Abstract
Neuro-immune alterations in the peripheral and central nervous system play a role in the pathophysiology of chronic pain, and non-coding RNAs – and microRNAs (miRNAs) in particular – regulate both immune and neuronal processes. Specifically, miRNAs control macromolecular complexes in neurons, glia and immune cells and regulate signals used for neuro-immune communication in the pain pathway. Therefore, miRNAs may be hypothesized as critically important master switches modulating chronic pain. In particular, understanding the concerted function of miRNA in the regulation of nociception and endogenous analgesia and defining the importance of miRNAs in the circuitries and cognitive, emotional and behavioral components involved in pain is expected to shed new light on the enigmatic pathophysiology of neuropathic pain, migraine and complex regional pain syndrome. Specific miRNAs may evolve as new druggable molecular targets for pain prevention and relief. Furthermore, predisposing miRNA expression patterns and inter-individual variations and polymorphisms in miRNAs and/or their binding sites may serve as biomarkers for pain and help to predict individual risks for certain types of pain and responsiveness to analgesic drugs. miRNA-based diagnostics are expected to develop into hands-on tools that allow better patient stratification, improved mechanism-based treatment, and targeted prevention strategies for high risk individuals.
Keywords: chronic pain, biomarker, polymorphism, miRNA-based diagnostics, miRNA expression patterns, miRNA polymorphisms, antagomir, miRNA-based analgesic
INTRODUCTION
Human chronic pain disorders are bio-psycho-social diseases, which are difficult to treat due to their diversity. Chronic pain syndromes that develop after nerve damage, trauma or surgery are characterized by persistent and severe pain; they induce anxiety and depression and greatly impair patients’ quality of life. One out of five Europeans suffers from chronic pain with most reporting that they endure it for more than two years (Breivik et al., 2006; Baker et al., 2010). Due to direct and follow-up costs they constitute a heavy burden for the health system (Phillips, 2006).
Of the painful neuropathies, the most frequent, painful diabetic polyneuropathy is a common complication of diabetes mellitus occurring in up to 20% of patients (Sommer, 2003; Sadosky et al., 2008). Good glycemic control can reduce the incidence of diabetic polyneuropathy but not painful diabetic polyneuropathy (PDPN) for which only symptomatic therapy of low to moderate efficacy is available to date (Vincent et al., 2011). Cellular mechanisms are emerging that include the classical changes of the diabetic milieu (Bierhaus and Nawroth, 2012; Bierhaus et al., 2012) however various studies have also identified signatures of neuroinflammation as critical components of painful diabetic polyneuropathy (Pabreja et al., 2011; Vincent et al., 2011). Pathological neuro-immune communication has also been associated with painful neuropathy that occurs in up to 50% of patients with traumatic peripheral nerve injury as a consequence of accidents, warfare or surgical procedures (Myers et al., 2006; Ciaramitaro et al., 2010; Birch et al., 2012). Also the neurogenic complex regional pain syndrome (CRPS) occurring as a complication of bone fracture, tissue injury or surgical interventions has a neuro-inflammatory component (Parkitny et al., 2013). In the majority of cases symptoms grossly resolve, however in 30% of patients pain symptoms persist or even intensify (Marinus et al., 2011). The beneficial effect of therapy with glucocorticosteroids in the acute phase of CRPS supports pathophysiological mechanisms associated with neuro-immune dysfunction (Üceyler et al., 2007a; Fischer et al., 2010; Marinus et al., 2011). Thus, converging evidence suggests that neuro-immune alterations in the peripheral and central nervous system play a major role in the general pathophysiology of neurogenic and neuropathic pain (McMahon and Malcangio, 2009; Kuner, 2010). Non-coding RNAs (ncRNAs), including microRNAs (miRNAs) and Piwi-binding piRNAs, are intimately associated with normal cellular as well as pathological processes (Mattick, 2004; Hüttenhofer et al., 2005; Hüttenhofer and Schattner, 2006). In this review we will focus on miRNAs since they are most extensively studied so far.
Various diseases, including neuropathic pain disorders, reveal unique miRNA expression signatures that can be exploited as diagnostic and prognostic markers. Recent reports on miRNA modulation of both neuronal and immune processes further predict therapeutic potential for manipulating disease-modified miRNAs in diseases affecting both the immune system and brain function, such as neuropathic pain disorders, Alzheimer’s disease, Parkinson’s disease, multiple sclerosis, and anxiety-related disorders (Soreq and Wolf, 2011; O’Connor et al., 2012).
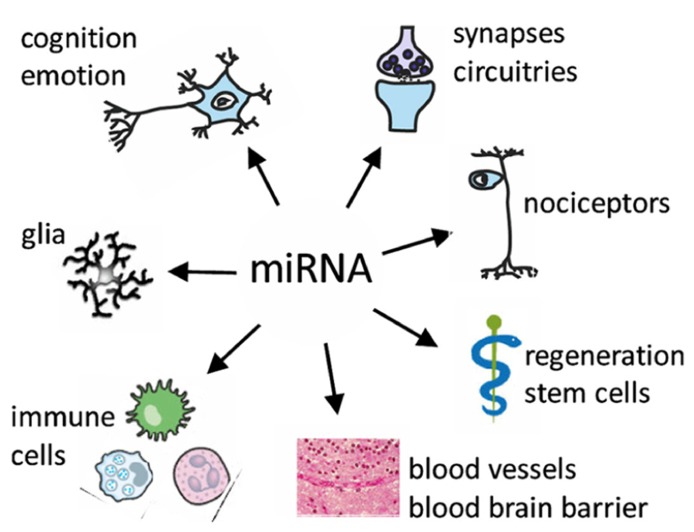
miRNAs that function within both the nervous and the immune systems possibly act as “negotiators” between these two interacting compartments (Figure 1). These “neurimmiRs” primarily target transcription factor genes or other regulatory genes, which enables simultaneous modulation of both immune and neuronal processes including cognition through direct or indirect alterations of neuron–glia or brain-to-body signaling (Soreq and Wolf, 2011). Thus, a given miRNA controls multiple cellular pathways, and miRNAs can act as “master switches” of the transcriptome or proteome, regulating multiple gene products and orchestrating multiple pathways including genes that encode cellular enzymes, trophic factors, receptor proteins, and ion channels many of which are individually pursued as drug targets.
FIGURE 1.
Targets of miRNA regulation networks.
Pain conditions have been suggested to deregulate the expression of miRNAs in pain pathways from primary afferent nociceptors to brain areas associated with emotional components of pain perception (Bai et al., 2007; Aldrich et al., 2009; Kusuda et al., 2011; Imai et al., 2011; Poh et al., 2011; von Schack et al., 2011). miRNAs are frequently deregulated and expressed at aberrant levels in diseased tissue, and first evidence suggests that this applies to neurogenic pain in CRPS (Orlova et al., 2011). Altered miRNA expression is frequently a consequence of genetic mutations, which may also cause loss or gain of function (Mishra and Bertino, 2009). This may account for inter-individual variation of pain sensitivity. However, the functional consequences of polymorphisms in miRNA genes and/or their binding sites, the downstream targets of miRNAs and the mechanisms by which miRNAs regulate circuitries and processes modulating nociception and endogenous analgesia are as yet unresolved.
Therapeutic miRNA regulation has been thoroughly studied and widely established in cancer research but its impact and the therapeutic prospects of miRNAs in the pain field are largely unexplored. Manipulation of miRNAs offers the possibility to control multiple targets including neuro-immune interactions, nociceptive processing and cognitive pathways. Both miRNAs and their isomiRNA versions are likely to each interact with many different targets, which may lead to downstream changes either due to the direct suppression of these targets or because of regulatory effects of those targets. Such downstream effects may be rather elaborate and are defined by some researchers “off-target” effects. However, we find that this definition may be misleading as it assumes that the physiological role of each miRNA is limited to the suppression of its direct targets. It is expected that miRNAs and miRNA derivatives will have few, if any, sequence-specific “off-target” effects. Thus, miRNA based diagnostics and therapeutics may have superior advantages by targeting multiple pain-associated genes and miRNA-based drugs may be the most appropriate therapy for the prevention or treatment of neuropathic pain.
BIOMARKERS FOR NEUROPATHIC AND NEUROGENIC PAIN SYNDROMES
Painful diabetic polyneuropathy is the most frequent painful neuropathy occurring in up to 20% of diabetic patients (Sommer, 2003; Sadosky et al., 2008). CRPS is an extremely painful condition that occurs in some patients after bone or tissue injury and peripheral nerve injury (traumatic neuropathy) and results in chronic neuropathic pain in many of these patients. These well-characterized albeit aetiologically diverse (metabolic, inflammatory, traumatic) neuropathic/neurogenic pain syndromes cover a spectrum of mechanisms underlying chronic pain. Nevertheless, the medical need for these syndromes is prevalent, and each of them is prototypic for an entire group of pain disorders.
It is unclear why diabetic neuropathy or traumatic neuropathy are painful in some instances and painless in others or why some patients develop CRPS after bone fracture, and why some recover from CRPS and others do not (Marinus et al., 2011). Thus, as yet unknown factors determine whether a given disorder entails chronic neuropathic pain. A first approach to be able to predict the individual risk of pain chronification was to use sensory phenotypes as surrogate markers for possible underlying mechanisms. Quantitative sensory testing (QST) is now well established but is still insufficient to disentangle specific pathophysiological mechanisms of chronic pain (Baron et al., 2012). One of the major hindrances in translating such findings into better therapy of neuropathic and neurogenic pain syndromes is the complexity of their pathophysiology. It is well known that alterations in many processes including ion channels, inflammatory mediators, neurotrophic factors, synaptic plasticity, and de- and regeneration, are involved, and that they even change during the course of the disease (Hehn et al., 2012). Therefore, a search for better and more specific diagnostic trait and state markers is one of the prerequisites for successful treatment in the future. Circulating miRNAs are detectable in body fluids including blood and cerebrospinal fluid and may be useful as novel biomarkers amenable to clinical diagnostic applications for various types of disease (Cogswell et al., 2008; Orlova et al., 2011; Ajit, 2012; Weiland et al., 2012; Machida et al., 2013). Therefore, it should be likewise promising to carefully assess which circulating miRNAs and novel ncRNAs are associated with neurogenic and neuropathic pain syndromes and may emerge as reliable diagnostic biomarkers for painful diabetic polyneuropathy, nerve injury pain, CRPS, headache and migraine.
NEW DRUGGABLE MOLECULAR TARGETS FOR PAIN TREATMENT
Treatment of painful diabetic polyneuropathy is far from satisfactory in many patients although this is the most intensely studied painful neuropathy in randomized controlled trials (RCTs). National and international guidelines differ in their recommendations about first and second line treatment choices. While pregabalin is favored by some (Bril et al., 2013), duloxetine or even tricyclic antidepressants are first choice in others (NICE-guideline; Attal et al., 2010; Dworkin et al., 2007). All of these drugs have adverse effects on diabetes. Furthermore, mean treatment effects comprise only two points of pain reduction on a 11-point Likert scale. In other types of neuropathy, like traumatic neuropathy or the frequent inflammatory types, there is little or no data at all from RCTs on pain treatment. Even worse, treatment of CRPS is neither standardized, nor satisfactory, nor based on multicentre RCTs. From single center studies with very limited patient numbers some evidence exists for anti-inflammatory treatment by corticosteroids or bisphosphonates in acute but not chronic stages, and for behavioral therapy for selected patients in chronic stages (de Tran et al., 2010). For the most frequently used invasive treatment modalities such as sympathetic blockers no RCT evidence of efficacy is available (Straube et al., 2010). Thus, more efficacious and specific medications are needed for both neurogenic and neuropathic pain syndromes.
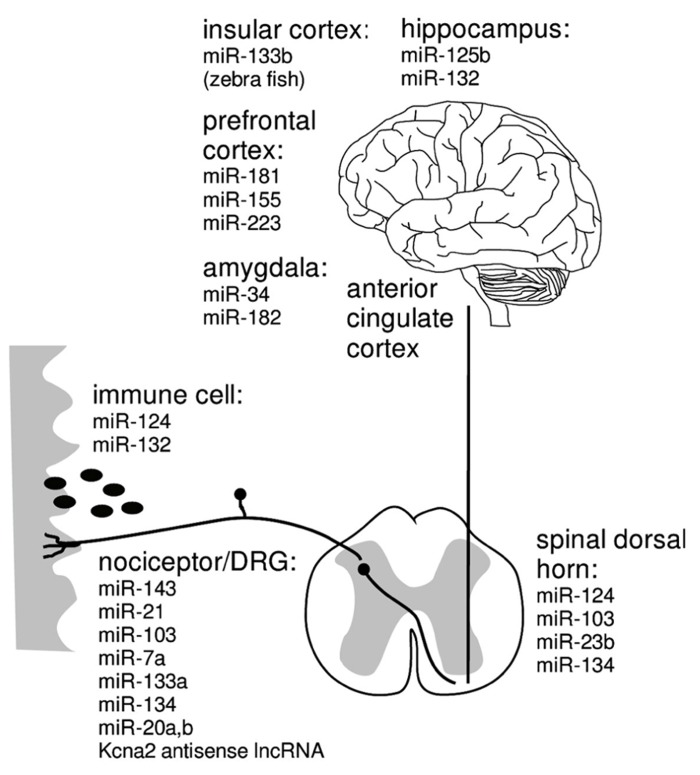
Both, the novel and specific mode of action and the ability to function as master switches of entire signaling networks has triggered enthusiasm for miRNAs as promising therapeutic targets although relatively little is known about the mechanisms of cellular uptake, storage and mode of action of miRNA modulators (van Rooji and Olson, 2012). In several rodent pain models, deregulated expression of miRNAs was found in pain pathways from primary afferent nociceptors to brain areas associated with emotional components of pain perception (Figure 2; Bai et al., 2007; Aldrich et al., 2009; Kusuda et al., 2011; Imai et al., 2011; Poh et al., 2011; von Schack et al., 2011). First evidence supporting a future for analgesic miRNA treatment comes from mice intrathecally receiving miR-124, miR-103 or miR-23b which are reported to prevent and treat persistent inflammatory and neuropathic pain (Favereaux et al., 2011; Imai et al., 2011; Willemen et al., 2012). Despite the fact that these miRNA treatments reduced signatures of synaptic modification, neuroinflammation and microglial response, the full extent and the mechanisms of the analgesic effect are not understood to date (Favereaux et al., 2011; Willemen et al., 2012).
FIGURE 2.
miRNA that may be causally associated with maintained neuropathic pain in immune cells (Shaked et al., 2009; Soreq and Wolf, 2011; Ponomarev et al., 2013), nociceptors/DRG (Ni et al., 2012; Sakai and Suzuki, 2013), spinal cord (Brandenburger et al., 2012; Favereaux et al., 2011; Im et al., 2012; Ni et al., 2012; Willemen et al., 2012), insular cortex (Sanchez-Simon et al., 2010), amygdala (Meerson et al., 2010; Haramati et al., 2011; Griggs et al., 2013), prefrontal cortex (Poh et al., 2011), hippocampus (Edbauer et al., 2010).
CIRCUITRIES AND PROCESSES MODULATING NOCICEPTION AND ENDOGENOUS ANALGESIA
Various studies have identified signatures of neuroinflammation as critical components of diabetic polyneuropathy (Pabreja et al., 2011; Vincent et al., 2011) in addition to the cellular mechanisms that include the classical changes of the diabetic milieu (Bierhaus et al., 2012; Bierhaus and Nawroth, 2012). Pathological neuro-immune communication has been associated with painful neuropathy following traumatic peripheral nerve injury (Myers et al., 2006; Ciaramitaro et al., 2010; Birch et al., 2012). Moreover, CRPS occurring as a complication of bone fracture or tissue injury results from neurogenic inflammatory processes (Goebel, 2011). In humans, a systemic pro-inflammatory profile distinguishes painful from painless neuropathy, and a local pro-inflammatory profile is part of the pathophysiology of small fiber neuropathy (Üceyler et al., 2007b, 2010). Specialized peripheral neurons, the nociceptors sense inflammatory or neuropathic conditions and respond with increased excitability and sensitivity leading to persisting pain and hyperalgesia (Costigan and Woolf, 2000; Sommer and Kress, 2004; Berta et al., 2008; Üceyler et al., 2009). However, mice lacking receptors for pro-inflammatory mediators in their nociceptor neurons are frequently protected from certain signatures of pathological pain (Andratsch et al., 2009; Schweizerhof et al., 2009; Mair et al., 2011; Quarta et al., 2011). The deficiency in anti-inflammatory cytokines in patients with CRPS (Üceyler et al., 2007a) together with beneficial effect of therapy with glucocorticosteroids support pathophysiological mechanisms associated with neuro-immune dysfunction (Fischer et al., 2010).
Inflammatory processes are also activated in the spinal cord upon peripheral nerve injury and involve microglia activation and leakage at the blood nerve barrier along the entire neuraxis (McMahon and Malcangio, 2009; Beggs et al., 2010, 2012). Microglia activation occurs in diabetic neuropathy in rodents (Wodarski et al., 2009; Beggs et al., 2012; Talbot and Couture, 2012) and has been recognized to be critical for the maintenance of neuropathic pain via the release of pro-nociceptive mediators (Clark et al., 2007). Leakage of the blood nerve barrier or the blood spinal barrier is just emerging in the pathophysiology of neuropathic pain accompanied by changes in tight junction proteins (Echeverry et al., 2011). Tight junction proteins which are critically involved in maintaining the blood–brain barrier like claudin-1 are also new targets, e.g., of miR-155 (Qin et al., 2013).
Deregulated miRNAs can be a consequence or cause of local inflammatory processes such as regulation of nociceptor sensitisation by controlling phospholipase A2 activation (Sun et al., 2012). Analyses of expression profiles of dorsal root ganglia (DRG) containing nociceptor cell bodies reveal that particular miRNAs are deregulated in rodent pain models giving rise to deregulation of miRNA-targeted ion channel expression patterns and metabotropic receptor transcripts in peripheral neurons which presumably cause nociceptor dysfunction (Zhao et al., 2010; von Schack et al., 2011). miRNAs are universal regulators of differentiation, activation and polarization of microglia in normal and inflammatory conditions (Ponomarev et al., 2013). Microglia and macrophage activity is suppressed by specific miRNAs, e.g., miR-124, and it is therefore anticipated that miRNA regulation is critically involved in endogenous inhibition and resolution of inflammation by e.g., resolvins (Ponomarev et al., 2011; Recchiuti et al., 2011). Certain miRNAs are substantially suppressed in glucocorticoid-treated thymocytes by reduced expression of the key miRNA processing enzymes Dicer, Drosha, and DGBR8/Pasha (Smith et al., 2010). This observation is of great relevance since CRPS for example is regarded a prototype disorder of failed termination of inflammation (Birklein and Kingery, 2009). The spinal release of immune modulators affects both spinal synaptic processes and local inhibitory circuits, possibly by classical cytokine-prostaglandin signaling and dys-inhibition of e.g., glycinergic spinal control (Samad et al., 2001; Harvey et al., 2004). Plastic changes at synapses in the spinal dorsal horn promote neuropathic and neurogenic pain via mechanisms involving enhanced nociceptive transmission but also inhibition of spinal endogenous analgesic circuits (Hartmann et al., 2004; Harvey et al., 2004; Fossat et al., 2007; Sandkühler, 2007, 2009; Pernïa-Andrade et al., 2009; Zeilhofer et al., 2009; Fossat et al., 2010; Laffray et al., 2012).
For a few miRNAs and long ncRNAs, downstream target proteins have been reported. For example, a conserved long ncRNA seems to modulate sensory neuron excitability by activation of a transcription factor MZF and downregulation of Kcna2 potassium channel expression and this has been causally associated with neuropathic pain (Zhao et al., 2013). In addition, the functional consequences of miR-103 regulation of voltage-gated Cav1.2 calcium channels and intrinsic excitability of spinal projection neurons have been demonstrated (Favereaux et al., 2011). It is well accepted that certain hereditary forms of migraine are associated with polymorphisms of voltage-gated calcium channels Cav2.1 and Cav2.2 (Pietrobon and Striessnig, 2003). Novel evidence suggests that in particular endogenous pain control systems including GABAergic and opioidergic synaptic signals are down-regulated by miRNAs such as miR-134 or miR-181a (Ni et al., 2012; Sengupta et al., 2013). Some of them link miRNAs like let-7 or miR-339 to opioid tolerance (He et al., 2010; He and Wang, 2012; Wu et al., 2013). In analogy, miRNA neuronal dys-regulation should not only apply to neurogenic or neuropathic pain but very likely the same principles and pathways should apply to other pain syndromes like headaches and in particular hereditary and other forms of migraine.
COGNITIVE, EMOTIONAL AND BEHAVIORAL COMPONENTS OF PAIN
Neuropsychological alterations are present in 65 % of CRPS patients and in particular cognitive impairment and deficits of emotional decision-making may impact their quality of life especially in risky, emotional situations (Apkarian et al., 2004). Emotional deficits and functional alterations in corresponding brain regions are reported in chronic CRPS patients and pain-related fear is one of the strongest predictors of disability in chronic pain disorders (Geha et al., 2008; de Jong et al., 2011).
Specific areas in the brain are actively involved in pain perception and behavior in humans and rodents and structural brain changes are associated with sensory and emotional function in rodent long-term neuropathic pain. In particular, decreased volumes of primary somatosensory and frontal cortex, retrosplenial and entorhinal cortex, anterior cingulate cortex and insula are maintained for months (Seminowicz et al., 2009). Specifically, abnormalities in hippocampus volume are observed in human CRPS and the mouse spared nerve injury (SNI) model. Similar to CRPS patients, SNI mice show increased anxiety like behavior and abnormal contextual fear extinction and this is associated with reduced extracellular signal-regulated kinase (ERK) expression, decreased neurogenesis and altered synaptic plasticity (Kodama et al., 2007; Mutso et al., 2012). Mice with experimental neuropathic pain also show cognitive deficits in novel object recognition and this is associated with deregulation of glycinergic neurotransmission in the hippocampus (Kodama et al., 2011), and may relate to reported enhanced quantal neurotransmitter release in the anterior cingulate cortex of mice with neuropathic pain (Toyoda et al., 2009). Dopaminergic and glutamatergic inputs from amygdala, hippocampus and prefrontal cortex to the nucleus accumbens participate in the putative emotional control circuits and recent human brain activity studies have examined the nucleus accumbens in the emotional aspects of pain processing (Baliki et al., 2010). These reports further link chronic pain with emotional dysfunction, and maladaptive responses of the nucleus accumbens in neuropathic pain have recently been associated with deregulated miRNAs in this region (Imai et al., 2011).
Brain-specific miRNAs are emerging as regulators of cognition, neuronal plasticity and memory by manipulating synapse structure and function, and specific miRNAs not only control cognition and emotional processes but also neuro-immune communication in the brain (Bredy et al., 2011; Soreq and Wolf, 2011). Mental retardation has been associated with miR-125b, miR-132 and other miRNAs and this arises from effects on dendritic spine morphology and synaptic physiology at hippocampal neurons. AMPA-mediated miniature mEPSC amplitude and frequency are reduced by neuronal over-expression of miR-125b and increased by miR-132 and this is due to differential regulation of glutamate NR2A and NR2B receptor mRNA levels (Edbauer et al., 2010). Other glutamate receptor subunits in the brain are regulated by dopamine through miR-181a which has recently been associated with the pain system (Saba et al., 2012). miR-132 is a highly interesting brain specific miRNA since it is up-regulated by brain derived neurotrophic factor (BDNF) and other growth factors in cortical neurons and this results in an increased expression of synaptic proteins including glutamate receptors (NR2A, NR2B and GluA1), an effect that is attenuated by glucocorticoids (Kawashima et al., 2010; Numakawa et al., 2011). Hippocampal miR-132 mediates stress-inducible cognitive deficits through acetylcholinesterase as a downstream target and specifically in the amygdala miR-34 is associated with the repression of stress-induced anxiety (Haramati et al., 2011; Shaltiel et al., 2013). More generally, happiness, anxiety and depression seem to depend on miRNA expression levels. Specific miRNAs are deregulated in patients suffering from depression and anxiety, and in pre-clinical models of psychological stress (Meerson et al., 2010). Moreover, psychoactive agents, including antidepressants and mood stabilizers, utilize miRNAs as downstream effectors (O’Connor et al., 2012). This further links neuropathic pain to emotional disorders and to the clinical benefit of antidepressants for pain treatment (Dworkin et al., 2007).
PAIN PREDISPOSING GENETIC POLYMORPHISMS
There is evidence that chronic pain, pain sensitivity and responsiveness to analgesic opioids show a sufficient heritability to make these phenotypes highly interesting sources for genetic variability which has an influence on pain (Angst et al., 2012; Hocking et al., 2012; Nielsen et al., 2012). Altered miRNA expression is frequently a consequence of genetic mutations, which may also cause loss or gain of function (Mishra and Bertino, 2009). This may account for significant inter-individual variation in the response to painful stimuli and analgesic drugs. Polymorphisms of specific molecular targets may be associated with certain pain phenotypes and this has emerged for example for a specific calcium channel subunit in a Drosophila screen that is conserved in mice and humans (Mogil, 2012; Neely et al., 2010). Several meta-analyses are available of the genetics of pain and associated specific loss- or gain of function polymorphisms with altered pain perception (LaCroix-Fralish et al., 2011; Mogil, 2012). A recent genome-wide association (GWA) study revealed three susceptibility loci for common migraine in the general population, however, systematic association studies are unavailable for DPN and CRPS to date (Chasman et al., 2011). In general, genetic studies have helped to understand the role and downstream mechanisms of individual proteins in pain processing, but specific single nucleotide polymorphism (SNP) related pain disorders apply to small numbers of individuals only and so far do not explain the large variability regarding susceptibility to distinct pain disorders or the responsiveness to different pain therapies in the general population (Dworkin et al., 2007; Attal et al., 2010).
The functional consequences of polymorphisms in miRNA genes and/or their binding sites, the downstream targets of miRNAs and the mechanisms by which miRNAs regulate circuitries and processes modulating nociception and endogenous analgesia are entirely unaddressed. SNPs in miRNAs or their target sites are not only bioinformatically predicted to be associated with the pathogenesis of diseases but are also experimentally validated (Wu et al., 2008; Coassin et al., 2010). It is known that SNPs are less common in miRNAs or their target sites than in other parts of the genome which points to the importance of miRNAs for cellular processes. However, on the other hand SNPs in these sites can affect the expression of a large number of genes when the production of the miRNA is influenced by that particular SNP. Moreover, SNPs in target sites of miRNAs can either modulate/disrupt existing binding sites or create new binding sites for the miRNAs that may then influence gene expression. SNPs in these regions have become a major focus of research and some of them are expected to explain pathogenetic mechanisms in disease development (Glinsky, 2008; Haas et al., 2012). For example, miRNA expression is markedly different between normal tissues and tumor tissues although otherwise miRNA expression is strictly controlled. This might be explained by somatic mutations that are introduced during carcinogenesis. The investigation of genetic variants at miRNAs or their target sites and their association with various diseases is only in its infancy. Initial studies show that these RNA chains might also be involved in neurological diseases such as Parkinson’s disease (Martins et al., 2011), Alzheimer’s disease (Serpente et al., 2011) or frontotemporal lobar degeneration (Villa et al., 2011). The identification of SNPs in miRNA related regions of the genome might be advantageous over classical GWA study since individual ncRNAs may control and regulate whole networks and pathways involving a multitude of functional proteins. This may open a new avenue that may potentially improve our understanding of extensive inter-individual differences in patients.
TRANSLATION OF PRE-CLINICAL AND CLINICAL RESULTS INTO SOLUTIONS FOR THE BENEFIT OF PATIENTS
As stated above, one of the major hindrances in the way of translating such findings into better therapy of neuropathic and neurogenic pain syndromes is the complexity of their pathophysiology, which even changes during the course of disease. Based on and in analogy to recent developments in the oncology field, an improved understanding of the role of miRNAs in neuropathic pain might be highly useful for diagnostic and prognostic assessments. For example, aberrant expression or functional deregulation of miRNAs has been associated with the risk for and progression of malignancies and this knowledge is expected to advance the management of certain cancer types through the development of novel personalized miRNA-based diagnostics and therapies (Dreussi et al., 2012; Rossi and Calin, 2013). Increasing evidence indicates that certain miRNAs may be aberrantly expressed or deregulated in certain individuals after tissue injury or with diabetes. This may be associated with increased risk of pain chronification or even responsiveness to analgesic drugs (Ivanov et al., 2012). Therefore, miRNAs are expected to have potential for personalized pain medicine as biomarkers for risk assessment, drug selection and novel therapies.
Therapeutic miRNA regulation has been thoroughly studied and begins to be established in different types of cancer, and the first miRNA targeted drug has entered phase II clinical trials (Lindow and Kauppinen, 2012). In contrast, the potential therapeutic impact of miRNAs in the pain field is as yet largely unexplored. To date, therapeutic approaches have been restricted to rodent models and intrathecal administration and some inconsistencies have emerged; thus miRNA increases in a disease may be either a cause or a feedback reaction to the observed symptoms. For example, although miR-124 is up-regulated after chronic constrictive nerve injury (CCI), intrathecal administration of miR-124 can prevent and treat persistent inflammatory and neuropathic pain (Willemen et al., 2012). Likewise, miR-132 levels are increased in colon biopsies from patients with intestinal bowel disease which should predictably limit inflammation (Maharshak et al., 2013). Importantly, manipulation of miRNAs offers the possibility to control multiple targets including neuro-immune interactions, nociceptive processing and cognitive and affective pathways. Thus, miRNA based therapeutics may have superior advantages by targeting multiple pain-associated genes and miRNA-based drugs may be the most appropriate therapy for the prevention or treatment of neuropathic and neurogenic pain. At least, recent developments provide an optimistic perspective on the evolution of therapeutic ncRNAs despite the drawback of unresolved obstacles for successful delivery and unknown, however unlikely, off-target effects (Cho, 2012).
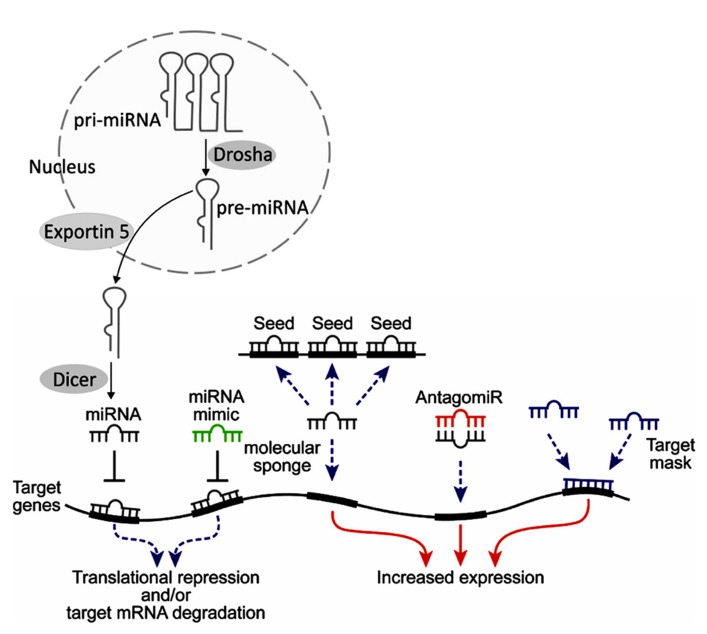
Manipulating miRNAs as a therapeutic tool presents significant theoretical and practical challenges that must be overcome before this approach becomes a reality. Specific examples involve two of the more straightforward approaches for miRNA modulation, miRNA mimics and antagomirs (Figure 3). miRNA mimics consist of over-expressing specific miRNAs that are reduced in the disease state. This mimic approach could be done by introducing synthetic oligonucleotides (natural or modified) or involve over-expression of such miRNAs from an introduced viral vector. Antagomirs are synthetic oligonucleotide sequences that are designed to be inversely oriented (antisense) to miRNAs that are over-expressed in the disease state and which can form Watson-Crick base pairing with the target miRNA. This can either inactivate a miRNA or result in its degradation. Similar to the miRNA mimics, these therapeutic and research tools can also consist of synthetic or modified nucleic acid sequences or be overexpressed from viral vectors.
FIGURE 3.
Endogenous miRNAs are generated from primary (pri-) miRNAs via cleavage by the RNAse Drosha into pre-miRNAs in the nucleus. They are exported into the cytosol by Exportin 5 and there are cleaved into active miRNAs by the RNAse Dicer. Depending on the degree of homology, miRNAs trigger translational repression or degradation of target mRNAs (for review see He and Hannon, 2004; Bartel, 2009). Therapeutic manipulations of miRNAs may involve various methods. Host tissue miRNAs (gray) bind to complementary sequences, which are often located in the 3′-untranslated region (3′-UTR) of the target genes. This leads to translational repression, often accompanied by degradation. Mimicking this process, miRNA mimics (green) with similar sequences to those of miRNAs may be designed to target the same mRNAs. Such mimics are synthetic oligonucleotides that are chemically protected against nucleolytic degradation. Alternative routes include molecular “sponges,” with several binding sites of a certain miRNA; antagomiRs (red), complementary oligonucleotides to the host miRNA which bind to it and limit its function, and target masks, which bind to part of target miRNAs and compete with their function. Thus, tools exist both for inducing gain of function (red arrows) or loss of function (dashed blue arrows).
Alternative methodologies used in experimental settings include miRNA sponges, which are exogenous DNA repeats of the target sequence and can serve to soak up excess copies of the excess miRNA (Ebert et al., 2007). The miRNA sponges may be produced under the regulation of RNA Polymerase III promoters and can generate high amounts of specific target sequences. Another novel yet promising approach involves target protection. In this application, modified antisense oligonucleotides such as LNA or morpholinos are prepared that will be complementary to a specific sequence in the target gene messenger RNA. These are added to the cells, where they bind to the target sequence, block its down-regulation by the miRNA complex and ensure sufficient expression of the target mRNA (Choi et al., 2007). Enhanced and prolonged miRNA suppression and simultaneous targeting of multiple miRNAs can be achieved by inhibitors carrying clustered hairpins based on the “Tough decoy” (TuD) design which offer the advantage of standardized suppression of families or clusters of miRNAs and can be combined with recombinant adenovirus vectors (Haraguchi et al., 2009; Xie et al., 2012; Bak et al., 2013; Hollensen et al., 2013).
An important difficulty that may be predicted for developing neuronal miRNA therapeutics is delivery, since targeting to the brain involves the significant hurdle of crossing the blood–brain barrier. Nevertheless, therapeutic efficacy of certain approaches such as the use of LNA antagomirs has been demonstrated even in primate models, and certain neuronal miRNA therapeutic approaches are now in preclinical development. These studies cover several creative approaches that have been developed to overcome the delivery problem. Thus, ~20-mer miRNA-size oligonucleotides are indeed unlikely to cross the blood–brain barrier. However, peripheral administration of oligonucleotide controllers of inflammation-regulating miRNAs would change the levels of cytokines, and cytokines can penetrate and affect the brain. Such effects have been demonstrated for miR-132 (Shaked et al., 2009) and miR-212 (Hollander et al., 2010). Other means include direct infection of cerebral neurons with viral vectors that may be adapted for better tropism to neuronal cells (Barbash et al., 2013). Direct introduction of antisense oligonucleotides can alternatively be performed by intracerebroventricular or local stereotactic injection though these would be extremely problematic in pain syndromes. Yet more recent work described the use of rabies virus glycoprotein labeled nanoparticles to enable direct delivery of a miRNA mimic to neuronal cells (Hwang do et al., 2011).
CONCLUSION
Recently, specific miRNAs have been associated with pathological pain and the deregulation of ion channel expression in sensory neurons in rodent pain models (Zhao et al., 2010; Favereaux et al., 2011; Li et al., 2011). Pain conditions have been suggested to deregulate the expression of miRNAs in pain pathways from primary afferent nociceptors to brain areas associated with emotional components of pain perception (Bai et al., 2007; Aldrich et al., 2009; Imai et al., 2011; Kusuda et al., 2011; Poh et al., 2011; von Schack et al., 2011; Arai et al., 2013; Genda et al., 2013; Sakai and Suzuki, 2013). Unique signatures of miRNAs are associated with altered innate immune signaling and secreted miRNAs are even considered a new form of neuroimmune communication and control immune cell activity as well as neuron function (Peng et al., 2010; Bredy et al., 2011; Chen et al., 2012; Ponomarev et al., 2013). miRNAs act at the neuro-immune interface which controls neuronal plasticity and memory but also are linked to the etiology of anxiety and mood disorders (Bredy et al., 2011; Soreq and Wolf, 2011; O’Connor et al., 2012; Shaltiel et al., 2013). Such deficits in the interaction of immune cells and neurons together with cognitive and emotional alterations in patients with neuropathic or neurogenic pain syndromes are hypothesized to converge on miRNA deregulated mechanisms along the entire neuraxis, and alterations in miRNA expression may account for the variation of susceptibility to certain types of pain or even for the responsiveness to analgesics and opioid tolerance (Parsons et al., 2008). Understanding the role of miRNAs in pain mechanisms is suggested to provide great benefit for clinical diagnostic and therapeutic applications.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work is supported by the European Commission (GA N 602133 - ncRNAPain).
REFERENCES
- Ajit S. K. (2012). Circulating microRNAs as biomarkers, therapeutic targets, and signaling molecules. Sensors (Basel) 12 3359–3369 10.3390/s120303359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aldrich B. T., Frakes E. P., Kasuya J., Hammond D. L., Kitamoto T. (2009). Changes in expression of sensory organ-specific microRNAs in rat dorsal root ganglia in association with mechanical hypersensitivity induced by spinal nerve ligation. Neuroscience 164 711–723 10.1016/j.neuroscience.2009.08.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andratsch M., Mair N., Constantin C. E., Scherbakov N., Benetti C., Quarta S., et al. (2009). A key role for gp130 expressed on peripheral sensory nerves in pathological pain. J. Neurosci. 29 13473–13483 10.1523/JNEUROSCI.1822-09.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angst M. S., Phillips N. G., Drover D. R., Tingl M., Ray A., Swan G. E., et al. (2012). Pain sensitivity and opioid analgesia: a pharmacogenomic twin study. Pain 153 1397–1409 10.1016/j.pain.2012.02.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apkarian A. V., Sossa Y., Krauss B. R., Thomas P. S., Fredrickson B. E., Levy R. E., et al. (2004). Chronic pain patients are impaired on an emotional decision-making task. Pain 108 129–136 10.1016/j.pain.2003.12.015 [DOI] [PubMed] [Google Scholar]
- Arai M., Genda Y., Ishikawa M., Shunsuke T., Okabe T., Sakamoto A. (2013). The miRNA and mRNA changes in rat hippocampi after chronic constriction injury. Pain Med. 14 720–729 10.1111/pme.12066 [DOI] [PubMed] [Google Scholar]
- Attal N., Cruccu G., Baron R., Haanpää M., Hansson P., Jensen T. S., et al. (2010). EFNS guidelines on the pharmacologcial treatment of neuropathic pain. Eur. J. Neurol. 17 1113–e88 10.1111/j.1468-1331.2010.02999.x [DOI] [PubMed] [Google Scholar]
- Bai G., Ambalavanar R., Wei D., Dessem D. (2007). Downregulation of selective microRNAs in trigeminal ganglion neurons following inflammatory muscle pain. Mol. Pain 3 15 10.1186/1744-8069-3-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bak R. O., Hollensen A. K., Primo M. N., Sørensen C. D., Mikkelsen J. G. (2013). Potent microRNA suppression by RNA Pol II-transcribed ‘Tough Decoy’ inhibitors. RNA 19 280–293 10.1261/rna.034850.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker M., Collet B., Fischer A., Hermann V., Huygen F. J. P., Trueman P., et al. (2010). Improving the Current and Future Management of Chronic Pain. A European Consensus Report. Brussels: Pfizer [Google Scholar]
- Baliki M. N., Geha P. Y., Fields H. L., Apkarian A. V. (2010). Predicting value of pain and analgesia: nucleus accumbens response to noxious stimuli changes in the presence of chronic pain. Neuron 66 149–160 10.1016/j.neuron.2010.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbash S., Hanin G., Soreq H. (2013). Stereotactic injection of microRNA-expressing lentiviruses to the mouse hippocampus CA1 region and assessment of the behavioral outcome. J. Vis. Exp. 10.3791/50170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baron R., Forster M., Binder A. (2012). Subgrouping of patients with neuropathic pain according to pain-related sensory abnormalities: a first step to a stratified treatment approach. Lancet Neurol. 11 999–1005 10.1016/S1474-4422(12)70189-8 [DOI] [PubMed] [Google Scholar]
- Bartel D. P. (2009). MicroRNAs: target recognition and regulatory functions. Cell 136 2015–2233 10.1016/j.cell.2009.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beggs S., Liu X. J., Kwan C., Salter M. W. (2010). Peripheral nerve injury and TRPV1-expressing primary afferent C-fibres cause opening of hte blood–brain barrier. Mol. Pain 6 74 10.1186/1744-8069-6-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beggs S., Trang T., Salter M. W. (2012). P2X4R(+) microglia drive neuropathic pain. Nat. Neurosci. 15 1068–1073 10.1038/nn.3155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berta T., Poirot O., Pertin M., Ji R. R., Kellenberger S., Decosterd I. (2008). Transcriptional and functional profiles of voltage-gated Na+ channels in injured and non-injured DRG neurons in the SNI model of neuropathic pain. Mol. Cell. Neurosci. 37 196–208 10.1016/j.mcn.2007.09.007 [DOI] [PubMed] [Google Scholar]
- Bierhaus A., Fleming T., Stoyanov S., Leffler A., Babes A., Neacsu C., et al. (2012). Methylglyoxal modification of Na(v)1.8 facilitates nociceptive neuron firing and causes hyperalgesia in diabetic neuropathy. Nat. Med. 18 926–933 10.1038/nm.2750 [DOI] [PubMed] [Google Scholar]
- Bierhaus A., Nawroth P. P. (2012). Critical evaluation of mouse models used to study pain and loss of pain perception in diabetic neuropathy. Exp. Clin. Endocrinol. Diabetes 120 188–190 10.1055/s-0032-1304567 [DOI] [PubMed] [Google Scholar]
- Birch R., Misra P., Stewart M. P., Eardley W. G., Ramasamy A., Brown K., et al. (2012). Nerve injuries sustained during warfare: part I – epidemiology. J. Bone Joint Surg. Br. 94 523–528 10.1302/0301-620X.94B4.28483 [DOI] [PubMed] [Google Scholar]
- Birklein F., Kingery W. S. (2009). Complex regional pain syndrome: a loss of inhibition? Pain 142 177–178 10.1016/j.pain.2009.01.029 [DOI] [PubMed] [Google Scholar]
- Brandenburger T., Castoldi M., Brendel M., Grievink H., Schlösser L., Werdehausen R., et al. (2012). Expression of spinal cord microRNAs in a rat model of chronic neuropathic pain. Neurosci. Lett. 506 281–286 10.1016/j.neulet.2011.11.023 [DOI] [PubMed] [Google Scholar]
- Bredy T. W., Lin Q., Wei W., Baker-Andresen D., Mattick J. S. (2011). MicroRNA regulation of neural plasticity and memory. Neurobiol. Learn. Mem. 96 89–94 10.1016/j.nlm.2011.04.004 [DOI] [PubMed] [Google Scholar]
- Breivik H., Collett B., Ventafridda V., Cohen R., Gallacher D. (2006). Survey of chronic pain in Europe: prevalence, impact on daily life, and treatment. Eur. J. Pain 10 287–333 10.1016/j.ejpain.2005.06.009 [DOI] [PubMed] [Google Scholar]
- Bril V., England J., Franklin G. M., Backonja M., Cohen J., Del Toro D., et al. (2013). Evidence-based guidelines: treatment of painful diabetic neuropathy: report of the American Academy of Neurology, the American Association of Neuromuscular and Electrodiagnostic Medicine and the American Academy of Physical Medicine and Rehabilitation. Neurology 76 1758–1765 10.1212/WNL.0b013e3182166ebe [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chasman D. I., Schürks M., Anttila V., de Vries B., Schminke U., Launer L. J., et al. (2011). Genome-wide association study reveals thress susceptibility loci for common migraine in the general population. Nat. Genet. 43 695–698 10.1038/ng.856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho W. C. (2012). Exploiting the therapeutic potential of microRNAs in human cancer. Expert Opin. Ther. Targets 16 345–350 10.1517/14728222.2012.663354 [DOI] [PubMed] [Google Scholar]
- Choi W. Y., Giraldez A. J., Schier A. F. (2007). Target protectors reveal dampening and balancing of Nodal agonist and antagonist by miR-430. Science 318 271–274 10.1126/science.1147535 [DOI] [PubMed] [Google Scholar]
- Ciaramitaro P., Mondelli M., Logullo F., Grimaldi S., Battiston B., Sard A., et al. (2010). Traumatic peripheral nerve injuries: epidemiological findings, neuropathic pain and quality of life in 158 patients. J. Peripher. Nerv. Syst. 15 120–127 10.1111/j.1529-8027.2010.00260.x [DOI] [PubMed] [Google Scholar]
- Clark A. K., Yip P. K., Grist J., Gentry C., Staniland A. A., Marchand F., et al. (2007). Inhibition of spinal microglial cathepsin S for the reversal of neuropathic pain. Proc. Natl. Acad. Sci. U.S.A. 104 10655–10660 10.1073/pnas.0610811104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coassin S., Brandstätter A., Kronenberg F. (2010). Lost in the space of bioinformatic tools: a constantly updated survival guide for genetic epidemiology. The GenEpi Toolbox. Atherosclerosis 209 321–335 10.1016/j.atherosclerosis.2009.10.026 [DOI] [PubMed] [Google Scholar]
- Cogswell J. P., Ward J., Taylor I. A., Waters M., Shi Y., Cannon B., et al. (2008). Identification of miRNA changes in Alzheimer’s disease brain and CSF yields putative biomarkers and insights into disease pathways. Alzheimers Dis. 14 27–41 [DOI] [PubMed] [Google Scholar]
- Costigan M., Woolf C. J. (2000). Pain: molecular mechanisms. J. Pain 3 35–44 10.1054/jpai.2000.9818 [DOI] [PubMed] [Google Scholar]
- de Jong J. R., Vlaeyen J. W., de Gelder J. M., Patijn J. (2011). Pain-related fear, perceived harmfulness of activities, and functional limitations in complex regional pain syndrome type I. J. Pain 12 1209–1218 10.1016/j.jpain.2011.06.010 [DOI] [PubMed] [Google Scholar]
- de Tran Q. H., Duon G., Bertini P., Finlayson R. J. (2010). Treatment of complex regional pain syndrome: a review of the evidence. Can. J. Anaesth. 57 149–166 10.1007/s12630-009-9237-0 [DOI] [PubMed] [Google Scholar]
- Dreussi E., Biason P., Toffoli G., Ceccin E. (2012). miRNA pharmacogenomics: the new frontier for personalized medicine in cancer? Pharmacogenomics 13 1635–1650 10.2217/pgs.12.147 [DOI] [PubMed] [Google Scholar]
- Dworkin R. H., O’Connor A. B., Backonja M., Farrar J. T., Finnerup N. B., Jensen T. S., et al. (2007). Pharmacological management of neuropathic pain: evidence-based recommendations. Pain 132 237–251 10.1016/j.pain.2007.08.033 [DOI] [PubMed] [Google Scholar]
- Ebert M. S., Neilson J. R., Sharp P. A. (2007). MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells. Nat. Methods 4 721–726 10.1038/nmeth1079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Echeverry S., Shi X. Q., Rivest S., Zhang J. (2011). Peripheral nerve injury alters blood-spinal cord barrier functional and molecular integrity through a selective inflammatory pathway. J. Neurosci. 31 10819–10828 10.1523/JNEUROSCI.1642-11.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edbauer D., Neilson J. R., Foster K. A., Wang C.-F., Seeburg D. P., Batterton M. N., et al. (2010). Regulation of synaptic structure and function by FMRP-associated microRNAs miR-125b and miR-132. Neuron 65 373–384 10.1016/j.neuron.2010.01.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favereaux A., Thoumine O., Bouali-Benazzouz R., Roques V., Papon M. A., Salam S. A., et al. (2011). Bidirectional integrative regulation of Cav1.2 calcium channel by microRNA miR-103: role in pain. EMBO J. 30 3830–3841 10.1038/emboj.2011.249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer S. G. L., Zuumond W. W. A., Birklein F., Loer S. A, Perez R. S. G. M. (2010). Anti-inflammatory treatment of complex regional pain syndrome. Pain 151 251–256 10.1016/j.pain.2010.07.020 [DOI] [PubMed] [Google Scholar]
- Fossat P., Dobremez E., Bouali-Benazzouz R., Favereaux A., Bertrand S., Kilk K., et al. (2010). Knock-down of L calcium channel subtypes: differential effects on neuropathic pain. J. Neurosci. 30 1073–1085 10.1523/JNEUROSCI.3145-09.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fossat P., Sibon I., LeMasson G., Landry M., Nagy F. (2007). L-type calcium channels and NMDA receptors: a determinant duo for short-term nociceptive plasticity. Eur. J. Neurosci. 25 127–135 10.1111/j.1460-9568.2006.05256.x [DOI] [PubMed] [Google Scholar]
- Geha P. Y., Baliki M. N., Harden R. N., Bauer W. R., Parrish T. B., Apkarian A. V. (2008). The brain in chronic CRPS pain: abnormal gray-white matter interactions in emotional and authnomic regions. Neuron 60 570–581 10.1016/j.neuron.2008.08.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genda Y., Arai M., Ishikawa M., Tanaka S., Okabe T., Sakamoto A. (2013). microRNA changes in the dorsal horn of the spinal cord of rats with chronic constriction injury: A TaqMan® Low Density Array study. Int. J. Mol. Med. 31 129–137 10.3892/ijmm.2012.1163 [DOI] [PubMed] [Google Scholar]
- Glinsky G. V. (2008). An SNP-guided microRNA map of fifteen common human disorders identifies a consensus disease phenocode aiming at prinicpal components of the nuclear import pathway. Cell Cycle 7 2570–2583 10.4161/cc.7.16.6524 [DOI] [PubMed] [Google Scholar]
- Goebel A. (2011). Complex regional pain syndrome in adults. Rheumatology 50 1739–1750 10.1093/rheumatology/ker202 [DOI] [PubMed] [Google Scholar]
- Griggs E. M., Young E. J., Rumbaugh G., Miller C. A. (2013). MicroRNA-182 regulates amygdala-dependent memory formation. J. Neurosci. 33 1734–1740 10.1523/JNEUROSCI.2873-12.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas U., Sczakiel G., Laufer S. D. (2012). MicroRNA-mediated regulation of gene expression is affected by disease-asssociated SNPs within the 3’UTR via altered RNA structure. RNA Biol. 9 924–937 10.4161/rna.20497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haraguchi T., Ozaki Y., Iba H. (2009). Vectors expressing efficient RNA decoys achieve the long-term suppression of specific microRNA activity in mammalian cells. Nucleic Acid Res. 37 e43 10.1093/nar/gkp040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haramati S., Navon I., Issler O., Ezra-Nevo G., Gil S., Zwang R., et al. (2011). microRNAs as repressors of stress-induced anxiety: the case of amygdalar miR-34. J. Neurosci. 31 14191–14203 10.1523/JNEUROSCI.1673-11.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann B., Ahmadi S., Heppenstall P. A., Lewin G. R., Schott C., Borchardt T., et al. (2004). The AMPA receptor subunits GluR-A and GluR-B reciprocally modulate spinal synaptic plasticity and inflammatory pain. Neuron 44 637–650 10.1016/j.neuron.2004.10.029 [DOI] [PubMed] [Google Scholar]
- Harvey R. J., Depner U. B., Wassle H., Ahmadi S., Heindl C., Reinold H., et al. (2004). GlyR alpha3: an essential target for spinal PGE2-mediated inflammatory pain sensitization. Science 304 884–887 10.1126/science.1094925 [DOI] [PubMed] [Google Scholar]
- He L., Hannon G. J. (2004). MicroRNAs: small RNAs with a big role in gene regulation. Nat. Rev. Genet. 5 522–531 10.1038/nrg1379 [DOI] [PubMed] [Google Scholar]
- He Y., Wang Z. J. (2012). Let-7 microRNAs and opoid tolerance. Front. Genet. 3:110 10.3389/fgene.2012.000110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y., Yang C., Kirkmire C. M., Wang Z. J. (2010). Regulation of opoid tolerace by let-7 family microRNA targeting the mu opioid receptor. J. Neurosci. 30 10251–10258 10.1523/JNEUROSCI.2419-10.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hehn C. A., Baron R., Woolf C. J. (2012). Deconstructing the neuropathic pain phenotype to reveal neural mechanisms. Neuron 73 638–652 10.1016/j.neuron.2012.02.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hocking L. J., Generation S., Morris A. D., Dominiczak A. F., Porteous D. J., Smith B. H. (2012). Heritability of chronic pain in 2195 extended families. Eur. J. Pain 16 1053–1063 10.1002/j.1532-2149.2011.00095.x [DOI] [PubMed] [Google Scholar]
- Hollander J. A., Im H. I., Amelio A. L., Kocerha J., Bali P., Lu Q., et al. (2010). Striatal microRNA controls cocaine intake through CREB signalling. Nature 466 197–202 10.1038/nature09202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollensen A. K., Bak R. O., Haslund D., Mikkelsen J. G. (2013). Suppression of microRNAs by dual targeting and clustered Tough Decoy inhibitors. RNA Biol. 10 406–414 10.4161/rna.23543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hüttenhofer A., Schattner P. (2006). The principles of guiding by RNA: chimeric RNA–protein enzymes. Nat. Rev. Genet. 7 475–482 10.1038/nrg1855 [DOI] [PubMed] [Google Scholar]
- Hüttenhofer A., Schattner P., Polacek N. (2005). Non-coding RNAs: hope or hype? Trends Genet. 21 289–297 10.1016/j.tig.2005.03.007 [DOI] [PubMed] [Google Scholar]
- Hwang do W., Son S., Jang J., Youn H., Lee S., Lee D., et al. (2011). A brain-targeted rabies virus glycoprotein-disulfide linked PEI nanocarrier for delivery of neurogenic microRNA. Biomaterials 32 4968–4975 10.1016/j.biomaterials.2011.03.047 [DOI] [PubMed] [Google Scholar]
- Im Y. B., Choi J. I., Cho H. T., Kwon O. H., Kang S. K. (2012). Molecular targeting of NOX4 for neuropathic pain after traumatic injury of the spinal cord. Cell Death Dis. 3 e426 10.1038/cddis.2012.168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai S., Saeki M., Yanase M., Horiuchi H., Abe M., Narita M., et al. (2011). Change in microRNAs associated with neuronal adaptive responses in the nucleus accumbens under neuropathic pain. J. Neurosci. 31 15294–15299 10.1523/JNEUROSCI.0921-11.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov M., Kacevska M., Ingelman-Sundberg M. (2012). Epigenomics and interindividual differences in drug response. Clin. Pharmacol. Ther. 92 727–736 10.1038/clpt.2012.152 [DOI] [PubMed] [Google Scholar]
- Kawashima H., Numakawa T., Kumumaru E., Adachi N., Mizuno H., Ninomiya M., Kunugi H., Hashido K. (2010). Glucocorticoid attenuates brain-derived neurotrophic factor-dependent upregulation of glutamate receptors via the suppression of microRNA-132 expression. Neuroscience 165 1301–1311 10.1016/j.neuroscience.2009.11.057 [DOI] [PubMed] [Google Scholar]
- Kodama D., Ono H., Tanabe M. (2007). Altered hippocampal long-term potentiation after peripheral nerve injury in mice. Eur. J. Pharmacol. 574 127–132 10.1016/j.ejphar.2007.07.054 [DOI] [PubMed] [Google Scholar]
- Kodama D., Ono H., Tanabe M. (2011). Increased hippocampal glycine uptake and cognitive dysfunction after peripheral nerve injury. Pain 152 809–817 10.1016/j.pain.2010.12.029 [DOI] [PubMed] [Google Scholar]
- Kuner R. (2010). Central mechanisms fo pathological pain. Nat. Med. 16 1258–1266 10.1038/nm.2231 [DOI] [PubMed] [Google Scholar]
- Kusuda R., Cadetti F., Ravanelli M. I., Sousa T. A., Zanon S., De Lucca F. L., et al. (2011). Differential expression of microRNAs in mouse pain models. Mol. Pain 7 17 10.1186/1744-8069-7-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaCroix-Fralish M. L., Austin J.-S., Zheng F. Y., Levitin D. J., Mogil J. S. (2011). Patterns of pain: meta-analysis of microarray studies of pain. Pain 152 1888–1898 10.1016/j.pain.2011.04.014 [DOI] [PubMed] [Google Scholar]
- Laffray S., Bouali-Benazzouz R., Papon M. A., Favereaux A., Jiang Y., Holm T., et al. (2012). Impairment of GABAB receptor dimer by endogenous 14-3-z in chronic pain conditions. EMBO J. 31 3239–3251 10.1038/emboj.2012.161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindow M., Kauppinen S. (2012). Discovering the first micrRNA-targeted drug. J. Cell Biol. 199 407–412 10.1083/jcb.201208082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machida A., Ohkubo T., Yokota T. (2013). Circulating microRNAs in the cerebrospinal fluid of patients with brain diseases. Methods Mol. Biol. 1024 203–209 10.1007/978-1-62703-453-1_16 [DOI] [PubMed] [Google Scholar]
- Maharshak N., Shenhar-Tsarfaty S., Aroyo N., Orpaz N., Guberman I., Canaani J., et al. (2013). Micro-RNA-132 modulates cholinergic signaling and inflammation in human inflammatory bowel disease. Inflamm. Bowel Dis. 19 1346–1353 10.1097/MIB.0b013e318281f47d [DOI] [PubMed] [Google Scholar]
- Mair N., Benetti C., Andratsch M., Leitner M. G., Constantin C. E., Camprubï-Robles M., et al. (2011). Genetic evidence for involvement of neuronally expressed S1P1receptor in nociceptor sensitization and inflammatory pain. PLoS ONE 6:e17268 10.1371/journal.pone.0017268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marinus J., Moseley G. L., Birklein F. B., Baron R., Maihöfner C., Kingery W. S., et al. (2011). Clinical features and pathophysiology of complex regional pain syndrome. Lancet Neurol. 10 637–648 10.1016/S1474-4422(11)70106-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins M., Rosa A., Guedes L. C., Fonseca B. V., Gotovac K., Violante S., et al. (2011). Convergence of miRNA expression profiling, a-synuclein interacton and GWAS in Parkinson’s disease. PLoS ONE 6:e25443 10.1371/journal.pone.0025443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattick J. S. (2004). RNA regulation: a new genetics? Nat. Rev. Genet. 5 316–323 10.1038/nrg1321 [DOI] [PubMed] [Google Scholar]
- McMahon S. B., Malcangio M. (2009). Current challenges in glia-pain biology. Neuron 64 46–54 10.1016/j.neuron.2009.09.033 [DOI] [PubMed] [Google Scholar]
- Meerson A., Cacheaux L., Goosens K. A., Sapolsky R. M., Soreq H., Kaufer D. (2010). Changes in brain microRNAs contribute to cholinergic stress reactions. J. Mol. Neurosci. 40 47–55 10.1007/s12031-009-9252-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra P. J., Bertino J. R. (2009). MicroRNA polymorphisms: the future of pharmacogenomics, molecular epidemiology and individualized medicine. Pharmacogenomics 10 399–416 10.2217/14622416.10.3.399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogil J. S. (2012). Pain genetics: past, present and future. Trends Genet. 28 258–266 10.1016/j.tig.2012.02.004 [DOI] [PubMed] [Google Scholar]
- Mutso A. A., Radzicki D., Baliki M. N., Huang L., Banisadr G., Centeno M. V., et al. (2012). Abnormalities in hippocampal functioning with persistent pain. J. Neurosci. 32 5747–5756 10.1523/JNEUROSCI.0587-12.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers R. R., Campana W. M., Shubayev V. I. (2006) The role of neuroinflammation in neuropathic pain: mechanisms and therapeutic targets. Drug Discov. Today 11 8–20 10.1016/S1359-6446(05)03637-8 [DOI] [PubMed] [Google Scholar]
- Neely G. G., Hess A., Costigan M., Keene A. C., Goulas S., Langeslag M., et al. (2010). A genome-wide Drosophila screen for heat nociception identifies a2d3 as evolutionarily conserved pain gene. Cell 143 628–638 10.1016/j.cell.2010.09.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni J., Gao Y., Gong S., Guo S., Hisamitsu T., Jiang X. (2012). Regulation of m-opioid type 1 receptors my microRNA134 in dorsal root gnaglion neurons following peripheral inflammation. Eur. J. Pain 10.1002/j.1532-2149.2012.00197.x [DOI] [PubMed] [Google Scholar]
- Nielsen C. C., Knudsen G. P., Steingrimsdottir O. A. (2012). Twin studies of pain. Clin. Genet. 82 331–340 10.1111/j.1399-0004.2012.01938.x [DOI] [PubMed] [Google Scholar]
- Numakawa T., Yamamoto N., Chiba S., Richards M., Ooshima Y., Kishi S., et al. (2011). Growth factors stimulate expression of neuronal and glial miR-132. Neurosci. Lett. 505 242–247 10.1016/j.neulet.2011.10.025 [DOI] [PubMed] [Google Scholar]
- O’Connor R. M., Dinan T. G., Cryan J. F. (2012). Little things on which happiness depends: micrRNAs as novel therapeutic targets for the treatment of axiety and depression. Mol. Psychiatry 17 359–376 10.1038/mp.2011.162 [DOI] [PubMed] [Google Scholar]
- Orlova I. A., Alexander G. M., Qureshi R. A., Sacan A., Graziano A., Barret J. E., et al. (2011). MicroRNA modulation in complex regional pain syndrome. J. Transl. Med. 9 195–204 10.1186/1479-5876-9-195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pabreja K., Dua K., Sharma S., Padi S. S. V., Kulkarni S. K. (2011). Minocycline attenuates the development of diabetic neuropathic pain: possible anti-inflammatory and anti-oxidant mechanisms. Eur. J. Pharmacol. 661 15–21 10.1016/j.ejphar.2011.04.014 [DOI] [PubMed] [Google Scholar]
- Parkitny L., McAuley J. H., Di Pietro F., Stanton T. R., O’Connell N. E., Marinus J., et al. (2013). Inflammation in complex regional pain syndrome: a systematic review and meta-analysis. Neurology 80 106–117 10.1212/WNL.0b013e31827b1aa1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons M. J., Grimm C. H., Paya-Cano J. L., Sugden K., Nietfeld W., Lehrach H., et al. (2008). Using hippocampal microRNA expression diffeences between mouse inbred strains to characterise miRNA function. Mamm. Genome 19 552–560 10.1007/s00335-008-9116-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng X., Gralinski L., Armour C. D., Ferris M. T., Thomas M. J, Proll S., et al. (2010). Unique signatures of long noncoding RNA expression in response to virus infection and altered innated immune signaling. mBio. 1 e00206–e00210 10.1128/mBio.00206-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pernïa-Andrade A. J., Kato A., Witschi R., Nyilas R., Katona I., Freund T. F., et al. (2009). Spinal endocannabinoids and CB1 receptors mediate C-fiber-induced heterosynaptic pain sensitization. Science 325 764 10.1126/science.1171870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips C. J. (2006). Economic burden of chronic pain. Exp. Rev. Pharmacoecon. Outcomes Res. 6 591–601 10.1586/14737167.6.5.591 [DOI] [PubMed] [Google Scholar]
- Pietrobon D., Striessnig J. (2003). Neurobiology of migraine. Nat. Rev. Neurosci. 4 386–398 10.1038/nrn1102 [DOI] [PubMed] [Google Scholar]
- Poh K. W., Yeo J. F., Ong W. Y. (2011). MicroRNA changes in the mouse prefrontal cortex after inflammatory pain. Eur. J. Pain 801 e1–e12 [DOI] [PubMed] [Google Scholar]
- Ponomarev E. D., Veremeyko T., Barteneva N., Krichevsky A. M., Weiner H. L. (2011). MicroRNA-124 promotes microglia quiescence and suppresses EAE by deactivating macrophages via the C/EPB-a-PU.1 pathway. Nat. Med. 17 64–70 10.1038/nm.2266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponomarev E. D., Veremeyko T., Weiner H. L. (2013). MicroRNAs are universal regulators of differentiation, activation, and polarization of microglia and macrophages in normal and diseased CNS. Glia 61 91–103 10.1002/glia.22363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin W., Ren Q., Liu T., Huang Y., Wang J. (2013). MicroRNA-155 is a novel suppressor of ovarian cancer-initiating cells that targets CLDN1. FEBS Lett. 587 1434–1439 10.1016/j.febslet.2013.03.023 [DOI] [PubMed] [Google Scholar]
- Quarta S., Vogl C., Constantin C. E., üceyler N., Sommer C., Kress M. (2011). Genetic evidence for an essential role of neuronally expressed IL-6 signal transducer gp130 in the induction and maintenance of experimentally induced mechanical hypersensitivity in vivo and in vitro. Mol. Pain 7 73 10.1186/1744-8069-7-73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Recchiuti A., Krishnamoorthy S., Fredman G., Chiang N., Serhan C. N. (2011). MicroRNAs in resolution of acute inflammation: identification of novel resolvin D1-miRNA circuits. FASEB J. 25 544–560 10.1096/fj.10-169599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi S., Calin G. A. (2013). Bioinformatics, non-coding RNAs and its possible application in personalized medicine. Adv. Exp. Med. Biol. 774 21–37 10.1007/978-94-007-5590-1_2 [DOI] [PubMed] [Google Scholar]
- Saba R., Störchel P. H., Aksoy-Aksel A., Kepura F., Lippi G., Plant T. D., et al. (2012). Dopamine-regulated microRNA miR-181a controls GluA2 surface expression in hippocampal neurons. Mol. Cell. Biol. 32 619–632 10.1128/MCB.05896-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadosky A., McDermott A. M., Brandenburg N. A., Strauss M. (2008). A review of the epidemiology of painful diabetic peripheral neuropathy, postherpetic neuroalgia, and less commonly studied neuropathic pain conditions. Pain Pract. 8 56 10.1111/j.1533-2500.2007.00164.x [DOI] [PubMed] [Google Scholar]
- Sakai A., Suzuki H. (2013). Nerve injury-induced upregulation of miR-21 in the primary sensory neurons contributes to neuropathic pain in rats. Biochem. Biophys. Res. Commun. 435 176–181 10.1016/j.bbrc.2013.04.089 [DOI] [PubMed] [Google Scholar]
- Samad T. A., Moore K. A., Sapirstein A., Billet S., Allchorne A., Poole S., et al. (2001). Interleukin-1b-mediated induction of Cox-2 in the CNS contributes to inflammatory pain hypersensitivty. Nature 410 471–475 10.1038/35068566 [DOI] [PubMed] [Google Scholar]
- Sanchez-Simon F. M., Zhang X. X., Loh H. H., Law P. Y., Rodriguez R. E. (2010). Morphine regulates dopaminergic neuron differentiation via miR-133b. Mol. Pharmacol. 78 942 10.1124/mol.110.066837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandkühler J. (2007). Understanding LTP in pain pathways. Mol. Pain 3 9 10.1186/1744-8069-3-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandkühler J. (2009). Models and mechanisms of hyperalgesia and allodynia. Physiol. Rev. 89 707–758 10.1152/physrev.00025.2008 [DOI] [PubMed] [Google Scholar]
- Schweizerhof M. Stösser S., Kurejova M., Njoo C, Gangadharan V., Agarwal N., et al. (2009). Hematopoietic colony-stimulating factors mediate tumor-nerve interactions and bone cancer pain. Nat. Med. 15 802–807 10.1038/nm.1976 [DOI] [PubMed] [Google Scholar]
- Seminowicz D. A., Laferriere A. L., Millecamps M., Yu J. S. C., Codderre T. J., Bushnell M. C. (2009). MRI structural brain changes associated with sensory and emotional function in a rat model of long-term neuropathic pain. Neuroimage 47 1007–1014 10.1016/j.neuroimage.2009.05.068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengupta J. N., Pochiraju S., Kannampalli P., Bruckert M., Addya S., Yadav P., et al. (2013). MicroRNA-mediated GABA(Aalpha-1) receptor subunit downregulation in adult spinal cord following neonatal cystitis-induced chronic visceral pain in rats. Pain 154 59–70 10.1016/j.pain.2012.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serpente M., Fenoglio C., Villa C., Cortini F., Cantoni C., Ridolfi E., et al. (2011). Role of OLR1 and its regulating hsa-miR369-3p in Alzheimer’s disease: genetics and expression analysis. J. Alzheimers Dis. 26 787–793 [DOI] [PubMed] [Google Scholar]
- Shaked I., Meerson A., Wolf Y., Avni R., Greenberg D., Gilboa-Geffen A., et al. (2009). MicroRNA-132 potentiates cholinergic anti-inflammatory signaling by targeting acetylcholinesterase. Immunity 31 965–973 10.1016/j.immuni.2009.09.019 [DOI] [PubMed] [Google Scholar]
- Shaltiel G., Hanan M., Wolf Y., Barbash S., Kovalev E., Shoham S., et al. (2013). Hippocampal microRNA-132 mediates stress-inducible cognitive deficits through its acetylcholinesterase target. Brain Struct. Funct. 218 59–72 10.1007/s00429-011-0376-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith L. K., Shah R. R., Cidlowski J. A. (2010). Glucocorticoids modulate microRNA expression and processing during lymphocyte apoptosis. J. Biol. Chem. 285 36698–36708 10.1074/jbc.M110.162123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer C. (2003). Painful neuropathies. Curr. Opin. Neurol. 16 623–628 10.1097/00019052-200310000-00009 [DOI] [PubMed] [Google Scholar]
- Sommer C., Kress M. (2004). Recent findings on how proinflammatory cytokines cause pain: peripheral mechanisms in inflammatory and neuropathic hyperalgesia. Neurosci. Lett. 361 184–187 10.1016/j.neulet.2003.12.007 [DOI] [PubMed] [Google Scholar]
- Soreq H., Wolf Y. (2011). NeurimmiRs: microRNAs in the neuroimmune interface. Trends Mol. Med. 17 548–555 10.1016/j.molmed.2011.06.009 [DOI] [PubMed] [Google Scholar]
- Straube S., Derry S., Moore R. A., McQuay H. J. (2010). Cervico-thoracic or lumbar sympathectomy for neuropathic pain and complex regional pain syndrome. Cochrane Database Syst. Rev. CD002918 10.1002/14651858.CD002918.pub2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y., Li X. Q., Sahbaie P., Shi X. Y., Li W. W., Liang D. Y., et al. (2012). miR-203 regulates nociceptive sensitization after incision by controlling phospholipase A2 activating protein expression. Anesthesiology 117 626–638 10.1097/ALN.0b013e31826571aa [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbot S., Couture R. (2012). Emerging role of microglial kinin B1 receptor in diabetic pain neuropathy. Exp. Neurol. 234 373–381 10.1016/j.expneurol.2011.11.032 [DOI] [PubMed] [Google Scholar]
- Tam Tam S., Bastian I., Zhou X. F., Vander Hoek M., Michael M. Z., Gibbins I. L., et al. (2011). MicroRNA-143 expression in dorsal root ganglion neurons. Cell Tissue Res. 346 163–173 10.1007/s00441-011-1263-x [DOI] [PubMed] [Google Scholar]
- Toyoda H., Zhao M. G., Zhou M. (2009). Enhnaced quantal release of excitatory transmitter in anterior cingulate cortex of adult mice with chronic pain. Mol. Pain 5 4 10.1186/1744-8069-5-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- üceyler N., Eberle T., Rolke R., Birklein F., Sommer C. (2007a). Differential expression patterns of cytokines in complex regional pain syndrome. Pain 132 195–205 10.1016/j.pain.2007.07.031 [DOI] [PubMed] [Google Scholar]
- üceyler N., Rogausch J. P., Toyka K. V., Sommer C. (2007b). Differential expression of cytokines in painful and painless neuropathies. Neurology 69 42–49 10.1212/01.wnl.0000265062.92340.a5 [DOI] [PubMed] [Google Scholar]
- üceyler N., Göbel K., Meuth S. G., Ortler S., Stoll G., Sommer C., et al. (2010). Deficiency of the negative immune regulator B7-H1 enhances inflammation and neuropathicpain after chronic constriction injury of mouse sciatic nerve. Exp. Neurol. 222 153–160 10.1016/j.expneurol.2009.12.026 [DOI] [PubMed] [Google Scholar]
- üceyler N., Schäfers M., Sommer C. (2009). Modeof action of cytokines on nociceptive neurons. Exp. Brain Res. 196 67–78 10.1007/s00221-009-1755-z [DOI] [PubMed] [Google Scholar]
- van Rooji E., Olson E. N. (2012). MicroRNA therapeutics for cardiovascular disease: opportunities and obstacles. Nat. Rev. Drug Discov. 11 860–872 10.1038/nrd3864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villa C., Fenoglio C., De Riz M., Clerici F., Marcone A., Benussi L., et al. (2011). Role of hnRNP-A1 and miR-590-3p in neuronal death: genetics and expression analysis in patients with Alzheimer disease and frontotemporal lobar degeneration. Rejuvenation Res. 14 275–281 10.1089/rej.2010.1123 [DOI] [PubMed] [Google Scholar]
- Vincent A. M., Callaghan B. C., Smith A. L., Feldman E. L. (2011). Diabetic neuropathy: cellular mechanisms as therapeutic targets. Nat. Rev. Neurol. 7 573–583 10.1038/nrneurol.2011.137 [DOI] [PubMed] [Google Scholar]
- von Schack D., Agostino M. J., Murray B. S., Li Y., Reddy P. S., Chen J., et al. (2011). Dynamic changes in the microRNA expression profile reveal mutliple regulatory mechanisms in the spinal nerve ligation model of neuropathic pain. PLoS ONE 6:e17670 10.1371/journal.pone.0017670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiland M., Gao X. H., Zhou L., Mi Q. S. (2012). Small RNAs have a large impact: circulating microRNAs as biomarkers for human diseases. RNA Biol. 9 850–859 10.4161/rna.20378 [DOI] [PubMed] [Google Scholar]
- Willemen H. L., Huo X. J., Mao-Ying Q. L., Zijlstra J., Heijnen C. J., Kavelaars A. (2012). MicroRNA-124 as a novel treatment for persistent hyperalgesia. J. Neuroinflammation 9 143 10.1186/1742-2094-9-143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wodarski R., Clark A. K., Grist J., Marchand F., Malcangio M. (2009). Gabapentin reverses microglial activation in the spinal cord of streptozotocin-induced diabetic rats. Eur. J. Pain 13 807–811 10.1016/j.ejpain.2008.09.010 [DOI] [PubMed] [Google Scholar]
- Wu M., Jolicoeur N., Li Z., Zhang L., Fortin Y., L’Abbe D., et al. (2008). Genetic variations of microRNAs in human cancer and their effects on teh expression of miRNAs. Carcinogenesis 29 1710–1716 10.1093/carcin/bgn073 [DOI] [PubMed] [Google Scholar]
- Wu Q., Hwang C. K., Zheng H., Wagley Y., Lin H. Y., Kim D. K., et al. (2013). MicroRNA 339 down-regulates m-opioid receptor at the post-transcriptional level in response to opioid treatment. FASEB J. 27 522–535 10.1096/fj.12-213439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie J., Ameres S. L., Friedline R., Hung J.-H., Zhang J., Zhang Y., et al. (2012). Long-term, efficient inhibition of microRNA function in mice using rAAV vectors. Nat. Methods 9 403–409 10.1038/nmeth.1903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeilhofer H. U., Witschi R, Hösl K. (2009). Subtype-selective GABAA receptor mimetics - novel antihyperalgesic agents? J. Mol. Med. 87 465–469 10.1007/s00109-009-0454-3 [DOI] [PubMed] [Google Scholar]
- Zhao J., Lee M. C., Momin A., Cendan C. M., Shepperd S. T., Baker M. D., et al. (2010). Small RNAscontrol sodium channel expression, nociceptor excitability and pain thresholds. J. Neurosci. 30 10860–10871 10.1523/JNEUROSCI.1980-10.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X., Tan Z., Zhang H., Atianjoh F. E., Zhao J. Y., Liang L., et al. (2013). A long noncoding RNA contributes to neurpathic pain by silencing Kcna2 in primary afferent neurons. Nat. Neurosci. 16 1024–1031 10.1038/nn.3438 [DOI] [PMC free article] [PubMed] [Google Scholar]