Abstract
The mechanistic interconnectivity between circadian regulation and the genotoxic stress response remains poorly understood. Here we show that the expression of Period 2 (Per2), a circadian regulator, is directly regulated by p53 binding to a response element in the Per2 promoter. This p53 response element is evolutionarily conserved and overlaps with the E-Box element critical for BMAL1/CLOCK binding and its transcriptional activation of Per2 expression. Our studies reveal that p53 blocks BMAL1/CLOCK binding to the Per2 promoter leading to repression of Per2 expression. In the suprachiasmatic nucleus (SCN), p53 expression and its binding to the Per2 promoter are under circadian control. Per2 expression in the SCN is altered by p53 deficiency or stabilization of p53 by Nutlin-3. Behaviorally, p53−/− mice have a shorter period length that lacks stability and they exhibit impaired photo-entrainment to a light pulse under a free-running state. Our studies demonstrate that p53 modulates mouse circadian behavior.
Introduction
It is recognized that circadian function and the genotoxic stress response are interconnected. This is thought to be an evolutionary adaption of the organism to sunlight irradiation on earth. The transcription factor p53 is known as the “guardian of the genome”1,2. The majority of all human tumors have mutations in the TP53 gene which encodes the p53 protein3. The roles of stress-induced p53 in cellular responses are well studied. For example, cell cycle arrest and the DNA damage response are among the biological processes that are regulated by stress-induced p534. Circadian rhythms are the daily oscillations of biological processes driven by endogenous molecular clocks5. The known core clock genes include Clock, Npas2, Bmal1, Per1, Per2, Cry1, Cry2, CK1ε, CK1δ, Rev-Erbα, and RORα. Typically, the CLOCK/BMAL1 heterodimer binds to the E-box in the promoter region of their target genes, leading to activation of the expression of these genes, including Per1, Per2, Cry1 and Cry2. The protein products of these core clock genes regulate each other’s expression in feedback loops. Recently, we showed that the promyelocytic leukemia (PML) protein regulates the circadian clock by interacting with PER2 and enhancing BMAL1/CLOCK mediated transcription6. PML is essential for formation of PML nuclear bodies (NB’s)7. It is thought that PML-NBs recruit specific proteins and regulate their activity by modulating their post-translational modification. To date, a growing list of PML interacting proteins has been reported8. Among them are both positive and negative regulators involved in the circadian clock mechanism including PER2, BMAL1, SIRT1, CBP, WDR5 and SIN3A6,9–12. These findings led us to screen other PML associated proteins for a possible role in modulating BMAL1/CLOCK mediated Per2 transcription. Among the PML associated proteins examined was p53, which is known to localize with the PML nuclear body. Studies have shown that acetylation and deacetylation of p53 by CBP and SIRT1, respectively, are PML-NB dependent13–15.
Here, we show that p53 modulates the circadian behavior of mice by modulating Per2 expression, acting as a direct competitor of BMAL1/CLOCK binding to the Per2 promoter. This mechanistic interaction may represent a key regulatory link between the p53-mediated cellular stress response and the circadian clock-regulated cellular pathways.
Results
p53 represses BMAL1/CLOCK-mediated transcription of Per2
A number of PML-associated proteins were screened for their potential ability to modulate BMAL1/CLOCK mediated Per2 promoter transcription. Expression constructs for p53, WRN, RB, and RARα were individually examined using a Per2-Luc reporter assay for their ability to up or down regulate BMAL1/CLOCK mediated transcription. Among these expression constructs, we observed that the one encoding p53 strongly repressed BMAL1/CLOCK mediated transcription of the Per2 promoter (Figure 1a). Therefore, we asked whether this apparent repression of Per2 transcription by p53 was mediated through BMAL1/CLOCK. High levels of p53 can induce apoptosis of tumor cell lines and this could result in an apparent non-specific transcriptional repression16. To rule out this possibility, we compared wildtype mouse embryonic fibroblasts (MEF) with HeLa cells using a Luc-reporter assay driven by either the Bmal1 or Per2 promoter. In both wildtype MEF and HeLa cells, the expression of p53 did not repress RORγ mediated transcription of the Bmal1 promoter but repressed Per2 promoter transcription mediated by BMAL1/CLOCK (Figure 1b and Supplementary Figure S1).
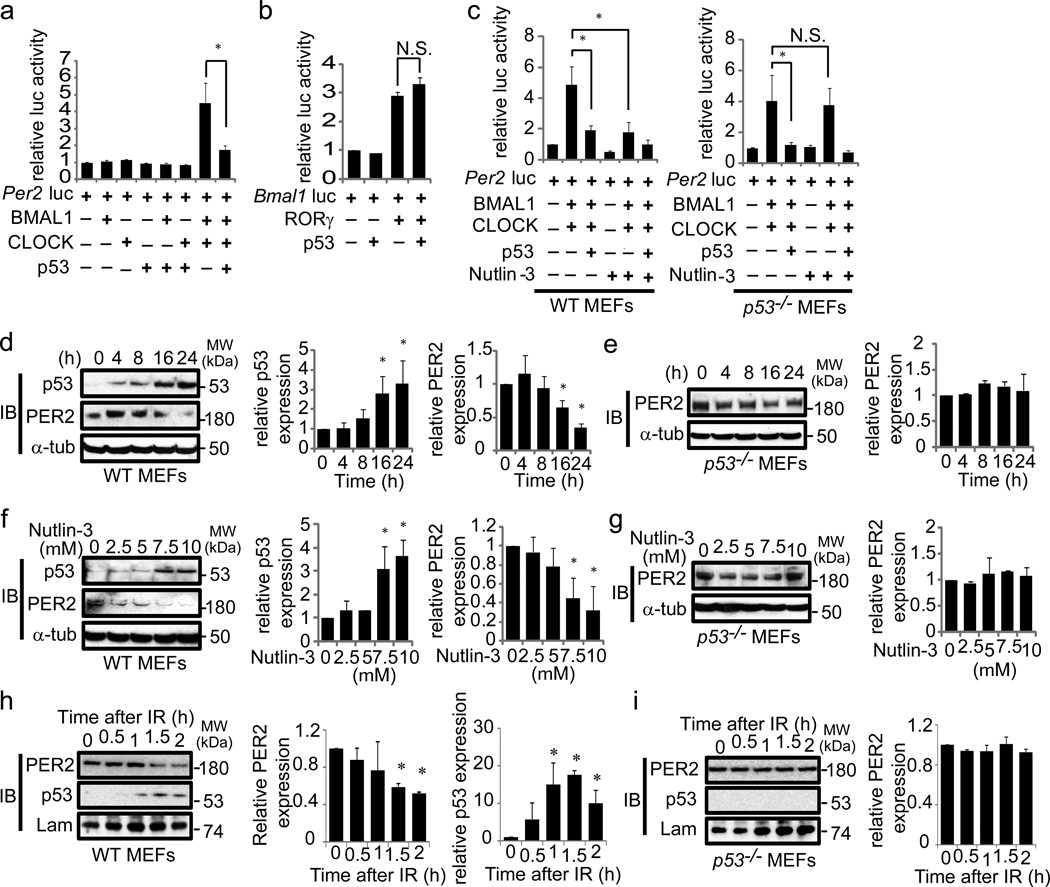
Figure 1. p53 modulates Per2 expression.
(a) Per2 or (b) Bmal1 promoter luciferase assay in wildtype (WT) MEF cells transfected with the indicated expression vectors, and harvested 24 h after transfection. (c) Per2 promoter luciferase assays in the absence or presence of Nutlin-3 (10 µM) using WT or p53−/− MEF cells. Western blotting using the indicated antibodies; (d) WT or (e) p53−/− MEF cells were treated with 10 µM Nutlin-3 and harvested at the indicated time points; (f) WT or p53−/− (g) MEF cells were treated with the indicated concentration of Nutlin-3 for 16 h. (h) WT or (i) p53−/− MEF cells were treated with γ-irradiation (10Gy) and harvested at indicated time points (Full-length immunoblots images for 1d and 1h are shown in Supplementary Figure S8). Error bars; mean ± SEM (n>3). T-test: *p< 0.05. N.S., not significant.
The endogenous p53 level is stabilized and enhanced by Nutlin-3, a small molecule inhibitor of MDM217. MDM2 is the major negative regulator of p53 and facilitates p53 degradation18. BMAL1/CLOCK-mediated transcription of the Per2 promoter was repressed in wildtype, but not in p53−/− MEF cells when treated with Nutlin-3 (Figure 1c). Transfection of a p53 expression construct into p53−/− MEF cells restored the repression of BMAL1/CLOCK-mediated transcription of the Per2 promoter in the presence of Nutlin-3. Western analysis with anti-p53 and anti-PER2 antibodies showed a time-dependent increase in the levels of endogenous p53, accompanied by a reciprocal decrease in endogenous PER2 in wildtype MEF cells following Nutlin-3 treatment (Figure 1d). No decrease in PER2 levels by Nutlin-3 treatment was observed in p53−/− MEF cells (Figure 1e). This inverse relationship between p53 and PER2 levels was further examined in a Nutlin-3 titration experiment. After a 16 h treatment, endogenous p53 levels correlated with increases in Nutlin-3 concentration, and were inversely correlated with PER2 levels in wildtype MEF cells (Figure 1f). No change in PER2 levels was observed when p53−/− MEF cells were treated with Nutlin-3 at a similar concentration (Figure 1g). Genotoxic stress such as γ-irradiation is known to rapidly stabilize p53 protein in some cells, including MEF and human osteosarcoma U2OS cells19. Western analysis of human U2OS cells and wildtype MEF treated with γ-irradiation displayed a rapid rise in p53 and a reciprocal decline in PER2 levels (Figure 1h, Supplementary Figure S2). This decline in PER2 level was not observed in p53−/− MEF cells that were similarly treated with γ-irradiation (Figure 1i). Together, these studies show that the cellular level of PER2 is inversely correlated with that of p53.
The Per2 promoter contains a conserved p53 response element
The inverse correlation between p53 and PER2 suggests that p53 is repressing BMAL1/CLOCK mediated transcription of the Per2 promoter. To map the putative response element for p53, BMAL1/CLOCK mediated transcription, reporter assays were carried out using serial deletion constructs of the mouse Per2 promoter at −2811 to −105 bp from the transcription start site (Figure 2a). The repression by p53 of BMAL1/CLOCK mediated transcription of the Per2 promoter was intact in all of these deletion constructs suggesting that the putative p53 response element is within the −105 bp region of the Per2 promoter. Using the NBLAST program, comparison of this 105 bp mouse sequence with Per2 promoter sequences of human, dog and rat from Genebank revealed a highly conserved 48 bp sequence (Figure 2b). Sequence analysis with the TFBIND program revealed a putative p53 response element. The p53 response element is a motif of two tandem decamers separated by 0–13 nucleotides. The consensus sequences can be depicted as 5'-RRXCXXGXYX-N(0–13)-XRXCXXGXYY-3', where R= A/G, Y= C/T and X= A/G/C/T 20. This putative p53 response element is upstream to the transcription start site of the Per2 promoter, and its proximal decamer overlaps with the critical E-box sequence for BMAL1/CLOCK binding. Mutation of nucleotides adjacent to the E-box severely dampened BMAL1/CLOCK mediated transcription of the Per2 promoter. These observations are consistent with previous findings that BMAL1/CLOCK mediated transcription of the Per2 promoter requires a segment about 20 bases from the transcription start site21. Interestingly, mutation of nucleotides in the distal decamer of the putative p53 response element did not significantly affect BMAL1/CLOCK mediated transcription while the strong repression by p53 was relieved (Figure 2b, 2c).
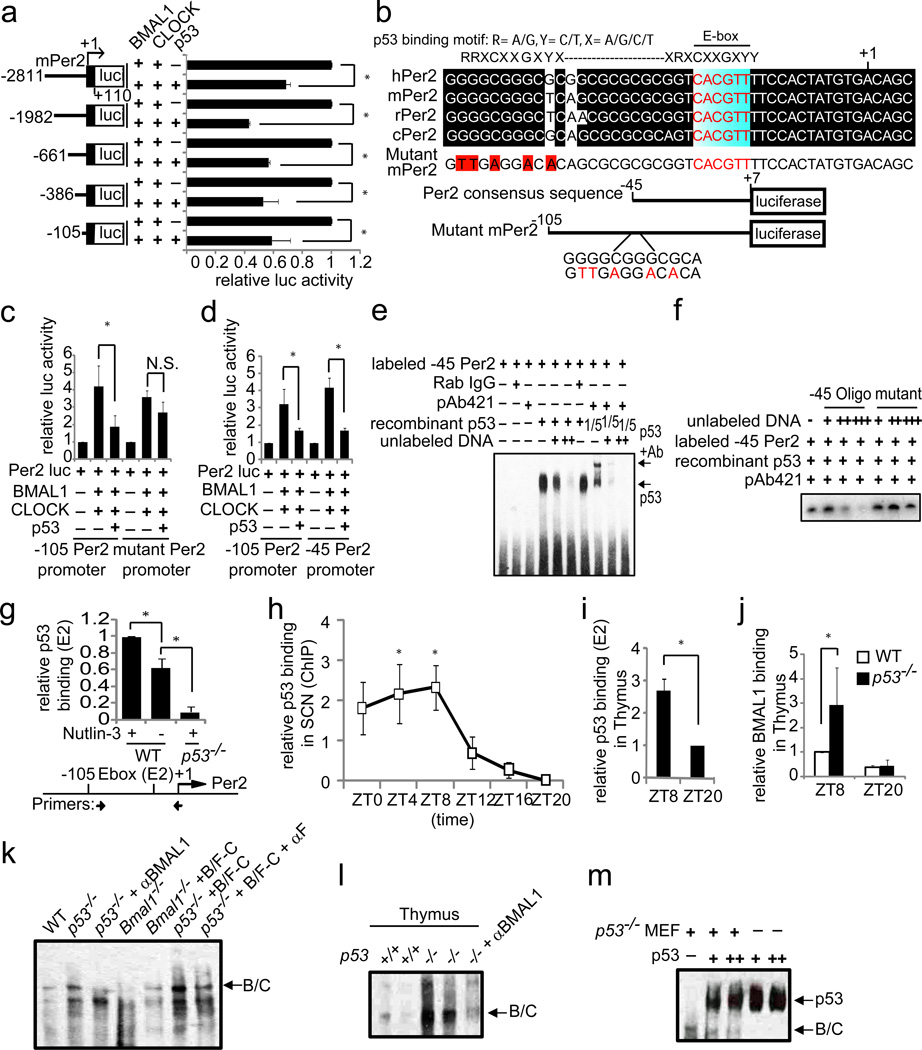
Figure 2. p53 binds to a Per2 promoter consensus region.
(a) Per2 promoter deletion luciferase assay. (b) Comparison of Per2 promoter among species (upper). Images of −45 bp WT and mutant Per2 promoter luciferase construct (lower). Luciferase assays were performed using: (c) −105 bp WT and mutant Per2 or (d) −105 bp and −45 bp Per2 promoter luciferase. (e) p53 binding to the Per2 promoter consensus region examined by EMSA. (f) Competition assay for p53 binding by EMSA. (g) Relative p53 binding to the Per2 promoter (E2) was examined by ChIP-qPCR assay. (h) ChIP-qPCR assay for p53 in mouse SCN. ANOVA: p< 0.05, post hoc t-test: *p< 0.05. (i) ChIP-qPCR assay (E2) for p53 in mouse Thymus. (j) ChIP-qPCR assay (E2) for BMAL1 in wildtype (clear) or p53−/− (shaded) mouse Thymus obtained at ZT8 and ZT20. (k) BMAL1/FLAG-CLOCK (B/F-C) binding to the Per2 promoter consensus region examined by EMSA. Nuclear extracts from the indicated genotype of fibroblast cells were used. (l) EMSA of BMAL1/CLOCK (B/C) binding using nuclear extract obtained from thymus. (m) Competitive binding assay to the Per2 promoter by EMSA using recombinant p53 and BMAL1/CLOCK from nuclear extract of p53−/− MEF cells. Error bars: mean ± SEM (n>3). T-test: *p< 0.05. N.S., not significant.
To further characterize the region upstream of the Per2 promoter initiation site, sense and antisense oligonucleotides corresponding to the mouse Per2 promoter sequence from +7 to −45 bp were synthesized, annealed and cloned into a luciferase reporter vector to generate −45bpPer2-Luc (Figure 2b). Transcriptional activation by BMAL1/CLOCK and repression by p53 of expression from the −45bpPer2-Luc and −105bpPer2-Luc reporter constructs were comparable, indicating that the −45bpPer2 DNA promoter sequence contained both the p53 response element and the E-box of BMAL1/CLOCK (Figure 2d).
Using an electrophoretic mobility shift assay (EMSA) we observed that recombinant p53 protein binds to −45bpPer2 DNA (Figure 2e). The binding of p53 to biotin-labeled −45bpPer2 DNA was competitively reduced by unlabeled −45bpPer2 DNA. Some antibodies to p53 are known to induce a super-shift of the p53/DNA complex. We tested two commercial p53 antibodies and they both generated the expected super-shifts (Figure 2e, Supplementary Figure S3). Further, the super-shift was competitively reduced by unlabeled −45bpPer2 DNA. However, when the unlabeled −45bpPer2 DNA contained a mutated sequence in the distal decamer, the super-shift binding was not reduced (Figure 2f).
The Per2 genomic locus has five described E-box elements (E1-E5), and the one most critical for BMAL1/CLOCK binding is E2 located in the promoter region22. Chromatin immuno-precipitation (ChIP) assays with anti-p53 antibodies were performed using genomic DNA that had been fragmented to 0.5 Kb on average. ChIP-qPCR analysis revealed that p53 pull-down of E2 was significantly enhanced in wildtype when cells were treated with Nutlin-3 (Figure 2g). In contrast, p53 pull-down of E5, about 3 Kb away from E2, was not significant (Supplementary Figure S4). Next, we determined whether BMAL1/CLOCK binding to the E2 region of the Per2 promoter was disrupted by p53. Transfection of p53−/− MEF with expression constructs for FLAG-tagged Clock, Bmal1 in the presence or absence of a p53 expression construct was carried out. ChIP-qPCR analysis showed that in the absence of p53, pull down of FLAG-CLOCK by the anti-FLAG antibody was significantly higher (Supplementary Figure S5).
We then investigated whether p53 binding to the E2 region of the Per2 promoter also occurs in the SCN in vivo. SCN from wildtype mouse brains were dissected at six zeitgeber time (ZT) points over 24 h for ChIP analysis. ChIP-qPCR with anti-p53 antibodies showed a peak level of endogenous p53 binding to the Per2 promoter in the SCN between ZT4-ZT8 and a nadir at ZT16-ZT20 (Figure 2h). These studies demonstrate that p53 is expressed in the SCN and its interaction with the Per2 promoter is under temporal control in vivo. Next, the level of p53 or BMAL1/CLOCK binding to the E2 region of the Per2 promoter was examined using wild type and p53−/− mouse thymus obtained at ZT8 and ZT20. Thymus was chosen since it expresses detectable levels of p53 under normal physiological conditions. ChIP-qPCR analysis revealed that p53 binding to the E2 region was about 2.5X higher at ZT8 than ZT20 (Figure 2i). BMAL1 binding to the E2 region in p53−/− MEF was significantly elevated compared with wildtype (Figure 2j).
To investigate whether p53 directly competes with BMAL1/CLOCK for binding to −45bpPer2 DNA, nuclear extracts from WT, p53−/− MEF and Bmal1−/− fibroblasts were prepared for EMSA. Unlike nuclear extracts from WT and p53−/− MEF, those from Bmal1−/− fibroblasts did not display a prominent protein/DNA complex on EMSA with −45bpPer2 DNA indicating that this complex is BMAL1 dependent (Figure 2k). Further, transfection of expression plasmids for Bmal1 and Flag-Clock into Bmal1−/− fibroblasts restored the BMAL1/CLOCK:-45bpPer2 protein/DNA complex in nuclear extracts. Treatment with either anti-BMAL1 or anti-FLAG antibodies significantly reduced this protein/DNA complex. Consistent with results from ChIP-qPCR analysis, nuclear extracts from p53−/− MEF and thymus showed an increased level of the BMAL1/CLOCK:-45bpPer2 protein/DNA complex when compared to wildtype samples (Figure 2k, 2l). Adding recombinant p53 to the p53−/− nuclear extract reduced the level of the BMAL1/CLOCK:-45bpPer2 DNA complex (Figure 2m). This observation shows that p53 competes with BMAL1/CLOCK for binding to the DNA probe. Collectively, these studies show that the Per2 promoter contains a functional p53 response element that overlaps with the E-box for BMAL1/CLOCK binding that is critical for transcription of the Per2 gene.
p53 levels regulate Per2 transcription in the SCN
To gain insight into the relevance of the p53 response element in the Per2 promoter to clock function, p53 levels in the SCN, the master clock structure in mammals, were investigated under temporal conditions. Previous studies have shown that in peripheral tissues, the p53 protein level is under temporal control23–25. Immunofluorescence analysis with anti-p53 antibodies showed that the level of p53 in the SCN from wildtype mice peaks at ZT4-ZT8 and reaches a nadir at ZT16-ZT20 (Figure 3a, 3b). There was no p53 staining in the SCN of p53−/− mice. This observation is consistent with results from the ChIP assay that showed peak levels of p53 binding to the Per2 promoter occurred at about ZT4-ZT8.
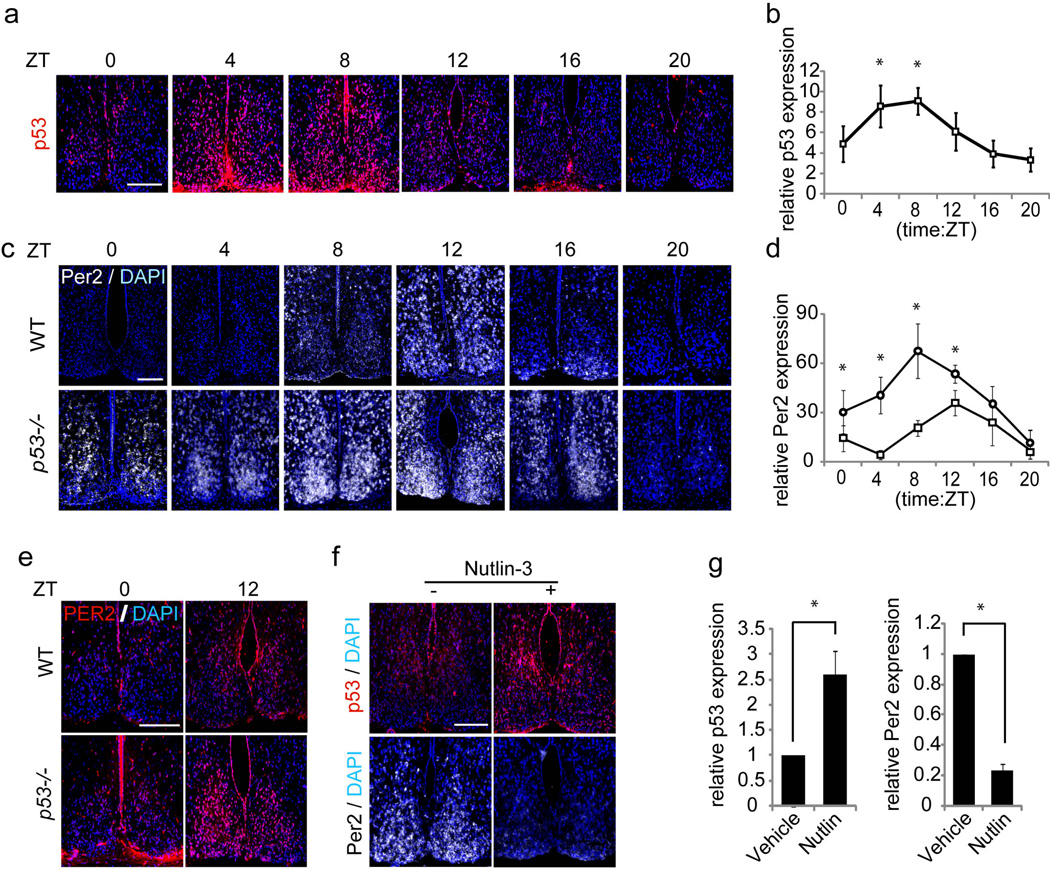
Figure 3. p53 oscillates and modulates Per2 expression in the SCN.
(a) Immunofluorescence analysis of p53 expression in WT mouse SCN. (b) Quantification of p53 expression in the SCN (n=3). ANOVA: p< 0.05. post hoc t-test: *p< 0.05. (c) In situ hybridization detection of Per2 mRNA in SCN of WT and p53−/− mice. (d) Quantification of Per2 expression in the WT (square) and p53−/− (circle) mouse SCN (n=3). ANOVA: p< 0.05. post hoc t-test: *p< 0.05. (e) Immunofluorescence analysis of PER2 expression in SCNs from WT or p53−/− mice (f) Correlation between p53 protein levels, shown by immunofluorescence, and Per2 expression, shown by in situ hybridization, in Nutlin-3 (200 mg/kg) or DMSO injected SCN. (g) Statistical analysis of (f) (n=2). Scale bars, 200 µm. Error bars: mean ± SEM. T-test:.*p< 0.05.
Next, we examined whether the loss of p53 elevates Per2 expression in the SCN as predicted by our in vitro studies. In situ hybridization studies showed that Per2 transcript levels in the SCN of p53−/− mice were significantly elevated compared to wildtype mice at most sampling times (Figure 3c, 3d). Interestingly, the expression of Per2 transcripts in p53−/− mice remained under temporal control and was apparently phase advanced, reaching peak levels at about ZT8 and a nadir at about ZT20. Consistent with results from in situ analysis, immunofluorescence studies with anti-PER2 antibodies showed that the level of PER2 was significantly elevated at ZT0 and ZT12 in the SCN of p53−/− mice compared to wildtype mice (Figure 3e).
Next, we examined whether increasing p53 protein levels by Nutlin-3 treatment would have the converse effect of repressing Per2 transcription in the SCN. First, we investigated whether Nutlin-3 could stabilize p53 levels in the SCN. Wildtype mice were administered Nutlin-3 or vehicle (DMSO) at ZT9 and then sacrificed at ZT14. We selected ZT14 for these studies since endogenous SCN Per2 and p53 protein levels are close to their peak and nadir, respectively at this time point. Immunofluorescence analysis with anti-p53 antibodies showed increased p53 protein in the SCN after treatment with Nutlin-3 but not vehicle (Figure 3f, 3g). In situ hybridization with an antisense Per2 probe showed that endogenous Per2 expression in the SCN of mice treated at ZT14 was significantly dampened after treatment with Nutlin-3 compared with vehicle (Figure 3f, 3g). Thus, in the SCN, increased p53 levels lead to repression of Per2 expression, consistent with the finding of elevated Per2 expression in p53−/− mice.
p53 modulates circadian behavior of mice
Next, we examined whether p53 regulates circadian rhythm behavior of mice. The circadian behavior of inbred male p53−/− and wildtype (C57BL/6) mice was analyzed by monitoring their wheel running activity. Both p53−/− and wildtype mice entrained to an LD cycle and were rhythmic throughout our studies. The p53−/− mouse period lengths were 22.8 ± 0.1 h (mean ± SEM, n=12) and were significantly shorter than wildtype period lengths of 23.3 ± 0.1 h (mean, ± SEM, n=10) (Figure 4a, 4b). In addition, the p53−/− mice had a more widely distributed period that exhibited sudden and unpredictable changes during free-running conditions (Figure 4a, 4c). To assess this phenotype further, the same p53−/− and wildtype mice were re-entrained to an LD cycle followed by a second release into free-running conditions. For wildtype mice, the changes in period measured from the second release into DD were all within 0.06 ± 0.01 h (mean ± SEM, n=8) of their first measurements. In contrast, p53−/− mice had periods that differed significantly from the first measurement (Figure 4c, 4d). Both lengthening and shortening of periods were observed. Comparing the longest and shortest period length of an individual mouse measured by linear regression sampled over at least 10 days revealed an average difference of 0.43 ± 0.1 h (mean ±SEM, n=7) for p53−/− mice.
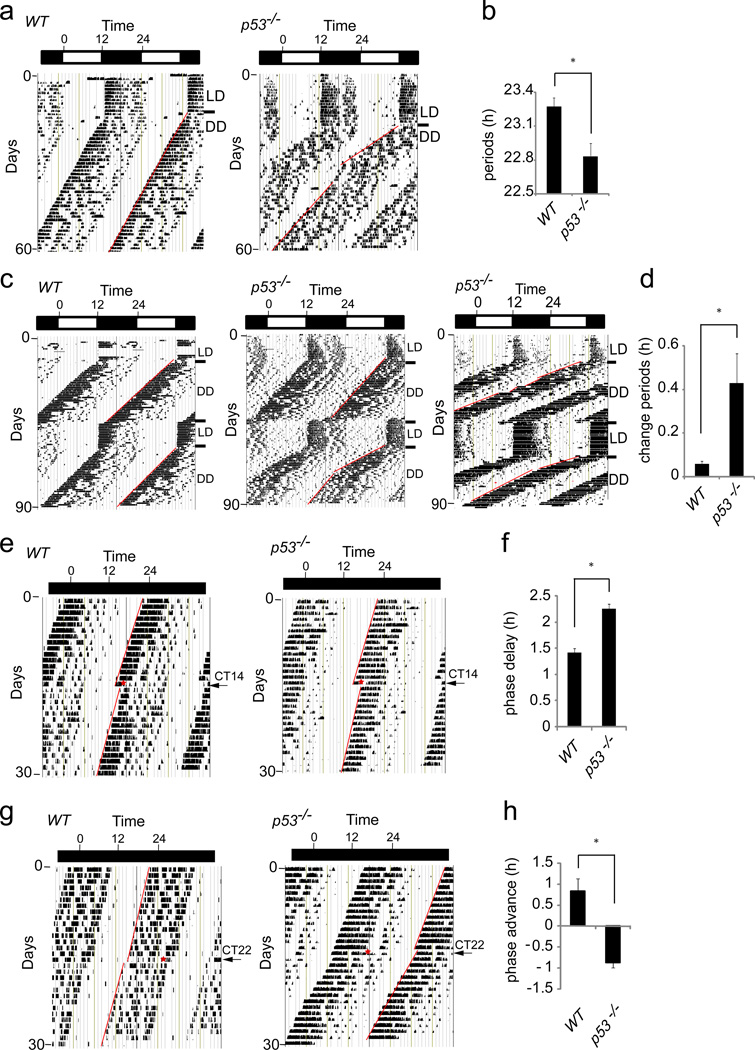
Figure 4. p53 controls circadian behavior of mice.
(a, c) Wheel running activity of WT and p53−/− mice. (b) Summary of periods for WT (n=10) and p53−/− mice (n=13). (d) Average difference in observed period length after re-entrainment of individual WT (n=8) and p53−/− (n=7) mice. (e) A 15 min light pulse applied at CT14 to free-running mice. (f) Summary of (e): WT (n=3) and p53−/− (n=4). (g) A 15 min light pulse applied at CT22 to free-running mice. (h) Summary of (g): WT (n=3) and p53−/− (n=5). Error bars, mean ± SEM. T-test: *p< 0.05.
Phase shift behavior in response to a light pulse of a specific circadian time (CT) is an intrinsic clock control behavior26. Mice in the free-running state were given a 15 min light pulse at either CT14 or CT22 to measure their phase delay or phase advance response, respectively. The level of phase delay in response to a CT14 light pulse averaged −1.4 ± 0.1 h (mean ± SEM n=3) in wildtype, whereas p53−/− mice displayed a significantly enhanced phase delay response that averaged −2.3 ± 0.1 h (mean ± SEM n=4) (Figure 4e, 4f). When the light pulse was given at CT22, wildtype mice displayed a phase advance response that averaged +0.8 h± 0.3 h (mean ± SEM n=3). Surprisingly, the p53−/− mice displayed no phase advance but instead exhibited a phase delay response that averaged −0.9 h ± 0.1 h (mean ± SEM n=5) (Figure 4g, 4h). Thus, the circadian period of p53−/− compared with wildtype mice is shorter, unstable and its phase shift response to a light cue displays an enhanced phase delay response during free running conditions.
Nutlin-3 phase shifts the circadian behavior of mice
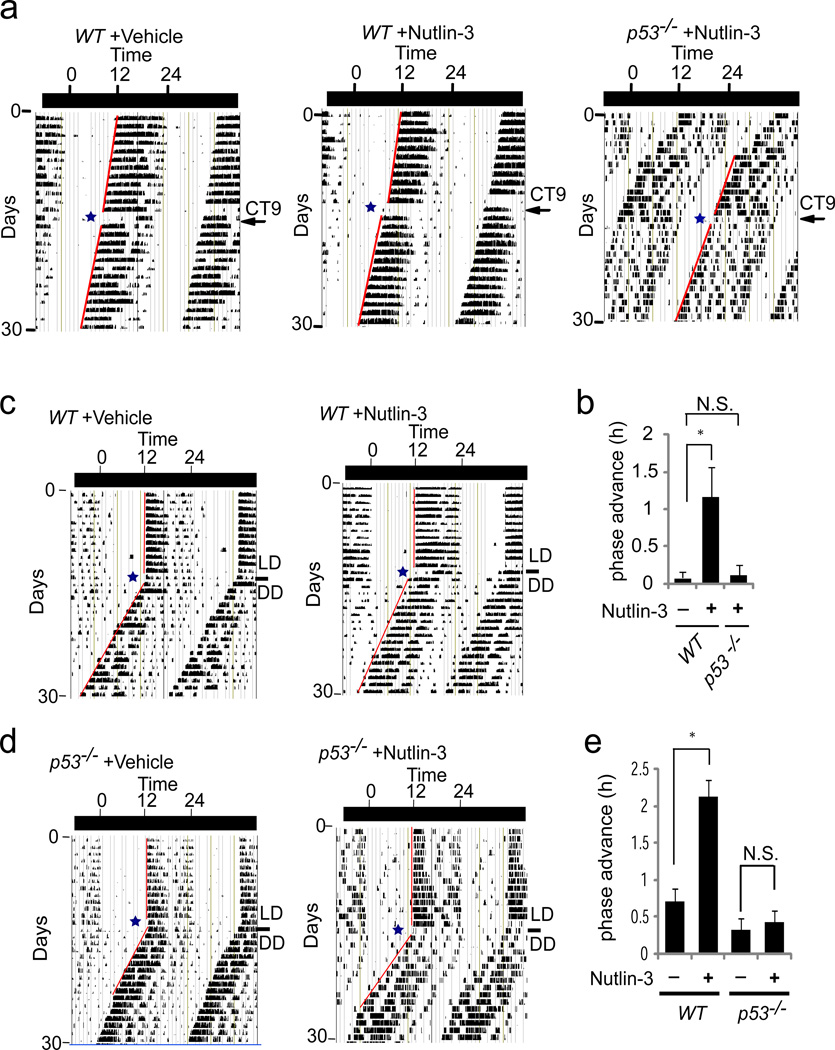
Nutlin-3 is currently in clinical trials as an anti-cancer drug27. Preclinical studies showed that Nutlin-3 has apparent low toxicity and can be given daily to mice17. Our molecular studies raised the possibility that Nutlin-3 can alter Per2 expression via stabilization of p53 and thus reset the circadian clock. To investigate this possibility, we examined whether Nutlin-3 or vehicle (DMSO) can phase shift circadian rhythm in wildtype mice during free-running DD conditions. CT9 and CT21 were chosen due to the approximate peak and nadir levels of endogenous p53 in the SCN, respectively. Both vehicle and Nutlin-3 treated mice displayed a temporary reduction of locomotor activity although the effect of Nutlin-3 persisted longer in some animals. However, no phase shift response was observed in vehicle treated wildtype or Nutlin-3 treated p53−/− mice whether treated at CT9 or CT21 (Figure 5a, 5b and Supplementary Figure S6). In contrast, a phase advance response was observed when wildtype mice were treated with Nutlin-3 at CT9 or CT21. The effect of Nutlin-3 was also examined using a different protocol for phase shift response in the presence of a lighting cue. In this protocol, delivery of vehicle or Nutlin-3 was carried out on the last day of the LD cycle entrainment followed by free-running conditions28. This protocol showed that vehicle treated mice did not display phase shift behavior while Nutlin-3 given at ZT9 induced a robust phase advance response (Figure 5c, 5d, 5e). In contrast, p53−/− mice treated with vehicle or Nutlin-3 displayed no phase shift response. These results show that the clock phase advance induced by Nutlin-3 treatment is mediated through stabilization of p53 and is consistent with light pulse studies of p53−/− mice showing enhanced phase delay response.
Figure 5. Actograms of phase shift response induced by Nutlin-3.
(a) WT or p53−/− mice were kept in a DD cycle and either vehicle (DMSO) (n=3) or Nutlin-3 (n=6) was injected at CT9. (b) Summary of phase advance (CT9). (c) Mice were kept in a 12 h/12 h LD cycle for 2 weeks, vehicle (n=6) or Nutlin-3 (n=4) were injected, and then mice were released into a DD cycle. (d) p53−/− mice given vehicle (n=5) or Nutlin-3 (n=6), as in (c). (e) Summary of phase advance of vehicle (-) or Nutlin-3 (+) injected mice. Error bars, mean ± SEM. T-test: *p< 0.05. N.S., not significant.
Discussion
The circadian clock mechanism governs the rhythmic activities that allow a living organism to anticipate and respond to opportunities and challenges presented in a daily diurnal rhythmic fashion. For this coordinated control to happen at all levels, from physiological processes to molecular signaling pathways, there must be a pervasive and effective mechanistic connection between the clock network and other cellular networks. It is now recognized that the clock mechanism or specific components of the clock are involved in responses to cellular stress including DNA damage response, checkpoint arrest and apoptosis29. p53 regulated genes are involved in diverse biological pathways including the cell cycle, DNA damage response, apoptosis and glucose metabolism4. However, the link between p53 controlled pathways such as the cell cycle and the circadian clock regulators is thought to be indirect or “opportunistic”30.
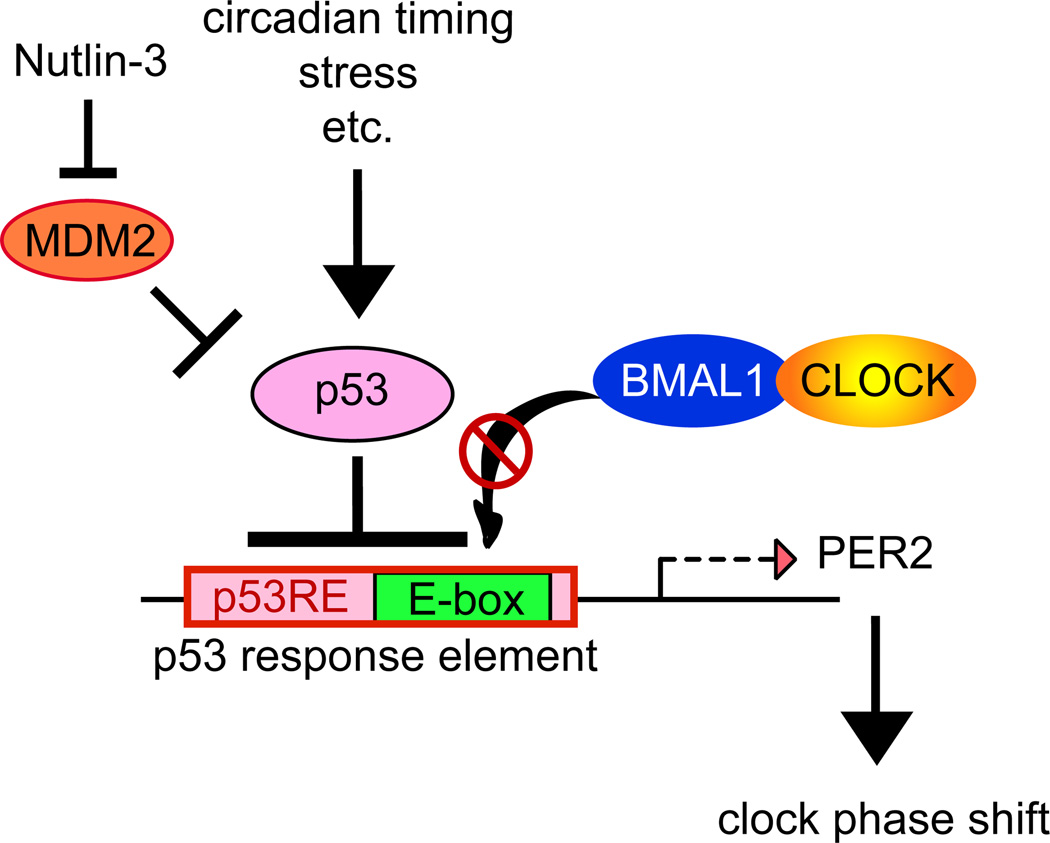
Our present study has shown that p53 directly binds to the Per2 promoter and represses its transcriptional activation by BMAL1/CLOCK. We mapped a p53 response element with two characteristic decamers to an evolutionarily conserved region within −45 bp of the Per2 promoter transcription start site. This p53 response element overlaps with the major E-box element necessary for BMAL1/CLOCK mediated transcription. Both EMSA and reporter assay with −45bpPer2DNA demonstrate p53 binding and repression of BMAL1/CLOCK mediated transcription. Independently, by ChIP-qPCR analysis, we showed that CLOCK/BMAL1 binding to the Per2 promoter is inhibited by the expression of p53. Four modes of p53 transcriptional repression are now recognized20. One of these modes is the overlap of the p53 response element with the binding site of another transcription factor, allowing p53 to compete with that transcription factor for DNA binding (Figure 6). Consistent with this mode of transcriptional repression, EMSA results show that the formation of a BMAL1/CLOCK:-45bpPer2 DNA complex was significantly reduced by addition of recombinant p53.
Figure 6. Model of p53 repression of Per2 expression and modulation of the circadian clock.
p53 levels are regulated by many factors such as circadian timing, cellular stress, and MDM2. Both physiological and stress induced p53 binds to the p53 response element of Per2 promoter which overlaps with the BMAL1 / CLOCK binding site (E-box) and suppresses BMAL1 / CLOCK mediated transcription of Per2 thereby modulating the circadian clock.
Our studies show that the physiological p53 level oscillates in the SCN of wildtype mice with peak expression around ZT8. In the SCN of p53−/− mice, Per2 expression was significantly elevated but remained under temporal control. These observations suggest that p53 regulates the amplitude of Per2 transcription. Indeed, the p53 response element of the Per2 promoter is within a region previously delineated to be important for the generation of amplitude21. The p53−/− mice display abnormal circadian behavior, exhibiting a shorter period length with reduced precision and altered phase shift response to a light pulse. Interestingly, transgenic mice that overexpress Per2 also display a shorter period length than wildtype mice31. Period length is also controlled by PER2 phosphorylation state since it regulates PER2 turnover32. Our current findings show that when the levels of p53 are elevated by Nutlin-3 treatment, γ-irradiation or by expression construct transfection in cultured cells, Per2 expression is significantly dampened. In the in vivo study, Nutlin-3 injection into mice produced elevated p53 protein and repressed Per2 expression in the SCN, resulting in phase advancement of the circadian clock. These findings suggest that both physiological and elevated levels of p53 regulate Per2 expression. The importance of physiological levels of p53 for other biological processes has been recently recognized. For example, physiological levels of p53 regulate CD44 expression and epithelial proliferation33. In addition, under non-stress conditions, p53 regulates adult neural stem cell proliferation34. Under stress conditions, stabilized p53 represses expression of Per2 that in turn could modulate the expression of other clock genes to help reset the clock. BMAL1/CLOCK mediated Per1 and Cry1 expressions also appear to be repressed by p53 (Supplementary Figure S7), though it is unclear if these genes are direct targets of p53 inhibition or if this is secondary to Per2 repression by p53. It was previously shown that loss of PER2 leads to highly dampened Per1 and Cry1 expression in vivo35.
This study shows that p53−/− mice displayed an increased phase delay and an absence of phase advance response to a light pulse at CT14 and CT22, respectively. Mice deficient in PER2 function exhibited enhanced phase advance and a weak phase delay response indicating that PER2 plays a major role in phase shift control36–38. Per2 expression is differentially induced by light over the span of the phase response curve. A light pulse given during early, but not late subjective night, induced Per2 expression in the SCN36,37. These observations suggest that relative to the phase response curve, increased induction of PER2 is associated with phase delay while a lack of induction of PER2 is associated with the phase advance response. This correlation of rapid reduction of PER2 levels with phase shift advance is supported by cell culture studies showing that DNA damage induced by methyl methane sulfonate (MMS) or UV-irradiation caused clock phase advancement as a result of a rapid decrease in PER2 levels39. Genotoxic agents such as UV or γ-irradiation rapidly induce p5319,24,40. Our studies show that γ-irradiation caused rapid p53 stabilization that was associated with decreased PER2 levels. This decline in PER2 was not observed in p53−/− MEF treated with γ-irradiation, indicating that it is a p53 dependent effect. Consistent with these findings, mice treated with γ-irradiation also displayed phase advancement of their circadian clock41. Conversely, it has been observed that known casein kinase 1 (CK1) inhibitors when given during free-running conditions induce a phase delay response in mice42. This phase shift effect of CK1 inhibitors is thought to be caused by increased stabilization of PERIOD proteins particularly PER242,43. Our observation that Nutlin-3 induced p53 mediated repression of Per2 expression and phase advancement of the clock is consistent with a phase advance response resulting from a reduced induction of PER2 levels by a light pulse at ZT22.
Previously, it had been observed that mice heterozygous for constitutively active p53 displayed an enhanced aging phenotype that included enhanced loss of soft tissues and development of spinal kyphosis44. Further, overactive p53 activities caused by the expression of the p53 isoform ΔN-p53 (p44) also resulted in a similar premature aging phenotype45,46. Interestingly, similar premature aging phenotypes, including progressive loss of soft tissues and spinal kyphosis, were observed in Per2 deficient mice47,48. These premature aging phenotypes in hyperactive p53 and Per2−/− mice are supportive of our findings that p53 suppresses Per2 expression.
Current evidence suggests that there is an inter-regulatory loop between p53, PML and Per2. PML is involved in the regulation a p53 acetylation and function14,15. However, PML is a p53 target49. Both p53 and PML are under circadian control6,25. Our studies showed PML and p53 can directly regulate the circadian clock via Per2. Clearly, the mechanistic detail of their inter-regulation is complex and remains to be fully deciphered.
In summary, our earlier study suggested that functional PER2 is important for p53-mediated stress signals to reach the circadian clock network. Here we show that p53 acts as a transcription factor that regulates the circadian clock by direct control of Per2 expression.
Methods
Animals
p53 −/− (C57BL/6) mice were obtained from The Jackson Laboratory (#002101). Animals were housed in a standard animal maintenance facility under a 12 h light : 12 h dark cycle. All animal studies were carried under institutional approved animal protocols (HSC-AWC-09–139).
Cells and Reagents
WT MEF cells and Trp53−/− MEF cells were obtained from Dr. C. Takahashi (Kanazawa University, Japan) and maintained in DMEM/10 % FCS. Nutlin-3 was purchased from Cayman Chemical (#10009816). The mouse p53 expression vector was purchased from ORIGENE. A mouse anti-α-tubulin monoclonal antibody was purchased from COVANCE (B-5-1–2). The mPer2 antibody was raised as described previously35. Anti-p53 antibodies are from Santa Cruz (FL-393, #sc-6243, DO-1, #sc-126) and from Calbiochem (PAb421, #OP03). The anti-BMAL1 antibody is from Abcam (ab3350). The anti-CLOCK antibody is from Thermo Scientific (PA1–520). The anti-Lamin A antibody is from BD Transduction Laboratories (#612162). mPer2-promoter Luc reporter constructs including −2811, −1982, −661, −386, −105 were kind gifts from Dr. T. Takumi21. The −105 bp Per2-luc reporter construct containing five point mutations was generated by 2-step site directed mutagenesis using QuikChange Site-Directed Mutagenesis Kit (#200519, Stratagene). An intermediate mutant was created by introducing 2 mutations to the p53 binding motif in the Per2 promoter using an oligonucleotide with the sequence 5’-GCGCGCGCAGGGGAGGACTCAGCGCGCGC-3’. Then, an additional 3 mutations were introduced to the same motif using an oligonucleotide with a sequence of 5’-CCAATGGCGCGCGCAGTTGAGGACACAGCGCGCGCGG-3’.
Luciferase reporter assay
Luciferase reporter assays were performed as previously described50. Briefly, MEF cells were seeded into 12-well plates and transfected with the Per2 luciferase reporter construct or the Bmal1 luciferase reporter construct using the Lipofectamine 2000 (Invitrogen). Co-transfections were performed with the respective expression constructs for Clock, Bmal1, Rorα and p53 Cells were harvested 24 h post-transfection, and luciferase assay was performed with an assay kit obtained from Promega using a TD-20/201 luminometer (Turner Designs).
Immunoblotting
Cells were lysed in ice-cold lysis buffer containing 50 mM Tris-HCl, pH7.4, 0.1% SDS, 0.25 mM deoxycholate, 150 mM NaCl, 2 mM EGTA, 0.1 mM Na3VO4, 10 mM NaF, 1 mM PMSF, and complete Protease Inhibitor (Roche). The cell lysates were resolved by SDS-PAGE and used for immunoblotting as described previously51. Briefly, the 1st antibody incubation for immunoblotting was performed at 4°C overnight in the blocking buffer (5% milk, 1% goat serum, TBS-T). The antibodies dilution we used were anti-mPer2 (x150,000), anti-p53 (x10,000), anti-alpha-tubulin (x10,000) and anti-Lamin A (x2,500).
Electric mobility shift assay
Electric mobility shift assay (EMSA) was carried out using a Light-Shift Chemiluminescent EMSA kit (#20148, PIERCE) as described previously52. Briefly, the following double stranded 5’-biotinylated or unlabeled DNA’s were synthesized (SIGMA Aldrich)., Per2 consensus sequence DNA: 5’-CAGGGGCGGGCTCAGCGCGCGCGGTCACGTTTTCCACTATGTGACAGCGGAGG-3’, Per2 mutant promoter DNA: 5’-CAGTTGAGGACACAGCGCGCGCGGTCACGTTTTCCACTATGTGACAGCGGAGG-3’. Per2 consensus −55bp oligo: 5’-ATGGCGCGCGCAGGGGCGGGCTCAGCGCGCGCGGTCACGTTTTCCACTATGTGACAGCGGAGG-3’, Per2 mutant consensus −55bp oligo: 5’- ATGGCGCGCGCAGTTGAGGACACAGCGCGCGCGGTCACGTTTTCCACTATGTGACAGCGGAGG-3’. Recombinant mouse p53 was purchased from Active Motif (#31103) and 2 ng of recombinant p53 was used per reaction. Binding assays were performed in a buffer containing 20 mM HEPES (pH7.6), 30 mM KCl, 1 mM EDTA, 10 mM (NH4)2SO4, 1 mM DTT, 0.2% Tween 20, 1 µg poly [d(A–T)], 0.1 µg poly L- lysine. Anti-p53 antibodies (pAb421, Calbiochem or DO-1, Santa Cruz) were used for supershift assay. For BMAL1/CLOCK binding assay, nuclear extracts were prepared as described previously53. Briefly, cells were washed once with Buffer A containing 10 mM HEPES (pH 7.3), 1.5 mM MgCl2, 10 mM KCl, and 0.5 mM DTT, and resuspended in Buffer A. Cells were homogenized, centrifuged (200rpm, 10min), and nuclei were washed again with Buffer A. Cell pellets were resuspended in Buffer C containing 20 mM HEPES (pH 7.3), 20 % glycerol, 0.42 M NaCl, 1.5 mM MgCl2, 0.2 mM EDTA, 0.5 mM DTT, 0.5 mM phenylmethylsulfonyl fluoride and protease inhibitor cocktail (Roche). Lysates were incubated for 30 min with constant rotation at 4 °C and centrifuged (8000 rpm, 10 min). Supernatants were used as nuclear extracts. Binding assays were performed in a buffer containing 50 mM Tris-HCl (pH 8.0), 100 mM KCl, 1 mM EDTA, 6 mM DTT and 1 µg poly [d(A–T)]54. The anti-BMAL1 antibody (N-20, 2 µg, Santa Cruz) and anti-FLAG M2 antibody (5 µg, SIGMA) were used to detect the specific BMAL1/CLOCK band. The anti-BMAL1 antibody was previously validated by other investigators55. Protein binding reactions were performed in 10 µl scale.
Chromatin immunoprecipitation assay
Chromatin immunoprecipitation (ChIP) assay was performed as described previously56. Briefly, cells or tissues were fixed with 1% formalin for 10 min. Crosslinking was stopped by adding 125 mM glycine. Cells were washed and homogenized using a homogenizer. Homogenized cells were collected by brief centrifugation and lysed with swelling buffer containing 5 mM PIPES (pH8.0), 85 mM KCl, 1% NP40, and protease inhibitor cocktail (# 11697498001, Roche). Nuclear fractions were collected by centrifugation and lysed with nuclear lysis buffer containing 50 mM Tris-HCl (pH 8.0), 10 mM EDTA, 1% SDS, and protease inhibitor cocktail. Sonication was performed to fragment DNA to about 500 bp. After centrifugation, lysates were precleared with protein A agarose (Invitrogen). Pre-cleared samples were diluted with the same volume of IP dilution buffer containing 16.7 mM Tris-HCl (pH 8.0), 0.01% SDS, 1.1% Triton X-100, 1.2 mM EDTA, 167 mM NaCl, and protease inhibitor cocktail. Then, p53 and BMAL1 were immunoprecipitated with anti-p53 antibody (2 µg, sc-6243) or anti-BMAL1 antibody (2 µg, ab3350, abcam) and collected on protein A agarose. After serial washing with dialysis buffer containing 50 mM Tris-HCl (pH8.0) 2 mM EDTA, and 0.2% sarkosyl and IP wash buffer containing 100 mM Tris-HCl (pH9.0), 500 mM LiCl, 1% NP40, 1% Deoxycholate, and protease inhibitor cocktail. p53-DNA complexes were eluted by elution buffer containing 50 mM NaHCO3 and 1% SDS. Eluted samples were mixed with 0.2 M NaCl and incubated at 67 °C overnight, then RNase A treated, and purified. PCR and quantitative real-time PCR were performed using Per2-Ebox2 forward primers: 5’-CAGGTTCCGCCCCGCCAGTAT-3’, reverse primer:5’-GTCGCCCTCCGCTGTCACATAG-3’ and Per2-Ebox5 forward primer: 5’-CTCTGTAGGGTGGAGCGGCGA-3’, reverse primer: 5’-ATCCCCACTGCTCCTTCGCAC-3’. ChIP samples obtained from SCN and thymus were analyzed by real-time PCR (ABI PRISM 7700 system, Applied Biosystems) using Maxima SYBR Green qPCR Master Mix (#0252, Fermentas). Real-time PCR data were analyzed as previously reported57. IgG control antibody was used as a negative control. ΔCt was obtained by Ct experiment sample–., Ct negative control and fold differences were calculated by [2(−ΔCt)].
Immunofluorescence staining
8–10 week old WT and Trp53−/− mouse brains were collected at the indicated time point, fixed with 4% PFA, and embedded in paraffin. Then 8 µm sections were obtained and stained with anti-p53 antibody (#sc-6243, x50, SantaCruz), or anti-mPER2 antibody (x4,000) and visualized by secondary anti-rabbit IgG Alexa Fluor 555 antibody (#A21429, x1000, Invitrogen).
In situ hybridization
8–10 week old WT and Trp53−/− mouse brains were collected at the indicated time point, fixed with 4% PFA, and embedded in paraffin. Then 8 µm sections were obtained and in situ hybridization was performed as described previously58 with some modifications. Briefly, paraffin sections were treated with 1 µg/ml protease K in PBS-T (0.1% Tween 20) at 37 °C for 15 min, washed, fixed with 4% performic acid at room temperature for 15 min, washed, and then incubated with digoxigenin-labeled cRNA probes in buffer containing 50% formamide, 1% SDS, 5 x SSC, 50 µg/ml tRNA, and 20 mg/ml heparin at 55°C overnight. The probe for Per2 was described previously59. Hybridized sections were washed with Buffer A containing 50% formamide, 1% SDS, and 5 x SSC twice at 55 °C for 30 min each and then washed with Buffer B containing 50% formamide and 2 x SSC three times at 55 °C for 30 min each. After washing with TBS-T (pH7.6, 0.1% Tween 20) three times, sections were blocked with 0.5% Blocking Reagent (#11096176001, Roche) for 1 h and incubated with anti-alkaline phosphatase-conjugated anti-DIG antibody (#11093274910, x1000, Roche) at 4 °C overnight. Sections were washed three times with TBS-T and immersed in NTMT buffer containing 100 mM Tris-HCl (pH 9.5), 100 mM NaCl, 50 mM MgCl2, 0.1% Tween 20, and 2 mM levamisol. Sections were then developed with NBT/BCIP reagents.
Locomoter activity
Wheel-running activity was monitored as described60. Male wild type and Trp53 −/− mice aged about 4 weeks were used. Nutlin-3 (200mg/kg) or vehicle (DMSO) was injected i.p. at the indicated time point.
Supplementary Material
Acknowledgement
This work was supported in part by a grant from NIH to CCL. TM was also supported by The Yasuda Medical Foundation. We thank Dr J. Lever and Jake Chen for comments in the preparation of the manuscript.
Footnotes
Author contribution: Takao Miki performed all the experiments in this study and contributed to the writing of the manuscript; Dr. Zhaoyang Zhao designed all the Per2 promoter mutants and contributed to the preparation of the manuscript; Tomoko Matsumoto generated the Per2 promoter mutants; Cheng Chi Lee directed the studies and wrote the manuscript.
Conflict of interest statement: The authors declare no competing financial interests.
References
- 1.Donehower LA, et al. Mice deficient for p53 are developmentally normal but susceptible to spontaneous tumours. Nature. 1992;356:215–221. doi: 10.1038/356215a0. [DOI] [PubMed] [Google Scholar]
- 2.Li F, Fraumeni J., Jr Rhabdomyosarcoma in children: epidemiologic study and identification of a familial cancer syndrome. J. Natl. Cancer. I. 1969;43:1365–1373. [PubMed] [Google Scholar]
- 3.Hollstein M, Sidransky D, Vogelstein B. p53 mutations in human cancers. Science. 1991;253:49–53. doi: 10.1126/science.1905840. [DOI] [PubMed] [Google Scholar]
- 4.Vousden KH, Prives C. Blinded by the Light: The Growing Complexity of p53. Cell. 2009;137:413–431. doi: 10.1016/j.cell.2009.04.037. [DOI] [PubMed] [Google Scholar]
- 5.Takahashi JS. Molecular neurobiology and genetics of circadian rhythms in mammals. Annu. rev. Neurosci. 1995;18:531–553. doi: 10.1146/annurev.ne.18.030195.002531. [DOI] [PubMed] [Google Scholar]
- 6.Miki T, et al. PML regulates PER2 nuclear localization and circadian function. The EMBO J. 2012;31:1427–1439. doi: 10.1038/emboj.2012.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hofmann TG, Will H. Body language: the function of PML nuclear bodies in apoptosis regulation. Cell Death Differ. 2003;10:1290–1299. doi: 10.1038/sj.cdd.4401313. [DOI] [PubMed] [Google Scholar]
- 8.Reineke EL, Kao H-Y. Targeting promyelocytic leukemia protein: a means to regulating PML nuclear bodies. Int. J. Biol. Sci. 2009;5:366–376. doi: 10.7150/ijbs.5.366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nakahata Y, et al. The NAD+-dependent deacetylase SIRT1 modulates CLOCK-mediated chromatin remodeling and circadian control. Cell. 2008;134:329–340. doi: 10.1016/j.cell.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee Y, et al. Coactivation of the CLOCK-BMAL1 complex by CBP mediates resetting of the circadian clock. J. Cell Sci. 2010;123:3547–3557. doi: 10.1242/jcs.070300. [DOI] [PubMed] [Google Scholar]
- 11.Duong Ha, Robles MS, Knutti D, Weitz CJA. A molecular mechanism for circadian clock negative feedback. Science. 2011;332:1436–1439. doi: 10.1126/science.1196766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brown Sa, et al. PERIOD1-associated proteins modulate the negative limb of the mammalian circadian oscillator. Science. 2005;308:693–696. doi: 10.1126/science.1107373. [DOI] [PubMed] [Google Scholar]
- 13.Cheng H-L, et al. Developmental defects and p53 hyperacetylation in Sir2 homolog (SIRT1)-deficient mice. Proc. Natl. Acad. Sci. 2003;100:10794–10799. doi: 10.1073/pnas.1934713100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pearson M, et al. PML regulates p53 acetylation and premature senescence induced by oncogenic Ras. Nature. 2000;406:207–210. doi: 10.1038/35018127. [DOI] [PubMed] [Google Scholar]
- 15.Langley E, et al. Human SIR2 deacetylates p53 and antagonizes PML/p53-induced cellular senescence. EMBO J. 2002;21:2383–2396. doi: 10.1093/emboj/21.10.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vousden KH, Ryan KM. P53 and Metabolism. Nat. rev. Cancer. 2009;9:691–700. doi: 10.1038/nrc2715. [DOI] [PubMed] [Google Scholar]
- 17.Vassilev LT, et al. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science. 2004;303:844–848. doi: 10.1126/science.1092472. [DOI] [PubMed] [Google Scholar]
- 18.Haupt Y, Rowan S, Shaulian E, Vousden HK, Oren M. Mdm2 promotes the rapid degradation of p53. Nature. 1997;387:296–299. doi: 10.1038/387296a0. [DOI] [PubMed] [Google Scholar]
- 19.Joseph TW, Zaika A, Moll UM. Nuclear and cytoplasmic degradation of endogenous p53 and HDM2 occurs during down-regulation of the p53 response after multiple types of DNA damage. FASEB J. 2003;17:1622–1630. doi: 10.1096/fj.02-0931com. [DOI] [PubMed] [Google Scholar]
- 20.Wang B, Xiao Z, Ko HL, Ren EC. The p53 response element and transcriptional repression. Cell cycle. 2010;9:870–879. doi: 10.4161/cc.9.5.10825. [DOI] [PubMed] [Google Scholar]
- 21.Akashi M, Ichise T, Mamine T, Takumi T. Molecular Mechanism of Cell-autonomous Circadian Gene Expression of Period2, a Crucial Regulator of the Mammalian Circadian Clock. Mol. Biol. Cell. 2006;17:555–565. doi: 10.1091/mbc.E05-05-0396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yoo S-H, et al. A noncanonical E-box enhancer drives mouse Period2 circadian oscillations in vivo. Proc. Natl. Acad. Sci. 2005;102:2608–2613. doi: 10.1073/pnas.0409763102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gréchez-Cassiau A, Rayet B, Guillaumond F, Teboul M, Delaunay F. The circadian clock component BMAL1 is a critical regulator of p21WAF1/CIP1 expression and hepatocyte proliferation. J. Biol. Chem. 2008;283:4535–4542. doi: 10.1074/jbc.M705576200. [DOI] [PubMed] [Google Scholar]
- 24.Fu L, Pelicano H, Liu J, Huang P, Lee C. The circadian gene Period2 plays an important role in tumor suppression and DNA damage response in vivo. Cell. 2002;111:41–50. doi: 10.1016/s0092-8674(02)00961-3. [DOI] [PubMed] [Google Scholar]
- 25.Bjarnason GA, Jordan RC, Sothern RB. Circadian variation in the expression of cell-cycle proteins in human oral epithelium. Am. J. Pathol. 1999;154:613–622. doi: 10.1016/S0002-9440(10)65306-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moore RY. Circadian rhythms: basic neurobiology and clinical applications. Annu. rev. Med. 1997;48:253–266. doi: 10.1146/annurev.med.48.1.253. [DOI] [PubMed] [Google Scholar]
- 27.Secchiero P, Bosco R, Celeghini C, Zauli G. Recent Advances in the Therapeutic Perspectives of Nutlin-3. Curr. Pharm. Design. 2011;17:569–577. doi: 10.2174/138161211795222586. [DOI] [PubMed] [Google Scholar]
- 28.Spoelstra K, Albrecht U, Van der Horst GTJ, Brauer V, Daan S. Phase responses to light pulses in mice lacking functional per or cry genes. J. Biol Rhythms. 2004;19:518–529. doi: 10.1177/0748730404268122. [DOI] [PubMed] [Google Scholar]
- 29.Sancar A, et al. Circadian clock control of the cellular response to DNA damage. FEBS lett. 2010;584:2618–2625. doi: 10.1016/j.febslet.2010.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hunt T, Sassone-Corsi P. Riding tandem: circadian clocks and the cell cycle. Cell. 2007;129:461–464. doi: 10.1016/j.cell.2007.04.015. [DOI] [PubMed] [Google Scholar]
- 31.Miyazaki K, Wakabayashi M, Chikahisa S, Sei H, Ishida N. PER2 controls circadian periods through nuclear localization in the suprachiasmatic nucleus. Genes cells. 2007;12:1225–1234. doi: 10.1111/j.1365-2443.2007.01129.x. [DOI] [PubMed] [Google Scholar]
- 32.Vanselow K, et al. Differential effects of PER2 phosphorylation: molecular basis for the human familial advanced sleep phase syndrome (FASPS) Genes. dev. 2006;20:2660–2672. doi: 10.1101/gad.397006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Godar S, et al. Growth-inhibitory and tumor- suppressive functions of p53 depend on its repression of CD44 expression. Cell. 2008;134:62–73. doi: 10.1016/j.cell.2008.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Meletis K, et al. P53 Suppresses the Self-Renewal of Adult Neural Stem Cells. Development. 2006;133:363–369. doi: 10.1242/dev.02208. [DOI] [PubMed] [Google Scholar]
- 35.Zheng B, et al. Nonredundant roles of the mPer1 and mPer2 genes in the mammalian circadian clock. Cell. 2001;105:683–694. doi: 10.1016/s0092-8674(01)00380-4. [DOI] [PubMed] [Google Scholar]
- 36.Yan L, Silver R. Differential induction and localization of mPer1 and mPer2 during advancing and delaying phase shifts. Eur. J. Neurosci. 2002;16:1531–1540. doi: 10.1046/j.1460-9568.2002.02224.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nagano M, Adachi A, Masumoto K, Meyer-Bernstein E, Shigeyoshi Y. rPer1 and rPer2 induction during phases of the circadian cycle critical for light resetting of the circadian clock. Brain Res. 2009;1289:37–48. doi: 10.1016/j.brainres.2009.06.051. [DOI] [PubMed] [Google Scholar]
- 38.Albrecht U, Zheng B, Larkin D, Sun ZS, Lee CC. mPer1 and mPer2 Are Essential for Normal Resetting of the Circadian Clock. J. Biol. rhythms. 2001;16:100–104. doi: 10.1177/074873001129001791. [DOI] [PubMed] [Google Scholar]
- 39.Gamsby J, Loros J, Dunlap JC. A phylogenetically conserved DNA damage response resets the circadian clock. J. biol. Rhythms. 2009;24:193–202. doi: 10.1177/0748730409334748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Latonen L, Taya Y, Laiho M. UV-radiation induces dose-dependent regulation of p53 response and modulates p53-HDM2 interaction in human fibroblasts. Oncogene. 2001;20:6784–6793. doi: 10.1038/sj.onc.1204883. [DOI] [PubMed] [Google Scholar]
- 41.Oklejewicz M, et al. Phase resetting of the mammalian circadian clock by DNA damage. Curr. Biol. 2008;18:286–291. doi: 10.1016/j.cub.2008.01.047. [DOI] [PubMed] [Google Scholar]
- 42.Badura L, et al. An Inhibitor of Casein Kinase I Induces Phase Delays in Circadian Rhythms under Free-Running and Entrained Conditions. Pharmacology. 2007;322:730–738. doi: 10.1124/jpet.107.122846. [DOI] [PubMed] [Google Scholar]
- 43.Eide EJ, et al. Control of Mammalian Circadian Rhythm by CKIε-Regulated Proteasome-Mediated PER2 Degradation. Mol. Cell. Biol. 2005;25:2795–2807. doi: 10.1128/MCB.25.7.2795-2807.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tyner SD, et al. P53 Mutant Mice That Display Early Ageing-Associated Phenotypes. Nature. 2002;415:45–53. doi: 10.1038/415045a. [DOI] [PubMed] [Google Scholar]
- 45.Maier B, et al. Modulation of mammalian life span by the short isoform of p53. Genes Dev. 2004;18:306–319. doi: 10.1101/gad.1162404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pehar M, et al. Altered longevity-assurance activity of p53:p44 in the mouse causes memory loss, neurodegeneration and premature death. Aging cell. 2010;9:174–190. doi: 10.1111/j.1474-9726.2010.00547.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lee CC. Tumor suppression by the mammalian Period genes. Cancer causes control: CCC. 2006;17:525–530. doi: 10.1007/s10552-005-9003-8. [DOI] [PubMed] [Google Scholar]
- 48.Pilorz V, Steinlechner S. Low reproductive success in Per1 and Per2 mutant mouse females due to accelerated ageing? Reproduction. 2008;135:559–568. doi: 10.1530/REP-07-0434. [DOI] [PubMed] [Google Scholar]
- 49.De Stanchina E, et al. PML is a direct p53 target that modulates p53 effector functions. Mol. Cell. 2004;13:523–535. doi: 10.1016/s1097-2765(04)00062-0. [DOI] [PubMed] [Google Scholar]
- 50.Kaasik K, Lee CC. Reciprocal regulation of haem biosynthesis and the circadian clock in mammals. Nature. 2004;430:467–471. doi: 10.1038/nature02724. [DOI] [PubMed] [Google Scholar]
- 51.Miki T, et al. The reversion-inducing cysteine-rich protein with Kazal motifs (RECK) interacts with membrane type 1 matrix metalloproteinase and CD13/aminopeptidase N and modulates their endocytic pathways. J. Biol. Chem. 2007;282:12341–12352. doi: 10.1074/jbc.M610948200. [DOI] [PubMed] [Google Scholar]
- 52.Yoshitane H, et al. Roles of CLOCK phosphorylation in suppression of E-box-dependent transcription. Mol. Cell Biol. 2009;29:3675–3686. doi: 10.1128/MCB.01864-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kiyama R, Camerini-Otero RD. A triplex DNA-binding protein from human cells: purification and characterization. Proc. Natl. Acad. Sci. 1991;88:10450–10454. doi: 10.1073/pnas.88.23.10450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ye R, Selby CP, Ozturk N, Annayev Y, Sancar A. Biochemical analysis of the canonical model for the mammalian circadian clock. J. Biol. Chem. 2011;286:25891–25902. doi: 10.1074/jbc.M111.254680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Oishi K, Shirai H, Ishida N. CLOCK is involved in the circadian transactivation of peroxisome-proliferator-activated receptor α ( PPAR α ) in mice. Biochem. J. 2005;386:575–581. doi: 10.1042/BJ20041150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bieda M, Xu X, Singer MA, Green R, Farnham PJ. Unbiased location analysis of E2F1-binding sites suggests a widespread role for E2F1 in the human genome. Genome Res. 2006;16:595–605. doi: 10.1101/gr.4887606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mukhopadhyay A, Deplancke B, Walhout AJM, Tissenbaum H. a Chromatin immunoprecipitation (ChIP) coupled to detection by quantitative real-time PCR to study transcription factor binding to DNA in Caenorhabditis elegans. Nat. Protoc. 2008;3:698–709. doi: 10.1038/nprot.2008.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hasegawa H, et al. Laminar patterning in the developing neocortex by temporally coordinated fibroblast growth factor signaling. J. Neurosci. 2004;24:8711–8719. doi: 10.1523/JNEUROSCI.3070-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Albrecht U, Sun ZS, Eichele G, Lee CC. A differential response of two putative mammalian circadian regulators, mper1 and mper2, to light. Cell. 1997;91:1055–1064. doi: 10.1016/s0092-8674(00)80495-x. [DOI] [PubMed] [Google Scholar]
- 60.Zheng B, et al. The mPer2 gene encodes a functional component of the mammalian circadian clock. Nature. 1999;400:169–173. doi: 10.1038/22118. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.