Abstract
Across amniotes, sex-determining mechanisms exhibit great variation yet the genes that govern sexual differentiation are largely conserved. Studies of evolution of sex-determining and sex-differentiating genes require an exhaustive characterization of functions of those genes such as FOXL2 and FGF9. FOXL2 is associated with ovarian development and FGF9 is known to play a role in testicular organogenesis in mammals and other amniotes. As a step toward characterization of the evolutionary history of sexual development, we measured expression of FOXL2 and FGF9 across three developmental stages and eight juvenile tissue types in male and female American alligators, Alligator mississippiensis. We report surprisingly high expression of FOXL2 before the stage of embryonic development when sex is determined in response to temperature and sustained and variable expression of FGF9 in juvenile male but not female tissue types. Novel characterization of gene expression in reptiles with temperature-dependent sex determination such as American alligators may inform the evolution of sex-determining and sex-differentiating gene networks as they suggest alternative functions from which the genes may have been exapted. Future functional profiling of sex-differentiating genes should similarly follow other genes and other species to enable a broad comparison across sex-determining mechanisms.
Introduction
A complex network of genes involved with sexual development is moderately conserved across amniotes [Graves and Peichel 2010]. Several studies have shown similar actions of sex-differentiating genes in different species and yet some amniotes determine sex based on chromosomal inheritance while others respond more directly to environmental cues such as incubation temperature [Bull 1983]. Still other groups appear to be susceptible to environmental influences, despite having differentiated sex chromosomes that largely dictate sex of offspring [Quinn et al., 2007; Radder et al., 2008]. Despite the conserved nature of the network of sex-differentiating genes, differences in timing and tissues of expression of certain genes are expected to shed light on the mechanistic differences among clades exhibiting genotypic sex determination (GSD) and temperature-dependent sex determination (TSD).
In GSD species, a sex-determining gene initiates male or female embryonic development whereas sex-differentiating genes continue to shape sexual development subsequent to the action of a sex-determining gene. While sex-differentiating genes are conserved across Amniota, the initial determinant or trigger of sex determination varies within and between GSD and TSD amniotes [Bull 1983; Organ and Janes 2008; Sarre et al., 2004]. Among GSD species, a sex-determining gene initiates the functional cascade of sex-differentiating genes as a result of presence (as in SRY in humans) or dosage (as in DMRT1 in chickens) [Sinclair et al., 1990; Smith et al., 2009]. However, it should be noted that DMRT1 dosage alone may not entirely direct sex determination in chickens or other birds [Clinton et al., 2012; Kuepper et al., 2012]. The male- or female-specific actions of sex-differentiating genes follow that of a sex-determining gene in GSD or a thermally responsive element in TSD, although no such element has been discovered. Functional differences of sex-determining and sex-differentiating genes likely arose from a long history of shifts from one sex-determining mechanism to another, or perhaps from gradual changes in sensitivity to environmental variables such as temperature [Sarre et al., 2004; Radder et al., 2008; Grossen et al., 2011]. Clearly, sex-determining mechanisms and the action of one or a series of sex-determining and/or sex-differentiating genes have changed in amniote history [Janzen and Phillips 2006; Organ and Janes 2008; Pokorna and Kratochvil 2009]. To study the effect of these shifts on the function of sex-differentiating genes or vice versa, a complete characterization of gene function and expression patterns across a range of taxa and developmental stages is essential.
To improve our understanding of the functional profiles of sex-differentiating genes, we characterized for the first time the relative expression of two sex-differentiating genes (Forkhead box protein L2 (FOXL2) and Fibroblast growth factor 9 (FGF9)) in American alligators (Alligator mississippiensis). American alligators exhibit TSD and lack gross chromosomal heteromorphy between males and females [Lang and Andrews 1994]. We chose to examine these two genes because they are widely conserved across amniotes and amplifiable in American alligators, a model species for studies of sex-determining mechanisms [Lang and Andrews 1994].
FOXL2 is closely associated with ovarian development in mammals [Crisponi et al., 2001]. It is expressed in ovarian tissue before folliculogenesis and is thought to be the earliest marker of ovarian development in mammals [Cocquet et al., 2002]. FOXL2-knockout mouse ovaries exhibited abnormal development across four developmental stages, suggesting a sustained effect on ovarian function [Garcia-Ortiz et al., 2009]. FOXL2 has also been implicated in ovarian development in fish [Nakamoto et al., 2006; Yamaguchi et al., 2007] and adult ovarian function in chicken [Govoroun et al., 2004]. Also, expression of FOXL2 was studied within the thermosensitive period during incubation (TSP) in two species of TSD turtles [Rhen et al., 2007; Shoemaker-Daly et al., 2010]. In both turtle species, FOXL2 expression was restricted to ovarian tissue near the end of the TSP.
The other gene, FGF9, plays a role in testicular organogenesis and has been shown to play a greater role across tissue types in mammalian embryos [Colvin et al., 2001]. Specifically, FGF9 is associated with development of Leydig cells within testes and testicular steroidogenesis after birth in mice, such that mice lacking FGF9 exhibit male-to-female sex reversal [Hiramatsu et al., 2009; Lin et al., 2010]. These observations might suggest an expectation of male-biased expression of FGF9 in alligators but the gene has also been shown to play a role in ovarian progesterone production in rats and sex-differential FGF9 expression was not reported in the frog, Rana rugosa, during the sex-determining stage of embryogenesis [Yamamura et al., 2005; Drummond et al., 2007]. Alligators might be expected to exhibit an expression profile of sex-differentiating genes that resembles that of mammals rather than that of amphibians since mammals and reptiles share a more recent common ancestor, despite their differences in mode of sex determination.
Patterns of gene expression across developmental stages contribute important information to the functions of genes uncharacterized in non-model species. For example, how do expression patterns of sex-differentiating genes before and after the TSP compare to patterns of expression within the TSP in which they have been more commonly studied [Valenzuela, 2008; Valenzuela, 2010]? We hypothesize that changes in FGF9 expression will demonstrate greater variability across stages in males than in females and across juvenile tissues as this gene is known to function specifically within testes. Evidence collected so far suggests that the gene has a function in tissues other than testes [Lin et al., 2010] but its expression has not yet been investigated comprehensively and the present study helps to fill that gap. Our experimental design permitted examination of FGF9 and FOXL2 expression in somatic tissues such as kidney and heart for the first time. Garcia-Ortiz et al. [2009] reported expression of FOXL2 at multiple stages of mice but not in multiple tissues whereas our study tests the effects of both stage and tissue type on expression of sex-differentiating genes. Furthermore, here we test the hypothesis that changes in expression levels are detectable in embryos collected before and after the TSP during which sex determination occurs in TSD species (Bull, 1983). The TSP marks the period during which FOXL2 and FGF9 are known to act in a sex-differential pattern in other studies.
Materials and Methods
Animals
Tissues were collected from twelve juvenile (six male & six female) American alligators (Alligator mississippiensis), according to instructions from veterinary staff at Harvard University. Alligators, collected in Rockefeller Refuge, LA in 2007, were selected as juveniles (~61cm in total length); representative of a developmental stage after hatching but before sexual maturity. From each juvenile, ~300mg of brain, heart, lung, liver, muscle, kidney, gastro-intestinal (GI) tract, and gonad-adrenal-mesonephros (GAM) were sampled. In addition, 24 alligator eggs were collected from wild nests and developmental stages of embryos were verified at the moment of collection by visual inspection according to Moore et al. (2008). Samples were collected from alligator embryos that visually approximated stages known to occur before and after the TSP during which sex determination occurs (Bull, 1983). Sexing of juveniles was conducted by external observation of genitalia and confirmed by macroscopic gonadal inspection. Early-stage embryos were not sexed as they were collected before the thermally sensitive sex-determining stage of incubation. Twenty late-stage embryos were incubated at either the female-producing temperature (28°C) of A. mississippiensis for 60 d or the male-producing temperature (33°C) for 44 d. This sampling scheme ensured that embryos developed past their TSP at their respective incubation temperature [Lang and Andrews 1994]. According to Derveaux et al. [2010] these sample sizes should suffice for establishing expression stability in representative samples of different tissues. Incubators were checked for heterogeneity in temperature and humidity and eggs were rotated daily within each incubator to avoid spatial differences in incubation environment. During tissue collection from late-stage alligator embryos, gonadal sex was confirmed by macroscopic inspection (Moore et al., 2008). From each embryo, ~300 mg of gonad and surrounding adrenal and kidney tissue were collected. Tissue samples were macerated with a scalpel, resuspended individually in 1.5ml tubes with 400ul of RNAlater (Sigma-Aldrich Cat. No. R0901), and stored at -80°C.
CDNAs
Samples (250mg) were later thawed on ice, homogenized with a rotor stator, and filtered for total RNA extraction, using an RNeasy Midikit (Qiagen Cat. No. 75144). Concentrations of extracted RNAs were measured with a NanoDrop ND-1000 Spectrophotometer and extracts were visualized on a gel for confirmation of quality by eye. Samples were diluted to equal concentrations of 4.2 ng/ul in order to use 50 ng of RNA as a template for reverse transcription of cDNA using an Omniscript kit (Qiagen Cat. No. 205113). If initial concentrations were lower than 4.2 ng/ul (less than optimal concentration for the Omniscript kit), then RNAs were reverse-transcribed using a Sensiscript kit (Qiagen Cat. No. 205211). Chemistries of these kits are similar except for a polymerase in the Sensiscript kit that is more effective under lower concentrations of template.
Target amplification
Primers were designed using Sequence Alignment Editor software v. 2.0a11 [Rambaut 2007] for housekeeping gene lactate dehydrogenase A (LDHA) (Genbank Accession # L79951) [Mannen et al., 1997] and sex-differentiating genes (Fibroblast growth factor 9 (FGF9) (To be submitted to Genbank with this study) and Forkhead box l2 (FOXL2) (Genbank Accession # EU848473)) (table 1). LDHA has been identified as a common housekeeping reference in mammal studies [Lee et al., 2002; Ren et al., 2010] and exhibits an expression profile in alligators that is similar to ß-tubulin, a housekeeping gene frequently used as a control in qPCR studies [Merchant et al., 2009]. We attempted to use ß-tubulin as a control in our study, but available primers failed to amplify the gene consistently. RNAs were treated with DNA-Free DNAse (Ambion Cat. No. AM1906) before reverse transcription using Qiagen’s One-Step RT-PCR kit (Qiagen Cat. No. 210212). Reverse-transcribed, DNAse-treated samples served as template for PCR to validate primers and confirm amplification of a single clear band as seen in comparison with the same amplification from genomic DNA. Target loci were amplified and quantified by a Stratagene MX3000p thermal cycler using an initial activation step at 95°C for 15 min, followed by 40 cycles of 94°C for 15 s, varying annealing temperatures depending on target (table 1) for 30 s, and 72°C for 30 s. The 40 cycles are followed by one cycle of 95°C for 1 min, 55°C for 30 s, and 95°C for 30 s. Ct values were downloaded from MxPro software v. 4.10. An effort was made to maximize assessment of expression of each gene by measuring expression in as many samples as possible on the same plate in the same qPCR run [Derveaux et al., 2010].
Table 1.
Primers and annealing temperatures used to amplify housekeeping and sex-differentiating genes in American alligators, Alligator mississippiensis
| Locus | Forward | Reverse | Annealing temperature, °C |
|---|---|---|---|
| LDHA | TGTGACTGCAAACTCCAAGC | GCCGCTACCAATAACACGAT | 57.0 |
| FGF9 | CCCAGGAGTGTGTATTCAGAG | CAGCATGTTCCATCCAAGCC | 60.8 |
| FOXL2 | TACTCSTACGTGGCSCTSAT | TTCTCGAACATGTCCTCGCAGG | 57.0 |
Analyses
Dissociation curves estimated the number of different transcripts amplified during one qPCR reaction and thus it provides a measure of the PCR specificity. Sample datapoints were discarded if either the targeted housekeeping gene, the targeted sex-differentiating gene or both produced more than a single product during qPCR as detected by the dissociation data. Ct expression data for sex-differentiating genes were normalized using LDHA Ct data. Sex and developmental stage differences in expression data (delta Ct) for sex-differentiating genes were tested using a t-test for pairwise comparisons or one-way ANOVA for stage comparisons, as implemented in SPSS Statistics 17.0 software [SPSS, 2008]. Sex and developmental stage differences in LDHA Ct data were also tested using the same t-test.
Results
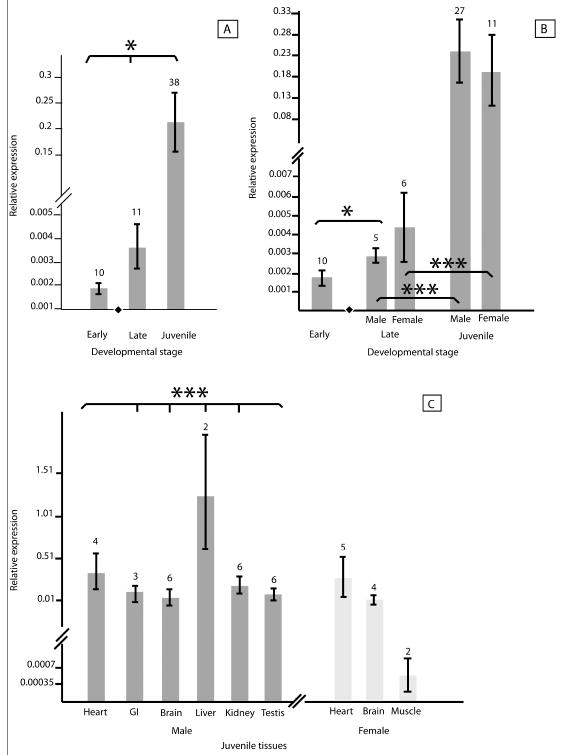
Datapoints were compared between sexes, across developmental stages, and among juvenile tissue types. Relevant values and levels of significance were recorded for each comparison with and without normalization of raw Ct values for FGF9 and FOXL2 with LDHA Ct values to demonstrate the effect of normalization on results (supplementary table 1). Log2-transformed FGF9 expression varied significantly across developmental stages (mean pre-TSP: 1.8 × 10-3; mean post-TSP: 3.7 × 10-3; mean juvenile: 2.2 × 10-1; p < 0.05) (fig. 1a). After the thermosensitive period (TSP), increased expression of FGF9 was less variable in males and significant increases were seen between post-TSP samples and juvenile tissues in both males and females (p < 0.001). Although mean FGF9 expression in late-stage embryos was higher in females (male (M) = 2.9 × 10-3; female (F) = 4.4 × 10-3), the variation (standard error) was also higher across the queried female embryos (fig. 1b), rendering the pattern non-significant. FGF9 expression varied significantly across juvenile tissues in males (p < 0.001) but not in females (fig. 1c).
Fig. 1.
Relative expression in male and female alligators of FGF9 at both early and late stages of embryogenesis and at a juvenile stage. (a) Analysis of FGF9 expression in all queried samples indicates significant variation across developmental stages. (b) Comparison of FGF9 expression before (termed early) and after (termed late) the thermally sensitive period of sex determination indicates a significant increase in FGF9 expression after sex determination among male but not female embryos. Expression increased significantly from late embryos to juvenile tissues in both males and females. (c) Comparing male and female tissue types from juvenile alligators indicates significant variation among male but not female tissues. Means were log2-transformed for ease of visual interpretation. Lines on bars represent standard error. Numbers above bars indicate the number of biological replicates. The black diamond in figures a and b indicates the developmental stage in which sex is determined. Asterisks represent significance (*: p < 0.05; ***: p < 0.001).
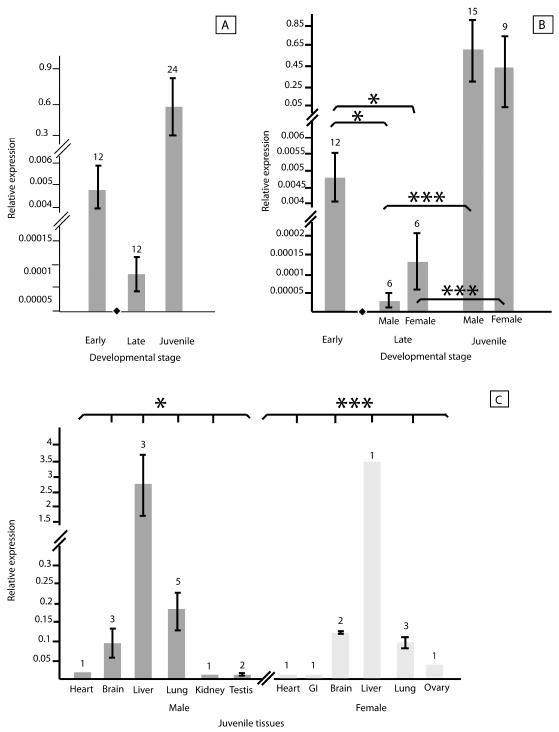
Overall, between-sex and interstage differences in Log2-transformed expression of FOXL2 were not significant across early, late and juvenile stages (mean pre-TSP: 4.8 × 10-3; mean post-TSP: 7.7 × 10-5; mean juvenile: 5.6 × 10-1; fig. 2a) but FOXL2 expression significantly decreased after the TSP (p < 0.05) and then increased in both male and female juveniles (p < 0.0001; fig. 2b). Although late-stage embryonic FOXL2 expression was higher, as expected, in females than in males, the difference was not significant. FOXL2 expression varied significantly among tissues within each sex (M: p < 0.05; F: p < 0.001) but not between male and female juveniles (fig. 2c). Log2-transformed LDHA expression varied significantly across developmental stages overall (mean pre-TSP: 20.0; mean post-TSP: 20.3; mean juvenile: 25.7; p < 0.001) and specifically for both males and females (M & F: p < 0.001; fig. 3a-b), but within stages, expression of LDHA did not differ between males and females (fig. 3b). Samples sizes vary due to conservative inclusion of expression data only of samples in which both amplifications of the sex-differentiating gene and the housekeeping gene yielded clear dissociation curves.
Fig. 2.
Relative expression in male and female alligators of FOXL2 at both early and late stages of embryogenesis and at a juvenile stage. (a) Analysis of FOXL2 expression in all queried samples does not indicate significant variation across developmental stages. (b) Comparison of FOXL2 expression before (termed early) and after (termed late) the thermally sensitive period of sex determination indicates a significant decrease in FOXL2 expression after sex determination among both male and female embryos. Expression increased significantly from late embryos to juvenile tissues in both males and females. (c) Comparing male and female tissue types from juvenile alligators indicates significant variation among both male and female tissues. Means were log2-transformed for ease of visual interpretation. Lines on bars represent standard error. Numbers above bars indicate the number of biological replicates. The black diamond in figures a and b indicates the developmental stage in which sex is determined. Asterisks represent significance (*: p < 0.05; ***: p < 0.001).
Fig. 3.
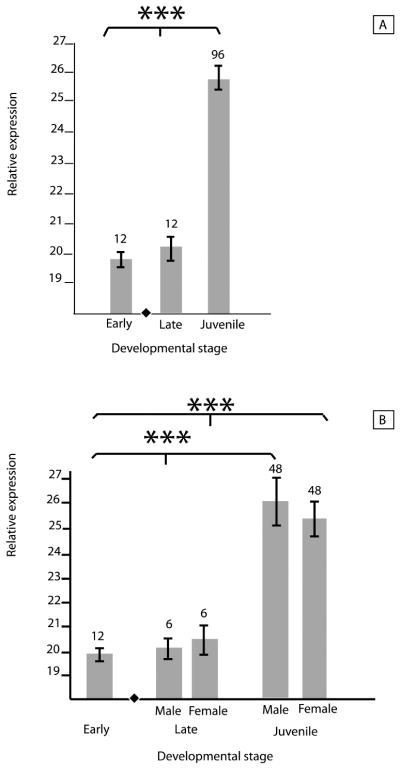
The housekeeping gene LDHA amplified from each queried tissue. (a) LDHA amplification varies by stage, suggestive of ontogenetic differences in cell proliferation (Merchant et al., 2009). (b) Subdivided by sex, the dataset shows stage-differential but not sex-differential expression of LDHA, suggesting the utility of the marker as a normalizer of expression data for genes of interest. Means were log2-transformed for ease of visual interpretation. Lines on bars represent standard error. Numbers above bars indicate the number of biological replicates. The black diamond in each figure indicates the developmental stage in which sex is determined. Asterisks represent significance (***: p < 0.001).
Discussion
Our study revealed sex differences of expression in FGF9 and FOXL2 in both embryonic and juvenile American alligators. FGF9 expression increased in a sex-specific manner after the TSP and differed significantly across juvenile male but not female tissue types. Our data revealed evidence of juvenile FGF9 expression in testes but not ovaries. FGF9 appeared to be acting in a sex-specific manner in both late-stage embryos and juveniles, suggesting a sustained role of FGF9 throughout sexual development of male but not female alligators. By contrast, FOXL2 was expressed at higher levels before the TSP than in either male or female late-stage embryos. This suggested an alternative function of FOXL2, distinct from its role specifically in ovarian development. The dataset for FGF9 expression in juveniles appears to be unbalanced in that more tissues are represented for males than for females (fig 1c). However, the absence of expression data for tissues such as female liver does not represent a lack of attempted amplification. Rather, these tissues were queried equally in males and females but FGF9 was not amplifiable from all samples. We consider absence of expression meaningful in this context.
It should be noted that differences in gene expression between sexes and developmental stages may not necessarily mean differences in function or distinct consequences for phenotypic variation. Gene expression is a complex culmination of many molecular factors, and the level of gene expression differences between species or sexes may have a basis in neutral, stochastic events rather than functional differences driven by natural selection [Bedford and Hartl 2009; Romero et al., 2012]. However, in comparative studies of gene expression, genes that exhibit strong differences in average expression in particular lineages or species are considered candidates for selection-driven differences with functional consequences for development [Perry et al., 2012; Romero et al., 2012]. In this study, the expression levels of FGF9 and FOXL2 in male and female embryos might be considered to exhibit strong differences in average expression and hence are our top candidates for expression differences that may have functional, selection-driven consequences for sex determination in alligators.
If juvenile or non-TSP embryonic patterns of expression are similar to expression patterns within the TSP, it would support the hypothesis that a similar function of the genes as reflected in their expression patterns, is sustained over a longer developmental phase than previously thought. By contrast, expression patterns that are dissimilar to those observed within the TSP would suggest potential alternative or additional functions for those genes during ontogeny. Identification of alternative functions of sex-differentiating genes is especially relevant to the evolution of sex determination because it is commonly accepted that sex-determining and sex-differentiating genes have been repeatedly recruited from different functions [Graves, 2001].
Identifying other functions that are suggested by time-, stage- or tissue-specific expression patterns may illuminate pathways by which sex-related genes are recruited, thereby informing the evolution of transitions between sex-determining mechanisms. For example, the alternative pre-TSP function(s) of FOXL2 has relevance to the study of transitions between mechanisms of sex determination. If genes are co-opted into the sex-differentiating cascade as sex-determining mechanisms evolve, then our result may suggest a function from which FOXL2 was co-opted into the function of sex differentiation. As another example, unlike the two studied sex-differentiating genes, LDHA was found to be expressed in every queried tissue. Relative expression levels of LDHA varied among developmental stages but not between sexes, possibly explained by stage differences in cell proliferation [Merchant et al., 2009]. This pattern supports the characterization of LDHA as a housekeeping gene and validates the use of LDHA as a normalizing factor with which to study relative expression levels of FGF9, FOXL2, and other genes in American alligators. Further, Merchant et al. [2009] reported expression levels of LDHA in American alligator similar to those of ß-tubulin, a confirmed housekeeping gene in this species.
Characterizing the expression of discrete markers under different conditions (i.e., male or female background) is only one approach by which functional genetics can be studied. Microarrays and sequencing of total transcriptomes are also increasingly popular tools for characterizing the functional network of sex-differentiating genes in both TSD and GSD species. Further research is needed encompassing a broader investigation of male and female transcriptomes of American alligators before, during and after the TSP. This research will benefit from whole genome sequencing of American alligators and other crocodilians [St. John et al., 2012]. We expect future transcriptomic studies to further support an early embryonic function of FOXL2 and a sustaining function of FGF9 throughout male development. These characterizations will serve as points of comparison between species and sex-determining mechanisms. By this approach, the evolutionary pathways and frequencies of change in mechanisms can be determined.
Supplementary Material
Acknowledgments
Alligators were donated by Rockefeller State Wildlife Refuge in Grand Chenier, LA. All animal work was approved under Institutional Animal Care and Use Protocol #24-06. David Ottenheimer, David Gonzalez Arroyo, Portia Botchway, Randi Griffin, Chris Organ, Andrew Shedlock, Jae Hur, Courtney Stern, and Lewis Ward assisted with sample collection and benchwork. Funding was provided by a National Institutes of Health National Research Service Award to DEJ (5F32GM072494) and by National Science Foundation Grant MCB-0817687 to NV and SVE.
Literature Cited
- Bedford T, Hartl DL. Optimization of gene expression by natural selection. Proc Natl Acad Sci. 2009;106:1133–1138. doi: 10.1073/pnas.0812009106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bull JJ. Evolution of sex-determining mechanisms. Benjamin/Cummings; California, USA: 1983. [Google Scholar]
- Clinton M, Zhao D, Nandi S, McBride D. Evidence for avian cell autonomous sex identity (CASI) and implications for the sex-determination process? Chromosome Res. 2012;20:177–190. doi: 10.1007/s10577-011-9257-9. [DOI] [PubMed] [Google Scholar]
- Cocquet J, Pailhoux E, Jaubert F, Servel N, Xia X, et al. Evolution and expression of FOXL2. J Med Genet. 2002;39:916–921. doi: 10.1136/jmg.39.12.916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colvin JS, Green RP, Schmahl J, Capel B, Ornitz DM. Male-to-female sex reversal in mice lacking fibroblast growth factor 9. Cell. 2001;104:875–889. doi: 10.1016/s0092-8674(01)00284-7. [DOI] [PubMed] [Google Scholar]
- Crisponi L, Deiana M, Loi A, Chiappe F, Uda M, et al. The putative forkhead transcription factor FOXL2 is mutated in blepharophimosis/ptosis/epicanthus inversus syndrome. Nat Genet. 2001;27:159–166. doi: 10.1038/84781. [DOI] [PubMed] [Google Scholar]
- Derveaux S, Vandesompele J, Hellemens J. How to do successful gene expression analysis using real-time PCR. Methods. 2010;50:227–230. doi: 10.1016/j.ymeth.2009.11.001. [DOI] [PubMed] [Google Scholar]
- Drummond AE, Tellbach M, Dyson M, Findlay JK. Fibroblast growth factor-9, a local regulator of ovarian function. Endocrinology. 2007;148:3711–3721. doi: 10.1210/en.2006-1668. [DOI] [PubMed] [Google Scholar]
- Garcia-Ortiz JE, Pelosi E, Omari S, Nedorezov T, Piao Y, et al. FOXL2 functions in sex determination and histogenesis throughout mouse ovary development. BMC Dev Biol. 2009;9 doi: 10.1186/1471-213X-9-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govoroun MS, Pannetier M, Pailhoux E, Cocquet J, Brillard JP, et al. Isolation of chicken homolog of the FOXL2 gene and comparison of its expression patterns with those of aromatase during ovarian development. Dev Dynam. 2004;231:859–870. doi: 10.1002/dvdy.20189. [DOI] [PubMed] [Google Scholar]
- Graves JAM. From brain determination to testis determination: evolution of the mammalian sex-determining gene. Reprod Fert Develop. 2001;13:665–672. doi: 10.1071/rd01093. [DOI] [PubMed] [Google Scholar]
- Graves JAM, Peichel CL. Are homologies in vertebrate sex determination due to shared ancestry or to limited options? Genome Biol. 2010;11:205. doi: 10.1186/gb-2010-11-4-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossen C, Neuenschwander S, Perrin N. Temperature-dependent turnovers in sex-determination mechanisms: a quantitative tool. Evolution. 2011;65:64–78. doi: 10.1111/j.1558-5646.2010.01098.x. [DOI] [PubMed] [Google Scholar]
- Hiramatsu R, Matoba S, Kanai-Azuma M, Tsunekawa N, Katoh-Fukui Y, et al. A critical time window of Sry action in gonadal sex determination in mice. Development. 2009;136:129–138. doi: 10.1242/dev.029587. [DOI] [PubMed] [Google Scholar]
- Janzen FJ, Phillips PC. Exploring the evolution of environmental sex determination, especially in reptiles. J Evolution Biol. 2006;19:1775–1784. doi: 10.1111/j.1420-9101.2006.01138.x. [DOI] [PubMed] [Google Scholar]
- Kuepper C, Augustin J, Edwards SV, Szekely T, Kosztolanyi A, Burke T, Janes DE. Triploid plover female provides support for a role of the W chromosome in avian sex determination. Biol Lett. 2012;8:787–789. doi: 10.1098/rsbl.2012.0329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang JW, Andrews HV. Temperature-dependent sex determination in crocodilians. J Exp Zool. 1994;270:28–44. [Google Scholar]
- Lee PD, Sladek R, Greenwod CMT, Hudson TJ. Control genes and variability: Absence of ubiquitous reference transcripts in diverse mammalian expression studies. Genome Res. 2002;12:292–297. doi: 10.1101/gr.217802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin YM, Tsai CC, Chung CL, Chen PR, Sun HS, et al. Fibroblast growth factor 9 stimulates steroidogenesis in postnatal Leydig cells. Int J Androl. 2010;33:545–553. doi: 10.1111/j.1365-2605.2009.00966.x. [DOI] [PubMed] [Google Scholar]
- Mannen H, Tsoi SCM, Krushkal JS, Li WH, Li SSL. The cDNA cloning and molecular evolution of reptile and pigeon lactate dehydrogenase isozymes. Mol Biol Evol. 1997;14:1081–1087. doi: 10.1093/oxfordjournals.molbev.a025717. [DOI] [PubMed] [Google Scholar]
- Merchant M, Kinney C, Sanders P. Differential protein expression in alligator leukocytes in response to bacterial lipopolysaccharide injection. Comp Biochem Phys D. 2009;4:300–304. doi: 10.1016/j.cbd.2009.08.001. [DOI] [PubMed] [Google Scholar]
- Moore BC, Uribe-Aranzabal MC, Boggs ASP, Guillette LJ. Developmental morphology of the neonatal alligator (Alligator mississippiensis) J Morphol. 2008;269:302–312. doi: 10.1002/jmor.10583. [DOI] [PubMed] [Google Scholar]
- Nakamoto M, Matsuda M, Wang DS, Nagahama Y, Shibata N. Molecular cloning and analysis of gonadal expression of FOXL2 in the medaka, Oryzias latipes. Biochem Bioph Res Co. 2006;344:353–361. doi: 10.1016/j.bbrc.2006.03.137. [DOI] [PubMed] [Google Scholar]
- Organ CL, Janes DE. Evolution of sex chromosomes in Sauropsida. Integr Comp Biol. 2008;48:512–519. doi: 10.1093/icb/icn041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pokorna M, Kratochvil L. Phylogeny of sex-determining mechanisms in squamate reptiles: are sex chromosomes an evolutionary trap? Zool J Linn Soc-Lond. 2009;156:168–183. [Google Scholar]
- Perry GH, Melsted P, Marioni JC, Wang Y, Bainer R, Pickrell JK, Michelini K, Zehr S, Yoder AD, Stephens M, Pritchard JK, Gilad Y. Comparative RNA sequencing reveals substantial genetic variation in endangered primates. Genome Res. 2012;22:602–610. doi: 10.1101/gr.130468.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinn AE, Georges A, Sarre SD, Guarino F, Ezaz T, Graves JAM. Temperature sex reversal implies sex gene dosage in a reptile. Science. 2007;316:411. doi: 10.1126/science.1135925. [DOI] [PubMed] [Google Scholar]
- Radder RS, Quinn AE, Georges A, Sarre SD, Shine R. Genetic evidence for co-occurrence of chromosomal and thermal sex-determining systems in a lizard. Biol Letters. 2008;4:176–178. doi: 10.1098/rsbl.2007.0583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambaut A. Sequence Alignment Editor v2.0a11. 2007 [Google Scholar]
- Ren S, Zhang F, Li D, Jia C, Li S, et al. Selection of housekeeping genes for use in quantitative reverse transcription PCR assays on the murine cornea. Mol Vis. 2010;16:1076–1086. [PMC free article] [PubMed] [Google Scholar]
- Rhen T, Metzger K, Schroeder A, Woodward R. Expression of putative sex-determining genes during the thermosensitive period of gonad development in the snapping turtle, Chelydra serpentina. Sex Dev. 2007;1:255–270. doi: 10.1159/000104775. [DOI] [PubMed] [Google Scholar]
- Romero IG, Ruvinsky I, Gilad Y. Comparative studies of gene expression and the evolution of gene regulation. Nat Genet. 2012;13:505–516. doi: 10.1038/nrg3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarre SD, Georges A, Quinn A. The ends of a continuum: genetic and temperature-dependent sex determination in reptiles. Bioessays. 2004;26:639–645. doi: 10.1002/bies.20050. [DOI] [PubMed] [Google Scholar]
- Shoemaker-Daly CM, Jackson K, Yatsu R, Matsumoto Y, Crews D. Genetic Network Underlying Temperature-Dependent Sex Determination Is Endogenously Regulated by Temperature in Isolated Cultured Trachemys scripta Gonads. Dev Dynam. 2010;239:1061–1075. doi: 10.1002/dvdy.22266. [DOI] [PubMed] [Google Scholar]
- Sinclair AH, Berta P, Palmer MS, Hawkins JR, Griffiths BL, et al. A gene from the human sex-determining region encodes a protein with homology to a conserved DNA-binding motif. Nature. 1990;346:240–244. doi: 10.1038/346240a0. [DOI] [PubMed] [Google Scholar]
- Smith CA, Roeszler KN, Ohnesorg T, Cummins DM, Farlie PG, et al. The avian Z-linked gene Dmrt1 is required for male sex determination in the chicken. Nature. 2009;461:267–271. doi: 10.1038/nature08298. [DOI] [PubMed] [Google Scholar]
- SPSS, I. SPSS Statistics 17.0. Chicago: 2008. [Google Scholar]
- St John JA, Braun EL, Isberg SR, Miles LG, Chong AY, et al. Sequencing three crocodilian genomes to illuminate the evolution of archosaurs and amniotes. Genome Biol. 2012;13:415. doi: 10.1186/gb-2012-13-1-415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valenzuela N. Evolution of the gene network underlying gonadogenesis in turtles with temperature-dependent and genotypic sex determination. Integr Comp Biol. 2008;48:476–485. doi: 10.1093/icb/icn031. [DOI] [PubMed] [Google Scholar]
- Valenzuela N. Multivariate expression analysis of the gene network underlying sexual development in turtle embryos with temperature-dependent and genotypic sex determination. Sex Dev. 2010;4:39–49. doi: 10.1159/000277935. [DOI] [PubMed] [Google Scholar]
- Yamaguchi T, Yamaguchi S, Hirai T, Kitano T. Follicle-stimulating hormone signaling and FOXL2 are involved in transcriptional regulation of aromatase gene during gonadal sex differentiation in Japanese flounder, Paralichthys olivaceus. Biochem Bioph Res Co. 2007;359:935–940. doi: 10.1016/j.bbrc.2007.05.208. [DOI] [PubMed] [Google Scholar]
- Yamamura Y, Aoyama S, Oshima Y, Kato T, Osawa N, et al. Molecular cloning and expression in gonad of Rana rugosa WT1 and FGF9. Zool Sci. 2005;22:1045–1050. doi: 10.2108/zsj.22.1045. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.