Abstract
Cystic fibrosis (CF) is an inherited disorder characterized by chronic airway inflammation. microRNAs (miRNAs) are endogenous small RNAs which act on messenger (m) RNA at a post transcriptional level, and there is a growing understanding that altered expression of miRNA is involved in the CF phenotype. Modulation of miRNA by replacement using miRNA mimics (premiRs) presents a new therapeutic paradigm for CF, but effective and safe methods of delivery to the CF epithelium are limiting clinical translation. Herein, polymeric nanoparticles are investigated for delivery of miRNA mimics into CF airway epithelial cells, using miR-126 as a proof-of-concept premiR cargo to determine efficiency. Two polymers, polyethyleneimine (PEI) and chitosan, were used to prepare miRNA nanomedicines, characterized for their size, surface (zeta) potential, and RNA complexation efficiency, and screened for delivery and cytotoxicity in CFBE41o- (human F508del cystic fibrosis transmembrane conductance regulator bronchial epithelial) cells using a novel high content analysis method. RNA extraction was carried out 24 hours post transfection, and miR-126 and TOM1 (target of Myb1) expression (a validated miR-126 target) was assessed. Manufacture was optimized to produce small nanoparticles that effectively complexed miRNA. Using high content analysis, PEI-based nanoparticles were more effective than chitosan-based nanoparticles in facilitating uptake of miRNA into CFBE41o- cells and this was confirmed in miR-126 assays. PEI-premiR-126 nanoparticles at low nitrogen/phosphate (N/P) ratios resulted in significant knockdown of TOM1 in CFBE41o- cells, with the most significant reduction of 66% in TOM1 expression elicited at an N/P ratio of 1:1 while chitosan-based miR-126 nanomedicines failed to facilitate statistically significant knockdown of TOM1 and both nanoparticles appeared relatively nontoxic. miRNA nanomedicine uptake can be qualitatively and quantitatively assessed rapidly by high content analysis and is highly polymer-dependent but, interestingly, there is not a direct correlation between the levels of miRNA uptake and the downstream gene knockdown. Polymeric nanoparticles can deliver premiRs effectively to CFBEs in order to modulate gene expression but must be tailored specifically for miRNA delivery.
Keywords: miR-126, nanotechnology, cystic fibrosis, TOM1, high content analysis, inflammation
Introduction
Cystic fibrosis (CF), an autosomal recessive disorder characterized by chronic airway inflammation,1 occurs due to mutations in the cystic fibrosis transmembrane conductance regulator (CFTR) gene involved in Cl− transport.2 A deletion of phenylalanine at position 508 of the CFTR protein (F508del) is the most frequent mutation, and accounts for approximately 70% of CFTR alleles.3 People with CF typically develop obstructive lung disease and disease in other organ systems, including pancreatic insufficiency, sweat electrolyte imbalance, and male infertility.3 Bronchial epithelial cells contribute significantly to the airway inflammation evident in the CF lung by responding to host-derived and pathogen-derived agonists, such as neutrophil elastase and Pseudomonas aeruginosa lipopolysaccharide1,4–9 that can signal via Toll-like receptors to augment interleukin-8 expression, leading to neutrophil-dominated inflammation. Therefore, components of these pathways may provide therapeutic targets for CF.
microRNAs (miRNAs) are 21–24 nucleotide duplex RNAs involved in the translational regulation of gene expression.10 RNA interference (RNAi) involving mature miRNAs occurs through the RNA induced silencing complex, where miRNA can bind to target messenger (m)RNA and induce cleavage degradation or translational repression of the mRNA target.10–12 Aberrant levels of miRNA are associated with many human diseases. miR-126, the first miRNA shown to be associated with CF, is downregulated in CF airway epithelial cells in vivo.1 TOM1 (target of Myb1) is a known target of miR-126, and is reciprocally upregulated in vivo in CF bronchial brushings.11 Other studies have also looked at miRNA expression in the CF airway and intestinal epithelial cells in humans and mice,13,14 and these support the concept that miRNAs have an important role in CF.15 Indeed, expression of wild-type and F508del CFTR are also known to be regulated by miRNAs.16–20
The use of RNAi in the targeted therapy of disease may prove very useful. Unlike DNA-based approaches, which require nuclear delivery, miRNAs and other RNAs, such as small interfering RNA (siRNA), only need to be delivered to the cytoplasm, and may be more benign to cells in terms of eliciting innate immune responses.21 Often miRNA has multiple targets, and this is of great benefit in terms of using replacement miRNA mimics.22 An added advantage of using miRNA over siRNA in regulation of aberrant mRNA expression is the reduced need for very high strand complementarity.
The systemic applications of naked miRNAs are restricted, because these and other small RNAs are polyanionic and highly susceptible to destruction by serum nucleases.23 Therefore, vectors are generally utilized to enhance in vivo stability as well as anatomic and cellular targeting. The use of nanoparticles and other nonviral vectors in the delivery of DNA and RNA into cells may be preferred therapeutically over viral vector-based delivery, due to the complications associated with viral delivery, including patient immune responses.21 Cationic polymers are now widely used to form RNA-containing nanoparticles, termed polyplexes. Examples of such polymers are polyethylenimine (PEI) and chitosan, and these are commercially available. PEI has a high cationic charge density, is of synthetic origin, and is available in various molecular weights and degrees of branching.24 Chitosan is a cationic polysaccharide polymer obtained by deacetylation of chitin. It can be sourced in many forms depending on molecular weight and degree of deacetylation.24,25 The physicochemical properties and subsequent biointeraction of RNA-cationic nanoparticles (polyplexes) is controlled by the ratio of amines on the cationic polymer to phosphates on the nucleic acid, and is known as the N/P ratio.
In the area of drug delivery, the cationic polymers described above have been used extensively to complex DNA and siRNA. However, very little work has been done using these polymers in the complexation of miRNA. Herein, we describe the preparation and characterization of miRNA nanomedicines using PEI and chitosan, and determine their levels of toxicity and miRNA uptake into a CFBE41o-(human F508del CFTR bronchial epithelial) cell line by harnessing state-of-the-art high content analysis, and using miR-126 as a proof-of-concept miRNA mimic (premiR) cargo to screen their efficiency by examining miR-126 and TOM1 expression.
Materials and methods
Materials
Chitosan glutamate (Protasan® UP G 113, molecular weight 160 kDa, degree of deacetylation 75%–90%), branched PEI (25 kDa, dialyzed), sodium tripolyphosphate pentabasic, Hoechst 33342, and phalloidin fluorescein isothiocyanate (FITC) were sourced from Sigma-Aldrich (St Louis, MO, USA). hsa-miR-126 (mature miRNA sequence UCGUACCGUGAGUAAUAAUGCG), hsa-miR-145 (sequence GUCCAGUUUUCCCAGGAAUCCCU), and Scrambled Silencer® Negative Control 1 siRNA (AM4611) were obtained from Applied Biosystems (Foster City, CA, USA). RiboJuice™ (Novagen, Billerica, MA, USA) transfection agent was used as the positive control in all transfections. Fluorescently labeled miRNA (Dharmacon Miridian miRNA-Dy547) and a Cellomics® multiparameter cytotoxicity 3 kit were obtained from Thermo Scientific (Waltham, MA, USA). All other chemicals used were of reagent grade and commercially available.
Preparation of miRNA nanomedicine
miRNA-PEI nanoparticles
Varying amounts of PEI (from 131 ng to 1.31 μg in distilled H2O) were added to 1 μg of miRNA (in 6.8 μL RNase-free H2O). This was diluted to 40 μL with phosphate-buffered saline (PBS) or 5% (w/v) glucose solution, mixed gently by pipette, and left on ice for 30 minutes.
miRNA-chitosan nanoparticles
For simple complexation, varying amounts of chitosan in 2% acetic acid were added to 1 μg of miRNA to produce N/P ratios of 50–200. This was diluted with PBS, mixed gently by pipette, and left on ice for 30 minutes. Chitosan-tripolyphosphate (TPP)-miRNA nanoparticles were prepared using various amounts of chitosan depending on the N/P ratio used. A weight ratio of 6:1 was used for all chitosan:TPP nanoparticles described here. Briefly, TPP solution was added to 0.5 μg miRNA and diluted to 100 μL with distilled water. This was left for 2 minutes at room temperature and then added dropwise to the relevant concentration of chitosan solution (100 μL) to produce N/P ratios of 50–200. The solution was then mixed gently by pipette and left on ice for 30 minutes.
Size and zeta potential
The sizes and zeta potential of all the miRNA nanomedicines were measured using the Zetasizer Nano ZS (Malvern Instruments, Malvern, UK). All complexes were diluted to 1 mL in 10 mM NaCl solution immediately before measurement on the Zetasizer instrument.
CFBE41o- uptake of miRNA nanomedicines: high content analysis
CFBE41o- cells (human F508del CFTR bronchial epithelial cells)26 were maintained in a 37°C, humidified 5% CO2 incubator in Minimal Essential Medium/GlutaMAX™ medium (Gibco®, Life Technologies, Carlsbad CA, USA). Cells were seeded at 3 × 104 cells/well in a 96-well plate. miRNA-nanomedicines were prepared as described with the fluorescently labeled miRNA (Dharmacon Miridian miRNA-Dy547, ThermoScientific). Thirty nM miRNA-Dy547 or equivalent in nanoparticles was added per well and following a 20.5-hour incubation, the cells were washed with PBS and fixed in 4% paraformaldehyde. Cells were stained for F-actin using phalloidin FITC and for the nucleus using Hoechst 33342. Cells were washed three times with Dulbecco’s PBS and stored in 150 μL of this solution. High content analysis was carried out using an IN Cell Analyzer 1,000 (GE Health/Amersham Biosciences, Little Chalfont, UK).
High content analysis of nanoparticle toxicity
CFBE41o- cells were seeded at 3 × 104 cells/well in a 96-well plate. miRNA nanomedicines were formed as described earlier, using premiR-126 (30 nM per well). Control wells were treated with 120 μM valinomycin for 18 hours as a positive control of toxicity prior to analysis. Cells were fixed and stained for the nucleus using Hoechst 33342 and the cell count was determined using the IN Cell Analyzer 1,000.
Transfection with miRNA nanomedicines
CFBE41o- cells were seeded on 24-well plates at 8 × 104 cells/well and were treated with premiR-126:PEI nanomedicines at N/P ratios of 1:1, 3:1, 5:1, or 10:1; premiR-126:chitosan nanomedicines at N/P ratios of 5:1, 50:1, 15:1, or 200:1; and compared with relevant controls including scrambled premiR control (scr), premiR-126 only; empty chitosan nanoparticles and PEI nanoparticles; premiR-126 using RiboJuice, or untreated. All miRNA treatments were at a concentration of 30 nM premiR per well. Twenty-four hours post-transfection, cells were lysed and RNA isolated for further analysis using TRI Reagent® (Sigma-Aldrich) according to the manufacturer’s specifications.
Complementary DNA (cDNA) synthesis and quantitative reverse transcription polymerase chain reaction (qRT-PCR) for miR-126
miRNA expression was measured using Taqman miRNA assays (Applied Biosystems) according to the manufacturer’s instructions, and qRT-PCR was performed on an LC480 Lightcycler (Roche, Basel, Switzerland). Expression of miRNA relative to β-actin was determined using the 2(−ΔΔCt) method.27 All qRT-PCR experiments were performed in triplicate, including no-template controls.
cDNA synthesis and qRT-PCR for TOM1
Equal quantities of RNA were reverse transcribed into cDNA using a Quantitect reverse transcription kit (Qiagen, Valencia, CA, USA). The cDNA resulting from this reaction was used as a template for qRT-PCR using SYBR Green (Roche) on the LightCycler 480 PCR system (Roche). The expression of TOM1 relative to β-actin was determined using the 2(−ΔΔCt) method.27 Primers for TOM1 and β-actin were obtained from MWG Eurofins Genetics (Ebersberg, Germany) (TOM1-F 5′-ATTCTGTGGGCACTGACTCC-3′ and TOM1-R 5′-CACTCACCATCTCCAGCTCA-3′, β-actin-F 5′-GGACTTCGAGCAAGAGATGG-3′ and β-actin-R 5′-AGGAAGGAAGGCTGGAAGAG-3′).
Statistical analysis
All analyses were performed using GraphPad Prism 4.0 software package (San Diego, CA, USA). Results are expressed as the mean ± standard error of the mean and were compared using the Student’s t-test (nonparametric, one-tailed). Differences were considered significant at P ≤ 0.05.
Results
Characterization of miRNA nanoparticles
Characteristics such as size, zeta potential (surface charge), and degree of complexation or encapsulation all affect the delivery, efficiency, and toxicity of nanoparticles, and the preparation methods were optimized to produce small miRNA-loaded, PEI, and chitosan nanoparticles that complexed and condensed miRNA.
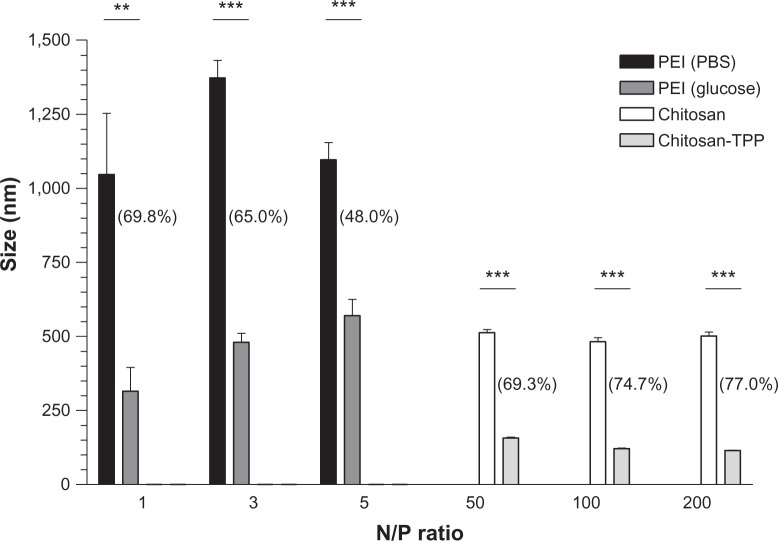
Large particles, with sizes greater than 1,000 nm, were obtained when PEI was complexed with premiR-126 at N/P ratios of 1:1, 3:1, and 5:1 using PBS as the diluent (Figure 1). These sizes are likely due to aggregation of nanoparticles. Using a higher N/P ratio of 10:1, a reduced size of 588 ± 34 nm was obtained. Using 5% (w/v) glucose as the complexation diluent for preparation of PEI:miRNA nanoparticles instead of PBS, much smaller complexes were obtained. The reductions in diameter are in the order of a 48%–70% decrease in size, with PEI:miRNA nanoparticles prepared at an N/P ratio of 1:1 in glucose having a size around 300 nm. No significant effect between diluents was observed with respect to zeta potential. Figure 1 also illustrates the effect of N/P ratio on the size of chitosan-miRNA nanoparticles, with mean diameters of between 480 nm and 590 nm for N/P ratios of 50:1, 100:1, 150:1, and 200:1. The inclusion of a crosslinking agent, TPP, in the manufacturing process significantly reduced the miRNA nanoparticles to as low as 115 ± 1.7 nm in diameter.
Figure 1.
Mean size (nm) of premiR-126-poly(ethyleneimine) (PEI) complexes prepared using phosphate-buffered saline (PBS) and 5% glucose, or premiR-126 chitosan complexes and premiR-126 chitosan-TPP nanoparticles. The percentage reduction in nanoparticle size using glucose or tripolyphosphate (TPP) for PEI and chitosan respectively at each N/P ratio, are shown in brackets.
Notes: Data are represented as mean ± standard error of the mean and were compared by t-test (non-parametric, one-tailed). Differences were considered significant at P ≤ 0.05. **P ≤ 0.01. ***P ≤ 0.001.
Abbreviations: miR, microribonucleic acid; N/P, nitrogen/phosphate ratio.
The zeta potential of free miRNA was found to be −15.98 ± 3.9 mV. PEI-miRNA nanoparticles were positively charged above an N/P ratio of 5, while chitosan-miRNA nanoparticles had a net positive charge at all N/P ratios over 50:1 (data not shown).
miRNA nanomedicine uptake into CFBE41o- cells: high content analysis
High content analysis allows for analysis and quantification of multiple parameters of both cellular uptake of nanoparticles and cytotoxicity induced as a result of treatment. The IN Cell Analyzer 1,000 algorithm detects the presence of whole fixed cells by the presence of both the nucleus (stained with Hoechst 33342) and the cytoplasm (stained with F-actin using phalloidin FITC). Harnessing high content analysis, PEI-based nanoparticles at N/P ratios of 5:1 and 10:1 appear to be more effective at delivery of miRNA to the cell than chitosan-based or chitosan-TPP-based nanoparticles, or transfection using the commercially available RNA transfection agent, RiboJuice (Figure 2). Figure 2B clearly indicates differences in the nature of the intracellular distribution of the miRNA within CFBE41o- cells when delivered using different carriers. Highly defined punctae are seen in PEI-miRNA-treated cells, while those treated with chitosan-miRNA nanoparticles show a diffuse distribution of miRNA in the cell. When TPP is used to prepare smaller and more defined miRNA nanoparticles, this can be seen to impact clearly the distribution of miRNA, with more defined areas of miRNA within the cell. These distribution differences can have a very significant effect on molecular kinetics and ultimately on the efficacy of the miRNA nanoparticles.
Figure 2.
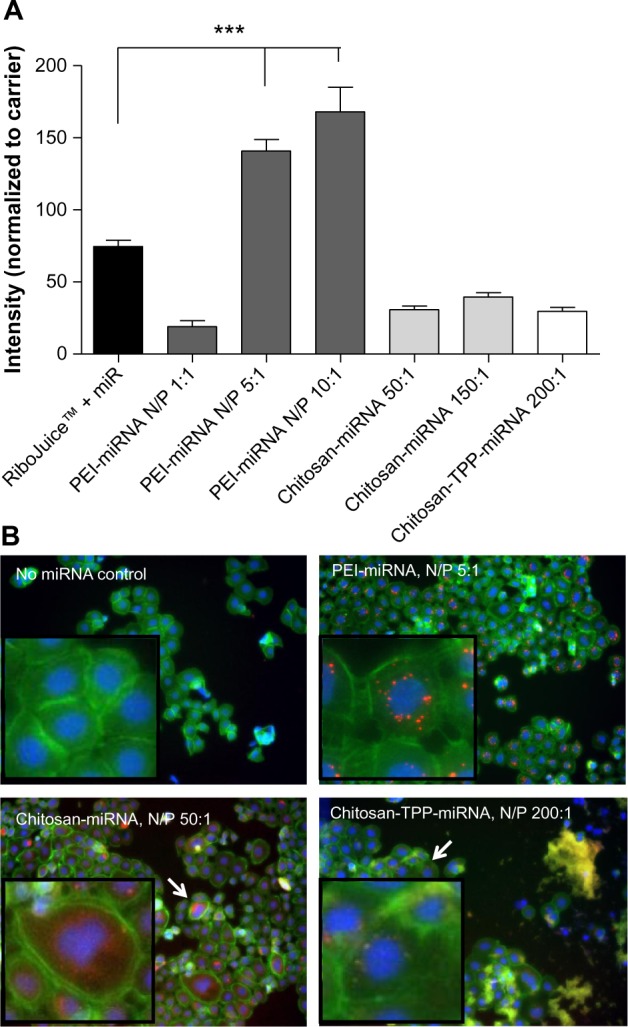
High content analysis of miRNA-Dy547 loaded nanoparticles association with CFBE41o- cells at 20.5 hours post transfection. (A) Comparative quantification of fluorescent miRNA delivered to CFBE cells by PEI- and chitosan-based nanoparticles at different N/P ratios and compared to commercial transfection agent, RiboJuice™. Data are represented as mean fluorescence intensity normalized to relative carrier, (ie, PEI, Chitosan or Chitosan-TPP uncomplexed) ± standard error of the mean and were compared by one-way analysis of variance, followed by Bonferroni’s multiple comparison test. Differences were considered significant at P ≤ 0.05. ***P ≤ 0.001. (B) Images of miRNA nanoparticles (red) internalized by CFBE cells (blue: nucleus, green: f-actin, arrow: area shown in higher zoom box) visualized using IN Cell analyzer 1,000.
Abbreviations: miRNA/miR, microribonucleic acid; PEI, poly(ethyleneimine); TPP, tripolyphosphate; N/P, nitrogen/phosphate ratio; CFBE41o-, human F508del cystic fibrosis transmembrane conductance regulator bronchial epithelial cells.
High content analysis of nanoparticle toxicity
Cell count is the most obvious marker of toxicity. miRNA-PEI-based and miRNA-chitosan-based nanoparticles induced little or no toxicity at the N/P ratios tested (Figure 3).
Figure 3.
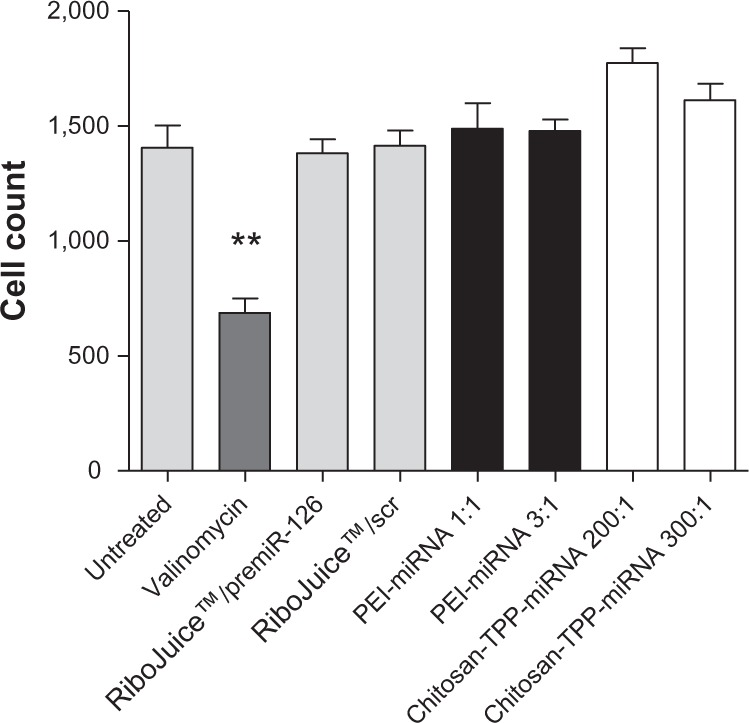
Cell count after treatment of CFBE41o- cells with miRNA-nanomedicines for 18 hours. Cells were subsequently stained and visualized using IN Cell Analyzer 1,000 (n = 3).
Note: Differences were considered significant at P ≤ 0.05. **P ≤ 0.01.
Abbreviations: miRNA/miR, microribonucleic acid; PEI, poly(ethyleneimine); TPP, tripolyphosphate; scr, scrambled control; CFBE41o-, human F508del cystic fibrosis transmembrane conductance regulator bronchial epithelial cells.
Modulation of miRNA in CFBE41o-cells using premiR-126 nanomedicines
miR-126 levels were quantified in CFBE41o- cells post transfection (Figure 4). Cells treated with miRNA-PEI nanoparticles at N/P ratios of 3:1 and higher had over 10,000-fold increases in miR-126 compared with untreated cells, similar to that seen using the commercially available transfection reagent, RiboJuice (P = 0.0378 and P = 0.0115 for PEI 3:1 and 5:1 versus scr). Chitosan-miR126-transfected cells also showed an increase in miR-126, most evident at N/P ratios of 200:1 and 300:1; however, these values were not statistically significant. The PEI-miRNA nanomedicines were found to be significantly more efficient than chitosan-miRNA nanomedicines at increasing miR-126 levels (PEI 3:1 versus chitosan-TPP 200:1 and 300:1, P = 0.0389 and P = 0.0381, respectively; PEI 5:1 versus chitosan-TPP 200:1 and 300:1, P = 0.0117 and P = 0.0116, respectively).
Figure 4.
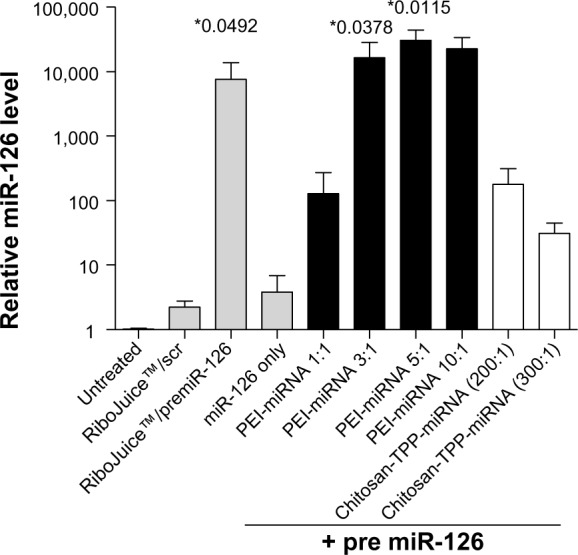
Effect of premiR-126 nanoparticles (as indicated) on miR-126 expression in CFBE41o- cells was assessed using qRT-PCR.
Notes: Data are represented as mean (n = 3) ± standard error of the mean and were compared by an unpaired t-test (one-tailed) to scrambled negative control complexed with RiboJuice™. Differences were considered significant at *P ≤ 0.05.
Abbreviations: scr, scrambled control; miRNA/miR, microribonucleic acid; PEI, poly(ethyleneimine); TPP, tripolyphosphate; qRT-PCR, quantitative reverse transcription polymerase chain reaction; CFBE41o-, human F508del cystic fibrosis transmembrane conductance regulator bronchial epithelial cells.
No significant reduction in TOM1 expression was seen in CFBE41o- cells after transfection with premiR-126 alone (Figure 5). However, TOM1 expression was significantly reduced in cells treated with PEI:premiR-126 at N/P ratios of 1:1, 3:1, and 5:1, although not at the highest N/P ratio studied of 10:1. The most significant reduction in TOM1 of 66% was seen using a PEI N/P ratio of 1:1, which had led to the lowest increase in miR-126 of any of the PEI nanoparticles in the miRNA assay (Figure 4). Interestingly, while RiboJuice:premiR-126 and chitosan:premiR-126 led to increased miR-126 levels, this did not translate into a statistically significant knockdown of TOM1 expression.
Figure 5.
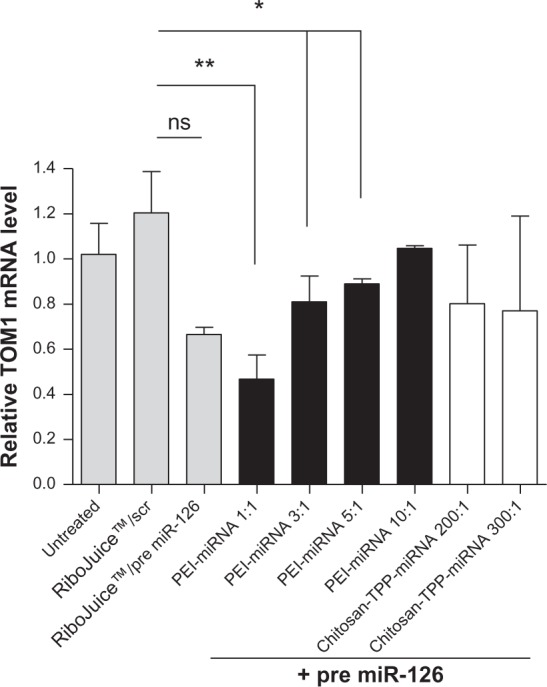
Effect of premiR-126 nanoparticles (as indicated) on TOM1 expression in CFBE41o- cells was assessed using qRT-PCR.
Notes: Data are represented as mean (n = 3) ± standard error of the mean and were compared by t-test (non-parametric, one-tailed) to scrambled negative control complexed with RiboJuice™. Differences were considered significant at P ≤ 0.05; *P ≤ 0.05, **P ≤ 0.01, ns = non statistically significant.
Abbreviations: scr, scrambled control; miRNA/miR, microribonucleic acid; PEI, poly(ethyleneimine); TPP, tripolyphosphate; qRT-PCR, quantitative reverse transcription polymerase chain reaction; TOM1, target of Myb1; CFBE41o-, human F508del cystic fibrosis transmembrane conductance regulator bronchial epithelial cells.
Discussion
The physiology and anatomy of the lungs makes the respiratory tract an ideal target for noninvasive local treatment of respiratory diseases using nanotechnology,28 including the respiratory component of CF. It is now well known that aberrant miRNA expression is involved in a range of diseases, and in the case of conditions involving overexpression of miRNA, various strategies are currently being investigated, including the use of modified antisense oligonucleotides (specifically antagomirs).29 For those conditions where underexpression of particular miRNAs is involved, these may be introduced into affected cells as premiRs. As for other nucleic acid-based therapeutics, effective delivery of antagomir and premiRs is a major obstacle to their clinical and commercial development. Therefore, it is critical that the relevant delivery technology is developed in parallel with progress in the field of epigenetics. Because nucleic acids are too large, too negatively charged, and targeted by many endogenous nucleases, polymeric nanoparticles can facilitate intracellular delivery, enhance in vivo stability, and target their delivery.30 Thus, microRNA nanomedicines may provide new therapeutic options for the treatment of many disease states, including cystic fibrosis. Herein we explored the use of cationic nanoparticles in the delivery of miRNA into CF cell lines to determine if they could modulate miR-126 levels in CFBE41o- cells and impact on the associated expression of the miR-126 validated target, TOM1.
There are limited data available on the physicochemical characteristics of miRNA nanoparticles. The sizes of PEI-miRNA complexes appear to be buffer-dependent, as size was significantly reduced when the complexes were prepared in a 5% (w/v) glucose solution instead of PBS (Figure 1). Others have found similar reductions in size of PEI-DNA complexes by using 5% glucose instead of salt-based solutions.31 As for other nucleic acids, including plasmid DNA and siRNA, PEI appears to be an extremely effective complexation agent for miRNA mimics, complexing the premiR completely at low N/P ratios to create small, positively charged nanoparticles. While the chitosan-miRNA nanoparticles prepared herein were slightly larger than the chitosan-siRNA polyplexes reported in the literature,32 use of the TPP crosslinker significantly reduced the size of the miRNA nanoparticles and altered the intracellular distribution of the miRNA once internalized by the CFBE cells (Figure 2B).
High content analysis (Figure 2A) confirms the effective uptake of miRNA nanomedicines into CFBE41o- cells. This technique has been successfully applied to screen delivery of plasmid DNA33 and siRNA,34 and herein we apply it for the first time to the screening of intracellular delivery for miRNA nanoparticles. The advantages of the technique include the ability to screen a range of delivery systems in parallel and generate both qualitative images and quantitative uptake data. The high content analysis data indicated that delivery of premiRNA-PEI nanomedicines to CFBE cells was twice as effective as miRNA transfection using RiboJuice, a commercial transfection agent often used for miRNA transfection in molecular biology applications. The high content analysis uptake data also indicated that PEI was a more efficient miRNA carrier than chitosan. The findings of the high content analysis uptake study are borne out by the miR-126 assay (Figure 4) in which PEI-miRNA nanomedicines were found to be significantly more efficient than chitosan-miRNA nanomedicines at increasing miR-126 levels. This may be explained by both the higher binding efficiency of premiRs to PEI than chitosan (data not shown) and its highly cationic nature that increases its interaction with the cell membrane, thereby enhancing transfection. This builds on and supports previous work harnessing PEI and chitosan for siRNA delivery that found PEI was significantly better at both complexing siRNA and transfecting cells with short RNA sequences.35,36
There is an interesting dichotomy in the miR-126 assay (Figure 4) and the TOM1 levels (Figure 5), wherein the formulation of PEI:premiR-126 N/P ratio of 1:1 that produces a relatively small increase in miR-126 compared with some of the other formulations of PEI leads to the most significant decrease in TOM1 expression. High N/P ratio nanoparticles composed of nucleic acids and PEI are often associated with cytotoxicity, which can then be associated with diminished gene expression or gene knockdown, depending on the cargo encapsulated therein. However, our cell toxicity studies (Figure 3) would indicate that overall cell viability is not compromised at the N/P ratios used in this study. Therefore, our hypothesis is that while PEI:premiR-126 N/P 3:1, 5:1 and 10:1 facilitate efficient uptake of the miRNA, greater than that facilitated by RiboJuice or PEI:premiR-126 N/P 1:1, this huge increase in fact saturates the miRNA induced silencing complex machinery, especially at a very high N/P ratio of 10:1, and interferes with its function. Previous work has found that high levels of artificial RNAi substrates delivered to cells can cause cellular toxicity and may compete for endogenous RNAi machinery, leading to disruption of natural miRNA function.39,40 Therefore, at high N/P ratios, the PEI-based nanomedicines may in essence be too effective at delivery and thereby negate the therapeutic benefits of the cargo. While RiboJuice appears to facilitate significantly more uptake and miR-126 expression than PEI 1:1, the downstream effects are almost comparable, although the RiboJuice fails to elicit a statistically significant decrease in TOM1 expression. This difference may relate to differences in intracellular trafficking of the two nanomedicines, with PEI capable of superior delivery than that of RiboJuice, of the internalized premiR-126 from the endolysosomal system. This material-dependent effect on gene expression, independent of simple cell uptake, can also be seen when comparing the chitosan-TPP nanoparticles. In Figure 2, the premiR uptake facilitated by the chitosan-TPP nanoparticles is slightly greater than PEI 1:1, while in Figure 4, the miR-126 expression level for chitosan-TPP 200:1 is approximately that of PEI 1:1, yet this fails to translate into significant knockdown of TOM1 expression. Again this may relate to differences in the molecular pharmacokinetics of these nanomedicines, with PEI’s “proton sponge” capacity enabling a small amount of miR-126 to effect significant knockdown of target gene expression. We would contend that this is an important finding in the context of miRNA nanomedicine development that differs from the development of other nucleic acid-based therapies, eg, plasmid DNA, where significant uptake is generally required to elicit gene expression. The more complex miRNA pathways may in fact respond to relatively small quantities of premiR delivered within a carrier that facilitates the appropriate intracellular trafficking and therefore development of these delivery systems must have an endpoint expression assay as their primary screening tool.
The PEI-miRNA nanomedicines (N/P ratio 1:1) produced a 66% knockdown in TOM1 compared with scrambled controls. This is a significantly lower N/P ratio than would be used for PEI transfections for plasmid DNA or siRNA, which are generally optimal at N/P ratios of 5:1 to 10:1.25,30 This would indicate that the preparation of miRNA nanomedicines needs to be optimized for each polymeric system and cannot be directly extrapolated from previous siRNA or plasmid DNA work. Of the miRNA nanomedicines assessed, PEI-miRNA nanomedicines (N/P ratio 1:1) offer the greatest potential, leading to a significant modulation in the target gene, ie, 66% knockdown of TOM1, with the advantage of a low N/P ratio, which limits toxicity issues associated with the polymer. These data would indicate that caution needs to be taken in the use of cell uptake studies to screen for miRNA delivery systems. Our understanding of their cellular role is slowly being elucidated and the complexities of this process might well mean that the simple correlation between increased miR-126 levels and knockdown of the target gene is not as straightforward a relationship as that seen, for example, with the highly sequence-specific siRNA molecules.
Local delivery of miRNA to the CF lung is one of the most promising approaches for bringing miRNA nanotechnologies targeting CF to the clinic. Inhalation offers tissue-specific targeting of the miRNA and minimal systemic exposure, thereby diminishing the risk of off-target effects. However, CF lungs represent both significant anatomic and pathologic barriers to inhaled nanomedicines, including obstructed airways covered with thickened mucus and mucus plugs. In order to develop therapeutics such as these for local aerosolized delivery to the CF lung, the next steps will be to evaluate their efficacy in mucus-producing air-liquid interface cultures, primary CF airway epithelial cell cultures, and ultimately in animal models of CF. The complex branched anatomy of the airways means inhaled nanomedicines require an effective device to deliver them to their site of action. The recent development of advanced nebulizers, eg, vibrating mesh devices, enables much more efficient delivery of nanomedicines to the lungs.
Prior to clinical testing, these miRNA nanomedicines will need to be screened in suitable in vivo CF models to examine efficacy, toxicity, and immunogenicity. Preclinical studies of this next generation of gene medicines represented by miRNA nanomedicine will benefit from novel animal models such as the CF pig and ferret models. The most common routes for pulmonary drug administration particularly in rodents are intratracheal and intranasal inhalation. Intratracheal administration works best for proof-of-concept work, and due to its invasive nature, it is not routinely used in humans and also does not require the assessment of aerosol droplet or particle size. However, inhaled delivery in animals cannot be extrapolated to humans due to differences in anatomy and immunology.
Conclusion
The degree of miRNA uptake facilitated by a range of nanomedicines can be qualitatively and quantitatively assessed rapidly by high content analysis and is highly carrier-dependent. High content analysis data indicate that miRNA-PEI nanomedicines facilitated greater uptake than miRNA-TPP-chitosan nanoparticles and the commercial transfection agent, RiboJuice. This superior delivery efficiency for PEI nanomedicines translated into modulation of TOM1 expression with PEI-miRNA nanomedicines (N/P ratio 1:1, 3:1, and 5:1), the only systems that could significantly knockdown TOM1 expression in CFBE41o-cells (P < 0.05). Interestingly, relative premiR-126 delivery efficiency for different PEI nanomedicines did not directly correlate with ability to modulate TOM1 expression in CFBE41o- cells with PEI-miRNA nanomedicines (N/P ratio 1:1, P < 0.05) offering the greatest knockdown of the formulations tested but not the most effective cellular uptake or miR-126 levels. Polymeric nanoparticles offer a biocompatible and efficient means of delivering premiRs effectively to CFBE41o- cells in order to modulate gene expression, thereby facilitating clinical translation. However, caution in extrapolation of uptake studies is required and downstream functional assays are ultimately required to determine the efficacy of each system.
Acknowledgments
This work was supported in part by a Science Foundation Ireland Strategic Research Cluster grant (07/SRC/B1154) and a Health Research Board grant (PhD/2007/11).
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Oglesby IK, Bray IM, Chotirmall SH, et al. miR-126 is downregulated in cystic fibrosis airway epithelial cells and regulates TOM1 expression. J Immunol. 2010;184:1702–1709. doi: 10.4049/jimmunol.0902669. [DOI] [PubMed] [Google Scholar]
- 2.Rommens J, Kerem B, Alon N, et al. Identification of the cystic fibrosis gene: cloning and characterization of complementary DNA. Science. 1989;245:1066–1073. doi: 10.1126/science.2475911. [DOI] [PubMed] [Google Scholar]
- 3.Sheridan MB, Hefferon TW, Wang N, et al. CFTR transcription defects in pancreatic sufficient cystic fibrosis patients with only one mutation in the coding region of CFTR. J Med Genet. 2011;48:235–241. doi: 10.1136/jmg.2010.083287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McNally P, Coughlan C, Bergsson G, et al. Vitamin D receptor agonists inhibit pro-inflammatory cytokine production from the respiratory epithelium in cystic fibrosis. J Cyst Fibros. 2011;10:428–434. doi: 10.1016/j.jcf.2011.06.013. [DOI] [PubMed] [Google Scholar]
- 5.Cosgrove S, Chotirmall SH, Greene CM, McElvaney NG. Pulmonary proteases in the cystic fibrosis lung induce interleukin 8 expression from bronchial epithelial cells via a heme/meprin/epidermal growth factor receptor/Toll-like receptor pathway. J Biol Chem. 2011;286:7692–7704. doi: 10.1074/jbc.M110.183863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Greene CM, Ramsay H, Wells RJ, O’Neill SJ, McElvaney NG. Inhibition of Toll-like receptor 2-mediated interleukin-8 production in cystic fibrosis airway epithelial cells via the alpha7-nicotinic acetylcholine receptor. Mediators Inflamm. 2010;2010:423241. doi: 10.1155/2010/423241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chotirmall SH, Greene CM, Oglesby IK, et al. 17Beta-estradiol inhibits IL-8 in cystic fibrosis by up-regulating secretory leucoprotease inhibitor. Am J Respir Crit Care Med. 2010;182:62–72. doi: 10.1164/rccm.201001-0053OC. [DOI] [PubMed] [Google Scholar]
- 8.Bergin DA, Greene CM, Sterchi EE, et al. Activation of the epidermal growth factor receptor (EGFR) by a novel metalloprotease pathway. J Biol Chem. 2008;283:31736–31744. doi: 10.1074/jbc.M803732200. [DOI] [PubMed] [Google Scholar]
- 9.Greene CM, Carroll TP, Smith SG, et al. TLR-induced inflammation in cystic fibrosis and non-cystic fibrosis airway epithelial cells. J Immunol. 2005;174:1638–1646. doi: 10.4049/jimmunol.174.3.1638. [DOI] [PubMed] [Google Scholar]
- 10.Rana TM. Illuminating the silence: understanding the structure and function of small RNAs. Nat Rev Mol Cell Biol. 2007;8:23–36. doi: 10.1038/nrm2085. [DOI] [PubMed] [Google Scholar]
- 11.Oglesby IK, McElvaney NG, Greene CM. MicroRNAs in inflammatory lung disease – master regulators or target practice? Respir Res. 2010;11:148. doi: 10.1186/1465-9921-11-148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bartel D. MicroRNAs genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 13.Bhattacharyya S, Balakathiresan NS, Dalgard C, et al. Elevated miR-155 promotes inflammation in cystic fibrosis by driving hyperexpression of interleukin-8. J Biol Chem. 2011;286:11604–11615. doi: 10.1074/jbc.M110.198390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bazett M, Paun A, Haston CK. MicroRNA profiling of cystic fibrosis intestinal disease in mice. Mol Genet Metab. 2011;103:38–43. doi: 10.1016/j.ymgme.2011.01.012. [DOI] [PubMed] [Google Scholar]
- 15.Xu W, Hui C, Yu SS, Jing C, Chan HC. MicroRNAs and cystic fibrosis – an epigenetic perspective. Cell Biol Int. 2011;35:463–466. doi: 10.1042/CBI20100664. [DOI] [PubMed] [Google Scholar]
- 16.Gillen AE, Gosalia N, Leir H, Harris A. MicroRNA regulation of expression of the cystic fibrosis transmembrane conductance regulator gene. Biochem J. 2011;438:25–32. doi: 10.1042/BJ20110672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Megiorni F, Cialfi S, Dominici C, Quattrucci S, Pizzuti A. Synergistic post-transcriptional regulation of the cystic fibrosis transmembrane conductance regulator (CFTR) by miR-101 and miR-494 specific binding. PLoS One. 2011;6:e26601. doi: 10.1371/journal.pone.0026601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ramachandran S, Karp PH, Jiang P, et al. A microRNA network regulates expression and biosynthesis of wild-type and deltaF508 mutant cystic fibrosis transmembrane conductance regulator. Proc Natl Acad Sci U S A. 2012;109:13362–13367. doi: 10.1073/pnas.1210906109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hassan F, Nuovo GJ, Crawford M, et al. miR-101 and miR-144 regulate the expression of the CFTR chloride channel in the lung. PLoS One. 2012;7:e50837. doi: 10.1371/journal.pone.0050837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Oglesby IK, Chotirmall SH, McElvaney NG, Greene CM. Regulation of CFTR by microRNA-145, -223 and -494 is altered in ΔF508 cystic fibrosis airway epithelium. J Immunol. 2013;190:3354–3362. doi: 10.4049/jimmunol.1202960. [DOI] [PubMed] [Google Scholar]
- 21.Durcan N, Murphy C, Cryan S-A. Inhalable siRNA: potential as a therapeutic agent in the lungs. Mol Pharm. 2008;5:559–566. doi: 10.1021/mp070048k. [DOI] [PubMed] [Google Scholar]
- 22.Esquela-Kerscher A, Slack FJ. Oncomirs – microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 23.Howard KA. Delivery of RNA interference therapeutics using polycation-based nanoparticles. Adv Drug Deliv Rev. 2009;61:710–720. doi: 10.1016/j.addr.2009.04.001. [DOI] [PubMed] [Google Scholar]
- 24.Guo P, Coban O, Snead NM, et al. Engineering RNA for targeted siRNA delivery and medical application. Adv Drug Deliv Rev. 2010;62:650–666. doi: 10.1016/j.addr.2010.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mao S, Sun W, Kissel T. Chitosan-based formulations for delivery of DNA and siRNA. Adv Drug Deliv Rev. 2010;62:12–27. doi: 10.1016/j.addr.2009.08.004. [DOI] [PubMed] [Google Scholar]
- 26.Bebok Z, Collawn JF, Wakefield J, et al. Failure of cAMP agonists to activate rescued deltaF508 CFTR in CFBE41o- airway epithelial monolayers. J Physiol. 2005;569(Pt 2):601–615. doi: 10.1113/jphysiol.2005.096669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-[delta][delta]CT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 28.Yang W, Peters JI, Williams RO. Inhaled nanoparticles – a current review. Int J Pharm. 2008;356:239–247. doi: 10.1016/j.ijpharm.2008.02.011. [DOI] [PubMed] [Google Scholar]
- 29.Soifer HS, Rossi JJ, Saetrom P. MicroRNAs in disease and potential therapeutic applications. Mol Ther. 2007;15:2070–2079. doi: 10.1038/sj.mt.6300311. [DOI] [PubMed] [Google Scholar]
- 30.Whitehead KA, Langer R, Anderson DG. Knocking down barriers: advances in siRNA delivery. Nat Rev Drug Discov. 2009;8:129–138. doi: 10.1038/nrd2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brissault B, Leborgne C, Guis C, Danos O, Cheradame H, Kichler A. Linear topology confers in vivo gene transfer activity to polyethylenimines. Bioconjug Chem. 2006;17:759–765. doi: 10.1021/bc050287v. [DOI] [PubMed] [Google Scholar]
- 32.Katas H, Alpar HO. Development and characterisation of chitosan nanoparticles for siRNA delivery. J Control Release. 2006;115:216–225. doi: 10.1016/j.jconrel.2006.07.021. [DOI] [PubMed] [Google Scholar]
- 33.de Raad M, Teunissen EA, Lelieveld D, Egan DA, Mastrobattista E. High-content screening of peptide-based non-viral gene delivery systems. J Control Release. 2012;158:433–442. doi: 10.1016/j.jconrel.2011.09.078. [DOI] [PubMed] [Google Scholar]
- 34.Hibbitts A, Lieggi N, McCabe O, et al. Screening of siRNA nanoparticles for delivery to airway epithelial cells using high content analysis. Ther Deliv. 2011;2:987–999. doi: 10.4155/tde.11.73. [DOI] [PubMed] [Google Scholar]
- 35.Laroui H, Theiss AL, Yan Y, et al. Functional TNFα gene silencing mediated by polyethyleneimine/TNFα siRNA nanocomplexes in inflamed colon. Biomaterials. 2011;32:1218–1228. doi: 10.1016/j.biomaterials.2010.09.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang J, Feng SS, Wang S, Chen ZY. Evaluation of cationic nanoparticles of biodegradable copolymers as siRNA delivery system for hepatitis B treatment. Int J Pharm. 2010;400:194–200. doi: 10.1016/j.ijpharm.2010.08.026. [DOI] [PubMed] [Google Scholar]
- 37.Günther M, Lipka J, Malek A, Gutsch D, Kreyling W, Aigner A. Polyethylenimines for RNAi-mediated gene targeting in vivo and siRNA delivery to the lung. Eur J Pharm Biopharm. 2011;77:438–449. doi: 10.1016/j.ejpb.2010.11.007. [DOI] [PubMed] [Google Scholar]
- 38.Gautam A, Densmore CL, Xu B, Waldrep JC. Enhanced gene expression in mouse lung after PEI-DNA aerosol delivery. Mol Ther. 2000;2:63–70. doi: 10.1006/mthe.2000.0087. [DOI] [PubMed] [Google Scholar]
- 39.Grimm D, Streetz KL, Jopling CL, et al. Fatality in mice due to oversaturation of cellular microRNA/short hairpin RNA pathways. Nature. 2006;441:537–541. doi: 10.1038/nature04791. [DOI] [PubMed] [Google Scholar]
- 40.Castanotto D, Sakurai K, Lingeman R, et al. Combinatorial delivery of small interfering RNAs reduces RNAi efficacy by selective incorporation into RISC. Nucleic Acids Res. 2007;35:5154–5164. doi: 10.1093/nar/gkm543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Conese M, Ascenzioni F, Boyd AC, et al. Gene and cell therapy for cystic fibrosis: from bench to bedside. J Cyst Fibros. 2011;10:S114–S128. doi: 10.1016/S1569-1993(11)60017-9. [DOI] [PubMed] [Google Scholar]