Summary
Natural epigenetic variation provides a source for the generation of phenotypic diversity, but to understand its contribution to phenotypic diversity, its interaction with genetic variation requires further investigation. Here, we report population-wide DNA sequencing of genomes, transcriptomes, and methylomes of wild Arabidopsis thaliana accessions. Single cytosine methylation polymorphisms are unlinked to genotype. However, the rate of linkage disequilibrium decay amongst differentially methylated regions targeted by RNA-directed DNA methylation is similar to the rate for single nucleotide polymorphisms. Association analyses of these RNA-directed DNA methylation regions with genetic variants identified thousands of methylQTL, which revealed the first population estimate of genetically dependent methylation variation. Analysis of invariably methylated transposons and genes across this population indicates that loci targeted by RNA-directed DNA methylation are epigenetically activated in pollen and seeds, which facilitates proper development of these structures.
Introduction
DNA methylation is a covalent base modification of plant nuclear genomes that is accurately inherited through both mitotic and meiotic1 cell divisions. However, similar to spontaneous mutations in DNA, errors in the maintenance of methylation states result in the accumulation of single methylation polymorphisms (SMPs) over an evolutionary timescale2,3. The rates of SMP formation are orders of magnitude greater than spontaneous mutations, which are in part, likely due to the fidelity of maintenance DNA methyltransferases and accompanying silencing machinery2,3,4,5. Epiallele formation in the absence of genetic variation can result in phenotypic variation, which is most evident in the plant kingdom as exemplified by the peloric and colorless non-ripening variants from Linaria vulgaris and Solanum lycopersicum, respectively6,7. Although rates of spontaneous variation in DNA methylation and mutation can be decoupled in the laboratory8,9,10,11, in natural settings, these two features of genomes co-evolve to create phenotypic diversity upon which natural selection can act. In plant genomes, DNA methylation is present in the symmetrical CG and CHG contexts (where H = A, C, or T ) as well as the asymmetrical CHH context. CG gene-body methylation is a common feature of animal and plant genomes12,13. Regions of plant genomes that contain methylation in the CG, CHG and CHH contexts are indicative of loci that are under control of RNA-directed DNA methylation (RdDM)14.
Similar to the limited examples of pure epialleles (methylation variants that form independent of genetic variation), few examples of DNA methylation variants linked to genetic variants are known15,16,17. Previous studies between two accessions of Arabidopsis thaliana or Zea mays revealed genome-wide natural variation in DNA methylation18,19,20,21, but the dependence of these methylation variants on genetic variants at the population level remains unaddressed. To understand the types and extent of natural DNA methylation variants in Arabidopsis thaliana, epigenomes for genotypically distinct, wild accessions, isolated from throughout the northern hemisphere were determined using MethylC-seq18, (152 methylomes, Supplementary Table 1), RNA-seq (144 transcriptomes, Supplementary Table 2) and gDNA-seq (217 genomes, Supplementary Table 3)18. Integration of genomic and epigenomic data allowed investigation into variable methylation states of both CG gene-body methylation and loci targeted by RdDM along with their interactions with genetic variants at the population level.
Population-wide patterns of SMPs
Recent reports of SMPs in a population of essentially isogenic plants indicated that they are major contributors to epigenomic variation2,3. Therefore, we assessed SMP diversity to understand their frequency and patterns throughout a population of genetically distinct accessions. A median of 390,255 SMPs ranging from 92,646 to 527,393 (Supplementary Table 4) were found in the sequenced accessions when compared to the Col-0 reference methylome. On average, CG-, CHG-, and CHH-SMPs accounted for 23%, 13%, and 64% of all SMPs, respectively. These newly identified SMPs were used to construct an epigenome-based phylogeny and then were compared to a genome-based (SNP) phylogeny (Supplementary Fig. 1–4). A high correlation in the tree structures was specifically observed between CG-SMPs and SNPs as compared to CHG-SMPs or CHH-SMPs and SNPs (Supplementary Table 5).
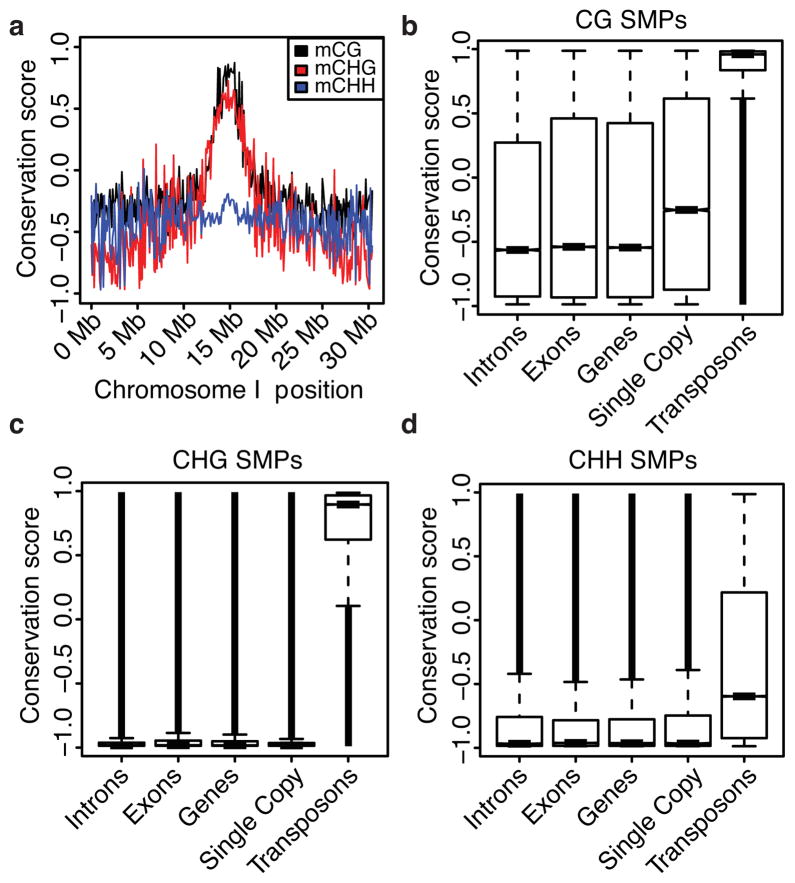
To determine patterns of SMP diversity, chromosome-wide conservation of methylation states at each SMP was examined by computing a conservation score (Fig. 1a and Supplementary Fig. 5). The methylation state of SMPs in the CG and CHG contexts is biased toward the methylated form at the pericentromere and biased toward the unmethylated form in gene-rich regions (Fig. 1a and Supplementary Fig. 5). Next, the distribution of conservation scores across different features and methylation contexts were plotted genome-wide (Fig. 1b–d). Like the pericentromeric regions, CG- and CHG-SMPs in transposable elements tend to be faithfully methylated throughout this population; whereas, CHH-SMPs are largely unmethylated. Unlike CHG and CHH-SMPs, CG-SMPs have a significantly larger amount of methylation at single-copy genes (Fig. 1b–d). Because CG gene-body methylation is associated with moderately expressed genes19, we postulated that these genes are more active because of the lack of other genes redundant in function. We tested this hypothesis by examining RNA-seq data for 144 of these accessions at these loci, which revealed the fraction of transcripts where expression was detected (i.e., FPKMs (Fragments Per Kilobase of exon per Million fragments mapped) > 0) was higher in single-copy genes than non-single-copy genes (85% vs. 71.8%). Moreover, the median expression level of single-copy genes was also significantly greater (361,814.50 FPKMs of single-copy genes vs. 56,107.85 FPKMs of non-single copy genes) supporting the finding that single-copy genes across the population are more transcriptionally active.
Fig. 1. Population-wide analyses of SMPs.
(a) A plot of the genome-wide distribution of methylation conservation across chromosome I. (b–d) The distributions of SMP conservation scores across various genomic features. Notches in the boxplots represent bootstrap estimated 95% confidence intervals around the medians.
Population-wide variation of DMRs
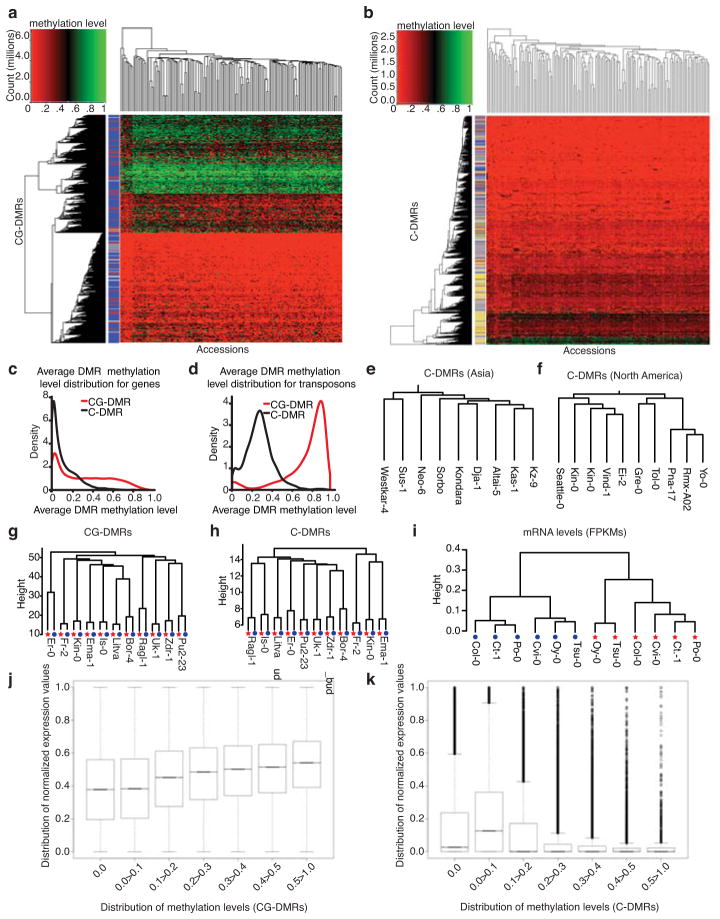
Spontaneous formation of SMPs represents one form of natural epigenetic variation, but variation also exists in the form of differentially methylated regions (DMRs)2,3. Therefore, we scanned this population for DMRs in the CG context (CG-DMRs) typically found in gene-bodies or in the CG, CHG and CHH contexts (C-DMRs) typical of regions targeted by RdDM. Hierarchical clustering of accessions based on weighted methylation levels20 (Supplementary Information), referred to as methylation levels throughout the rest of the paper, of CG-DMRs or C-DMRs revealed patterns across the population that were coincident with certain genomic features (Fig. 2a and b). For example, CG-DMRs are enriched in gene bodies and are present in both unmethylated and methylated states equally throughout the population (Fig. 2a), whereas C-DMRs occur in both gene bodies and transposons (Fig. 2b). Additionally, the C-DMRs in genes are largely unmethylated, which contrast to the heavy methylation levels that occur in transposons (Fig. 2b). In total, 40,269 CG-DMRs (Supplementary Table 6), with an average size of 321 bp, (Supplementary Fig. 6) were identified across the population that were enriched in gene bodies and depleted in transposons (Fig. 2a, Supplementary Figure 7 and Supplementary Table 7); whereas, 13,485 C-DMRs (Supplementary Table 8), with an average size of 221 bp (Supplementary Fig. 6) were identified that show enrichment in transposons and depletion in genes (Fig. 2b, Supplementary Figure 7 and Supplementary Table 7). The distribution of both CG- and C-DMRs reflects the distribution of genes and transposons along each chromosome and the type of DNA methylation primarily associated with these features, CG gene-body methylation versus RdDM. Furthermore, the distribution of methylation levels of CG-DMRs is skewed towards lower levels when the CG-DMR overlaps a gene and higher levels when it overlaps a transposon (Fig. 2c and d). The distribution of methylation levels in CG-DMRs resembles the patterns of CG-SMPs for genes versus transposons as the transposon sequences often contained highly methylated sites or DMRs when compared to genes, supporting the observation that these regions are faithfully repressed by methylation across the population. A comparison of the distribution of methylation levels of the C-DMRs revealed that genes are infrequently methylated at high levels in the population when compared to C-DMRs overlapping transposons (Fig. 2c and d). In this regard, C-DMRs overlapping genes are rare variants in the population; whereas, most transposon sequences are almost invariably methylated. Clustering these accessions based on their methylation levels of C-DMRs revealed that accessions that are geographically separated are less likely to cluster together indicating the potential for underlying genetic structure (Fig. 2e and f). Alternatively, these results could also be obtained for methylation variants that are not dependent on genetic variants if they are stable. Most likely, this result is due to a combination of both of these scenarios.
Fig. 2. Population-wide analyses of DMRs.
Heatmaps representing the methylation levels across CG-DMRs (a) and C-DMRs (b) with coverage in all 152 accessions. The colored row labels on the left of the heatmap indicate what feature a DMR falls in (blue = gene, gold = transposon, red = gene with a transposon inserted in an intron, grey = no feature). Rows indicate genomic locus of DMR and columns indicate accessions. The density (y-axis) and average methylation levels (x-axis) of CG-DMRs and C-DMRs in (c) genes and (d) transposons. Asian (e) and North American (f)) methylome profiles reflected geographical distribution. Dendrograms from hierarchical clustering of CG-DMRs (g), C-DMRs (h) and mRNA levels (i) from accessions that had samples from two different tissues. Red stars and blue circles indicate leaf and mixed stage inflorescence samples, respectively. (j) Boxplot representation of transcriptional variation reveals a positive association with higher levels of methylation levels of CG-DMRs. (k) Increasing methylation levels of C-DMRs is negatively associated with gene expression.
For a subset of accessions examined, methylation data were produced for two tissue types: leaf and mixed-stage inflorescence. Regardless of the tissue used for methylome analysis, when hierarchical clustering was performed using methylation levels of either CG-DMRs (Fig. 2g) or C-DMRs (Fig. 2h), these accessions grouped by their genotype not their tissue type. When the same analysis was applied to RNA-seq data from the same tissues of six accessions, samples clustered based on their tissue type not their genotype (Fig. 2i). Collectively, these data indicate that DNA methylation is less dynamic than gene expression patterns in plants and only plays a role during specific stages of development or cell types1,21,22. Although DNA methylation is more static than transcription, it varies appreciably over an evolutionary timescale, significantly affecting the transcriptional output of specific genes (Fig. 2j and k). Using CG-DMRs that overlap with genes, a positive correlation (Spearman correlation; P value < 2.2e−16) between their methylation levels and gene expression levels were found (Fig. 2j); whereas, the opposite was true for C-DMRs that overlapped genes, supporting a role for RdDM in transcriptionally silencing these loci (Spearman correlation; P value < 2.2e−16, Fig. 2k and Supplementary Fig. 8 and 9 and Supplementary Information). Although the role of CG gene-body methylation is still unclear, these data indicate that CG-DMRs that are heavily methylated are associated with higher gene expression levels and can possibly give rise to transcriptional variation.
Linking genetic and methylation variants
Genome sequencing was performed for 217 individuals of which 152 had a matching sequenced DNA methylome. We used the SHORE analysis pipeline23 to identify SNPs between each accession and the Col-0 genome (Supplementary Information). The identification of SMPs and SNPs that were variable between at least two accessions was used to determine the population-level frequency of these variants, which revealed approximately 70% of CG-SMPs and 41% of SNPs are present at <1% allele frequency (Supplementary Table 9). These results indicate that a large fraction of SMPs and SNPs are rare variants similar to the results observed for C-DMRs and further indicate that the high epimutation rate for SMPs results in greater numbers of rare alleles. Therefore, even though the spontaneous epimutation rate is at least four orders of magnitude greater than SNPs, the reversible nature of certain SMPs governs their accumulation within populations2,3,5.
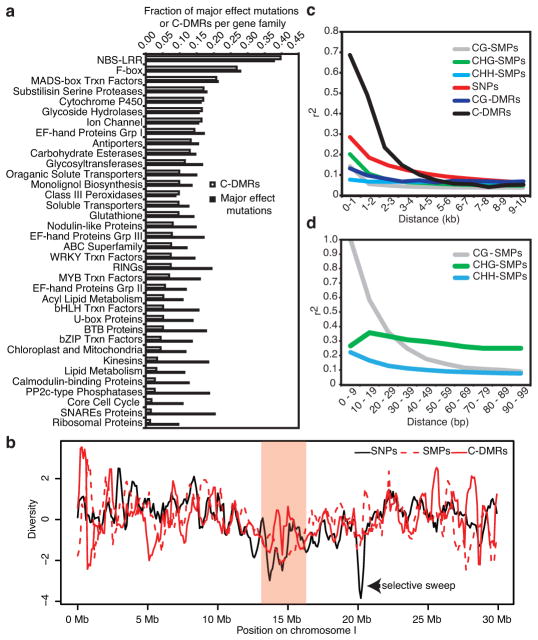
Analysis of gene families that contained the highest number of major effect mutations (Supplementary Information, NBS-LRR – defense response, F-box – protein degradation and MADS-box transcription factor - development) is consistent with previous studies24,25, and these gene families also contained the highest frequency of C-DMRs (Fig. 3a). Furthermore, gene ontology analysis for genes overlapping with C-DMRs identified terms enriched in protein degradation and immune response functions indicating that these three genes families are equally prone to hypervariable genetic and epigenetic states (hypermutable) (Supplementary Table 10). Although the frequency of major effect mutations and C-DMRs was similar for these hypermutable families, the remaining gene families tested revealed no such co-occurrence of genetic and methylation variation as the frequency of C-DMRs approached zero; whereas, the frequency of major effect mutations reached a background rate (Fig. 3a). Aside from the hypermutable families discussed above, there is little relationship between major effect mutations and frequency of C-DMRs. Furthermore, there is no correlation between methylation level and mutation rate in genes containing C-DMRs (Supplementary Table 11). Therefore, the majority of genes targeted by RdDM are functional and silencing by this pathway may limit their expression to specific stages of development similar to observations made for transposons26 and/or limit their expression until released from silencing by bacterial infection27, possibly explaining the high frequency of C-DMRs in members of the NBS-LRR family.
Fig. 3. Population-wide analyses of natural genetic and epigenetic variation.
(a) A comparison of genetic and epigenetic variation across gene families that have the highest incidence of C-DMRs. The y-axis indicates the gene family and the x-axis represents the relative fraction of major effect mutations or fraction of C-DMRs. Fractions were used to adjust for the size of the gene families. (b) A plot of SNP, SMP, and C-DMR diversity across chromosome I. The shaded pink region indicates the position of the pericentromere. (c and d) Linkage disequilibrium/positional association decay plots for genetic and epigenetic variants.
To determine the extent to which variation in both DNA methylation and genotype are linked, diversity estimates were calculated for SNPs, all forms of SMPs and C-DMRs (Fig. 3b and Supplementary Fig. 10). A known selective sweep on chromosome I26 was identified (Fig. 3b). However, no corresponding depletion was observed for either CG-SMPs or C-DMRs. At this resolution, no correlation between genotype and epigenotype was detected (Supplementary Table 12). Therefore, to understand the relationship and possible dependence of methylation variants on genotype, a higher resolution positional association and linkage disequilibrium (LD) decay analysis was performed using SNPs, CG-SMPs, CHG-SMPs, CHH-SMPs, CG-DMRs and C-DMRs (Fig. 3c and d). Similar to past reports for SNPs, LD decays within 10 kb reaching 50% of its initial value at ~2 kb25,28 (Fig. 3c). This value is similar to the rate of decay for the association amongst C-DMRs (~10 kb), which reaches 50% of its initial value at ~1 kb (Fig. 3c). Surprisingly, the rate of decay for association amongst methylation variants such as CG-SMPs and CG-DMRs occurs rapidly, within 100 bp, which is especially true for genes when compared to transposons (Fig. 3d and Supplementary Fig. 11 and 12). Collectively, these data indicate that SMPs and CG-DMRs are truly epigenetic in nature as they occur largely independent of genetic variation. In contrast, although spontaneous C-DMR formation can occur independent of genetic variation2,3, the LD and association decay analysis revealed that the presence of C-DMRs may be due, in part, to local genetic variants.
Association mapping methylation variants
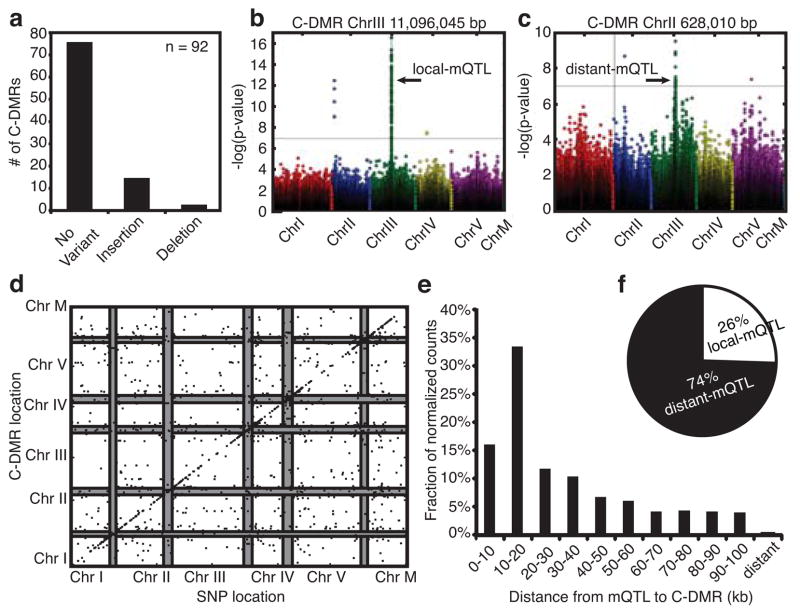
Although there are many mechanisms that can give rise to DNA methylation variation2,3,15,29, the extent to which each plays a role in the formation of the observable methylation variation is unknown. We noted that some sites of known transposition events possessed C-DMRs and posited that these structural variants could be responsible for these differences (Supplementary Fig. 13). To experimentally determine the proportion of C-DMRs with a local structural variant, regions surrounding 92 C-DMRs were PCR amplified and sequenced. Most of these C-DMRs failed to overlap with structural variants; however, structural variations were detected at ~17% (16 / 92) of the C-DMRs assayed (Fig. 4a, Supplementary Table 13). To better inspect any direct relationship between genetic variants and C-DMRs and to identify potential methylQTL (mQTL)30, we utilized a genome-wide association technique, EMMAX, as this methodology was successfully used in another similarly sized Arabidopsis population28,31 (Supplementary Information). Furthermore, we employed two different methodologies to control for false discoveries and found them highly concordant (Supplementary Information). To minimize the number of false positives, we used SNPs that were significant in both methodologies. Application of EMMAX to the 152 accessions with SNP and C-DMR data uncovered C-DMRs that associated with local (Fig. 4b) and distant genetic variants (Fig. 4c) and identified the well-characterized PAI epialleles (Supplementary Fig. 14)15. In total, 2,739 significant mQTL were associated with 1,045 of the 3,023 tested C-DMRs (~35%) (Supplementary Fig. 15–24).
Fig. 4. Association of natural genetic variants and methylation variants.
(a) A summary of the type and number of variants (non-SNPs and small indels) discovered at 92 C-DMRs. Manhattan plots with examples of (b) local and (c) distant mQTL. (d) Distribution of significant mQTL and the C-DMRs with which they associate. Each point represents a significant association between a C-DMR and a block of SNPs. The x-axis denotes the genomic location of the SNP block, and the y-axis indicates the position of the C-DMR. The pericentromeres on each chromosome are shown as grey bars. (e) The distribution of distances of mQTL from their C-DMRs normalized for the base space covered by each range of distances. (f) The ratio of distant mQTL to local mQTL.
Of the tested C-DMRs, 377 (~12%) overlap with a genomic locus with which they associate, which is a similar proportion to the number of experimentally determined local variants. We grouped significant mQTL into blocks and plotted the position of these blocks and the corresponding C-DMR in Fig. 4d (Supplementary Information). An enrichment of local mQTL is visible in particular at the pericentromeric regions (Fig. 4d). When corrected for the genome space in which local events can occur, local mQTL account for a larger fraction of the overall results; although, the raw number of distant mQTL exceeds the number of local mQTL (Fig. 4f). Furthermore, 61.3% of the local mQTL occur within 30 kb of the C-DMR (Fig. 4e). These association-mapping results also indicated that there were more than twice as many mQTL than C-DMRs. To address whether or not many of the C-DMRs are being controlled in a polygenic manner, we applied the tool MLMM32 to the 1,045 C-DMRs with at least one mQTL. Roughly half of the significant C-DMRs reported as polygenic by EMMAX were also reported as polygenic by MLMM (Supplementary Fig. 25). Given these results, there are polygenic C-DMRs, although it remains to be determined what types of mechanisms lead to the methylation variation of these C-DMRs. Lastly, applying EMMAX to CG-DMRs resulted in a much lower detection rate of mQTL (Supplementary Table 14 and Supplementary Fig. 26). Together, the above data demonstrate that a considerable fraction of C-DMRs and to a much lesser extent CG-DMRs exist as a result of genetic variation.
All C-DMRs randomly selected for Fig. 4a are rare in the population and had been filtered out prior to association mapping. Consequently, to determine potential causal variants that are associated with the methylation variants, we PCR amplified 96 C-DMRs associated with a local mQTL. Of these tested loci, 86 successfully amplified and revealed 16 structural variants (Supplementary Table 15), which are similar to the results from the randomly selected C-DMRs (16/92 versus 16/86). Alternative to structural variation, distant mQTL may result from SNPs as reported for the VIM1 variant in the Bor-4 accession33. Analysis of components with known involvements in DNA methylation within these distant mQTL regions (Supplementary Table 16) revealed VIM3 and AGO2 as possible causal loci. Potential causal variants for the remaining local and distant mQTL likely involve a combination of either SNPs or structural variations that will undoubtedly be uncovered with future whole-genome assemblies.
RdDM targets are activated in pollen
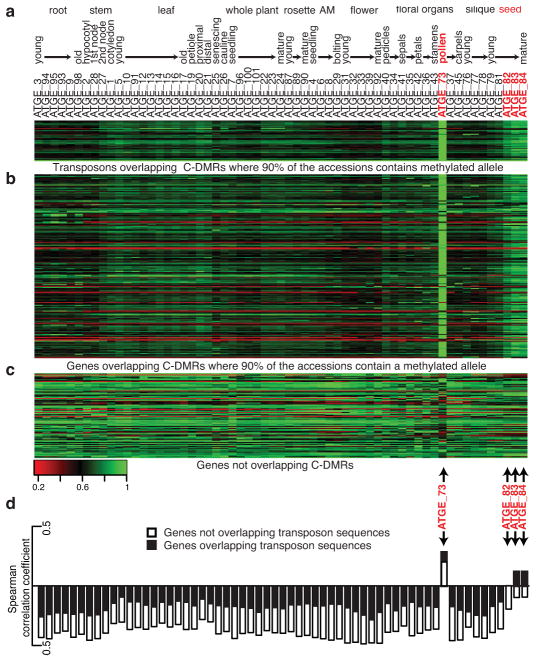
The mQTL identified revealed that there is an association among some genetic variants and DNA methylation variants, especially for C-DMRs. It is well established that other genetic features, such as repeats, are important for guiding RdDM to target loci. For example, the intergenic sub-telomeric repeats 3′ to the MEDEA locus and the repeated SINE elements and tandem repeats around the transcription start site of FWA are key regulatory sequences for controlling gene expression of these loci34,35. Although these loci are under transcriptional control by genetic elements, these specific elements are present and invariably methylated in every accession examined. Therefore, to understand the role of regions of the epigenome that are less prone to natural epigenetic variation we searched for loci that contained methylated alleles (methylation level ≥ 10%) in >90% of the accessions and identified 283 genes and 255 transposons. The expression of these loci was specifically activated in pollen (Fig. 5a and b). A previous study demonstrated that transposons are activated in the pollen vegetative nucleus, providing a substrate to generate mobile small RNAs, which can be transmitted to the sperm cells (germ line)26. This mechanism is not restricted to transposons as we identified protein-coding genes that are under control of RdDM and invariably methylated across this population are also activated in pollen (Fig. 5b). This activation is not a general feature of pollen, as a control set of genes that are not targeted by RdDM are not activated in pollen (Fig. 5c). A closer examination of these invariably methylated genes with gene ontology revealed a significant enrichment for two major categories, cell wall biology and translation (Supplementary Table 17), both related to major functions of pollen development.
Fig. 5. Epigenetic reprogramming of genes targeted by the RdDM pathway.
(a–c) A heatmap representation of mRNA expression levels throughout a developmental time course37 for (a) transposons and (b) genes that overlap with C-DMRs where > 90% of the alleles are methylated across the population and (c) genes not overlapping with C-DMRs. Each row represents a locus with mRNA expression values. Each column represents a different developmental stage. mRNA expression values range from low (red) to medium (black) to high (green). (d) Correlation test between the nonCG methylation levels and microarray gene expression values for genes targeted by RdDM in Col-0. Open rectangles are genes that do not overlap transposon sequences; whereas black rectangles represent genes overlapping transposons sequences.
Although these invariably methylated loci are under similar epigenetic control as transposons (Fig. 5a and b), it is likely that all RdDM-targeted loci are under control of this mechanism regardless of their variability within this population. In fact, Col-0 genes targeted by RdDM and their corresponding expression levels are positively correlated (Spearman correlation; P Value 5.81e−27) in pollen and seed development (Fig. 5d); whereas, all 55 other tissues tested revealed either a negative correlation or no correlation (Fig. 5d, Supplementary Table 18). Furthermore, categories of genes with positive correlations are stronger for loci that overlap transposon sequences (Fig. 5d). These data indicate that these loci have likely come under control of sequences that are evolutionarily silenced, which acts to restrict their expression to these specific stages of development (Fig. 5d – see expanded section and discussion).
Conclusion
Natural epigenomic variation is widespread within Arabidopsis thaliana and the population-based epigenomics presented here has uncovered features of the DNA methylome that are unlinked to underlying genetic variation such as all forms of SMPs and CG-DMRs. However, C-DMRs have positional association decay patterns similar to LD decay patterns for SNPs and in some cases are associated with genetic variants, but the majority of C-DMRs were not tested by association mapping due to low allele frequencies and could result from rare sequence variants. Our combined analyses of genetic and methylation variation did not uncover a correlation between major effect mutations and genes silenced by RdDM suggesting that this pathway may target these genes for another purpose. This purpose could be to restrict expression from vegetative tissues similar to transposons. Another possible purpose of being targeted by RdDM could be to coordinate expression specifically in pollen and in seed to ensure proper gametophytic and embryonic development. Animals also use small RNA-directed DNA methylation and heterochromatin formation mechanisms to maintain the epigenome of the germ line through the use of Piwi-interacting RNAs36. In both plants and animals these small RNAs are derived from the genome of companion cells, which are terminal in nature and can afford widespread reactivation of transposon and repeat sequences as they are not passed on to the next generation. Our study provides evidence that RdDM-targeted genes may have co-opted this transposon silencing mechanism to maintain their silenced state in vegetative tissues and transgenerationally as well as to ensure proper expression important for pollen, seed, and germ line development.
Materials and Methods
Plant material
Leaf and mixed stage inflorescence tissue were flash frozen in liquid nitrogen, and then the tissue was ground to a fine powder with a mortar and pestle. Leaf tissue was used for genomic and RNA-Seq, and the tissues used for each MethylC-Seq experiment is listed in Supplementary Table 1. DNA was isolated using a Qiagen Plant DNeasy kit (Qiagen, Valencia, CA) following the manufacturer’s recommendations. RNA was isolated using the Qiagen Plant RNeasy kit (Qiagen) following the manufacturer’s instructions.
Genomic DNA sequencing library construction
Approximately two micrograms of genomic DNA was sonicated to ~250 bp using the Covaris S2 System using the following parameters: cycle number = 2, duty cycle = 10%, intensity = 4, cycles/burst = 200 and time = 40 seconds. Sonicated DNA was purified with a PCR Purification Minielute column according to the manufacturer’s instructions (Qiagen). Purified DNA was end repaired at room temperature for 45 minutes using the End-It Repair Kit (Epicentre, Madison, WI) and purified with a minielute column (Qiagen). Purified samples were then A-tailed with dATP and Klenow 3′ – 5′ exo minus (New England Biolabs, Ipswich, MA) for 30 minutes at 37C and then purified with a minielute column (Qiagen). Purified DNA was then used for an overnight ligation to TruSeq barcoded adapters (Illumina, San Diego, CA) with T4 DNA ligase at 16C (New England Biolabs). Ligated fragments were purified twice using Ampure XP purification beads (Beckman, Brea, CA) at 1.3X ratio of beads to sample and then PCR amplified for 15 cycles using Phusion High Fidelity DNA Polymerase (New England Bioloabs).
MethylC-Seq library construction
Approximately one to three micrograms of genomic DNA was sonicated to ~100 bp using the Covaris S2 System using the following parameters: cycle number = 6, duty cycle = 20%, intensity = 5, cycles/burst = 200 and time = 60 seconds. Sonicated DNA was purified using Qiagen DNeasy minielute columns (Qiagen). Each sequencing library was constructed similar to genomic DNA libraries except the ligation was performed with methylated adapters provided by Illumina. Ligation products were purified with AMPure XP beads (Beckman) at a ratio of 1.8X of beads to sample. Up to 450 ng of ligated DNA was bisulfite treated using the MethylCode Kit (Invitrogen, Carlsbad, CA) following the manufacturer’s guidelines and then PCR amplified using Pfu Cx Turbo (Agilent, Santa Clara, CA) using the following PCR conditions (2 minutes at 95C, 4 cycles of 15 seconds at 98C, 30 seconds at 60C, 4 minutes at 72C and 10 minutes at 72C).
RNA-Seq library construction
RNA-Seq libraries were prepared according to the described methods in38 except for data collected for Fig. 2i. These libraries were prepared using a TruSeq RNA Sample Kit v2 (Illumina, CA).
Sequencing
Paired-end genomic DNA and single-end MethylC-Seq libraries were sequenced using the Illumina GAIIx (Illumina) as per manufacturer’s instructions. Sequencing of genomic DNA and MethylC-Seq libraries was performed up to 101 and 85 cycles, respectively. Image analysis and base calling were performed with the standard Illumina pipeline. Sequencing of RNA-Seq libraries was performed on the SOLiD4 platform (Life Technologies) for 50 bp according to the manufacturer’s instructions.
Variant identification
The SHORE package was used to call variants for all of our accessions23. The following is a list of each submodule and arguments that we ran for the strains: shore import -v Fastq -e Shore -a genomic -x <forward reads> -y <reverse reads>-o<output directory> -n 200, shore mapflowcell -i <TAIR10 Reference> -f <output directory> -v bwa -n 5% -g 3 -c 7 -b 500000, shore correct4pe –l <input directory> -x 250 -e 1001, shore merge -p <input directory> -d <output directory>, shore consensus -n <accession_name> -f <TAIR10 Reference> -o <output directory> -i <input directory> -g 4 -q 7 -a <Arabidopsis default scoring matrix> -b 0.51 -v –r. Any variant with a quality score of 25 or above was deemed significant. These variants were then substituted into the TAIR10 reference genome to create sample specific references (also referred to as SNP-substituted references) for the mapping of other data sets. In the case of the MethylC-seq mapping, we were able to map, on average, an additional 943,182 reads and allowed us to call an additional 225,894 methylated cytosines (Supplementary Table 19).
MethylC-Seq sequencing analysis
Fastq files were aligned to SNP-substituted reference genomes for each accession using Bowtie39, and custom algorithms were used for identification of mC sites as described previously40.
RNA-Seq data analysis
Bioscope version 1.3 was used to align .csfasta and .qual files to SNP-substituted reference genomes for each accession using default parameters which allows up to 10 locations per sequenced read. Cufflinks version 1.1 was used to quantify gene expression values using the following parameters: -F 0 –b –N –library-type fr-secondstrand –G TAIR10.gtf.
Identification of SMPs
We identified SMPs by looking for sites that either were called methylated by our pipeline, or were covered by at least five reads, which we defined as an unmethylated site. Any other site was listed as missing. A SMP was defined as any site with an accession that had a methylation state different between at least two accessions but contained the same sequence as the Col-0 reference genome.
Dendrogram construction
Throughout this work, we present various clustering results of SMPs, SNPs, and DMRs. In the cases where these dendrograms are presented with a heatmap, we used the R function heatmap.2 in the gplots package with the default clustering parameters to produce the figure. The dendrograms that lack heatmaps were produced by first generating a distance matrix with R’s dist function and passing this matrix to the hclust function, both with their default parameters.
Clustering comparison
To compare the results of the clustering of SMPs and SNPs, we generated distance matrices using R’s dist function with the methylation statuses of SMPs as well as the alleles of the SNPs and then compared the spearman correlation coefficients between the SNP distance matrix and each of the SMP distance matrices (Supplementary Table 5).
Identification of DMRs
All classes of DMRs were identified as previously reported3. CG-DMRs and C-DMRs are not mutually exclusive because C-DMRs are a subset of CG-DMRs. Consequently, for any CG-DMR analyses the subset of C-DMRs were removed.
Definition of methylation levels
Throughout this work, we refer to the level of methylation of genomic regions. To compute this level for a given region, we summed the number of sequenced C bases across all cytosines that were called statistically significantly methylated by our pipeline and divided that sum by the number of sequenced bases covering all cytosines in the given region.
Relationship between DNA methylation and mutation
In an attempt to look at the relationship between mutation and DNA methylation, we calculated the weighted average of DNA methylation and mutation rates across all genes. Genes were defined as entries in the TAIR10 reference GFF file having the word “gene” in the feature column. Methylation levels were calculated as described above, and SNP effects were determined using the SNPeff tool (Cingolani, P. “snpEff: Variant effect prediction”, http://snpeff.sourceforge.net, 2012.) and its athalianaTair10 reference file. We computed two mutation rates, the overall mutation rate and the major effect mutation rate, which we obtained by calculating the fraction of mutations in that gene out of the total number of mutations that were observed in that gene across all accessions. Major effect mutations were defined as mutations that introduced or removed a start or stop codon. The methylation level and mutation rates for each locus were normalized to the maximum value observed at that locus for each measurement type. This normalization yielded measurements on a scale from 0 to 1. We performed a correlation test on these measurements to try and detect a relationship between methylation level and either of the mutation types. As we had no reason to suspect a linear relationship between these variables, we chose to use a Kendall statistic to evaluate the correlation. We detected small but statistically significant relationships between all three of our measurements. Although these results are statistically significant given the small magnitude of the correlation coefficients, we believe that these relationships are at least difficult to interpret but probably not biologically meaningful (Supplementary Table 11).
Enrichment of DMRs in genes and transposons
To determine if CG- and C-DMRs were enriched or depleted in genes or transposons, we performed a binomial test based on these features proportions throughout the genome. The results of these tests can be found in Supplementary Table 7.
LD/positional association decay analysis
To determine the rate of decay for C-DMRs and CG-DMRs we computed a Pearson correlation coefficient between each pair of DMRs within 10 kb of one another. These coefficients were then separated into 1 kb or 200 bp bins based on the distances between the midpoints of the DMRs. We took the median correlation coefficient of each bin as the rate of decay at a particular distance. In the case of SMPs and SNPs, we utilized the software package PLINK to determine the association/LD between all pairs of sites with a minor allele frequency of 20% and that were within 10 kb of one another. In the case of DMRs, we computed the minor allele frequency by first scoring each accession’s DMR as methylated (methylation level >= 10%) or unmethylated (methylation level < 10%). These scores were binned as in the case of DMRs, and the median value of each bin was taken as the decay rate for a particular distance.
DMR saturation analysis
We estimated how close we are to saturating the discovery of DMRs by randomly subsetting our data and calling DMRs on those subsets (Supplementary Fig. 27). For each of the sample sizes, five random subsets were drawn from the samples and run using the same DMR calling pipeline previously outlined. Although the discovery of new CHH-DMRs seems to be saturated, DMRs in the other contexts remain to be found.
mQTL analysis
Given our small sample size, we made several efforts to control for the number of false positives we undoubtedly found. To this end, we only tested DMRs that had at least 75% (114 samples) of their observations present and at least 10% of their observations over a 10% methylation level (i.e., what we defined as a methylated allele). Additionally, we only tested phenotypes that had genomic inflation factors (GIFs) between .985 and 1.015. To obtain these GIFs, we calculated the 50th percentile of each tested C-DMR’s distribution of p-values as well as the 50th percentile of the distribution of p-values generated by randomly permuting the phenotypes of 20 randomly chosen C-DMRs 10 times (200 permutations in total). These filtering steps left us with 3,023 C-DMRs and 1,877 CG-DMRs to test. We then randomly sampled 1% of the p-values tested and input them to the R package Q-Value41. The p-value corresponding to a 1% false discovery rate was then used as a cutoff to determine the significance of each association test (we refer to this methodology as the “Q-Value method”). The results for significant SNPs are detailed in Supplementary Table 20. As further validation to ensure that this methodology was working, we compared it to the randomization method outlined in Breitling et al42 (we refer to the following methodology as the “randomization method”). To this end, we randomized the labels in our genotype matrix (i.e., so every sample now had genotypes from a different, randomly chosen sample) and ran EMMAX on the DMRs that had passed our quality control thresholds. Specifically, we ran those DMRs that had at least 10% of their DMRs in the “methylated” state, at least 75% of their observations present, and a GIF between .985 and 1.015. For each DMR tested, we attempted to find the largest P-Value that kept the false discovery rate (FDR) under 1%. In this case, we defined the FDR of a given P-Value cutoff as the fraction of significant (i.e., below the P-Value cutoff in question) hits found in the randomized set out of the total number of significant hits found in the randomized and non-randomized sets. The results for significant SNPs are detailed in Supplementary Table 21. We found that the methodology employing Q-Value discovered fewer mQTL than the randomization method (Supplementary Table 22), but both methods found a similar proportion of cis and trans mQTL (Supplementary Fig. 28). Furthermore, the Q-Value results are nearly a perfect subset of the randomization results (~93% overlap). Consequently, to be conservative, we utilized the SNPs that overlapped in both methodologies for the analysis in the paper. We grouped these significant SNPs into blocks with the following method. If a significant SNP lies within 10kb of another significant SNP combine these two SNPs into a block (i.e., the block’s start and end are now the positions of these two SNPs). Using this block as a starting point, look for other significant SNPs that are within 10kb of either end of the block. If such SNPs exist, add them to the existing block, update the block ends with the new SNP, and look for significant SNPs within 10kb of these new block ends. Repeat this procedure until no significant SNPs can be found within 10kb of the block ends. These blocks are what we refer to as mQTL throughout the paper. To prioritize candidate loci for follow up studies, we have listed all genes (i.e., protein-coding genes defined in the file here ftp://ftp.arabidopsis.org/home/tair/Genes/TAIR10_genome_release/TAIR10_gff3/TAIR10_GFF3_genes.gff) that fall within the mQTL blocks defined by these significant SNPs, the number of significant SNPs that directly overlap these genes, and whether or not they have been implicated in DNA methylation processes (Supplementary Tables 16 and 23). To better address the validity of mQTL that associated with more than one mQTL, we ran the 1,045 C-DMRs with at least one significant mQTL through the MLMM software provided in32. When evaluating results from this program, we chose the model that minimized the EBIC criterion reported. We used the same P-Value cutoff given by the Q-Value method above to determine which results were significant and collapsed them in the same fashion as mentioned above. We have included the individual results for the significant SNPs in Supplementary Table 24.
Expression of genes containing DMRs
The lists of C-DMRs and CG-DMRs were use to find the overlap between them and a list of protein coding genes (i.e., genes with the “protein-coding gene” descriptor in the TAIR10 reference annotation file found here ftp://ftp.arabidopsis.org/home/tair/Genes/TAIR10_genome_release/TAIR10_gff3/TAIR10_GFF3_genes.gff). We then compared the methylation level of these DMRs with the expression levels of the genes they overlapped. We created boxplots of the expression levels for various methylation levels (e.g., the expression values for all genes with a DMR that had a methylation level greater than 0.2 but less than 0.3). All the expression values of a locus were divided by the maximum observed value at that locus, so the expression values plotted are the fraction of the maximum expression level observed at a given locus. It is interesting to note that genes with no C methylation are expressed at a lower level than those that have a methylation level between (but excluding) 0 and 0.1. This dip is due to genes that have no gene body (i.e., CG methylation) as has been shown in20 and is also apparent in these loci (Supplementary Fig. 8). Consequently, we plotted these data again excluding those sites without gene body methylation (i.e., 0 now represents loci with no CHG or CHH methylation) and saw the median expression rise to match the median expression level at the 0 to 0.1 level (Supplementary Fig. 9). To make the differences in the medians clearer, we have plotted the median values for the boxplots in Figure 2k and 2j along with the bootstrap confidence intervals in Supplementary Fig. 29 and 30.
Developmental gene expression profiling
Microarray analysis was previously performed for a broad range of developmental stages throughout the plant life cycle37. These data were downloaded from http://www.weigelworld.org/resources/microarray/AtGenExpress/AtGE_dev_gcRMA.txt.zip/at_download/file. These lists of loci that are targeted by the RdDM pathway were matched against probe IDs and the resulting information was extracted. Triplicate data for each developmental time point was average and then row normalized according to the developmental time point that displayed the highest expression level and then plotted as a heatmap.
Analysis of local sequence variants at C-DMRs overlapping genes
Primer sets were designed and used for PCR amplification of 92 methylated C-DMRs and for amplification of 86 C-DMRs with local mQTL. Individual PCR products were purified with a PCR purification column (Qiagen) and then sequenced with Sanger sequencing technology. All primer sets can be found in Supplementary Table 13 and 15.
SMP conservation
To get a global look at the diversity of methylation across each chromosome, we binned cytosine positions into 10 kb windows. To examine the conservation of methylation state at cytosines throughout the genome, we computed a score for each site. Any cytosine that had less than five reads covering it was excluded. We used the following formula to estimate the amount of conservation at each site that was missing data from no more than 50 samples: (count(methylated accessions) −count(unmethylated accessions)) / (count(methylated accessions) + count(unmethylated accessions)). This score reaches its maximum value of 1 when all accessions are methylated and a minimum of −1 when all accessions are unmethylated. We computed this score for each site within a bin (Fig. 1a, Supplementary Fig. 5) and then averaged those statistics together. The distributions of these scores are plotted across features in Fig. 1b, 1d, and 1e.
Genome-wide running correlation of SMP, SNP, and C-DMR diversity measures
To evaluate how the correlation between the diversity measures calculated for SMPs, SNPs, and C-DMRs changed across the genome, we calculated diversity measures in the same way as in Figure 3b, but in 100kb windows offset by 20kb instead of 500kb windows offset by 100kb. We changed the window size and offset in order to generate more points with which to perform correlation tests. First, we calculated the percentiles of all the diversity measures. Next, we performed a Kendall Tau correlation test on these percentiles for all windows that started within 500kb (upstream or downstream) of a genomic coordinate (listed as the Window Center in Supplementary Table 25). The coefficients from these tests as well as their P-Values are listed in Supplementary Table 25.
Supplementary Material
Acknowledgments
We thank Chongyuan Luo, Robert Dowen, and Naden Krogan for critical reading of this manuscript; Brian Coullahan for technical assistance with SOLiD RNA-seq; and Stephan Ossowski, Korbinian Schneeberger, and Detlef Weigel for assistance in establishing a variant calling pipeline. RJS was supported by NIH postdoctoral fellowships F32HG004830 and K99GM100000. MDS was supported by an NSF IGERT training grant (DGE-0504645). OL and NJS are supported by NIH/NCRR Grant Number UL1 RR025774. This work was supported by the NSF (MCB-0929402 and MCB-1122246), the Howard Hughes Medical Institute and the Gordon and Betty Moore Foundation to JRE. JRE is a HHMI-GBMF Investigator.
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Author Contributions. RJS, MDS and JRE conceived and designed the study. RJS, MAU, JRN, AA, RBM performed experiments. RJS, MDS, MP, OL, HC and NJS performed data analysis. RJS, MDS and JRE wrote the paper.
Genome sequence data can be downloaded from NCBI SRA (SRA012474). Processed datasets can be viewed at http://neomorph.salk.edu/1001_epigenomes.html and http://signal.salk.edu/atg1001/index.php.
Reprints and permissions information is available at www.nature.com/reprints.
The authors declare no competing financial interests.
References
- 1.Calarco JP, et al. Reprogramming of DNA methylation in pollen guides epigenetic inheritance via small RNA. Cell. 2012;151:194–205. doi: 10.1016/j.cell.2012.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Becker C, et al. Spontaneous epigenetic variation in the Arabidopsis thaliana methylome. Nature. 2011;480:245–249. doi: 10.1038/nature10555. [DOI] [PubMed] [Google Scholar]
- 3.Schmitz RJ, et al. Transgenerational epigenetic instability is a source of novel methylation variants. Science. 2011;334:369–373. doi: 10.1126/science.1212959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Genereux DP, Miner BE, Bergstrom CT, Laird CD. A population-epigenetic model to infer site-specific methylation rates from double-stranded DNA methylation patterns. Proc Natl Acad Sci U S A. 2005;102:5802–5807. doi: 10.1073/pnas.0502036102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ossowski S, et al. The rate and molecular spectrum of spontaneous mutations in Arabidopsis thaliana. Science. 2010;327:92–94. doi: 10.1126/science.1180677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cubas P, Vincent C, Coen E. An epigenetic mutation responsible for natural variation in floral symmetry. Nature. 1999;401:157–161. doi: 10.1038/43657. [DOI] [PubMed] [Google Scholar]
- 7.Manning K, et al. A naturally occurring epigenetic mutation in a gene encoding an SBP-box transcription factor inhibits tomato fruit ripening. Nat Genet. 2006;38:948–952. doi: 10.1038/ng1841. [DOI] [PubMed] [Google Scholar]
- 8.Johannes F, et al. Assessing the impact of transgenerational epigenetic variation on complex traits. PLoS Genet. 2009;5:e1000530. doi: 10.1371/journal.pgen.1000530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mirouze M, et al. Selective epigenetic control of retrotransposition in Arabidopsis. Nature. 2009;461:427–430. doi: 10.1038/nature08328. [DOI] [PubMed] [Google Scholar]
- 10.Reinders J, et al. Compromised stability of DNA methylation and transposon immobilization in mosaic Arabidopsis epigenomes. Genes Dev. 2009;23:939–950. doi: 10.1101/gad.524609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Teixeira FK, et al. A role for RNAi in the selective correction of DNA methylation defects. Science. 2009;323:1600–1604. doi: 10.1126/science.1165313. [DOI] [PubMed] [Google Scholar]
- 12.Feng S, et al. Conservation and divergence of methylation patterning in plants and animals. Proc Natl Acad Sci U S A. 2010;107:8689–8694. doi: 10.1073/pnas.1002720107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zemach A, McDaniel IE, Silva P, Zilberman D. Genome-wide evolutionary analysis of eukaryotic DNA methylation. Science. 2010;328:916–919. doi: 10.1126/science.1186366. [DOI] [PubMed] [Google Scholar]
- 14.Law JA, Jacobsen SE. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat Rev Genet. 2010;11:204–220. doi: 10.1038/nrg2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bender J, Fink GR. Epigenetic control of an endogenous gene family is revealed by a novel blue fluorescent mutant of Arabidopsis. Cell. 1995;83:725–734. doi: 10.1016/0092-8674(95)90185-x. [DOI] [PubMed] [Google Scholar]
- 16.Woo HR, Richards EJ. Natural variation in DNA methylation in ribosomal RNA genes of Arabidopsis thaliana. BMC Plant Biol. 2008;8:92. doi: 10.1186/1471-2229-8-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martin A, et al. A transposon-induced epigenetic change leads to sex determination in melon. Nature. 2009;461:1135–1138. doi: 10.1038/nature08498. [DOI] [PubMed] [Google Scholar]
- 18.Lister R, et al. Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell. 2008;133:523–536. doi: 10.1016/j.cell.2008.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zilberman D, Gehring M, Tran RK, Ballinger T, Henikoff S. Genome-wide analysis of Arabidopsis thaliana DNA methylation uncovers an interdependence between methylation and transcription. Nat Genet. 2006;39:61–69. doi: 10.1038/ng1929. [DOI] [PubMed] [Google Scholar]
- 20.Schultz MD, Schmitz RJ, Ecker JR. ‘Leveling’ the playing field for analyses of single-base resolution DNA methylomes. Trends Genet. 2012;28:583–585. doi: 10.1016/j.tig.2012.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ibarra CA, et al. Active DNA demethylation in plant companion cells reinforces transposon methylation in gametes. Science. 2012;337:1360–1364. doi: 10.1126/science.1224839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gehring M, Bubb KL, Henikoff S. Extensive demethylation of repetitive elements during seed development underlies gene imprinting. Science. 2009;324:1447–1451. doi: 10.1126/science.1171609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ossowski S, et al. Sequencing of natural strains of Arabidopsis thaliana with short reads. Genome Res. 2008 doi: 10.1101/gr.080200.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cao J, et al. Whole-genome sequencing of multiple Arabidopsis thaliana populations. Nat Genet. 2011;43:956–963. doi: 10.1038/ng.911. [DOI] [PubMed] [Google Scholar]
- 25.Clark RM, et al. Common sequence polymorphisms shaping genetic diversity in Arabidopsis thaliana. Science. 2007;317:338–342. doi: 10.1126/science.1138632. [DOI] [PubMed] [Google Scholar]
- 26.Slotkin RK, et al. Epigenetic reprogramming and small RNA silencing of transposable elements in pollen. Cell. 2009;136:461–472. doi: 10.1016/j.cell.2008.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dowen RH, et al. Widespread dynamic DNA methylation in response to biotic stress. Proc Natl Acad Sci U S A. 2012 doi: 10.1073/pnas.1209329109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Atwell S, et al. Genome-wide association study of 107 phenotypes in Arabidopsis thaliana inbred lines. Nature. 2010;465:627–631. doi: 10.1038/nature08800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ahmed I, Sarazin A, Bowler C, Colot V, Quesneville H. Genome-wide evidence for local DNA methylation spreading from small RNA-targeted sequences in Arabidopsis. Nucleic Acids Res. 2011;39:6919–6931. doi: 10.1093/nar/gkr324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Johannes F, Colot V, Jansen RC. Epigenome dynamics: a quantitative genetics perspective. Nat Rev Genet. 2008;9:883–890. doi: 10.1038/nrg2467. [DOI] [PubMed] [Google Scholar]
- 31.Kang HM, et al. Variance component model to account for sample structure in genome-wide association studies. Nat Genet. 2010;42:348–354. doi: 10.1038/ng.548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Segura V, et al. An efficient multi-locus mixed-model approach for genome-wide association studies in structured populations. Nat Genet. 2012;44:825–830. doi: 10.1038/ng.2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Woo HR, Pontes O, Pikaard CS, Richards EJ. VIM1, a methylcytosine-binding protein required for centromeric heterochromatinization. Genes Dev. 2007;21:267–277. doi: 10.1101/gad.1512007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kinoshita T, et al. One-way control of FWA imprinting in Arabidopsis endosperm by DNA methylation. Science. 2004;303:521–523. doi: 10.1126/science.1089835. [DOI] [PubMed] [Google Scholar]
- 35.Xiao W, et al. Imprinting of the MEA Polycomb gene is controlled by antagonism between MET1 methyltransferase and DME glycosylase. Dev Cell. 2003;5:891–901. doi: 10.1016/s1534-5807(03)00361-7. [DOI] [PubMed] [Google Scholar]
- 36.Vagin VV, et al. A distinct small RNA pathway silences selfish genetic elements in the germline. Science. 2006;313:320–324. doi: 10.1126/science.1129333. [DOI] [PubMed] [Google Scholar]
- 37.Schmid M, et al. A gene expression map of Arabidopsis thaliana development. Nature Genetics. 2005 doi: 10.1038/ng1543. [DOI] [PubMed] [Google Scholar]
Methods References
- 38.Li L, et al. Linking photoreceptor excitation to changes in plant architecture. Genes Dev. 2012;26:785–790. doi: 10.1101/gad.187849.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lister R, et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462:315–322. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Storey JD. A direct approach to false discovery rates. Journal of the Royal Statistical Society. 2002;64:479–498. [Google Scholar]
- 42.Breitling R, et al. Genetical genomics: spotlight on QTL hotspots. PLoS Genet. 2008;4:e1000232. doi: 10.1371/journal.pgen.1000232. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.