Summary
Understanding how interactions between extracellular signalling pathways and transcription factor networks influence cellular decision making will be crucial for understanding mammalian embryogenesis and for generating specialised cell types in vitro. To this end, pluripotent mouse Embryonic Stem (mES) cells have proven to be a useful model system. However, understanding how transcription factors and signalling pathways affect decisions made by individual cells is confounded by the fact that measurements are generally made on groups of cells, whilst individual mES cells differentiate at different rates and towards different lineages, even in conditions that favour a particular lineage. Here we have used single-cell measurements of transcription factor expression and Wnt/β-catenin signalling activity to investigate their effects on lineage commitment decisions made by individual cells. We find that pluripotent mES cells exhibit differing degrees of heterogeneity in their expression of important regulators from pluripotency, depending on the signalling environment to which they are exposed. As mES cells differentiate, downregulation of Nanog and Oct4 primes cells for neural commitment, whilst loss of Sox2 expression primes cells for primitive streak commitment. Furthermore, we find that Wnt signalling acts through Nanog to direct cells towards a primitive streak fate, but that transcriptionally active β-catenin is associated with both neural and primitive streak commitment. These observations confirm and extend previous suggestions that pluripotency genes influence lineage commitment and demonstrate how their dynamic expression affects the direction of lineage commitment, whilst illustrating two ways in which the Wnt signalling pathway acts on this network during cell fate assignment.
Keywords: Single cell analysis, Pluripotency, Lineage priming, Mouse embryonic stem cells, Transcription factor networks, Wnt/β-catenin signalling
Introduction
The mammalian embryo is derived from a small group of cells, the epiblast, which is first segregated from extraembryonic tissue just before implantation, at the blastocyst stage (Tam and Loebel, 2007; Rossant and Tam, 2009). This group of cells forms an epithelium (the postimplantation epiblast) that undergoes a proliferative expansion before giving rise to different lineages through the spatio-temporal segregation of patterns of gene expression. The first symmetry-breaking event in the embryonic epiblast occurs around the time of implantation (E4.5), with the definition of an anteroposterior axis, mediated by signalling between the extraembryonic endoderm and the epiblast (Takaoka et al., 2006). This event positions the primordium for the brain (ectoderm) anteriorly, and the primitive streak, that will give rise to the endoderm and the mesoderm, posteriorly (Kimura et al., 2000; Perea-Gomez et al., 2002; Yamamoto et al., 2004; Rivera-Pérez and Magnuson, 2005; Arkell and Tam, 2012). The transition from the blastocyst to the patterned epiblast is a continuous process that preludes the diversification and spatial organization of the embryo and the signalling pathways involved have been studied in detail (Tam and Loebel, 2007). However, the molecular events that initially specify a cell as anterior or posterior in the epiblast, have not been identified, as they coincide with implantation of the embryo. Embryonic Stem (ES) cells provide an alternative and complementary system to study these events.
Mouse Embryonic Stem (mES) cells are clonal populations of cultured cells derived from the blastocyst of the embryo that can give rise to all of the cell types that constitute the adult organism, i.e. they are pluripotent (Bradley et al., 1984; Smith, 2001). Several studies have revealed that pluripotent cells in vitro fluctuate between a ground state of pluripotency and one in which cells are primed for lineage commitment (Chambers et al., 2007; Hayashi et al., 2008; Toyooka et al., 2008; Kalmar et al., 2009; Canham et al., 2010; Martinez Arias and Brickman, 2011). Manipulation of different extracellular signalling pathways can shift the population as a whole towards ground state pluripotency or cause germ layer differentiation, depending on the combination of signals the cells are exposed to (Ying et al., 2008; Wray et al., 2010). Leukemia Inhibitory Factor (LIF) and Bone Morphogenetic Protein (BMP) are sufficient for self-renewal of pluripotency of an mES cell population in a chemically defined medium like N2B27 (Ying et al., 2003a). However, culturing cells in the presence of inhibitors of Extracellular-signal-Regulated Kinases (ERK) and Glycogen Synthase Kinase 3 (GSK3), hereafter referred to as “2i” conditions, shifts cells to ground state pluripotency (Ying et al., 2008). Conversely, in N2B27 basal medium alone, which contains the Retinoic Acid precursor Vitamin A and insulin, pluripotent cells adopt a neural fate, whilst activation of Wnt or Nodal signalling elicits a primitive streak like population that serves as a precursor for the endoderm and the mesoderm (Ying et al., 2003b; Gadue et al., 2006; Bakre et al., 2007; Nostro et al., 2008; Hansson et al., 2009). On the assumption that mES cells reflect an embryonic population, they can be used as a model to study cell fate assignment and maintenance as well as the early stages of mammalian development.
Here we have analyzed the controlled exit of mES cells from pluripotency under established neural differentiation conditions (N2B27) by measuring gene expression in single cells. This technique has previously been used to study mouse blastocysts (Guo et al., 2010), pluripotent mES cells (Hayashi et al., 2008; Tang et al., 2010; MacArthur et al., 2012; Trott et al., 2012) and haematopoietic stem cells (Pina et al., 2012; Moignard et al., 2013), but our study represents the first analysis of the early stages of differentiation of mES cells.
We find that initial differentiation of mES cells in N2B27 is not restricted to neural lineages, but that during the first four days of differentiation, individual cells express markers for a variety of lineages. Furthermore, our single cell analysis suggests that the expression of individual components of the core pluripotency network (Nanog, Oct4 and Sox2) restricts differentiation to specific lineages as cells exit pluripotency. Our findings lead us to suggest that during the first four days of differentiation in culture, cells undergo a transition that mimics events in vivo between the blastocyst and the postimplantation epiblast, by the end of which cells are biased towards commitment to different lineages. We also find that, during this period, ES cells display a widespread activation of canonical Wnt/β-catenin signalling which is not affiliated with specific lineages and discuss the possible implications of this observation.
Results
Pluripotency at the single cell level
Mouse ES cells self-renew under a variety of culture conditions and attempts to reveal key molecules supporting pluripotency revealed that LIF is central in the promotion of “naïve pluripotency” (Smith et al., 1988; Williams et al., 1988). Subsequently it was found that mES cells placed in synthetic medium (N2B27) in the presence of LIF and BMP4 maintained their pluripotency, which demonstrated that BMP is the component in Serum responsible for inhibition of differentiation (Ying et al., 2003a). A number of experiments have since established that heterogeneity in the expression of Rex1, Nanog, Klf4 and Stella is a hallmark of naïve pluripotent mES cells, with cells expressing high levels being pluripotent and cells expressing low levels being primed for differentiation, as reflected by their increased propensity to exit pluripotency permanently (Chambers et al., 2007; Hayashi et al., 2008; Toyooka et al., 2008; Kalmar et al., 2009). Furthermore, in Serum and LIF or LIF and BMP, cells can transit between different expression states. Culturing mES cells in 2i conditions eliminates differentiation-primed cells from the culture and leads to a robust state of pluripotency that has been termed “ground state pluripotency” and that can be propagated in these growth conditions (Ying et al., 2008; Wray et al., 2010). Accordingly, culturing cells in 2i increases colony formation and chimaera contribution rates.
To determine whether the enhanced pluripotency of mES cells cultured in 2i has a transcriptional basis at the level of single cells, expression of several key markers of pluripotency and lineage commitment was measured directly in individual cells by qPCR (Fig. 1A). In the presence of LIF and BMP4, it is possible to observe some cells expressing differentiation markers: Sox1 (neural) and T/Bra (Primitive Streak, the precursor of the mesoderm and the endoderm) (Herrmann et al., 1990; Wood and Episkopou, 1999; Smith, 2004). These cells express low levels of pluripotency markers and are likely to represent the differentiating population known to be present in these conditions. When cells are placed in 2i (in this case supplemented with LIF), the heterogeneities disappear and ∼80% of the cells express high and homogeneous levels of Oct4, Sox2 and Nanog alongside negligible levels of differentiation markers (Fig. 1B). This demonstrates that the enhanced pluripotency of cells cultured in 2i is associated with more stable expression of key pluripotency regulators at the level of single cells and consequently a lack of differentiation in such cultures.
Fig. 1. Gene expression in individual wild-type (E14Tg2A) pluripotent mES cells in N2B27 supplemented with BMP4+LIF or 2i+LIF.
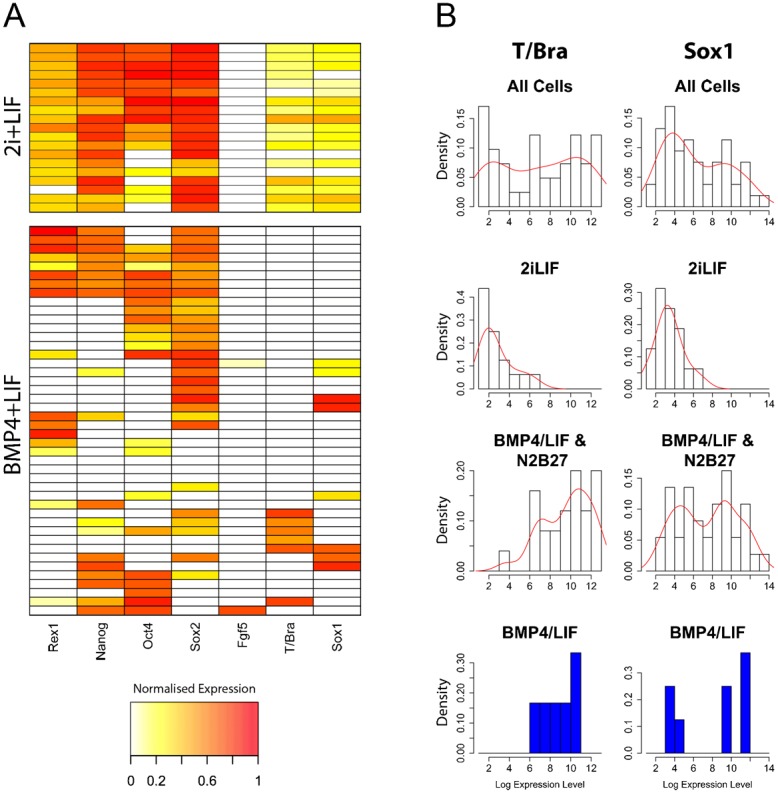
(A) Heat maps of pluripotency (Rex1, Nanog, Oct4 and Sox2) and differentiation (Fgf5, T/Bra and Sox1) marker gene expression in mES cells maintained in 2i+LIF or BMP4+LIF. Plotted values are natural logarithms of Gapdh-normalised expression levels, scaled such that cells with no expression of a particular gene have a value of 0 whilst cells that express the maximum detected level of a gene have a value of 1. (B) Histograms of primitive streak (T/Bra) and neural (Sox1) marker gene expression in different groups of mES cells. Natural logarithms of Gapdh-normalised expression levels are plotted as histograms (black outlined bars) with the corresponding probability density functions overlaid (red lines). The top row of histograms show data for all cells cultured in 2i+LIF, BMP4+LIF and after one to four days BMP4+LIF withdrawal (see Materials and Methods), whilst the lower three rows show data for subsets of the above. Only cells with detectable expression of the gene in question are included. The histograms show bimodal expression level distributions for both Sox1 and T/Bra, suggesting there are discrete groups of cells expressing high or low levels of these genes. These differentiation markers are only ever expressed at low levels by cells cultured in 2i+LIF, and are often coexpressed in individual cells (Fig. 1A). Conversely, in BMP4+LIF and after BMP4+LIF withdrawal (N2B27), these genes can be expressed at high or low levels, but tend not to be coexpressed (Fig. 1A). These observations suggest that cells expressing high levels of these genes are differentiating, whilst low-level expression of these genes is not indicative of commitment to a particular lineage. The fact that some cells in BMP4+LIF express high levels of Sox1 or T/Bra suggests these populations contain a proportion of differentiating cells.
The exit from the pluripotent state
Removal of LIF and BMP4 from an N2B27 based culture medium is often used to trigger loss of pluripotency and the differentiation of the culture towards neural lineages (Ying et al., 2003b); in these conditions, addition of Retinoic Acid (RA) is used to enhance this effect and we have observed that it accelerates differentiation, though it increases cell death (J.T. and A.M.A., unpublished observations). As mES cells begin to differentiate, they undergo two sequential transitions: firstly from a multi-layered to a monolayer epithelium – which presumably reflects the transition to an EpiSC state – and secondly, after five or six days, through an Epithelial Mesenchymal Transition (EMT), to a phenotypically heterogeneous population. While some cells form rosette like structures typical of neural cells, others have morphologies typical of mesenchymal or epithelial tissues (Fig. 2A). Prolonged culture in N2B27 appears to eliminate the latter and allows the expansion of a population with neural characteristics, such that by six days cells express high levels of Sox1 (Ying et al., 2003b; Abranches et al., 2009; Engberg et al., 2010; Stavridis et al., 2010). These observations raised the possibility that N2B27 might not provide an environment to direct differentiation strictly into neural fates during the exit from pluripotency and led us to monitor gene expression across different lineages as cells exit pluripotency in N2B27.
Fig. 2. Changes in morphology and gene expression as mES cells are differentiated over five days in N2B27 medium following BMP4+LIF withdrawal (see Materials and Methods).
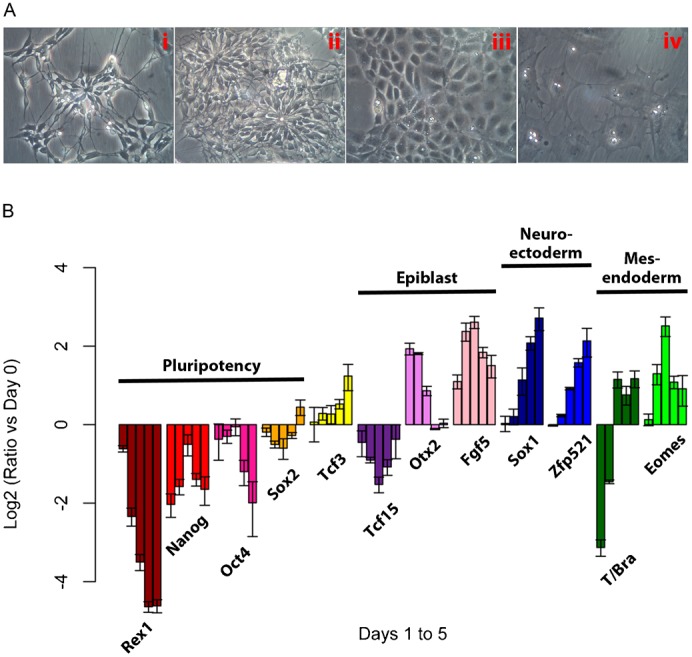
(A) Examples of cellular morphologies observed after five days in N2B27: neural rosettes (i,ii), epithelial (iii) and mesenchymal (iv). (B) Population level changes in gene expression over the course of five days of differentiation. Expression of markers of pluripotency (Rex1, Nanog, Oct4 and Sox2), epiblast (Tcf15, Otx2 and Fgf5), neuroectoderm (Sox1 and Zfp521) and mesendoderm (T/Bra and Eomes) was measured. For each gene on each of the days from day 1 (left most bar) to day 5 (right most bar), the value plotted is the log2 of the ratio on that day compared to the level in BMP4+LIF (both values normalised to the levels of the gene Ppia). Error bars above and below each bar represent the standard deviation of the mean of 3 or more qPCR replicates.
Cells that had been grown in the presence of LIF and BMP4 were placed in N2B27 and the expression of several genes, some associated with pluripotency and some with differentiation, was monitored daily over time (Fig. 2B). We observed the expected rapid decrease in Rex1, Nanog and Oct4 expression as cells exit pluripotency, as well as transient expression of the epiblast affiliated genes Fgf5 and Otx2. There is also an expected, steady increase in the expression of genes associated with neural fates (Sox1 and Zfp521) and an early decrease in the expression of Sox2 as cells exit pluripotency, followed by an increase as cells adopt neural fates. However, we also observed expression of T/Bra and Eomes, which are genes associated with Primitive Streak and endodermal fates (the sharp decline in T/Bra levels on day 1 most likely represents the failure of cells expressing this gene in BMP4+LIF conditions to re-adhere when reseeded). These profiles additionally reveal a small, but significant and reproducible, rise in the levels of Nanog and Oct4 on day 3 of differentiation, which can be observed, but is rarely remarked on, in many reports. Dynamic expression of Nanog and Oct4 probably reflects reactivation of these genes as cells pass through the epiblast state, where both are expressed (Brons et al., 2007). These results demonstrate that the early stages of differentiation in N2B27 involve expression of fates other than neural commitment.
The exit from pluripotency from the perspective of the pluripotency network
Population analyses, like the one described above, provide a useful guide to the behaviour of an ensemble of cells, but are less helpful when trying to determine how individual cells choose between different lineages during differentiation. Therefore, we used single cell qPCR to identify changes in gene expression that occur as cells exit pluripotency. Individual cells were selected at random from mES cell populations over four days of differentiation in N2B27 and processed for measuring gene expression. We were able to obtain 19 cells grown in 2i+LIF, 45 cells grown in LIF and BMP4, and 44, 42, 36 and 34 cells for each of one, two, three and four days following withdrawal of LIF and BMP4 (the cells were taken from the same populations for which bulk gene expression was presented in Fig. 2B). Expression of genes representative of pluripotency (Rex1, Nanog, Oct4 and Sox2), epiblast differentiation (Fgf5), neural commitment (Sox2 and Sox1) and primitive streak commitment (T/Bra) was measured to determine the differentiation state of individual cells over the course of the experiment (for details of the procedure, see Materials and Methods).
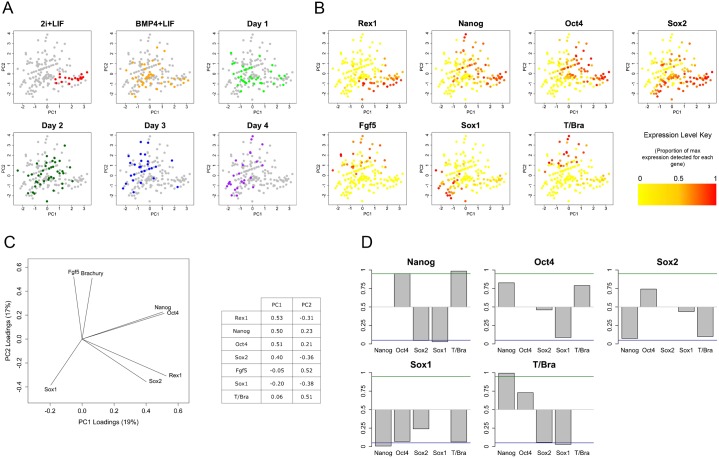
Principal Components Analysis (PCA) of the gene expression profiles of individual cells shows that in 2i conditions cells have very similar expression profiles, which gradually diverge as they differentiate (Fig. 3A). In 2i, cells cluster at the bottom right of the PCA plot alongside cells from other conditions that also express Rex1 (Fig. 3B), indicating that cells in this region of the plot are pluripotent. After four days differentiation, most cells are to be found towards the top- or bottom-left of the plot where cells that express the primitive streak marker T/Bra or the neural marker Sox1 are found. On days 1 and 2 following BMP4+LIF withdrawal, cells are dispersed between these two extremes, which suggests that, in vitro, individual cells proceed towards lineage commitment at different rates. Furthermore, coexpression of significant levels of Brachyury and Sox1 is observed very rarely suggesting multi-lineage priming does not occur frequently in this system.
Fig. 3. Relationships between the expression levels of markers of pluripotency and lineage commitment in mES cells exiting pluripotency.
The expression of markers of pluripotency (Rex1, Nanog, Oct4 and Sox2) and lineage commitment (Fgf5, Sox1 and T/Bra) was measured in mES cells maintained in 2i+LIF, BMP4+LIF or differentiated over four days in N2B27 following withdrawal of BMP4 and LIF. Natural logarithms of Gapdh-normalised expression levels were used for all calculations relating to this figure and for plotting. (A,B,C) Principal Components Analysis (PCA) of gene expression in individual mES cells. Each point in the plots in panels A and B represents a single cell, the position of which is a function of the expression of seven genes (C) in that cell. The positions of individual cells are identical in each of the plots shown. The cells analysed here, with the exception of those cultured in 2i+LIF, were taken from the same population as those used to produce the population level gene expression data in Fig. 2. Individual cells are coloured according to treatment/differentiation day (A) or their expression of a particular gene (B). In panel B the natural logarithms of the Gapdh-normalised expression levels (including cells in which expression was not detected) are scaled such that they range between 0 and 1. Colours are then mapped to these values as shown by the colour key. The principal component loading values used to calculate the positions of individual cells and the percentage of the variability in the dataset that is explained by each axis are shown in panel C. (D) Relationships between the expression of pluripotency and differentiation genes after three to four days differentiation in N2B27. Cells were segregated (in silico) according to whether or not they express a first gene (plot titles). The mean level of a second gene (horizontal labels below each bar) in cells that express the first gene was then calculated. Bootstrapping was used to determine the probability that the mean level of the second gene was higher in cells that express the first gene than would be expected by chance (P value). These P values were plotted as grey bars. Bootstrapping involves shuffling the levels of the second gene across the population and repeating the calculation described above 100,000 times; the P value is the proportion of the bootstrap means that are lower than the real mean. A high P value indicates that the levels of two genes are positively correlated, whilst a low P values indicates they are negatively correlated. The green and blue lines at P values of 0.95 and 0.05 denote statistically significant relationships.
Patterns of pluripotency gene expression across the plot suggest that individual pluripotency genes influence the direction of lineage commitment (Fig. 3B,C). Pluripotent cells at the bottom right of the plot coexpress Nanog, Oct4 and Sox2 homogeneously. However, differentiating cells express different combinations of these genes, suggesting that the sequence in which the expression of pluripotency genes is downregulated differs between differentiating cells. Interestingly, cells that retain high levels of Sox2 expression often coexpress Sox1, but very rarely express T/Bra, whilst the opposite is true of cells that retain high levels of Nanog and/or Oct4 expression. These observations suggest that retention of Sox2 expression predisposes cells towards neural commitment, whilst retention of Nanog or Oct4 expression biases cells towards primitive streak commitment.
To test these relationships more formally we looked at probabilities of coexpression after four days differentiation (Fig. 3D). This analysis confirmed the strong anti-correlation between expression of Nanog and T/Bra on the one hand, and Sox2 and Sox1 on the other, in differentiating cells. The relationships between expression of Oct4 and the other genes were similar to those of Nanog, but generally much weaker. These findings confirm earlier suggestions that different pluripotency genes specifically inhibit commitment to individual lineages, and additionally demonstrate how these interactions are relieved over time in order to guide cells towards different lineages.
β-catenin biases lineage commitment through the regulation of Nanog
Nanog acts as a gatekeeper for pluripotency but, as we have shown here and others have observed previously, it is also associated with the emergence of a primitive streak fate (Hart et al., 2004; Chambers et al., 2007; Osorno et al., 2012). Similarly, β-catenin stimulation has been shown to promote both pluripotency and primitive streak commitment (Liu et al., 1999; Huelsken et al., 2000; Wray et al., 2011; Yi et al., 2011). Since β-catenin has been shown to enhance Nanog expression (Pereira et al., 2006; Yi et al., 2008), we tested whether β-catenin regulates pluripotency and lineage commitment via upregulation of Nanog expression by assessing the effects of activating Wnt/β-catenin signalling in the absence of Nanog (Fig. 4). Nanog mutant mES cells exhibit an unstable pluripotency as shown by their high rates of differentiation. However, Chiron, which activates β-catenin signalling, is able to block upregulation of Fgf5 expression in the absence of Nanog, demonstrating that Chiron maintains pluripotency independently of Nanog.
Fig. 4. The effects of β-catenin stimulation on pluripotency and lineage marker gene expression in wild-type (E14Tg2A) and Nanog−/− (44Cre6) cells.
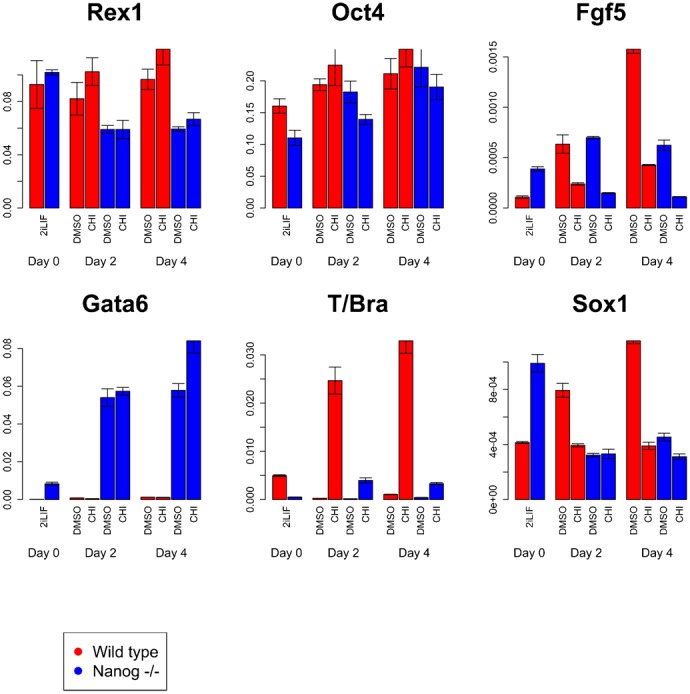
Wild-type (red bars) and Nanog−/− (blue bars) mES cells maintained in 2i+LIF on gelatin-coated tissue culture plastic were transferred to serum+LIF to allow a proportion of cells to exit pluripotency (a percentage of mES cells differentiate spontaneously in serum+LIF), in the presence or absence of the β-catenin agonist Chiron. The effects of Chiron stimulation of β-catenin on expression of markers of pluripotency (Rex1, Nanog and Oct4), epiblast (Fgf5), neuroectoderm (Sox1 and Zfp521), mesendoderm (T/Bra) and primitive endoderm (Gata6) were then assessed by qPCR after two and four days. Plotted are the levels of individual genes in each sample relative to the levels of the reference gene Gapdh. Error bars above and below each bar represent the standard deviation of the mean of at least 3 qPCR replicates. DMSO is used a control in this experiment as Chiron is dissolved in DMSO.
We notice that Nanog mutant cells have a strong tendency to upregulate expression of the endoderm marker Gata6, which most likely reflects increased primitive endoderm commitment, consistent with the established role of Nanog in blocking primitive endoderm commitment (Mitsui et al., 2003). Nonetheless, Nanog mutant cells are able to commit to both neural and primitive streak fates (Chambers et al., 2007). However, we observe that in the absence of Nanog, β-catenin is unable to block Sox1 expression or stimulate T/Bra expression significantly. These observations indicate that the ability of β-catenin to predispose cells towards primitive streak commitment is dependent on Nanog.
Wnt signalling as a signature for the exit from pluripotency
When LIF is substituted for Retinoic Acid (RA) in Serum, mES cells undergo preferential neural differentiation and we have observed that they activate a reporter of Wnt signalling (Faunes et al., 2013). This is surprising as Wnt signalling inhibits the neural fate (Aubert et al., 2002; Haegele et al., 2003; Engberg et al., 2010). Analysis of the reporter at the single cell level reveals that not every cell activates Wnt signalling, and therefore raises the possibility that only those cells that are negative for Wnt signalling will develop into neural precursors. If this were the case, there should be a negative correlation between reporter expression and neural differentiation. To test this, we made use of the fact that, in a population of cells self-renewing in Serum and LIF, some cells exhibit low levels of Wnt signalling. Using a fluorescent Wnt reporter, we selected cells in the top 10% of the distribution for analysis of gene expression (Faunes et al., 2013). These cells amount to ∼1% of the population (Fig. 5A) and Axin2 is only detectable in these cells, in agreement with their high levels of Wnt reporter activity.
Fig. 5. Wnt signalling marks the exit from pluripotency.
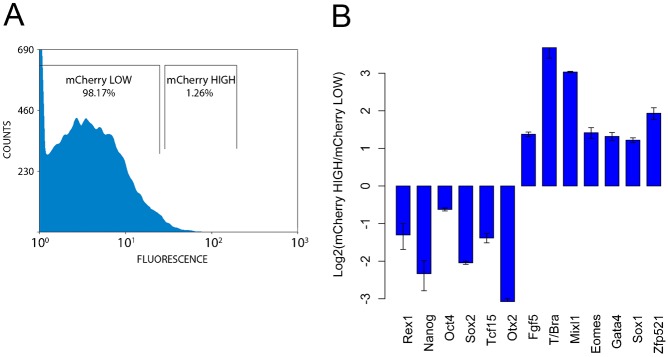
(A) Fluorescence profile of mES cells cultured in serum+LIF that express an mCherry reporter of transcriptionally active β-catenin (Ferrer-Vaquer et al., 2010). The proportion of cells in each of the gates is shown. (B) Gene expression in cells sorted for expression of mCherry using the gates shown in panel A. The two populations were assessed for their expression of markers of pluripotency (Rex1, Nanog, Oct4 and Sox2), epiblast differentiation (Fgf5), primitive streak and its derivatives (T/Bra, Mixl1, Eomes and Gata4) and neural differentiation (Sox1 and Zfp521) using qPCR. Each bar shows the log2 value of the ratio of gene expression in mCherry high cells versus mCherry low cells. All values were first normalised to Ppia expression. Error bars above and below each bar represent the standard deviation of the mean of 3 or more qPCR replicates. Axin2, a direct transcriptional target of β-catenin signalling, was also measured in both populations and, as expected, was only expressed in the mCherry high population. Increased levels of β-catenin-mediated transcription are associated with loss of pluripotency gene expression and upregulation of markers of multiple opposing lineages.
Cells that express the Wnt reporter exhibit hallmarks of differentiation (Fig. 5B): Fgf5 expression and low levels of the pluripotency genes Rex1, Nanog, Oct4, Sox2, Tcf15 and Otx2 relative to the rest of the population. These cells also express lineage associated genes, but without a bias for particular fates. Thus we observe expression of markers of primitive streak and its derivatives (T/Bra, Mixl1, Eomes and Gata4), as expected, and also genes associated with neural development (Sox1 and Zfp521). These results suggest that, as cells exit pluripotency in culture, they activate Wnt signalling and that this activation is not associated with specific lineages, but rather appears to be a feature of the early stages of differentiation. It is known that during self-renewal there is little expression of Wnt genes and that as cells differentiate they express many members of the Wnt family and increase expression of those expressed during self-renewal (Nordin et al., 2008). The increase in reporter expression is likely to be associated with this. The generalized expression of differentiation genes in this population is, at this moment, more difficult to explain.
Discussion
Here we have analysed gene expression in individual mES cells during self-renewal and the early stages of differentiation. Our results confirm suggestions from population studies that naïve pluripotency (mES cells grown in N2B27 supplemented with BMP4 and LIF) is associated with variable but generally high levels of expression of pluripotency genes (Nanog, Oct4, Sox2 and Rex1), as well as differentiation markers in a proportion of cells. In ground state pluripotency (2i conditions), these genes are expressed more homogeneously and the expression of differentiation markers is reduced, though we observe a small degree of heterogeneity, which is in agreement with previous observations (Canham et al., 2010) (C. L. Lim, Investigating the dynamics of Nanog heterogeneity in mouse embryonic stem cells, PhD thesis, University of Cambridge, 2011).
When cells exit pluripotency, the expression of the pluripotency markers, with the exception of Rex1, does not disappear completely but individual pluripotency genes become restricted to subsets of cells alongside genes affiliated with different lineages: Oct4 and Nanog become associated with the primitive streak gene T/Bra, whilst Sox2 becomes associated with Sox1, which marks the neural fate (Wood and Episkopou, 1999; Rivera-Pérez and Magnuson, 2005). Others have reported that individual pluripotency factors are associated with differentiation towards specific lineages (Avilion et al., 2003; Graham et al., 2003; Hart et al., 2004; Osorno et al., 2012). Furthermore, it was recently demonstrated that Oct4 and Sox2 specifically inhibit neural and primitive streak differentiation, respectively (Thomson et al., 2011). Our experiments extend these analyses to reveal how dynamic expression of the pluripotency genes during differentiation is used to guide individual cells towards particular lineages. In support of our model, we find that very few cells maintain high-level coexpression of Nanog, Oct4 and Sox2 during the differentiation process. In this context our experiments reveal that the ability of Wnt/β-catenin signalling to promote Primitive Streak differentiation is dependent on Nanog and that this interaction is independent of the roles of Nanog and β-catenin in the maintenance of pluripotency (Silva et al., 2009; Wray et al., 2011; Yi et al., 2011).
In the course of our experiments we were surprised to observe cells primed for endoderm and mesoderm in N2B27, which is commonly used as a base medium to generate neural tissue (Ying et al., 2003b; Engberg et al., 2010; Stavridis et al., 2010). Furthermore, after five days in culture most of the cells can be coaxed towards neural fates if treated with high levels of RA, whilst T/Bra protein is rarely detected under these differentiation conditions (A.M.A., unpublished observations). This suggests that the initial phases of differentiation involve a general priming of multiple lineages at the level of single cells, preceding a process of selection that, in N2B27, results in most cells eventually expressing Sox1 and undergoing neural commitment and differentiation (Abranches et al., 2009). We also notice a very low proportion of single cells coexpressing markers of different lineages, although the number of cells analyzed in our experiment is not very large. This would suggest that multilineage priming, in the manner that has been shown to occur in hematopoietic precursors (Cross and Enver, 1997; Hu et al., 1997) is a minor event during loss of pluripotency. Instead it appears that even the earliest stages of differentiation of mES cells are mediated by changes in gene expression that are specific to individual lineages.
Our experiments confirm previous observations that differentiation is associated with an increase in Wnt/β-catenin signalling (Faunes et al., 2013). Surprisingly, we observe that this activity is not associated with a particular fate and that cells with high levels of Wnt/β-catenin signalling express genes associated with neural as well as primitive streak fates. It may be that this is part of the ‘priming’ process and that in the same manner that cells are primed for particular fates by lineage specific transcription factors, they are also primed for signalling, i.e. signalling is activated indiscriminately and only later becomes restricted to the cells dependent on it, i.e. those fated to the become the Primitive Streak. Further studies should shed light on the meaning of this observation. We have shown that single cell analysis of the early stages of differentiation of mES cells in culture can reveal features of this process that are masked by population averaging. It will be important to extend these studies to include more genes and to carry out a comparative study of single cells from embryos at the equivalent stages.
Materials and Methods
Cell culture and differentiation
E14Tg2A wild-type (129/Ola), Nanog−/− (44Cre6) (Chambers et al., 2007) and Wnt reporter (Faunes et al., 2013) mES cells were routinely cultured on gelatin-coated tissue culture plastic in serum-containing medium (Glasgow Minimal Essential Medium (Sigma, G5154), Foetal Bovine Serum (10%), non-essential amino acids 0.1 mM (Life Technologies, 11140-050), GlutaMAX 2 mM (Life Technologies, 35050-038), sodium pyruvate 1 mM (Sigma, S8636), 2-mercaptoethanol 100 nM (Life Technologies, 31350-010)) supplemented with 100 U/mL LIF (Leukaemia Inhibitory Factor – Recombinant human, Department of Biochemistry, University of Cambridge). Prior to initiating experiments, cells were transferred to NDiff N2B27 (StemCells Inc, SCS-SF-NB-02) supplemented with 100 U/mL LIF and 10 ng/mL BMP4 (Department of Biochemistry, University of Cambridge) or 3 µM Chiron (CHIR99021, Division of signal transduction, University of Dundee) and 2 µM PD03 (PD0325901, Division of signal transduction, University of Dundee) and cultured for two passages. Cells were propagated by trypsinisation every two days with a split ratio of approximately 1 in 5. For differentiations, cells were trypsinised and reseeded in N2B27 alone at a density of 10,000 cells/cm2.
Quantitative Polymerase Chain Reaction (qPCR)
RNA was isolated from ∼5×105 trypsinised and pelleted mES cells using the RNeasy Mini kit (Qiagen, 74104) according to the manufacturer's instructions, and resuspended in 30 µL distilled water. RNA was reverse transcribed as follows. RNA samples (1 µg in 38 µL nuclease free water) were combined with 2 µL Oligo-dT anchored primers (Life Technologies, 12577-011) and incubated at 80°C for 2 minutes before transferring immediately to ice for 2 minutes. PCR master mix was then added to each sample: 1.5 µl dNTPs (Life Technologies, 18427-013), 12 µl 5× First Strand buffer (Life Technologies, 18080-400), 3 µl 0.1 M DTT (Life Technologies, D-1532), 1.5 µl RNaseOUT (Life Technologies, 10777-019) and 2 µl Superscript III Reverse Transcriptase (Life Technologies, 18080-400). Thermal cycling was carried out as follows: 25°C 10 minutes, 50°C 30 minutes, 70°C 15 minutes. To remove RNA from the resultant cDNA samples 1 µL RNaseH (Life Technologies, 18021071) was added and samples were incubated at 37°C for 20 minutes and then at 80°C for 20 minutes. Samples were diluted 1 in 10 before being analysed for gene expression using a Rotor-gene Q instrument. Reactions were set up using a Qiagility liquid-handling robot (12.5 µL 2× QuantiFast SYBR Green Master Mix (Qiagen, 204052), 0.5 µL primer mix (50 µM each), 9 µL PCR grade water and 3 µL diluted cDNA) and analysed using the 2-step protocol described in the Quantifast manual (95°C for 5 minutes, then 40 cycles of 95°C for 10 s + 60°C for 30 s, followed by melt curve analysis). Fluorescence curves were quantified using the MAK3 algorithm found in the R package qpcR. Primer sequences are listed in supplementary material Table S1.
Single cell qPCR
FACS was used to distribute individual mES cells into the wells of a 96-well plate containing lysis buffer (see below). The transcriptomes of isolated cells were then amplified according to a previously published protocol (up to step 30) (Tang et al., 2010). The resultant cDNA samples were purified using a PCR purification kit (Qiagen, 28104) and diluted 1 in 10 in PCR-grade water. Gene expression was measured as described above.
Flow cytometry/FACS
Cells were trypsinised and dissociated, spun down in a microcentrifuge, resuspended in serum-containing medium, then analysed and/or sorted on a Beckman–Coulter Cytomation MoFlo High Performance Cell Sorter.
Supplementary Material
Acknowledgments
J.T. was funded by the Wellcome Trust PhD programme. A.M.A. was funded by an ERC Senior Investigator Grant. The Nanog−/− (44Cre6) cells described were a gift from Prof. Austin Smith. We want to thank members of the Martinez Arias group for discussions throughout the course of this work.
Footnotes
Author contributions: J.T. and A.M.A. conceived and designed experiments and wrote the manuscript. J.T. performed experiments and analysed data.
Competing interests: The authors have no competing interests to declare.
References
- Abranches E., Silva M., Pradier L., Schulz H., Hummel O., Henrique D., Bekman E. (2009). Neural differentiation of embryonic stem cells in vitro: a road map to neurogenesis in the embryo. PLoS ONE 4, e6286 10.1371/journal.pone.0006286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arkell R. M., Tam P. P. (2012). Initiating head development in mouse embryos: integrating signalling and transcriptional activity. Open Biol 2, 120030 10.1098/rsob.120030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aubert J., Dunstan H., Chambers I., Smith A. (2002). Functional gene screening in embryonic stem cells implicates Wnt antagonism in neural differentiation. Nat. Biotechnol. 20, 1240–1245 10.1038/nbt763 [DOI] [PubMed] [Google Scholar]
- Avilion A. A., Nicolis S. K., Pevny L. H., Perez L., Vivian N., Lovell-Badge R. (2003). Multipotent cell lineages in early mouse development depend on SOX2 function. Genes Dev. 17, 126–140 10.1101/gad.224503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakre M. M., Hoi A., Mong J. C., Koh Y. Y., Wong K. Y., Stanton L. W. (2007). Generation of multipotential mesendodermal progenitors from mouse embryonic stem cells via sustained Wnt pathway activation. J. Biol. Chem. 282, 31703–31712 10.1074/jbc.M704287200 [DOI] [PubMed] [Google Scholar]
- Bradley A., Evans M., Kaufman M. H., Robertson E. (1984). Formation of germ-line chimaeras from embryo-derived teratocarcinoma cell lines. Nature 309, 255–256 10.1038/309255a0 [DOI] [PubMed] [Google Scholar]
- Brons I. G., Smithers L. E., Trotter M. W., Rugg-Gunn P., Sun B., Chuva de Sousa Lopes S. M., Howlett S. K., Clarkson A., Ahrlund-Richter L., Pedersen R. A. et al. (2007). Derivation of pluripotent epiblast stem cells from mammalian embryos. Nature 448, 191–195 10.1038/nature05950 [DOI] [PubMed] [Google Scholar]
- Canham M. A., Sharov A. A., Ko M. S., Brickman J. M. (2010). Functional heterogeneity of embryonic stem cells revealed through translational amplification of an early endodermal transcript. PLoS Biol. 8, e1000379 10.1371/journal.pbio.1000379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers I., Silva J., Colby D., Nichols J., Nijmeijer B., Robertson M., Vrana J., Jones K., Grotewold L., Smith A. (2007). Nanog safeguards pluripotency and mediates germline development. Nature 450, 1230–1234 10.1038/nature06403 [DOI] [PubMed] [Google Scholar]
- Cross M. A., Enver T. (1997). The lineage commitment of haemopoietic progenitor cells. Curr. Opin. Genet. Dev. 7, 609–613 10.1016/S0959-437X(97)80007-X [DOI] [PubMed] [Google Scholar]
- Engberg N., Kahn M., Petersen D. R., Hansson M., Serup P. (2010). Retinoic acid synthesis promotes development of neural progenitors from mouse embryonic stem cells by suppressing endogenous, Wnt-dependent nodal signaling. Stem Cells 28, 1498–1509 10.1002/stem.479 [DOI] [PubMed] [Google Scholar]
- Faunes F., Hayward P., Descalzo S. M., Chatterjee S. S., Balayo T., Trott J., Christoforou A., Ferrer-Vaquer A., Hadjantonakis A. K., Dasgupta R. et al. (2013). A membrane-associated β-catenin/Oct4 complex correlates with ground-state pluripotency in mouse embryonic stem cells. Development 140, 1171–1183 10.1242/dev.085654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrer-Vaquer A., Piliszek A., Tian G., Aho R. J., Dufort D., Hadjantonakis A. K. (2010). A sensitive and bright single-cell resolution live imaging reporter of Wnt/ß-catenin signaling in the mouse. BMC Dev. Biol. 10, 121 10.1186/1471-213X-10-121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadue P., Huber T. L., Paddison P. J., Keller G. M. (2006). Wnt and TGF-beta signaling are required for the induction of an in vitro model of primitive streak formation using embryonic stem cells. Proc. Natl. Acad. Sci. USA 103, 16806–16811 10.1073/pnas.0603916103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham V., Khudyakov J., Ellis P., Pevny L. (2003). SOX2 functions to maintain neural progenitor identity. Neuron 39, 749–765 10.1016/S0896-6273(03)00497-5 [DOI] [PubMed] [Google Scholar]
- Guo G., Huss M., Tong G. Q., Wang C., Li Sun L., Clarke N. D., Robson P. (2010). Resolution of cell fate decisions revealed by single-cell gene expression analysis from zygote to blastocyst. Dev. Cell 18, 675–685 10.1016/j.devcel.2010.02.012 [DOI] [PubMed] [Google Scholar]
- Haegele L., Ingold B., Naumann H., Tabatabai G., Ledermann B., Brandner S. (2003). Wnt signalling inhibits neural differentiation of embryonic stem cells by controlling bone morphogenetic protein expression. Mol. Cell. Neurosci. 24, 696–708 10.1016/S1044-7431(03)00232-X [DOI] [PubMed] [Google Scholar]
- Hansson M., Olesen D. R., Peterslund J. M., Engberg N., Kahn M., Winzi M., Klein T., Maddox-Hyttel P., Serup P. (2009). A late requirement for Wnt and FGF signaling during activin-induced formation of foregut endoderm from mouse embryonic stem cells. Dev. Biol. 330, 286–304 10.1016/j.ydbio.2009.03.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart A. H., Hartley L., Ibrahim M., Robb L. (2004). Identification, cloning and expression analysis of the pluripotency promoting Nanog genes in mouse and human. Dev. Dyn. 230, 187–198 10.1002/dvdy.20034 [DOI] [PubMed] [Google Scholar]
- Hayashi K., Lopes S. M., Tang F., Surani M. A. (2008). Dynamic equilibrium and heterogeneity of mouse pluripotent stem cells with distinct functional and epigenetic states. Cell Stem Cell 3, 391–401 10.1016/j.stem.2008.07.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmann B. G., Labeit S., Poustka A., King T. R., Lehrach H. (1990). Cloning of the T gene required in mesoderm formation in the mouse. Nature 343, 617–622 10.1038/343617a0 [DOI] [PubMed] [Google Scholar]
- Hu M., Krause D., Greaves M., Sharkis S., Dexter M., Heyworth C., Enver T. (1997). Multilineage gene expression precedes commitment in the hemopoietic system. Genes Dev. 11, 774–785 10.1101/gad.11.6.774 [DOI] [PubMed] [Google Scholar]
- Huelsken J., Vogel R., Brinkmann V., Erdmann B., Birchmeier C., Birchmeier W. (2000). Requirement for beta-catenin in anterior-posterior axis formation in mice. J. Cell Biol. 148, 567–578 10.1083/jcb.148.3.567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalmar T., Lim C., Hayward P., Muñoz-Descalzo S., Nichols J., Garcia-Ojalvo J., Martinez Arias A. (2009). Regulated fluctuations in nanog expression mediate cell fate decisions in embryonic stem cells. PLoS Biol. 7, e1000149 10.1371/journal.pbio.1000149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura C., Yoshinaga K., Tian E., Suzuki M., Aizawa S., Matsuo I. (2000). Visceral endoderm mediates forebrain development by suppressing posteriorizing signals. Dev. Biol. 225, 304–321 10.1006/dbio.2000.9835 [DOI] [PubMed] [Google Scholar]
- Liu P., Wakamiya M., Shea M. J., Albrecht U., Behringer R. R., Bradley A. (1999). Requirement for Wnt3 in vertebrate axis formation. Nat. Genet. 22, 361–365 10.1038/11932 [DOI] [PubMed] [Google Scholar]
- MacArthur B. D., Sevilla A., Lenz M., Müller F. J., Schuldt B. M., Schuppert A. A., Ridden S. J., Stumpf P. S., Fidalgo M., Ma'ayan A. et al. (2012). Nanog-dependent feedback loops regulate murine embryonic stem cell heterogeneity. Nat. Cell Biol. 14, 1139–1147 10.1038/ncb2603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez Arias A., Brickman J. M. (2011). Gene expression heterogeneities in embryonic stem cell populations: origin and function. Curr. Opin. Cell Biol. 23, 650–656 10.1016/j.ceb.2011.09.007 [DOI] [PubMed] [Google Scholar]
- Mitsui K., Tokuzawa Y., Itoh H., Segawa K., Murakami M., Takahashi K., Maruyama M., Maeda M., Yamanaka S. (2003). The homeoprotein Nanog is required for maintenance of pluripotency in mouse epiblast and ES cells. Cell 113, 631–642 10.1016/S0092-8674(03)00393-3 [DOI] [PubMed] [Google Scholar]
- Moignard V., Macaulay I. C., Swiers G., Buettner F., Schütte J., Calero-Nieto F. J., Kinston S., Joshi A., Hannah R., Theis F. J. et al. (2013). Characterization of transcriptional networks in blood stem and progenitor cells using high-throughput single-cell gene expression analysis. Nat. Cell Biol. 15, 363–372 10.1038/ncb2709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordin N., Li M., Mason J. O. (2008). Expression profiles of Wnt genes during neural differentiation of mouse embryonic stem cells. Cloning Stem Cells 10, 37–48 10.1089/clo.2007.0060 [DOI] [PubMed] [Google Scholar]
- Nostro M. C., Cheng X., Keller G. M., Gadue P. (2008). Wnt, activin, and BMP signaling regulate distinct stages in the developmental pathway from embryonic stem cells to blood. Cell Stem Cell 2, 60–71 10.1016/j.stem.2007.10.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osorno R., Tsakiridis A., Wong F., Cambray N., Economou C., Wilkie R., Blin G., Scotting P. J., Chambers I., Wilson V. (2012). The developmental dismantling of pluripotency is reversed by ectopic Oct4 expression. Development 139, 2288–2298 10.1242/dev.078071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perea-Gomez A., Vella F. D., Shawlot W., Oulad-Abdelghani M., Chazaud C., Meno C., Pfister V., Chen L., Robertson E., Hamada H. et al. (2002). Nodal antagonists in the anterior visceral endoderm prevent the formation of multiple primitive streaks. Dev. Cell 3, 745–756 10.1016/S1534-5807(02)00321-0 [DOI] [PubMed] [Google Scholar]
- Pereira L., Yi F., Merrill B. J. (2006). Repression of Nanog gene transcription by Tcf3 limits embryonic stem cell self-renewal. Mol. Cell. Biol. 26, 7479–7491 10.1128/MCB.00368-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pina C., Fugazza C., Tipping A. J., Brown J., Soneji S., Teles J., Peterson C., Enver T. (2012). Inferring rules of lineage commitment in haematopoiesis. Nat. Cell Biol. 14, 287–294 10.1038/ncb2442 [DOI] [PubMed] [Google Scholar]
- Rivera-Pérez J. A., Magnuson T. (2005). Primitive streak formation in mice is preceded by localized activation of Brachyury and Wnt3. Dev. Biol. 288, 363–371 10.1016/j.ydbio.2005.09.012 [DOI] [PubMed] [Google Scholar]
- Rossant J., Tam P. P. (2009). Blastocyst lineage formation, early embryonic asymmetries and axis patterning in the mouse. Development 136, 701–713 10.1242/dev.017178 [DOI] [PubMed] [Google Scholar]
- Silva J., Nichols J., Theunissen T. W., Guo G., van Oosten A. L., Barrandon O., Wray J., Yamanaka S., Chambers I., Smith A. (2009). Nanog is the gateway to the pluripotent ground state. Cell 138, 722–737 10.1016/j.cell.2009.07.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith A. G. (2001). Embryo-derived stem cells: of mice and men. Annu. Rev. Cell Dev. Biol. 17, 435–462 10.1146/annurev.cellbio.17.1.435 [DOI] [PubMed] [Google Scholar]
- Smith J. C. (2004). Role of T-box genes during gastrulation. Gastrulation: From Cells To Embryo Stern C D, ed731.Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press. [Google Scholar]
- Smith A. G., Heath J. K., Donaldson D. D., Wong G. G., Moreau J., Stahl M., Rogers D. (1988). Inhibition of pluripotential embryonic stem cell differentiation by purified polypeptides. Nature 336, 688–690 10.1038/336688a0 [DOI] [PubMed] [Google Scholar]
- Stavridis M. P., Collins B. J., Storey K. G. (2010). Retinoic acid orchestrates fibroblast growth factor signalling to drive embryonic stem cell differentiation. Development 137, 881–890 10.1242/dev.043117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takaoka K., Yamamoto M., Shiratori H., Meno C., Rossant J., Saijoh Y., Hamada H. (2006). The mouse embryo autonomously acquires anterior-posterior polarity at implantation. Dev. Cell 10, 451–459 10.1016/j.devcel.2006.02.017 [DOI] [PubMed] [Google Scholar]
- Tam P. P., Loebel D. A. (2007). Gene function in mouse embryogenesis: get set for gastrulation. Nat. Rev. Genet. 8, 368–381 10.1038/nrg2084 [DOI] [PubMed] [Google Scholar]
- Tang F., Barbacioru C., Nordman E., Li B., Xu N., Bashkirov V. I., Lao K., Surani M. A. (2010). RNA-Seq analysis to capture the transcriptome landscape of a single cell. Nat. Protoc. 5, 516–535 10.1038/nprot.2009.236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson M., Liu S. J., Zou L. N., Smith Z., Meissner A., Ramanathan S. (2011). Pluripotency factors in embryonic stem cells regulate differentiation into germ layers. Cell 145, 875–889 10.1016/j.cell.2011.05.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyooka Y., Shimosato D., Murakami K., Takahashi K., Niwa H. (2008). Identification and characterization of subpopulations in undifferentiated ES cell culture. Development 135, 909–918 10.1242/dev.017400 [DOI] [PubMed] [Google Scholar]
- Trott J., Hayashi K., Surani A., Babu M. M., Martinez-Arias A. (2012). Dissecting ensemble networks in ES cell populations reveals micro-heterogeneity underlying pluripotency. Mol. Biosyst. 8, 744–752 10.1039/c1mb05398a [DOI] [PubMed] [Google Scholar]
- Williams R. L., Hilton D. J., Pease S., Willson T. A., Stewart C. L., Gearing D. P., Wagner E. F., Metcalf D., Nicola N. A., Gough N. M. (1988). Myeloid leukaemia inhibitory factor maintains the developmental potential of embryonic stem cells. Nature 336, 684–687 10.1038/336684a0 [DOI] [PubMed] [Google Scholar]
- Wood H. B., Episkopou V. (1999). Comparative expression of the mouse Sox1, Sox2 and Sox3 genes from pre-gastrulation to early somite stages. Mech. Dev. 86, 197–201 10.1016/S0925-4773(99)00116-1 [DOI] [PubMed] [Google Scholar]
- Wray J., Kalkan T., Smith A. G. (2010). The ground state of pluripotency. Biochem. Soc. Trans. 38, 1027–1032 10.1042/BST0381027 [DOI] [PubMed] [Google Scholar]
- Wray J., Kalkan T., Gomez-Lopez S., Eckardt D., Cook A., Kemler R., Smith A. (2011). Inhibition of glycogen synthase kinase-3 alleviates Tcf3 repression of the pluripotency network and increases embryonic stem cell resistance to differentiation. Nat. Cell Biol. 13, 838–845 10.1038/ncb2267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto M., Saijoh Y., Perea-Gomez A., Shawlot W., Behringer R. R., Ang S. L., Hamada H., Meno C. (2004). Nodal antagonists regulate formation of the anteroposterior axis of the mouse embryo. Nature 428, 387–392 10.1038/nature02418 [DOI] [PubMed] [Google Scholar]
- Yi F., Pereira L., Merrill B. J. (2008). Tcf3 functions as a steady-state limiter of transcriptional programs of mouse embryonic stem cell self-renewal. Stem Cells 26, 1951–1960 10.1634/stemcells.2008-0229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi F., Pereira L., Hoffman J. A., Shy B. R., Yuen C. M., Liu D. R., Merrill B. J. (2011). Opposing effects of Tcf3 and Tcf1 control Wnt stimulation of embryonic stem cell self-renewal. Nat. Cell Biol. 13, 762–770 10.1038/ncb2283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying Q. L., Nichols J., Chambers I., Smith A. (2003a). BMP induction of Id proteins suppresses differentiation and sustains embryonic stem cell self-renewal in collaboration with STAT3. Cell 115, 281–292 10.1016/S0092-8674(03)00847-X [DOI] [PubMed] [Google Scholar]
- Ying Q. L., Stavridis M., Griffiths D., Li M., Smith A. (2003b). Conversion of embryonic stem cells into neuroectodermal precursors in adherent monoculture. Nat. Biotechnol. 21, 183–186 10.1038/nbt780 [DOI] [PubMed] [Google Scholar]
- Ying Q. L., Wray J., Nichols J., Batlle-Morera L., Doble B., Woodgett J., Cohen P., Smith A. (2008). The ground state of embryonic stem cell self-renewal. Nature 453, 519–523 10.1038/nature06968 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.