Abstract
An understanding of parameters that modulate gene transfer in 3-D will assist in the formation of gene delivery systems and scaffolds, which can mediate efficient non-viral delivery for guiding in-vivo tissue regeneration and therapy. We have previously demonstrated the cell area and length, integrin expression, and RhoGTPases mediated signalling to be pivotal parameters that guide gene transfer to mouse mesenchymal stem cells (mMSCs) cultured in 2-D and are modulated by ECM proteins. In this study, we were interested in determining if cationic polymer mediated gene transfer to cells seeded in 3-D would occur through different mechanisms as compared to 2-D. In particular, we examined the endocytosis pathways used to internalize polyplexes, and the role of cytoskeletal dynamics and RhoGTPases on non-viral gene transfer for cells seeded in 2-D and 3-D. Inhibition of clathrin- and caveolae- mediated endocytosis resulted in more drastic decrease in overall transgene expression for cells seeded in 3-D than those in 2-D. In addition, polyplex internalization was only significantly decreased in 3-D when clathrin-mediated endocytosis was inhibited, while caveolae-mediated endocytosis inhibition for cells seeded in 2-D resulted in the strongest polyplex internalization inhibition. Actin and microtubule polymerization affected 2-D and 3-D transfection differently. Microtubule depolymerization enhanced transgene expression in 2-D, but inhibited transgene expression in 3-D. Last, inhibition of RhoGTPases also affected 2-D and 3-D transfection differently. The inhibition of ROCK effector resulted in a decrease of transgene expression and internalization for cells seeded in 3-D, but not 2-D and the inhibition of effector PAK1 resulted in an increase of transgene expression for both 2-D and 3-D. Overall, our study suggests that the process of gene transfer occurs through different mechanisms for cells seeded in 2-D compared to those seeded in 3-D.
Introduction
Increasing the efficacy of non-viral gene delivery will mobilize its application in tissue regeneration and therapy. The design of the vector system 1-3 and characteristics of the cellular microenvironment such as composition 4, stiffness 5, surface chemistry 6 and topography 7 are able to influence the process of non-viral gene transfer. However, the underlying intracellular mechanisms guiding gene transfer have not been fully elucidated. Most of the studies have focused on identifying the gene transfer mechanisms in cells plated in 2-D. Cell surface receptors such as the integrins 8, 9 and syndecans 10 have been shown to participate in gene transfer and interact with non-viral delivery systems. The interaction of cell surface glycosaminoglycans (GAGs) with non-viral vectors has also been shown to influence the intranuclear uptake and post nuclear processes 11. Main parameters that are able to modulate gene transfer in 2-D include cell proliferation 12, 13, internalization 14 and nuclear area 15. We have previously demonstrated that extent of cell spreading and cell length, intracellular trafficking, endocytosis pathways and cytoskeletal dynamics influence gene transfer in mouse mesenchymal stem cells plated in 2-D 4, 16. For cells plated on fibronectin coated surfaces in 2-D, intracellular signalling mediated by RhoGTPases specifically RhoA, Rac1 and Cdc42, is instrumental in mediating efficient gene transfer 17. Furthermore, gene expression of RAP1A (member of RAS oncogene family) and HSP6 (heat shock protein 6) was shown to be upregulated in cells transfected with non-viral vectors in 2-D, using a microarray analysis 18. Transfection of cells in presence of activators of RAP1A and HSP6 resulted in enhanced transgene expression 18.
On the other hand, little is known about the intracellular mechanisms involved in gene transfer in cells seeded in 3-D. Recent studies have demonstrated that balancing cell migration with rate of matrix degradation is able to enhance gene transfer in 3-D 19. Cell-matrix interactions can also be manipulated to modulate gene transfer in 3-D 20. However, a more comprehensive understanding of mechanisms guiding gene transfer in 3-D is needed to effectively employ non-viral gene delivery in in-vivo experiments and regenerative therapies.
We believe that dimensionality influences non-viral gene transfer and studies conducted in 2-D do not simulate the gene transfer process in 3-D. In this study, we were interested in determining if cationic polymer mediated gene transfer to cells seeded in 2-D or 3-D would occur through different mechanisms. In particular, we examined the endocytosis pathways used to internalize polyplexes, and the role of cytoskeletal dynamics and RhoGTPases on non-viral gene transfer for cells seeded in 2-D and 3-D. Hyaluronic acid (HA) hydrogels, cross-linked with matrix metalloproteinase (MMP) degradable peptides and modified with RGD were used as our 3-D environment and conventional tissue culture plastic surface (TCP) as our 2-D environment.
Results
Synthesis and characterization of hyaluronic acid hydrogels
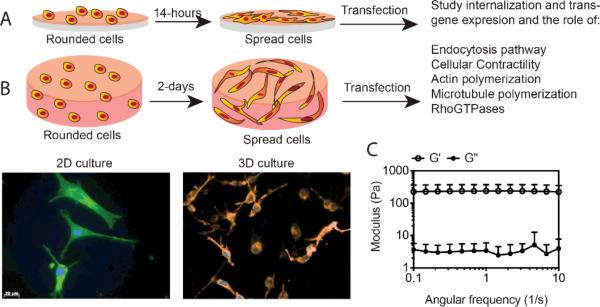
The goal of these studies was to study the process of non-viral gene transfer to mesenchymal stem cells cultured on a flat 2-D surface specifically tissue culture polystyrene (Fig. 1A) and compare that to cells cultured inside a hydrogel scaffold (3-D) (Fig. 1B). We choose to transfect cells seeded in 3-D after spreading to better mimic the 2-D situation and to avoid transfection that occurs during hydrogel formation if the polyplexes are added to the gel precursor solution as is commonly done for transfection inside hydrogels 19-22. After two days of culture, the cells inside of the hydrogel scaffold were spread (Fig. 1B). Thus, 2-D transfections were performed after 14-hours of plating and 3-D transfections after 2-days of plating (Fig. 1).
Fig. 1. Adhesion peptide functionalized hyaluronic acid hydrogel for 2-D and 3-D cell culture.
A) and B) represent a schematic for cells seeded on tissue culture polystyrene (TCP) and inside HA-RGD hydrogels (3-D). Cell spreading was analyzed through actin and DNA staining using Alexa488 conjugated phalloidin, and Hoechst dye, respectively. A) For cells plated in 2-D on tissue culture plastic surface (TCP), cell spreading was analyzed after 14 hours. B) For 3-D culture, 10 μl gel was placed per well in 96-well plates with 50,000 cells per well, and cell spreading was analyzed after 2 days. C) HA hydrogels were formed by Michael-type addition of biscysteine containing MMPxl peptides onto HA-AC functionalized with cell adhesion peptides (RGD). For cell culture and rheology, gelation was achieved by placing drops of 10 μl and 50 μl of the precursor solution, respectively, between sigmacote coated glass slides with 1 mm spacer, for 30 min at 37°C. The gel was then placed in media. The mechanical properties of the hydrogels were determined using plate-to-plate rheometry. The gel was cut to a size of 8.0 mm in diameter to fit the plate. The storage and loss modulus over a frequency range of 0.1-10 rad/s at a constant strain of 0.01 was plotted.
HA was cross-linked to form a hydrogel material using Michael-type addition of biscysteine containing MMPxl peptides onto HA-AC, functionalized with cell adhesion peptides (RGD). The storage (G’) and loss moduli (G”) of hydrogels were measured at 37°C using plate-to-plate rheology with 8 mm geometry. An evaporation blocker was utilized to avoid drying of the hydrogel sample. It was observed that G’ and G” did not cross at any measured frequency (0.1 to 10 Hz) and were frequency-independent (Fig. 1C), both of which are consistent with typical hydrogel characteristics.
To ensure that polyplexes could diffuse inside the hydrogel scaffold mesh size calculations were performed using Flory Rehner and Rubber Elasticity theory 23-25 (see materials and methods). The mesh size of HA hydrogel was determined to be in the range of ~330 nm to ~800 nm from day 1 to day 4 of culture in the presence of cells. Since the average size of the LPEI-DNA complexes used in this study was determined to be 75.86 ± 1.09 nm and 73.48 ± 0.04 nm for N/P of 6.7 and 12, respectively we expect that the polyplexes can diffuse into the hydrogel scaffold some distance and transfect the embedded cells.
Polyplex internalization pathways differ for cells cultured inside hydrogel scaffolds
Gene transfer in 2-D was studied by plating 20,000 cells onto 48-well plates and allowing the cells to attach and spread overnight (14-15 hours) before incubating the cells with the polyplex solution. 0.5 μg DNA was added as bolus per 20,000 cells per well, as this was the same DNA amount used in our previous studies where we had shown that cell area, intracellular trafficking, endocytosis pathways and cytoskeletal dynamics modulate gene transfer in cells plated on ECM proteins in 2-D 4, 16. In this study we wanted to know if the mechanisms of gene transfer we had observed for cells seeded in 2-D on ECM proteins, was same for cells seeded in 2-D on TCPS and in 3-D in HA hydrogels.
The transgene expression and polyplex internalization in 3-D was studied by culturing 50,000 cells in 10 μl hydrogels in 96-well plates and allowing the cells to attach and spread for 2-days before incubating the hydrogels with the polyplex solution. 5 μg DNA was added as a bolus per well for 3-D transfection. The 3-D transfection protocol used was based on studies conducted by our lab demonstrating and optimizing 3-D non-viral gene transfer in HA hydrogels 20, 21 . While internalization of polyplexes by flowcytometry in 2-D was observed after 2-hours, negligible internalization signal was observed in 3-D after 2-6 hours (Supplementary figure S1), indicating that as expected diffusion of the polyplexes in the hydrogel scaffold is slower than diffusion in free media. However, after 12-hours of polyplex exposure of cells to polyplexes in 3D comparable internalization was observed. Hence, for 2-D transfection 4- hours was used to transfect cells in presence of the inhibitor or activator used and for 3-D 12-hours was used. At the time of analyzing transfection in untreated cells we observed an average of about 2.4×106 RLUs per 20,000 cells for cells seeded in 2-D and 3.7×105 RLU per 50,000 cells for cells seeded in 3-D (Supplementary figure S1).
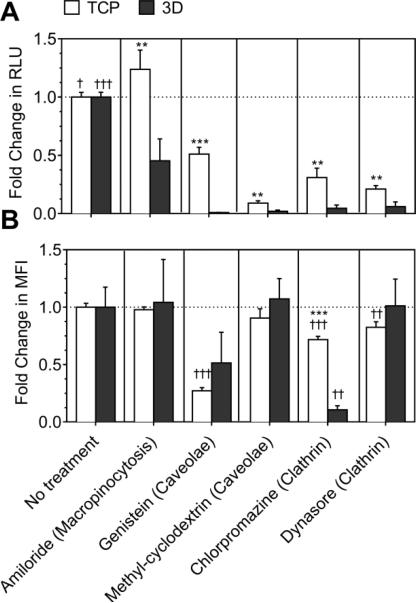
Amiloride was used to inhibit macropinocytosis, genistein and methyl-cyclodextrin were used to inhibit caveolae-mediated endocytosis, and chlorpromazine and dynasore were used to inhibit clathrin-mediated endocytosis. Inhibition of macropinocytosis significantly decreased the transgene expression in 2-D (p<0.001, Fig. 2A), and significantly increased the transgene expression in 3-D (p<0.05). Inhibition of caveolae using genistein significantly decreased the transgene expression by 49% in 2-D, and >99% in 3-D (p<0.001, Fig. 2A). Inhibition of caveolae using methyl-cyclodextrin decreased the transgene expression by >90% in cells seeded in both 2-D and 3-D.
Fig. 2. Effect of endocytic inhibitors on LPEI/DNA complex mediated gene transfer.
Macropinocytosis was inhibited using 100 μM amiloride (-macropinocytosis). Caveolae mediated pathway was inhibited using chlorpromazine and cyclodextrin. Clathrin-mediated endocytosis was inhibited using genestein, and indirectly inhibited using dynasore which inhibits dynamin. Cells were cultured on tissue culture surface (TCP) for 13-14 hours before pretreatment and for 2-days in hydrogels (3-D) before pretreatment and incubation with or without YOYO-1 labelled polyplexes for 4 hours and 12-13 hours in 2-D and 3-D, respectively. A) The transgene expression was analyzed using gaussia luciferase assay for cells seeded in 2-D and 3-D. B) Internalization was analyzed using flow-cytometry. A total of 7000 events for 2-D and 5000 events for 3-D were analyzed per sample and the mean fluorescence of events was plotted. Statistical analysis was done using tukey Multiple Comparison test, which compares all columns with each other for 2-D and 3-D, respectively. The symbols +,++ and +++ represents a significant change to the level of p<0.05, p<0.01 and p<0.001, respectively, as compared to untreated sample for 2-D or 3-D. For each treatment, transgene expression and internalization in 2-D was statistically compared with 3-D using the unpaired t-test (two tail p value). The symbols *, ** and *** represents a significant change to the level of p<0.05, p<0.01 and p<0.001, respectively, between 2-D and 3-D reading.
Inhibition of clathrin using either chlorpromazine or dynasore significantly decreased the transgene expression by ≥ 69% and ≥94% in 2-D and 3-D, respectively (at least p<0.05, Fig. 2A). Moreover, the decrease in transgene expression on inhibiting caveolae- and clathrin-mediated endocytosis was significantly more in 3-D as compared to in 2-D (at least p<0.01). Therefore, all three pathways are involved in the overall non-viral gene transfer process in 2-D, while only the clathrin- and caveolae-mediated endocytosis are involved in 3-D.
The level of internalization did not correspond with transgene expression in all conditions. Internalization was not significantly affected by inhibition of macropinocytosis in both 2-D and 3-D (Fig. 2B). Inhibition of caveolae using genistein significantly decreased polyplex uptake by 73% in 2-D, and 49% in 3-D (p<0.001, Fig. 2B). Inhibition of caveolae using methyl-cyclodextrin did not significantly influence polyplex uptake in cells seeded in 2-D and 3-D.
Inhibition of clathrin using chlorpromazine significantly reduced polyplex internalization by 28% and 89% in 2-D and 3-D, respectively (at least p<0.01, Fig. 2B). Inhibition of clathrin using chlorpromazine reduced the internalization significantly more in cells seeded in 3-D as compared to 2-D (p<0.001). However, inhibition of clathrin using dynasore did not significantly influence polyplex internalization in 2-D as well as in 3-D.
The cell viability in 3-D was not influenced by treatment with endocytic inhibitors at the time of transgene expression analysis, which is 24 hours post removal of polyplexes after overnight transfection (Supplementary figure S3).
Role of cytoskeletal dynamics in gene transfer in hydrogels
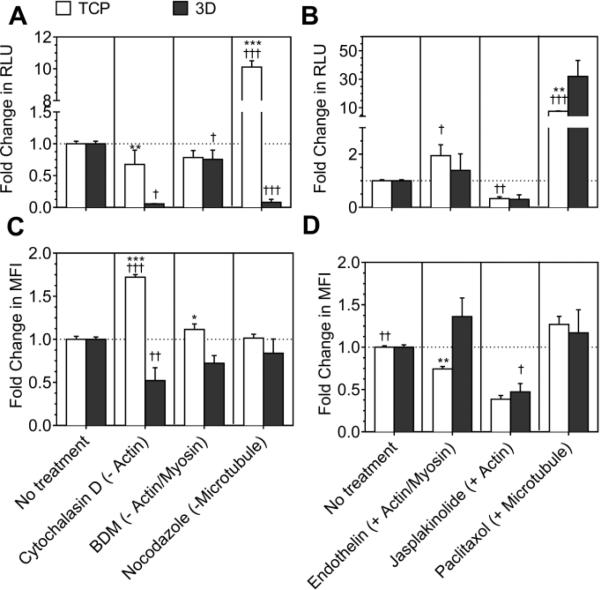
Cytochalasin-D (-actin), nocodazole (-microtubule) and butanedione-monoxime (-actin-myosin) were used to inhibit polymerization, polymerization of microtubules, and actinmyosin interactions, respectively. Jasplakinolide (+actin), paclitaxel (+microtubule) and endothelin I (+actin-myosin) were used to induce actin polymerization, microtubule polymerization and actin myosin interactions, respectively.
For cells cultured in 2-D, inhibition of actin polymerization and actin-myosin interactions did not significantly affect transgene expression (Fig. 3A). For cells cultured in 3-D, inhibition of either actin or actin-myosin interactions significantly decreased the transgene expression (at least >0.05, Fig. 3A). Inhibition of microtubule polymerization increased transgene expression by 10-fold in cells plated in 2-D, but inhibited the transgene expression by >90% in cells plated in 3-D (Fig. 3A).
Fig. 3. Effect of cytoskeletal inhibitors and activators on LPEI/DNA complex mediated gene transfer.
Actin polymerization, actin-myosin interactions and microtubule polymerization were inhibited using cytochalasin D (-actin), BDM (-actin-myosin) and nocodazole (-microtubule). Treatment using jasplakinolide (+actin), paclitaxol (+microtubule) and endothelin I (+actin-myosin) was given for enhancing actin polymerization, microtubule polymerization, and actin-myosin interactions, respectively. Before treatment the cells were cultured for 14-15 hours on tissue culture plastic (TCP) and for 2 days in hydrogels (3-D). A) and B) Transgene expression was analyzed using gaussia luciferase assay for cells seeded in 2-D and 3-D. C) and D) Internalization was analyzed using flowcytometry. A total of 7000 events for 2-D and 5000 events for 3-D were analyzed per sample and the mean fluorescence of events was plotted. Statistical analysis was done using tukey Multiple Comparison test, which compares all columns with each other for 2-D and 3-D, respectively. The symbols +,++ and +++ represents a significant change to the level of p<0.05, p<0.01 and p<0.001, respectively, as compared to untreated sample for 2-D or 3-D. For each treatment, transgene expression and internalization in 2-D was statistically compared with 3-D using the unpaired t-test (two tail p value). The symbols *, ** and *** represents a significant change to the level of p<0.05, p<0.01 and p<0.001, respectively, between 2-D and 3-D reading.
Activation of actin polymerization and stabilization significantly decreased the transgene expression in both 2-D and 3-D (Fig. 3B). Activation of actin-myosin interactions significantly increased the transgene expression in 2-D (P<0.01), but did not significantly influence transgene expression in 3-D (Fig. 3B). Interesting, inducing microtubule polymerization increased the transgene expression by 7.5-fold in 2-D and by 35-fold in 3-D (Fig. 3B).
As was seen with endocytic inhibitors, the level of polyplex internalization did not always correspond to transgene expression observed after using inhibitors and activators of cytoskeletal dynamics. Inhibition of actin polymerization significantly increased internalization in 2-D (p<0.001) and significantly decreased internalization in 3-D (p<0.05, Fig. 3C). Inhibition of actin-myosin interactions and microtubule polymerization had no significant effect on polyplex uptake in cells plated in 2-D as well as 3-D (Fig. 3C).
Induction of actin polymerization significantly decreased internalization for cells seeded in 2-D and 3-D (at least p<0.05, Fig. 3D). Activation of actin-myosin interactions significantly reduced internalization in 2-D (p<0.01) and had no effect on internalization in 3-D (Fig. 3D). Induction of microtubule polymerization significantly increased internalization in 2-D (p<0.001), while no significant effect was observed in polyplex internalization in 3-D (Fig. 3D).
At the time of transgene expression analysis same level of proliferation was observed for all conditions as compared to untreated cells (Supplementary figure S3).
Role of RhoGTPases in gene transfer in hydrogels
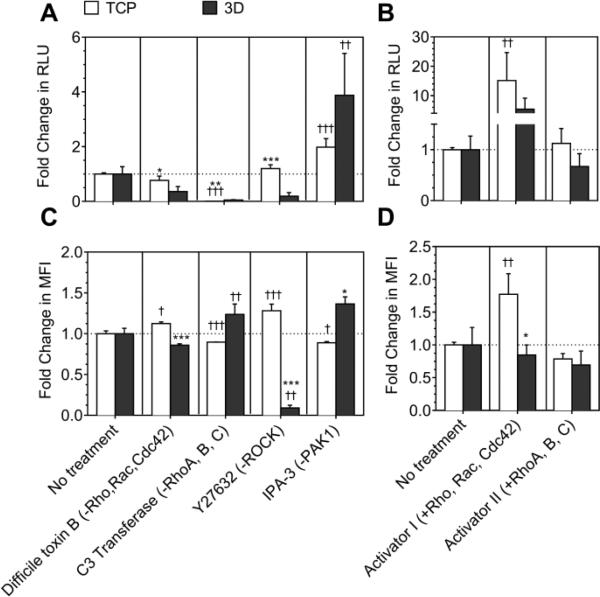
Inhibition of Rho, Rac and Cdc42 using TcdB (-Rho,Rac,Cdc42) decreased the transgene expression to a significantly greater extent in cells seeded in 3-D in HA gels (64%), as compared to cells seeded in 2-D (Fig. 4A, Supplementary table 3). Inhibition of RhoA,B,C using C3 transferase (-RhoA,B,C) drastically reduced transgene expression by ≥95% in both 2-D and 3-D. Inhibition of ROCK which is a downstream effector of RhoA, using Y27632 (-ROCK) drastically decreased transgene expression in cells seeded 3-D by 81%, in contrast to cells seeded in 2-D (Fig. 4A). Importantly, inhibition of PAK1, which is a downstream effector of Rac and Cdc42, using IPA3 (-PAK1) significantly increased transgene expression in 2-D and 3-D by 2- and 3.9-fold, respectively (at least p<0.05, Fig. 4A). Activation of Rho, Rac and Cdc42 using Rho/Rac/Cdc42 activator I (+Rho,Rac,Cdc42) significantly increased the transgene expression in 2-D by 15-fold (p<0.05), while a 5.4-fold increase in transgene expression was observed in 3-D (Fig. 4B, Supplementary table 3). Activation of RhoA,B,C using Rho direct activator II (+RhoA,B,C) did not influence the transgene expression significantly in 2-D and 3-D (Fig. 4B).
Fig. 4. Effect of RhoGTPase mediated signaling on LPEI/DNA complex mediated gene transfer.
Rac, Cdc42 and Rho were inhibited using difficile toxin B (-Rho,rac,cdc42) and Rho was inhibited using C3 transferase (-RhoA,B,C). PAK1, downstream effector of Rac and Cdc42 was inhibited using IPA3 (-PAK1), and ROCK, downstream effector of RhoA was inhibited using Y27632 (-ROCK). The effect of activation of RhoGTPases was studied by activation of Rho, Rac and Cdc42 with Rho,Rac,Cdc42 activator I (+Rho,Rac,Cdc42) and by specific activation of Rho using Rho direct activator II (+RhoA,B,C). For inhibitor treatment, the cells were cultured for 14-15 hours and for 2 days in 2-D and 3-D, respectively, before pretreatment. For activator treatment cells were cultured for 1 day in 2-D and for 2 days in 3-D, and then serum starved for 8 hours in 2-D and for 1 day in 3-D before pretreatment. After pre-treatment cells were transfected with or without YOYO-1 labelled polyplexes. A) and B) Transgene expression was analyzed using gaussia luciferase assay for cells seeded in 2-D and 3-D. C) and D) Internalization was analyzed 2 hours post transfection in 2-D, and after overnight transfection in 3-D using flow-cytometry. A total of 7000 events for 2-D and 5000 events for 3-D were analyzed per sample and the mean fluorescence of events was plotted. Statistical analysis was done using Tukey Multiple Comparison test, which compares all columns with each other for 2-D and 3-D, respectively. The symbols +,++ and +++ represents a significant change to the level of p<0.05, p<0.01 and p<0.001, respectively, as compared to untreated sample for cells cultured in 2-D or 3-D. For each treatment, transgene expression and internalization in 2-D was statistically compared with 3-D using the unpaired t-test (two tail p value). The symbols *, ** and *** represents a significant change to the level of p<0.05, p<0.01 and p<0.001, respectively, between 2-D and 3-D reading.
Next the effect of RhoGTPase inhibitors and activators was studied on internalization of polyplexes. Treatment with -Rho,Rac,Cdc42 did not statistically influence internalization in cells cultured in 2-D and 3-D (Fig. 4C). Inhibition of RhoA,B,C resulted in a statistically significant increase in internalization in cells seeded in 2-D on TCPS (p<0.05), but not in 3-D. Treatment with -ROCK significantly increased the internalization in 2-D and significantly decreased internalization by >90% in 3-D (p<0.001, Fig. 4C). The effect of inhibition of ROCK on internalization corresponded with transgene expression. On the other hand, treatment with -PAK1 significantly decreased internalization in 2-D (p<0.001) and did not significantly influence internalization in 3-D (Fig. 4C).
Activation of Rho, Rac and Cdc42-GTPases significantly increased internalization in 2-D but not in 3-D (Fig. 4D). Activation of only Rho (+RhoA,B,C) did not significantly influence internalization in 2-D as well as in 3-D (Fig. 4D). The cell viability was not influenced significantly by treatment with RhoGTPase inhibitors and activators at the time of transgene expression analysis (Supplementary figure S3).
Discussion
Non-viral gene delivery is mainly limited by its efficiency. In addition to the type of polymer used in the vector system 26, 27, the cell and its microenvironment also modulate gene transfer efficiency. In relation to the cell microenvironment, gene transfer efficiency has been shown to be dependent on the constitution of the matrix 4, 14, stiffness of the matrix 5 and presentation of ligands in cellular microenvironment. In relation to the cell itself, cell proliferation, cell area, internalization, intracellular trafficking and integrin expression modulate gene transfer efficiency. Furthermore, for cells plated in 2-D on ECM proteins, the gene transfer is modulated by internalization pathways, cytoskeletal dynamics and RhoGTPases mediated signalling 16, 17. Most of the studies have focused on studying the parameters influencing non-viral gene delivery in cells plated in 2-D, while little is known about 3-D.
3-D matrices provide a better model system for studying cellular physiological processes, as cells in tissues exist in a 3-D system. Polymeric nano-particles have been shown to have different transfection efficiencies in mammary epithelial cells cultured in 2-D and 3-D 28. We believe that the gene transfer mechanism is also dependent on the dimensionality in which cells are cultured. If so, an understanding of these differences would assist in the development of efficient gene delivery systems for 3-D cell culture. Here the role of Internalization pathways, cytoskeletal dynamics and RhoGTPases in non-viral gene transfer in cells cultured in 3-D hydrogels has been studied and compared with gene transfer mechanism in cells cultured in 2-D on conventional tissue culture plastic surface (TCP). Ideally we wanted to compare cells seeded on tissue culture polystyrene, to those seeded on top of HA-RGD hydrogels, and to those seeded inside HA-RGD hydrogels. However, cells did not spread well at the 2-day time point when seeded on top of HA-RGD hydrogels (Supplementary figure S1) and resulted in clumping and detachment during the duration of our study (4-days). Spreading on top of gels is typically observed in stiffer 26-28, 29 and higher RGD concentration hydrogels (0.5-5.4 mM range, for example see ref 30-32). However, culture of cells inside the hydrogel is achieved in hydrogels with less than 1 kPa bulk modulus 20, 33 and can be achieved with lower integrin ligand density (10-500 μM range, see ref 20, 34, 35). We decided to compare transfection for cells seeded on the traditional polystyrene surface used for transfection compared to cells seeded inside hydrogel scaffolds. Pathways for internalization of non-viral gene delivery systems depend on the type of polymer as vector system 36, cell type 37, size of the particle 38, particle design/shape 39 and composition of the extra-cellular matrix 4, 16. On studying the role of endocytic pathways on gene transfer, our results indicated that the internalization pathways are modulated by dimensionality of cell culture system. It was observed that only caveolae- and clathrin-mediated endocytosis contribute to overall gene transfer in 2-D (49% and 67%, respectively, supplementary table 1). And all three endocytic pathways namely macropinocytosis, caveolae-mediated endocytosis and clathrin-mediated endocytosis mediate efficient gene transfer in 3-D. Moreover, inhibition of caveolaeand clathrin-mediated endocytosis resulted in a more drastic decrease in overall transgene expression in 3-D (>98%), as compared to in 2-D.
Inhibition of caveolae mediated endocytosis using genistein resulted in maximal polyplex uptake inhibition in 2-D by 73%, while the polyplex uptake in 3-D was reduced by 49% (supplementary table 1). However, caveolae inhibition using methyl-cyclodextrin did not affect polyplex uptake in 2-D and 3-D. Caveolae mediated endocytosis is dependent on tyrosine phosphorylation 40. The differences in polyplex uptake inhibition by the two caveolae-pathway inhibitors could be because genestein inhibits caveolae pathway by inhibiting tyrosine kinase, while cyclodextrin inhibits by depleting cholesterol. After extraction of cholesterol, caveolin remains clustered in the plasma membrane, as if caveolae (invaginations) were not flattened or disappeared. The influence of inhibition using methyl-cyclodextrin on internalization might not be fully detected as polyplexes possibly remained associated with caveolae giving a false positive reading.
Unlike caveolae pathway inhibition, inhibition of clathrin-medicated endocytosis using chlorpromazine resulted in maximal inhibition of polyplex uptake in 3-D by 89%, while internalization was reduced by 28% in 2-D (supplementary table 1). Yet, inhibition with dynasore did not influence the internalization in 3-D while a low significant decrease of 17.5% was observed in internalization in 2-D (p < 0.05, supplementary table 1). This could be because dynasore inhibits clathrin-mediated endocytosis which depends on dynamin by rapidly blocking coated vesicle formation 41. Two types of coated pit intermediates accumulate during dynasore treatment, U-shaped, half formed pits and O-shaped, fully formed pits, captured while pinching off 41. Inhibition of polyplex internalization using dynasore is not fully detected as a positive reading is observed from polyplexes that possibly remain attached to the pits as the clathrin lattice is not disrupted while endocytosis is inhibited.
Next, cytoskeletal dynamics were observed to differentially influence non-viral gene transfer in cells seeded in 2-D and 3-D. Inhibition of actin polymerization decreased transgene expression by ≥90% in cells seeded in 3-D, but not in 2-D. In a recent study, primary fibroblast were shown to assemble actin cytoskeleton when cultured on flat 2-D substrates but did not assemble a detectable cytoskeleton when cultured in soft 3-D microwells 42. Thus, the decrease in overall gene transfer on inhibiting actin in 3-D, could be because cells in 3-D have limited actin cytoskeleton. Our results also show that enhancement of actin polymerization decreased transgene expression by 67% and 70% in 2-D and 3-D, respectively (supplementary table 2). This correlated with significant decrease in internalization in both 2-D and 3-D (at least p<0.01). Therefore, internalization and trafficking in 2-D and 3-D is significantly inhibited on enhancing actin polymerization, which increases cellular tension.
Inhibition of microtubule polymerization increased transgene expression by 10-fold in 2-D, while the same treatment decreased transgene expression to almost background levels in cells seeded inside our hydrogel scaffolds. As was observed in this study (Fig. 3A), disruption of the microtubular network with nocodazole has been shown to enhance transgene expression for cells seeded in tissue culture polystyrene 43-45. Microtubules play a role in vesicular transport and depolymerization of microtubules likely disrupts the vesicular transport. The >90% decrease observed in transgene expression in 3-D indicates that microtubules positively regulate polyplex trafficking in cells seeded in 3-D. On the other hand, for cells seeded in 2-D, inhibition of microtubule polymerization increases contractile force and promotes focal adhesions 46. The increase in transgene expression on inhibiting microtubule dynamics in 2-D could be a result of increased actin-dynamics that aid in efficient trafficking of the polyplexes.
Furthermore, a 7.5- and 35-fold increase in transgene expression was observed after enhancement of microtubule polymerization in 2-D and 3-D, respectively (supplementary table 2). This can be attributed to increased vesicular transport. Our results agree with the study showing that co-delivery of paclitaxel with PEI-DNA complexes enhances transfection in-vitro in HEK293, HepG2 and 4T1 mouse breast tumor cells, as well as in-vivo in 4T1 mouse breast tumor cells 47.
Actin-myosin interactions were observed to not influence overall transgene expression in 2-D. Contrarily for cells plated in 3-D, inhibition and enhancement of actin-myosin interactions significantly decreased and increased transgene expression, respectively. This indicates that actin-myosin interactions mediate efficient gene transfer leading to transgene expression in 3-D, but not in 2-D. It has been shown that cytoskeletal elements regulate each other 48. It is important to note that even though specific functions have been assigned to different cytoskeletal elements, they are overlapping and not separate. Actin plays a role in membrane trafficking and microtubules are involved in the control of protrusive and contractile forces 48.
Lastly, the role of RhoGTPases specifically Rho, Rac and Cdc42, in non-viral gene transfer in cells cultured in 2-D and 3-D was analyzed using specific inhibitors and activators. Inhibition of ROCK, a downstream effector of RhoA drastically reduced the overall transgene expression by 81% and polyplex uptake by 91% in cells plated in 3-D, but not in 2-D (supplementary table 3). This indicates that cell contractility is necessary to mediate efficient polyplex uptake and transfer in 3-D. This result corresponds with the significant decrease in transgene expression observed on inhibition of actin-myosin interactions in 3-D.
PAK1, a downstream effector of Rac and Cdc42 also modulates gene transfer differentially in 2-D and 3-D. Inhibition of PAK1 increased the transgene expression by 2-fold but significantly reduced polyplex internalization in 2-D (p<0.05, supplementary table 3). The transgene expression increased by 3.9-fold, and the internalization was not significantly effected in 3-D, after inhibition of PAK1. This could be because cells employ different internalization pathways in 2-D and 3-D leading to overall gene transfer. Decreased internalization leading to overall increased transgene expression occurs in mMSCs when they are plated on Fn coated surfaces in 2-D 16. Additionally, activation of Rho,Rac,Cdc42 (+Rho,Rac,Cdc42) increased transgene expression by 15-fold and internalization by 1.8-fold in cells plated in 2-D. For cells plated in 3-D, transgene expression increased by 5.4-fold while internalization was not significantly influenced (supplementary table 3). Taken together, our results indicate the RhoGTPases also differentially modulate non-viral gene delivery in 2-D and 3-D.
Just as the mechanism of gene-transfer is not universal in 2-D, the mechanism of gene transfer is not universal in 3-D. Mechanism of gene transfer in 3-D would be influenced by the type of adhesions. The adhesions of the cell with the matrix in which it is seeded, mediates the cross talk between the cell and its microenvironment, affecting RhoGTPase activation and cytoskeletal dynamics. The type of adhesions formed depends on the stiffness of the matrix, expression level of focal adhesion component, fibrillar and non-fibrillar organization of the matrix, and bidirectional interaction between matrix stiffness and cytoskeletal contractility 49, 50.
Henceforth, the cell behaviour pertaining to non-viral gene transfer in conventional 2-D surface cannot be compared with the cell behaviour in 3-D. It is important to develop and assess the efficacy of a transfection protocol in 3-D cell culture systems before using it to transfect cells in 3-D culture systems for invitro applications 51, 52, or before using it for in-vivo transfection 53.
Experimental
Materials
Peptides Ac-GCRDGPQGIWGQDRCG-NH2 (MMPxl) and Ac-GCGWGRGDSPG-NH2 (RGD) were obtained from Genescript (Piscataway, NJ). Sodium hyaluronan was a gift from Genzyme Corp. (Boston,MA). Gaussia luciferase expression vector (pGluc, New England BioLabs, Ipswich, MA) was expanded using an endotoxin-free Giga Prep kit from Qiagen following the manufacturer's instructions. Linear PEI (25 kg/mol) was purchased from Polysciences (Warrington, PA). Amiloride hydrochloride hydrate, Methyl-β-cyclodextrin, Cytochalasin D, Nocodazole, Butanedione monoxime (BD), IPA-3 and Y27632 were purchased from Sigma Aldrich (St Louis, MO). Genistein, Chlorpromazine hydrochloride and were purchased from Fisher scientific. Jasplikinolide was purchased from Invitrogen (Grand Island, NY) and Endothelin I was purchased from Calbiochem (EMD biosciences, San Diego, CA). Difficile toxin B (TcdB) was purchased from List Biologicals (Campbell, CA). C3 transferase (C3), Paclitaxel, , Rho/Rac/Cdc42 activator I and Rho activator II were purchased from Cytoskeleton Inc (Denver, CO). All other products were purchased from Fisher Scientific unless noted otherwise.
Cell culture
Mouse bone marrow cloned mesenchymal stem cells (D1, CRL12424) were purchased from ATCC (Manassas, VA, USA). Cells were maintained in Dulbecco's modified eagle's medium (Sigma-Aldrich) containing 10% bovine growth serum (BGS, Hyclone, Logan, Utah) and 1% penicillin/streptomycin antibiotics (Invitrogen, Grand Island, NY) and cultured at 37°C and 5% CO2.
Modification of HA
Acrylated hyaluronic acid (HA-AC) was prepared using a two-step synthesis as previously described 20, 21. Briefly, HA (60,000 Da, Genzyme Corporation, Cambridge, MA) (2.0 g, 5.28 mmol, 60 kDa) was reacted with 18.0 g (105.5 mmol) of adipic dihydrazide (ADH) at pH 4.75 in the presence of 4.0 g (20 mmol) of 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide hydrochloride (EDC) overnight and purified through dialysis (8000 MWCO) in deionized (DI) water for 2 days. The purified intermediate (HA-ADH) was lyophilized and stored at 20 °C until used. A portion of 29% of the carboxyl groups was modified with ADH based on the trinitrobenzene sulfonic acid (TNBSA, Pierce, Rockford, Illinois) assay. HA-ADH was then reacted with 5 molar excess of N-acryloxysuccinimide (NHS-Ac) in HEPES buffer (pH 7.2) overnight, and purified through dialysis in DI water for 2-3 days. All the primary amines were acrylated based on the 2,4,6-trinitrobenzene sulfonic acid (TNBSA, Thermo Scientific) assay performed following the product manual.
HA Hydrogel Synthesis
HA hydrogels were formed by Michael-type addition of biscysteine containing MMPxl peptides onto HA-AC functionalized with cell adhesion peptides (RGD). Hydrogels were prepared with 3% HA and 0.4-0.44 ratio of thiols to acrylates (r ratio), containing 100 μM RGD with a 0.53 degree of clustering (mmol RGD/mmol HA-RGD) 34, 20. A lyophilized aliquot of HA-Ac was first dissolved in 0.3 M TEOA buffer (pH = 8.4) to a concentration of 0.08 mg/μl. 4.3% of total HA-Ac (0.08 mg/μl) was added to lyophilized aliquot of RGD peptides and allowed to react for 15-20 min at 37°C. The cell solution (500,000 cells per 100 μl of final gel volume) and the required amount of non-RGD functionalized HA-AC were then added. A lyophilized aliquot of the cross-linker (0.91 mg MMPxl) was then diluted in 18.2 μl of 0.3 M TEOA buffer (pH = 8.4) and immediately added to the gel precursor solution. The final gel solution had a pH = 8-8.1. Gelation was achieved by placing drops of 10 μl of the precursor solution between sigmacote coated glass slides for 30 min at 37°C. The final gel was placed inside 96-well plates for culture. Thorough mixing was used to ensure the homogeneity in cell and crosslinker distribution in the precursor solution.
Characterization of HA Hydrogel Mechanical Properties
The storage and loss modulus were measured with a plate-to-plate rheometer (Physica MCR, Anton Par, Ashland, VA) using a 8 mm plate under a constant strain of 0.01 and frequency ranging from 0.1 to 10 rad/s. Hydrogels were made as detailed above and cut to a size of 8.0 mm in diameter to fit the plate. A humid hood was used to prevent the hydrogel from drying, and the temperature was kept at 37°C.
Calculations for determining the mesh size of HA Hydrogel
The mesh size was determined using the following equation 24,25 derived from Flory-Rehner theory, and modified by Peppas et.al 2006 24 :
Correlation distance between crosslinks
| (1) |
where ξ is the correlation distance between crosslinks or the corresponding mesh size, is the polymer volume fraction and (ro2)1/2 is the root mean square distance.
Swelling experiments and Flory-Rehner calculations conducted by Leach et.al. for photocrosslinked hyaluronic hydrogels (using glycidyl methacrylate-HA) provided the following three equations23 :
| (2a) |
| (2b) |
From (2) and (3) and substituting Ṁc for Mn
| (2c) |
where Ṁc is the average molecular weight between crosslinks and Qv is the volumetric swelling ratio.
| (3) |
The average molecular weight between crosslinks was calculated using the following equation derived from the Rubber Elasticity theory:
| (4) |
where ρ is the polymer density in the hydrogel, R is the universal gas constant, T is the absolute temperature and G is the average storage modulus
The average storage modulus for 3% HA gels was determined to be ~ 225 Pa (Fig. 1c) on day 1 after formation and before culturing cells. The corresponding Ṁc from eq (4) was 343650 g/mol and the corresponding mesh size, from eq (3) was ξ = 330 85 nm.
Study conducted by Lam et.al, 201334 showed that the storage modulus for 3% HA gels decreases from ~ 200 Pa on day 1 to ~ 40 Pa on day 4 of cell culture due to gel degradation by MMPs produced by the growing cells. On day 4, for G = 40 Pa, the corresponding Ṁc from eq (4) was 1933000 g/mol and the corresponding mesh size, from eq (3) was ξ = 777 nm.
Therefore, the mesh size of the HA gels is in the range of ~330 nm to ~800 nm, from day 1 to day 4 of cell culture. The values determined here are considered approximations, as approximations were made while deriving equation (2c) by Leach et.al.23 and (3).
Cell morphology
For 2-D cell culture, wells were placed on a plastic coverslip using an 8-well flexiperm and the assembly was placed inside a sterile tissue culture petri dish. 20,000 cells were seeded per well on the coverslip and cultured for 14 hours before analyzing cell morphology. For 3-D cell culture, HA-RGD hydrogels containing mMSCs (5,000 cells/μl) were synthesized and cultured in 96 well plates for 2 days before analyzing cell morphology. For analyzing cell morphology in 2-D and 3-D, cells were then fixed and stained for actin and nuclear DNA using Alexa488-phalloidin and Hoechst dye following the manufacturer's instructions and standard staining protocols. Briefly, after two PBS washes the cells were fixed with 4% paraformaldehyde (PFA) for 15-30 minutes at room temperature and the cell membrane was weakened with 0.1% tritonX100 in PBS for 3-6 minutes. Hoechst dye and Alexa488-phalloidin were then added and left in the dark for 60 minutes at room temperature followed by three washes with 0.05% tween-20. The samples were imaged using fluorescence microscopy at 40× magnification for 2-D and using confocal microscopy at 20× magnification for 3-D.
Cell proliferation
50,000 cells per 10 μl hydrogel per well were cultured in 3-D in a 96 well plate. Cells were treated as required by the experiments mentioned. To determine the cytotoxicity and proliferation of the cells, Cell Titer 96®Aqueous One Solution Cell Proliferation Assay (Promega) was performed. Media was replaced with fresh media, and the Aqueous One Solution (20 μl) was added in to the media in each well to be assayed and incubated for 2 hours. SDS (25 μl of a 10% solution) was added to each well after the incubation. The fluorescence was measured using a plate reader at 490 nm. Three gels for each condition were analyzed at each time point.
Transfection in 2-D
20,000 cells were allowed to attach per well of a 48 well plate and cultured under various conditions as required for the experiments mentioned. For gene transfer studies, DNA/PEI polyplexes were formed by mixing equal volumes of plasmid DNA with PEI. For every 1 μg of DNA, 1.65 μg of PEI was added to the DNA solution to get N/P ratio of 12, vortexed for 15 seconds and incubated at room temperature for 15 minutes. Polyplexes were added directly to the medium of the plated cells at a final DNA concentration of 0.5 μg per 20,000 cells per well in a 48-well plates. Salt was added directly to the wells post addition of transfection solution to get a final concentration of 150 mM NaCl. Transfection was quantified at 48 hours post transfection using the Gaussia Luciferase Assay System following the manufacturer's instructions.
Internalization of polyplexes in 2-D
20,000 cells were plated per well of a 48 well plate and cultured under various conditions as required for the experiments mentioned. For analyzing internalization, plasmid DNA (pGluc) and the fluorescent DNA-intercalator YOYO-1 were mixed at a ratio of 1 YOYO-1 molecule per 50 base pairs and were allowed to complex for 60 minutes at room temperature. YOYO-1 labelled DNA was then used to prepare PEI/DNA complexes at a N/P of 12 (as mentioned above) and bolus transfection was performed. Two hours after exposure to the polyplexes, cells were washed with PBS, trysinized with 50 μl of 0.25% trypsin-EDTA and finally suspended in 350 μl of 0.04% tryphan blue in 1% BGS in PBS. Fluorescent cells were detected by flow-cytometry with a FACScan X and data was analyzed with CELLQuest (Beckton Dickinson). Experiments were performed in triplicates analyzing 5000 total events per sample and mean fluorescence intensity of total events was plotted.
Transfection in hydrogels
HA-RGD hydrogels containing mMSCs (5,000 cells/μl) were synthesized and cultured in 96 well plates for 2 days to allow the cells to spread. Prior to transfection the cells were treated with the different activators or inhibitors. DNA/PEI polyplexes were formed by mixing equal volumes of plasmid DNA with 25 kDa-Linear PEI. For every 1 μg of DNA, 0.913 μg of PEI was added to the DNA solution to get N/P of 7, vortexed for 15 seconds and incubated at room temperature for 15 minutes. Subsequently salt was added to the polyplex solution to get a final concentration of 150 mM NaCl. After addition of salt, polyplexes were immediately added as bolus directly to the medium of the plated cells at a final DNA concentration of 5 μg per 50,000 cells per gel (10 μl) per well of 96 well low binding plate. Transfection was quantified at 24 hours post overnight transfection using the Gaussia Luciferase Assay System following the manufacturer's instructions.
Internalization in hydrogels
HA-RGD hydrogels containing mMSCs (5,000 cells/μl) were synthesized and cultured in 96 well plates for 2 days to allow the cells to spread. Prior to transfection the cells were treated with the different activators or inhibitors. Plasmid DNA (pSEAP) and the fluorescent DNA-intercalator YOYO-1 were mixed at a ratio of 1 YOYO-1 molecule per 50 base pairs and were allowed to complex for 60 minutes at room temperature. YOYO-1 labelled DNA was then used to prepare PEI/DNA complexes at a N/P of 7 (as mentioned above for transfection in hydrogels) and bolus transfection was performed. After overnight (~16 hours) exposure to the polyplexes, gels were washed with PBS, degraded by trysinization with 50 μl of 0.25%trypsin-EDTA for 5 minutes, and finally suspended in 250 μl of 0.04% tryphan blue in 1% BGS in PBS. Fluorescent cells were detected by flow cytometry with a FACScanX and data was analyzed with CELLQuest (Beckton Dickinson). Experiments were performed in triplicates analyzing 5000 total events per sample and mean fluorescence intensity of total events was plotted.
Measuring polyplex size
The size of the polyplexes formed was determined using dynamic light scattering (DLS). The Nano ZS (Malvern zetasizer) instrument and ZEN0040 Disposable micro cuvette was used. The measurements were performed in triplicate.
Analyzing endocytic pathways
Clathrin mediated endocytosis was inhibited by using 10 μg/ml chlorpromazine and indirectly inhibited using 50 μM dynasore, caveolae mediated endocytosis was inhibited using 200 μM genestein and 0.5-1 mM cyclodextrin, and macropinocytosis was inhibited using 100 μM amiloride. For 2-D cell culture 20,000 cells were cultured per well in a 48 well plate for 13-14 hours before pretreatment with inhibitors for endocytosis to determine the endocytic pathways active in cells. For 3-D cell culture 50,000 cells were cultured per 10 μl hydrogel per well in a 96 well plate for 2 days before pretreatment. For genistein, chlorpromazine and amiloride pretreatment was given for 0.5 and 1.5 hours for cells cultured in 2-D and 3-D, respectively. For cyclodextrin and dynasore 1 hour pretreatment was given in 2-D and 3-D. Subsequently, cells were transfected using polyplexes labelled with or without YOYO-1, in presence of inhibitors for 4 hours and overnight (12-13 hours) in 2-D and 3-D, respectively. The media was then replaced with fresh media. Internalization was analyzed 2 hours post transfection in 2-D, and after overnight transfection in 3-D. Transgene expression was analyzed 48 hours after 4 hour transfection for cells plated in 2-D and 24 hours after overnight transfection for cells plated in 3-D, respectively. Cell proliferation was analyzed 24 hours post overnight transfection for cells plated in 3-D.
Studying the role of cytoskeleton in gene transfer
20 μM cytochalasin D (-actin) treatment was given to inhibit actin polymerization and the resultant tension, 10 μM nocodazole (-microtubulin) treatment was given to depolymerize microtubules and 10 mM butanedione monoxime (-actin-myosin) treatment was given to inhibit myosin ATPase. For activation, 500 nM jasplakinolide (+actin), 10 μM paclitaxel (+microtubule) and 20 nM endothelin I (+actin-myosin) was given for actin, microtubule, and actin-myosin interactions, respectively. For 2-D cell culture 20,000 cells were cultured per well in a 48 well plate for 13-14 hours before pretreatment. For 3-D cell culture 50,000 cells were cultured per 10 μl hydrogel per well in a 96 well plate for 2 days before pretreatment. For pretreatment, the media in the wells was replaced with media containing the inhibitor for 0.5 and 1.5 hours for cells cultured in 2-D and 3-D respectively, or with media containing the activator for 2 hours for cells cultured in 2-D and 3-D. For endothelin I, a 2.5 minute pretreatment was given in 2-D and 3-D. For cells cultured in 2-D, immediately after pretreatment media was replaced with fresh media, and cells were transfected using with or without YOYO-1 labelled polyplexes. For cells cultured in 3-D, immediately after pretreatment overnight transfection was done with or without YOYO-1 labelled polyplexes, in presence of inhibitor or activator. And after overnight transfection the media was replaced with fresh media. Internalization was analyzed 2 hours post transfection in 2-D, and after overnight transfection in 3-D. Transgene expression was analyzed 48 hours post transfection for cells plated in 2-D and 24 hours post overnight transfection for cells plated in 3-D. Cell proliferation was also analyzed 24 hours after overnight transfection for cells plated in 3-D.
Inhibition of RhoGTPases in hydrogels
200 ng/ml difficile toxin B (-Rho,Rac,Cdc42, List Biologicals, Campbell, CA) was used to pharmacologically inhibit Rho, Rac and Cdc42, Rho was specifically inhibited using cell permeable 1 μg/ml C3 transferase (-RhoA,B,C, Cytoskeleton, Denver, CO) and Rac and Cdc42 were both inhibited using 10 μM IPA3 (-PAK1). 20,000 cells were cultured per well in a 48 well plate for 14-15 hours in 2-D and 50,000 cells were cultured per 10 μl hydrogel per well in a 96 well plate in 3-D for 2 days. Cells cultured in 2-D and 3-D were then pretreated by replacing the media in the wells with serum free media containing -Rho,Rac,Cdc42 or -ROCK for 4 hours or -PAK1 for 0.5 hours. Immediately after pretreatment, media was changed with fresh media and cells were transfected with or without YOYO-1 labelled polyplexes, for cells seeded in 2-D, For cells seeded in 3-D, immediately after pretreatment overnight transfection was done with or without YOYO-1 labelled polyplexes, in presence of inhibitor. Media was replaced with fresh media after overnight transfection in 3-D. Internalization was analyzed 2 hours post transfection in 2-D, and after overnight transfection in 3-D. Transgene expression was analyzed 48 hours after transfection for cells plated in 2-D. For cells plated in 3-D, transgene expression and cell proliferation was analyzed 24 hours after overnight transfection.
Activation of RhoGTPases
1 μg/ml Rho/Rac/Cdc42 activator I (+Rho, Rac,Cdc42) and 1 μg/ml Rho direct activator II (+RhoA,B,C) were used to activate all three Rho, Rac and Cdc42, and to activate only Rho respectively. In 2-D, 20,000 cells were cultured per well in a 48 well plate for 1 day, and then serum starved for 8 hours by culturing them in serum free media. In 3-D, 50,000 cells were cultured per 10 μl hydrogel per well in a 96 well plate for 2 days and then serum starved for 1 day using serum free media. Subsequently, pre-treatment was given for 4 hours with activators in serum free media for cells cultured in 2-D and 3-D. Immediately, after pretreatment bolus transfection was done in presence of activators in serum free media, with or without YOYO-1 labelled polyplexes for 4 hours and overnight (12-13 hours) in cells plated in 2-D and 3-D, respectively. The media was then replaced with fresh media. Internalization was analyzed 2 hours post transfection in 2-D, and after overnight transfection in 3-D. Transgene expression was analyzed 48 hours post transfecting cells for 4 hours for cells plated in 2-D and post overnight transfection, for cells plated in 3-D, respectively. Cell proliferation was analyzed 24 hours post overnight transfection for cells plated in 3-D.
Statistics
All statistical analysis were performed using the computer program Instat (GraphPad, San Diego, CA). Experiments were statistically analyzed using the Tukey test, which compares all pairs of columns, using a 95% confidence interval or using unpaired t-test (two tail p value) which compared any two columns.
Conclusions
To conclude, we have shown that endocytic pathways, cytoskeletal dynamics and RhoGTPase mediated signalling differentially modulate non-viral gene transfer in cells cultured in 2-D and 3-D. Caveolae- and clathrin- mediated endocytosis contribute to efficient gene transfer in 2-D. For cells seeded in 3-D all three pathways namely macropinocytosis, caveolae and clathrin mediated endocytosis play a role in mediating gene transfer. Regarding cytoskeletal dynamics, actin and microtubule polymerization is necessary to mediate efficient gene transfer in 3-D, but not 2-D. Moreover, inhibition of microtubule polymerization increases transgene expression by 10-fold in 2-D. The enhancement of microtubule polymerization results in a 7.5- and 35-fold increase in transgene expression in 2-D and 3-D, respectively. Lastly, with respect to RhoGTPases, it was observed that activation of ROCK, downstream effector of Rho, is necessary in mediating efficient gene transfer in 3-D but not in 2-D. Furthermore, activation of Rho, Rac and Cdc42 increases transgene expression by 15-fold in 2-D and by 5.4-fold in 3-D. This is an initial study comparing mechanism of non-viral gene delivery in cells cultured in 2-D and 3-D. A lot of work still needs to be done to gain a complete understanding of gene transfer mechanism in 3-D. For example, future studies about the role of integrins and other cell surface receptors, effect of ECM protein identity and density, and intracellular trafficking of gene delivery vectors in cells cultured in 3-D systems would be helpful.
Supplementary Material
Acknowledgement
We sincerely appreciate the co-operation and valuable advice rendered by all the members of the Segura Lab. We would also like to thank undergraduate researchers Clayton Lin from UCLA and Yining Zhan from MIT, for their time and effort during this project. Flow cytometry was performed in the UCLA Jonsson Comprehensive Cancer Center (JCCC) and Center for AIDS Research Flow Cytometry Core Facility that is supported by National Institutes of Health awards CA-16042 and AI-28697, and by the JCCC, the UCLA AIDS Institute, and the David Geffen School of Medicine at UCLA.
Footnotes
¶ Electronic supplementary information (ESI) available: Supplementary figure 1 showing a schematic for cells plated on top of HA-RGD hydrogels. Supplementary figure 2 showing transgene expression for untreated cells seeded in 2-D and 3-D, and internalization in 3-D at various time intervals. Supplementary figure 3 showing the effect of various inhibitors and activators on cell viability. Supplementary tables showing and comparing the statistical analysis for transgene expression and internalization in cells plated in 2-D and 3-D, after the various treatments.
Notes and references
- 1.Jager M, Schubert S, Ochrimenko S, Fischer D, Schubert US. Chem Soc Rev. 2012;41:4755–4767. doi: 10.1039/c2cs35146c. [DOI] [PubMed] [Google Scholar]
- 2.Mairhofer J, Grabherr R. Mol Biotechnol. 2008;39:97–104. doi: 10.1007/s12033-008-9046-7. [DOI] [PubMed] [Google Scholar]
- 3.Rekha MR, Sharma CP. Recent Pat DNA Gene Seq. 2012;6:98–107. doi: 10.2174/187221512801327389. [DOI] [PubMed] [Google Scholar]
- 4.Dhaliwal A LJ, Maldonado M, Lin C, Segura T. Soft Matter. 2012:1451–1459. [Google Scholar]
- 5.Kong HJ, Liu J, Riddle K, Matsumoto T, Leach K, Mooney DJ. Nature materials. 2005;4:460–464. doi: 10.1038/nmat1392. [DOI] [PubMed] [Google Scholar]
- 6.Kasputis T, Pannier AK. J Biol Eng. 2012;6:17. doi: 10.1186/1754-1611-6-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Teo BK, Goh SH, Kustandi TS, Loh WW, Low HY, Yim EK. Biomaterials. 32:9866–9875. doi: 10.1016/j.biomaterials.2011.08.088. [DOI] [PubMed] [Google Scholar]
- 8.Chowdhury EH, Nagaoka M, Ogiwara K, Zohra FT, Kutsuzawa K, Tada S, Kitamura C, Akaike T. Biochemistry. 2005;44:12273–12278. doi: 10.1021/bi050595g. [DOI] [PubMed] [Google Scholar]
- 9.Zuhorn IS, Kalicharan D, Robillard GT, Hoekstra D. Mol Ther. 2007;15:946–953. doi: 10.1038/mt.sj.6300139. [DOI] [PubMed] [Google Scholar]
- 10.Kopatz I, Remy JS, Behr JP. The journal of gene medicine. 2004;6:769–776. doi: 10.1002/jgm.558. [DOI] [PubMed] [Google Scholar]
- 11.Ziraksaz Z, Nomani A, Ruponen M, Soleimani M, Tabbakhian M, Haririan I. Eur J Pharm Sci. 2013;48:55–63. doi: 10.1016/j.ejps.2012.10.016. [DOI] [PubMed] [Google Scholar]
- 12.Mortimer I, Tam P, MacLachlan I, Graham RW, Saravolac EG, Joshi PB. Gene therapy. 1999;6:403–411. doi: 10.1038/sj.gt.3300837. [DOI] [PubMed] [Google Scholar]
- 13.Brunner S, Sauer T, Carotta S, Cotten M, Saltik M, Wagner E. Gene therapy. 2000;7:401–407. doi: 10.1038/sj.gt.3301102. [DOI] [PubMed] [Google Scholar]
- 14.Bengali Z, Rea JC, Shea LD. J Gene Med. 2007;9:668–678. doi: 10.1002/jgm.1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Uchimura E, Yamada S, Uebersax L, Yoshikawa T, Matsumoto K, Kishi M, Funeriu DP, Miyake M, Miyake J. Neuroscience Letters. 2005;378:40–43. doi: 10.1016/j.neulet.2004.12.018. [DOI] [PubMed] [Google Scholar]
- 16.Dhaliwal A, Maldonado M, Han Z, Segura T. Acta Biomater. 2010;6:3436–3447. doi: 10.1016/j.actbio.2010.03.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dhaliwal MMA, Lin C, Segura T. PLoS One. 2012;7:e35046. doi: 10.1371/journal.pone.0035046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Plautz SA, Boanca G, Riethoven JJ, Pannier AK. Mol Ther. 2011;19:2144–2151. doi: 10.1038/mt.2011.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shepard JA, Huang A, Shikanov A, Shea LD. J Control Release. 2010;146:128–135. doi: 10.1016/j.jconrel.2010.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gojgini S, Tokatlian T, Segura T. Mol Pharm. 2011;8:1582–1591. doi: 10.1021/mp200171d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lei Y, Rahim M, Ng Q, Segura T. J Control Release. 2011;153:255–261. doi: 10.1016/j.jconrel.2011.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kidd ME, Shin S, Shea LD. J Control Release. 2012;157:80–85. doi: 10.1016/j.jconrel.2011.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baier Leach J, Bivens KA, Patrick CW, Jr., Schmidt CE. Biotechnol Bioeng. 2003;82:578–589. doi: 10.1002/bit.10605. [DOI] [PubMed] [Google Scholar]
- 24.Peppas NA, Hilt Z, Khademhosseini A, Langer R. Adv Mater. 2006;18:1345–1360. [Google Scholar]
- 25.Ferreira L, Figueiredo MM, Gil MH, Ramos MA. J Biomed Mater Res B Appl Biomater. 2006;77:55–64. doi: 10.1002/jbm.b.30394. [DOI] [PubMed] [Google Scholar]
- 26.Loh XJ, Ong SJ, Tung YT, Choo HT. Macromol Biosci. doi: 10.1002/mabi.201300050. [DOI] [PubMed] [Google Scholar]
- 27.Wightman L, Kircheis R, Rossler V, Carotta S, Ruzicka R, Kursa M, Wagner E. J Gene Med. 2001;3:362–372. doi: 10.1002/jgm.187. [DOI] [PubMed] [Google Scholar]
- 28.Bhise NS, Gray RS, Sunshine JC, Htet S, Ewald AJ, Green JJ. Biomaterials. 2010;31:8088–8096. doi: 10.1016/j.biomaterials.2010.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nie T, Akins RE, Jr., Kiick KL. Acta Biomater. 2009;5:865–875. doi: 10.1016/j.actbio.2008.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rizzi SC, Ehrbar M, Halstenberg S, Raeber GP, Schmoekel HG, Hagenmuller H, Muller R, Weber FE, Hubbell JA. Biomacromolecules. 2006;7:3019–3029. doi: 10.1021/bm060504a. [DOI] [PubMed] [Google Scholar]
- 31.Liu SQ, Ee PL, Ke CY, Hedrick JL, Yang YY. Biomaterials. 2009;30:1453–1461. doi: 10.1016/j.biomaterials.2008.11.023. [DOI] [PubMed] [Google Scholar]
- 32.Zhang Z, Lai Y, Yu L, Ding J. Biomaterials. 2010;31:7873–7882. doi: 10.1016/j.biomaterials.2010.07.014. [DOI] [PubMed] [Google Scholar]
- 33.Kim J, Kim IS, Cho TH, Kim HC, Yoon SJ, Choi J, Park Y, Sun K, Hwang SJ. J Biomed Mater Res A. 2010;95:673–681. doi: 10.1002/jbm.a.32884. [DOI] [PubMed] [Google Scholar]
- 34.Lam J, Segura T. Biomaterials. 2013;34:3938–3947. doi: 10.1016/j.biomaterials.2013.01.091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lei Y, Segura T. Biomaterials. 2009;30:254–265. doi: 10.1016/j.biomaterials.2008.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rejman J, Bragonzi A, Conese M. Mol Ther. 2005;12:468–474. doi: 10.1016/j.ymthe.2005.03.038. [DOI] [PubMed] [Google Scholar]
- 37.Douglas KL, Piccirillo CA, Tabrizian M. European Journal of Pharmaceutics and Biopharmaceutics. 2008;68:676–687. doi: 10.1016/j.ejpb.2007.09.002. [DOI] [PubMed] [Google Scholar]
- 38.Rejman J, Oberle V, Zuhorn IS, Hoekstra D. The Biochemical journal. 2004;377:159–169. doi: 10.1042/BJ20031253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gratton SE, Ropp PA, Pohlhaus PD, Luft JC, Madden VJ, Napier ME, DeSimone JM. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:11613–11618. doi: 10.1073/pnas.0801763105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sverdlov SAM, Minshall RD. J Cell Mol Med. 2007;11:1239–1250. doi: 10.1111/j.1582-4934.2007.00127.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Macia E, Ehrlich M, Massol R, Boucrot E, Brunner C, Kirchhausen T. Dev Cell. 2006;10:839–850. doi: 10.1016/j.devcel.2006.04.002. [DOI] [PubMed] [Google Scholar]
- 42.Mirjam MT. Ochsner1¤b, Viola Vogel2, Michael L. Smith2¤a*. PLoS One. 2010;5:e9445. doi: 10.1371/journal.pone.0009445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang L, MacDonald RC. Mol Ther. 2004;9:729–737. doi: 10.1016/j.ymthe.2004.02.009. [DOI] [PubMed] [Google Scholar]
- 44.Brisson M, Tseng WC, Almonte C, Watkins S, Huang L. Human gene therapy. 1999;10:2601–2613. doi: 10.1089/10430349950016645. [DOI] [PubMed] [Google Scholar]
- 45.Lindberg J, Fernandez MA, Ropp JD, Hamm-Alvarez SF. Pharmaceutical research. 2001;18:246–249. doi: 10.1023/a:1011001022570. [DOI] [PubMed] [Google Scholar]
- 46.Elbaum M CA, Levy ET, Shtutman M, Bershadsky AD. Biochem Soc Symp. 1999;65:147–172. [PubMed] [Google Scholar]
- 47.Wang Y, Gao S, Ye WH, Yoon HS, Yang YY. Nature materials. 2006;5:791–796. doi: 10.1038/nmat1737. [DOI] [PubMed] [Google Scholar]
- 48.S E-M. Traffic. 2004;5:470–477. doi: 10.1111/j.1600-0854.2004.00196.x. [DOI] [PubMed] [Google Scholar]
- 49.Yamada A. B. a. K. M. J.Cell.Physiol. 2007;213:565–573. doi: 10.1002/jcp.21237. [DOI] [PubMed] [Google Scholar]
- 50.Harunaga JS, Yamada KM. Matrix Biol. 2011;30:363–368. doi: 10.1016/j.matbio.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Benzoni E, Torre ML, Faustini M, Stacchezzini S, Cremonesi F, Conte U, Villani S, Russo V, Ricevuti G, Vigo D. Int J Immunopathol Pharmacol. 2005;18:677–682. doi: 10.1177/039463200501800409. [DOI] [PubMed] [Google Scholar]
- 52.Zhang H, Lee MY, Hogg MG, Dordick JS, Sharfstein ST. ACS Nano. 2010;4:4733–4743. doi: 10.1021/nn9018812. [DOI] [PubMed] [Google Scholar]
- 53.Marrero B, Heller R. Biomaterials. 2012;33:3036–3046. doi: 10.1016/j.biomaterials.2011.12.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.