The prolyl isomerase Pin1 stimulates the dephosphorylation of histone H1, stabilizing its binding to chromatin at transcriptionally active chromatin.
Abstract
Histone H1 plays a crucial role in stabilizing higher order chromatin structure. Transcriptional activation, DNA replication, and chromosome condensation all require changes in chromatin structure and are correlated with the phosphorylation of histone H1. In this study, we describe a novel interaction between Pin1, a phosphorylation-specific prolyl isomerase, and phosphorylated histone H1. A sub-stoichiometric amount of Pin1 stimulated the dephosphorylation of H1 in vitro and modulated the structure of the C-terminal domain of H1 in a phosphorylation-dependent manner. Depletion of Pin1 destabilized H1 binding to chromatin only when Pin1 binding sites on H1 were present. Pin1 recruitment and localized histone H1 phosphorylation were associated with transcriptional activation independent of RNA polymerase II. We thus identify a novel form of histone H1 regulation through phosphorylation-dependent proline isomerization, which has consequences on overall H1 phosphorylation levels and the stability of H1 binding to chromatin.
Introduction
Histone H1 has an important role in the formation and mechanical stability of the 30-nm chromatin fiber by facilitating folding and increasing internucleosomal contacts (Thoma and Koller, 1977; Thoma et al., 1979; Bednar et al., 1998; Carruthers et al., 1998; Hansen, 2002; Robinson and Rhodes, 2006; Kruithof et al., 2009). Reversible phosphorylation of H1 is the most extensively studied post-translational modification in a wide range of cellular processes. It is maintained by the antagonistic actions of protein phosphatases and CDC2/CDK2 kinase activities (Roth et al., 1991; Herrera et al., 1996; Paulson et al., 1996; Swank et al., 1997). The kinases require the presence of a consensus sequence (T/S)PXZ, where X can be any amino acid and Z represents a basic amino acid (Moreno and Nurse, 1990). Different variants of H1 have different numbers of these motifs. For example, H1.1 has two T/SPKK sites, whereas H1.5 has five (Parseghian and Hamkalo, 2001). Additionally, whereas interphase phosphorylation of H1 is restricted to Ser residues, both Thr and Ser residues are phosphorylated in mitosis (Sarg et al., 2006; Zheng et al., 2010), resulting in a maximally phosphorylated state at the G2–M transition (Bradbury, 1992; Roth and Allis, 1992; Th’ng et al., 1994; Talasz et al., 1996).
Increased levels of H1 phosphorylation are observed in cells that express several oncogenes and this correlates with a relaxed chromatin structure (Chadee et al., 1995; Taylor et al., 1995). H1 phosphorylation promotes chromatin decondensation at transcriptionally active sites to allow access to other DNA-binding proteins (Hohmann, 1983; Roth and Allis, 1992; Chadee et al., 1995; Lu et al., 1995; Koop et al., 2003; Vicent et al., 2011).
Although our interpretation of the function of histone H1 phosphorylation has largely been dominated by the assumption that such phosphorylations are regulated by electrostatic processes, the recent recognition of the C-terminal domain (CTD) of histone H1 as an intrinsically disordered structure that adopts a more structured state when it interacts with DNA or nucleosomes (Clark et al., 1988; Roque et al., 2005; Caterino et al., 2011; Fang et al., 2012) necessitates other considerations. Proline isomerization is a mechanism to significantly alter the structure of a protein in a single enzymatic step. Interestingly, the phosphorylation sites within the CTD of H1 are all adjacent to prolines and match the known target sequence of the phosphorylation-directed proline isomerase activity of Pin1.
In this study, we examined whether or not phosphorylated S/T-Pro residues on H1 act as substrates for Pin1, a peptidyl-prolyl isomerase (PPIase). Pin1 recognizes and catalyzes the interconversion between the cis and trans conformations of the peptidyl-prolyl bond (Lu et al., 1996). Pin1 is a highly abundant nuclear protein that is essential for progression through the cell cycle, and has been shown to interact with a host of proteins, including RNA polymerase II and Cdc25 (Lu et al., 1996; Albert et al., 1999; Stukenberg and Kirschner, 2001). Pin1 has two domains, an N-terminal WW domain that recognizes and binds phosphorylated Ser/Thr-Pro residues and a C-terminal PPIase domain (Lu et al., 1999; Lu et al., 2007). Isomerization can induce a conformational change in the protein backbone of a substrate, which has been shown to alter the catalytic activity, localization, and stability, as well as the kinetics of phosphorylation and dephosphorylation events (Zhou et al., 2000; Stukenberg and Kirschner, 2001).
In this study, we found that Pin1 binds to histone H1 in a phosphorylation-dependent manner. Using fluorescence resonance energy transfer (FRET), we determined that Pin1 could directly alter the conformation of the phosphorylated but not the nonphosphorylated H1 CTD when bound to nucleosomes in vitro. Furthermore, sub-stoichiometric levels of Pin1 were found to promote H1 dephosphorylation in vitro, consistent with an isomer preference for H1 phosphatase activity. Pin1 stabilized the binding of H1 on chromatin by increasing its residence time. Pin1 and H1 phosphorylation levels were found to increase early after transcriptional activation, which is consistent with H1 phosphorylation playing a crucial role in transcription (Langan, 1969; Lamy et al., 1977; Koop et al., 2003; Zheng et al., 2010). In the absence of Pin1, transcriptionally active and inactive sites are further decondensed and associated with increased H1 mobility. Together, our results implicate Pin1 and phosphorylation-dependent proline isomerization as a chromatin regulatory mechanism that promotes a more compact chromatin state. As the only histone protein containing Pin1 target sites, Pin1-dependent regulation of histone H1, stimulating its dephosphorylation and promoting its binding to chromatin, is a promising mechanism to explain this function.
Results
Pin1 interacts with histone H1
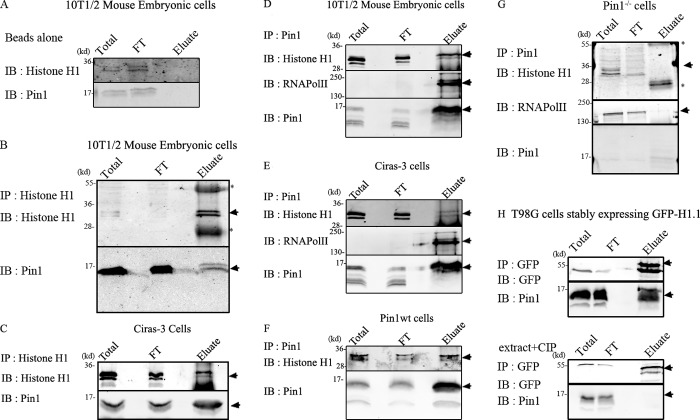
Interaction between Pin1 and H1 was analyzed using antibodies specific to H1 or Pin1 bound to magnetic Dynabeads to immunoprecipitate proteins from nuclear extracts. The beads did not bind any detectable amount of either H1 or Pin1 in the absence of antibodies (Fig. 1 A). Anti-H1 antibodies coimmunoprecipitated Pin1 in 10T1/2 cells (Fig. 1 B) and Ciras-3 cells (Fig. 1 C). In a reciprocal experiment, H1 coimmunoprecipitated with Pin1 in 10T1/2, Ciras-3, and Pin1wt cells (Fig. 1, D–F). Pin1 antibody did not pull down H1 in Pin1−/− cells (Fig. 1 G; Fujimori et al., 1999; Liou et al., 2002). We confirmed that Pin1 was physically bound to GFP-H1.1, using extracts from T98G cells that stably expressed GFP-H1.1. This interaction was dependent upon the phosphorylation status of H1 because treatment of the nuclear extracts with calf intestinal phosphatases before immunoprecipitation prevented Pin1 from binding to GFP-H1.1 (Fig. 1 H). These observations are consistent with previous reports using Pin-1 pull-down experiments to show that H1 is a substrate of Pin1 (Tatara et al., 2010; Steger et al., 2013).
Figure 1.
Pin1 interacts with histone H1. Coimmunoprecipitation experiments were performed to test whether Pin1 and H1 interacted with each other in vivo. “Total” refers to the total nuclear extract before the addition of the antibody, and “FT” refers to flow-through (∼3–6% of the total volume). The entire contents of the eluate were run on the gel. Black lines indicate that intervening lanes were spliced out, and arrows indicate bands that correspond to the protein being IB. Asterisks indicate heavy/light chain IgG antibodies that form part of the eluate. (A) Under the conditions used, both histone H1 and Pin1 did not bind beads nonspecifically. Histone H1 antibodies were used to immunoprecipitate (IP) H1 from mouse embryonic cells (B) and from Ciras-3 cells (C). Immunoblots (IB) reveal pull-down of Pin1 along with histone H1 demonstrating their association in vitro. Reciprocal experiments were performed using Pin1 antibody to pull down Pin1 from extracts prepared from 10T1/2 mouse embryonic cells (D), Ciras-3 cells (E), and Pin1wt cells (F) and Pin1−/− cells (G). RNA polymerase II, which is an established substrate for Pin1, was used as a positive control. Both H1 and RNA polymerase II form a part of the eluate in 10T1/2 and Ciras-3 cells, but not in Pin1−/− cells, demonstrating specific interactions mediated by Pin1. (H, top) Interaction between Pin1 and GFP-H1.1 in extracts prepared from T98G cells stably expressing GFP-H1.1. GFP-H1.1 was immunoprecipitated using GFP antibody coupled to magnetic particles (GFP-Trap). This interaction is dependent on the phosphorylation status of proteins (H, bottom) as treatment of the extracts with calf intestinal phosphatase (CIP), a general nonspecific protein phosphatase, abrogated the interaction between H1 and Pin1.
Pin1 promotes the dephosphorylation of H1
Pin1 has been shown to promote the dephosphorylation of substrates, such as Cdc25 and Tau proteins (Zhou et al., 2000). Elegant in vitro experiments found that the major proline-directed protein phosphatase PP2A specifically dephosphorylates the trans-isomer of the pSer/Thr-Pro bond (Zhou et al., 2000), thereby imparting a post-phosphorylation, conformation-specific regulatory step to Pin1 substrates. The cis isomer has to achieve the right conformation either through slow spontaneous isomerization, or through Pin1-mediated isomerization in order to be a substrate of phosphatases. We hypothesized that a similar Pin1-mediated regulatory mechanism for H1 may also exist.
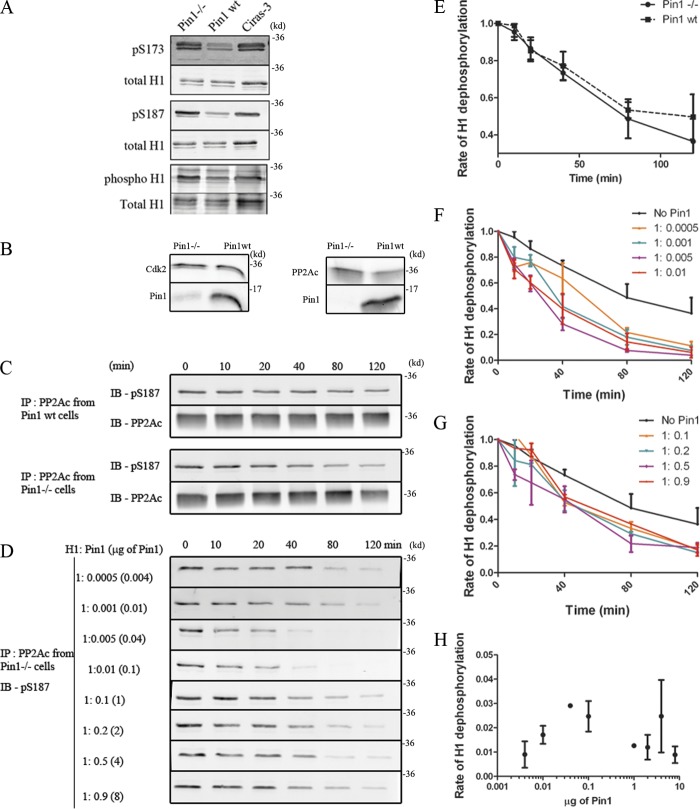
We first determined whether or not steady-state H1 phosphorylation levels were altered in Pin1−/− cells compared with their wild-type counterparts. We probed both total H1 phosphorylation levels using a general phospho-specific stain, and site/variant-specific (H1.2/H1.5 pS173, H1.4 pS187; Zheng et al., 2010) phosphorylation levels in Pin1wt and Pin1−/− cells (Fig. 2 A). We found that Pin1−/− cells had higher total levels of phosphorylated H1 histones including pS173 and pS187 levels compared with the Pin1wt cells. Ciras-3 cells, mouse embryonic fibroblasts with an activated ras pathway, were used as positive control.
Figure 2.
Pin1 promotes H1 dephosphorylation. (A) Histones were extracted from Pin1−/−, Pin1wt, and Ciras-3 cells, and were then probed with either pS173(H1.2/H1.5), pS187(H1.4), a phospho-specific stain that labels all phosphorylated proteins, or with a stain that labels total protein. Levels of pS173, pS187, and net H1 phosphorylation levels were found to be higher in Pin1−/− cells as compared with Pin1wt cells, similar to those observed in Ciras-3 cells (positive control). (B) Nuclear extracts from Pin1−/− cells and Pin1wt cells revealed that the levels of Cdk2 and PP2Ac were similar in both cells. (C) The dephosphorylation activity of PP2Ac activity was analyzed using purified H1 as a substrate. PP2Ac was immunoprecipitated from either Pin1wt cells or Pin1−/− cells and assessed for its ability to dephosphorylate pS187. The kinetics of this dephosphorylation reaction are plotted in E with each dot/square representing the average H1 phosphorylation level obtained from at least three independent experiments. The average intensity from the zero-minute time point is set as the maximum, against which all other time points are compared. (D) PP2Ac was immunoprecipitated from Pin1−/− cells and was mixed with a constant amount of H1, while levels of purified Pin1 were varied from 0.004 to 8 µg. The former corresponds to a molar stoichiometry of H1/Pin1 = 1:0.0005, whereas the latter corresponds to H1/Pin1 = 1:0.9. The kinetics of dephosphorylation is plotted in F and G, with the average intensity at the zero-minute time point set to 1. These curves were then submitted to a one-phase decay curve analysis and the rate obtained was plotted as a function of the amount of Pin1 added to the reaction (H).
The higher levels of phosphorylated H1 molecules in Pin1−/− cells compared with Pin1wt cells were not due to increased levels (Fig. 2 B) or altered activity of Cdk2 (Fig. S1, A–D). The levels (Fig. 2 B) and activity of PP2Ac were also found to be similar in both Pin1wt and Pin1−/− cells (Fig. 2, C and E). Upon modeling the curves based on a one-phase decay, we found the rate of dephosphorylation in PP2Ac extracted from Pin1−/− cells to be 0.019 ± 0.006 min−1, whereas the extract from Pin1wt cells registered a dephosphorylation rate of 0.014 ± 0.003 min−1. This suggested that PP2A activity was not compromised in Pin1−/− cells.
To test if Pin1 was able to impose a post-phosphorylation regulatory step, we performed the H1 dephosphorylation assay in the presence of increasing amounts of Pin1 (Fig. 2, D, F, and G). We found that at sub-stoichiometric concentrations (molar ratio of H1/Pin1 = 1:0.0005 to 1:0.1) Pin1 increased the rate of H1 dephosphorylation. The rate of H1 dephosphorylation increased from 0.019 ± 0.006 min−1 observed in the absence of Pin1 to 0.0291 ± 0.0003 min−1 at a molar stoichiometry of H1/Pin1 = 1:0.005 (0.04 µg of Pin1; Fig. 2 H). This suggested that sub-stoichiometric levels of Pin1 are able to promote H1 dephosphorylation in vitro. The increased rate of H1 dephosphorylation was abrogated in the presence of in vitro inhibitor of Pin1, Juglone (Juglone/Pin1 = 10:1; Hennig et al., 1998), alluding toward the requirement of the catalytic activity of Pin1 for such an effect (Fig. S1 E). At higher levels of Pin1, with the molar stoichiometry approaching 1:1 (H1/Pin1), we noticed that the rate of H1 dephosphorylation approached that seen in the absence of Pin1. This was probably due to competition between Pin1 and PP2A for binding to the H1 substrate.
To investigate whether or not Pin1 promoted H1 dephosphorylation in living cells, we used roscovitine, a competitive inhibitor of Cdk2, and measured the rate of in vivo pS187 dephosphorylation kinetics in both Pin1−/− and Pin1wt cells. Consistent with the role of Pin1 in regulating the kinetics of Cdc25 and tau protein phosphorylation, we observed the rate of H1 dephosphorylation of pS187 was faster in Pin1wt cells, compared with that of Pin1−/− cells (Fig. S1, F and G). Fitting these curves using one-phase decay kinetics, we found a higher half-life for H1 phosphorylation of 72.81 min in Pin1−/− cells compared with 58.93 min in Pin1wt cells, explaining the higher steady-state levels of pS187 observed in Pin1−/− cells. This suggested that Pin1 promoted the dephosphorylation of these Ser residues on H1. Similarly, the level of pT146 was higher in Pin1−/− cells but, in contrast to pS187, displayed a similar apparent rate of dephosphorylation compared with wild-type cells (Fig. S1, H and I). However, it was difficult to assess the initial rate of dephosphorylation given the rapid rate at which Thr residues are dephosphorylated (an estimated half-life of 7–8 min compared with 59–72 min for Ser residues).
Pin1 and phosphorylation of H1 cause conformational changes in H1 CTD
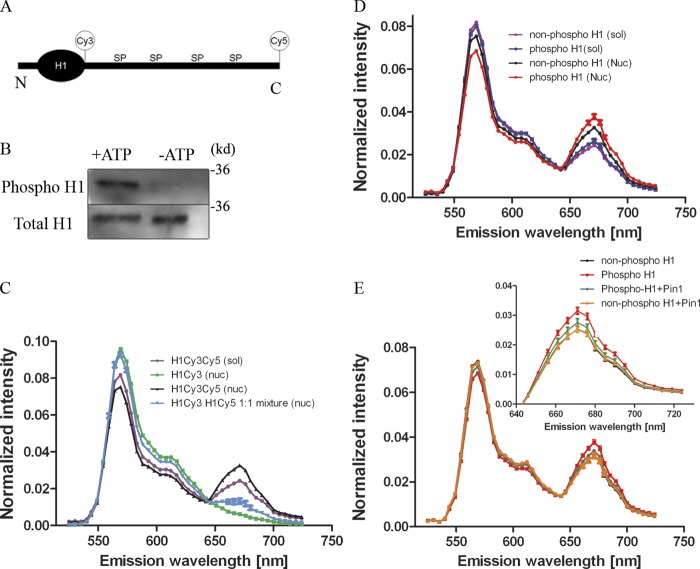
The H1 CTD adopts a random disordered structure in solution, whereas in the presence of DNA and nucleosomes it is thought to assume characteristics of classical secondary structures, such as α-helices and β-sheets (Clark et al., 1988; Roque et al., 2005, 2007; Caterino et al., 2011). Phosphorylation of H1 increases the proportion of β-sheets at the expense of α-helices (Roque et al., 2008). Recently, H1 labeled with Cy3 and Cy5 on either end of the CTD in the same molecule was used to show that H1 binding to nucleosomes brings the two ends of the CTD into close proximity, resulting in significant FRET (Fang et al., 2012). The same level of FRET is not attained with H1 in solution, confirming the change in conformation only upon binding nucleosomes. Given these studies, we tested whether or not phosphorylation of labeled H1 and Pin1 would alter FRET levels, reflecting a further change in the conformation of the CTD.
We used purified Xenopus H1 that was directly labeled with either Cy3 or Cy5 or both on either end of the CTD, as shown in Fig. 3 A. Fluorescence emission spectra were recorded with excitation at 514 nm and 633 nm with 5-nm slit width. Minimal excitation of H1-Cy5 with 514 nm was observed, whereas H1-Cy3 produced a spectra characteristic of a Cy3 signal at this wavelength (Fig. S2 B). Excitation of H1 Cy3Cy5 with 633 nm also produced an emission spectra characteristic of H1Cy5, indicating that the addition of a Cy3 tag did not compromise the emission spectra of Cy5 (Fig. S2 C). This signal increased dramatically when both Cy3 and Cy5 were present on the same molecule, as seen in the emission spectra for free H1Cy3Cy5 (Fig. 3 C). There is a further increase in FRET when the same molecules are mixed in the presence of reconstituted nucleosomes (1:1 stoichiometry), suggesting a change in the conformation of the CTD upon binding nucleosomes (Fig. 3 C). Consistent with previous results, H1Cy3 and H1Cy5 mixed together at a net 1:1 stoichiometry with nucleosomes and showed minimal inter-molecular FRET, as judged by the small amount of Cy5 emission at 671 nm (Fig. 3 C).
Figure 3.
Pin1 and H1 phosphorylation change the structure of the CTD. (A) The position of the Cy3 and Cy5 label are indicated in relation to the whole H1 molecule, not to scale (N, N-terminal; C, C-terminal; SP, Ser-Pro). (B) H1 labeled with Cy3 and Cy5 were treated with Cdk2 immunoprecipitated from Pin1−/− cells in the presence or absence of ATP and probed with a phospho-specific stain. These blots reveal successful phosphorylation of H1 in the presence of ATP (now referred to as phosphoH1), whereas Cdk2 was unable to phosphorylate labeled H1 molecules in the absence of ATP (now referred to as nonphospho H1). (C) Labeled phospho H1 molecules were then diluted either in solution (sol) or with reconstituted nucleosomes (nuc). A 514-nm laser was then used to excite the molecules and fluorescence emission spectra was obtained from 525–724nm (5-nm slit width). Fluorescence intensity was normalized to the total fluorescence intensity obtained from each spectrum. The spectra show a slight increase in FRET signal (peak at 671 nm) in the mono-labeled H1s (either Cy3 or Cy5) mixed with each other in 1:1 stoichiometry together with nucleosomes, indicating inter-molecular FRET, whereas this signal increases dramatically when both Cy3 and Cy5 are on the same H1 molecule. (D) FRET signal was compared between phosphorylated H1 and nonphosphorylated H1 in solution versus these molecules added to reconstituted nucleosomes. Although FRET signal remains the same when H1 is in solution, FRET signal is dependent on the phosphorylation status of H1 in the presence of reconstituted nucleosomes. (E) FRET signal was compared between phosphorylated H1 and nonphosphorylated H1 with reconstituted nucleosomes in the presence or absence of Pin1. Although phosphorylation alone increases the FRET signal, addition of Pin1 reduces this signal toward that of the nonphosphorylated H1 molecules.
Next, H1Cy3Cy5 was treated with Cdk2 immunoprecipitated from Pin1−/− cells in the presence or absence of ATP (Fig. 3 B). Addition of labeled in vitro–phosphorylated H1 to reconstituted nucleosomes led to higher levels of FRET signal when compared with labeled H1 that was not phosphorylated (incubated in buffer containing Cdk2 devoid of ATP; Fig. 4 D). This change in FRET signal between phosphorylated and nonphosphorylated H1 was not observed when H1 molecules are in solution devoid of nucleosomes, indicating that changes in the conformation of the H1 CTD induced by H1 phosphorylation occur only in the nucleosomal bound H1 state. The difference in Cy5 emission at 671 nm between phosphorylated H1 molecules and nonphosphorylated H1 molecules, upon binding of nucleosomes, was significantly reduced in the presence of stoichiometric levels of Pin1 (0.1 µg of Pin1 or 1:1.4 H1/Pin1 molar ratio; Fig. 3 E). In fact, FRET levels of phosphorylated H1 molecules were restored to the levels seen in nonphosphorylated H1 molecules in the presence of Pin1 (Fig. 3 E). For example, the FRET efficiency calculated using the RatioA method (Clegg, 1992; Poirier et al., 2009; Fang et al., 2012) increases from 0.222 ± 0.008 to 0.27 ± 0.01 upon phosphorylation of H1 (Fig. S2 D). This translates to a decrease in the separation between the Cy3 and Cy5 tags from 6.18 ± 0.05 nm in the nonphosphorylated state to 5.94 ± 0.06 nm in the phosphorylated state. Upon addition of Pin1, the FRET efficiency of phosphorylated H1 molecules bound to nucleosome was reduced to 0.24 ± 0.01, increasing the apparent separation between Cy3 and Cy5 to 6.09 ± 0.08 nm. Adding Pin1 did not change the FRET levels of nonphosphorylated H1, suggesting once again that the interaction of H1 and Pin1 was dependent on H1 phosphorylation. Furthermore, the change in FRET levels was dependent upon the catalytic activity of Pin1 because addition of the in vitro Pin1 inhibitor Juglone abrogated this effect (Fig. S2 E).
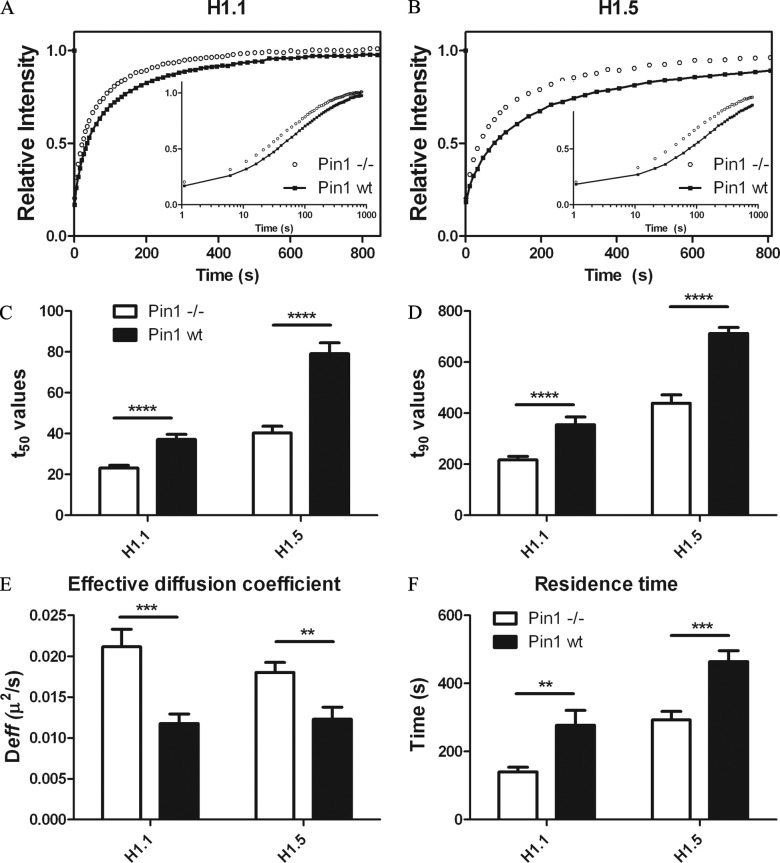
Figure 4.
Pin1 stabilizes GFP-H1.1 and GFP-H1.5 dynamics. GFP H1.1 (A) or GFP-H1.5 (B) was expressed either in Pin1−/− cells or Pin1wt cells. FRAP experiments were performed to measure the dynamics of H1; each curve represents an average of ∼20 cells (total) in three independent experiments. The inset represents the same FRAP curve with the x-axis in log (time) to highlight changes in the earlier phases of the FRAP curve. Both H1.1 and H1.5 recover much faster in Pin1−/− cells as compared with Pin1wt cells. This trend was affirmed with a statistically significant increase in both t50 (C) and t90 (D) values in the presence of Pin1. Mathematical modeling of FRAP curves show that Pin1 causes a decrease in effective diffusion coefficient (E), a measure of the freely diffusing and low-affinity population, while at the same time causes increases residence time (F) of the high-affinity H1 population. Significance between Pin1−/− vs. Pin1wt was analyzed using unpaired t test (95% confidence interval). Notation for significance: *** if P value is < 0.001; ** if P is between 0.001 and 0.01; * if P is between 0.01 and 0.05.
Pin1 depletion destabilizes H1 binding
To determine whether or not Pin1 regulates H1 binding in vivo, we tested the contribution of Pin1 to H1 binding by performing fluorescence recovery after photobleaching (FRAP) experiments with N-terminally GFP-tagged H1.1 (enriched in euchromatin) and H1.5 (enriched in heterochromatin; Th’ng et al., 2005). Human H1.1 has one Thr-Pro site (TP) and one Ser-Pro site (SP) on its CTD at positions 152 and 183, respectively. In contrast, H1.5 has the highest number of T/SP sites among known human H1 variants, having three TP and three SP sites.
The dynamics of H1.1 and H1.5 were markedly different when compared in Pin1wt versus Pin1−/− cells (Fig. 4, A and B). For instance, the t50 values of H1.1 decreased from 37 ± 2 s in Pin1wt cells to 23 ± 1 s in Pin1−/− cells (Fig. 4, C and D). In Pin1wt cells expressing GFP H1.5, a t50 value of 80 ± 5 s was recorded, which was half of that observed in Pin1−/− cells, where a t50 value of 40 ± 3 s was observed. Thus, both H1.1 and H1.5 molecules are more mobile in the absence of Pin1.
Upon mathematical modeling of the FRAP curves (Carrero et al., 2004a,b), we observed that Pin1 induces a significant increase in the residence time of H1.1, from 140 ± 13 s in Pin1−/− cells to 277 ± 44 s in Pin1wt cells, and of H1.5 molecules, 464 ± 32 s in Pin1wt cells to that of 292 ± 24 s in Pin1−/− cells (Fig. 4, E and F). This suggests that the H1.1 molecules were able to engage in high-affinity interactions for longer durations in the presence of Pin1. Furthermore, a steep reduction in the effective diffusion coefficient of H1.1 and H1.5 in the presence of Pin1 was observed. Given that the diffusion rate is unlikely to change, the reduction in effective diffusion coefficient implies either a greater proportion of H1 pool that is bound to chromatin, or alternatively, although not mutually exclusive, that there is an increase in the affinity of the weakly bound H1 fraction in the presence of Pin1.
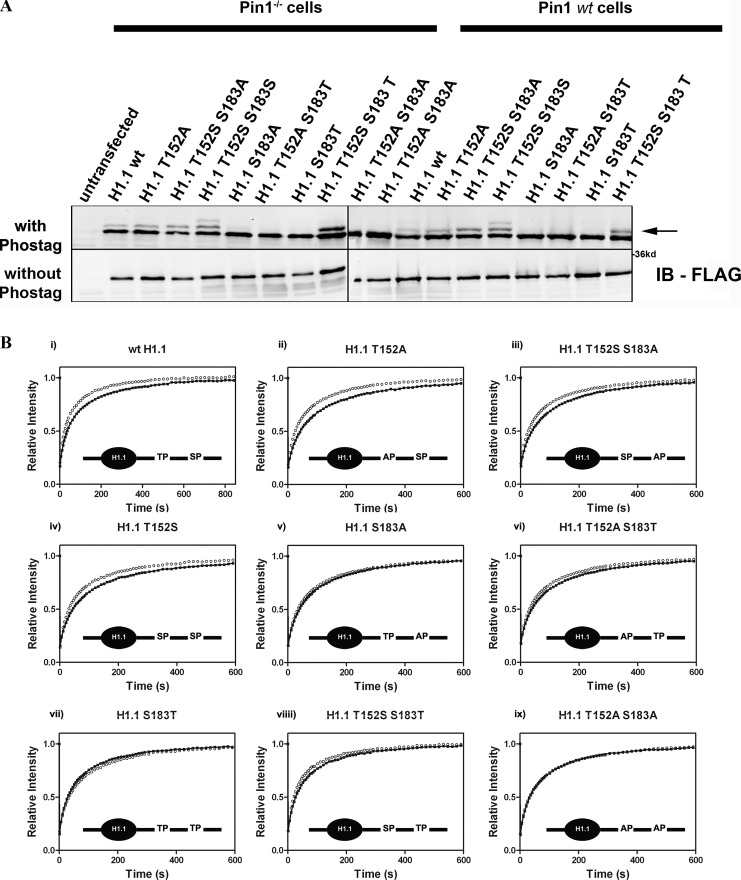
To test the contribution of specific residues on H1.1 that are involved in interacting with Pin1, we created a series of point mutations in H1.1, where Thr152 and Ser183 were mutated and/or switched to Ala/Ser/Thr. We first confirmed that these mutants can be phosphorylated in vivo by expressing FLAG-tagged versions of these mutants in either Pin1wt or Pin1−/− cells and then separating the extracted histones on a 10% SDS-gel ± Phos-tag (Fig. 5 A). Phos-tag is a phosphate-binding molecule that, when supplemented in an acrylamide gel, retards phospho-proteins, thus decreasing the mobility of any phosphorylated species of a protein (Kinoshita et al., 2008, 2009). We found that in the absence of Phos-tag all FLAG-H1.1 mutants migrate as single band; however, in the presence of Phos-tag there is a mutant-specific reduction in the migration. Wild-type H1.1, for example, migrates as two distinct species, a lower nonphosphorylated band and an upper mono-phosphorylated band. Consistent with previous reports (Sarg et al., 2006; Zheng et al., 2010), Ser was the primary residue to be phosphorylated in H1 extracted from primarily interphase cells and, interestingly, the phosphorylation status of Ser was independent of its position on the H1 molecule (152 or 183). There was no evidence for significant Thr phosphorylations in our extracts. Consistent with these data, only residues that were phosphorylated were subject to Pin1 mediated stabilization of H1 binding in FRAP experiments. In other words, for wild-type H1.1, most of the changes induced by Pin1 were mediated through its interaction with Ser at position 183 (and not Thr 152) in H1.1 (Fig. 5 B). Furthermore, substituting these residues with Ala abrogated the Pin1-mediated stabilization of H1, suggesting that the effect of Pin1 on H1 dynamics was a direct result of Pin1 interacting with phosphorylation-compatible residues on H1.
Figure 5.
Mobility shift assay for detecting phosphorylated H1 and FRAP analysis of H1.1 mutants. (A) FLAG-tagged H1.1 wt and H1.1 mutants were transfected in Pin1−/− and Pin1wt cells. Histones were then extracted using 0.4 N H2SO4 and the extracts were then run in a 10% acrylamide gel ± Phos-tag. Phos-tag is a ligand that interacts with phosphate molecules imparting shifts in mobility. H1.1wt migrates as two distinct species in the presence of Phos-tag, whereas H1.1T152S migrates as three distinct species (shown by arrows). In the absence of Phos-tag, all mutants migrate as a single band. (B) GFP H1.1 (i) or GFP H1.1 mutants (ii–ix) were expressed either in Pin1wt (black filled circles) or Pin1−/− (open circles) cells. FRAP experiments were performed to measure the dynamics of the H1 molecules. Each curve represents an average of ∼20 cells (total), three independent experiments. The inset is a diagrammatic representation of the genetic alteration and relative position of serines (S), theonines (T), prolines (P), and alanines (A). (ii–iv) Role played by serine at either position 183 or 152 in contributing toward Pin1 mediated changes in H1 dynamics. (v–vii) Role played by altering the Thr residue on H1.1 in Pin1-mediated changes in H1 dynamics. (viii) Recovery of H1.1 when the ser and thr positions are switched. (ix) Lack of any change in H1 dynamics when both the ser and thr residues are changed to ala.
We further tested the impact of other known peptidyl-prolyl cis-trans isomerases, the cyclophilins and FKBP, on H1 mobility, by selectively inhibiting these classes of proteins using cyclosporine A and rapamycin, respectively. A 1-h incubation of 10T1/2 cells in the presence of either drug produced no change in H1.1 dynamics (Fig. S3 A). These results show that among the classes of prolyl-isomerases, only Pin1 affects the mobility of H1 in a significant manner.
To determine if the changes in H1 mobility in Pin1−/− and Pin1wt cells could be due to other chromatin modifications that have the potential to modify H1 mobility, we analyzed the composition of core histones and the level of histone acetylation by acetic acid-urea-Triton X-100 PAGE and found each to be similar in both Pin1−/− and Pin1wt cells (Fig. S3 B). These results help confirm that the changes seen in H1 mobility are a direct consequence of an interaction between H1 and Pin1.
Role of Pin1 in transcription
Phosphorylated histone H1 molecules have been shown to be enriched at sites of active transcription, and in vitro studies have shown that phosphorylation of H1 is one of the prerequisite steps for gene induction (Koop et al., 2003; Zheng et al., 2010; Vicent et al., 2011). In addition, Pin1 has also been shown to interact with proteins involved in transcription, most notably RNA polymerase II (Xu and Manley, 2007). Furthermore, chromatin has been shown to decondense after transcriptional activation (Tumbar et al., 1999). We quantified chromatin condensation, levels of Pin1, H1 phosphorylation, and the dynamics of H1 at transcriptionally silent versus active chromatin to determine how Pin1 modulated these events associated with transcription.
We first analyzed the changes in H1 phosphorylation and Pin1 levels at transcriptionally active regions in vivo. We used the lac array system developed by the Belmont laboratory and, more specifically, an array system constructed by the Spector and Janicki laboratories (Robinett et al., 1996; Tsukamoto et al., 2000; Janicki et al., 2004; Rafalska-Metcalf et al., 2010). The system is comprised of 256 repeats of the lac operon, followed by 96 repeats of tet-responsive elements that are upstream of a minimal CMV promoter, which drives the transcription of a CFP-SKL gene. The CFP-SKL gene also has MS2 repeats, which when transcribed bind transfected YFP-MS2 proteins and is used as readout for transcriptional elongation (Rafalska-Metcalf et al., 2010). Transcription is activated by adding Tamoxifen, which binds mcherry-ER-tTA, causing it to relocalize to the nucleus and bind the tet-responsive elements, allowing increased accumulation of RNA polymerase II (Fig. S4 B) and YFP-MS2 enrichment (Fig. S4 C) at these sites (Rafalska-Metcalf et al., 2010). Enrichment of YFP-MS2 and accumulation of RNA polymerase II were used as surrogate readouts for the level of transcriptional activity. To visualize transcriptionally inactive chromatin, we transfected cells with mcherry-LacR alone (Rafalska-Metcalf et al., 2010).
Upon activation of transcription, we found that there was a twofold increase in the levels of H1 phosphorylation at the lac arrays (Fig. 6 B; Fig. S4 D; Dou et al., 1999; Koop et al., 2003; Zheng et al., 2010). This increase was significantly higher than H1 phosphorylation levels seen at transcriptionally inactive sites, using mcherry LacR as a marker to generate a mask for measurements. Surprisingly, we found that the increase in H1 phosphorylation levels was independent of transcript elongation by RNA polymerase II because overnight pretreatment with α-amanitin before the activation of transcription by Tamoxifen led to the accumulation of high levels of H1 phosphorylation at the lac arrays. Note that this increase at the lac arrays is in contrast to the reduction in total H1 phosphorylation levels after inhibition of transcription by α-amanitin treatment (Fig. S4 G; Chadee et al., 1997).
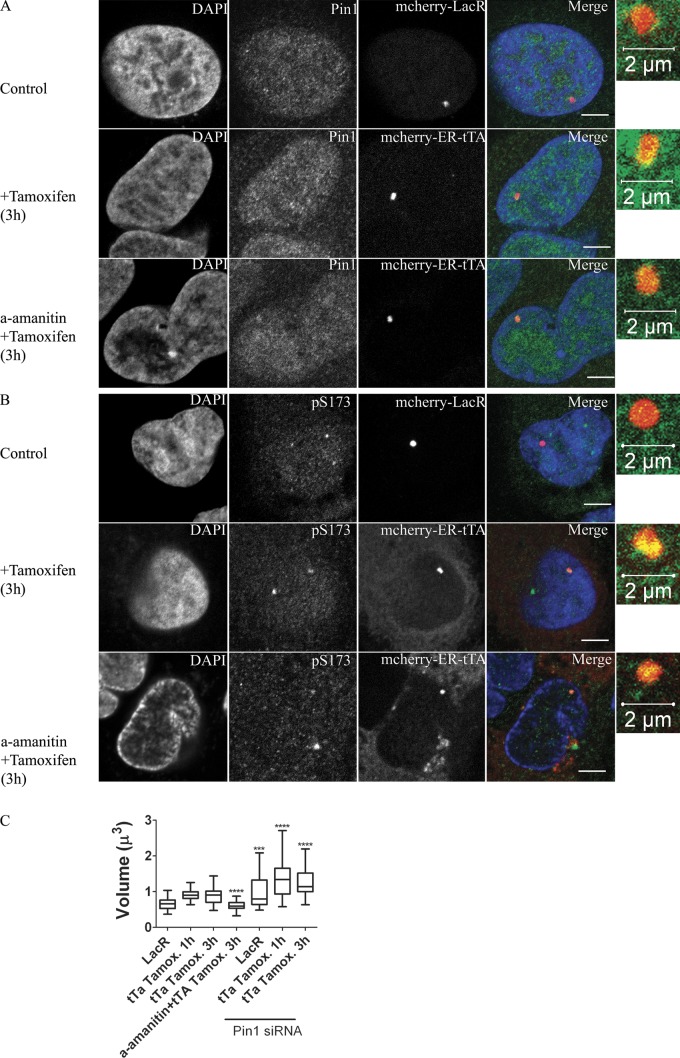
Figure 6.
Pin1 and H1 phosphorylation are marks of transcriptionally competent chromatin. U2Os 263 cells harboring lac arrays followed by TRE, CMV promoter, and CFP-SKL gene were either transfected with mCherry LacR or mCherry ER-tTA. The former represented the transcriptionally inactive state while addition of tamoxifen (3 h) to the latter represented the transcriptionally active state of chromatin. Pin1 (A) and pS173H1.2 (B) levels were measured using immunofluorescence. Both Pin1 and pS173H1.2 levels were found to increase at sites of active transcription. α-Amanitin, was used to deplete RNA polymerase II levels in the cells. When transcription was activated in these competent, yet transcriptionally silent cells, Pin1 and H1 phosphorylation levels were elevated, suggesting that these were early events in the initiation of transcription. Bar, 5 µm (unless otherwise specified). (C) The volume occupied by the arrays was measured in both the transcriptionally inactive state (mCherry LacR alone) and in the transcriptionally active state (addition of tamoxifen to cells expressing mCherry ER-tTA for either 1 h or 3 h). Volume was measured through rapid acquisition of z-stacks in living cells. Whereas transcription caused an increase in the volume occupied by the arrays, treatment of cells with α-amanitin led to compact arrays. Both transcriptionally active and inactive arrays were found to occupy larger volumes when Pin1 was depleted by Pin1siRNA treatment. Significance between control vs. treated (amanitin or Pin1si-RNA) was analyzed using unpaired t test (95% confidence interval). Notation for significance: *** if P value is < 0.001; ** if P is between 0.001 and 0.01; * if P is between 0.01 and 0.05.
Consistent with the increase in H1 phosphorylation, we observed an increase in the relative amounts of Pin1 at sites of transcription (Fig. 6 A; Fig. S4 E). The modest increase in Pin1 is significant given the high abundance of Pin1 in the nucleus. The increase in Pin1 was also independent of RNA polymerase II because overnight treatment with α-amanitin before the addition of Tamoxifen led to high levels of Pin1 being targeted to the mcherry-ER-tTA sites. This suggests that Pin1 accumulation and an increase in H1 phosphorylation are marks of transcriptionally competent chromatin and not dependent on transcriptional elongation.
In accordance with published literature, we observed that transcriptionally elongating chromatin were more decondensed, occupying 40% more nuclear volume compared with chromatin in a transcriptionally inactive state (Fig. 6 C; Fig. S4 F; Robinett et al., 1996; Tumbar et al., 1999; Müller et al., 2001; Janicki et al., 2004; Hu et al., 2009; Rafalska-Metcalf et al., 2010). Chromatin decondensation was strictly dependent on elongation induced by RNA polymerase II because an overnight treatment with α-amanitin completely abolished the decondensation induced by targeting mcherry-ER-tTA to the lac arrays (Fig. 6 C; Müller et al., 2001). Pin1 knockdown had a maximal effect on the nuclear volume occupied by the lac arrays (Fig. 6 C). Under reduced Pin1 levels, the lac arrays occupied ∼40% greater volume compared with Pin1-proficient cells. Similarly, after induction of transcription, the nuclear volume occupied by the arrays in Pin1-deficient cells was 40% higher than the transcriptionally active locus in Pin1-proficient cells.
To analyze H1 mobility at sites of transcription, we cotransfected GFP-H1.5 and mcherry-ER-tTA in cells housing the lac arrays (Fig. 7 A). After addition of Tamoxifen (1 h/3 h), a circular spot of 0.7-µm diameter was photobleached and the intensity of GFP-H1.5 was monitored over time. The region photobleached coincided with the mcherry-ER-tTA region within the nucleus, while a second circular spot of the same dimension and y-axis position was simultaneously photo-bleached and served as an internal control. To control for transcription dependence, parallel experiments were done in cells cotransfected with mcherry-LacR and GFP-H1.5, conditions where the array is not transcriptionally active. H1 kinetics, as measured by FRAP is shown in Fig. 7 C, whereas the t50 values obtained from these curves are shown in Fig. 7 B. Note the t50 values from these spot bleaches are considerably lower compared with linear bleaches (Fig. 4). This reflects the smaller size of the photobleached region.
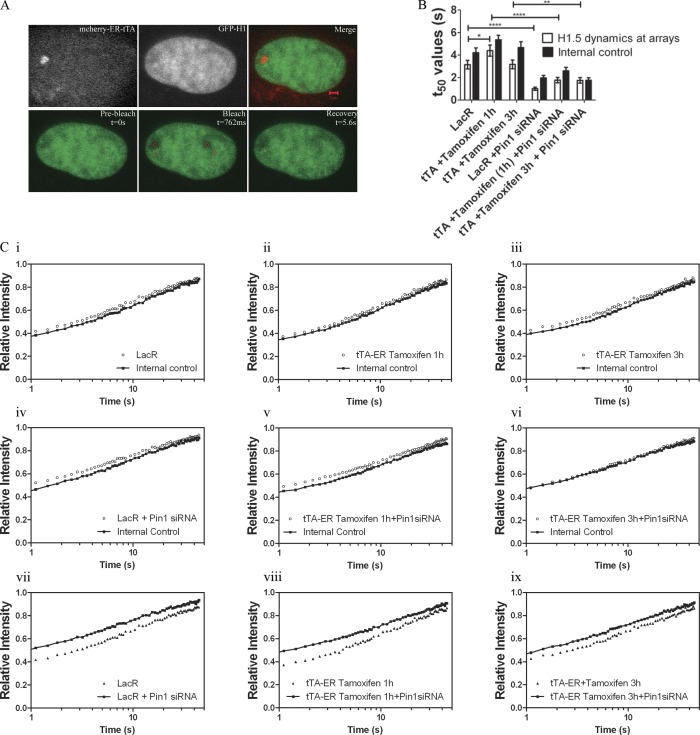
Figure 7.
Pin1 stabilizes H1 binding at sites of transcription. (A) GFP-H1.5 was cotransfected with either mCherry LacR or mCherry-ER-tTA in U2OS 263 cells harboring the arrays. H1 dynamics were monitored using FRAP with two separate regions in the nucleus being photo-bleached. One bleached region corresponded to either the mCherry LacR (transcriptionally inactive site) or mCherry-ER-tTA (transcriptionally active site), and photo-bleached region 2 corresponded to a random site within the nucleus in the same horizontal plane. (B) T50 values of the FRAP curves (C) show that H1.5 dynamics at the lac arrays is fairly similar to those of internal controls, in the transcriptionally uninduced state (Ci). The same trend is seen even when transcription is stimulated by transfection of mCherry-ER-tTA and tamoxifen is added for either 1 h (Cii) or 3 h (Ciii). Similar experiments were performed in cells treated with Pin1siRNA (Civ–Cvi). Major differences in H1 mobility can be observed when comparing the recovery rate in Pin1-proficient cells vs. those seen in Pin1-deficient cells (Cvii–ix). The increase in H1.5 dynamics upon Pin1 depletion is independent of transcriptional activity. Significance between control vs. Pin1siRNA treated t50 values was analyzed using unpaired t test (95% confidence interval). Notation for significance: *** if P value is < 0.001; ** if P is between 0.001 and 0.01; * if P is between 0.01 and 0.05. Each FRAP curve represents an average of ∼30 unique sites of transcription from three independent experiments.
H1.5 kinetics was only slightly faster at the arrays compared with internal control (Fig. 7 C, i). We next stimulated transcription at the arrays by transfecting mcherry-ER-tTA and incubating these cells in the presence of Tamoxifen for 1 h or 3 h. Despite the increase in levels of Pin1, H1 phosphorylation, and decondensed chromatin after the addition of Tamoxifen (1 h/3 h), we found H1 mobility to be similar to that observed in transcriptionally inactive chromatin (Fig. 7 C, ii and iii). For example, after 1 h of transcriptional activation at the lac arrays, H1.5 was found to bind more stably than that observed at transcriptionally silent chromatin (Fig. 7, B and C ii). However, after a longer treatment with Tamoxifen (3 h) there was no difference in the FRAP recovery profiles of H1.5 observed at transcriptionally inactive versus transcriptionally elongating chromatin (Fig. 7, B and C iii). This trend was seen with other variants of H1, such as GFP-H1.1 and GFP-H1.2, which are enriched in euchromatin (Th’ng et al., 2005). The mobility of these molecules remained unchanged after activation of transcription (Fig. S5).
We next wished to determine whether reducing the level of Pin1 would destabilize H1 at sites of transcription. Using siRNA directed against Pin1, we reduced Pin1 to 50–60% of its original level. Under these conditions, there was a statistically significant increase in the phosphorylation levels of H1 at transcriptionally inactive chromatin (Fig. S4 D). Note that this increase was in the absence of any transfected transcriptional activators, such as mcherry-ER-tTA. This suggested that Pin1 depletion was able to increase the steady-state levels of H1 phosphorylation, consistent with our earlier observation in Pin1−/− cells. When transcription was induced in Pin1 siRNA-treated cells through transfection of mcherry-ER-tTA and Tamoxifen, H1 phosphorylation was found to further increase to levels seen in Pin1-proficient cells. This suggested that Pin1 does not inhibit the activity of the Cdk2 enzyme that phosphorylates H1 during transcriptional induction.
We then analyzed the dynamics of H1 at transcriptionally inactive versus active sites in these Pin1-depleted cells (Fig. 7, B and C iv–ix). We found H1.5 mobility at the arrays to be similar to the internal controls (chromatin sites away from the lac arrays), irrespective of whether the site was transcriptionally active or not (Fig. 7, B and C iv–vi). A major difference in H1 mobility was seen when the kinetics were compared in Pin1-proficient cells compared with Pin1-deficient cells (Fig. 7, B and C vii–ix). This difference was maintained irrespective of transcriptional status. Our results imply that Pin1 plays a role in stabilizing H1 binding at both transcriptionally active and inactive chromatin sites.
Discussion
In this study, we have shown an interaction between phosphorylated H1 molecules and Pin1 and have examined the implications of this interaction. Lack of Pin1 leads to reduced H1 retention on chromatin and reduced turnover of the phosphorylated state of H1. At transcriptionally active sites, where both H1 phosphorylation and Pin1 levels are elevated, Pin1 serves to stabilize the binding of H1 and this correlated with a reduction in chromatin decondensation.
Histone H1 interacts with Pin1
Our current understanding of how H1 phosphorylation affects its binding centers around electrostatic repulsions between negatively charged phosphorylation residues and the high degree of negative charge associated with the DNA backbone (Dou and Gorovsky, 2000; Dou et al., 2002). Our data favor a model that is a product of several biophysical studies describing the CTD of H1 as intrinsically disordered (Clark et al., 1988; Roque et al., 2005, 2007, 2008; Hansen et al., 2006). Upon interaction with DNA or nucleosomes, the CTD condenses and acquires classical secondary structures such as α-helices and β-sheets (Roque et al., 2005; Caterino et al., 2011; Fang et al., 2012). Interestingly, the proportions of CTD secondary structures appear to be dependent upon the phosphorylation status of H1 (Roque et al., 2008). The acquisition of structure is thought to further determine the strength of H1 binding to chromatin, with high affinity H1 particles having a more folded structure compared with freely diffusing molecules (Misteli et al., 2000; Raghuram et al., 2009; Stasevich et al., 2010). In this study, we have shown that Pin1, through its interaction with the phosphorylated residues on H1, can modulate the conformation of the CTD, which may critically influence the binding dynamics of H1. We previously showed that variants of H1 bind chromatin with differing affinity, where the affinity roughly correlates with the length of the CTD (Hendzel et al., 2004). This correlation can be further extended to residues that are phosphorylated on H1. For example, in interphase, H1.1 is mono-phosphorylated, while H1.5 can exist as a tri-phosphorylated species (Sarg et al., 2006). This implies that the higher the number of phosphorylated residues on H1, greater is the amount of stabilization mediated by Pin1. This argument is consistent with the fact that H1.5 has a much higher residence time compared with H1.1.
Histone H1 phosphorylation, Pin1, and transcription
A well-characterized target of Pin1 in transcription is the CTD of RNA polymerase II (Xu and Manley, 2007). Here we show that Pin1 recruitment to sites of transcription is independent of RNA polymerase II and parallels the increase in H1 phosphorylation at these transcriptionally competent sites. Increases in H1 phosphorylation may act to recruit Pin1 to these sites, establishing a dynamic cycle of H1 phosphorylation and dephosphorylation at sites of transcription.
The rapid mobility of H1 in the nucleus is well documented (Lever et al., 2000; Misteli et al., 2000). However, experiments from our laboratory and others demonstrate a localized increase in H1 phosphorylation, such as upon induction of transcription (Zheng et al., 2010; see Fig. 6 B), or upon targeting LacR-Cdk2 to lac arrays (Alexandrow and Hamlin, 2005), wherein H1 phosphorylation increases locally as opposed to the spreading of H1 phosphorylation from a focal point. These data collectively confirm the stringent spatial regulation of H1 phosphorylation and dephosphorylation in vivo. The early recruitment of Pin1 to sites of transcription helps to promote H1 dephosphorylation and stabilize its binding, which may act as a mechanism designed to prevent inappropriate transcriptional activation of adjacent genes.
The lack of any real change in H1 dynamics when chromatin is subject to strong transcriptional activators contradicts in vitro experiments that suggest H1 to be displaced upon initiation of transcription. This, however, could be due to the limitations of in vitro systems to replicate in vivo complexity. In recent in vitro transcription assays where such complexity was established using reconstituted chromatin assembled from purified core histones, H1, and histone chaperone, where the level of compaction approximated that of the 30-nm fiber (Li et al., 2010), H1 molecules were found to be present through a complete cycle of elongation, including the preceding changes to chromatin that are associated with transcriptional activation. These results complement electron microscopy data showing H1 to be present in all stages of transcription from the Balbiani ring genes (Ericsson et al., 1990). The presence of H1 from initiation to elongation, even at very high frequencies of transcription (1 Pol II enzyme/100 bp of DNA; Ericsson et al., 1990) suggests a role for H1 in the process of transcription. Furthermore, in vivo transcription of a model DHFR gene has been shown to occur in chromatin structures that were much more condensed than a 30-nm fiber (Tumbar et al., 1999; Hu et al., 2009). Induction of transcription did lead to decondensation; however, the resulting chromatin was significantly more compact than expected of a 30-nm fiber conformation (Hu et al., 2009).
Pin1 could play a pivotal role in stabilizing H1 at transcriptionally active sites. Reduction in the level of Pin1 led to an increase in H1 mobility that was accompanied by chromatin decondensation. Transcriptionally active regions undergo an additional increase in chromatin decondensation that parallels a further increase in H1 phosphorylation. Furthermore, transcriptionally active regions in Pin1-competent cells bind H1 with a higher affinity compared with Pin1-deficient cells, suggesting that Pin1 could play a role in stabilizing H1 at sites of transcription. However, it remains to be seen if additional factors play a role in this process, or if it is a direct consequence of Pin1 acting on H1.
Materials and methods
Cell culture and transfections
Pin1−/− and Pin1wt cells (MEF, E13.5) were cultured in DMEM (Gibco). Pin1−/− cells were generated by using a targeting vector designed to delete all the exons of Pin1 (GenBank/EMBL/DDBJ accession no. AB009691; Fujimori et al., 1999). U2Os 263 cells were grown in high-glucose DMEM supplemented with 100 µg/ml hygromycin B. Transfections were done using Effectene (QIAGEN) transfection reagents, according to the manufacturer’s protocol. Cells transfected with mCherry-tTA-ER were treated with 1 µM tamoxifen (Sigma-Aldrich; dissolved in ethanol) for either 1 h or 3 h to activate transcription. For α-amanitin analysis, the 263 cells were transfected overnight with mCherry-tTA-ER and then pretreated with α-amanitin (100 µg/ml, overnight) after which they were treated with tamoxifen.
Coimmunoprecipitation
Approximately 106 cells were centrifuged at 1,300 g and resuspended in ice-cold nuclei isolation buffer (250 mM sucrose, 150 mM NaCl, 20 mM Tris, pH 8, 1.5 mM MgCl2, 0.2 mM CaCl2, and 0.1% IGEPAL CA630) supplemented with PhosSTOP (Roche) and Complete protease inhibitor (Roche). These were then centrifuged at 3,200 g, and the nuclei were resuspended in modified RIPA buffer (Tris, pH 7.4, 1% NP-40, and 150 mM NaCl) along with PhosSTOP, protease inhibitors, and an endonuclease, benzonase (EMD Millipore). After 4 h, the reaction was stopped by adding 1 mM EDTA. After 30 min incubation at 4°C, the extract was spun down at 13,000 g to remove aggregates. The extract was then treated with antibody prebound to dynabeads (Invitrogen) overnight. For coimmunoprecipitation with T98G cells stably expressing GFP-H1.1, extracts were treated with 50 µl GFP-TRAP (ChromoTek). The antibody–beads mixture was then separated using a magnetic rack and washed three times, after which 3× SDS loading buffer was added directly to the beads. Samples were then run on a standard 15% or 18% acrylamide gel, transferred onto a nitrocellulose membrane. Pin1 (G-8) antibody (Santa Cruz Biotechnology, Inc.) was used at a dilution of 1:1,000, anti-H1 (Novus Biologicals) was used at 1:250 dilution, anti-GFP (Ab290; Abcam) was used at 1:10,000 dilution, and anti-Pol II (8WG16; Promega) was used at 1:1,000 dilution. Secondary antibodies were conjugated with infrared-specific dyes (Alexa Flour 680, Alexa Fluor 750), and all blots were scanned on the Odyssey Infrared Imaging System (LI-COR Biosciences).
H1 phosphorylation/dephosphorylation assays
Approximately 1.5 × 107 cells were centrifuged at 1,300 g for 4 min at 4°C, resuspended in RIPA buffer and 1 mM EDTA along with PhosTOP protease inhibitors and phosphatase inhibitors (the latter was excluded in dephosphorylation assays). The extract was spun at 14,000 g for 10 min at 4°C, and the supernatant was treated with either anti-Cdk2 antibody (M2, sc-163, 2.4 µg; Santa Cruz Biotechnology, Inc.) or anti-PP2Ac (1D6, 4 µg; EMD Millipore) overnight at 4°C. Dynabeads were then added the next day for 2 h at 4°C, after which the beads were separated magnetically, washed three times with fresh RIPA buffer, once with 40 mM Tris, pH 7.6, and then resuspended either in phosphorylation buffer (40 mM, Tris 7.6, 2 mM DTT, and 10 mM MgCl2) or in dephosphorylation buffer (40 mM Tris, pH 7.6, 2 mM DTT, and 1.5 mM MgCl2). Purified calf-thymus H1 (EMD Millipore; resuspended in water at a concentration of 1 mg/ml, 3 µg of H1/reaction) was added to the reaction mixture with or without ATP (8.5 mM). The reaction mixture was then incubated at either 30°C (for kinase reaction) or 37°C (dephosphorylation) for the given time. Adding 3×SDS loading buffer stopped the reaction and H1 was resolved on a denaturing 18% acrylamide gel. Cy3/Cy5-labeled H1 was phosphorylated in a similar manner, except the reaction was allowed to progress for 90 min, after which the reaction was stopped with the addition of EDTA. To verify phosphorylation, labeled H1 was resolved on a denaturing 18% acrylamide gel, transferred to nitrocellulose, and stained with ProQ Diamond Phosphoprotein Blot Stain (Molecular Probes) to detect phosphorylated H1 molecules or SyproRuby Protein blot stain to detect total H1 protein. The blots were visualized using 302-nm UV light.
Fluorescence resonance energy transfer (FRET)
FRET was performed on H1Cy3Cy5 either in solution or when added to reconstituted nucleosomes that were placed on live-cell imaging dishes (total volume of 150 µl). H1 proteins were diluted to a concentration of 26 nM and the solution was spread on a glass-bottomed dish. A glass coverslip was placed on top to prevent evaporation. The sample was then imaged on a laser scanning microscope (LSM 710; Carl Zeiss) equipped with a Plan-Apochromat 40×/1.3 NA oil DIC M27 objective and a heat stage that was maintained at 37°C. The sample was excited with a 514- or 633-nm laser, both operating at 5% laser output. Emission spectra (5-nm slit width) were obtained using a 523–727-nm filter when excited with 514 nm and a 639–727-nm filter when excited with the 633-nm laser. Pixel dwell time was maintained at 2.55 µs and pinhole was set at 600 µm. FRET was calculated using the RatioA method (Clegg, 1992; Poirier et al., 2009), using peak heights and with extinction coefficient εA (630) = 150,000 (Cy5); εA (514) = 5,000 (Cy5); εD (514) = 75,000 (Cy3); and d+ = 1. R0 was set at 5.4 nm (Fang et al., 2012).
Immunofluorescence microscopy
Cells were grown on Fisherbrand coverglass (18 × 18–1.5) overnight. Cells were fixed with 4% paraformaldehyde in 1× PBS for 10 min, permeabilized with 0.5% Triton X-100 for 5 min. For pS173 H1 antibody, cells were fixed with 1% paraformaldehyde for 10 min. Coverslips were then washed with PBS, inverted onto 50–100 µl of primary antibody in PBS, and incubated for at least 30 min. Coverslips were then washed with 0.1% Triton X-100 and then with PBS, before incubation with secondary antibody coupled with a flourophore. Cells were then mounted on slides using a 90% glycerol/PBS-based medium containing 1 mg of paraphenylenediamine/ml and 0.5 µg DAPI/ml. Pin1 was detected using a monoclonal antibody (G-8; Santa Cruz Biotechnology, Inc.) at 1:500 dilution; pS173 antibodies were used at 1:400 dilution. The slides were then imaged using a laser-scanning confocal microscope (LSM 710; Carl Zeiss). Confocal sections were obtained with a pinhole aperture setting of 1 Airy unit. DAPI was excited with a 405-nm laser, GFP with a 488-nm laser, and Cy3 with 514-nm laser. For images that show a field of cells, typically a 1.3 NA oil DIC M27 Plan-Apochromat 40× objective lens (Carl Zeiss) was used, whereas a higher magnification lens (63× 1.4 NA oil DIC M27 Plan-Apochromat) objective lens was used for most images that were further used for quantifying protein accumulation. The confocal sections were then analyzed using ImageJ software (National Institutes of Health) to draw a mask around regions of interest and intensity measurements were obtained. Similarly, a mask was drawn around the nucleus to provide nuclear intensity. A ratio of the intensity in the region of interest to that of the entire nucleus provided values for fold enrichment.
Nucleosome reconstitution
Nucleosomes were reconstituted using the EpiMark nucleosome assembly kit (New England Biolabs, Inc.) with some minor modifications (Steger and Workman, 1999). 50 pmol of nucleosomes were incubated with 50 pmol of DNA (208 bp of containing the Lytechinus variegatus 5SrDNA) at 2 M NaCl, 1 µg BSA in a final volume of 20 µl at 37°C for 15 min. The reaction mixture was serially diluted to 1.5, 1, 0.8, 0.7, 0.6, 0.5, 0.4, 0.25, and 0.2 M NaCl using 50 mM Hepes, pH 7.5, 1 mM EDTA, 5 mM DTT, and Complete protease inhibitors (Roche) for 30°C (15 min for each dilution). One final dilution was performed in Tris, pH 7.5, 1 mM EDTA, 0.1% NP-40, 5 mM DTT, Complete protease inhibitors, 20% glycerol, and 100 µg/ml BSA to bring the final salt concentration to 0.1 M NaCl. Reconstitutions were analyzed by electrophoresis on 5% acrylamide nucleoprotein gels.
Roscovitine treatment/acid extraction of histones
Pin1−/− and wt cells were treated with roscovitine (30 µM) for the times indicated. Cold nuclei isolation buffer was added directly to cells, and incubated at 4°C for ∼10 min. The cells were then washed, and spun at 3,200 g to isolate the nuclei. Histones were extracted using 0.4 N H2SO4 + protease inhibitors, DTT, and PhoSTOP (1 h, 4°C). Extracts were then spun at 10,000 g and the supernatant was then precipitated with −20°C acetone (overnight). The precipitated histones were then resuspended in SDS loading buffer and separated on 15% acrylamide gels. H1 was detected using pS173, pS187, and pT146 (1:250; Abcam). For AUT gels, acid-extracted histones were separated on a 15% separating gel at 200 V for 3.5 h at 4°C. Gels were then stained with Coomassie brilliant blue (Panyim and Chalkley, 1969).
Mobility shift assay for detecting phosphorylated H1
Pin1−/− and Pin1wt cells were transfected with either H1.1-FLAG or the appropriate H1.1mut FLAG. The following day, histones were extracted using 0.4 N H2SO4, as described above. Phos-tag ligands bind with strong affinity to phosphate molecules, and when cross-linked in an acrylamide gel, can provide specific mobility shifts based on the presence/absence of phosphorylation (Kinoshita et al., 2009). Phos-tag SDS-PAGE gels (100 µM Phos-tag, 10% acrylamide gels) were prepared as per the manufacture’s protocol. Gels were run at constant current (20 mA). The gels were then washed extensively with transfer buffer + 4 mM EDTA to chelate the Mn2+, rinsed with water, and then washed with transfer buffer without EDTA. Proteins were then transferred to a nitrocellulose membrane (110 V, 0.37 mA, 90 min) and then probed with anti-FLAG (M2; Sigma-Aldrich) at 1:50,000 dilution.
Volumetric measurements of lac arrays
Cells expressing either mCherry LacR or mCherry-ER-tTA were plated on live-cell imaging dishes and where then imaged using a 63× Plan-Apochromatic 1.4 oil DIC M27 objective (LSM 710; Carl Zeiss) with an objective warmer maintained at 37°C. A Piezo stage (Piezostem Jena; Carl Zeiss) was used, allowing rapid acquisition of z-stacks at a rate of 50 images (9.8 µm in z-direction)/9 s (total acquisition time), with a pixel dwell time of 0.79 µs and pinhole set at 48 µm. A 561-nm laser operating at 1–2% laser output was used to excite the mCherry signal. The images were then analyzed on Imaris x64 7.3.0 surface-rendering algorithm. The images were thresholded based on 30% of the maximum absolute intensity recorded (approximately equal to one standard deviation), and the volume of the surface generated was then recorded.
FRAP and kinetic modeling
FRAP was performed as described in Raghuram et al. (2010). In brief, cells were grown on #1.5 glass coverslips in tissue culture media along with transfection reagents. The following day, the coverslips were mounted on glass slides with media enclosed by vacuum grease. FRAP was performed using a laser-scanning confocal microscope (LSM 510 NLO; Carl Zeiss) using a 488-nm laser operating at 100% for bleaching (30 iterations) and 0.5% for acquiring images. Pixel dwell time was maintained at 1.26 µs. A 40× 1.3 NA oil objective lens equipped with a heated stage and objective warmer maintained at 37°C was used. For global H1 analysis, a 1.5-µm rectangular region was photobleached. Only cells with low GFP background fluorescence were picked for photobleaching. All images were corrected for cell/nuclear movement using ImageJ software and the StackReg plugin. Intensity measurements were performed using MetaMorph, while statistical tests for t50 and t90 values were done using GraphPad software. Kinetic modeling of the FRAP curves were done essentially as described in Carrero et al. (2004a,b). For H1 FRAP at sites of transcription, cells were cotransfected with GFP H1 and mCherry-ER-tTA or mCherry-LacR. After the addition of tamoxifen, the mCherry signal was used as a guide to locate the arrays. This was used as a mark to photobleach a spot (0.07 µm in diameter) that corresponded to the arrays. Another nonarray spot was simultaneously photobleached, and served as an internal control. The recovery of both the photobleached spots was monitored at regular time intervals.
Online supplemental material
Fig. S1 shows the similar kinetics of H1 phosphorylation mediated by Cdk2 extracted from Pin1wt and Pin1−/− cells. Fig. S2 shows the controls of the in vitro FRET experiments as well as the changes in FRET efficiencies upon H1 phosphorylation and proline isomerization induced by Pin1. Fig. S3 shows the lack of any real change in H1 dynamics after addition of cyclosporine and rapamycin. Also shown in Fig. S3 is an AUT gel comparing histone composition and histone acetylation status in Pin1wt and Pin1−/− cells. Fig. S4 shows the accumulation of RNA polymerase II and YFP-MS2 after the addition of tamoxifen and the effects of α-amanitin on RNA polymerase II. The figure also shows the increases in H1 phosphorylation and Pin1 after transcriptional activation and a representative example of the decondensation after such activation. Fig. S5 shows the lack of any change in H1.1 and H1.2 at sites of active transcription. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201305159/DC1.
Supplementary Material
Acknowledgments
We thank K.P. Lu and S. Janicki for supplying valuable reagents. We also thank L. Schang, G. Chan, and R. Gieni for their input and C. Andrin, X. Sun, G. Barron, and the Cell Imaging Facility for their technical assistance.
This work was supported by an operating grant from the Canadian Cancer Society Research Institute. N. Raghuram was supported by graduate studentships from the Alberta Innovates Health Solutions, Alberta Cancer Foundation, and the CIHR studentship. H. Strickfaden was supported by an ACF Research (PDF) fellowship. M.J. Hendzel is supported by an Alberta Innovates Health Solutions Senior Scholarship.
The authors declare they have no conflict of interest.
Author contributions: N. Raghuram designed and performed experiments, analyzed data, and wrote the paper. D. McDonald performed experiments and analyzed data. K. Williams and H. Fang produced valuable reagents. J. Hayes and C. Mizzen provided valuable reagents and critically read and wrote the manuscript. H. Strickfaden, J. Th’ng, and M.J. Hendzel designed experiments, analyzed the data, and wrote the paper.
Footnotes
Abbreviations used in this paper:
- CTD
- C-terminal domain
- FRET
- fluorescence resonance energy transfer
- PPIase
- peptidyl-prolyl isomerase
References
- Albert A., Lavoie S., Vincent M. 1999. A hyperphosphorylated form of RNA polymerase II is the major interphase antigen of the phosphoprotein antibody MPM-2 and interacts with the peptidyl-prolyl isomerase Pin1. J. Cell Sci. 112:2493–2500 [DOI] [PubMed] [Google Scholar]
- Alexandrow M.G., Hamlin J.L. 2005. Chromatin decondensation in S-phase involves recruitment of Cdk2 by Cdc45 and histone H1 phosphorylation. J. Cell Biol. 168:875–886 10.1083/jcb.200409055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bednar J., Horowitz R.A., Grigoryev S.A., Carruthers L.M., Hansen J.C., Koster A.J., Woodcock C.L. 1998. Nucleosomes, linker DNA, and linker histone form a unique structural motif that directs the higher-order folding and compaction of chromatin. Proc. Natl. Acad. Sci. USA. 95:14173–14178 10.1073/pnas.95.24.14173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradbury E.M. 1992. Reversible histone modifications and the chromosome cell cycle. Bioessays. 14:9–16 10.1002/bies.950140103 [DOI] [PubMed] [Google Scholar]
- Carrero G., Crawford E., Hendzel M.J., de Vries G. 2004a. Characterizing fluorescence recovery curves for nuclear proteins undergoing binding events. Bull. Math. Biol. 66:1515–1545 10.1016/j.bulm.2004.02.005 [DOI] [PubMed] [Google Scholar]
- Carrero G., Crawford E., Th’ng J., de Vries G., Hendzel M.J. 2004b. Quantification of protein-protein and protein-DNA interactions in vivo, using fluorescence recovery after photobleaching. Methods Enzymol. 375:415–442 10.1016/S0076-6879(03)75026-5 [DOI] [PubMed] [Google Scholar]
- Carruthers L.M., Bednar J., Woodcock C.L., Hansen J.C. 1998. Linker histones stabilize the intrinsic salt-dependent folding of nucleosomal arrays: mechanistic ramifications for higher-order chromatin folding. Biochemistry. 37:14776–14787 10.1021/bi981684e [DOI] [PubMed] [Google Scholar]
- Caterino T.L., Fang H., Hayes J.J. 2011. Nucleosome linker DNA contacts and induces specific folding of the intrinsically disordered H1 carboxyl-terminal domain. Mol. Cell. Biol. 31:2341–2348 10.1128/MCB.05145-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chadee D.N., Taylor W.R., Hurta R.A., Allis C.D., Wright J.A., Davie J.R. 1995. Increased phosphorylation of histone H1 in mouse fibroblasts transformed with oncogenes or constitutively active mitogen-activated protein kinase kinase. J. Biol. Chem. 270:20098–20105 10.1074/jbc.270.34.20098 [DOI] [PubMed] [Google Scholar]
- Chadee D.N., Allis C.D., Wright J.A., Davie J.R. 1997. Histone H1b phosphorylation is dependent upon ongoing transcription and replication in normal and ras-transformed mouse fibroblasts. J. Biol. Chem. 272:8113–8116 10.1074/jbc.272.13.8113 [DOI] [PubMed] [Google Scholar]
- Clark D.J., Hill C.S., Martin S.R., Thomas J.O. 1988. Alpha-helix in the carboxy-terminal domains of histones H1 and H5. EMBO J. 7:69–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clegg R.M. 1992. Fluorescence resonance energy transfer and nucleic acids. Methods Enzymol. 211:353–388 10.1016/0076-6879(92)11020-J [DOI] [PubMed] [Google Scholar]
- Dou Y., Gorovsky M.A. 2000. Phosphorylation of linker histone H1 regulates gene expression in vivo by creating a charge patch. Cell. 103:263 10.1016/S0092-8674(00)00118-5 [DOI] [PubMed] [Google Scholar]
- Dou Y., Mizzen C.A., Abrams M., Allis C.D., Gorovsky M.A. 1999. Phosphorylation of linker histone H1 regulates gene expression in vivo by mimicking H1 removal. Mol. Cell. 4:641–647 10.1016/S1097-2765(00)80215-4 [DOI] [PubMed] [Google Scholar]
- Dou Y., Bowen J., Liu Y., Gorovsky M.A. 2002. Phosphorylation and an ATP-dependent process increase the dynamic exchange of H1 in chromatin. J. Cell Biol. 158:1161–1170 10.1083/jcb.200202131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ericsson C., Grossbach U., Björkroth B., Daneholt B. 1990. Presence of histone H1 on an active Balbiani ring gene. Cell. 60:73–83 10.1016/0092-8674(90)90717-S [DOI] [PubMed] [Google Scholar]
- Fang H., Clark D.J., Hayes J.J. 2012. DNA and nucleosomes direct distinct folding of a linker histone H1 C-terminal domain. Nucleic Acids Res. 40:1475–1484 10.1093/nar/gkr866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimori F., Takahashi K., Uchida C., Uchida T. 1999. Mice lacking Pin1 develop normally, but are defective in entering cell cycle from G(0) arrest. Biochem. Biophys. Res. Commun. 265:658–663 10.1006/bbrc.1999.1736 [DOI] [PubMed] [Google Scholar]
- Hansen J.C. 2002. Conformational dynamics of the chromatin fiber in solution: determinants, mechanisms, and functions. Annu. Rev. Biophys. Biomol. Struct. 31:361–392 10.1146/annurev.biophys.31.101101.140858 [DOI] [PubMed] [Google Scholar]
- Hansen J.C., Lu X., Ross E.D., Woody R.W. 2006. Intrinsic protein disorder, amino acid composition, and histone terminal domains. J. Biol. Chem. 281:1853–1856 10.1074/jbc.R500022200 [DOI] [PubMed] [Google Scholar]
- Hendzel M.J., Lever M.A., Crawford E., Th’ng J.P. 2004. The C-terminal domain is the primary determinant of histone H1 binding to chromatin in vivo. J. Biol. Chem. 279:20028–20034 10.1074/jbc.M400070200 [DOI] [PubMed] [Google Scholar]
- Hennig L., Christner C., Kipping M., Schelbert B., Rücknagel K.P., Grabley S., Küllertz G., Fischer G. 1998. Selective inactivation of parvulin-like peptidyl-prolyl cis/trans isomerases by juglone. Biochemistry. 37:5953–5960 10.1021/bi973162p [DOI] [PubMed] [Google Scholar]
- Herrera R.E., Chen F., Weinberg R.A. 1996. Increased histone H1 phosphorylation and relaxed chromatin structure in Rb-deficient fibroblasts. Proc. Natl. Acad. Sci. USA. 93:11510–11515 10.1073/pnas.93.21.11510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohmann P. 1983. Phosphorylation of H1 histones. Mol. Cell. Biochem. 57:81–92 10.1007/BF00223526 [DOI] [PubMed] [Google Scholar]
- Hu Y., Kireev I., Plutz M., Ashourian N., Belmont A.S. 2009. Large-scale chromatin structure of inducible genes: transcription on a condensed, linear template. J. Cell Biol. 185:87–100 10.1083/jcb.200809196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janicki S.M., Tsukamoto T., Salghetti S.E., Tansey W.P., Sachidanandam R., Prasanth K.V., Ried T., Shav-Tal Y., Bertrand E., Singer R.H., Spector D.L. 2004. From silencing to gene expression: real-time analysis in single cells. Cell. 116:683–698 10.1016/S0092-8674(04)00171-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita E., Kinoshita-Kikuta E., Matsubara M., Yamada S., Nakamura H., Shiro Y., Aoki Y., Okita K., Koike T. 2008. Separation of phosphoprotein isotypes having the same number of phosphate groups using phosphate-affinity SDS-PAGE. Proteomics. 8:2994–3003 10.1002/pmic.200800243 [DOI] [PubMed] [Google Scholar]
- Kinoshita E., Kinoshita-Kikuta E., Koike T. 2009. Separation and detection of large phosphoproteins using Phos-tag SDS-PAGE. Nat. Protoc. 4:1513–1521 10.1038/nprot.2009.154 [DOI] [PubMed] [Google Scholar]
- Koop R., Di Croce L., Beato M. 2003. Histone H1 enhances synergistic activation of the MMTV promoter in chromatin. EMBO J. 22:588–599 10.1093/emboj/cdg052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruithof M., Chien F.T., Routh A., Logie C., Rhodes D., van Noort J. 2009. Single-molecule force spectroscopy reveals a highly compliant helical folding for the 30-nm chromatin fiber. Nat. Struct. Mol. Biol. 16:534–540 10.1038/nsmb.1590 [DOI] [PubMed] [Google Scholar]
- Lamy F., Lecocq R., Dumont J.E. 1977. Thyrotropin stimulation of the phosphorylation of serine in the N-terminal of thyroid H1 histones. Eur. J. Biochem. 73:529–535 10.1111/j.1432-1033.1977.tb11347.x [DOI] [PubMed] [Google Scholar]
- Langan T.A. 1969. Phosphorylation of liver histone following the administration of glucagon and insulin. Proc. Natl. Acad. Sci. USA. 64:1276–1283 10.1073/pnas.64.4.1276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lever M.A., Th’ng J.P., Sun X., Hendzel M.J. 2000. Rapid exchange of histone H1.1 on chromatin in living human cells. Nature. 408:873–876 10.1038/35048603 [DOI] [PubMed] [Google Scholar]
- Li G., Margueron R., Hu G., Stokes D., Wang Y.H., Reinberg D. 2010. Highly compacted chromatin formed in vitro reflects the dynamics of transcription activation in vivo. Mol. Cell. 38:41–53 10.1016/j.molcel.2010.01.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liou Y.C., Ryo A., Huang H.K., Lu P.J., Bronson R., Fujimori F., Uchida T., Hunter T., Lu K.P. 2002. Loss of Pin1 function in the mouse causes phenotypes resembling cyclin D1-null phenotypes. Proc. Natl. Acad. Sci. USA. 99:1335–1340 10.1073/pnas.032404099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu K.P., Hanes S.D., Hunter T. 1996. A human peptidyl-prolyl isomerase essential for regulation of mitosis. Nature. 380:544–547 10.1038/380544a0 [DOI] [PubMed] [Google Scholar]
- Lu K.P., Finn G., Lee T.H., Nicholson L.K. 2007. Prolyl cis-trans isomerization as a molecular timer. Nat. Chem. Biol. 3:619–629 10.1038/nchembio.2007.35 [DOI] [PubMed] [Google Scholar]
- Lu M.J., Mpoke S.S., Dadd C.A., Allis C.D. 1995. Phosphorylated and dephosphorylated linker histone H1 reside in distinct chromatin domains in Tetrahymena macronuclei. Mol. Biol. Cell. 6:1077–1087 10.1091/mbc.6.8.1077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu P.J., Zhou X.Z., Shen M., Lu K.P. 1999. Function of WW domains as phosphoserine- or phosphothreonine-binding modules. Science. 283:1325–1328 10.1126/science.283.5406.1325 [DOI] [PubMed] [Google Scholar]
- Misteli T., Gunjan A., Hock R., Bustin M., Brown D.T. 2000. Dynamic binding of histone H1 to chromatin in living cells. Nature. 408:877–881 10.1038/35048610 [DOI] [PubMed] [Google Scholar]
- Moreno S., Nurse P. 1990. Substrates for p34cdc2: in vivo veritas? Cell. 61:549–551 10.1016/0092-8674(90)90463-O [DOI] [PubMed] [Google Scholar]
- Müller W.G., Walker D., Hager G.L., McNally J.G. 2001. Large-scale chromatin decondensation and recondensation regulated by transcription from a natural promoter. J. Cell Biol. 154:33–48 10.1083/jcb.200011069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panyim S., Chalkley R. 1969. High resolution acrylamide gel electrophoresis of histones. Arch. Biochem. Biophys. 130:337–346 10.1016/0003-9861(69)90042-3 [DOI] [PubMed] [Google Scholar]
- Parseghian M.H., Hamkalo B.A. 2001. A compendium of the histone H1 family of somatic subtypes: an elusive cast of characters and their characteristics. Biochem. Cell Biol. 79:289–304 10.1139/o01-099 [DOI] [PubMed] [Google Scholar]
- Paulson J.R., Patzlaff J.S., Vallis A.J. 1996. Evidence that the endogenous histone H1 phosphatase in HeLa mitotic chromosomes is protein phosphatase 1, not protein phosphatase 2A. J. Cell Sci. 109:1437–1447 [DOI] [PubMed] [Google Scholar]
- Poirier M.G., Oh E., Tims H.S., Widom J. 2009. Dynamics and function of compact nucleosome arrays. Nat. Struct. Mol. Biol. 16:938–944 10.1038/nsmb.1650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rafalska-Metcalf I.U., Powers S.L., Joo L.M., LeRoy G., Janicki S.M. 2010. Single cell analysis of transcriptional activation dynamics. PLoS ONE. 5:e10272 10.1371/journal.pone.0010272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghuram N., Carrero G., Th’ng J., Hendzel M.J. 2009. Molecular dynamics of histone H1. Biochem. Cell Biol. 87:189–206 10.1139/O08-127 [DOI] [PubMed] [Google Scholar]
- Raghuram N., Carrero G., Stasevich T.J., McNally J.G., Th’ng J., Hendzel M.J. 2010. Core histone hyperacetylation impacts cooperative behavior and high-affinity binding of histone H1 to chromatin. Biochemistry. 49:4420–4431 10.1021/bi100296z [DOI] [PubMed] [Google Scholar]
- Robinett C.C., Straight A., Li G., Willhelm C., Sudlow G., Murray A., Belmont A.S. 1996. In vivo localization of DNA sequences and visualization of large-scale chromatin organization using lac operator/repressor recognition. J. Cell Biol. 135:1685–1700 10.1083/jcb.135.6.1685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson P.J., Rhodes D. 2006. Structure of the ‘30 nm’ chromatin fibre: a key role for the linker histone. Curr. Opin. Struct. Biol. 16:336–343 10.1016/j.sbi.2006.05.007 [DOI] [PubMed] [Google Scholar]
- Roque A., Iloro I., Ponte I., Arrondo J.L., Suau P. 2005. DNA-induced secondary structure of the carboxyl-terminal domain of histone H1. J. Biol. Chem. 280:32141–32147 10.1074/jbc.M505636200 [DOI] [PubMed] [Google Scholar]
- Roque A., Ponte I., Suau P. 2007. Macromolecular crowding induces a molten globule state in the C-terminal domain of histone H1. Biophys. J. 93:2170–2177 10.1529/biophysj.107.104513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roque A., Ponte I., Arrondo J.L., Suau P. 2008. Phosphorylation of the carboxy-terminal domain of histone H1: effects on secondary structure and DNA condensation. Nucleic Acids Res. 36:4719–4726 10.1093/nar/gkn440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth S.Y., Allis C.D. 1992. Chromatin condensation: does histone H1 dephosphorylation play a role? Trends Biochem. Sci. 17:93–98 10.1016/0968-0004(92)90243-3 [DOI] [PubMed] [Google Scholar]
- Roth S.Y., Collini M.P., Draetta G., Beach D., Allis C.D. 1991. A cdc2-like kinase phosphorylates histone H1 in the amitotic macronucleus of Tetrahymena. EMBO J. 10:2069–2075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarg B., Helliger W., Talasz H., Förg B., Lindner H.H. 2006. Histone H1 phosphorylation occurs site-specifically during interphase and mitosis: identification of a novel phosphorylation site on histone H1. J. Biol. Chem. 281:6573–6580 10.1074/jbc.M508957200 [DOI] [PubMed] [Google Scholar]
- Stasevich T.J., Mueller F., Brown D.T., McNally J.G. 2010. Dissecting the binding mechanism of the linker histone in live cells: an integrated FRAP analysis. EMBO J. 29:1225–1234 10.1038/emboj.2010.24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steger D.J., Workman J.L. 1999. Transcriptional analysis of purified histone acetyltransferase complexes. Methods. 19:410–416 10.1006/meth.1999.0877 [DOI] [PubMed] [Google Scholar]
- Steger M., Murina O., Hühn D., Ferretti L.P., Walser R., Hänggi K., Lafranchi L., Neugebauer C., Paliwal S., Janscak P., et al. 2013. Prolyl isomerase PIN1 regulates DNA double-strand break repair by counteracting DNA end resection. Mol. Cell. 50:333–343 10.1016/j.molcel.2013.03.023 [DOI] [PubMed] [Google Scholar]
- Stukenberg P.T., Kirschner M.W. 2001. Pin1 acts catalytically to promote a conformational change in Cdc25. Mol. Cell. 7:1071–1083 10.1016/S1097-2765(01)00245-3 [DOI] [PubMed] [Google Scholar]
- Swank R.A., Th’ng J.P., Guo X.W., Valdez J., Bradbury E.M., Gurley L.R. 1997. Four distinct cyclin-dependent kinases phosphorylate histone H1 at all of its growth-related phosphorylation sites. Biochemistry. 36:13761–13768 10.1021/bi9714363 [DOI] [PubMed] [Google Scholar]
- Talasz H., Helliger W., Puschendorf B., Lindner H. 1996. In vivo phosphorylation of histone H1 variants during the cell cycle. Biochemistry. 35:1761–1767 10.1021/bi951914e [DOI] [PubMed] [Google Scholar]
- Tatara Y., Terakawa T., Uchida T. 2010. Identification of Pin1-binding phosphorylated proteins in the mouse brain. Biosci. Biotechnol. Biochem. 74:2480–2483 10.1271/bbb.100580 [DOI] [PubMed] [Google Scholar]
- Taylor W.R., Chadee D.N., Allis C.D., Wright J.A., Davie J.R. 1995. Fibroblasts transformed by combinations of ras, myc and mutant p53 exhibit increased phosphorylation of histone H1 that is independent of metastatic potential. FEBS Lett. 377:51–53 10.1016/0014-5793(95)01314-8 [DOI] [PubMed] [Google Scholar]
- Th’ng J.P., Guo X.W., Swank R.A., Crissman H.A., Bradbury E.M. 1994. Inhibition of histone phosphorylation by staurosporine leads to chromosome decondensation. J. Biol. Chem. 269:9568–9573 [PubMed] [Google Scholar]
- Th’ng J.P., Sung R., Ye M., Hendzel M.J. 2005. H1 family histones in the nucleus. Control of binding and localization by the C-terminal domain. J. Biol. Chem. 280:27809–27814 10.1074/jbc.M501627200 [DOI] [PubMed] [Google Scholar]
- Thoma F., Koller T. 1977. Influence of histone H1 on chromatin structure. Cell. 12:101–107 10.1016/0092-8674(77)90188-X [DOI] [PubMed] [Google Scholar]
- Thoma F., Koller T., Klug A. 1979. Involvement of histone H1 in the organization of the nucleosome and of the salt-dependent superstructures of chromatin. J. Cell Biol. 83:403–427 10.1083/jcb.83.2.403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukamoto T., Hashiguchi N., Janicki S.M., Tumbar T., Belmont A.S., Spector D.L. 2000. Visualization of gene activity in living cells. Nat. Cell Biol. 2:871–878 10.1038/35046510 [DOI] [PubMed] [Google Scholar]
- Tumbar T., Sudlow G., Belmont A.S. 1999. Large-scale chromatin unfolding and remodeling induced by VP16 acidic activation domain. J. Cell Biol. 145:1341–1354 10.1083/jcb.145.7.1341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicent G.P., Nacht A.S., Font-Mateu J., Castellano G., Gaveglia L., Ballaré C., Beato M. 2011. Four enzymes cooperate to displace histone H1 during the first minute of hormonal gene activation. Genes Dev. 25:845–862 10.1101/gad.621811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y.X., Manley J.L. 2007. Pin1 modulates RNA polymerase II activity during the transcription cycle. Genes Dev. 21:2950–2962 10.1101/gad.1592807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y., John S., Pesavento J.J., Schultz-Norton J.R., Schiltz R.L., Baek S., Nardulli A.M., Hager G.L., Kelleher N.L., Mizzen C.A. 2010. Histone H1 phosphorylation is associated with transcription by RNA polymerases I and II. J. Cell Biol. 189:407–415 10.1083/jcb.201001148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X.Z., Kops O., Werner A., Lu P.J., Shen M., Stoller G., Küllertz G., Stark M., Fischer G., Lu K.P. 2000. Pin1-dependent prolyl isomerization regulates dephosphorylation of Cdc25C and tau proteins. Mol. Cell. 6:873–883 10.1016/S1097-2765(05)00083-3 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.