MAD2L2 is rapidly degraded by APC/CCDC20 at the onset of anaphase, allowing release of sequestered CDH1 to activate the dephosphorylated APC/C.
Abstract
The switch from activation of the anaphase-promoting complex/cyclosome (APC/C) by CDC20 to CDH1 during anaphase is crucial for accurate mitosis. APC/CCDC20 ubiquitinates a limited set of substrates for subsequent degradation, including Cyclin B1 and Securin, whereas APC/CCDH1 has a broader specificity. This switch depends on dephosphorylation of CDH1 and the APC/C, and on the degradation of CDC20. Here we show, in human cells, that the APC/C inhibitor MAD2L2 also contributes to ensuring the sequential activation of the APC/C by CDC20 and CDH1. In prometaphase, MAD2L2 sequestered free CDH1 away from the APC/C. At the onset of anaphase, MAD2L2 was rapidly degraded by APC/CCDC20, releasing CDH1 to activate the dephosphorylated APC/C. Loss of MAD2L2 led to premature association of CDH1 with the APC/C, early destruction of APC/CCDH1 substrates, and accelerated mitosis with frequent mitotic aberrations. Thus, MAD2L2 helps to ensure a robustly bistable switch between APC/CCDC20 and APC/CCDH1 during the metaphase-to-anaphase transition, thereby contributing to mitotic fidelity.
Introduction
Ubiquitination of key mitotic regulatory proteins by the anaphase-promoting complex/cyclosome (APC/C) and their subsequent degradation by the proteasome is crucial for the correct progression of cells through mitosis. The APC/C is a large multi-subunit ubiquitin ligase whose activity is controlled and directed by two activator subunits, CDC20 and CDH1 (Sullivan and Morgan, 2007; Pines, 2011). The APC/C is activated sequentially, first by CDC20 and then by CDH1. This switch of activators broadens the substrate specificity of the APC/C and consequently regulates the timing of substrate degradation (Hagting et al., 2002; Lindon and Pines, 2004).
APC/CCDC20 has a limited set of substrates, most notably Securin and Cyclin B1, whose rapid degradation is essential for the initiation of anaphase (Cohen-Fix et al., 1996; Lim et al., 1998; Shirayama et al., 1999). Securin inhibits the protease Separase, hence degradation of Securin permits Separase to cleave the cohesin rings that hold the sister chromatids together at metaphase (Ciosk et al., 1998). Proteolysis of Cyclin B1 inactivates the main mitotic kinase CDK1, favoring the dephosphorylation of Cyclin B1–CDK1 substrates necessary for exit from mitosis (Visintin et al., 1998; Wäsch and Cross, 2002). CDC20 first associates with the APC/C during prometaphase, promoted by the phosphorylation of the APC/C, mainly by CDK1 (Shteinberg et al., 1999; Kramer et al., 2000). However, APC/CCDC20 is kept inactive by the spindle assembly checkpoint (SAC), which monitors the attachment of spindle microtubules to kinetochores. The SAC is mediated by the mitotic checkpoint complex, made up of MAD2, MAD3 (BUBR1), and BUB3 (Musacchio and Salmon, 2007), which blocks the recognition of the degron motifs on substrates of APC/CCDC20 (Chao et al., 2012). When the SAC is satisfied, APC/CCDC20 is activated by the dissociation of MAD2 and anaphase is initiated.
Once cells enter anaphase, the switch from CDC20 to CDH1 is driven by degradation of Cyclin B1. In early mitosis, phosphorylation of CDH1 inhibits its association with APC/C (Zachariae et al., 1998; Jaspersen et al., 1999; Kramer et al., 2000). As CDK1 activity declines during anaphase, APC/C and CDH1 are dephosphorylated. Further CDC20 is degraded, promoted initially by APC/CCDC20 autoubiquitination (Foe et al., 2011) and then by APC/CCDH1 (Prinz et al., 1998; Shirayama et al., 1998; Pfleger and Kirschner, 2000). Together, these events lead to a switch from activation of the APC/C by CDC20 to CDH1. APC/CCDH1 is responsible for the destruction of a wide range of mitotic regulators including the kinases Aurora A (AURKA), Aurora B (AURKB), and polo-like kinase (PLK1), the ordered proteolysis of which is important for proper mitotic exit (Zur and Brandeis, 2001; Lindon and Pines, 2004; Stewart and Fang, 2005).
Inhibitor proteins also help regulate the activity of APC/CCDH1 at different points in the cell cycle. APC/CCDH1 remains active during G1, which restrains the levels of mitotic Cyclin/CDK complexes (Pines, 2011). To allow entry into S phase, APC/CCDH1 is deactivated by the binding of EMI1 (Reimann et al., 2001; Hsu et al., 2002), phosphorylation by Cyclin E/Cyclin A–CDK complexes (Reber et al., 2006) and by JNK (Gutierrez et al., 2010), and by its own degradation (Listovsky et al., 2004). In budding yeast, a second inhibitor, Acm1, provides additional restriction of APC/CCDH1 activity in prometaphase by acting as a pseudosubstrate inhibitor until the onset of anaphase when it is ubiquitinated by APC/CCDC20 and degraded (Enquist-Newman et al., 2008).
MAD2L2 was first identified as the small subunit of DNA polymerase ζ, an enzyme involved in the replication of damaged DNA, and named REV7 (Lawrence et al., 1985; Nelson et al., 1996). It is also a paralogue of the mitotic checkpoint complex component MAD2. Although not a component of the SAC (Chen and Fang, 2001), MAD2L2 has been shown to be an APC/C inhibitor in vitro (Chen and Fang, 2001; Pfleger et al., 2001). However, there exists uncertainty about its precise mechanism, in particular whether it inhibits solely APC/CCDH1 or both APC/CCDH1 and APC/CCDC20 and whether it acts by binding to and inhibiting the APC/C itself or whether it inhibits the activator proteins off the APC/C. Further, the in vivo significance of its role as an APC/C inhibitor has not been explored in detail.
Here we show in human cells that MAD2L2 ensures accurate mitosis by preventing premature activation of APC/CCDH1 during prometaphase and early anaphase. Loss of MAD2L2 results in dysregulation of APC/C substrate degradation and unscheduled mitotic exit. During normal mitosis, the CDH1-inhibitory effect of MAD2L2 is rapidly overcome at anaphase by APC/CCDC20-mediated degradation of MAD2L2. Thus, both the sequestration of CDH1 by MAD2L2 and the rapid degradation of MAD2L2 at anaphase help ensure a robustly stable switch between activation of the APC/C by CDC20 and then by CDH1.
Results
Loss of MAD2L2 results rapid and unfaithful mitosis
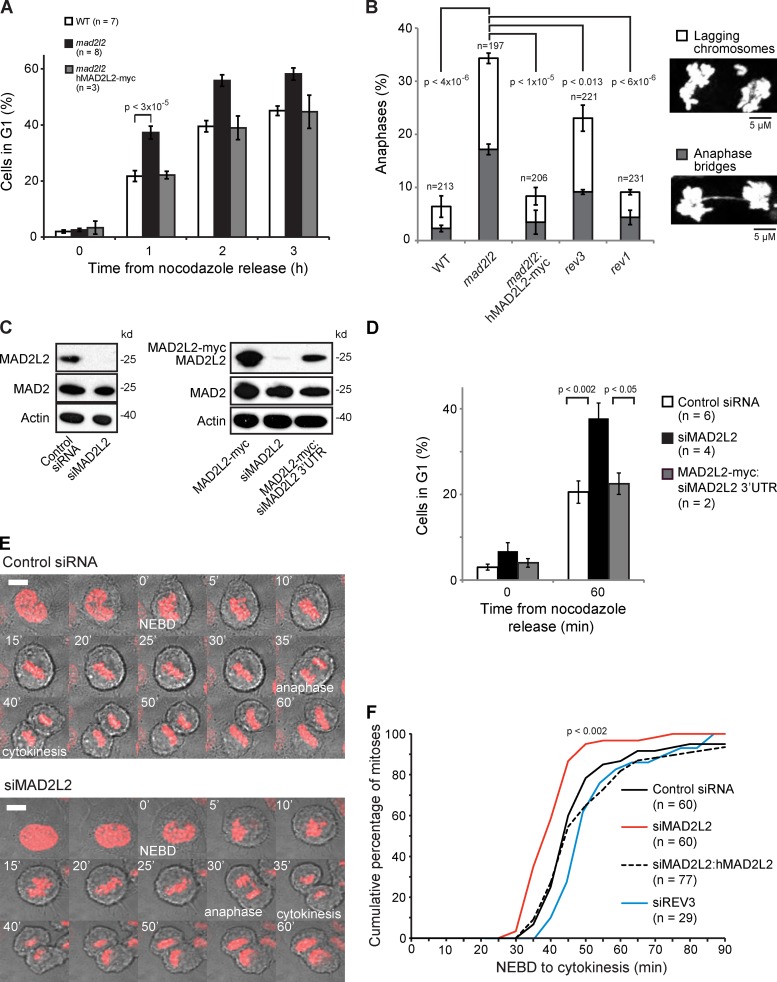
We initially observed that MAD2L2-deficient chicken DT40 cells progressed more rapidly to G1 than wild-type controls when released from nocodazole, a microtubule poison that arrests cells in prometaphase. This effect was reversed by ectopic expression of human MAD2L2 (Fig. 1 A). MAD2L2-deficient DT40 also exhibited a high rate of mitotic aberrations, most notably lagging chromosomes and anaphase bridges (Fig. 1 B). Such mitotic aberrations can result from problems with DNA replication and repair or from defects in mitotic regulation (Fenech et al., 2011). MAD2L2 has an established role as the REV7 subunit of DNA polymerase ζ, an enzyme involved in both translesion synthesis and homologous recombination (Sale, 2013). Hence, we next examined the frequency of mitotic aberrations in isogenic DT40 cells lacking REV3 (the catalytic subunit of Pol ζ) and REV1, a key regulator of translesion synthesis, which interacts with Pol ζ (Sale, 2013). The frequency of aberrations was significantly higher in MAD2L2-deficient cells than either rev1 or rev3 cells, suggesting that defective Pol ζ function may not explain all the mitotic aberrations in MAD2L2-deficient cells (Fig. 1 B).
Figure 1.
Accelerated mitosis in MAD2L2-deficient cells. (A) MAD2L2 DT40 cells release more rapidly from nocodazole block into G1 than wild-type cells. “n” represents the number of independent experiments. Error bars = SEM; p, unpaired t test. (B) Mitotic aberrations in mad2l2(rev7), rev3, and rev1 DT40 cells. Lagging chromosomes, white segments; anaphase bridges, gray segments. The total number of metaphases scores is indicated. These data were derived from two independent experiments in each of which at least 85 metaphases were counted. Error bars = 1 SD for the independently determined percentages from the two experiments; p, unpaired t test. (C) Confirmation of the effectiveness of the MAD2L2 siRNAs used in this study, showing silencing of only MAD2L2 but not MAD2. The right-hand panel shows the effect of the siRNA against the 3′UTR of human MAD2L2 that does not deplete ectopically expressed human MAD2L2 cDNA carrying a C-terminal myc tag. (D) MAD2L2-depleted U2OS cells release more rapidly than controls from nocodazole block into G1. The percentage of cells in G1 was assessed 60 min after nocodazole release. “n” represents the number of independent experiments. Error bars = SEM; p, unpaired t test. (E) Frames from time-lapse movies of control and siMAD2L2 U2OS cells. NEBD, nuclear envelope breakdown. Note the lagging chromosome at 30 min in the siMAD2L2 frames. Example movies can be found online (Videos 1–4). The white bar in the first frame represents 5 µm. (F) Quantification of the time taken for control (Control siRNA; solid black line), MAD2L2-depleted (siMAD2L2; solid red line), complemented (siMAD2L2 + hMAD2L2; dashed black line), and REV3-depleted (siREV3; solid blue line) U2OS cells expressing mCherry-H2B to complete mitosis, assessed by time-lapse video microscopy (1 frame every 5 min). The plot shows the cumulative percentage of cells that completed mitosis, measured from NEBD to cytokinesis. “n” represents the number of cells examined, collected from at least three independent experiments. The P-value to test whether the distribution of times in siMAD2L2-treated cells is distinct from controls was calculated by the Kolmogorov-Smirnov test.
We were able to observe similar rapid release from nocodazole in human U2OS cells treated with an siRNA against MAD2L2 (Fig. 1, C and D). To examine the effect of depletion of MAD2L2 in the absence of spindle poisons, we tracked mitosis in live U2OS cells stably expressing mCherry-tagged histone H2B to allow direct visualization of chromosome dynamics (Fig. 1 E; Videos 1–4). After silencing of MAD2L2, cells completed mitosis more quickly than controls (Fig. 1 F). In control cells, the average time taken from nuclear envelope breakdown to cytokinesis was 55 ± 6 min. After depletion of MAD2L2 by siRNA this was reduced to 42 ± 1 min (P = 0.032), whereas expression of siRNA-resistant hMAD2L2 restored mitosis to an average of 60 ± 2 min. Importantly, neither of the siRNAs that we used had an effect on the levels of MAD2 (Fig. 1 C), excluding dysfunction of the spindle assembly checkpoint due to a nonspecific effect on MAD2 levels.
MAD2L2 depletion results in premature degradation of APC/C substrates from prometaphase
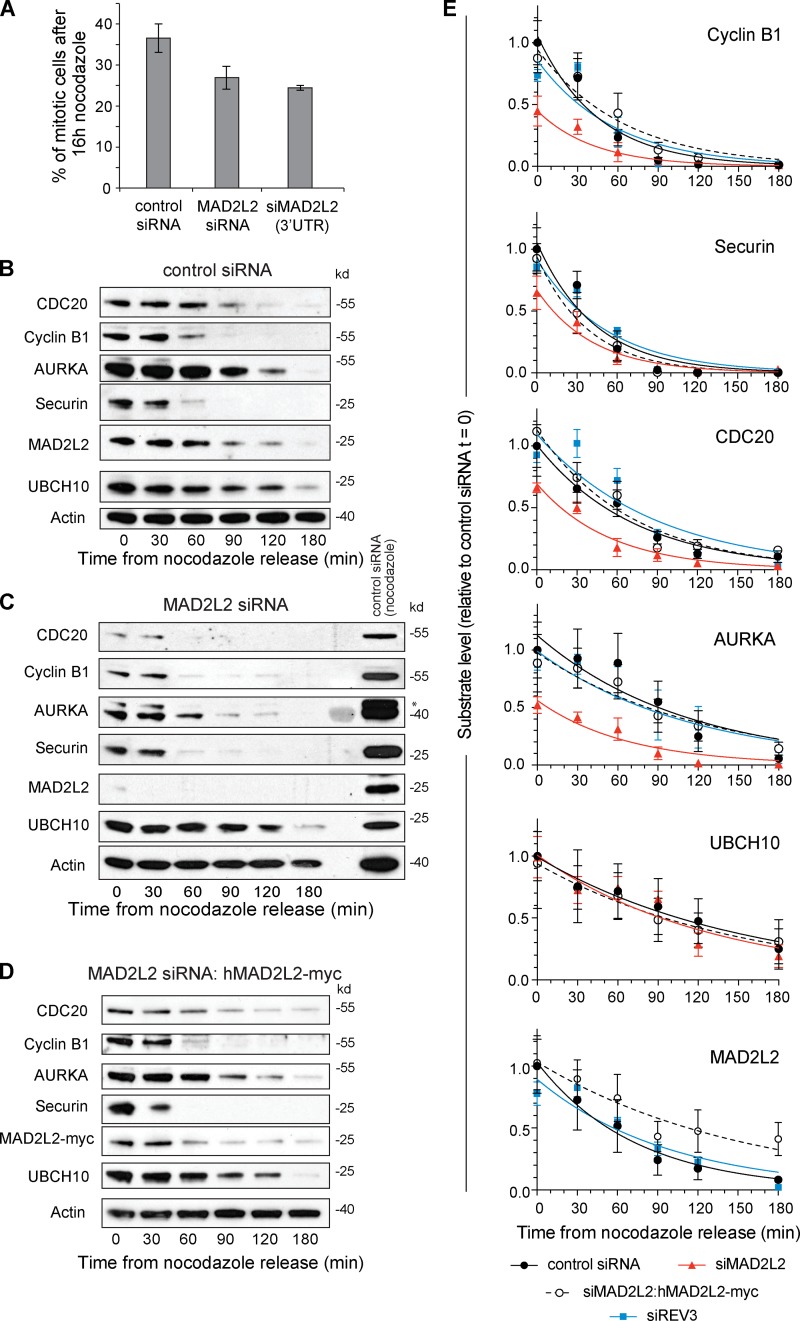
These observations suggested that cells lacking MAD2L2 may initiate anaphase prematurely. We therefore examined the ability of these cells to maintain prometaphase arrest in nocodazole. Cells depleted of MAD2L2 did arrest on nocodazole treatment, suggesting that the SAC is active, but with a slightly lower mitotic index than control cells. This is consistent with idea that loss of MAD2L2 could cause “mitotic slippage,” in which degradation of substrates by the APC/C leads to escape from mitosis despite an active SAC (Fig. 2 A; Lee et al., 2010). We therefore examined the stability of key APC/C substrates in the first 180 min after release from prometaphase arrest in nocodazole. Cells treated with MAD2L2 siRNA exhibited lower levels of APC/C substrates than control or complemented cells after 16 h of nocodazole treatment (Fig. 2, B–E). This was most marked for the substrates normally degraded early in anaphase, Cyclin B1, Securin, CDC20, and AURKA, which is an obligate APC/CCDH1 substrate (Floyd et al., 2008). However, UBCH10, which is degraded later in G1, thereby allowing Cyclin A accumulation before S-phase entry (Rape and Kirschner, 2004), was not significantly affected. Once anaphase was initiated, Cyclin B1, Securin, CDC20, and AURKA reached a low, almost undetectable, basal level at earlier time points in MAD2L2-deficient cells than controls (Fig. 2, B–E; Table 1). This was, in part, due to lower substrate levels as cells enter anaphase (Fig. 2 E), but also, in the case of CDC20 and AURKA, to a shorter half-life of degradation once anaphase is initiated (Table 2). Together, these data suggesting that there is a degree of premature APC/C activation in cells lacking MAD2L2. This effect was independent of the role played by MAD2L2 in DNA polymerase ζ as cells depleted for REV3 exhibited normal kinetics of APC/C substrate degradation (Fig. 2 E; and Fig. S4, A and B).
Figure 2.
Premature degradation of APC/C substrates in cells depleted of MAD2L2. (A) Silencing of MAD2L2 results in a lower mitotic index in nocodazole. The percentage of mitotic U2OS cells was calculated by flow cytometry monitoring histone H3 phospho-serine 10 in cells treated with nocodazole for 16 h. Error bars = 1 SD. (B) APC/C substrate degradation in control siRNA-treated U2OS cells. (C) APC/C substrate degradation in siMAD2L2-treated cells. The asterisk indicates remnant Cyclin B1 signal in the AURKA blot. (D) APC/C substrate degradation in cells complemented with siRNA-resistant hMAD2L2-myc. (E) Summary of substrate degradation in control and siMAD2L2-treated cells in the 180 min after nocodazole release. Substrate levels at each time point are normalized to actin and then shown as a fraction of the level of the substrate at t = 0 in control siRNA-treated cells. The curve fit is an exponential decay. The t1/2 and fitting statistics are presented in Table 2. Error bars = SEM. Control siRNA: solid black circle/solid black line, n = 5. siMAD2L2: solid red triangles/solid red line, n = 3. siMAD2L2 complemented with hMAD2L2-myc: open circles/dashed black line, n = 4. siREV3: solid blue square/solid blue line, n = 3. “n” represents the number of independent experiments. The additional blots that contribute to this analysis are shown in Fig. S1 (control, siMAD2L2, and complemented) and Fig. S4 (siREV3).
Table 1.
Time taken from nocodazole release for substrate levels to reach 20% of the starting level in control cells
| Substrate | Control siRNA | siMAD2L2 | siMAD2L2:hMAD2L2-myc |
| Cyclin B1 | 73.6 ± 2 | 32.1 ± 10.2 | 95.7 ± 20 |
| Securin | 69.7 ± 2.2 | 48.5 ± 12.6 | 57.9 ± 12 |
| Cdc20 | 118 ± 1.5 | 67.9 ± 12.2 | 122 ± 44.3 |
| AURKA | 179 ± 14.8 | 61.3 ± 15.3 | 180 ± 60.3 |
| UBCH10 | 248 ± 4.2 | 214 ± 55.8 | 234 ± 102 |
| MAD2L2 | 119 ± 1 | – | 251 ± 75 |
Calculated from fitted exponential decay curves (Fig. 2 E).
Table 2.
Half-lives of APC/C substrates
| Substrate | Parameter | Control siRNA | siMAD2L2 | siMAD2L2:hMAD2L2-myc |
| Cyclin B1 | T1/2 | 31.7 ± 0.9 | 29.6 ± 0.01 | 44.3 ± 1.5 |
| Cyclin B1 | R2 | 0.947 | 0.999 | 0.932 |
| Securin | T1/2 | 30 ± 0.9 | 29.1 ± 0.50 | 26.8 ± 0.2 |
| Securin | R2 | 0.938 | 0.965 | 0.982 |
| CDC20 | T1/2 | 50.6 ± 0.6 | 38.1 ± 0.8 | 49.2 ± 1.3 |
| CDC20 | R2 | 0.975 | 0.958 | 0.950 |
| AURKA | T1/2 | 77.0 ± 6.4 | 46.5 ± 2.1 | 85.2 ± 3.8 |
| AURKA | R2 | 0.842 | 0.913 | 0.914 |
| UBCH10 | T1/2 | 106 ± 1.8 | 91.4 ± 5.8 | 105 ± 0.9 |
| UBCH10 | R2 | 0.967 | 0.876 | 0.983 |
| MAD2L2 | T1/2 | 51 ± 0.4 | – | 108 ± 5.3 |
| MAD2L2 | R2 | 0.983 | – | 0.904 |
Calculated from fitted exponential decay curves (Fig. 2 E).
MAD2L2 degradation in early anaphase is mediated by APC/CCDC20
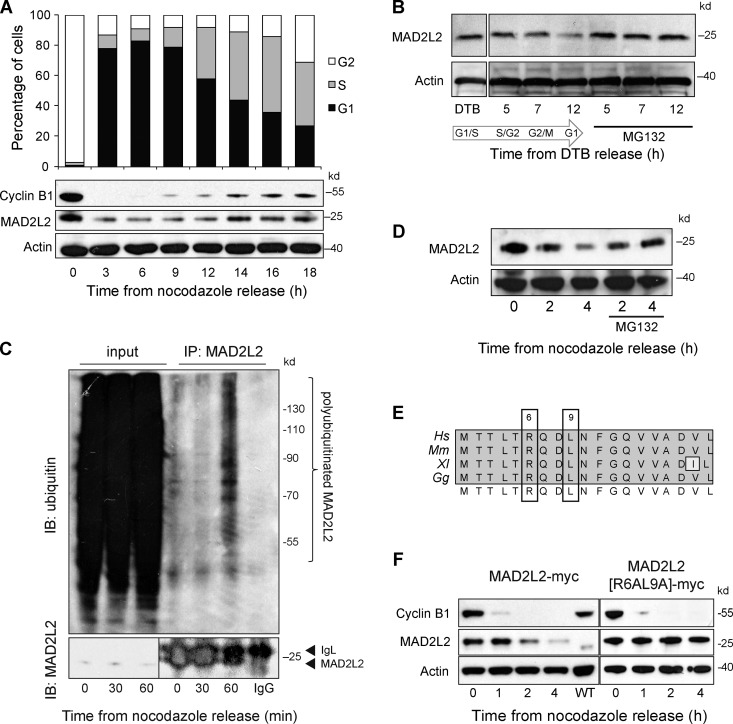
MAD2L2 itself was also degraded rapidly as cells progress into anaphase, with kinetics similar to CDC20, whose degradation is initiated by APC/CCDC20-dependent autoubiquitination (Fig. 2, B and E; Foe et al., 2011). A longer time course, monitoring MAD2L2 levels after release of cells from nocodazole, confirmed that MAD2L2 exhibits cyclical behavior through the cell cycle (Fig. 3 A). This pattern of degradation was also seen after release from double thymidine block, which arrests cells at the G1/S boundary (Fig. 3 B). MAD2L2 polyubiquitination could be detected by 60 min after release from nocodazole arrest (Fig. 3 C) and degradation of MAD2L2 was prevented by treatment of cells with the proteasome inhibitor MG132 (Fig. 3, B and D). Together, these observations suggested that MAD2L2 protein levels might be regulated by APC/C-mediated proteasomal degradation. APC/C substrates are usually characterized by the presence of either a D-box (RXXL) (Glotzer et al., 1991) or KEN-box (Pfleger and Kirschner, 2000). Examination of the sequence of MAD2L2 from a number of vertebrate species revealed a potential D-box within the N terminus of the protein (Fig. 4 E). This D-box motif is seen in both substrates of APC/CCDC20 and APC/CCDH1, although not all RXXL motifs are D-boxes (Simpson-Lavy et al., 2010). To test whether the RXXL motif in MAD2L2 is required for its degradation, we transfected U2OS cells with MAD2L2 tagged at its C terminus with a myc epitope. MAD2L2-myc was degraded on release of the cells from nocodazole (Fig. 4 F). Importantly, mutation of the D-box resulted in stabilization of the protein, suggesting that the RXXL motif in MAD2L2 is a genuine D-box (Fig. 4 F).
Figure 3.
MAD2L2 degradation in early anaphase is dependent on a destruction box and the proteasome. (A) Long time-course of MAD2L2 expression in U2OS after release from nocodazole with a representative cell cycle profile (of three repeats). (B) Stabilization of MAD2L2 levels by MG132 after release from double thymidine block. (C) In vivo ubiquitination of MAD2L2 on release from nocodazole. Left-hand three lanes: input blotted with anti-MAD2L2 (bottom) and anti-ubiquitin (top). Right-hand lanes: the anti-MAD2L2 immunoprecipitate blotted with anti-ubiquitin with IgG only control on the far right. (D) Stabilization of MAD2L2 by proteasome inhibition with MG132 after release from nocodazole. (E) The RXXL motif in vertebrate MAD2L2. Alignments of the first 19 amino acids of human (Hs), mouse (Mm), Xenopus (Xl), and chicken (Gg) MAD2L2. (F) MAD2L2 levels are stabilized by mutation of the D-box. Ectopically expressed myc-tagged MAD2L2 is degraded on release into G1 (left) and this is prevented by mutation of R6 and L9 to A (right).
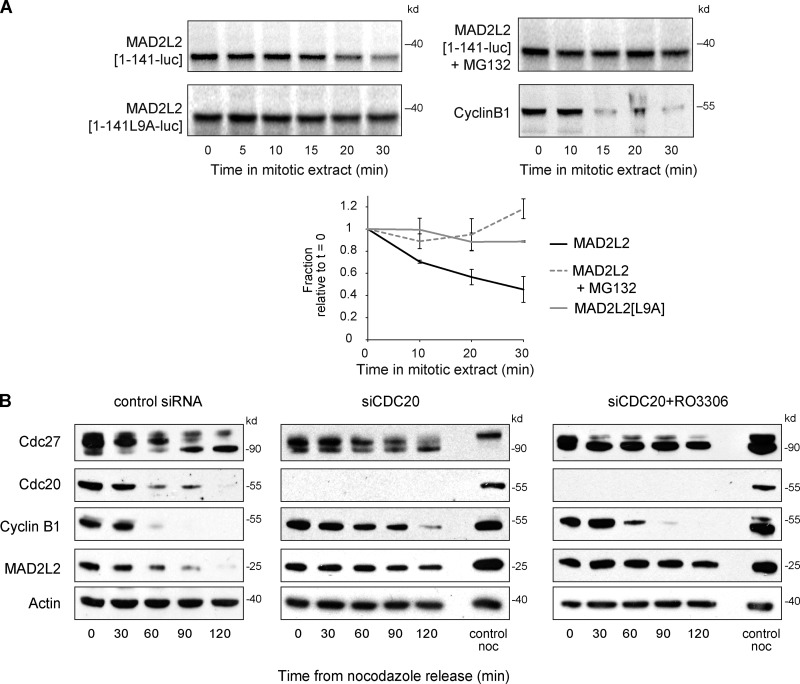
Figure 4.
MAD2L2 degradation in anaphase is mediated by APC/CCDC20. (A) D-box and proteasome-dependent destruction of MAD2L2 in extracts of mitotic Xenopus oocytes. The first 141 amino acids of MAD2L2, and the first 200 amino acids of Cyclin B1, were fused to luciferase, in vitro translated, and added to mitotic Xenopus oocyte extracts. Degradation is stabilized by mutation of the D-box or by addition of MG132. The graphs shows quantification of three repeats of the experiment with the level of translated MAD2L2 being normalized to time 0. Error bars = 1 SD. (B) The degradation of MAD2L2 in early anaphase is delayed by depletion of CDC20. Depletion of CDC20 alone results in prolonged delay in mitotic exit and hence also a delay in MAD2L2 and Cyclin B1 degradation and CDC27 dephosphorylation. Addition of the CDK1 inhibitor RO3306 permits mitotic exit in the absence of CDC20, but MAD2L2 degradation remains delayed.
To examine whether the D-box in MAD2L2 is indeed recognized by the APC/C, and in particular by APC/CCDC20, we examined the stability of MAD2L2 fused to Renilla luciferase in mitotic extracts of Xenopus laevis oocytes, in which activation of the APC/C is dependent only on CDC20 (Lorca et al., 1998). Full-length MAD2L2 is not degraded in vitro (Pfleger et al., 2001), consistent with it being an APC/C inhibitor (Chen and Fang, 2001; Pfleger et al., 2001). We therefore asked whether a C-terminally truncated form of the protein, which retains the RXXL destruction box, could be recognized as an APC/C substrate. hMAD2L2[1–141] was degraded over the course of 30 min, but with slower kinetics than Cyclin B1[1–200] (Fig. 4 A). Degradation of hMAD2L2[1–141] was blocked either by the addition of MG132 or by mutation of the D-box (Fig. 4 A), confirming that the destruction box can be recognized by APC/CCDC20. To examine whether MAD2L2 is a substrate of APC/CCDC20 in vivo, we assessed the stability of the protein after nocodazole release in cells in which CDC20 was silenced and mitotic exit forced by treatment with the CDK inhibitor RO3306 (Vassilev et al., 2006). Under these conditions the degradation of MAD2L2 was retarded despite rapid Cyclin B1 degradation and dephosphorylation of the APC/C subunit CDC27 (Fig. 4 B). Together, these data support the degradation of MAD2L2 in early anaphase as being mediated by APC/CCDC20.
MAD2L2 interacts with a pool of free CDH1
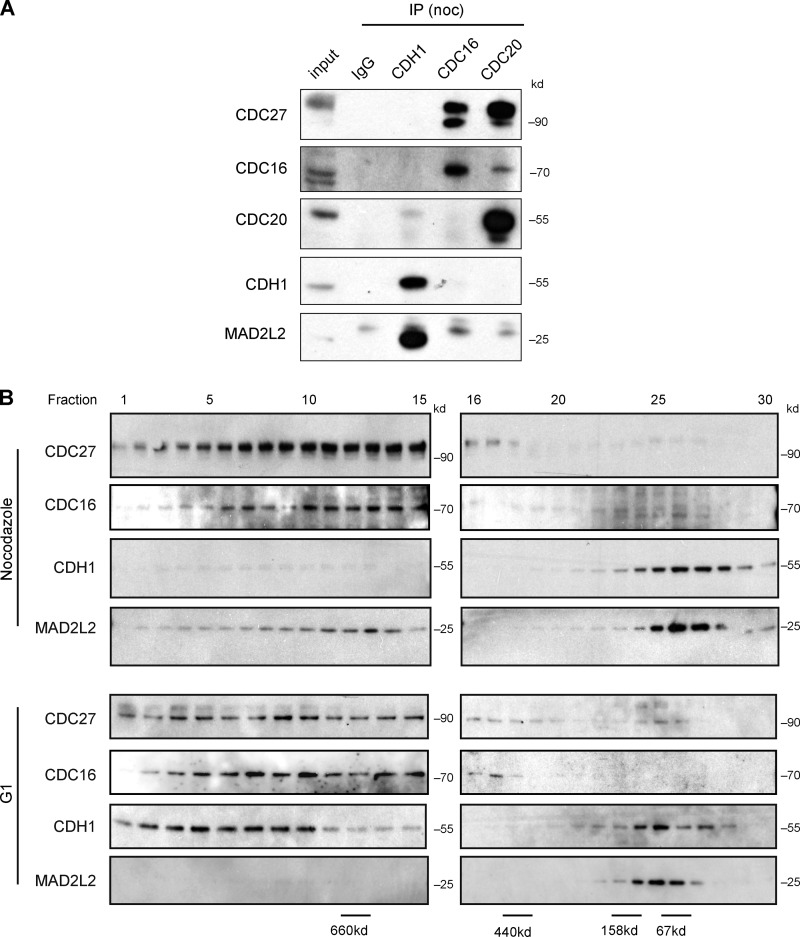
Thus far, our data suggested that MAD2L2 exerts an inhibitory effect on the APC/C during prometaphase and early anaphase. Because it has been proposed that APC/C inhibition by MAD2L2 in vitro is directed toward the activator proteins (Chen and Fang, 2001; Pfleger et al., 2001), we examined the in vivo binding of the two APC/C activators to MAD2L2 in cells arrested in prometaphase with nocodazole. CDH1 interacted with MAD2L2, but, as expected, did not precipitate either the CDC27 or CDC16 subunits of the APC/C (Fig. 5 A and Fig. S2 A). Equally, MAD2L2 immunoprecipitated CDH1 but not CDC27 in asynchronous cells (Fig. S2 A), confirming that the interaction between MAD2L2 and CDH1 takes place independently of the APC/C throughout the cell cycle. Gel filtration analysis of purified recombinant MAD2L2 and CDH1 indicated that the interaction between the two proteins is likely to be direct (Fig. S2, B and C). In contrast to CDH1, we detected no interaction between MAD2L2 and CDC20 in prometaphase-arrested cells (Fig. 5 A).
Figure 5.
In vivo MAD2L2 interacts with a pool of CDH1 that is not bound to the APC/C. (A) MAD2L2 binds CDH1 but not the APC/C or CDC20 in nocodazole-arrested cells. (B) Gel filtration of synchronized U2OS cells. In nocodazole, the bulk of CDH1 and MAD2L2 co-elute in a low molecular weight fraction, while in G1 CDH1 is present in a higher molecular weight fractions containing the APC/C subunits CDC27 and CDC16.
These data suggest that CDH1 can interact with free MAD2L2 that is not bound to the APC/C, and thus that CDH1 exists in two pools during the cell cycle. In one pool it is bound to CDC27 as part of the APC/CCDH1 complex, and in the other to MAD2L2. To examine the dynamics of these CDH1 pools we performed gel filtration on whole-cell extracts made from cells arrested in nocodazole and from cells released into G1. In extracts from nocodazole-treated cells, CDH1 is found in low molecular weight fractions together with MAD2L2, quite distinct from the higher molecular weight fractions containing the CDC27 and CDC16 subunits of the APC/C (Fig. 5 B). Although a small amount of MAD2L2 was seen at higher molecular weight, the immunoprecipitation experiments demonstrate that this is not due to binding the APC/C (Figs. 5 A and S2 A). It may reflect a remaining fraction binding the REV3 subunit of DNA polymerase ζ, which has a predicted monomeric molecular weight of more than 350 kD. In contrast, the majority of CDH1 in G1 is found in higher molecular weight fractions with CDC27. We could not detect any significant fraction of free MAD2L2 in either extract.
Depletion of MAD2L2 results in premature association of CDH1 with the APC/C and activation of APC/CCDH1
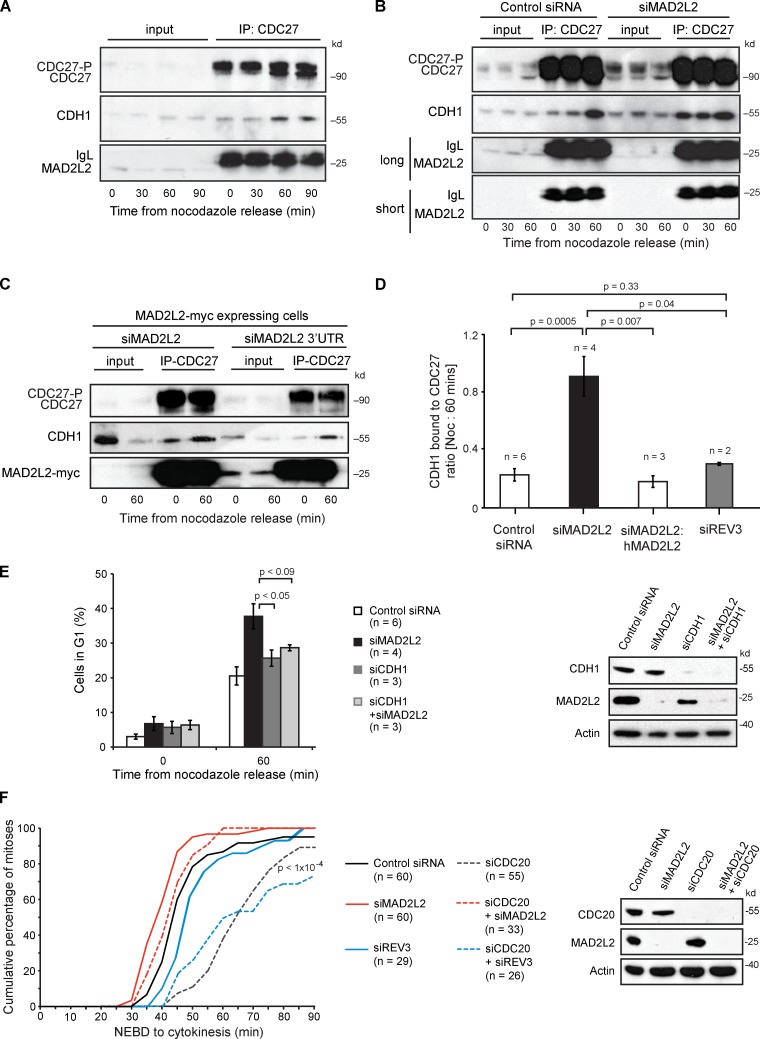
Together, these observations suggest a model in which MAD2L2 helps to sequester CDH1 away from the APC/C during prometaphase and early anaphase. Such a model predicts that loss of MAD2L2 would allow CDH1 to gain premature access to the APC/C, explaining the premature APC/C substrate degradation seen in MAD2L2-deficient cells. To test this idea we monitored the interaction between CDC27 and CDH1 after release of cells from nocodazole arrest. In control cells the association of CDH1 with the CDC27 subunit of the APC/C is seen by 60 min after release from nocodazole block (Fig. 6 A). However, in cells depleted for MAD2L2 there is constitutive association of CDH1 with the APC/C even in nocodazole, a situation that is reversed by stably expressing an siRNA-resistant MAD2L2 (Fig. 6, B–D; Fig. S1 D). Increased association of CDH1 with the APC/C in nocodazole-arrested cells is not observed after depletion of REV3 (Figs. 6 D and S4 C).
Figure 6.
Depletion of MAD2L2 leads to premature binding of CDH1 to the APC/C in prometaphase. (A) CDH1 interacts with the APC/C within 60 min after nocodazole release. Immunoprecipitation with anti-CDC27 from U2OS cells arrested in nocodazole and at 30, 60, and 90 min after release blotted for CDH1 to monitor the association of CDH1 with the APC/C. (B) Silencing of MAD2L2 leads to premature association of CDH1 with CDC27. (C) Complementation of the premature association of CDH1 with CDC27. The cells in this experiment all stably express hMAD2L2-myc. siMAD2L2 silences both the endogenous MAD2L2 and the transgenic MAD2L2-myc. The siMAD2L2 3′UTR only silences the endogenous MAD2L2, leaving the MAD2L2-myc expressed. See also Fig. 1 C. (D) Quantification of the premature association of CDH1 with CDC27 in MAD2L2-depleted cells. The amount of CDH1 immunoprecipitated with CDC27 was normalized to the IgG signal for each immunoprecipitation and the ratio of the amount of CDH1 pulled down in nocodazole (time = 0) and at 60 min after release was calculated. The constitutive association of CDH1 with CDC27 in prometaphase in MAD2L2-silenced cells results in an increase in this ratio, which approaches 1. “n” represents the number of independent experiments. Error bars = SEM; P-value calculated with unpaired, two-tailed t test assuming equal variance. The additional blots contributing to this analysis are shown in Fig. S1 D. (E) Depletion of CDH1 prevents the rapid mitotic exit seen in cells lacking MAD2L2. The percentage of cells in G1 was assessed 60 min after nocodazole release. “n” represents the number of independent experiments. Error bars = SEM; p, unpaired t test. The control siRNA and siMAD2L2 data are reproduced from Fig. 1 D for comparison. The effectiveness of the siRNA protocol is illustrated in the panel on the right. (F) Silencing of MAD2L2, but not REV3, can rescue the mitotic delay in CDC20-depleted cells. Quantification of the time taken for control (Control siRNA; solid black line), MAD2L2-depleted (siMAD2L2; solid red line), REV3-depleted (siREV3; solid blue line), CDC20-depleted (siCDC20, dashed dark gray line), CDC20- and MAD2L2-depleted (siCDC20 + siMAD2L2, dashed red line), and CDC20- and REV3-depleted (siCDC20 + siREV3, dashed blue line) U2OS cells expressing mCherry-H2B to complete mitosis, assessed by time-lapse video microscopy (1 frame every 5 min). The plot shows the cumulative percentage of cells that completed mitosis, measured from NEBD to cytokinesis. “n” represents the number of cells examined, collected from at least three independent experiments. P-value for siCDC20 and siCDC20 + siREV3 vs. Control siRNA, siMAD2L2, and siMAD2L2 + siCDC20 < 1 × 10−4.
Interestingly, overexpression of stable MAD2L2[R6AL9A] caused only a subtle delay in the association of CDH1 with the APC/C but did not prevent it (Fig. S3). Consistent with this, there was at best a minor delay in APC/C substrate degradation (Fig. S3), although this might simply represent the overexpression of myc-tagged MAD2L2, as we also detected some delay in substrate degradation after expression of the myc-tagged wild-type protein, whose levels are also higher than endogenous MAD2L2 (Fig. 2 E). Thus, although MAD2L2 acts as an APC/C inhibitor in prometaphase and early anaphase, it cannot prevent full activation of APC/CCDH1 in G1. Because CDH1 is not thought to be essential for mitotic exit, which relies initially on CDC20, this observation predicted that removing CDH1 would prevent rapid exit of MAD2L2-depleted cells from mitosis on nocodazole release. This is indeed observed (Fig. 6 E). Additionally, the premature activation of APC/CCDH1 in cells lacking MAD2L2, but not REV3, appears to be sufficiently strong to bypass the delay in mitotic exit caused by depletion of CDC20. Thus, cells lacking both CDC20 and MAD2L2 execute mitosis with practically normal kinetics (Fig. 6 F).
Discussion
Despite being shown to act as an in vitro APC/C inhibitor over ten years ago (Chen and Fang, 2001; Pfleger et al., 2001), the in vivo significance of the APC/C inhibitory activity of MAD2L2 has remained unclear. Further, its mechanism of action as an APC/C inhibitor is also unresolved. Pfleger et al. (2001) proposed a model in which MAD2L2 acts directly on the APC/C by binding to and inhibiting APC/CCDH1 by preventing substrate release. The data of Chen and Fang (2001), on the other hand, support a model in which MAD2L2 is able to inhibit both APC/CCDH1 and APC/CCDC20 by binding directly to free CDH1 or CDC20.
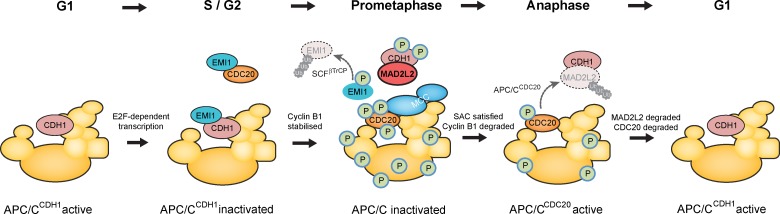
Here we address the mechanism by which MAD2L2 acts to inhibit the APC/C in vivo and demonstrate the functional consequence of its loss. We show that in vivo MAD2L2 is not targeted directly to the APC/C but instead to the pool of free CDH1, not associated with the APC/C. We suggest that this binding helps to sequester CDH1 away from the APC/C during prometaphase, preventing it from binding and prematurely activating the APC/C (Fig. 7). This inhibitory effect is rapidly relieved as cells enter anaphase by the APC/CCDC20-mediated degradation of MAD2L2, thereby contributing to the orderly handover from APC/CCDC20 to APC/CCDH1 necessary for the correct execution of mitosis (Sullivan and Morgan, 2007).
Figure 7.
The role played by MAD2L2 in the control of APC/C activation. As cells enter S phase, APC/CCDH1, which has been active in G1, is inhibited by Emi1 and by degradation of CDH1. Emi1 can also inhibit newly produced CDC20 by sequestering it from the APC/C. Loss of APC/C activity results in Cyclin B1 stabilization and consequent phosphorylation of the APC/C, the activators CDC20 and CDH1 and of Emi1. Phosphorylated Emi1 is degraded via ubiquitination by the SCFβTrCP. APC/C phosphorylation promotes binding of CDC20, but the complex is kept inactive by MAD2 and the mitotic checkpoint complex. Meanwhile, APC/C and CDH1 phosphorylation antagonizes activation of the APC/C by CDH1. CDH1 is additionally bound by MAD2L2, contributing to its sequestration away from the APC/C. Once the spindle assembly checkpoint (SAC) is satisfied and anaphase is initiated, APC/CCDC20 degrades MAD2L2 and releases CDH1. CDH1 and the APC/C are also dephosphorylated, allowing CDH1 to bind and activate the APC/C. The sequestration of CDH1 by MAD2L2 acts to prevent premature activation of the APC/C by preventing binding of CDH1 to hypophosphorylated forms of the complex, thus ensuring the timing and bistability of the APC/CCDC20 to APC/CCDH1 switch. Model adapted from Pines, 2011.
Although we show that MAD2L2 degradation is dependent on APC/CCDC20, full-length MAD2L2 inhibits APC/CCDC20 in mitotic extracts in vitro (Chen and Fang, 2001). This apparent paradox may be explained by a combination of the inability of MAD2L2 to inactivate an active APC/CCDC20 (Chen and Fang, 2001) and the failure of MAD2L2 and CDC20 to interact during prometaphase in vivo (Fig. 5 A). This latter observation may reflect the physical separation of the two proteins during prometaphase: CDC20 is concentrated on the kinetochores (Howell et al., 2004) while MAD2L2 is excluded from condensed chromosomes (Medendorp et al., 2009, 2010; Bhat et al., 2013). Thus, in vivo, APC/CCDC20 would be in a position to trigger the degradation of MAD2L2 on release of the SAC.
Consistent with the model we propose (Fig. 7), depletion of MAD2L2 results in more CDH1 bound to the APC/C during prometaphase. Several lines of evidence suggest that this results in premature activation of the APC/C by CDH1. The constitutive association of APC/C and CDH1 that we see during prometaphase in cells depleted of MAD2L2 correlates with lower levels of key early APC/C substrates, and with more rapid mitotic exit. Interestingly, of the substrates we examined after MAD2L2 depletion, AUKRA, which has been previously shown to be specifically degraded by APC/CCDH1 (Floyd et al., 2008), was most significantly affected, providing further support to the idea that the CDH1-dependent activation of the APC/C is functionally relevant. In agreement with our hypothesis, silencing of CDH1 simultaneously with MAD2L2 prevented the rapid release of cells from nocodazole arrest (Fig. 6 E). Finally, and perhaps most strikingly, silencing of both MAD2L2 and CDC20 reveals that the APC/C activation by CDH1 appears to be sufficiently robust to prevent the prolonged metaphase delay caused by lack of CDC20 (Fig. 6 F).
Our data suggest that the phosphorylation of CDH1 (Zachariae et al., 1998; Jaspersen et al., 1999) and of the APC/C during prometaphase may not be sufficient to fully prevent activation of APC/CCDH1. Indeed, it is well established that addition of exogenous CDH1 can activate the APC/C in any cell cycle stage (Schwab et al., 1997; Visintin et al., 1997; Listovsky et al., 2000). We suggest that MAD2L2 contributes a second layer of regulation to ensure robust but rapidly reversible suppression of APC/CCDH1 activity. Indeed, there are other examples that suggest that APC/C activation is more generally regulated by an interplay of phosphorylation and specific inhibitor proteins, including MAD2, Emi1, and Acm1. It is instructive to consider the role of MAD2L2 in the context of these mechanisms (Fig. 7).
As cells enter S phase, residual APC/CCDH1 is inactivated by binding of the inhibitor protein Emi1, which is able to inactivate APC/C already bound by CDH1 (Reimann et al., 2001; Hsu et al., 2002), allowing the accumulation of Cyclin B1. During G2 and M phases, Cyclin B1/CDK1 phosphorylates CDH1 and the APC/C itself, which together reduces the ability of CDH1 to bind and activate the APC/C (Zachariae et al., 1998; Jaspersen et al., 1999). Although inhibitory to CDH1 binding, APC/C phosphorylation promotes binding of CDC20 (Shteinberg et al., 1999; Kramer et al., 2000). However, CDC20 does not activate that APC/C in prometaphase, as it is inhibited by MAD2 and the mitotic checkpoint complex (Musacchio and Salmon, 2007). Emi1, meanwhile, is degraded during prometaphase by the SCFβTrCP/Slimb ubiquitin ligase in response CDK1/Cyclin B–dependent phosphorylation (Margottin-Goguet et al., 2003). In the preexisting scheme this leaves phosphorylation of APC/C and CDH1 as the sole mechanism to prevent activation of the APC/C by CDH1 during prometaphase in vertebrate cells. We propose that it is at this point that MAD2L2 contributes to the inhibition of APC/CCDH1 activation by sequestering CDH1 (Fig. 7). Interestingly, in budding yeast, Acm1 (Martinez et al., 2006; Dial et al., 2007) has also been shown to provide a second mechanism to control APC/CCdh1 activity during prometaphase by binding to Cdh1 (Enquist-Newman et al., 2008). Like MAD2L2, Acm1 is itself targeted by APC/CCdc20, resulting in its destruction at the onset of anaphase, thereby releasing the inhibition of APC/CCdh1 activation (Enquist-Newman et al., 2008). However, no clear homologue of Acm1 has yet been identified in higher eukaryotes. It is therefore tempting to speculate that MAD2L2 is performing an analogous role in vertebrate cells. Interestingly, although Rev7/Mad2l2 is present in budding yeast, it is not well conserved when compared with the vertebrate protein and its levels have been reported not to fluctuate during the cell cycle (Waters and Walker, 2006). It is also worth noting that, perhaps contrary to expectations, expression of a destruction box mutant of either Acm1 in yeast (Enquist-Newman et al., 2008) or MAD2L2 in human cells (Fig. 6 F) does not significantly retard the degradation of APC/C substrates. This suggests that the role of both proteins is likely to be most important before and during the switch between APC/CCDC20 and APC/CCDH1.
Finally, it is likely that the origin of the mitotic defects in cells depleted of MAD2L2 is complex. MAD2L2 has a well-documented function with REV3 as a subunit of DNA polymerase ζ, a specialized DNA polymerase important in translesion synthesis, homologous recombination, and the replication of fragile sites (Sonoda et al., 2003; Bhat et al., 2013; Sale, 2013). Deficiencies in these processes are well known to increase the rate of mitotic errors, likely because of failure to complete replication or resolve recombination intermediates before anaphase (Fenech et al., 2011). Although loss of either gene results in identical sensitivity to DNA damage, MAD2L2-deficient cells exhibit more chromosome defects in metaphase spreads than REV3-deficient cells (Okada et al., 2005) and, as we report, more mitotic aberrations in anaphase (Fig. 1 B). We suggest that this increased frequency of mitotic aberration in MAD2L2-deficient cells results from a combination of defective replication/recombination and premature mitotic exit, which shortens the time for late resolution of unreplicated segments and recombination intermediates.
Although other reports have also shown that loss of MAD2L2 function leads to an increase in the total level of mitotic aberrations, the level of anaphase bridges and lagging chromosomes varies between studies and cell lines (Medendorp et al., 2009, 2010; Bhat et al., 2013). Moreover, interpretation of the pattern of mitotic aberrations in terms of specific mechanisms is difficult because both anaphase bridges and lagging chromosomes can result from defective replication/recombination and from defects in mitotic control and timing (Fenech et al., 2011). Thus, it is currently not possible to accurately determine the relative contribution of these two functions of MAD2L2 in preventing mitotic aberrations.
Indeed, aside from the direct effects of MAD2L2 loss, there are also likely to be indirect consequences caused by the lower levels of APC/C substrates seen during prometaphase in MAD2L2-deficient cells. For instance, loss of AURKA function can precipitate chromosome segregation errors (Hégarat et al., 2011). Additionally, interactions between MAD2L2 and the GTPase RAN (Medendorp et al., 2009) and the clathrin light chain A (Medendorp et al., 2010) have also been proposed to contribute toward normal mitosis. Future studies should help untangle the multi-faceted roles of MAD2L2 in maintaining mitotic fidelity.
Materials and methods
Cell culture, synchronization, and drug treatment
The experiments in this study were performed using the human osteosarcoma cell line U2OS, unless otherwise stated. Mammalian cell lines were grown in DMEM + GlutaMAX-I (Life Technologies) supplemented with penicillin/streptomycin and 10% fetal bovine serum (Hyclone) and maintained at 37°C in a humidified incubator with 5% CO2. For live-cell imaging, U2OS cells were grown in CO2-independent medium (18045-054; Life Technologies) supplemented with penicillin/streptomycin, 10% fetal bovine serum (Hyclone), and 2 mM l-Glutamine (25030–024; Life Technologies). The chicken avian leukosis virus–tranformed B cell lymphoma cell line DT40 (Baba et al., 1985) and its derivatives were propagated at 37°C in RPMI 1640 penicillin/streptomycin supplemented with 7% FBS, 3% chicken serum, and 50 mM 2-mercaptoethanol. The rev1 DT40 cell line has been described previously (Simpson and Sale, 2003). The mad2l2 (rev7) (Okada et al., 2005) and rev3 (Sonoda et al., 2003) DT40 mutants were a gift of S. Takeda (Kyoto University, Kyoto, Japan).
Cell synchronization in prometaphase was performed by treatment with 0.2 µM nocodazole (M1404; Sigma-Aldrich). U2OS cells were treated for 16 h and DT40 cells were treated for 6 h. Prometaphase-arrested mammalian cells were obtained by gentle shake-off and either collected for time 0 or washed once with PBS and plated in fresh media to resume cycling for later time points. MG-132 (474791; EMD Millipore) was used at 40 µM for 1–2 h before immunoblotting. The Cdk1 inhibitor RO-3306 (sc-358700; Santa Cruz Biotechnology, Inc.) was used at 9 µM, added at nocodazole release.
Double thymidine block (DTB) was used for early S-phase synchronization. Cells were treated for 18 h with 2 mM thymidine, washed, and then released for 9 h to allow reentry into the cell cycle. The second thymidine treatment was also for 18 h with 2 mM thymidine, after which cells were washed and released into fresh media. MG-132 (474791; EMD Millipore) was used at 40 µM for 1–2 h before immunoblotting.
Expression vectors, mutagenesis, and siRNAs
MAD2L2-myc was constructed in pcDNA3.1(+) (Life Technologies) by fusion of a myc tag to the C terminus of full-length human MAD2L2. The first 141 amino acids of MAD2L2 were amplified with primers: 5′MAD2L2, 5′-TCGAAGCTTATGACCACGCTCACACGACA-3′; 3′MADL2141, 5′-CGAATTCTTGCGTTTCTGGAAGATGCCCACGGGGTAGACC-3′ and fused to full-length Renilla luciferase to obtain the MAD2L2[1–141]-luc vector used in Fig. 2. Histone H2B pmCherry-N1 (Takara Bio Inc.) was a gift from the laboratory of Alan Warren (LMB, Cambridge, England, UK). The point mutations in MAD2L2, R6A, and L9A were generated by site-directed mutagenesis with primers: 5′R6A, 5′-ATGACCACGCTCACAGCACAAGACCTCAACTTTG-3′; 3′R6A, 5′-CAAAGTTGAGGTCTTGTGCTGTGAGCGTGGTCAT-3′; 5′L9A, 5′-CTCACACGACAAGACGCCAACTTTGGCCAAGTG-3′; 3′L9A, 5′-CACTTGGCCAAAGTTGGCGTCTTGTCGTGTGAG-3′. Small interfering RNAs (siRNA) used for targeting human MAD2L2 were: siMAD2L2, 5′-GAUGCAGCUUUACGUGGAAGA-3′ (IDT); siMAD2L2-3′UTR, 5′-CAACACUGUCUGUCUCAAAUA-3′ (QIAGEN); for Cdc20 targeting, 5′-CACCACCAUGAUGUUCGGGUA-3′ (QIAGEN); for REV3 targeting, 5′-CGGGAUGUAGUCAAACUGCAAA-3′; and for control siCTR, 5′-GCGUAUUGCCUACAUUAC-3′ (Thermo Fisher Scientific). Primers for qPCR of hREV3 were 5′-CTTTCTCAGATGGCATTCAG-3′ and 5′-TTTCGGAACTTGACAGCAGC-3′.
DNA and siRNA transfections
Stably transfected cell lines were used for all experiments. U2OS cells were transfected with G418-resistant vectors containing the cloned gene of interest, using calcium phosphate transfection kit (K2780-01; Life Technologies). Cells were selected using G418 at 400 µg/ml and single colonies collected. DT40 cells were transfected by electroporation at 250 V and 950 µF in 0.4-cm cuvettes in a Gene Pulser (Bio-Rad Laboratories). Cells were selected using G418 at 2 mg/ml and single colonies were collected. For gene-silencing experiments, U2OS cells were transfected with 25 nM control or indicated siRNA, using INTERFERin reagent (409-50; Polyplus-Transfection). In all experiments, 6-well plates with 50,000–100,000 cells/well were used. When mitotic cells were needed, nocodazole was added 48 h after INTERFERin transfection.
Cell cycle analysis
To analyze cell cycle stage and quality of mitotic release, cells were fixed in 70% ethanol at −20°C, washed in PBS, and stained with 25 µg/ml propidium iodide in PBS supplemented with 25 µg/ml RNaseA (R6513; Sigma-Aldrich). The percentage of M and G1 phase cells was determined by flow cytometry using a cytometer (model LSR II; BD) and the data analyzed with FlowJo software (Tree Star).
Mitotic index
To quantify cells arrested in mitosis during nocodazole treatment we used anti–phospho-histone (Ser10) 488 conjugate (3465; Cell Signaling Technology). In brief, cells were fixed in 70% EtOH, washed, and resuspended with PBS + 0.25% Triton X-100 and incubated for 10 min on ice. Cells were washed and incubated for 1 h with phospho-histone antibody in the dark, washed again in PBS, and stained with 25 µg/ml propidium iodide in PBS supplemented with 25 µg/ml RNaseA (R6513; Sigma-Aldrich). The percentage of mitotic cells was determined by flow cytometry using a cytometer (model LSR II; BD) with data analysis on FlowJo software (Tree Star).
Western blotting, immunoprecipitations, and antibodies
For immunoblotting and immunoprecipitations, cells were lysed in extraction buffer ([EB] 50 mM Tris-HCl, pH 8, 250 mM NaCl, 20 mM EGTA, 50 mM NaF, and 1% Triton X-100) supplemented with Complete protease inhibitor cocktail (11873580001; Roche) and Halt phosphatase inhibitor cocktail (78420; Thermo Fisher Scientific). Cells were lysed on ice for 10–30 min, and cleared by centrifugation at 20,000 g for 10–30 min at 4°C. For immunoblotting, extracts were boiled in Laemmli buffer for 5 min. Equal amounts of protein sample (30 µg) were loaded on NuPAGE gels (Life Technologies) and transferred to a nitrocellulose membrane (Whatman).
For immunoprecipitations, clarified lysates were supplemented with the appropriate primary antibody and incubated for 1–2 h at 4°C. Next, 30 µl of equilibrated protein G–Sepharose beads (PC-G5; Generon) was added for 1 h. Finally, beads were washed three times in TBS buffer (30 mM Tris-HCl, pH 7.5, and 150 mM NaCl) and boiled in Laemmli buffer for 5 min.
The following primary antibodies were used at 1:1,000 dilution: human IAK1 (Aurora A, 610936; BD), CDC16 (sc-6395; Santa Cruz Biotechnology, Inc.), CDC20 H-175 (sc-8358; Santa Cruz Biotechnology, Inc.), CDC27 (610454; BD), CDH1 (Clone DH01, MS-116-P; Thermo Fisher Scientific), MAD2 (610678; BD), mouse MAD2L2 (612266; BD), rabbit MAD2L2 (12683-1-AP; ProteinTech), Myc-Tag (2272; Cell Signaling Technology), Securin (3305; Abcam), UBCH10 (AB3681; EMD Millipore), and ubiquitin FK2 (04-263; EMD Millipore). β-Actin (ab8227-50; Abcam) was used at 1:5,000 dilution. To avoid background signal form the IgG heavy chain in the immunoprecipitation experiments, anti–mouse (115-035-174; Jackson ImmunoResearch Laboratories, Inc.) and –rabbit (211-032-171; Jackson ImmunoResearch Laboratories, Inc.) IgG light chain–specific secondary antibodies were used at 1:10,000 dilution.
Western blots were scanned to 8-bit TIFF images using a flatbed scanner (9950F; Canon). Scanned images were processed using the “Levels” command in Adobe Photoshop to ensure that >95% pixels mapped to the full range of 256 output levels of an 8-bit grayscale image. Band densitometry was performed using the ImageJ (National Institutes of Health) Gel Analyzer function. No bands were saturated, as indicated by a plateau on the lane densitometry plot. Substrate intensities were normalized to the actin signal and then to time t = 0 of control siRNA cells. The nocodazole release experiments were performed at least three times for each condition and the average fraction remaining at each time point plotted. Fitting of an exponential decay curve was performed with Prism v6 (GraphPad Software) using a least squares fit algorithm. The R2 for these fits is given in Table 2. The additional blots contributing to this analysis that are not in the main manuscript are shown in Fig. S1, A–C.
Degradation assays in Xenopus mitotic extracts
Activated interphase egg extracts were supplemented with 5 µg of recombinant nondegradable cyclin BΔ90 per 100 µl of extract (both gifts of A. Philpott, Hutchison/MRC Research Centre, Cambridge, UK). To enable extracts to enter mitosis, extracts were incubated at 21°C for 20 min. Mitotic extracts were then supplemented with 100 mg/ml cycloheximide (C4859; Sigma-Aldrich), 1.5 mg/ml ubiquitin (from bovine erythrocytes, U6253; Sigma-Aldrich), and when indicated 50 µM MG-132. 5 µl of 35S-labeled protein was added to 20 µl of mitotic extract and the reaction was incubated at 21°C. 5-µl samples were taken at the indicated time point and boiled in Laemmli buffer for 5 min. Labeled substrates for degradation assays were synthesized in TnT T7 quick-coupled transcription/translation reaction (L1171; Promega) containing [35S]methionine.
Protein purification and size-exclusion column chromatography
CDH1 and MAD2L2 were cloned into pGEX-6P-1 vector and transfected to BL21-competent Escherichia coli to allow protein overexpression. GST-tagged proteins were purified with glutathione Sepharose 4B (17-0756-01; GE Healthcare) and cleaved with 50 units/glutathione Sepharose bed of PreScission protease. Beads were incubated at 4°C overnight. Beads were centrifuged at 200 g for 2 min and the supernatant containing the purified protein was collected. The quality of proteins was verified with Western blots. For size-exclusion chromatography of recombinant proteins, samples of the different complexes were prepared in PBS and 100 µl injected onto a PC3.2/30 (2.4 ml) Superdex 200 gel filtration column (GE Healthcare) preequilibrated in 50 mM Hepes, pH 7.5, 150 mM NaCl, and 2 mM DTT. For size-exclusion chromatography of U2OS protein extracts, cells were lysed in EB buffer and the lysate cleared by centrifugation at 20,000 g for 30 min at 4°C. Samples were fractionated using an ÄKTApurifier (GE Life Sciences) on a Superose 6 prep grade XK16/70 column, (GE Life Sciences) preequilibrated with EB buffer without Triton X-100, at a flow rate of 0.5 ml/min with collection of 500 µl fractions.
Time-lapse imaging and analysis
Time-lapse imaging was performed on a confocal microscope (model C1-si; Nikon) fitted with a modified incubation chamber (Tokai-HIT) to maintain the specimen at 37°C. Cells were grown in CO2–independent medium (18045-054; Life Technologies) supplemented with penicillin/streptomycin, 10% fetal bovine serum (Hyclone), and 2 mM l-Glutamine (25030-024; Life Technologies). Focus was maintained using the Nikon PerfectFocus system. Images were acquired every 5 min using a 63×/NA 1.4 oil-immersion objective and Nikon EZ-C1 software. Videos were created using the “merge channels” function of ImageJ.
Online supplemental material
Fig. S1 documents the additional blots that contributed to the analysis of APC/C substrate degradation in Fig. 2 E and Tables 1 and 2, and additional examples of the CDH1 immunoprecipitations that contribute to Fig. 6 D. Fig. S2 provides further evidence of the interaction between CDH1 and MAD2L2 both in vivo and in vitro. Fig. S3 shows the effect of overexpression of nondegradable MAD2L2[R6AL9A] on the timing of CDH1 association with the APC/C and on the kinetics of APC/C substrate degradation. Fig. S4 provides supporting material for the evidence presented in Figs. 1 F, 2 E, and 6, D and F that the role of MAD2L2 in the control of APC/CCDH1 activation is independent of REV3. Videos 1 and 2 show time-lapse movies of mitosis in mCherry-H2B expressing U2OS treated with control siRNA. Videos 3 and 4 show movies of mitosis in cells treated with MAD2L2 siRNA. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201302060/DC1. Additional data are available in the JCB DataViewer at http://dx.doi.org/10.1083/jcb.201302060.dv.
Supplementary Material
Acknowledgments
We thank A. Philpott and G. McDowell for providing the Xenopus egg extracts and for advice on the in vitro degradation assay; S. Takeda for rev7 DT40; M. Hodgkinson for advice on gel filtration; and R. Fernandez-Leiro and M. Lamers for advice on size-exclusion chromatography and providing the polymerase III epsilon (exonuclease) subunit and PreScission protease. We also thank E. Rajendra and members of the laboratory for helpful discussions and comments on the manuscript; and M. Daly and F. Zhang of the LMB flow cytometry core facility for their help.
T. Listovsky was supported by an EMBO Long Term Fellowship and the Medical Research Council, and work in the laboratory is supported by a central grant to the LMB from the Medical Research Council (U105178808).
Footnotes
Abbreviations used in this paper:
- APC/C
- anaphase-promoting complex/cyclosome
- SAC
- spindle assembly checkpoint
References
- Baba T.W., Giroir B.P., Humphries E.H. 1985. Cell lines derived from avian lymphomas exhibit two distinct phenotypes. Virology. 144:139–151 10.1016/0042-6822(85)90312-5 [DOI] [PubMed] [Google Scholar]
- Bhat A., Andersen P.L., Qin Z., Xiao W. 2013. Rev3, the catalytic subunit of Polζ, is required for maintaining fragile site stability in human cells. Nucleic Acids Res. 41:2328–2339 10.1093/nar/gks1442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao W.C., Kulkarni K., Zhang Z., Kong E.H., Barford D. 2012. Structure of the mitotic checkpoint complex. Nature. 484:208–213 10.1038/nature10896 [DOI] [PubMed] [Google Scholar]
- Chen J., Fang G. 2001. MAD2B is an inhibitor of the anaphase-promoting complex. Genes Dev. 15:1765–1770 10.1101/gad.898701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciosk R., Zachariae W., Michaelis C., Shevchenko A., Mann M., Nasmyth K. 1998. An ESP1/PDS1 complex regulates loss of sister chromatid cohesion at the metaphase to anaphase transition in yeast. Cell. 93:1067–1076 10.1016/S0092-8674(00)81211-8 [DOI] [PubMed] [Google Scholar]
- Cohen-Fix O., Peters J.M., Kirschner M.W., Koshland D. 1996. Anaphase initiation in Saccharomyces cerevisiae is controlled by the APC-dependent degradation of the anaphase inhibitor Pds1p. Genes Dev. 10:3081–3093 10.1101/gad.10.24.3081 [DOI] [PubMed] [Google Scholar]
- Dial J.M., Petrotchenko E.V., Borchers C.H. 2007. Inhibition of APCCdh1 activity by Cdh1/Acm1/Bmh1 ternary complex formation. J. Biol. Chem. 282:5237–5248 10.1074/jbc.M606589200 [DOI] [PubMed] [Google Scholar]
- Enquist-Newman M., Sullivan M., Morgan D.O. 2008. Modulation of the mitotic regulatory network by APC-dependent destruction of the Cdh1 inhibitor Acm1. Mol. Cell. 30:437–446 10.1016/j.molcel.2008.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenech M., Kirsch-Volders M., Natarajan A.T., Surralles J., Crott J.W., Parry J., Norppa H., Eastmond D.A., Tucker J.D., Thomas P. 2011. Molecular mechanisms of micronucleus, nucleoplasmic bridge and nuclear bud formation in mammalian and human cells. Mutagenesis. 26:125–132 10.1093/mutage/geq052 [DOI] [PubMed] [Google Scholar]
- Floyd S., Pines J., Lindon C. 2008. APC/C Cdh1 targets aurora kinase to control reorganization of the mitotic spindle at anaphase. Curr. Biol. 18:1649–1658 10.1016/j.cub.2008.09.058 [DOI] [PubMed] [Google Scholar]
- Foe I.T., Foster S.A., Cheung S.K., DeLuca S.Z., Morgan D.O., Toczyski D.P. 2011. Ubiquitination of Cdc20 by the APC occurs through an intramolecular mechanism. Curr. Biol. 21:1870–1877 10.1016/j.cub.2011.09.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glotzer M., Murray A.W., Kirschner M.W. 1991. Cyclin is degraded by the ubiquitin pathway. Nature. 349:132–138 10.1038/349132a0 [DOI] [PubMed] [Google Scholar]
- Gutierrez G.J., Tsuji T., Chen M., Jiang W., Ronai Z.A. 2010. Interplay between Cdh1 and JNK activity during the cell cycle. Nat. Cell Biol. 12:686–695 10.1038/ncb2071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagting A., Den Elzen N., Vodermaier H.C., Waizenegger I.C., Peters J.M., Pines J. 2002. Human securin proteolysis is controlled by the spindle checkpoint and reveals when the APC/C switches from activation by Cdc20 to Cdh1. J. Cell Biol. 157:1125–1137 10.1083/jcb.200111001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hégarat N., Smith E., Nayak G., Takeda S., Eyers P.A., Hochegger H. 2011. Aurora A and Aurora B jointly coordinate chromosome segregation and anaphase microtubule dynamics. J. Cell Biol. 195:1103–1113 10.1083/jcb.201105058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell B.J., Moree B., Farrar E.M., Stewart S., Fang G., Salmon E.D. 2004. Spindle checkpoint protein dynamics at kinetochores in living cells. Curr. Biol. 14:953–964 10.1016/j.cub.2004.05.053 [DOI] [PubMed] [Google Scholar]
- Hsu J.Y., Reimann J.D., Sørensen C.S., Lukas J., Jackson P.K. 2002. E2F-dependent accumulation of hEmi1 regulates S phase entry by inhibiting APC(Cdh1). Nat. Cell Biol. 4:358–366 10.1038/ncb785 [DOI] [PubMed] [Google Scholar]
- Jaspersen S.L., Charles J.F., Morgan D.O. 1999. Inhibitory phosphorylation of the APC regulator Hct1 is controlled by the kinase Cdc28 and the phosphatase Cdc14. Curr. Biol. 9:227–236 10.1016/S0960-9822(99)80111-0 [DOI] [PubMed] [Google Scholar]
- Kramer E.R., Scheuringer N., Podtelejnikov A.V., Mann M., Peters J.M. 2000. Mitotic regulation of the APC activator proteins CDC20 and CDH1. Mol. Biol. Cell. 11:1555–1569 10.1091/mbc.11.5.1555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence C.W., Das G., Christensen R.B. 1985. REV7, a new gene concerned with UV mutagenesis in yeast. Mol. Gen. Genet. 200:80–85 10.1007/BF00383316 [DOI] [PubMed] [Google Scholar]
- Lee J., Kim J.A., Margolis R.L., Fotedar R. 2010. Substrate degradation by the anaphase promoting complex occurs during mitotic slippage. Cell Cycle. 9:1792–1801 10.4161/cc.9.9.11519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim H.H., Goh P.Y., Surana U. 1998. Cdc20 is essential for the cyclosome-mediated proteolysis of both Pds1 and Clb2 during M phase in budding yeast. Curr. Biol. 8:231–234 10.1016/S0960-9822(98)70088-0 [DOI] [PubMed] [Google Scholar]
- Lindon C., Pines J. 2004. Ordered proteolysis in anaphase inactivates Plk1 to contribute to proper mitotic exit in human cells. J. Cell Biol. 164:233–241 10.1083/jcb.200309035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Listovsky T., Zor A., Laronne A., Brandeis M. 2000. Cdk1 is essential for mammalian cyclosome/APC regulation. Exp. Cell Res. 255:184–191 10.1006/excr.1999.4788 [DOI] [PubMed] [Google Scholar]
- Listovsky T., Oren Y.S., Yudkovsky Y., Mahbubani H.M., Weiss A.M., Lebendiker M., Brandeis M. 2004. Mammalian Cdh1/Fzr mediates its own degradation. EMBO J. 23:1619–1626 10.1038/sj.emboj.7600149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorca T., Castro A., Martinez A.M., Vigneron S., Morin N., Sigrist S., Lehner C., Dorée M., Labbé J.C. 1998. Fizzy is required for activation of the APC/cyclosome in Xenopus egg extracts. EMBO J. 17:3565–3575 10.1093/emboj/17.13.3565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margottin-Goguet F., Hsu J.Y., Loktev A., Hsieh H.M., Reimann J.D., Jackson P.K. 2003. Prophase destruction of Emi1 by the SCF(betaTrCP/Slimb) ubiquitin ligase activates the anaphase promoting complex to allow progression beyond prometaphase. Dev. Cell. 4:813–826 10.1016/S1534-5807(03)00153-9 [DOI] [PubMed] [Google Scholar]
- Martinez J.S., Jeong D.E., Choi E., Billings B.M., Hall M.C. 2006. Acm1 is a negative regulator of the CDH1-dependent anaphase-promoting complex/cyclosome in budding yeast. Mol. Cell. Biol. 26:9162–9176 10.1128/MCB.00603-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medendorp K., van Groningen J.J., Vreede L., Hetterschijt L., van den Hurk W.H., de Bruijn D.R., Brugmans L., van Kessel A.G. 2009. The mitotic arrest deficient protein MAD2B interacts with the small GTPase RAN throughout the cell cycle. PLoS ONE. 4:e7020 10.1371/journal.pone.0007020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medendorp K., Vreede L., van Groningen J.J., Hetterschijt L., Brugmans L., Jansen P.A., van den Hurk W.H., de Bruijn D.R., van Kessel A.G. 2010. The mitotic arrest deficient protein MAD2B interacts with the clathrin light chain A during mitosis. PLoS ONE. 5:e15128 10.1371/journal.pone.0015128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musacchio A., Salmon E.D. 2007. The spindle-assembly checkpoint in space and time. Nat. Rev. Mol. Cell Biol. 8:379–393 10.1038/nrm2163 [DOI] [PubMed] [Google Scholar]
- Nelson J.R., Lawrence C.W., Hinkle D.C. 1996. Thymine-thymine dimer bypass by yeast DNA polymerase zeta. Science. 272:1646–1649 10.1126/science.272.5268.1646 [DOI] [PubMed] [Google Scholar]
- Okada T., Sonoda E., Yoshimura M., Kawano Y., Saya H., Kohzaki M., Takeda S. 2005. Multiple roles of vertebrate REV genes in DNA repair and recombination. Mol. Cell. Biol. 25:6103–6111 10.1128/MCB.25.14.6103-6111.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfleger C.M., Kirschner M.W. 2000. The KEN box: an APC recognition signal distinct from the D box targeted by Cdh1. Genes Dev. 14:655–665 [PMC free article] [PubMed] [Google Scholar]
- Pfleger C.M., Salic A., Lee E., Kirschner M.W. 2001. Inhibition of Cdh1-APC by the MAD2-related protein MAD2L2: a novel mechanism for regulating Cdh1. Genes Dev. 15:1759–1764 10.1101/gad.897901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pines J. 2011. Cubism and the cell cycle: the many faces of the APC/C. Nat. Rev. Mol. Cell Biol. 12:427–438 10.1038/nrm3132 [DOI] [PubMed] [Google Scholar]
- Prinz S., Hwang E.S., Visintin R., Amon A. 1998. The regulation of Cdc20 proteolysis reveals a role for APC components Cdc23 and Cdc27 during S phase and early mitosis. Curr. Biol. 8:750–760 10.1016/S0960-9822(98)70298-2 [DOI] [PubMed] [Google Scholar]
- Rape M., Kirschner M.W. 2004. Autonomous regulation of the anaphase-promoting complex couples mitosis to S-phase entry. Nature. 432:588–595 10.1038/nature03023 [DOI] [PubMed] [Google Scholar]
- Reber A., Lehner C.F., Jacobs H.W. 2006. Terminal mitoses require negative regulation of Fzr/Cdh1 by Cyclin A, preventing premature degradation of mitotic cyclins and String/Cdc25. Development. 133:3201–3211 10.1242/dev.02488 [DOI] [PubMed] [Google Scholar]
- Reimann J.D., Gardner B.E., Margottin-Goguet F., Jackson P.K. 2001. Emi1 regulates the anaphase-promoting complex by a different mechanism than Mad2 proteins. Genes Dev. 15:3278–3285 10.1101/gad.945701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sale J.E. 2013. Translesion DNA synthesis and mutagenesis in eukaryotes. Cold Spring Harb. Perspect. Biol. 5:a012708 10.1101/cshperspect.a012708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwab M., Lutum A.S., Seufert W. 1997. Yeast Hct1 is a regulator of Clb2 cyclin proteolysis. Cell. 90:683–693 10.1016/S0092-8674(00)80529-2 [DOI] [PubMed] [Google Scholar]
- Shirayama M., Zachariae W., Ciosk R., Nasmyth K. 1998. The Polo-like kinase Cdc5p and the WD-repeat protein Cdc20p/fizzy are regulators and substrates of the anaphase promoting complex in Saccharomyces cerevisiae. EMBO J. 17:1336–1349 10.1093/emboj/17.5.1336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirayama M., Tóth A., Gálová M., Nasmyth K. 1999. APC(Cdc20) promotes exit from mitosis by destroying the anaphase inhibitor Pds1 and cyclin Clb5. Nature. 402:203–207 10.1038/46080 [DOI] [PubMed] [Google Scholar]
- Shteinberg M., Protopopov Y., Listovsky T., Brandeis M., Hershko A. 1999. Phosphorylation of the cyclosome is required for its stimulation by Fizzy/cdc20. Biochem. Biophys. Res. Commun. 260:193–198 10.1006/bbrc.1999.0884 [DOI] [PubMed] [Google Scholar]
- Simpson L.J., Sale J.E. 2003. Rev1 is essential for DNA damage tolerance and non-templated immunoglobulin gene mutation in a vertebrate cell line. EMBO J. 22:1654–1664 10.1093/emboj/cdg161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson-Lavy K.J., Oren Y.S., Feine O., Sajman J., Listovsky T., Brandeis M. 2010. Fifteen years of APC/cyclosome: a short and impressive biography. Biochem. Soc. Trans. 38:78–82 10.1042/BST0380078 [DOI] [PubMed] [Google Scholar]
- Sonoda E., Okada T., Zhao G.Y., Tateishi S., Araki K., Yamaizumi M., Yagi T., Verkaik N.S., van Gent D.C., Takata M., Takeda S. 2003. Multiple roles of Rev3, the catalytic subunit of polzeta in maintaining genome stability in vertebrates. EMBO J. 22:3188–3197 10.1093/emboj/cdg308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart S., Fang G. 2005. Destruction box-dependent degradation of aurora B is mediated by the anaphase-promoting complex/cyclosome and Cdh1. Cancer Res. 65:8730–8735 10.1158/0008-5472.CAN-05-1500 [DOI] [PubMed] [Google Scholar]
- Sullivan M., Morgan D.O. 2007. Finishing mitosis, one step at a time. Nat. Rev. Mol. Cell Biol. 8:894–903 10.1038/nrm2276 [DOI] [PubMed] [Google Scholar]
- Vassilev L.T., Tovar C., Chen S., Knezevic D., Zhao X., Sun H., Heimbrook D.C., Chen L. 2006. Selective small-molecule inhibitor reveals critical mitotic functions of human CDK1. Proc. Natl. Acad. Sci. USA. 103:10660–10665 10.1073/pnas.0600447103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visintin R., Prinz S., Amon A. 1997. CDC20 and CDH1: a family of substrate-specific activators of APC-dependent proteolysis. Science. 278:460–463 10.1126/science.278.5337.460 [DOI] [PubMed] [Google Scholar]
- Visintin R., Craig K., Hwang E.S., Prinz S., Tyers M., Amon A. 1998. The phosphatase Cdc14 triggers mitotic exit by reversal of Cdk-dependent phosphorylation. Mol. Cell. 2:709–718 10.1016/S1097-2765(00)80286-5 [DOI] [PubMed] [Google Scholar]
- Wäsch R., Cross F.R. 2002. APC-dependent proteolysis of the mitotic cyclin Clb2 is essential for mitotic exit. Nature. 418:556–562 10.1038/nature00856 [DOI] [PubMed] [Google Scholar]
- Waters L.S., Walker G.C. 2006. The critical mutagenic translesion DNA polymerase Rev1 is highly expressed during G(2)/M phase rather than S phase. Proc. Natl. Acad. Sci. USA. 103:8971–8976 10.1073/pnas.0510167103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zachariae W., Schwab M., Nasmyth K., Seufert W. 1998. Control of cyclin ubiquitination by CDK-regulated binding of Hct1 to the anaphase promoting complex. Science. 282:1721–1724 10.1126/science.282.5394.1721 [DOI] [PubMed] [Google Scholar]
- Zur A., Brandeis M. 2001. Securin degradation is mediated by fzy and fzr, and is required for complete chromatid separation but not for cytokinesis. EMBO J. 20:792–801 10.1093/emboj/20.4.792 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.