hydrophobic region), AtNHX8 seemed to prefer Li+ over Na+ as its substrate (An et al., 2007).
It appears that the AtSOS1 C-terminus contains an activation and an auto-inhibitory domain that are physically associated to each other, and this interaction seems to suppress AtSOS1 activity at the resting state (Quintero et al., 2011). The activation of AtSOS1 occurs via the direct phosphorylation of the auto-inhibitory domain by the serine/threonine kinase AtSOS2, which requires a calcium binding protein, AtSOS3, or SCaBP8/CBL10, for its kinase activity (Figure 1; Qiu et al., 2002; Quan et al., 2007; Quintero et al., 2011). In addition, AtSOS2 and AtSOS3 have also shown to be required for the Na+-dependent induction of AtSOS1 gene (Shi et al., 2000).
The physiological significance of SOS1 transporters in saline environments has been well established in Arabidopsis, tomato, and T. salsuginea, as significant increases in salt sensitivity has been observed for the knockout/knockdown plants of corresponding SOS1 genes(Wu et al., 1996; Shi et al., 2000, 2002; Oh et al., 2009; Olías et al., 2009). All of these knockout/knockdown plants tend to accumulate higher amounts of Na+ than wild type plants under severe salt stress, revealing critical roles of SOS1 transporters in limiting Na+ entry by promoting Na+ efflux at the plasma membrane of the root cells (Shi et al., 2002; Oh et al., 2009; Olías et al., 2009). This is well in agreement with the observation that the AtSOS1 transcript is predominantly expressed in epidermal cells of the root tip (Shi et al., 2002), and that the overexpression of AtSOS1, T. salsuginea SOS1, and rice OsSOS1 in Arabidopsis successfully conferred resistance to salinity stress (Shi et al., 2003; Martínez-Atienza et al., 2007; Oh et al., 2009; Yang et al., 2009). Furthermore, SOS1 appears to play a role in protecting the root plasma membrane K+ uptake under salinity stress (Qi and Spalding, 2004). Involvement in xylem loading of Na+ for root to shoot transport has also been suggested for AtSOS1 under moderate salt stress and low transpiration conditions (Ding and Zhu, 1997; Shi et al., 2002). This is based on the preferential expression of AtSOS1 in xylem parenchyma cells and decreased shoot Na+ contents in the atsos1 mutant than in wild type under hydroponic or high humidity conditions (Shi et al., 2002). On the other hand, SlSOS1-silenced tomato (Solanum lycopersicum) exhibited increased Na+ content in roots and leaves as compared to wild type under salt stress, while stems of silenced plants accumulated less Na+ than the wild type (Olías et al., 2009). These results prompted a suggestion that SlSOS1 possibly functioned in Na+ extrusion from the leaf cells back to the xylem stream in order to protect photosynthetic organs (Olías et al., 2009). SOS1 also appears to have several other cellular functions that are not directly related to Na+-homeostasis. A recent study using fluorescent probes reported that the loss of the functional AtSOS1 gene resulted in altered pH homeostasis, both in the cytosol and the vacuole of root cells, probably due to the alteration in plasma membrane H+ flux (Shabala et al., 2005; Oh et al., 2010). In addition, the fragmentation of vacuoles and deficiencies in endocytosis have also been reported in atsos1 mutants under salinity stress (Oh et al., 2010).
HKT TRANSPORTERS
The gene encoding high-affinity K+ transporter, HKT1 was first identified in the wheat genome by functional complementation screening for genes that rescue the deficiency of TRK1 and TRK2-mediated K+ uptake activity in S. cerevisiae mutants (Schachtman and Schroeder, 1994). HKT1 belongs to the Trk/Ktr/HKT-type K+ transporter superfamily found in microorganisms, plant cells, and parasites (Gaber et al., 1988; Ko and Gaber, 1988; Dosch et al., 1991; Schlösser et al., 1991; Schachtman and Schroeder, 1994; Nakamura et al., 1998; Mosimann et al., 2010). The further study on HKT1 revealed that the wheat HKT1 (TaHKT2;1) showed Na+/K+ co-transport activity at high Na+ as compared to K+ in extracellular compartment (Rubio et al., 1995; Gassmann et al., 1996). The Na+-dependent activation of K+ uptake was later found in KtrAB in Vibrio alginolyticus (Nakamura et al., 1998), and KtrABE in cyanobacteria Synechosytis PCC 6803 as well (Matsuda et al., 2004). TbHKT1 in Trypanosoma brucei (Mosimann et al., 2010) and TsHKT1;2 in T. salsuginea (Ali et al., 2012) showed higher selectivity for K+ over Na+. On the other hand, a strong Na+ selective transport activity was found in AtHKT1;1 isolated from Arabidopsis (Uozumi et al., 2000). Na+ selectivity was also seen in HKTs from other species including Eucalyptus camaldulensis and rice (Fairbairn et al., 2000; Horie et al., 2001).
An array of biochemical approaches including bacterial alkaline phosphatase-fusion analysis, glycosylation scanning, and immunofluorescence detection, revealed that AtHKT1 contains eight transmembrane domains that correspond to fourfold bacterial simple K+ channels consisting of transmembrane-pore-transmembrane motifs, and N- and C-termini that face the cytoplasm (Kato et al., 2001). Trk/Ktr/HKT-type transporters appear to have evolutional correlation with the ion-conducting regions of K+ channels, Na+ channels, Ca2+ channels, and Kdp-type K+ pumps(Durell et al., 1999; Durell and Guy, 1999; Uozumi and Dreyer, 2012). The glycine of the K+ ion selective filter in the four pore domains of Trk/Ktr/HKT-type transporters is analogous to the first glycine in the GYG signature sequence (Mäser et al., 2002b; Tholema et al., 2005; Uozumi and Dreyer, 2012). In AtHKT1, the serine in the first pore region in AtHKT1 is substituted for glycine, while the following three glycine residues are conserved. Replacement of the serine by glycine in AtHKT1 led to the increase of K+ transport activity, which strongly suggested that AtHKT1 functions as a Na+ selective transporter rather than a K+ transport system. Based on the relationship of Gly/Ser and Na+/K+ selectivity in Trk/Ktr/HKT-type transporters in combination with sequence alignments, HKTs were categorized into two major groups (Platten et al., 2006). The conserved arginine in the middle of the last transmembrane domain of Trk/Ktr/HKT-type transporters, which is not conserved in K+ channels, is one of the determinants of “transporter” activity but not “channel” activity (Kato et al., 2007). The gating of the ion transport is controlled by the sixth transmembrane in KtrB of V. alginolyticus (Hänelt et al., 2010a,b). This structure–function relationship is supported by the crystal structure of a TrkH from Vibrio parahaemolyticus (Cao et al., 2011).
A long-term exposure of a plant to Na+ in soil will translocate Na+ from the root to above ground tissue (Mäser et al., 2002a). AtHKT1 may participate in the Na+ uptake and Na+ circulation in plant body. The analysis of AtHKT1 gene expression by the use of the GUS reporter system showed that AtHKT1 was expressed in vascular tubes (Mäser et al., 2002a; Berthomieu et al., 2003). This detailed observation, in combination with immunoelectron microscopic detection, showed that AtHKT1;1 resides at the plasma membrane of xylem parenchyma cells and phloem tissues (Berthomieu et al., 2003; Sunarpi et al., 2005). Likewise, one of the rice HKTs was present in xylem parenchyma cells (Ren et al., 2005). The sas2 (sodium over-accumulation in shoots) mutant exhibited increased Na+ accumulation in aerial organs and decreased it in roots (Berthomieu et al., 2003). Taken together, the hypothetical model proposed here to explain the physiological role of AtHKT1 is that AtHKT1 works by unloading Na+ from shoot xylem sap of Arabidopsis in the presence of high salinity and for recirculation of shoot to root (Figure 1; Mäser et al., 2002a; Berthomieu et al., 2003; Sunarpi et al., 2005).
Some transcriptional regulatory elements have been identified in the AtHKT1 promoter. The tandem repeat regions found far from the AtHKT1 initiation codon (approximately -3.9 to -5.3 kb) were responsible for the expression in roots (Rus et al., 2006). The second-half of the repeat sequence, which was located in the 3′ region of two repeat sequences, acted as enhancer elements for AtHKT1 expression (Baek et al., 2011). Reduction of the expression in the root by the inactivation of the repeat sequence induced overaccumulation of Na+ in the shoot (Rus et al., 2006). In accordance with this result, the engineered root stele-specific AtHKT1-overexpressor reduced salt content in the shoot, whereas AtHKT1 expression driven by the 35S promoter in whole plant resulted in salt hypersensitivity (Møller et al., 2009). The putative highly methylated sequence (250 bp) was found 2.6 kb upstream from the initiation codon of AtHKT1. It is believed that this region is a potential small RNA target site and the methylation in this region may control the expression of AtHKT1 at a low level (Baek et al., 2011).
CONCLUDING REMARKS
Almost two decades of extensive molecular studies have clearly established the involvement of NHX, SOS1, and HKT transporters in plant salt tolerance. The genes encoding some of these transporters have since been successfully utilized as genetic tools for enhancing salt tolerance of model and crop plants. Recent advancements have also provided evidence on the involvement of these transporters in other vital cellular processes. This indicates the possibility that further enhancement of salt tolerance by overexpression of any of the above-mentioned genes might reach its limit due to excessive perturbation of related cellular and physiological processes. A much broader and profound understanding of the molecular properties, regulations, and cellular/physiological functions of Na+-transporters, may provide further insight into the development of efficient strategies for the engineering of “super salt-tolerant” crop plants and therefore progress in this research field is highly anticipated.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Mario Fon for the proofreading and editing the manuscript. This work was supported by Grants-in-Aid for Scientific Research from Japan Society for the Promotion of Science (24102504, 24580135, and 25292055).
REFERENCES
- Agarwal P. K., Shukla P. S., Gupta K., Jha B. (2013). Bioengineering for salinity tolerance in plants: state of art. Mol. Biotechnol. 54 102–123 10.1007/s12033-012-9538-3 [DOI] [PubMed] [Google Scholar]
- Ali Z., Park H. C., Ali A., Oh D. H., Aman R., Kropornicka A., et al. (2012). TsHKT1;2, a HKT1 homolog from the extremophile Arabidopsis relative Thellungiella salsuginea, shows K+ specificity in the presence of NaCl. Plant Physiol. 158 1463–1474 10.1104/pp.111.193110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- An R., Chen Q. J., Chai M. F., Lu P. L., Su Z., Qin Z. X., et al. (2007). AtNHX8, a member of the monovalent cation: proton antiporter-1 family in Arabidopsis thaliana, encodes a putative Li/H antiporter. Plant J. 49 718–728 10.1111/j.1365-313X.2006.02990.x [DOI] [PubMed] [Google Scholar]
- Apse M. P., Aharon G. S., Snedden W. A., Blumwald E. (1999). Salt tolerance conferred by overexpression of a vacuolar Na+/H+ antiport in Arabidopsis. Science 285 1256–1258 10.1126/science.285.5431.1256 [DOI] [PubMed] [Google Scholar]
- Apse M. P., Blumwald E. (2007). Na+ transport in plants. FEBS Lett. 581 2247–2254 10.1016/j.febslet.2007.04.014 [DOI] [PubMed] [Google Scholar]
- Apse M. P., Sottosanto J. B., Blumwald E. (2003). Vacuolar cation/H+ exchange, ion homeostasis, and leaf development are altered in a T-DNA insertional mutant of AtNHX1, the Arabidopsis vacuolar Na+/H+ antiporter. Plant J. 36 229–239 10.1046/j.1365-313X.2003.01871.x [DOI] [PubMed] [Google Scholar]
- Barragan V., Leidi E. O., Andres Z., Rubio L., Luca A. D., Fernandez J. A., et al. (2012). Ion exchangers NHX1 and NHX2 mediate active potassium uptake into vacuoles to regulate cell turgor and stomatal function in Arabidopsis. Plant Cell 24 1127–1142 10.1105/tpc.111.095273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassil E., Coku A., Blumwald E. (2012). Cellular ion homeostasis: emerging roles of intracellular NHX Na+/H+ antiporters in plant growth and development. J. Exp. Bot. 63 5727–5740 10.1093/jxb/ers250 [DOI] [PubMed] [Google Scholar]
- Bassil E., Ohto M. A., Esumi T., Tajima H., Zhu Z., Cagnac O., et al. (2011a). The Arabidopsis intracellular Na+/H+ antiporters NHX5 and NHX6 are endosome associated and necessary for plant growth and development. Plant Cell 23 224–239 10.1105/tpc.110.079426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassil E., Tajima H., Liang Y. C., Ohto M. A., Ushijima K., Nakano R., et al. (2011b). The Arabidopsis Na+/H+ antiporters NHX1 and NHX2 control vacuolar pH and K+ homeostasis to regulate growth, flower development, and reproduction. Plant Cell 23 3482–3492 10.1105/tpc.111.089581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baek D., Jiang J., Chung J. S., Wang B., Chen J., Xin Z., et al. (2011). Regulated AtHKT1 gene expression by a distal enhancer element and DNA methylation in the promoter plays an important role in salt tolerance. Plant Cell Physiol. 52 149–161 10.1093/pcp/pcq182 [DOI] [PubMed] [Google Scholar]
- Berthomieu P., Conéjéro G., Nublat A., Brackenbury W. J., Lambert C., Savio C., et al. (2003). Functional analysis of AtHKT1 in Arabidopsis shows that Na+ recirculation by the phloem is crucial for salt tolerance. EMBO J. 22 2004–2014 10.1093/emboj/cdg207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumwald E. (2000). Sodium transport and salt tolerance in plants. Curr. Opin. Cell Biol. 12 431–434 10.1016/S0955-0674(00)00112-5 [DOI] [PubMed] [Google Scholar]
- Blumwald E., Aharon G. S., Apse M. P. (2000). Sodium transport in plant cells. Biochim. Biophys. Acta 1465 140–151 10.1016/S0005-2736(00)00135-8 [DOI] [PubMed] [Google Scholar]
- Brett C. L., Donowitz M., Rao R. (2005). Evolutionary origins of eukaryotic sodium/proton exchangers. Am. J. Physiol. Cell Physiol. 288 C223–C239 10.1152/ajpcell.00360.2004 [DOI] [PubMed] [Google Scholar]
- Cao Y., Jin X., Huang H., Derebe M. G., Levin E. J., Kabaleeswaran V., et al. (2011). Crystal structure of a potassium ion transporter, TrkH. Nature 471 336–340 10.1038/nature09731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding L., Zhu J. K. (1997). Reduced Na+ uptake in the NaCl-hypersensitive sos1 mutant of Arabidopsis thaliana. Plant Physiol. 113 795–799 10.1104/pp.113.3.795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dosch D. C., Helmer G. L., Sutton S. H., Salvacion F. F., Epstein W. (1991). Genetic analysis of potassium transport loci in Escherichia coli: evidence for three constitutive systems mediating uptake potassium. J. Bacteriol. 173 687–696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durell S. R., Guy H. R. (1999). Structural models of the KtrB, TrkH, and Trk1,2 symporters based on the structure of the KcsA K+ channel. Biophys. J. 77 789–807 10.1016/S0006-3495(99)76932-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durell S. R., Hao Y., Nakamura T., Bakker E. P., Guy H. R. (1999). Evolutionary relationship between K+ channels and symporters. Biophys. J. 77 775–788 10.1016/S0006-3495(99)76931-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairbairn D. J., Liu W., Schachtman D. P., Gomez-Gallego S., Day S. R., Teasdale R. D. (2000). Characterisation of two distinct HKT1-like potassium transporters from Eucalyptus camaldulensis. Plant Mol. Biol. 43 515–525 10.1023/A:1006496402463 [DOI] [PubMed] [Google Scholar]
- Flowers T. J, Läuchli A. (1983). “Sodium versus potassium: substitution and compartmentation,” in Encyclopedia of Plant Physiology 15B, New Series, Inorganic Plant Nutrition eds Läuchli A., Bieleski R. A. (Berlin: Springer; ) 651–681 [Google Scholar]
- Fukada-Tanaka S., Inagaki Y., Yamaguchi T., Saito N., Iida S. (2000). Colour-enhancing protein in blue petals. Nature 407 581 10.1038/35036683 [DOI] [PubMed] [Google Scholar]
- Gaber R. F., Styles C. A., Fink G. R. (1988). TRK1 encodes a plasma membrane protein required for high-affinity potassium transport in Saccharomyces cerevisiae. Mol. Cell. Biol. 8 2848–2859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gassmann W., Rubio F., Schroeder J. I. (1996). Alkali cation selectivity of the wheat root high-affinity potassium transporter HKT1. Plant J. 10 869–882 10.1046/j.1365-313X.1996.10050869.x [DOI] [PubMed] [Google Scholar]
- Gruwel M. L. H., Rauw V. L., Loewen M., Abrams S. R. (2001). Effects of sodium chloride on plant cells: a 31P and 23Na NMR system to study salt tolerance. Plant Sci. 160 785–794 10.1016/S0168-9452(00)00424-6 [DOI] [PubMed] [Google Scholar]
- Hall J. L., Flowers T. J. (1973). The effect of salt on protein synthesis in the halophyte Suaeda maritima. Planta 110 361–368 10.1007/BF00387064 [DOI] [PubMed] [Google Scholar]
- Hänelt I., Löchte S., Sundermann L., Elbers K., Vor der Brüggen M., Bakker E. P. (2010a). Gain of function mutations in membrane region M2C2 of KtrB open a gate controlling K+ transport by the KtrAB system from Vibrio alginolyticus. J. Biol. Chem. 285 10318–10327 10.1074/jbc.M109.089870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hänelt I., Wunnicke D., Müller-Trimbusch M., Vor der Brüggen M., Kraus I., Bakker E. P., et al. (2010b). Membrane region M2C2 in subunit KtrB of the K+ uptake system KtrAB from Vibrio alginolyticus forms a flexible gate controlling K+ flux: an electron paramagnetic resonance study. J. Biol. Chem. 285 28210–28219 10.1074/jbc.M110.139311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauser F., Horie T. (2010). A conserved primary salt tolerance mechanism mediated by HKT transporters: a mechanism for sodium exclusion and maintenance of high K+/Na+ ratio in leaves during salinity stress. Plant Cell Environ. 33 552–565 10.1111/j.1365-3040.2009.02056.x [DOI] [PubMed] [Google Scholar]
- Horie T., Yoshida K., Nakayama H., Yamada K., Oiki S., Shinmyo A. (2001). Two types of HKT transporters with different properties of Na+ and K+ transport in Oryza sativa. Plant J. 27 129–138 10.1046/j.1365-313x.2001.01077.x [DOI] [PubMed] [Google Scholar]
- Huertas R., Rubio L., Cagnac O., García-Sánchez M. J., Alché J. D., Venema K., et al. (2013). The K+/H+ antiporter LeNHX2 increases salt tolerance by improving K+ homeostasis in transgenic tomato. Plant Cell Environ. 10.1111/pce.12109 [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- Kato N., Akai M., Zulkifli L., Matsuda N., Kato Y., Goshima S., et al. (2007). Role of positively charged amino acids in the M2D transmembrane helix of Ktr/Trk/HKT-type cation transporters. Channels 1 161–171 [DOI] [PubMed] [Google Scholar]
- Kato Y., Sakaguchi M., Mori Y., Saito K., Nakamura T., Bakker E. P., et al. (2001). Evidence in support of a four transmembrane-pore-transmembrane topology model for the Arabidopsis thaliana Na+/K+ translocating AtHKT1 protein, a member of the superfamily of K+ transporters. Proc. Natl. Acad. Sci. USA. 98 6488–6493 10.1073/pnas.101556598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko C. H., Gaber R. F. (1988). TRK1 and TRK2 encode structurally related K+ transporters in Saccharomyces cerevisiae. Mol. Cell. Biol. 11 4266–4273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leidi E. O., Barragán V., Rubio L., El-Hamdaoui A., Ruiz M. T., Cubero B., et al. (2010). The AtNHX1 exchanger mediates potassium compartmentation in vacuoles of transgenic tomato. Plant J. 61 495–506 10.1111/j.1365-313X.2009.04073.x [DOI] [PubMed] [Google Scholar]
- Martínez-Atienza J., Jiang X., Garciadeblas B., Mendoza I., Zhu J. K., Pardo J. M., et al. (2007). Conservation of the salt overly sensitive pathway in rice. Plant Physiol. 143 1001–1012 10.1104/pp.106.092635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mäser P., Eckelman B., Vaidyanathan R., Fairbain D. J., Kubo M., Yamagami M., et al. (2002a). Altered shoot/root Na+ distribution and bifurcating salt sensitivity in Arabidopsis by genetic disruption of the Na+ transporter AtHKT1. FEBS Lett. 531 157–161 10.1016/S0014-5793(02)03488-9 [DOI] [PubMed] [Google Scholar]
- Mäser P., Hosoo Y., Goshima S., Horie T., Eckelman B., Yamada K., et al. (2002b). Glycine residues in potassium channel-like selectivity filters determine potassium selectivity in four-loop-per-subunit HKT transporters from plants. Proc. Natl. Acad. Sci. USA. 99 6428–6433 10.1073/pnas.082123799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda N., Kobayashi H., Katoh H., Ogawa T., Futatsugi L., Nakamura T., et al. (2004). Na+-dependent K+ uptake Ktr system from the cyanobacterium Synechocystis sp. PCC 6803 and its role in the early phases of cell adaptation to hyperosmotic shock. J. Biol. Chem. 279 54952–54962 10.1074/jbc.M407268200 [DOI] [PubMed] [Google Scholar]
- Møller I. S., Gilliham M., Jha D., Mayo G. M., Roy S. J., Coates J. C., et al. (2009). Shoot Na+ exclusion and increased salinity tolerance engineered by cell type-specific alteration of Na+ transport in Arabidopsis. Plant Cell 21 2163–2178 10.1105/tpc.108.064568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosimann M., Goshima S., Wenzler T., Lüscher A., Uozumi N, Mäser P. (2010). A Trk/HKT-type K+ transporter from Trypanosoma brucei. Eukaryot. Cell 9 539–546 10.1128/EC.00314-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munns R., Tester M. (2008). Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 59 651–681 10.1146/annurev.arplant.59.032607.092911 [DOI] [PubMed] [Google Scholar]
- Murguía J. R., Bellés J. M., Serrano R. (1995). A salt-sensitive 3′(2′),5′-bisphosphate nucleotidase involved in sulfate activation. Science 267 232–234 10.1126/science.7809627 [DOI] [PubMed] [Google Scholar]
- Nakamura T., Yamamuro N., Stumpe S., Tsutomu U., Bakker E. P. (1998). Cloning of the trkAH gene cluster and characterization of the Trk K+-uptake system of Vibrio alginolyticus. Microbiology 144 2281–2289 10.1099/00221287-144-8-2281 [DOI] [PubMed] [Google Scholar]
- Núñez-Ramírez R. Sánchez-Barrena M. J., Villalta I., Vega J. F., Pardo J. M., Quintero F. J., et al. (2012). Structural insights on the plant salt-overly-sensitive 1 (SOS1) Na+/H+ antiporter. J. Mol. Biol. 424 283–294 10.1016/j.jmb.2012.09.015 [DOI] [PubMed] [Google Scholar]
- Oh D. H., Leidi E., Zhang Q., Hwang S. M., Li Y., Quintero F. J., et al. (2009). Loss of halophytism by interference with SOS1 expression. Plant Physiol. 151 210–222 10.1104/pp.109.137802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh D. H., Lee S. Y., Bressan R. A., Yun D. J., Bohnert H. J. (2010). Intracellular consequences of SOS1 deficiency during salt stress. J. Exp. Bot. 61 1205–1213 10.1093/jxb/erp391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okazaki Y., Kikuyama M., Hiramoto Y., Iwasaki N. (1996). Short-term regulation of cytosolic Ca2+, cytosolic pH and vacuolar pH under NaCl stress in the charophyte alga Nitellopsis obtusa. Plant Cell Environ. 19 569–576 10.1111/j.1365-3040.1996.tb00390.x [DOI] [Google Scholar]
- Olías R., Eljakaoui Z., Li J., De Morales P. A., Marín-Manzano M. C., Pardo J. M., et al. (2009). The plasma membrane Na+/H+ antiporter SOS1 is essential for salt tolerance in tomato and affects the partitioning of Na+ between plant organs. Plant Cell Environ. 32 904–916 10.1111/j.1365-3040.2009.01971.x [DOI] [PubMed] [Google Scholar]
- Pardo J. M., Cubero B., Leidi E. O., Quintero F. J. (2006). Alkali cation exchangers: roles in cellular homeostasis and stress tolerance. J. Exp. Bot. 57 1181–1199 10.1093/jxb/erj114 [DOI] [PubMed] [Google Scholar]
- Platten J. D., Cotsaftis O., Berthomieu P., Bohnert H., Davenport R. J., Fairbairn D. J., et al. (2006). Nomenclature for HKT transporters, key determinants of plant salinity tolerance. Trends Plant Sci. 11 372–374 10.1016/j.tplants.2006.06.001 [DOI] [PubMed] [Google Scholar]
- Qi Z., Spalding E. P. (2004). Protection of plasma membrane K+ transport by the salt overly sensitive 1 Na+–H+ antiporter during salinity stress. Plant Physiol. 136 2548–2555 10.1104/pp.104.049213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu Q. S. (2012). Plant and yeast NHX antiporters: roles in membrane trafficking. J. Integr. Plant Biol. 54 66–72 10.1111/j.1744-7909.2012.01097.x [DOI] [PubMed] [Google Scholar]
- Qiu Q. S., Barkla B. J., Vera-Estrella R., Zhu J. K., Schumaker K. S. (2003). Na+/H+ exchange activity in the plasma membrane of Arabidopsis. Plant Physiol. 132 1041–1052 10.1104/pp.102.010421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu Q. S., Guo Y., Dietrich M. A., Schumaker K. S., Zhu J. K. (2002). Regulation of SOS1, a plasma membrane Na+/H+ exchanger in Arabidopsis thaliana, by SOS2 and SOS3. Proc. Natl. Acad. Sci. USA. 99 8436–8441 10.1073/pnas.122224699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quan R., Lin H., Mendoza I., Zhang Y., Cao W., Yang Y., et al. (2007). SCABP8/CBL10, a putative calcium sensor, interacts with the protein kinase SOS2 to protect Arabidopsis shoots from salt stress. Plant Cell 19 1415–1431 10.1105/tpc.106.042291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintero F. J., Martinez-Atienza J., Villalta I., Jiang X., Kim W. Y., Ali Z., et al. (2011). Activation of the plasma membrane Na/H antiporter salt-overly-sensitive 1 (SOS1) by phosphorylation of an auto-inhibitory C-terminal domain. Proc. Natl. Acad. Sci. USA. 108 2611–2616 10.1073/pnas.1018921108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren Z. H., Gao J. P., Li L. G., Cai X. L., Huang W., Chao D. Y., et al. (2005). A rice quantitative trait locus for salt tolerance encodes a sodium transporter. Nat. Genet. 37 1141–1146 10.1038/ng1643 [DOI] [PubMed] [Google Scholar]
- Rodríguez-Rosales M. P., Gálvez F. J., Huertas R., Aranda M. N., Baghour M., Cagnac O., et al. (2009). Plant NHX cation/proton antiporters. Plant Signal. Behav. 4 265–276 10.4161/psb.4.4.7919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Rosales M. P., Jiang X. J., Gálvez F. J., Aranda M. N., Cubero B., Venema K. (2008). Overexpression of the tomato K+/H+ antiporter LeNHX2 confers salt tolerance by improving potassium compartmentalization. New Phytol. 179 366–377 10.1111/j.1469-8137.2008.02461.x [DOI] [PubMed] [Google Scholar]
- Rubio F., Gassmann W., Schroeder J. I. (1995). Sodium-driven potassium uptake by the plant potassium transporter HKT1 and mutations conferring salt tolerance. Science 270 1660–1663 10.1126/science.270.5242.1660 [DOI] [PubMed] [Google Scholar]
- Rus A., Baxter I., Muthukumar B., Gustin J., Lahner B., Yakubova E., et al. (2006). Natural variants of AtHKT1 enhance Na+ accumulation in two wild populations of Arabidopsis. PLoS Genet. 2:e210. 10.1371/journal.pgen.0020210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Säier M. H., Jr., Tran C. V., Barabote R. D. (2006). TCDB: the transporter classification database for membrane transport protein analyses and information. Nucleic Acids Res. 34 D181–D186 10.1093/nar/gkj001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato Y., Sakaguchi M. (2005). Topogenic properties of transmembrane segments of Arabidopsis thaliana NHX1 reveal a common topology model of the Na+/H+ exchanger family. J. Biochem. (Tokyo) 138 425–431 10.1093/jb/mvi132 [DOI] [PubMed] [Google Scholar]
- Schachtman D. P., Schroeder J. I. (1994). Structure and transport mechanism of a high-affinity potassium uptake transporter from higher plants. Nature 370 655–658 10.1038/370655a0 [DOI] [PubMed] [Google Scholar]
- Schlösser A., Kluttig S., Hamann A., Bakker E. P. (1991). Subcloning, nucleotide sequence, and expression of trkG, a gene that encodes an integral membrane protein involved in potassium uptake via the Trk system of Escherichia coli. J. Bacteriol. 173 3170–3176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shabala L., Cui T. A., Newman I. A., Shabala S. (2005). Salinity-induced ion flux patterns from the excised roots of Arabidopsis sos mutants. Planta 222 1041–1050 10.1007/s00425-005-0074-2 [DOI] [PubMed] [Google Scholar]
- Shi H., Ishitani M., Kim C., Zhu J. K. (2000). The Arabidopsis thaliana salt tolerance gene SOS1 encodes a putative Na+/H+ antiporter. Proc. Natl. Acad. Sci. USA. 97 6896–6901 10.1073/pnas.120170197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H., Lee B. H., Wu S. J., Zhu J. K. (2003). Overexpression of a plasma membrane Na+/H+ antiporter gene improves salt tolerance in Arabidopsis thaliana. Nat. Biotechnol. 21 81–85 10.1038/nbt766 [DOI] [PubMed] [Google Scholar]
- Shi L. Y., Li H. Q., Pan X. P., Wu G. J., Li M. R. (2008). Improvement of Torenia fournieri salinity tolerance by expression of Arabidopsis AtNHX5. Funct. Plant Biol. 35 185–192 10.1071/FP07269 [DOI] [PubMed] [Google Scholar]
- Shi H., Quintero F. J., Pardo J. M., Zhu J. K. (2002). The putative plasma membrane Na+/H+ antiporter SOS1 controls long-distance Na+ transport in plants. Plant Cell 14 465–477 10.1105/tpc.010371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sottosanto J. B., Gelli A., Blumwald E. (2004). DNA array analyses of Arabidopsis thaliana lacking a vacuolar Na+/H+ antiporter: impact of AtNHX1 on gene expression. Plant J. 40 752–771 10.1111/j.1365-313X.2004.02253.x [DOI] [PubMed] [Google Scholar]
- Sunarpi H. T., Motoda J., Kubo M., Yang H., Yoda K., Horie R., et al. (2005). Enhanced salt tolerance mediated by AtHKT1 transporter-induced Na unloading from xylem vessels to xylem parenchyma cells. Plant J. 44 928–938 10.1111/j.1365-313X.2005.02595.x [DOI] [PubMed] [Google Scholar]
- Tholema N., Vor der Brüggen M., Mäser P., Nakamura T., Schroeder J. I., Kobayashi H., et al. (2005). All four putative selectivity filter glycine residues in KtrB are essential for high affinity and selective K+ uptake by the KtrAB system from Vibrio alginolyticus. J. Biol. Chem. 280 41146–41154 10.1074/jbc.M507647200 [DOI] [PubMed] [Google Scholar]
- Tsugane K., Kobayashi K., Niwa Y., Ohba Y., Wada K., Kobayashi H. (1999). A recessive Arabidopsis mutant that grows photoautotrophically under salt stress shows enhanced active oxygen detoxification. Plant Cell 11 1195–1206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsunekawa K, Shijuku T., Hayashimoto M., Kojima Y., Onai K., Morishita M., Ishiura M., et al. (2009). Identification and characterization of the Na+/H+ antiporter Nhas3 from the thylakoid membrane of Synechocystis sp. PCC 6803. J. Biol. Chem. 284 16513–16521 10.1074/jbc.M109.001875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uozumi N., Dreyer I. (2012). “Structure–function correlates in plant ion channels,” in Comprehensive Biophysics, Vol.6, Channel Proteins ed. Egelman E. (Amsterdam: Elsevier BV; ) 234–245 [Google Scholar]
- Uozumi N., Kim E. J., Rubio F., Yamaguchi T., Muto S., Tsuboi A., et al. (2000). The Arabidopsis HKT1 gene homolog mediates inward Na+ currents in Xenopus laevis oocytes and Na+ uptake in Saccharomyces cerevisiae. Plant Physiol. 122 1249–1259 10.1104/pp.122.4.1249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uozumi N., Schroeder J. I. (2010). “Ion channels and plant stress: past, present and future,” in Ion Channels and Plant Stress Responses eds Demidchik V., Maathuis F. (Berlin: Springer; ) 1–22 [Google Scholar]
- Vera-Estrella R., Barkla B. J., García-Ramírez L., Pantoja O. (2005). Salt stress in Thellungiella halophila activates Na+ transport mechanisms required for salinity tolerance. Plant Physiol. 139 1507–1517 10.1104/pp.105.067850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S. J., Ding L., Zhu J. K. (1996). SOS1, a genetic locus essential for salt tolerance and potassium acquisition. Plant Cell 8 617–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H., Jiang X., Zhan K., Cheng X., Chen X., Pardo J. M., et al. (2008). Functional characterization of a wheat plasma membrane Na+/H+ antiporter in yeast. Arch. Biochem. Biophys. 473 8–15 10.1016/j.abb.2008.02.018 [DOI] [PubMed] [Google Scholar]
- Yamaguchi T., Aharon G. S., Sottosanto J. B., Blumwald E. (2005). Vacuolar Na+/H+ antiporter cation selectivity is regulated by calmodulin from within the vacuole in Ca2+- and pH-dependent manner. Proc. Natl. Acad. Sci. U.S.A. 102 16107–16112 10.1073/pnas.0504437102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi T., Apse M. P., Shi H., Blumwald E. (2003). Topological analysis of a plant vacuolar Na+/H+ antiporter reveals a luminal C terminus that regulates antiporter cation selectivity. Proc. Natl. Acad. Sci. U.S.A. 100 12510–12515 10.1073/pnas.2034966100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi T., Blumwald E. (2005). Developing salt tolerant crop plants: challenges and opportunities. Trends Plant Sci. 10 615–620 10.1016/j.tplants.2005.10.002 [DOI] [PubMed] [Google Scholar]
- Yamaguchi T., Fukada-Tanaka S., Inagaki Y., Saito N., Yonekura-Sakakibara K., Tanaka Y., et al. (2001). Genes encoding the vacuolar Na+H+ exchanger and flower coloration. Plant Cell Physiol. 42 451–461 10.1093/pcp/pce080 [DOI] [PubMed] [Google Scholar]
- Yang Q., Chen Z. Z., Zhou X. F., Yin H. B., Li X. F., Xin X. F., et al. (2009). Overexpression of SOS (salt overly sensitive) genes increases salt tolerance in transgenic Arabidopsis. Mol. Plant 2 22–31 10.1093/mp/ssn058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo A. R. (1999). Predicting the interaction between the effects of salinity and climate change on crop plants. Sci. Hortic. (Amsterdam) 78 159–174 10.1016/S0304-4238(98)00193-9 [DOI] [Google Scholar]
- Zhang H. H., Hodson J. N., Williams J. P., Blumwald E. (2001). Engineering salt-tolerant Brassica plants: characterization of yield and seed oil quality in transgenic plants with increased vacuolar sodium accumulation. Proc. Natl. Acad. Sci. U.S.A. 98 12832–12836 10.1073/pnas.231476498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H. X., Blumwald E. (2001). Transgenic salt-tolerant tomato plants accumulate salt in foliage but not in fruit. Nat. Biotechnol. 19 765–768 10.1038/90824 [DOI] [PubMed] [Google Scholar]