Multiple formins cooperate during cytokinesis, but their functions in de novo actin assembly at the division site play the primary role in contractile ring assembly.
Abstract
Both de novo–assembled actin filaments at the division site and existing filaments recruited by directional cortical transport contribute to contractile ring formation during cytokinesis. However, it is unknown which source is more important. Here, we show that fission yeast formin For3 is responsible for node condensation into clumps in the absence of formin Cdc12. For3 localization at the division site depended on the F-BAR protein Cdc15, and for3 deletion was synthetic lethal with mutations that cause defects in contractile ring formation. For3 became essential in cells expressing N-terminal truncations of Cdc12, which were more active in actin assembly but depended on actin filaments for localization to the division site. In tetrad fluorescence microscopy, double mutants of for3 deletion and cdc12 truncations were severely defective in contractile ring assembly and constriction, although cortical transport of actin filaments was normal. Together, these data indicate that different formins cooperate in cytokinesis and that de novo actin assembly at the division site is predominant for contractile ring formation.
Introduction
Cytokinesis is the final step of the cell division cycle that partitions cellular components into two daughter cells. The contractile ring containing actin filaments, myosin II, and many other proteins is required for cytokinesis in fungi and animal cells (Balasubramanian et al., 2004; Barr and Gruneberg, 2007; Pollard and Wu, 2010). Both de novo actin assembly at the division site and cortical transport/flow contribute actin filaments to the contractile ring (White and Borisy, 1983; Bray and White, 1988; Cao and Wang, 1990; Lee et al., 1998; Pelham and Chang, 2002; Chen et al., 2008; Zhou and Wang, 2008; Alsop et al., 2009; Huang et al., 2012; Subramanian et al., 2013). However, the relative importance of these sources of actin filaments for the contractile ring is unknown in any cell type. Mechanisms of actin accumulation at the division site hold the key to understanding different models for cleavage site selection and contractile ring assembly. However, determining the contributions from de novo assembly and cortical transport has been difficult because of the overlap between the two mechanisms (Zhou and Wang, 2008; Huang et al., 2012).
The fission yeast Schizosaccharomyces pombe is an excellent model organism for investigating molecular mechanisms of cytokinesis (Roberts-Galbraith and Gould, 2008; Laporte et al., 2010). The anillin-like protein Mid1 is essential for division site specification (Chang et al., 1996; Sohrmann et al., 1996; Bähler et al., 1998a; Paoletti and Chang, 2000; Celton-Morizur et al., 2004; Almonacid et al., 2009, 2011). Mid1 acts as a scaffold and positional cue to assemble IQGAP Rng2, myosin-II Myo2 and its light chains Cdc4 and Rlc1, F-BAR protein Cdc15, and the formin Cdc12 into cytokinesis nodes and then the contractile ring (Coffman et al., 2009; Almonacid et al., 2011; Laporte et al., 2011; Padmanabhan et al., 2011; Lee and Wu, 2012). Search, capture, pull, and release (SCPR) is a stochastic ring assembly model whereby actin filaments nucleated in random directions by Cdc12 are captured by myosin-II motors in neighboring nodes to pull them together into the contractile ring (Vavylonis et al., 2008; Lee et al., 2012). The SCPR model describes ring assembly by de novo actin nucleation without considering cortical transport of actin filaments. The ring remains at a constant diameter as it matures by addition of more proteins during anaphase B (Wu et al., 2003). After anaphase, the contractile ring begins to disassemble as it constricts.
Formins are a family of conserved proteins that nucleate and elongate linear actin filaments (Castrillon and Wasserman, 1994; Evangelista et al., 2002; Pruyne et al., 2002; Sagot et al., 2002a,b; Kovar et al., 2003; Pring et al., 2003). All formins contain a highly conserved formin homology (FH) 2 domain that forms a stable homodimer with an actin binding surface for nucleation and processive barbed end association (Moseley et al., 2004; Xu et al., 2004) and an FH1 domain with proline-rich tracts to bind and rapidly elongate profilin-actin (Wasserman, 1998; Kovar et al., 2003; Li and Higgs, 2003; Romero et al., 2004; Kovar, 2006; Vavylonis et al., 2006; Neidt et al., 2009; Courtemanche and Pollard, 2012). GTPase binding domains and FH3 domains existing in some formins are involved in localization or activation (Petersen et al., 1998; Carnahan and Gould, 2003; Gorelik et al., 2011; Liu et al., 2012). Some formins remain bound to barbed ends of growing actin filaments for >1,000 s in vitro (Kovar and Pollard, 2004; Kovar, 2006), sufficient to make a >30-µm filament, whereas actin filaments average only ∼1 µm in vivo (Karpova et al., 1998; Kamasaki et al., 2005; Coffman et al., 2009), suggesting that formin activity must be tightly regulated. Many formins are regulated by autoinhibition when the two ends of the protein interact to prevent actin nucleation. Inhibition is relieved by Rho GTPase binding, phosphorylation, or localization to cortical nucleation sites (Takeya et al., 2008; Wang et al., 2009; Ramalingam et al., 2010; Buttery et al., 2012; H. Chen et al., 2012; Maiti et al., 2012). In addition, inhibitors regulate nucleation activity or processivity of some formins by displacing them from barbed ends of actin filaments (Eisenmann et al., 2007; Quinlan et al., 2007; Chesarone et al., 2009).
S. pombe uses three formins for distinct actin structures: Fus1 assembles actin filaments for the mating projection (Petersen et al., 1998); For3 assembles actin filaments within long actin cables for cellular transport and polarization (Feierbach and Chang, 2001b); and Cdc12 is essential to assemble the contractile ring for cytokinesis (Chang et al., 1997). For3 has established roles in cell polarity (Feierbach and Chang, 2001b; Martin and Chang, 2006; Martin et al., 2007; Wang and Vavylonis, 2008; Scott et al., 2011), but it has not been studied in the contractile ring. For3 localizes to the division site but is not essential for cytokinesis (Pelham and Chang, 2002; Martin and Chang, 2006). The prevailing assumption is that For3 localization to the division site is to mark the new cell end (Feierbach and Chang, 2001a; Kamasaki et al., 2005). Cdc12 assembles actin filaments both from cytokinesis nodes at the division site (Coffman et al., 2009) and from nonmedial locations (Huang et al., 2012). Most knowledge of Cdc12 targeting and regulation stems from structure-function analyses of Cdc12 and F-BAR protein Cdc15. An N-terminal fragment of the first ∼500 aa localizes normally to the contractile ring in the presence of native full-length Cdc12 and binds to Cdc15 (Carnahan and Gould, 2003; Yonetani et al., 2008). The regulation of Cdc12 activity remains a mystery because mutational analyses provided no obvious evidence of autoinhibition (Yonetani et al., 2008). However, the formation of interphase contractile rings when the C terminus is deleted indicates an activated allele, suggesting autoinhibition may occur by an unknown mechanism (Laporte et al., 2010; Yonetani and Chang, 2010).
Here we investigate the relative importance of different sources of actin filaments for the contractile ring in fission yeast cytokinesis by studying the formin For3 and N-terminal truncations of Cdc12. The Cdc12 truncations depend on actin and For3 for localization to the division site. Despite the truncations nucleating more actin filaments throughout the cell, without proper Cdc12 localization for de novo actin assembly at the division site, contractile ring assembly is inefficient and unreliable. Collectively, these results reveal a novel role for For3 at the division site and the cooperation between two formins for successful cytokinesis.
Results
Condensation of cytokinesis nodes without the essential formin Cdc12
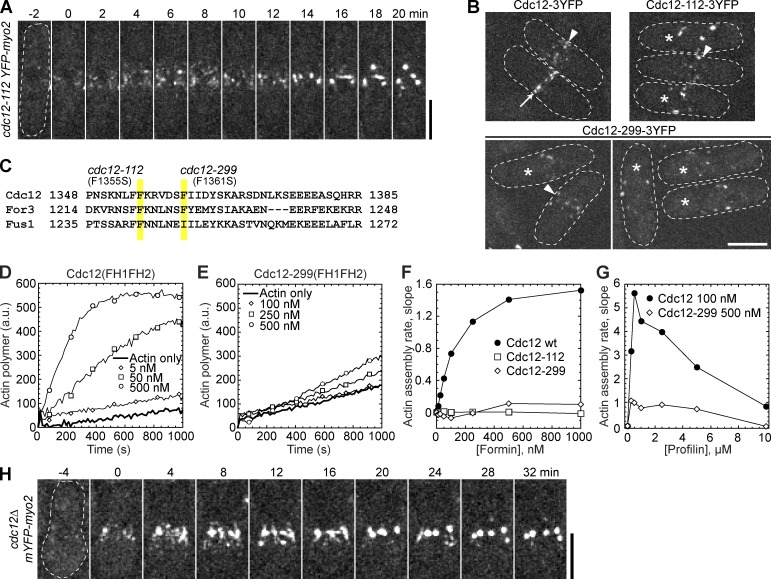
Actin filaments are required for condensation of cytokinesis nodes (defined as 20–65 punctate structures at the cell equator containing formin Cdc12, myosin-II heavy chain Myo2, regulatory light chain Rlc1, and other proteins) into the contractile ring during early mitosis (Wu et al., 2006; Huang et al., 2008; Saha and Pollard, 2012). The formin Cdc12, a nucleator of linear actin filaments, is essential for cytokinesis (Chang et al., 1996, 1997; Kovar et al., 2003). Instead of remaining dispersed, Myo2 nodes condensed into clumps (hereafter we refer to partially condensed nodes as clumps) but not a contractile ring in temperature-sensitive (ts) cdc12-112 cells at the restrictive temperature of 36°C (Fig. 1 A), similar to Rlc1 (Hachet and Simanis, 2008). It was suggested that clump formation might arise from the ts mutants being less efficient at actin filament nucleation or elongation at the restrictive temperature, resulting in fewer or shorter actin filaments at the division site (Ojkic and Vavylonis, 2010). To investigate the mechanism of node movement in the mutants, we first determined if the ts formins are defective in cortical localization. After 2 h at 36°C, Cdc12-112 and Cdc12-299 concentrated to the nodes or clumps, whereas wild-type (wt) Cdc12 localized to nodes or rings (Fig. 1 B). Thus, improper Cdc12 localization is not the main contributor to defective node condensation.
Figure 1.
Condensation of cytokinesis nodes without functional formin Cdc12. (A and B) Cells were grown at 36°C for 2 h before imaging at 36°C. (A) Time course of Myo2 nodes condensing into clumps in cdc12-112 mutant. Time 0 is when nodes appear. (B) Localization of Cdc12, Cdc12-112, and Cdc12-299. Nodes (arrowheads), contractile rings (arrows), and cells with scattered clumps (asterisks) are marked. (C) Alignment of the last α-helix of the FH2 domains (αT; Xu et al., 2004) of S. pombe formins. Positions of point mutations in cdc12-112 and cdc12-299 are highlighted. (D–G) In vitro spontaneous assembly of 2.5 µM Mg-ATP actin monomers (10% pyrene labeled) using purified FH1-FH2 domains from wt and mutant Cdc12, in the absence (D–F) and presence (G) of profilin. The data are single representative experiments out of at least two repeats. (D and E) Time course of actin polymerization with the increasing concentrations of wt (D) or mutant (E) Cdc12. (F) Dependence of the actin assembly rate on formin concentration. (G) Dependence of Cdc12-mediated actin assembly rate on profilin concentration. (H) Time course of condensation of Myo2 nodes in cdc12Δ cells. Spores from cdc12Δ/cdc12+ diploid strain were germinated and grown at 25°C for 12 h before imaging at 25°C. Time 0 is when nodes appear. Bars, 5 µm.
The cdc12-112 mutant (Chang et al., 1997) has a point mutation resulting in a substitution of phenylalanine to serine at residue 1355 (Fig. 1 C) in the last α helix of the FH2 domain (Xu et al., 2004). The cdc12-299 mutant (Chang et al., 1996) has a similar mutation, F1361S (Fig. 1 C), as well as a deletion of three proline residues (948–950) in the FH1 domain. Both point mutations in the FH2 domain of Cdc12 alter residues that are highly conserved in evolutionarily diverse formins (Xu et al., 2004). To determine if the mutations affect actin nucleation/elongation activities, we purified wt and mutant Cdc12 FH1FH2 from Escherichia coli and tested their actin assembly properties in vitro. Unlike wt FH1FH2, the mutants were not capable of stimulating the assembly of actin or profilin-actin (Fig. 1, D–G). Although these assays were performed at a permissive temperature of 25°C, the purified proteins’ inability to function is probably reflective of their state at the restrictive temperature because cells are normally viable at room temperature. To rule out the possibility that residual activity of the mutant Cdc12 at 36°C is responsible to condense the nodes into clumps, we deleted one copy of cdc12 in a diploid strain (Laporte et al., 2011). Surprisingly, we found that germinated cdc12Δ spores also condensed nodes into clumps instead of a contractile ring (Fig. 1 H). These data from cdc12 mutants reveal that other actin nucleators also function at the division site during contractile ring assembly.
Involvement of the formin For3 in cytokinesis
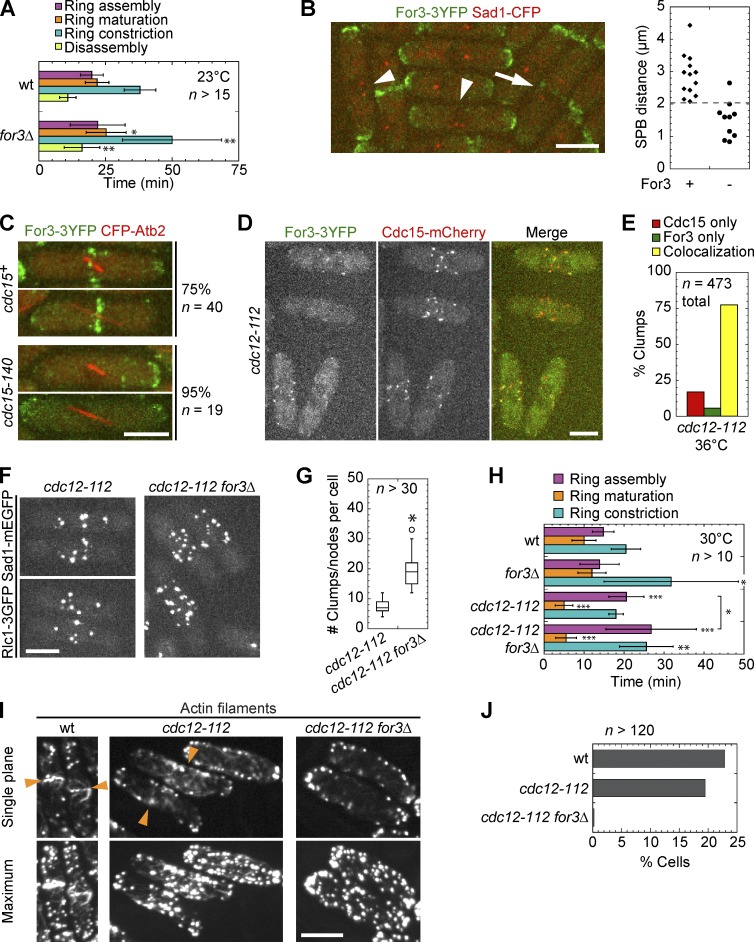
The formin For3 is the nucleator of linear actin filaments in actin cables (Feierbach and Chang, 2001b). It has been reported that the Arp2/3 complex, the nucleator of branched actin filaments, participates in the formation of actin rings at the division site and that the actin ring is not detected in fixed arp3-c1 for3Δ double mutant cells (Pelham and Chang, 2002). However, YFP-actin, which is used by the Arp2/3 complex but not by formins in fission yeast, is not visible in the contractile ring (Wu et al., 2006; Q. Chen et al., 2012). We examined live cells expressing Rlc1-3GFP and spindle pole body (SPB) protein Sad1-mEGFP in for3Δ, arp3-c1, or the for3Δ arp3-c1 double mutant. After shifting cells to 19°C for 6–16 h, the restrictive temperature for arp3-c1, both single and double mutant cells formed rings normally, despite polarity defects in the mutants (Fig. S1, A and B). These data suggest that neither the Arp2/3 complex nor For3 is required for contractile ring assembly in a wt cdc12 background.
Although the timing of ring assembly was not significantly different in for3Δ and wt cells, later stages of ring constriction and disassembly were slowed in for3Δ (Fig. 2 A), consistent with its relocalization to the septation site during ring constriction (Fig. S1 C). However, we found that for3Δ was synthetic lethal with rng2 and cdc4 mutants, two genes essential for contractile ring assembly (Table 1), suggesting that For3 might play an overlapping role in contractile ring assembly with other proteins. Consistently, For3 arrived at the division site shortly after SPB separation during ring assembly and usually before the spindle began to elongate during anaphase B (Fig. 2 B). This localization did not depend on actin (Fig. S2 A) because For3 still localized to the division site during ring assembly after Latrunculin A (Lat-A) treatment or in the actin-binding mutant for3-I930A that is defective for actin assembly (Martin and Chang, 2006; Scott et al., 2011). Of the cytokinesis mutants tested, only cdc15-140 affected For3 localization to the division site (Fig. 2 C and Fig. S2, B and C). Proteins that affect For3 localization to cell tips or interact with For3 at tips were not required for its localization to the division site (Fig. S2 D). For3 localized to the division site later than F-BAR protein Cdc15 in wt cells (Wu et al., 2003), but it colocalized with Cdc15 in clumps in cdc12-112 cells at 36°C, although at the elevated temperature 3YFP is more difficult to detect (Fig. 2, D and E). Double mutant cdc12-112 for3Δ cells had significantly more but dimmer node-like structures at the division site in cells with two nuclei than in single mutant cdc12-112 cells (Fig. 2, F and G), suggesting that actin filaments nucleated by For3 might be responsible for node movement and node condensation into clumps. Indeed, at a semipermissive temperature of 30°C, ring assembly was significantly delayed in cdc12-112 for3Δ compared with cdc12-112 (Fig. 2 H). Consistently, meshwork-like actin filaments existed at the cell equator in cdc12-112 cells, similar to a previous study (Arai and Mabuchi, 2002), but not in cdc12-112 for3Δ double mutants (Fig. 2, I and J). Together, these data suggest that For3 and Cdc12 may work together to nucleate actin filaments for the contractile ring during cytokinesis.
Figure 2.
The formin For3 localization at the division site and its involvement in cytokinesis. (A) Timing (mean ± SD) of cytokinesis in for3Δ and wt at 23°C. *, P < 0.05; **, P < 0.005 compared with wt. This and subsequent p-values were determined using a two-tailed t test. Here and in later figures, ring assembly is from Rlc1 node appearance until a compact ring forms; ring maturation is from a compact ring to the start of ring constriction; ring constriction is from the start of constriction until the ring becomes a dot; and disassembly is from a dot until all the signal disappears from the division site. (B) For3 (green) localization to the division site. Arrowheads indicate cells with two SPBs (red) but no For3, whereas the arrow indicates a cell with two SPBs and For3. Graph (right) depicts 3D SPB distance in cells with short (<5 µm) spindles with (+) and without (−) For3 at the division site. (C–G) Cells were grown at 36°C for 2 h before imaging at 36°C. (C) For3 (green) localization in wt and cdc15-140 cells expressing CFP-Atb2 (red). Cells with similar spindle lengths are shown. Percentage of cells (with 2–7-µm spindles) with (wt) or without (cdc15-140) For3 at the division site is given. (D) For3 and Cdc15 localization in clumps in cdc12-112 mutants. (E) Quantification of clumps/nodes in (D) with Cdc15 only, For3 only, or colocalization of For3 and Cdc15. (F) Node condensation is impaired in cdc12-112 for3Δ double mutants compared with cdc12-112. (G) Quantification of clumps/nodes per cell in images as in F, subtracting two SPBs from the total. *, P < 0.0001 compared with cdc12-112. Box plots include 50% of the data inside the box, the line is the median, the whiskers indicate upper and lower bounds, and the circles outside the whiskers are outliers. (H) Timing of contractile ring stages (mean ± SD) in indicated Rlc1-3GFP labeled strains grown at 30°C for 2 h before imaging at 30°C. *, P < 0.05; **, P < 0.005; ***, P < 0.0005 in t tests compared with wt (except where the bracket indicates). (I) Staining of actin filaments in wt, cdc12-112, and cdc12-112 for3Δ cells. Cells were fixed at 36°C after growing at 36°C for 2 h. Single planes near the top of the cells and maximum intensity projections are shown. The meshwork/ring of filaments at the division site is marked (arrowheads). (J) Quantification of cells as in I. Percent cells with actin meshwork and rings (wt only) at the division site. Bars, 5 µm.
Table 1.
Genetic interactions of for3Δ with cytokinesis mutants
| Mutanta | Viable double mutants at 25°C | Viable single mutants at 25°Cb | Total number of tetrads |
| % | % | ||
| cdc12-299 | 100 | 100 | 11 |
| rng2-D5 | 0 | 100 | 10 |
| cdc4-8 | 0 | 100 | 9 |
| rlc1Δ | 36 | 100 | 8 |
| cdc15-140 | 100 | 100 | 11 |
| mid1-366 | 100 | 100 | 11 |
Parent strains are given in Table S1.
Single mutants of for3Δ were 100% viable on tetrad plates in this group of crosses. Some ts mutants had been backcrossed before these crosses.
N-terminal truncations of Cdc12 affect actin cytoskeleton organization
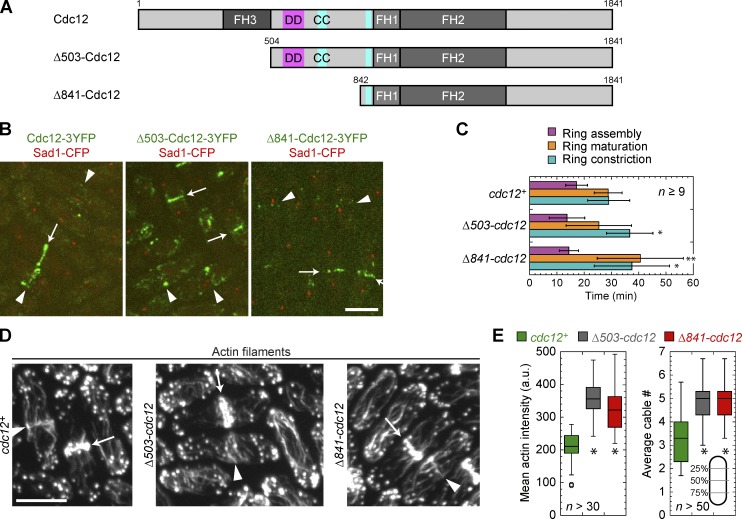
To further explore the relationship between the formins Cdc12 and For3 during cytokinesis, we tested whether altering Cdc12 activity or localization by truncating Cdc12 would make For3 essential. We truncated the first 503 aa of Cdc12 (Δ503-Cdc12; Fig. 3 A) to eliminate the binding site for Cdc15 (Carnahan and Gould, 2003) and another localization domain (Yonetani et al., 2008). We also made a truncation that only contained FH1, FH2, and the C terminus (Δ841-Cdc12; Fig. 3 A). The Δ841-Cdc12 fragment can rescue cdc12-112 or cdc12Δ when overexpressed from a high-copy plasmid (Yonetani et al., 2008). Both N-terminal truncations were expressed from the cdc12 promoter at its native locus replacing the wt Cdc12. Cells with the truncations resembled wt cells. The truncations localized to node-like structures and contractile rings at the division site, although fewer nodes were detectable than in wt (Fig. 3 B and Video 1). We used fluorescence intensity to estimate the expression of the truncations (Wu and Pollard, 2005; Coffman et al., 2011; Laporte et al., 2011). Δ503-Cdc12 levels in the whole cell and the contractile ring were not significantly different from those in wt cells; however, the levels of Δ841-Cdc12 were reduced to ∼60% in the whole cell and ∼15% at the contractile ring (Fig. S3 A). The contractile ring assembled slightly faster in both truncations, whereas ring constriction was delayed (Fig. 3 C and Video 2).
Figure 3.
N-terminal truncations of Cdc12 affect actin cytoskeleton organization. (A) Domain architecture of full-length Cdc12 and two truncations, approximately to scale. Δ503-Cdc12 and Δ841-Cdc12 delete the first 503 aa and the first 841 aa, respectively. FH domains 1, 2, and 3 are in darker shades of gray. Putative dimerization domain (DD, pink) and coiled-coil domains (CC, teal) are shown (Yonetani et al., 2008). (B) Localization of N-terminal truncations of Cdc12 (green) and SPB protein Sad1 (red) as a cell cycle marker. Localizations to ring precursors (arrowheads) and rings (arrows) are shown. (C) Rlc1-3GFP ring assembly, maturation, and constriction (mean ± SD) in Cdc12 N-terminal truncations and wt. *, P < 0.05; **, P < 0.005. (D and E) Staining of actin filaments in wt, Δ503-cdc12, and Δ841-cdc12 cells to reveal an actin meshwork at the division site (arrowheads), actin rings (arrows), and more and brighter actin cables in the mutants. (E, left) Box plots of cell size–corrected integrated actin intensity from the sum intensity projection of 0.4-µm-spaced sections. (right) Box plots of actin cable numbers per cross section in interphase cells. Cables were counted in three line scans of the maximum intensity projection at the positions indicated (gray lines) and the mean number from those three positions for each cell is plotted. *, P < 0.0001 compared with wt. Bars, 5 µm.
Next we asked if their actin assembly activities are altered. Actin staining revealed a significant increase in total actin filaments (Fig. 3, D and E), whereas actin patch (Arp2/3 complex-nucleated, branched actin filaments) intensities were unchanged (mean actin patch intensities were ∼97% of wt in both truncations; n ≥ 50 patches each). In addition, actin cables per cross section increased from 3.3 ± 1.3 in wt to 5.0 ± 1.1 and 4.9 ± 1.2 in Δ503-cdc12 and Δ841-cdc12 cells (mean ± SD; Fig. 3 E). Thus, Δ841-Cdc12 even at the reduced level is sufficient to form excess actin cables to the same extent as Δ503-Cdc12. Together, these data show that the Cdc12 truncations are activated alleles for actin assembly and are sufficient for contractile ring formation.
For3 is essential for contractile ring assembly and constriction in Cdc12 truncations
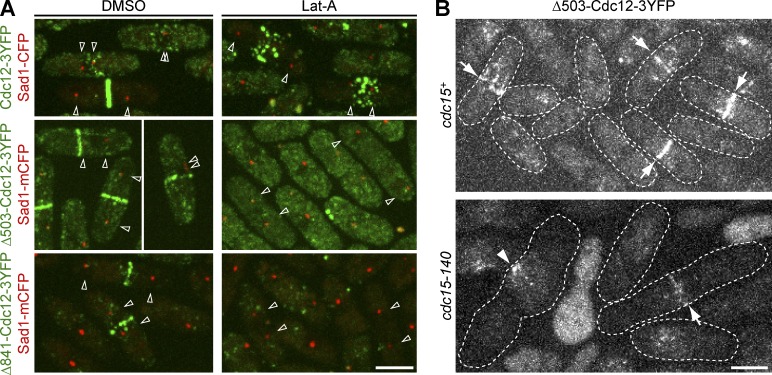
Surprisingly, unlike full-length Cdc12 (Wu et al., 2003, 2006), localization of both truncations to the division site was abolished after Lat-A treatment (Fig. 4 A). We hypothesized that For3-nucleated actin filaments are crucial for the localization of the Cdc12 truncations. To test this hypothesis, we abolished For3 localization to the division site using cdc15-140 (Fig. 2 C). The cdc15-140 Δ503-cdc12 cells were viable at 25°C but synthetic lethal from 30 to 36°C (Table 2). After 4 h at 36°C, Δ503-Cdc12-3YFP localized weakly to a contractile ring in 1% of living cells in cdc15-140, compared with 18% in wt (Fig. 4 B; n > 100 cells each). Some cells also had large Δ503-Cdc12 clumps at the division site. The perturbed localization is not likely to be a direct effect of the cdc15 mutation because the Cdc12 truncation lacks the Cdc15 binding site.
Figure 4.
N-terminal truncations of Cdc12 depend on actin and Cdc15 for localization. (A) Cdc12 N-terminal truncations or wt were treated with DMSO or Lat-A for 30 min. Cells with similar SPB distances are marked by open arrowheads for comparison. (B) Localization of Δ503-Cdc12 to the contractile ring/forming ring (arrows) or clump (arrowhead) in wt or cdc15-140 after growth at 36°C for 4 h and then imaged at 36°C. Bars, 5 µm.
Table 2.
Genetic interactions of Δ503-cdc12 with cytokinesis mutants
| Mutanta | Viable double mutants at 25°C | Viable single mutants at 25°Cb | Total number of tetrads | Lowest synthetic lethal temperaturec |
| % | % | °C | ||
| for3Δ | 0 | 90 | 42 | N/A |
| for3-I930A | 0 | 42 | 43 | N/A |
| mid1Δ | 0 | 64 | 18 | N/A |
| rng2-D5 | 7 | 89 | 11 | 30 |
| rng2-346 | 0 | 88 | 11 | N/A |
| cdc4-8 | 0 | 64 | 10 | N/A |
| myo2-E1 | 5d | 82 | 19 | N/A |
| rlc1Δ | 0 | 93 | 11 | N/A |
| rng3-65 | 100 | 100 | 10 | 30 |
| cdc15-140 | 60 | 80 | 10 | 30 |
| mid1-6 | 50 | 86 | 14 | ND |
| mid1-366 | 67 | 100 | 9 | ND |
| blt1Δ | 93 | 100 | 10 | ND |
Parent strains are given in Table S1.
Single mutants of Δ503-cdc12 were 93% viable on tetrad plates in this group of crosses. Ts single mutants were more viable after backcrossing; see Table 1.
Tested by streaking wt, single mutants, and double mutant on YE5S plates and incubating at temperatures from 25 to 36°C. The Δ503-cdc12 single mutants grew normally from 25 to 36°C.
The only viable double mutant isolated likely had a suppressor based on cell morphology at 36°C.
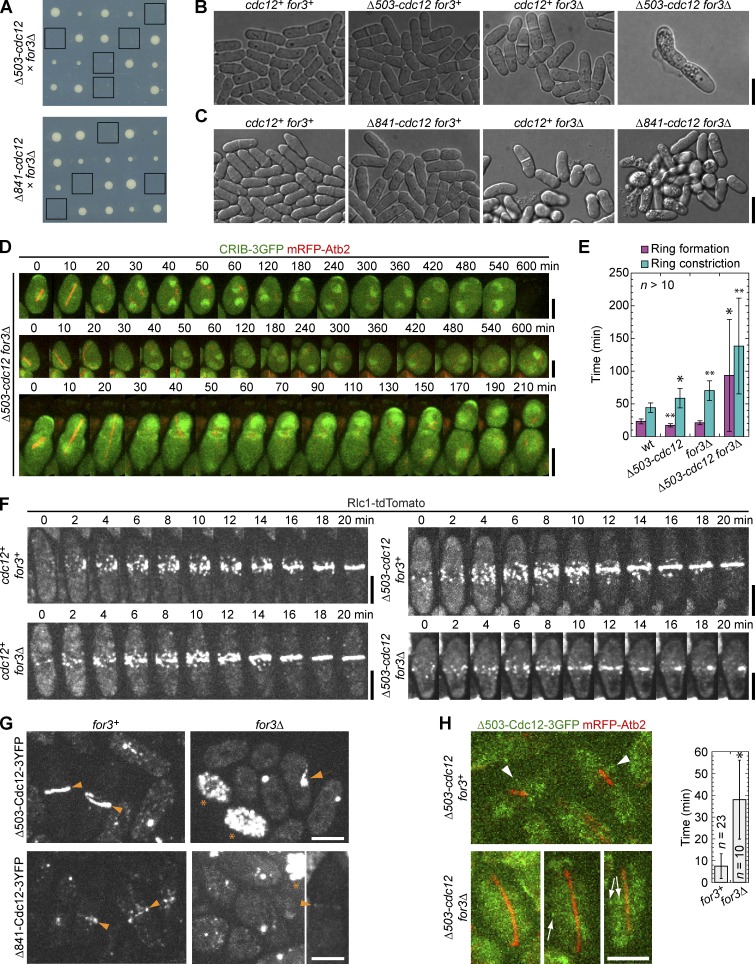
Crossing the Cdc12 truncations to for3Δ yielded no visible double mutant colonies despite 80–100% single mutant and wt spores forming colonies (Fig. 5 A and Table 2; n > 40 tetrads for each cross), indicating that For3 is essential in these Cdc12 truncations. Because neither Δ503-cdc12 for3Δ nor Δ841-cdc12 for3Δ double mutants could be recovered from the crosses, we observed tetrads 2–4 d after dissection to determine the lethal defects (Fig. 5, B–H), using a confocal microscope with an automated stage (tetrad fluorescence microscopy; see Materials and methods). Many double mutants died with only one or two elongated cells (∼30%; Fig. 5 B), suggesting a cytokinesis defect. Although we found colonies of up to 32 cells, most had 3–8 cells (∼52%; Fig. 5 C). We mainly imaged the double mutants with 3–8 cells because the long cells had likely failed cytokinesis. Because for3Δ cells have polarity and spindle orientation defects (Feierbach and Chang, 2001b), we used CRIB-3GFP and mRFP-Atb2 as markers for polarized growth and the mitotic spindle, respectively, to observe tetrad colonies. The CRIB domain from Saccharomyces cerevisiae Cdc42 effector Gic2 is a marker for active Cdc42 (Tatebe et al., 2008; H. Chen et al., 2012). Although cell polarization was affected in the double mutants based on abnormal cell shapes, the majority of interphase cells had one or two cell tips with CRIB localization (Fig. 5 D and Video 3). Mitosis occurred in ∼40% of double mutants during the movies, but less than half of those succeeded in cytokinesis. The low rate of mitosis in 12 h is likely caused by the polarity and growth defects. However, these defects alone are not sufficient to explain the cytokinesis defect, because arp3-c1 for3Δ cells were not well polarized but assembled contractile rings normally (Fig. S1 A).
Figure 5.
Tetrad fluorescence microscopy reveals that for3Δ cdc12 truncation double mutants are defective in contractile ring assembly and constriction. (A) Sections of tetrad plates from crosses between for3Δ and cdc12 truncations grown at 25°C. Double mutants from each tetrad (vertical) are boxed. (B) Differential interference contrast images of a tetratype tetrad from the for3Δ × Δ503-cdc12 cross (JW1665 × JW5124) 3 d after tetrad dissection. The entire colony of the double mutant is shown, while the wt and single mutants are focused at the edge of the colonies where the cells are in a single layer. (C) Same as B for for3Δ × Δ841-cdc12 cross (JW5281 × JW5283). (D) Time courses showing three Δ503-cdc12 for3Δ CRIB-3GFP mRFP-atb2 cells that undergo mitosis during the movies. Time 0 is the presence of a short spindle. Tetrads were dissected from a cross JW5281 × JW5282-2 and imaged on YE5S agar (see Materials and methods) at 23°C. CRIB-3GFP is a marker for cell polarization and mRFP-Atb2 is a marker for MTs and the spindle. (E and F) for3Δ Δ503-cdc12 cells are defective in contractile ring assembly and constriction. Tetrad fluorescence microscopy of a cross between for3Δ rlc1-tdTomato and Δ503-cdc12 rlc1-tdTomato. (E) Timing (mean ± SD) of ring assembly and constriction as shown in F from tetrad colonies. Ring constriction here is from the appearance of a compact ring to ring constriction to a dot (including the ring maturation time). *, P < 0.05; **, P < 0.005 compared with wt. (F) Time courses of four representative cells from the same tetratype tetrad. (G) For3 is crucial for the localization of Cdc12 N-terminal truncations. Tetrad fluorescence microscopy of crosses between Δ503-cdc12-3YFP or Δ841-cdc12-3YFP with for3Δ. The localizations to rings (arrowhead) and clumps (arrow) are marked. Dead cells with high autofluorescence in for3Δ are marked with asterisks. (H) Micrographs (left) of mRFP-Atb2 spindles and Δ503-Cdc12-3GFP localization to faint rings (arrowhead) or clumps (arrow). Graph (right) shows timing (mean ± SD) of Δ503-cdc12-3GFP localization to the medial region of the cell after the appearance of a spindle using a 5–10-min delay in tetrad fluorescence microscopy. *, P < 0.001. Bars: (D and F–H) 5 µm; (B and C) 10 µm.
To observe contractile ring assembly directly, we crossed for3Δ rlc1-tdTomato to Δ503-cdc12 rlc1-tdTomato cells and observed them 2–3 d after tetrad dissection (Fig. 5, E and F). Of the cells that attempted contractile ring formation during imaging, 39% of Δ503-cdc12 for3Δ cells successfully assembled a compact ring (Fig. 5 F), but it took approximately three times as long to assemble and constrict the ring compared with wt cells (Fig. 5 E). The single mutants were only slightly delayed in ring constriction, similar to our previous results from liquid cultures (Figs. 2 A and 3 C). In the double mutants, 51% of cytokinesis attempts (n = 37) resulted in formation of large Rlc1 clumps at the division site instead of a compact ring (Fig. 5 F and Video 4). The large variation in times to condense the nodes into a compact ring and to constrict the ring in the double mutants suggests stochastic and unreliable cytokinesis pathways without normal formin function or localization. Because Rlc1 localizes to nodes and other upstream node proteins are independent of Cdc12 for localization (Laporte et al., 2011; Padmanabhan et al., 2011), we expect that the nodes only lack Cdc12.
To test whether ring assembly might fail in the double mutants because of defective localization of the Cdc12 truncations to the division site, we crossed tagged truncations to for3Δ and observed their localization in the double mutants. Both truncations localized to the division site mainly at a large spot or clump in for3Δ cells in contrast to a ring in for3+ cells (Fig. 5 G). The time from spindle appearance to spindle breakdown was similar in wt (38.0 ± 5.6 min, n = 15 cells) and for3Δ Δ503-cdc12 cells (37.9 ± 10.5 min, n = 14), indicating that the duration of mitosis is normal. However, in cells that formed spindles, 83% cells formed a Δ503-Cdc12 spot/clumps at the division site 38.0 ± 18.1 min after spindle appearance in for3Δ (no signal in other cells) compared with 100% cells forming a Δ503-Cdc12 ring after 7.5 ± 5.7 min in for3+ (Fig. 5 H). In two cells that managed to form formin rings out of >40 cells (Fig. 5 G), the ring assembly took >2.5 h. The actin-binding mutant for3-I930A was also synthetic lethal with cdc12 truncations, indicating that actin filament assembly by For3 is essential for the localization of the truncations. The expression level and localization of For3 in cdc12 truncation strains were similar to wt (Fig. S3, B–D). Thus, the dependence on For3 was not an artifact of abnormal For3 behavior. Together, these data indicate that Cdc12 lacking its known N-terminal localization domains (Carnahan and Gould, 2003; Yonetani et al., 2008) can be recruited to the division site by actin filaments nucleated by For3.
Genetic interactions further support For3’s roles in cytokinesis. Both Δ503-cdc12 and for3Δ exhibited synthetic lethal interactions with several cytokinesis mutants involved in contractile ring assembly and constriction including rng2, cdc4, myo2, rlc1Δ, and mid1Δ (Tables 1 and 2). Thus, For3 has a direct role in contractile ring assembly in cooperation with Cdc12.
Contributions of actin filaments assembled at or transported to the division site for contractile ring assembly
Cdc12 concentrates to cytokinesis nodes from scattered cytoplasmic speckles (Coffman et al., 2009) and For3 relocates from cell tips to the division site (Fig. 2 A) during contractile ring assembly in wt cells. Cdc12 and For3 assemble actin filaments at the division site and at other places in the cell (Fig. 3 D; Martin and Chang, 2006; Coffman et al., 2009; Huang et al., 2012). One open question is the relative contribution of de novo actin filament assembly at the division site and cortical transport/flow of actin filaments assembled elsewhere to the contractile ring (Huang et al., 2012). The actin binding peptide, Lifeact (Riedl et al., 2008), is reported to label actin structures with the least impact on actin dynamics and cell growth (Huang et al., 2012). We compared the timing of ring assembly using Rlc1 as a marker in wt and strains expressing tagged Lifeact or GFP alone from the strong actin promoter. The Lifeact tagged with a GFP that has a tendency to dimerize at high concentration (Yang et al., 1996) doubled the times needed for ring assembly, unlike the monomeric GFP (mGFP L221K)-tagged Lifeact (Zacharias et al., 2002; Huang et al., 2012) or overexpressed GFP alone (Fig. S4 A). We also compared the intensity of the actin meshwork at the cell equator to the regions just outside the broad band (to avoid actin patches concentrated at cell tips) in fixed wt cells stained for actin filaments and in live cells expressing Lifeact-GFP or Lifeact-mGFP during early contractile ring assembly (Fig. S4 B). All three strains were similar with approximately three times more actin filaments at the cell equator than at the adjacent regions. In later stages, cells expressing Lifeact-GFP usually had thick, wavy bundles outside of the compact ring; whereas in wt cells (Fig. S4 C) or Lifeact-mGFP cells (Fig. S4 D and Video 5), we rarely detected prominent nonmedial cables after a compact actin ring had formed. Together these data suggest that the excess cables in Lifeact-GFP–expressing cells are not crucial for contractile ring assembly or maturation (see Discussion).
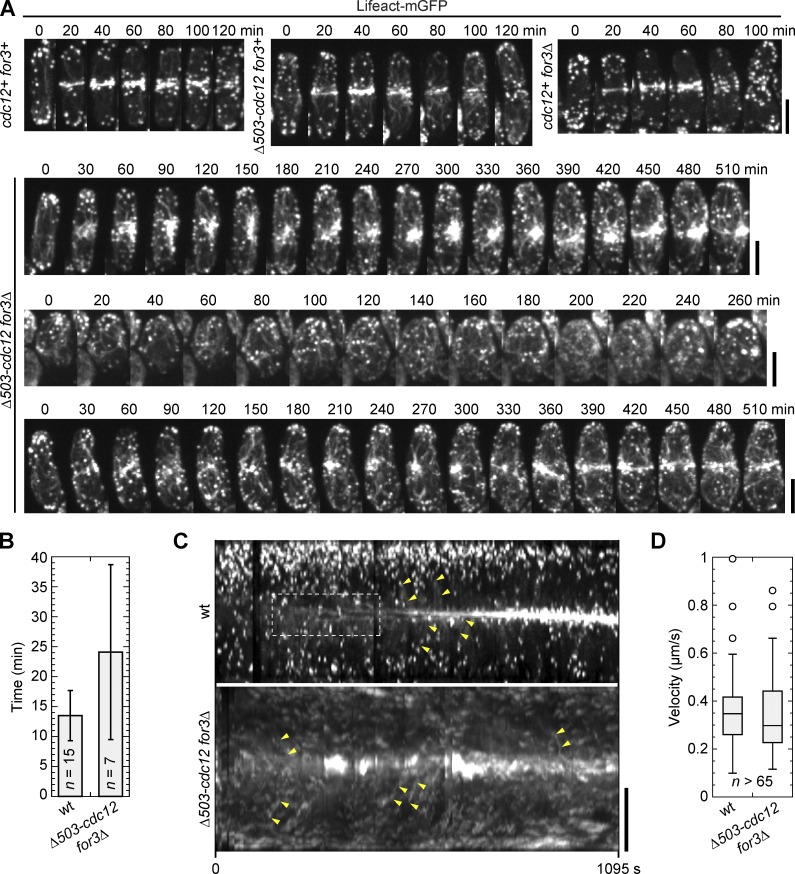
We detected more actin filaments at the cell equator than at the adjacent regions during contractile ring assembly (Fig. S4 B), but it is challenging to tell whether these filaments are assembled de novo at the division site or transported from elsewhere by cortical flow. We predicted that double mutants of for3Δ and cdc12 truncations are ideal for testing the importance of de novo actin assembly at the division site because no formins could localize to the cell equator efficiently at early mitosis (Fig. 5, G and H). We crossed for3Δ and Δ503-cdc12 cells both expressing Lifeact-mGFP for tetrad fluorescence microscopy. Consistent with previous experiments (Fig. 5), 100% of wt and single mutant cells assembled and constricted the contractile ring with normal timing (Fig. 6 A and Video 6). However, only ∼25% of double mutant cells assembled actin rings (n = 28 cells), and the timing of assembly and constriction were severely delayed, despite these cells having an excess of actin filaments throughout the cells (Fig. 6 A and Video 6). Because myosin-II marked with Rlc1 still localized to the division site (Fig. 5 F), cortical transport might be normal. Indeed, in tetrad fluorescence microscopy, the timing of actin accumulation at the division site was not significantly different in Δ503-cdc12 and for3Δ Δ503-cdc12 cells (P = 0.10; Fig. 6 B). Of those double mutant cells that formed an actin aster, 43% eventually formed rings. Because Δ503-Cdc12 accumulation was delayed (Fig. 5 H), this result suggests that actin accumulates at the division site by cortical transport before Δ503-Cdc12 does. The velocity of actin filaments moving toward the division site in wt and for3Δ Δ503-cdc12 was also similar (Fig. 6, C and D; and Video 7). Thus, cortical transport is normal, whereas de novo actin filament assembly at the division site could not occur until after Δ503-Cdc12 localized to the clump in for3Δ Δ503-cdc12. These results suggest that although the Cdc12 truncation assembles actin filaments efficiently in for3Δ cells, cortical transport alone is not effective and reliable for contractile ring formation and constriction. Together, these data indicate that de novo assembly of actin at the medial cortex is crucial for contractile ring assembly and constriction in fission yeast cytokinesis.
Figure 6.
Medial actin filaments are predominant for contractile ring assembly. (A) Time courses of contractile ring assembly and constriction by tetrad fluorescence microscopy in cells expressing Lifeact-mGFP from the cross JW5463 × JW5481. The interval of the time-lapse movie is 10 min and time 0 is the time point just before actin filaments concentrate near the division site (except for the double mutant with no accumulation at the division site). Three classes of double mutant cells are shown: large clump at the division site (top); no specific structures (middle); and severely delayed ring assembly (bottom). (B) Timing of actin accumulation in a ring/meshwork (wt) or aster (Δ503-cdc12 for3Δ) after Rlc1-tdTomato node appearance in tetrad fluorescence microscopy. Many double mutants could not be included in this quantification because they did not attempt division during the imaging. Error bars show SDs. (C and D) Actin filament movements toward the division site. (C) Kymographs of wt and double mutant (Δ503-cdc12 for3Δ) cells expressing Lifeact-mGFP. De novo actin filament nucleation (dashed box, wt only) and cortical flow (examples are marked by pairs of yellow arrowheads) are evident in tetrad fluorescence microscopy from the same cross as B. (D) Box plots of velocity of actin movements toward the division site as shown in C. Bars, 5 µm.
Discussion
The formin For3 participates in assembly of actin filaments for the contractile ring and becomes essential in the absence of other pathways to efficiently localize the formin Cdc12 to the division site. In addition, cells that cannot anchor the activated Cdc12 to the division site assemble contractile rings inefficiently, indicating that de novo nucleated actin filaments at the cleavage site are crucial for contractile ring assembly and constriction.
Cooperation between the formins Cdc12 and For3 in fission yeast cytokinesis
Our data support a previously uncharacterized role of For3 at the division site that is redundant with Cdc12. In the presence of normal Cdc12 function, For3 has little effect on the fidelity of contractile ring assembly and only affects ring constriction. However, when the function of Cdc12 is affected by point mutations or N-terminal truncations, actin assembly by For3 is needed for node movement and cell survival. The lower efficiency of actin nucleation by For3 (Scott et al., 2011) might explain node clumping in cdc12 mutants, based on the predictions of the SCPR model (Vavylonis et al., 2008; Ojkic and Vavylonis, 2010).
The localization of Cdc12 N-terminal truncations depends on actin and For3, but the mechanism is unclear. There are at least four possibilities for the recruitment of Cdc12 truncations to the division site: (1) Cdc12 displaces For3 from the barbed end of actin filaments at the division site. This is likely because the barbed end affinity of Cdc12 is ∼10× higher than that of For3 (Scott et al., 2011) and capping protein localizes to the division site after a compact ring is formed (Wu et al., 2003). (2) Cdc12 travels on actin filaments. Both For3 and the formin Bni1 are released from actin nucleation sites and incorporate into actin cables by remaining bound to the barbed ends of short bundled filaments (Kamasaki et al., 2005; Martin and Chang, 2006; Buttery et al., 2007). Thus, Cdc12 might travel to the division site by displacing For3 within actin cables. (3) The actin filaments nucleated by Cdc12 truncations could be bundled with those assembled by For3 at the division site, which recruits Cdc12 to the division site. (4) Actin nucleated by the Cdc12 truncations might be captured and pulled into the division site by Myo2 or Rng2 in the nodes, which accumulate there independently of actin filaments and Cdc12 (Wu et al., 2006; Laporte et al., 2011; Padmanabhan et al., 2011). This mechanism could also explain the limited localization of Cdc12 truncations in for3Δ to the contractile ring or large clump (Fig. 5, G and H) and might be similar to myosin-II–dependent retrograde flow of actin filaments in budding yeast (Huckaba et al., 2006; Gao and Bretscher, 2009). These four possibilities are not mutually exclusive, but further experiments are needed to distinguish them. We cannot exclude the possibility that For3 has additional roles at the division site beyond localizing Cdc12.
The increase of actin filaments in the Cdc12 N-terminal truncations is consistent with these being activated Cdc12 alleles. A Cdc12 fragment missing the C terminus (downstream of the FH2 domain) is not sufficient for viability, but induces the formation of contractile rings during interphase when overexpressed in the presence of native protein (Yonetani and Chang, 2010). These data suggest that the N and C termini of Cdc12 might be involved in an autoinhibitory interaction, but the regulation of such an interaction remains a mystery.
Although formins often have distinct functions (Kovar et al., 2011; Scott et al., 2011), cooperation or redundancy between them is also common. Mouse formins mDia1 and mDia2 have both been implicated in cytokinesis (Tominaga et al., 2000; Watanabe et al., 2008). In budding yeast, both formins Bni1 and Bnr1 localize to the bud neck and are involved in cytokinesis (Vallen et al., 2000). In addition, cells can survive with only one formin (Pruyne et al., 2004; Gao and Bretscher, 2009), suggesting functional redundancy. Most remarkably, budding yeast can polarize and divide when only the FH1FH2 fragment of one formin is expressed in bni1Δ bnrΔ (Gao and Bretscher, 2009). We find some differences in fission yeast, where Cdc12 N-terminal truncations alone are not viable in for3Δ and Cdc12 is essential for cytokinesis, which might reflect the fact that the contractile ring is more important for cytokinesis in cells with a larger cleavage site diameter (Bi et al., 1998; Tolliday et al., 2003; Laporte et al., 2010).
Sources of actin filaments for contractile ring assembly during cytokinesis
One of the outstanding and challenging questions in fungal and animal cytokinesis is the relative importance of the sources of actin filaments for the contractile ring (Zhou and Wang, 2008; Huang et al., 2012). The defect in for3Δ cdc12 truncation double mutants and our analysis of actin dynamics labeled by Lifeact can address this important question. Our data reveal for the first time the effect of mislocalizing Cdc12 but not upstream proteins in node and contractile ring assembly pathways (Almonacid et al., 2011; Laporte et al., 2011; Padmanabhan et al., 2011). These activated alleles of Cdc12 are efficient at nucleating actin filaments throughout the cell (Figs. 3 D and 6 A), but when not anchored properly to the division site, severe cytokinesis defects emerge. These data indicate that actin filaments assembled at nonmedial sites are not efficient for contractile ring assembly even when the cortical transport is normal. It is important to note that the lethality of the for3Δ cdc12 truncation double mutant might not arise solely from the contractile ring assembly defect, because polarity defects, slow growth, and cell lysis were also observed.
An interesting question is whether de novo actin assembly or cortical transport alone is sufficient for contractile ring formation. Cdc12 truncation cells have more actin cables (∼3–7 per cross section) than wt cells (Fig. 3 E) but most cdc12 truncation for3Δ mutants form large aggregates of actin and other ring components instead of a contractile ring (Fig. 5, F and G; and Fig. 6 A). The inefficiency of cortical transport for ring assembly might be a result of the asymmetric distribution of actin cables in contrast to symmetric distribution of cytokinesis nodes, the sites of de novo assembly at the division site. Collectively, eliminating de novo actin assembly in the presence of normal cortical transport during early stages of contractile ring formation results in failure of contractile ring assembly in most cells, suggesting that de novo actin assembly at the division site is more crucial than cortical transport in fission yeast cytokinesis. Indeed, after Cdc12 accumulates at the clump by cortical transport, de novo actin nucleation might also contribute to the successful ring assembly in some cells.
The actin promoter used for the Lifeact constructs is a strong constitutive promoter (Huang et al., 2012). High levels of Lifeact expression in plants altered actin filament dynamics (van der Honing et al., 2011). Although Huang et al. (2012) demonstrated that Lifeact-GFP colocalized with actin staining in the same cells, our data suggest that Lifeact-GFP might promote the formation of aberrant actin structures that inhibit contractile ring assembly (Fig. S4). Using the concentration of actin in fission yeast (63.2 µM; Wu and Pollard, 2005) and the dissociation constants measured for YFP and mYFP dimers (0.11 mM and 9.7 mM, respectively; Zacharias et al., 2002), we estimate that the concentration of Lifeact-GFP dimer may be as high as 12.8 µM, whereas Lifeact-mGFP dimer is only 0.4 µM in the cells with the respective Lifeact constructs. The range of dissociation constants given by different measurements for Lifeact and F-actin is 1.3 to 2.3 µM and for G-actin is 40 to 280 nM (Riedl et al., 2008). The combination of an actin-binding motif and a dimerization motif is a feature shared by actin filament nucleators such as formins and bundlers such as α-actinin. Thus, dimerization of GFP at high concentrations might transform the Lifeact-GFP peptide into an actin filament nucleator or bundler because of its high affinity with actin, which may result in thick actin cable formation throughout the cell.
In conclusion, our results revealed the contributions of different sources of actin to the contractile ring. Both formins Cdc12 and For3 nucleate actin filaments at the division site, with the essential Cdc12 playing the major role. Nonmedial filaments nucleated by activated Cdc12 truncation alleles are unable to efficiently assemble a contractile ring when formin localization to the division site is impaired. Together, these data suggest that de novo actin nucleation at the division site by Cdc12 and For3 is the most important source of actin for contractile ring assembly in fission yeast. It will be interesting to know if de novo actin nucleation at the cleavage furrow is also the predominant source of actin filaments for the contractile ring in other cell types.
Materials and methods
Strains, growing conditions, and genetic, molecular, and cellular methods
Table S1 lists the S. pombe strains used in this study. All tagged genes are integrated at their native chromosomal loci under the control of endogenous promoters unless noted otherwise (Bähler et al., 1998b). Cells were grown in exponential phase for 36–48 h before microscopy as described previously (Wu et al., 2006). Spore germination for cdc12 deletion was performed as described previously (Laporte et al., 2011). In brief, one copy of cdc12 was deleted using the KS-ura4+ marker (Bähler et al., 1998b) in a diploid strain with two copies of the mYFP-myo2. The resulting strain was grown for 2 d on SPA5S to induce sporulation, and then tetrads were incubated 12 h in 2% glusulase at 25°C to kill remaining diploids and release the spores. Spores were washed 5× in 1.7 g/liter of rinsing medium (Yeast Nitrogen Base; BD) and germinated in YE5S-ura liquid medium 12–24 h before imaging. Because fission yeast diploid cells are not stable for long-term storage, only the parental strains are listed in Table S1. Lat-A treatments were performed at a final concentration of 100 µM (Wu et al., 2001) and imaging was done on bare slides to maintain the drug concentration.
The N-terminal truncations of cdc12 at the native locus were made by first cloning the cdc12 promoter (−800 to +3 bp from the ATG) into the KS-ura4 vector at the multicloning sites upstream of the ura4 promoter (Bähler et al., 1998b) to obtain plasmid JQW213. A cdc12 truncation strain was constructed with an mYFP tag at the N terminus under control of the 81nmt1 promoter and selected by G418 resistance (Bähler et al., 1998b). Then the ura4+-Pcdc12 region of plasmid JQW213 was amplified for targeting to the cdc12 locus to replace the kanMX6-P81nmt1-mYFP portion, selected for growth on EMM-uracil, and screened for loss of YFP signal. The resulting ura4+-Pcdc12 truncation strains were then tagged at the C terminus with 3YFP. In some truncation strains, the ura4+ marker was looped out during crosses because it resided between 800 bp of identical cdc12 promoter sequence. Control strains were matched to the same auxotrophic state of the truncations for quantifications. Sequencing of the mutants was done by cloning Cdc12 ORF into plasmids and sequencing the plasmids in both directions for duplicate coverage.
For actin staining, 300 U BODIPY-phallacidin 488 (Molecular Probes) was dissolved in methanol at a final concentration of 0.2 U/µl, and then dispensed in 25-µl aliquots and dried in a vacuum in the dark and stored at −20°C. 1 ml of log phase cells was fixed by adding 333 µl of 16% paraformaldehyde (Electron Microscopy Sciences) at the growing temperature for 5–15 min, washed in 1 ml PEM buffer (0.1 M Na Pipes, pH 6.8, 1 mM EGTA, and 1 mM MgCl2) 3×, permeabilized with 1% Triton X-100 in PEM buffer for 2 min, and washed in 1 ml PEM buffer 3×. Each phallacidin aliquot was resuspended in 50 µl PEM buffer and 10 µl was combined with 10 µl of permeabilized cells. After ∼30 min the stained cells were washed once in 1 ml PEM buffer and then centrifuged at 7,000 rpm for 30 s to collect cells. Most of the supernatant was removed and ∼5 µl of cell suspension was added to a gelatin slide for imaging.
Plasmid constructs, protein purification, and actin assembly assays
wt and mutant FH1FH2 domain constructs (residues 882–1390) were cloned between BamHI and XhoI (wt and Cdc12-112) or BamHI and NotI (Cdc12-299) restriction sites into the bacterial expression vector pET21a, under control of the IPTG-inducible T7 promoter (EMD Millipore; Kovar and Pollard, 2004). Cdc12-112 (F1355S) was cloned by QuikChange site-directed mutagenesis (Agilent Technologies). Cdc12-299 (Δ948–950; F1361S) was cloned by amplification of genomic DNA and Infusion (Takara Bio Inc.). wt and mutant Cdc12(FH1FH2)-HIS proteins were purified as described previously (Kovar and Pollard, 2004; Scott et al., 2011). Constructs were expressed in E. coli strain BL21-Codon Plus (DE3)-RP (Agilent Technologies) with 0.5 mM IPTG for 16 h at 16°C, broken with an EmulsiFlex-C3 (Avestin) in extraction buffer (50 mM NaH2PO4, pH 8.0, 500 mM NaCl, 10% glycerol, 10 mM imidazole, and 10 mM β-mercaptoethanol supplemented with protease inhibitors), clarified by spinning at 30,000 and 50,000 g for 15 and 30 min, incubated with Talon Metal Affinity Resin (Takara Bio Inc.) for 2 h at 4°C, and then loaded onto a disposable column. After a 50-ml wash with extraction buffer, formin was eluted with 250 mM imidazole in extraction buffer. Pure protein was dialyzed with formin buffer (20 mM Hepes, pH 7.4, 1 mM EDTA, 200 mM KCl, 0.01% NaN3, and 1 mM DTT), concentrated, flash frozen in liquid nitrogen, and stored at −80°C. Ca-ATP actin was purified from chicken skeletal muscle acetone powder by extraction with G Buffer (2 mM Tris, pH 8.0, 0.2 mM ATP, 0.1 mM CaCl2, and 0.5 mM DTT) and a cycle of polymerization and depolymerization (Spudich and Watt, 1971), gel filtered on Sephacryl S-300 in G Buffer, and labeled on Cys-374 with pyrenyliodoacetamide (Life Technologies). S. pombe profilin Cdc3 was expressed and purified from bacteria by binding to poly-l-proline Sepharose, elution with 7.0 M urea, and dialysis in profilin buffer (20 mM Tris, 150 mM KCl, and 0.2 mM DTT, pH 7.5; Kovar et al., 2003). The spontaneous assembly of actin was measured from the fluorescence of 10% pyrene-actin with a Safire 2 fluorescence plate reader (Tecan) as described in detail previously (Scott et al., 2011). In brief, pyrene-labeled Mg-ATP-actin monomers were placed in an upper row of a 96-well black plate (Corning). Cdc12, profilin, and 10× KMEI (500 mM KCl, 10 mM MgCl2, 10 mM EGTA, and 100 mM imidazole, pH 7.0) were placed in a lower row. Reactions were initiated by mixing contents of the lower wells with actin monomers in the upper wells.
Microscopy and data analysis
Cells for microscopy were collected from liquid cultures as described previously (Wu et al., 2006; Coffman et al., 2009). In brief, they were centrifuged at 5,000 rpm, washed into EMM5S, placed on a thin layer of EMM5S liquid medium with 20% gelatin (Sigma-Aldrich) and 25 nM n-propyl-gallate, and observed at 23–24°C except where noted. Figs. 4 A and 5 D were taken from single stacks or time-lapse imaging at 23°C in a Delta TPG dish (Bioptechs) with YE5S agar on top of the cells to maintain a single cell layer. For imaging of ts mutants at 36°C, cells were collected and placed on EMM5S + 2% agar pads prewarmed for 10 min at 36°C. Precautions were taken to maintain cells at 36°C using a benchtop incubator (Boekel) and an Objective Heater (Bioptechs; Laporte et al., 2011). Imaging in Figs. 2 C and 4 B was done in a prewarmed Delta TPG dish from a small incubator with prewarmed YE5S agar placed on top of the cells. The dish was transferred to the preheated imaging chamber (Stage Top Incubator [INUB-PPZI2-F1] equipped with dish holder [UNIV2-D350; Tokai Hit) on the microscope. Microscopy at 19°C was done by cooling the entire room to 19°C. Cells were grown at 19°C in a heated water bath in a 4°C cold room and centrifuged in the cold room before imaging.
We imaged cells using 100×/1.4 NA Plan-Apo objective lenses (Nikon) on two microscopes. For most imaging, we used a spinning disk confocal microscope (UltraVIEW ERS; PerkinElmer) with 440, 568-nm solid state lasers and 488, 514-nm argon ion lasers and a cooled charge-coupled device camera (ORCA-AG; Hamamatsu Photonics) on an Eclipse (TE2000-U; Nikon) with 2 × 2 binning. Actin staining was imaged with 1 × 1 binning. Some imaging (Fig. 2, C and D; Figs. 4–6; and Fig. S4, A, B, and D) was performed on another spinning disk confocal microscope (UltraVIEW Vox CSUX1 system; PerkinElmer) with 440-, 488-, 515-, and 561-nm solid state lasers and back-thinned EMCCD camera (Hamamatsu Photonics) on a Ti-E inverted microscope (Nikon) without binning.
Tetrad fluorescence microscopy was performed as follows on a Ti-E inverted microscope equipped with the UltraVIEW Vox spinning disk confocal microscope system. The cross was performed as normal on SPA5S for 2 d, followed by tetrad dissection on YE5S agar plate during which spores are placed 5 mm apart in both X and Y directions. Imaging was done after 2–4 d of colony growth at 25°C and genotypes were determined by cell morphology compared with previous crosses for synthetic interaction. Before imaging, the tetrad dissection needle was used to mark the agar around the double mutant colonies to enable us to find them using the 100× objective lens. For tetrad imaging before even the wt colonies were visible to the naked eye, the plate and the agar were marked at the first and last colony in each tetrad as a guide. A section of the agar with two to four tetrads was then cut out of the tetrad plate and inverted onto a large coverslip (24×60-1.5; Thermo Fisher Scientific) that was clamped into place on an automated stage on the microscope (Nikon). The marks in the agar were used to focus and to assist in finding the first colony. Subsequent colonies were found using the X-Y stage position based on the 5-mm distance of colony positions. Each imaging position was saved in the Volocity software so that they could be revisited for imaging at each time point. Tetrads were imaged with 2–10-min delays, and the autofocus drive in the software was used to ensure that the focus was maintained for ∼12 h of imaging. For viable genotypes, cells at the edges of the colonies were imaged, whereas the entire colony was imaged for the double mutants. Time-lapse images of fluorescence and differential interference contrast were acquired at each time point for each colony. The parent strains are listed in the strain table, because the double mutants could not be isolated and kept. The tetrad fluorescence microscopy we developed here is a promising new approach to determine the cause of cell death in synthetic lethal double mutants by following the progeny of a meiotic event. Using both the tetrad microscope and a sensitive fluorescence microscope, this imaging technique enabled several possible causes of cell death to be distinguished with much greater certainty and resolution than traditional random spore assays.
Maximum-intensity projections and other image analyses were performed using UltraVIEW, Volocity, and/or ImageJ. Images in figures are maximum-intensity projections of z-sections spaced at 0.2–0.8 µm except where noted. Two tagged proteins were considered in the same nodes if their centers were within one radius of the nodes. Images in Fig. 5 H are of 3GFP-tagged truncation instead of 3YFP, and thus the GFP signal is not as brightly visible above the higher autofluorescence at 488 nm. Rotated regions in Fig. S1 C were scaled to 400% in ImageJ using interpolation and resized in Adobe Illustrator to match the scale bar of the maximum intensity projection. In Fig. S4 B, cells with partially condensed actin meshworks at the division site were excluded from our analysis. Statistical analysis was done using Microsoft Excel, and t tests were performed using the “t test” function in Excel, with two tails and type 2 (homoscedastic) or 3 (heteroscedastic) as appropriate. Given numbers (n) in the figures are for each bar/box unless otherwise noted.
Analysis of actin filament movement was done using ImageJ as described previously (Huang et al., 2012) on ∼20-min movies taken with 5-s delay, visualizing only a single slice through the cell. Up to half the cell width was used to make a kymograph along the long axis of the cell, and the angle and the distance traveled by actin filaments that moved toward the division site was measured. The velocity was calculated from these measurements based on the distance traveled and the time between frames. The kymographs in Fig. 6 C cover the entire cell width.
Supplemental videos were made with the QuickTime movie writer plug-in in ImageJ with Video Compression and Best Quality, with key frame every frame (Video 1) or every 5–10 frames. Videos 1 and 7 were scaled to 400% in ImageJ using interpolation. Side-by-side images were automatically adjusted for brightness and contrast the same way so the intensities are comparable for data collected on the same day (Videos 1–4, 6, and 7), whereas the left and right portions of Video 5 were taken on different days and might not be comparable.
Counting numbers of molecules in whole cells and at the division site
Cells were imaged at 0.4-µm z spacing, the full width at half maximum of the point spread function on our system (Coffman et al., 2011; Laporte et al., 2011; Coffman and Wu, 2012), and measurements were taken using the sum intensity of all sections with signal (Wu and Pollard, 2005; Wu et al., 2008). wt cells in the same field were measured the same way and intensity was subtracted to remove autofluorescence and background from the measurement. Global intensity of Cdc12 truncations tagged with 3YFP was compared with global intensity of Cdc12-3YFP, setting Cdc12 to 600 molecules per cell on average and then adjusting for cell size (Wu and Pollard, 2005). The division site intensity was measured using a rectangle region of interest over the ring area and a 2× larger rectangle along the cell’s long axis to determine the background (Wu et al., 2008).
Online supplemental material
Table S1 lists the S. pombe strains used in this study. Fig. S1 shows contractile ring formation in arp3-c1 for3Δ cells at 19°C. Fig. S2 shows For3 localization to the division site independent of the cytokinesis and polarity proteins. Fig. S3 shows Cdc12 truncations and For3 expression levels and For3 localization in cdc12 truncation strains. Fig. S4 shows the characterization of Lifeact-mGFP and Lifeact-GFP. Video 1 shows contractile ring assembly from a band of Cdc12 nodes or node-like structures in wt and Δ503-cdc12. Video 2 shows contractile ring assembly from a broad band of Rlc1-3GFP nodes in wt and cdc12 truncations. Video 3 shows tetrad fluorescence microscopy from a cross between for3Δ and Δ503-cdc12 cells expressing CRIB-3GFP and mRFP-Atb2 as markers for active Cdc42 and microtubules (MTs). Video 4 shows tetrad fluorescence microscopy from a cross between for3Δ and Δ503-cdc12 cells expressing Rlc1-tdTomato as a marker for cytokinesis nodes and the contractile ring. Video 5 shows the visualization of Lifeact-GFP and Lifeact-mGFP during contractile ring assembly. Video 6 shows Lifeact-mGFP in tetrad fluorescence microscopy from a cross between for3Δ and Δ503-cdc12. Video 7 shows actin filaments moving toward the division site in cells attempting cell division. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201305022/DC1.
Supplementary Material
Acknowledgments
We thank I-Ju Lee, Stephen Osmani, Paul Herman, and Michael Ostrowski for critical reading of the manuscript; Damien Laporte for some data collection; and Mohan Balasubramanian, Fred Chang, Kathleen Gould, Sophie Martin, Dannel McCollum, Jürg Bähler, Matt Lord, and Kaz Shiozaki for strains.
This work is supported by a predoctoral fellowship from the American Heart Association and Elizabeth Clay Howald Presidential Fellowship to V.C. Coffman and the National Institutes of Health grants R01GM079265 to D.R. Kovar and R01GM086546 to J.-Q. Wu.
Footnotes
Abbreviations used in this paper:
- FH
- formin homology
- fps
- frames per second
- Lat-A
- Latrunculin A
- MT
- microtubule
- SCPR
- search, capture, pull, and release
- SPB
- spindle pole body
- ts
- temperature-sensitive
- wt
- wild type
References
- Almonacid M., Moseley J.B., Janvore J., Mayeux A., Fraisier V., Nurse P., Paoletti A. 2009. Spatial control of cytokinesis by Cdr2 kinase and Mid1/anillin nuclear export. Curr. Biol. 19:961–966 10.1016/j.cub.2009.04.024 [DOI] [PubMed] [Google Scholar]
- Almonacid M., Celton-Morizur S., Jakubowski J.L., Dingli F., Loew D., Mayeux A., Chen J.S., Gould K.L., Clifford D.M., Paoletti A. 2011. Temporal control of contractile ring assembly by Plo1 regulation of myosin II recruitment by Mid1/anillin. Curr. Biol. 21:473–479 10.1016/j.cub.2011.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alsop G.B., Chen W., Foss M., Tseng K.-F., Zhang D. 2009. Redistribution of actin during assembly and reassembly of the contractile ring in grasshopper spermatocytes. PLoS ONE. 4:e4892 10.1371/journal.pone.0004892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arai R., Mabuchi I. 2002. F-actin ring formation and the role of F-actin cables in the fission yeast Schizosaccharomyces pombe. J. Cell Sci. 115:887–898 [DOI] [PubMed] [Google Scholar]
- Bähler J., Steever A.B., Wheatley S., Wang Y.-L., Pringle J.R., Gould K.L., McCollum D. 1998a. Role of polo kinase and Mid1p in determining the site of cell division in fission yeast. J. Cell Biol. 143:1603–1616 10.1083/jcb.143.6.1603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bähler J., Wu J.-Q., Longtine M.S., Shah N.G., McKenzie A., III, Steever A.B., Wach A., Philippsen P., Pringle J.R. 1998b. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast. 14:943–951 [DOI] [PubMed] [Google Scholar]
- Balasubramanian M.K., Bi E., Glotzer M. 2004. Comparative analysis of cytokinesis in budding yeast, fission yeast and animal cells. Curr. Biol. 14:R806–R818 10.1016/j.cub.2004.09.022 [DOI] [PubMed] [Google Scholar]
- Barr F.A., Gruneberg U. 2007. Cytokinesis: placing and making the final cut. Cell. 131:847–860 10.1016/j.cell.2007.11.011 [DOI] [PubMed] [Google Scholar]
- Bi E., Maddox P., Lew D.J., Salmon E.D., McMillan J.N., Yeh E., Pringle J.R. 1998. Involvement of an actomyosin contractile ring in Saccharomyces cerevisiae cytokinesis. J. Cell Biol. 142:1301–1312 10.1083/jcb.142.5.1301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray D., White J.G. 1988. Cortical flow in animal cells. Science. 239:883–888 10.1126/science.3277283 [DOI] [PubMed] [Google Scholar]
- Buttery S.M., Yoshida S., Pellman D. 2007. Yeast formins Bni1 and Bnr1 utilize different modes of cortical interaction during the assembly of actin cables. Mol. Biol. Cell. 18:1826–1838 10.1091/mbc.E06-09-0820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buttery S.M., Kono K., Stokasimov E., Pellman D. 2012. Regulation of the formin Bnr1 by septins anda MARK/Par1-family septin-associated kinase. Mol. Biol. Cell. 23:4041–4053 10.1091/mbc.E12-05-0395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao L.G., Wang Y.L. 1990. Mechanism of the formation of contractile ring in dividing cultured animal cells. I. Recruitment of preexisting actin filaments into the cleavage furrow. J. Cell Biol. 110:1089–1095 10.1083/jcb.110.4.1089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carnahan R.H., Gould K.L. 2003. The PCH family protein, Cdc15p, recruits two F-actin nucleation pathways to coordinate cytokinetic actin ring formation in Schizosaccharomyces pombe. J. Cell Biol. 162:851–862 10.1083/jcb.200305012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castrillon D.H., Wasserman S.A. 1994. Diaphanous is required for cytokinesis in Drosophila and shares domains of similarity with the products of the limb deformity gene. Development. 120:3367–3377 [DOI] [PubMed] [Google Scholar]
- Celton-Morizur S., Bordes N., Fraisier V., Tran P.T., Paoletti A. 2004. C-terminal anchoring of mid1p to membranes stabilizes cytokinetic ring position in early mitosis in fission yeast. Mol. Cell. Biol. 24:10621–10635 10.1128/MCB.24.24.10621-10635.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang F., Woollard A., Nurse P. 1996. Isolation and characterization of fission yeast mutants defective in the assembly and placement of the contractile actin ring. J. Cell Sci. 109:131–142 [DOI] [PubMed] [Google Scholar]
- Chang F., Drubin D., Nurse P. 1997. cdc12p, a protein required for cytokinesis in fission yeast, is a component of the cell division ring and interacts with profilin. J. Cell Biol. 137:169–182 10.1083/jcb.137.1.169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W., Foss M., Tseng K.-F., Zhang D. 2008. Redundant mechanisms recruit actin into the contractile ring in silkworm spermatocytes. PLoS Biol. 6:e209 10.1371/journal.pbio.0060209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Kuo C.-C., Kang H., Howell A.S., Zyla T.R., Jin M., Lew D.J. 2012. Cdc42p regulation of the yeast formin Bni1p mediated by the effector Gic2p. Mol. Biol. Cell. 23:3814–3826 10.1091/mbc.E12-05-0400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q., Nag S., Pollard T.D. 2012. Formins filter modified actin subunits during processive elongation. J. Struct. Biol. 177:32–39 10.1016/j.jsb.2011.10.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesarone M., Gould C.J., Moseley J.B., Goode B.L. 2009. Displacement of formins from growing barbed ends by bud14 is critical for actin cable architecture and function. Dev. Cell. 16:292–302 10.1016/j.devcel.2008.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffman V.C., Wu J.-Q. 2012. Counting protein molecules using quantitative fluorescence microscopy. Trends Biochem. Sci. 37:499–506 10.1016/j.tibs.2012.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffman V.C., Nile A.H., Lee I.-J., Liu H., Wu J.-Q. 2009. Roles of formin nodes and myosin motor activity in Mid1p-dependent contractile-ring assembly during fission yeast cytokinesis. Mol. Biol. Cell. 20:5195–5210 10.1091/mbc.E09-05-0428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffman V.C., Wu P., Parthun M.R., Wu J.-Q. 2011. CENP-A exceeds microtubule attachment sites in centromere clusters of both budding and fission yeast. J. Cell Biol. 195:563–572 10.1083/jcb.201106078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courtemanche N., Pollard T.D. 2012. Determinants of Formin Homology 1 (FH1) domain function in actin filament elongation by formins. J. Biol. Chem. 287:7812–7820 10.1074/jbc.M111.322958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenmann K.M., Harris E.S., Kitchen S.M., Holman H.A., Higgs H.N., Alberts A.S. 2007. Dia-interacting protein modulates formin-mediated actin assembly at the cell cortex. Curr. Biol. 17:579–591 10.1016/j.cub.2007.03.024 [DOI] [PubMed] [Google Scholar]
- Evangelista M., Pruyne D., Amberg D.C., Boone C., Bretscher A. 2002. Formins direct Arp2/3-independent actin filament assembly to polarize cell growth in yeast. Nat. Cell Biol. 4:260–269 10.1038/ncb718 [DOI] [PubMed] [Google Scholar]
- Feierbach B., Chang F. 2001a. Cytokinesis and the contractile ring in fission yeast. Curr. Opin. Microbiol. 4:713–719 10.1016/S1369-5274(01)00273-9 [DOI] [PubMed] [Google Scholar]
- Feierbach B., Chang F. 2001b. Roles of the fission yeast formin for3p in cell polarity, actin cable formation and symmetric cell division. Curr. Biol. 11:1656–1665 10.1016/S0960-9822(01)00525-5 [DOI] [PubMed] [Google Scholar]
- Gao L., Bretscher A. 2009. Polarized growth in budding yeast in the absence of a localized formin. Mol. Biol. Cell. 20:2540–2548 10.1091/mbc.E09-03-0194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorelik R., Yang C., Kameswaran V., Dominguez R., Svitkina T. 2011. Mechanisms of plasma membrane targeting of formin mDia2 through its amino terminal domains. Mol. Biol. Cell. 22:189–201 10.1091/mbc.E10-03-0256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hachet O., Simanis V. 2008. Mid1p/anillin and the septation initiation network orchestrate contractile ring assembly for cytokinesis. Genes Dev. 22:3205–3216 10.1101/gad.1697208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y., Yan H., Balasubramanian M.K. 2008. Assembly of normal actomyosin rings in the absence of Mid1p and cortical nodes in fission yeast. J. Cell Biol. 183:979–988 10.1083/jcb.200806151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J., Huang Y., Yu H., Subramanian D., Padmanabhan A., Thadani R., Tao Y., Tang X., Wedlich-Soldner R., Balasubramanian M.K. 2012. Nonmedially assembled F-actin cables incorporate into the actomyosin ring in fission yeast. J. Cell Biol. 199:831–847 10.1083/jcb.201209044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huckaba T.M., Lipkin T., Pon L.A. 2006. Roles of type II myosin and a tropomyosin isoform in retrograde actin flow in budding yeast. J. Cell Biol. 175:957–969 10.1083/jcb.200609155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamasaki T., Arai R., Osumi M., Mabuchi I. 2005. Directionality of F-actin cables changes during the fission yeast cell cycle. Nat. Cell Biol. 7:916–917 10.1038/ncb1295 [DOI] [PubMed] [Google Scholar]
- Karpova T.S., McNally J.G., Moltz S.L., Cooper J.A. 1998. Assembly and function of the actin cytoskeleton of yeast: relationships between cables and patches. J. Cell Biol. 142:1501–1517 10.1083/jcb.142.6.1501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovar D.R. 2006. Molecular details of formin-mediated actin assembly. Curr. Opin. Cell Biol. 18:11–17 10.1016/j.ceb.2005.12.011 [DOI] [PubMed] [Google Scholar]
- Kovar D.R., Pollard T.D. 2004. Insertional assembly of actin filament barbed ends in association with formins produces piconewton forces. Proc. Natl. Acad. Sci. USA. 101:14725–14730 10.1073/pnas.0405902101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovar D.R., Kuhn J.R., Tichy A.L., Pollard T.D. 2003. The fission yeast cytokinesis formin Cdc12p is a barbed end actin filament capping protein gated by profilin. J. Cell Biol. 161:875–887 10.1083/jcb.200211078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovar D.R., Sirotkin V., Lord M. 2011. Three’s company: the fission yeast actin cytoskeleton. Trends Cell Biol. 21:177–187 10.1016/j.tcb.2010.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laporte D., Zhao R., Wu J.-Q. 2010. Mechanisms of contractile-ring assembly in fission yeast and beyond. Semin. Cell Dev. Biol. 21:892–898 10.1016/j.semcdb.2010.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laporte D., Coffman V.C., Lee I.-J., Wu J.-Q. 2011. Assembly and architecture of precursor nodes during fission yeast cytokinesis. J. Cell Biol. 192:1005–1021 10.1083/jcb.201008171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee I.-J., Wu J.-Q. 2012. Characterization of Mid1 domains for targeting and scaffolding in fission yeast cytokinesis. J. Cell Sci. 125:2973–2985 10.1242/jcs.102574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee E., Shelden E.A., Knecht D.A. 1998. Formation of F-actin aggregates in cells treated with actin stabilizing drugs. Cell Motil. Cytoskeleton. 39:122–133 [DOI] [PubMed] [Google Scholar]
- Lee I.-J., Coffman V.C., Wu J.-Q. 2012. Contractile-ring assembly in fission yeast cytokinesis: Recent advances and new perspectives. Cytoskeleton (Hoboken). 69:751–763 10.1002/cm.21052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F., Higgs H.N. 2003. The mouse Formin mDia1 is a potent actin nucleation factor regulated by autoinhibition. Curr. Biol. 13:1335–1340 10.1016/S0960-9822(03)00540-2 [DOI] [PubMed] [Google Scholar]
- Liu W., Santiago-Tirado F.H., Bretscher A. 2012. Yeast formin Bni1p has multiple localization regions that function in polarized growth and spindle orientation. Mol. Biol. Cell. 23:412–422 10.1091/mbc.E11-07-0631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiti S., Michelot A., Gould C., Blanchoin L., Sokolova O., Goode B.L. 2012. Structure and activity of full-length formin mDia1. Cytoskeleton (Hoboken). 69:393–405 10.1002/cm.21033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin S.G., Chang F. 2006. Dynamics of the formin for3p in actin cable assembly. Curr. Biol. 16:1161–1170 10.1016/j.cub.2006.04.040 [DOI] [PubMed] [Google Scholar]
- Martin S.G., Rincón S.A., Basu R., Pérez P., Chang F. 2007. Regulation of the formin for3p by cdc42p and bud6p. Mol. Biol. Cell. 18:4155–4167 10.1091/mbc.E07-02-0094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moseley J.B., Sagot I., Manning A.L., Xu Y., Eck M.J., Pellman D., Goode B.L. 2004. A conserved mechanism for Bni1- and mDia1-induced actin assembly and dual regulation of Bni1 by Bud6 and profilin. Mol. Biol. Cell. 15:896–907 10.1091/mbc.E03-08-0621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neidt E.M., Scott B.J., Kovar D.R. 2009. Formin differentially utilizes profilin isoforms to rapidly assemble actin filaments. J. Biol. Chem. 284:673–684 10.1074/jbc.M804201200 [DOI] [PubMed] [Google Scholar]
- Ojkic N., Vavylonis D. 2010. Kinetics of myosin node aggregation into a contractile ring. Phys. Rev. Lett. 105:048102 10.1103/PhysRevLett.105.048102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padmanabhan A., Bakka K., Sevugan M., Naqvi N.I., D’souza V., Tang X., Mishra M., Balasubramanian M.K. 2011. IQGAP-related Rng2p organizes cortical nodes and ensures position of cell division in fission yeast. Curr. Biol. 21:467–472 10.1016/j.cub.2011.01.059 [DOI] [PubMed] [Google Scholar]
- Paoletti A., Chang F. 2000. Analysis of mid1p, a protein required for placement of the cell division site, reveals a link between the nucleus and the cell surface in fission yeast. Mol. Biol. Cell. 11:2757–2773 10.1091/mbc.11.8.2757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelham R.J., Chang F. 2002. Actin dynamics in the contractile ring during cytokinesis in fission yeast. Nature. 419:82–86 10.1038/nature00999 [DOI] [PubMed] [Google Scholar]
- Petersen J., Nielsen O., Egel R., Hagan I.M. 1998. FH3, a domain found in formins, targets the fission yeast formin Fus1 to the projection tip during conjugation. J. Cell Biol. 141:1217–1228 10.1083/jcb.141.5.1217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard T.D., Wu J.-Q. 2010. Understanding cytokinesis: lessons from fission yeast. Nat. Rev. Mol. Cell Biol. 11:149–155 10.1038/nrm2834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pring M., Evangelista M., Boone C., Yang C., Zigmond S.H. 2003. Mechanism of formin-induced nucleation of actin filaments. Biochemistry. 42:486–496 10.1021/bi026520j [DOI] [PubMed] [Google Scholar]
- Pruyne D., Evangelista M., Yang C., Bi E., Zigmond S., Bretscher A., Boone C. 2002. Role of formins in actin assembly: nucleation and barbed-end association. Science. 297:612–615 10.1126/science.1072309 [DOI] [PubMed] [Google Scholar]
- Pruyne D., Gao L., Bi E., Bretscher A. 2004. Stable and dynamic axes of polarity use distinct formin isoforms in budding yeast. Mol. Biol. Cell. 15:4971–4989 10.1091/mbc.E04-04-0296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan M.E., Hilgert S., Bedrossian A., Mullins R.D., Kerkhoff E. 2007. Regulatory interactions between two actin nucleators, Spire and Cappuccino. J. Cell Biol. 179:117–128 10.1083/jcb.200706196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramalingam N., Zhao H., Breitsprecher D., Lappalainen P., Faix J., Schleicher M. 2010. Phospholipids regulate localization and activity of mDia1 formin. Eur. J. Cell Biol. 89:723–732 10.1016/j.ejcb.2010.06.001 [DOI] [PubMed] [Google Scholar]
- Riedl J., Crevenna A.H., Kessenbrock K., Yu J.H., Neukirchen D., Bista M., Bradke F., Jenne D., Holak T.A., Werb Z., et al. 2008. Lifeact: a versatile marker to visualize F-actin. Nat. Methods. 5:605–607 10.1038/nmeth.1220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts-Galbraith R.H., Gould K.L. 2008. Stepping into the ring: the SIN takes on contractile ring assembly. Genes Dev. 22:3082–3088 10.1101/gad.1748908 [DOI] [PubMed] [Google Scholar]
- Romero S., Le Clainche C., Didry D., Egile C., Pantaloni D., Carlier M.-F. 2004. Formin is a processive motor that requires profilin to accelerate actin assembly and associated ATP hydrolysis. Cell. 119:419–429 10.1016/j.cell.2004.09.039 [DOI] [PubMed] [Google Scholar]
- Sagot I., Klee S.K., Pellman D. 2002a. Yeast formins regulate cell polarity by controlling the assembly of actin cables. Nat. Cell Biol. 4:42–50 [DOI] [PubMed] [Google Scholar]
- Sagot I., Rodal A.A., Moseley J., Goode B.L., Pellman D. 2002b. An actin nucleation mechanism mediated by Bni1 and profilin. Nat. Cell Biol. 4:626–631 [DOI] [PubMed] [Google Scholar]
- Saha S., Pollard T.D. 2012. Anillin-related protein Mid1p coordinates the assembly of the cytokinetic contractile ring in fission yeast. Mol. Biol. Cell. 23:3982–3992 10.1091/mbc.E12-07-0535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott B.J., Neidt E.M., Kovar D.R. 2011. The functionally distinct fission yeast formins have specific actin-assembly properties. Mol. Biol. Cell. 22:3826–3839 10.1091/mbc.E11-06-0492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohrmann M., Fankhauser C., Brodbeck C., Simanis V. 1996. The dmf1/mid1 gene is essential for correct positioning of the division septum in fission yeast. Genes Dev. 10:2707–2719 10.1101/gad.10.21.2707 [DOI] [PubMed] [Google Scholar]
- Spudich J.A., Watt S. 1971. The regulation of rabbit skeletal muscle contraction. I. Biochemical studies of the interaction of the tropomyosin-troponin complex with actin and the proteolytic fragments of myosin. J. Biol. Chem. 246:4866–4871 [PubMed] [Google Scholar]
- Subramanian D., Huang J., Sevugan M., Robinson R.C., Balasubramanian M.K., Tang X. 2013. Insight into actin organization and function in cytokinesis from analysis of fission yeast mutants. Genetics. 194:435–446 10.1534/genetics.113.149716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeya R., Taniguchi K., Narumiya S., Sumimoto H. 2008. The mammalian formin FHOD1 is activated through phosphorylation by ROCK and mediates thrombin-induced stress fibre formation in endothelial cells. EMBO J. 27:618–628 10.1038/emboj.2008.7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatebe H., Nakano K., Maximo R., Shiozaki K. 2008. Pom1 DYRK regulates localization of the Rga4 GAP to ensure bipolar activation of Cdc42 in fission yeast. Curr. Biol. 18:322–330 10.1016/j.cub.2008.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolliday N., Pitcher M., Li R. 2003. Direct evidence for a critical role of myosin II in budding yeast cytokinesis and the evolvability of new cytokinetic mechanisms in the absence of myosin II. Mol. Biol. Cell. 14:798–809 10.1091/mbc.E02-09-0558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tominaga T., Sahai E., Chardin P., McCormick F., Courtneidge S.A., Alberts A.S. 2000. Diaphanous-related formins bridge Rho GTPase and Src tyrosine kinase signaling. Mol. Cell. 5:13–25 10.1016/S1097-2765(00)80399-8 [DOI] [PubMed] [Google Scholar]
- Vallen E.A., Caviston J., Bi E. 2000. Roles of Hof1p, Bni1p, Bnr1p, and Myo1p in cytokinesis in Saccharomyces cerevisiae. Mol. Biol. Cell. 11:593–611 10.1091/mbc.11.2.593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Honing H.S., van Bezouwen L.S., Emons A.M., Ketelaar T. 2011. High expression of Lifeact in Arabidopsis thaliana reduces dynamic reorganization of actin filaments but does not affect plant development. Cytoskeleton (Hoboken). 68:578–587 10.1002/cm.20534 [DOI] [PubMed] [Google Scholar]
- Vavylonis D., Kovar D.R., O’Shaughnessy B., Pollard T.D. 2006. Model of formin-associated actin filament elongation. Mol. Cell. 21:455–466 10.1016/j.molcel.2006.01.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vavylonis D., Wu J.-Q., Hao S., O’Shaughnessy B., Pollard T.D. 2008. Assembly mechanism of the contractile ring for cytokinesis by fission yeast. Science. 319:97–100 10.1126/science.1151086 [DOI] [PubMed] [Google Scholar]
- Wang H., Vavylonis D. 2008. Model of For3p-mediated actin cable assembly in fission yeast. PLoS ONE. 3:e4078 10.1371/journal.pone.0004078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Neo S.P., Cai M. 2009. Regulation of the yeast formin Bni1p by the actin-regulating kinase Prk1p. Traffic. 10:528–535 10.1111/j.1600-0854.2009.00893.x [DOI] [PubMed] [Google Scholar]
- Wasserman S. 1998. FH proteins as cytoskeletal organizers. Trends Cell Biol. 8:111–115 10.1016/S0962-8924(97)01217-8 [DOI] [PubMed] [Google Scholar]
- Watanabe S., Ando Y., Yasuda S., Hosoya H., Watanabe N., Ishizaki T., Narumiya S. 2008. mDia2 induces the actin scaffold for the contractile ring and stabilizes its position during cytokinesis in NIH 3T3 cells. Mol. Biol. Cell. 19:2328–2338 10.1091/mbc.E07-10-1086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White J.G., Borisy G.G. 1983. On the mechanisms of cytokinesis in animal cells. J. Theor. Biol. 101:289–316 10.1016/0022-5193(83)90342-9 [DOI] [PubMed] [Google Scholar]
- Wu J.-Q., Pollard T.D. 2005. Counting cytokinesis proteins globally and locally in fission yeast. Science. 310:310–314 10.1126/science.1113230 [DOI] [PubMed] [Google Scholar]
- Wu J.-Q., Bähler J., Pringle J.R. 2001. Roles of a fimbrin and an α-actinin-like protein in fission yeast cell polarization and cytokinesis. Mol. Biol. Cell. 12:1061–1077 10.1091/mbc.12.4.1061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J.-Q., Kuhn J.R., Kovar D.R., Pollard T.D. 2003. Spatial and temporal pathway for assembly and constriction of the contractile ring in fission yeast cytokinesis. Dev. Cell. 5:723–734 10.1016/S1534-5807(03)00324-1 [DOI] [PubMed] [Google Scholar]
- Wu J.-Q., Sirotkin V., Kovar D.R., Lord M., Beltzner C.C., Kuhn J.R., Pollard T.D. 2006. Assembly of the cytokinetic contractile ring from a broad band of nodes in fission yeast. J. Cell Biol. 174:391–402 10.1083/jcb.200602032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J.-Q., McCormick C.D., Pollard T.D. 2008. Chapter 9: Counting proteins in living cells by quantitative fluorescence microscopy with internal standards. Methods Cell Biol. 89:253–273 10.1016/S0091-679X(08)00609-2 [DOI] [PubMed] [Google Scholar]
- Xu Y., Moseley J.B., Sagot I., Poy F., Pellman D., Goode B.L., Eck M.J. 2004. Crystal structures of a Formin Homology-2 domain reveal a tethered dimer architecture. Cell. 116:711–723 10.1016/S0092-8674(04)00210-7 [DOI] [PubMed] [Google Scholar]
- Yang F., Moss L.G., Phillips G.N., Jr 1996. The molecular structure of green fluorescent protein. Nat. Biotechnol. 14:1246–1251 10.1038/nbt1096-1246 [DOI] [PubMed] [Google Scholar]
- Yonetani A., Chang F. 2010. Regulation of cytokinesis by the formin cdc12p. Curr. Biol. 20:561–566 10.1016/j.cub.2010.01.061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonetani A., Lustig R.J., Moseley J.B., Takeda T., Goode B.L., Chang F. 2008. Regulation and targeting of the fission yeast formin cdc12p in cytokinesis. Mol. Biol. Cell. 19:2208–2219 10.1091/mbc.E07-07-0731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zacharias D.A., Violin J.D., Newton A.C., Tsien R.Y. 2002. Partitioning of lipid-modified monomeric GFPs into membrane microdomains of live cells. Science. 296:913–916 10.1126/science.1068539 [DOI] [PubMed] [Google Scholar]
- Zhou M., Wang Y.-L. 2008. Distinct pathways for the early recruitment of myosin II and actin to the cytokinetic furrow. Mol. Biol. Cell. 19:318–326 10.1091/mbc.E07-08-0783 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.