Abstract
Biological networks with a structured syntax are a powerful way of representing biological information generated from high density data; however, they can become unwieldy to manage as their size and complexity increase. This article presents a crowd-verification approach for the visualization and expansion of biological networks.
Web-based graphical interfaces allow visualization of causal and correlative biological relationships represented using Biological Expression Language (BEL). Crowdsourcing principles enable participants to communally annotate these relationships based on literature evidences. Gamification principles are incorporated to further engage domain experts throughout biology to gather robust peer-reviewed information from which relationships can be identified and verified.
The resulting network models will represent the current status of biological knowledge within the defined boundaries, here processes related to human lung disease. These models are amenable to computational analysis. For some period following conclusion of the challenge, the published models will remain available for continuous use and expansion by the scientific community.
Keywords: community curation, biological network models, reputation system, Biological Expression Language
From Crowdsourcing to Crowd-verification
For nearly 20 years, crowdsourcing initiatives have been used to draw upon and focus the expertise of a broad, heterogeneous scientific community to address specific biological questions framed as ‘challenges’. These challenges have addressed topics as diverse and labor-intensive as knowledge discovery and data mining (KDD cup,1 see www.kdd.org/kddcup/), microarray and next-generation sequencing (MAQC,2 see www.fda.gov/MicroArrayQC/), and protein-folding (FoldIt,3 see www.fold.it). Crowd-based approaches have enabled the collection of scientific knowledge in common repositories such as BioCarta (see www.biocarta.com/) and WikiPathways4 (see www.wikipathways.org). Challenge-based verification processes may offer a unique way to leverage the explosive growth of scientific data and publications. Sophisticated computational methods that are used to analyze complex data are not easily evaluated through the classical peer review process.5 Crowdsourcing initiatives with the aim to solve a particular challenge are used to reach a better understanding of the strengths and weaknesses of methods that are used to handle “big data”. Such evaluation enables progress in their respective disciplines.5 sbv IMPROVER6 (see www.sbvimprover.com) is a challenge-based verification process with a specific focus on the validation of industrial research processes related to systems biology by decomposing a research workflow into individual components, termed building blocks, that can be independently verified via crowdsourcing.5
The first sbv IMPROVER initiative, the Diagnostic Signature Challenge (DSC), was designed to determine which computational approaches and types of transcriptomic data could be used for phenotype prediction.7 The second initiative, the Species Translation Challenge (STC), was designed to address whether or not biological events observed in rodents were “translatable” to humans (see www.sbvimprover.com). Each sbv IMPROVER challenge was developed under the hypothesis that “crowdsourcing . . . may be a fruitful strategy for assessing the quality of analyses and predictions from high-throughput data”5 and each was built on the lessons from other challenge-based processes such as CASP,8 CAPRI,9 BioCREATIVE,10 and DREAM.11 Crowdsourcing initiatives in systems biology can potentially complement the classical peer review process, by enabling a practical assessment of robustness for complex methodologies.
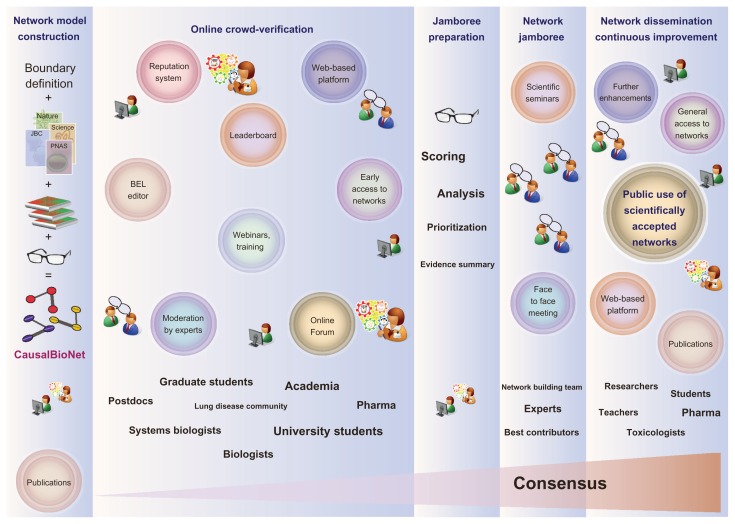
Crowdsourcing challenges aimed at verifying methodologies require the organizers to have a solution (“gold standard”) against which the predictions are assessed and that is available to participants after the challenge is closed.5 Obtaining a “gold standard” is difficult in the area of systems biology or when the challenge is focused on scientific content rather than the method, as is the case for the construction of representative biological networks. In this instance, the knowledge must be updated continuously to reflect the current state of the field.12 To verify and enhance previously built biological network models,12–15 the sbv IMPROVER Network Verification Challenge (NVC)-to be held between October 2013 and March 2014-will use a proven social networking approach to generate high-quality curation results.16 This crowd-verification process is designed to assemble the knowledge of domain experts and focus the critical minds of biologists from across multiple fields of biology to effectively and efficiently review the evidence available in the literature to improve biological network annotations, similar to other community-driven curation efforts like WikiPathways4 and TBCAP.17 The additions to and modifications of the provided biological networks will be evaluated using an online verification process (see Fig. 1) and an in-person scientific ‘jamboree’ session to resolve the final representation of the networks.
Figure 1.
Outline of the crowd-verification verification process that will be used in the Network Verification Challenge (NVC).
The NVC consists of five phases. In the first phase, network models are constructed based on the literature and data-driven hypothesis validation. The models are imported into a Web-based platform (CausalBioNet) for the second phase (online Crowd-verification). In phase 2, experts and biology students and researchers are encouraged to access and verify/enhance the network models directly on the platform. This process is set up as a reputation-based collaborative competition, where actions on the network are given points that are recorded in a leaderboard. After this online phase is closed, in phase 3, the results and actions are analyzed, and the organizers select a number of edges that appeared to be the most controversial for discussion in a jamboree (phase 4) that will gather together scientific experts and the best contributors in the online phase. After a wrap up of the conclusions and actions on the network discussed during the jamboree, in phase 5, verified versions of the networks will be released for the scientific community at large to use.
Network Models: From the Bigger Picture to Detailed Biological Mechanisms
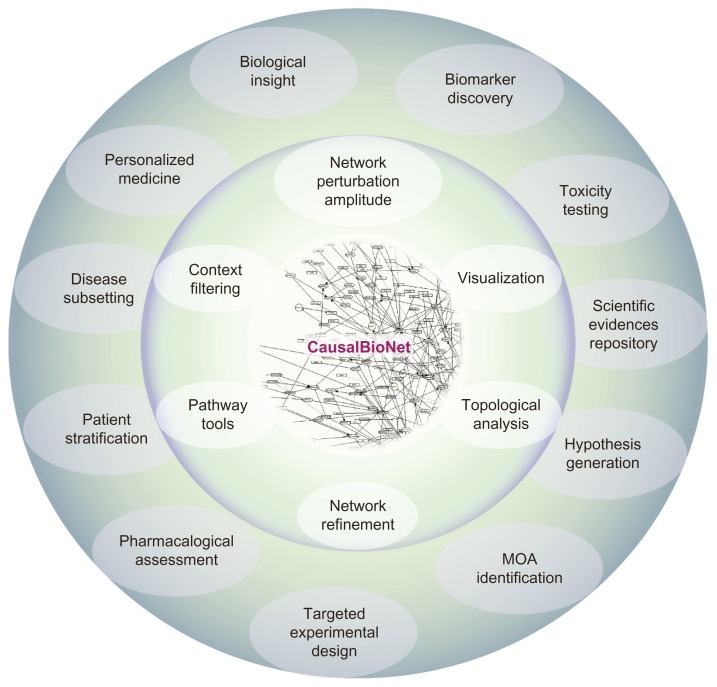
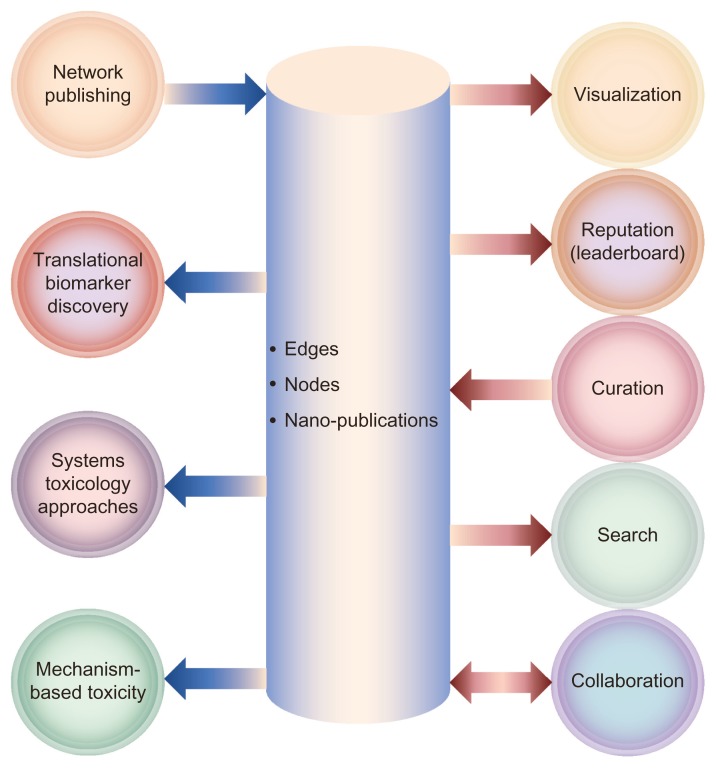
Over the last 10–20 years, the development of innovative tools for biological research has enabled the acquisition of high density data in a systems-wide approach. This has enabled the evaluation of gene expression changes in various settings to generate new hypotheses. Consequently, the size and number of datasets being reposited into databases are growing exponentially, as are the number of scientific articles being published. Despite the advantages brought by the rising number of online-accessible repositories, researchers could easily drown in this deluge of data and lose biological focus. For this reason, top-down approaches that first look at the bigger picture before investigating single pathways or gene changes and the combination of approaches (supervised and unsupervised) can provide a broad framework for a researcher to delve more effectively and efficiently into the vast amount of available data. Pathway signaling and network biology enable these types of approaches, contributing to a variety of research applications, including drug discovery, personalized medicine, and toxicological risk assessment (Fig. 2).18
Figure 2.
Potential applications of CausalBionet.
The networks provided in the NVC are potentially at the center of very diverse applications. They provide the ability to extract mechanistic insights from large datasets in toxicity or pharmacological testing. The networks will also provide teachers and students with a centralized repository of lung-related biology-relevant genes and pathways. Their usefulness in patient stratification, personalized medicine, and other areas of research is accomplished through a functional layer of available tools for visualization, refinement, and diverse types of quantitative and qualitative analyses.
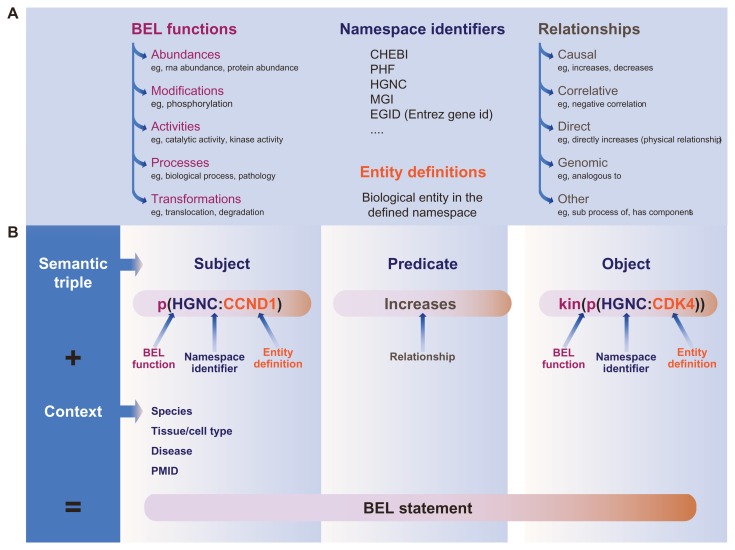
Biological network models that represent an up-to-date summary of known biology within defined boundaries (eg, species, tissue, and disease) offer a readily human-understandable metaphor for biological relationships and are amenable to computational analysis when encoded in the appropriate format. The curated network models can be encoded in the Biological Expression Language (BEL)19,20 (see www.openbel.org). BEL is a structured language that represents scientific findings by capturing causal and correlative relationships between biological entities in computable statements that are composed by functions and entity definitions expressed with a defined ontology (eg, HGNC, see www.genenames.org). A BEL Statement (see Fig. 3) is designed as a semantic triple (subject, predicate, object) to represent discrete scientific findings and their relevant contextual information as qualitative causal relationships.
Figure 3.
BEL Elements and an Example of a BEL Statement.
(A) In a BEL statement, functions and entity definitions are expressed with a defined ontology (namespace) and the relationships as causal or correlative (B) A BEL Statement is designed as a semantic triple (subject, predicate, object) to represent discrete scientific findings and their relevant contextual information as qualitative causal relationships. Functions and entity definitions are expressed with a defined ontology (namespace). For example p(HGNC:CCND1) => kin(p(HGNC:CDK4) is a statement equivalent to “Increased abundance of the protein designated by ‘CCND1’ in the HGNC namespace directly increases the kinase activity of the abundance of the protein designated by ‘CDK4’ in the HGNC namespace”. The rest of the BEL Statement consists of fields pertaining to the context of the statement: the literature reference from which the statement was derived, the tissue, cell line, organism, and disease context of the statement.
The main strength of BEL is that it is easily human-readable and machine-computable, making it an ideal language to capture literature evidences from manual curation as well as text mining pipelines. It also allows literature evidence to be displayed in the context of network visualization. Additionally, the OpenBEL community is continuously developing and assembling tools in an emerging open-platform technology named the BEL framework. In contrast, BioPAX,21 another language frequently used to model knowledge, focuses on the integration and data exchange of biological pathway information across a large array of existing pathway resources. With its unique representation of specific relationships between entities and their respective biological contexts, BEL was adopted as the syntactical structure of the network models in the NVC.
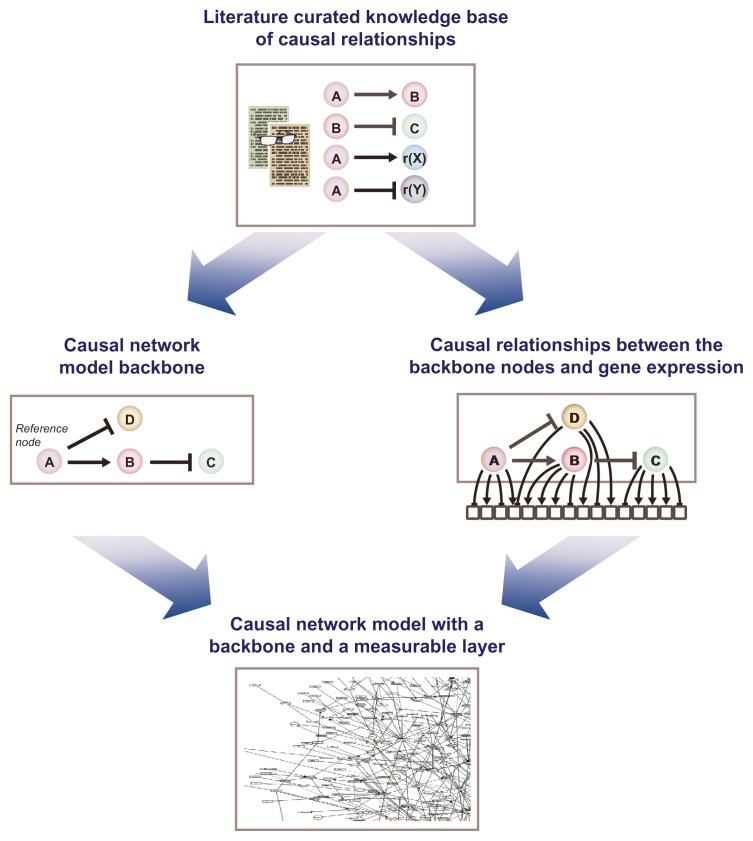
Sets of network models have been generated to score high-throughput data sets.12–15 The basics of the model building are shown in Figure 4. Network model construction is a multi-step, iterative process, which has been described in detail in previous publications. 12–15,22 Briefly, the construction of the network models starts with a careful selection of model boundaries, i.e., the selection of appropriate tissue/cell context and biological processes to be included in the model. Then, the relevant scientific literature is reviewed to extract causal relationships that comprise the nodes and edges in backbone of the literature model. The network is subsequently augmented based on gene expression data using reverse causal reasoning.23 Multiple data sets are used to verify the network content, ideally from experiments where the biological mechanisms captured by the network model under construction are perturbed.
Figure 4.
Building of CausalBioNet network model sets. The cause and effect relationships that are curated from the scientific literature form a knowledgebase. This information can then be used to build the model backbone as well as the downstream measurable layer, which together form the computable causal network model.
These models can be used in combination with an algorithm that can predict the activity of a backbone node based on gene expression changes attributable to a perturbation of the biological process.24 Proof-of-principle verification of possible applications of network models has been published previously.25 In this study, human bronchial epithelial (NHBE) cells were treated with TNF and the transcriptomic response was analyzed in the context of the TNF-NFkB signaling pathway. Dynamic changes were detected in the amplitude of network perturbation following TNF treatment. Importantly, the measured changes in network perturbation amplitude corresponded to the direct experimental measurement of NFkB nuclear translocation following TNF treatment. This result illustrates how the network models contained in CausalBioNet (or similar networks) can identify and quantitate chemically induced biological changes.26
A Challenge of Skills and Incentives
A structured crowdsourcing initiative requires a large number of highly engaged participants to gather scientific information from which new relationships can be. To this end, the NVC has incorporated both traditional and non-traditional incentives to promote user activity. The reputation gained by participating in a game of skills becomes part of the reward, as opposed to (or in addition to) material incentives such as financial awards, ie, traditional incentives. Reputation is measured by the points or badges accrued from different actions. Participants’ motivation can be further increased when their reputation is made visible to others on a leaderboard, instead of being provided solely at the conclusion. To complement the individual leaderboards, the NVC will also have team/institution leaderboards to encourage collaborative competition.
Beyond the reputation points, reputation badges, and leaderboard system, the NVC will offer a number of professional and scientific incentives to stimulate participation and engagement. Participating researchers will have an early access to curated network models of signaling pathways. Participants who performed a certain number of actions will also be able to download the networks. Because these models summarize biological knowledge and relationships that may have been very dispersed in the literature, they are likely to help scientists generate new hypotheses for their own research. Additionally, users will gain early expertise in BEL, which is becoming more widely adopted as a biological syntax conducive for computational manipulation.
Scientists will also be incented to contribute actively to the networks of interest and to develop new understanding through discourse with other domain experts. Communication between participants will be made possible via the commenting system, which allows users to provide remarks and responses specific to individual nodes and edges throughout the network. This social aspect of the NVC is an important feature because it encourages users to engage with academic peers to drive the approval or disapproval of network actions. It offers the users the opportunity not only to gain reputation but also to assign changes to the network that represents validated information from which new insights can be drawn. The push towards greater interaction naturally increases an individual user’s personal network, which is traditionally an important component of a scientific career.
Furthermore, a user’s visibility can be increased greatly should they rise to the top of a particular network’s leaderboard or gain prominence as a pathway expert, disease expert, or curation expert by participating in the challenge. Notably, top participants and performers in the NVC will be awarded a travel bursary and invited to a ‘jamboree’ session. The session allows participants to share knowledge, grow personal networks, strengthen the decisions made in the open online verification phase, and discuss items that arose as being of popular interest but lacked consensus from the crowd.
Crowd-verification Process
The crowd-verification of biological network models will be performed through the following steps:
Develop a high-performance platform for crowd-verification of biological network models and import created biological network models onto the platform;
Start the crowd-verification period by making the platform accessible to the research community, with associated incentives to stimulate online verification of nodes and edges supported by scientific findings;
Interpret the results after a previously set period in order to select questionable edges (eg, edges that did not obtain a consensus from the community);
Organize a ‘jamboree’ session where community members that contributed significantly to the online verification can meet recognized experts and analyze scientific evidence for selected questionable edges. Publish the verified and extended networks;
Assess the resulting networks and determine to what extent the biological mechanisms were further expanded, revised or invalidated. Disseminate the networks for public use.
These five steps are summarized in Figure 1 and are described below in more detail.
Network Verification Platform
Though biological networks are a powerful way of representing complex information, they can easily become unwieldy to navigate and manage as their size, complexity and density increases with additional data. Crowd-verification of networks can mitigate some of these difficulties because many individuals can work in parallel to manage large networks; however, currently, the tools required to collaboratively build, share, and maintain these networks are lacking. The NVC website (Fig. 5) will provide a collaborative, crowd-sourced, network building and verification application that is self-managing through the use of a social reputation engine. This application will offer network building, knowledge capture, verification, and extension features with a reputation system to aid in moderation of the network verification process. The site goes further by developing a framework where, in the future, new biological networks can be created de novo. It has been suggested that intuitive interfaces for biological network visualization and interaction may “one day replace printed biology textbooks as the primary resource for knowledge about cellular processes”.21 The platform and its components described below provide the research community with a high-performance environment for the crowd-verification of biological network models.
Figure 5.
Outline of the Network Verification Challenge (NVC) website at the interface between the scientific community and systems biology.
Database of networks encoded in BEL (CausalBioNet)
The CausalBioNet networks presented in the sbv IMPROVER project for verification and enhancement are expressed in BEL and represent qualitative biology in a scale-free representation. The nodes are BEL terms and are identified using biological databases such as SwissProt (www.uniprot.org), Entrez-Gene (www.ncbi.nlm.nih.gov/gene), Rat Genome Database (rgd.mcw.edu), and ChEBI (www.ebi.ac.uk/chebi/). The network edges are BEL Statements that connect two nodes, maintain the computability of the network, and are supported by evidence from the published and peer-reviewed scientific literature. Both the network structure and the supporting evidence are stored in a MongoDB database (www.mongodb.org).
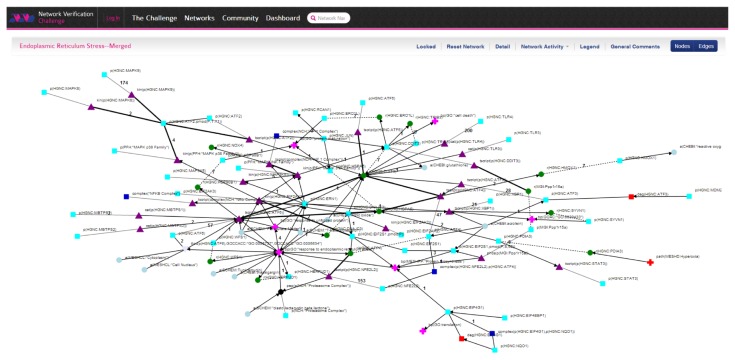
Website to facilitate visualization and the review process
The NVC website will display an overview of the CausalBioNet network representing the connections and relationships between several networks and provide the functionality to select one of these networks for review (Fig. 6). It will also be possible to see a list of the available networks for selection or to use a search function that will search across the network title, summary, individual nodes, and gene or protein synonyms. The website will support the full set of actions allowed in curating a network. For instance, participants will be able to add and remove edges or nodes as well as add and remove evidence supporting the network edges. However, all of these actions will be labeled as suggested changes that will need to be ratified by other participants through voting.
Figure 6.
Screenshot of the mock-up of a network as visualized on the NVC website.
Network visualization engine
The CausalBioNet networks will be visualized using a web-based network visualization engine powered by D3.js (www.d3js.org). The network viewer will allow participants to add and delete edges through the addition or deletion of evidence supporting those edges. Additional features such as adding comments to a network and providing different visualization filters for the networks will be made available. The filters will include the visualization of the original network, the current network updated with all recommended changes, or the original network with the recommended changes presented as layers on top of the original network.
BEL web-based statement editor
All network edges are represented by BEL Statements (see Fig. 2), which must be supported by at least one published literature reference. The BEL web-based statement editor supports several features that provide guidance to the participants on the functional syntax of the BEL Statement. An auto-complete terminology service provides support in entering protein names, chemical compound names, Gene Ontology terms,27 and other biological entities used in a BEL Statement. The editor will also suggest which statement functions and types of entities are allowed at the cursor position as the BEL Statement is being created.
At a later stage, it is also envisioned that model-building efforts will be fuelled by advances in text mining that can make possible the semi-automated assembly of BEL encoded knowledge bases. Text mining is the technology that is used to support the targeted retrieval of relevant terms28 and bring them into a structured relationship. Historically, the conversion of prior knowledge into BEL Statement has largely been a manual process that required many curators with expertise in the related fields and their full commitment to the curation task. The OpenBEL community envisions technologies that will use state-of-the-art algorithms that are specifically designed to extract biological facts from scientific literature and assemble them into BEL Statements based on their context and meaning.29 Text mining pipelines of this kind could provide potential BEL Statements for review and incorporation either into the current networks or into a biological knowledge base.
Curation system moderated by a reputation system
Another important aspect of the website will be the reputation engine that will provide self-management of curation tasks. Reputation engines have been used in other initiatives that involve game of skills principles, such as StackOverflow (stackoverflow.com), that reward “submitters” and “voters”. In the NVC, “Submitters” are participants who propose an action on the network website and “voters” are participants who vote to approve or disapprove an action. Depending on the type of action and the effort required to propose an action, a corresponding number of reputation points will be awarded to the submitter. Voters also gain reputation points. Once an action has obtained a minimum number of votes, the action is ‘locked’ to further voting. If a consensus is reached, additional points are given to the submitter if the action is approved or, if the action is disapproved, the points originally awarded points for the action will be removed. Voters are awarded bonus points only if the action reaches a consensus and their vote aligns with the consensus.
Reputation badges will also be awarded as users complete a pre-defined set of tasks. For example, a user may be given a badge if they create 10 approved network edges. Though these badges do not affect a user’s point total or leaderboard position, they are an important acknowledgment of their contributions to the network and the NVC.
To mitigate attempts to obtain reputation points not based on skilled scientific actions, several quality review checks will be introduced into the system. For example, the system will measure the co-occurrence of submission and voting activity between participants. If an abnormal amount of activity is measured between participants who seem to be supporting each other’s submissions, their activities will be reviewed by the site moderators to confirm the scientific rationale underpinning the actions. In addition, the system will allow only a limited number of actions per hour, to avoid automated scripts being used to create a high number of actions.
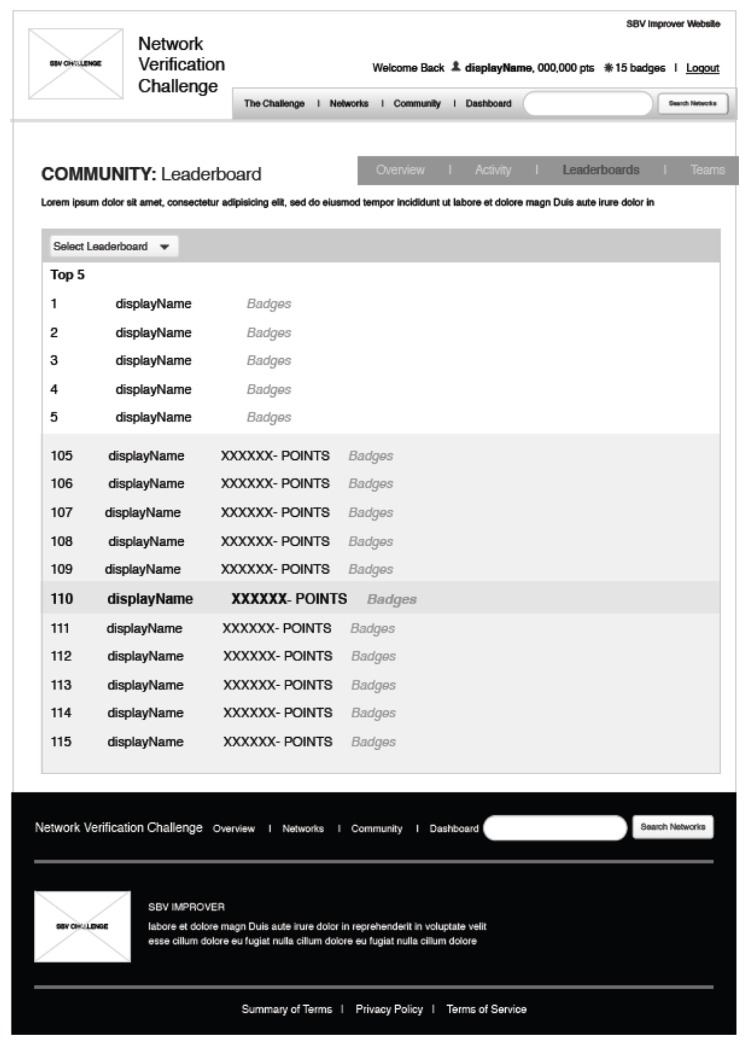
A leaderboard (see Fig. 7) will be generated to show participants and their reputation points. Users with the highest reputation points are likely to be those who were highly engaged, had strong biological knowledge, and gained the most experienced in biological knowledge representation. The NVC will help to determine the future role of crowd-sourced network creation and management in network biology. The highest- scoring participants overall and within the subnetworks will be invited to a ‘jamboree’ session (described below) to review or curate the networks that were created during the crowd-verification challenge.
Figure 7.
Leaderboard page as part of the NVC curation system.
Properties of the CausalBioNet Networks
The CausalBioNet networks that will be the subjects of NVC possess a unique set of features that distinguishes them from, and makes them complementary to, the collection of signaling pathways and networks already available to the scientific community12,15 (see Supplementary Table 1). Repositories such as STRING30 or HPRD31 try to create a genome-wide picture of protein-protein interactions in an almost context-free setting. Other signaling pathway repositories, such as KEGG32 and BioCarta (www.biocarta.com), employ manual curation of the literature but still do not offer significant biological context. The aim of the NVC is to provide curated networks constructed within precisely defined contextual boundaries for associated literature. The literature context for the networks is primarily human, although mouse and rat evidence was included when supporting literature from human context was not available. Most of the evidence is derived from non-diseased respiratory tissue biology augmented with chronic obstructive respiratory disease biology (i.e., excluding, for example, lung cancer context). The current CausalBioNet networks encode the exact relationships between entities at the highest level of granularity possible, and provide the associated literature evidence as captured in BEL. Hence, a wide range of biological information is represented in these networks, including proteins, DNA variants, coding and non-coding RNA, phenotypic or clinical observations, chemicals, lipids, methylation states, and other modifications (e.g., phosphorylation). Additionally, because the networks are encoded in BEL, they reflect the causal nature of relationships between nodes, allowing the biological intent of the network model to be easily digested by a scientist, and enabling inference and computation using the network as a whole (See Fig. 7). While most of the known biology is represented in the visible backbone of the networks, network computations can be performed using a downstream (hidden) layer of RNA abundance of genes regulated by the nodes in the backbone. Future development of the CausalBio- Net networks may accommodate other -omics datasets, such as proteomics, metabolomics, or lipidomics. The gene expression data that underlie these networks greatly facilitates the biological interpretation of the complex datasets in the search for explanations of the observations. Another important feature of the networks, as implemented on the NVC website, is that they are dynamic: they can be modified to represent specific species and/or tissue contexts by the application of appropriate boundaries and can be updated as new knowledge becomes available.
Lung physiology and pathophysiology networks
The above approach was used to build a set of networks representing important biological processes implicated in human lung physiology. These networks have been published previously as separate entities: cell proliferation,15 cellular stress,15 DNA damage and cell fates,12 pulmonary inflammation,13 tissue repair and angiogenesis.14
COPD is a common inflammatory lung disease in which the airways become narrowed, causing shortness of breath. COPD is a major and increasing global health problem. It is predicted by the World Health Organization to become the third most common cause of death and the fifth most common cause of disability in the world by 2020.33 The main risk factor for emphysema/COPD in the developed world is exposure to tobacco smoke.34
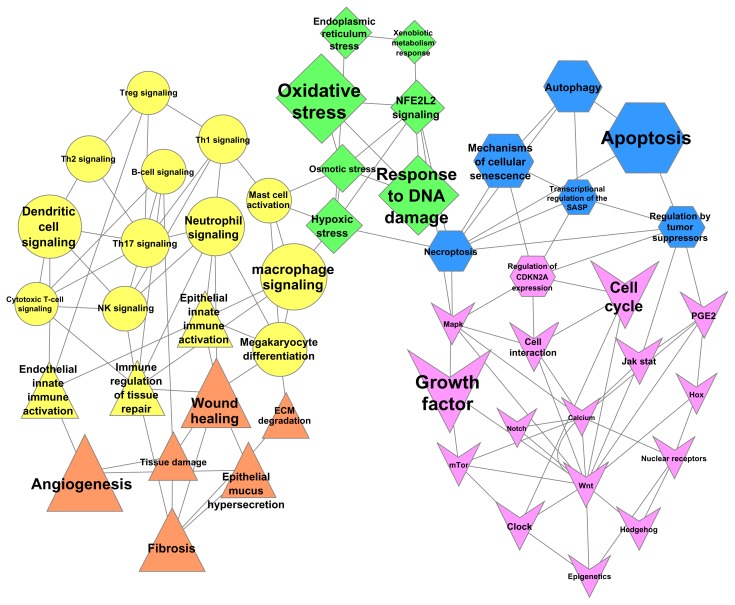
The non-disease networks described above were augmented with chronic obstructive pulmonary disease (COPD) pathophysiology-relevant connections to yield the CausalBioNet collection of networks. In addition, four networks were either built exclusively or modified extensively to suit the COPD context. B-cell Activation and T-cell Recruitment and Activation subnetworks were built to represent these immune processes and their role in COPD, and extra-cellular matrix (ECM) Degradation and Efferocytosis subnetworks were constructed by strongly modifying non diseased models to specifically comprise COPD-relevant mechanisms. The set of networks that describe the biological elements related to the COPD process in humans (Fig. 8) will be made available for the NVC.
Figure 8.
Biological networks included in CausalBioNet. Each node is labeled with the network name and the size of the node shows the sum of nodes in each network. We have split the 50 networks submitted for crowd-verification in five different tracks, namely cell fate (blue nodes), cell proliferation (pink), response to cell stress (green), immune response (yellow), and tissue response (orange). The separation is biologically not always as clear, so the shape of the nodes also reflects common areas. For example, the immune regulation of tissue repair is both an immune and a tissue response.
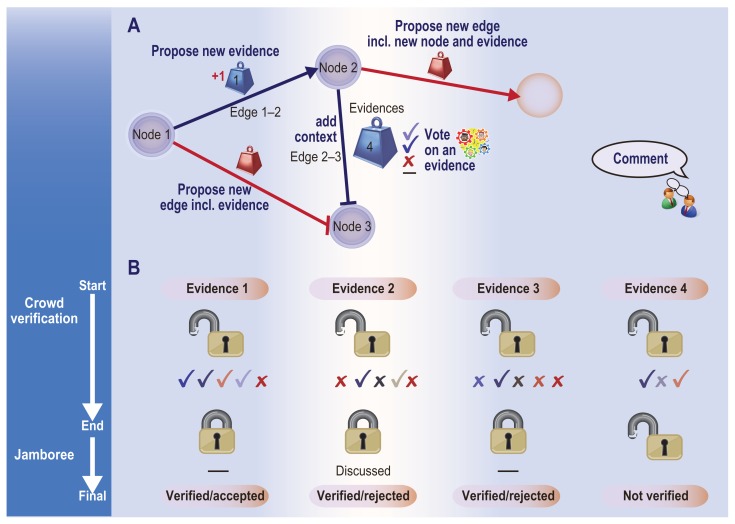
Crowd-verification within the scope of crowd curation of biological networks and the online verification of this curation, the NVC has implemented a submission, approval, and commenting system designed to encourage scientists to critically evaluate evidence supporting various network relationships (Fig. 9). When verifying edges and nodes, users are required to use controlled syntax in the form of a BEL Statement and must generally support their action with a reference to one or more peer-reviewed publications. The use of the BEL Statement with references ensures structural and logical correctness and addresses an important concern regarding knowledge curation platforms: consistency checking.35 BEL Statements enforce consistent input structures that allow evidence evaluation algorithmically or manually. The requirement for references allows other participants to judge the applicability and logical soundness of the comment or modification to the network, species, tissue, or process being verified.
Figure 9.
Outline of the crowd-verification process showing the actions available in the NVC online phase and verification outcomes. (A) Actions that will be available for verification of the networks. The “starting” network is shown as pink nodes and black edges. Each edge is supported by a number of evidences or BEL statements. The weight close to the edge shows the number of evidences available for each edge (ie, the weight of the evidences for that edge). Participants can vote for (approve/reject) evidences. If the evidence is for a new edge, that edge will then be added to the network. (B) Examples of possible verification outcomes. A limited number of votes will be allowed on each evidence after which the evidence will be locked and the points rewarded accordingly (see Table 1). If the majority consensus validates the evidence, then it is considered verified and accepted, and locked from further voting. Conversely, if the majority finds the evidence inappropriate, then it can be rejected. In cases where no clear consensus emerges, the evidence will be locked and considered for discussion in the jamboree. Evidences that do not accumulate a sufficient number of votes will be considered as not verified.
The NVC further stipulates that for any network action (such as creating or removing edges or evidences) to be approved or disapproved there must be a consensus among users as measured by their votes, here referred to as approvals and disapprovals. Users are awarded a baseline set of points when they create an action in the system. Bonus points are awarded following a specific reputation action being locked to further approvals or disapprovals, and the modification being approved or disapproved (Table 1). An action will be locked once a minimum number of approvals or disapprovals have been cast. The action will then be approved or disapproved if there is a clear consensus, as indicated by a pre-determined threshold ratio of approvals to disapprovals. If this threshold is not met, no bonus points will be awarded, and the action will remain as ambiguous and be prioritized for further study at the ‘jamboree’ session.
Table 1.
Points system used to rank participants based on their contributions to the NVC.
| Reputations actions | Points awarded initial/approval/rejection | Final score after community responses to the actions | |||
|---|---|---|---|---|---|
|
| |||||
| 4 votes approving | 4 votes disapproving | 5 total votes, but fewer than 4 for/against | Less than 5 total votes and fewer than 4 for/against | ||
| Network edge creation | 5/100/0 | 100 | 0 | 5 | 5† |
| Evidence creation | 5/50/0 | 50 | 0* | 5† | 5† |
| Peer approval | 1–3/11–13‡/0 | 11–13‡ | 0* | 1–3†,‡ | 1–3†,‡ |
| Peer disapproval | 1–3‡/11–13‡/0 | 0* | 11–13‡ | 1–3†,‡ | 1–5†,‡ |
Notes:
Loss of initial points;
Keep initial points;
Depending on how many fields were completed.
By implementing a system that rewards network modifications that are agreed upon by the wider set of participants, the NVC places greater emphasis and importance on high-quality curation and discourages indiscriminate actions. Furthermore, the system requires voters to offer evidence to support actions; thereby, the system discourages malicious or arbitrary down-voting of other participants. However, if the disapproval is appropriate and the action to which it is applied is subsequently disapproved, the voter will be awarded a bonus point to reward removal of incorrect actions. This system can be implemented with minimal moderator oversight, and in such a form, it becomes a nearly real-time online crowd-verification system for curation of biological networks.
Prior to the locking of an action, any user can view the votes or comments on that action but the usernames are kept anonymous to prevent unintended personal influence. However, after an action is locked, all usernames of submitters and voters will be viewable. Such transparency can be useful in generating a greater, persistent scientific dialog that may be carried over to other areas of the network, to the ‘jamboree’ session, and to areas outside the scope of the NVC. At the close of the open challenge, all edges that have been locked without clear consensus will be evaluated, sorted, and selected from for further analysis in the ‘jamboree’ session (Fig. 9). Criteria for edges selected for analysis at the ‘jamboree’ session will be determined by the total number of approvals and disapprovals.
For the NVC, crowd-verification will be mediated using a reputation point system that encourages high-quality scientific contributions and the development of a consensus network model. Users will be able to accrue reputation points and reputation badges as well as interact with the larger network of participants through a leaderboard system and commenting structure. Of particular importance, the sources of points and positive rewards are primarily conferred based on contributions of verified, biological knowledge, as opposed to more purely action-based reward systems like Foldit.36 By placing the emphasis on the scientific information provided, the NVC is more analogous to crowdsourcing efforts that aim to confine gamification elements principally to leaderboard systems to drive friendly competition and engagement, as exemplified by DREAM,37 the Netflix Challenge,38 and the challenges hosted by Kaggle (www.kaggle.com). The NVC point and badge system also has elements of the reputation systems seen in Q&A sites like StackOverflow (www.stackoverflow.com), where answers to questions are up-and down-voted by users, and participants who submit answers are rewarded with points attributed in relation to the crowd approval or disapproval of their answer.
The NVC will also implement a leaderboard system to offer participants an understanding of their relative performance in the overall challenge and in each specific subnetwork. The leaderboard system will be designed to encourage friendly competition and greater engagement within each of the networks. Leaderboards will indicate username, rank as determined by the total number of reputation points, and specific metrics such as quantity of edges created/rejected. These leaderboards will operate at a global level, including activity across all subnetworks, and will also run for each individual subnetwork. Importantly, to promote competition and continued engagement while avoiding discouragement because of large differences in point totals, users will be able to see only the ranks and points of the five participants above and below their rank within the global and specific subnetwork leaderboards. In addition, to reward top contributors without discouraging other participants, the top five usernames for all leaderboards will be shown, but without their point totals.
Users may participate in the NVC as individuals or as a team. Although users will ultimately be evaluated as individuals, self-identification with others as a team may encourage participation within and competition between groups. In addition, the NVC infrastructure will be maintained and available to the community for further action after the official close of the challenge.
‘Jamboree’ Session and Publication of Results
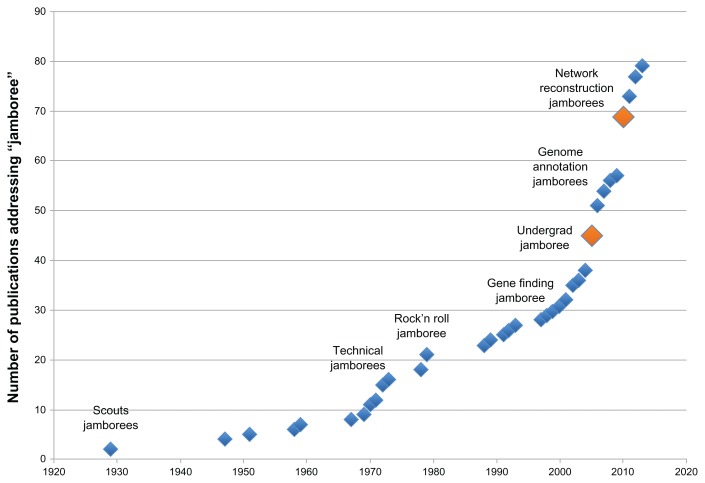
The analysis performed following the open phase of network verification/enhancement should pinpoint a number of edges and nodes that are ambiguous. The decision to include or exclude these edges or nodes from the finalized consensus network would benefit greatly from discussions between subject matter experts, the team of scientists who first generated the networks, and the “best” participants in the NVC. Though the web platform used during the open phase is built to facilitate interactions within the scientific community, face-to-face meetings and the efficiency of experts sitting together to discuss predefined topics are irreplaceable. Therefore, the NVC will include a 2–4 day ‘jamboree’ network verification meeting that will conclude the open phase of the challenge. Concepts surrounding the value of jamboree meetings in science have emerged only recently (see Fig. 10). In a scientific context, they may be defined as “focused events at which domain experts apply their knowledge to refine and consolidate biochemical knowledge from existing reconstructions and published literature”.39
Figure 10.
Numbers of publications that mention the word “jamboree”.
The Merriam-Webster definitions of “jamboree” are “a large festive gathering” or “a national or international camping assembly of boy Scouts”. The concept of “jamboree” meetings in the context of science has emerged only recently. One of its first appearances in the scientific literature was in a report about the outcome of a scientific jamboree for undergraduate students. The jamboree was held to conclude their summer work of trying to construct small biological devices.2 The jamboree allowed students to share data, experiences, and most importantly to realize how their contribution had been valuable. It also showed that coordinated efforts can lead to better results than the sum of individual efforts. Similarly, endeavors such as network reconstruction or genome annotation projects, which entail countless efforts that are best distributed among many teams, also benefit from jamboree sessions where results can be homogenized and validated. The reconstruction of a consensus yeast metabolic network project set the ground for the type of scientific ventures that benefit greatly from jamboree meetings.3 The goals set for the yeast jamboree meeting were threefold: (i) define the standards that should be used, (ii) annotate, and (iii) resolve discrepancies. What a reconstruction annotation jamboree entails and how it should be organized was later summarized very nicely by Thiele and Palsson.4 Jamborees have also been used in other projects, namely a community-based effort to construct a model of Salmonella typhimurium LT2,5 and the reconstruction of the human metabolic network.1 Another successful jamboree was held for the Little Skate genome project.40 The goal of these jamboree sessions was not only to provide a floor for discussion, but also to deliver training in various fields related to genome annotation and analysis,6 and to coordinate the efforts.
Community-based jamborees are commonly used in genome-based annotation projects34,40 and they have been shown to be valuable for (i) defining the standards that should be used, (ii) annotating, and (iii) resolving discrepancies. The ‘jamboree’ session constitutes an integral part of the network verification process, and scientists will be invited to give their opinion on the online verification phase and outcome as well as to adjudicate ambiguous edges and evidences.
After the ‘jamboree’ session, the discussions and decisions will be summarized and an updated version of the network models will be generated incorporating the feedback from the session. These updated network models, which will represent what is expected to be the most relevant knowledge on human lung biology, will be made available to the scientific community through the bionet.sbvimprover.com website.
Interpretation of Results
In the last phase of the NVC, the results of the challenge will be evaluated to determine to what extent the biological mechanisms were further expanded, revised or invalidated by the challenge. To address these questions, we will thoroughly review the changes to the network and the transactions performed by the participants based on defined metrics such as.
Number of statements supporting each edge, before and after the NVC.
The specificity of contextual annotations for each statement relative to the network’s intended context, before and after the NVC.
Ratio of positive and negative reviews of each evidence prior to locking.
Number of editing events for each remaining edge.
Number of evidence deletion events and number of edges removed when all supporting evidence is removed.
Number of locked vs. unlocked evidences.
Finally, we will review the transactions and the resulting network to make sure that all unproductive activities were flagged and removed by moderators. If there are any unusual patterns of success by individuals or groups, the science of the resulting statements and edges will be reviewed to determine whether the scientific content of the final network was in any way compromised for the sake of competition.
Outlook
The NVC aims to encourage continual user participation; therefore, the network will continually be refined and expanded as new evidence is added throughout the challenge and after the close of the official challenge time. In this way, the challenge is more than a simple verification, but is a curation and refinement process that looks to expand and maintain up-to-date biological knowledge as well as facilitate the inference of new relationships.
Challenges such as the NVC are expected to bring benefits to the scientific community that engages in the crowd-verification process. The benefits might include:
providing an accelerated mechanism for the dissemination and validation of knowledge;
providing better maps of disease and a forum for reproducible and re-usable data and analyses;
providing a platform that links model generators with researchers and clinicians who together are poised to validate modeling hypotheses and incorporate modeling results into research directed at understanding physiological or disease states, and therapeutic development efforts;
sharing and working as a social network of distributed teams with the needs and opportunities created by the genomics revolution and the desire to translate public and private investment into demonstrable human benefit;
supporting the scientific community through a combination of the network model and analytics25 in the endeavors to identify biomarkers of lung disease as well as biomarkers relevant to environmental or tobacco smoke exposure.41
Finally, researchers who choose to participate will gain early insights into the data and relationships that will be evaluated in the sbv IMPROVER’s Grand Challenge that will follow the NVC. The Grand Challenge will aim to leverage the wisdom of crowds to develop methodologies for predicting the prognostic impact of different compounds and substances on COPD. The network information verified by the NVC will be included as one of the inputs for the Grand Challenge. Thus, early involvement in the curation and validation of these data may offer opportunities for significant insights into the data that could enhance the methods developed for the Grand Challenge.
Supplementary Table
Table S1.
| CausalBioNet | KEGG30 (kyoto encyclopedia of genes and genomes) | Reactome | BioCarta | Wikipathways4 | SPIKE31 (signaling pathway integrated knowledge engine) | UCSD Signaling gateway | NCI pathway interaction database* | NetPath32 | |
|---|---|---|---|---|---|---|---|---|---|
| Species human (Hs); Mouse (Mm); Rat (Rn) | Hs, Mm, Rn | >20 species | Hs (curated) +20 species (inferred) |
Hs, Mm | >25 species | Hs | Hs, Mm | Hs | Hs |
| Literature support shown | At edge-level | At pathway-level | At pathway level | At pathway level | At pathway level | At edge level | At molecule/state/transition levels | At edge and pathway level | At edge level |
| Defined biological boundaries | Species Tissue Disease context Biological pathways |
Species Disease context (Hs) Biological pathways |
Biological pathways | Species Biological pathways |
Species Disease context Biological pathways |
Species Biological pathways |
Species Biological pathways |
Species Disease context (cancer) Biological pathways |
|
| Manual curation | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes* | Yes |
| Data-driven enhancement | Yes | No? | No? | No | No | Yes | No | Yes | |
| Crowd curation | Yes | No | No | Partial (comments) | Yes | No | No | No | Through Wikipathways |
| Causal | Yes | No | No | No | No | Yes | No | Yes | Yes |
| Directional edges | Yes | Yes | Partial | Yes | Yes | Yes | Yes | Yes | Yes |
| Multiple types of gene-centric entities | Yes | No | Yes | No | No | No | Yes | Yes | No |
| Interactive visualization | Yes | No | Yes | Yes | Yes | Yes | No | No | No |
| Computable | Yes | Yes/no | Yes (pathway enrichment) | No | No | No | No | No | No but include transcriptionally regulated genes in each pathway |
| Size | >30 networks | >400 signaling modules | 1402 pathways (Hs) | >350 pathways | >430 pathways | 28 curated pathways | ~3500 proteins and their proximal connections | 137 NCI-nature curated pathways (+Reactome + BioCarta) | 32 curated pathways (immune signaling/cancer) |
| Available for download | Yes | Yes | Yes | No | Yes | Yes | No | Yes | Yes |
Notes:
No longer actively curated. The website will be retired in September 2013.
Acknowledgements
The authors express their gratitude to Vincenzo Belcastro, Mark Ciapka, and Ryan Todd for their invaluable input while discussing the challenge and for the design of the website, Filipe Bonjour and Sylvain Gubian for their assistance in technical matters, and Tim Kilchenmann for his assistance in the challenge preparation.
Footnotes
Author Contributions
Developed overarching strategy and conceived the project: JH, MCP. Developed technical design and implementation: JB, SB, KR, DP, RK, RN, AI, CP, WKS, GS, WH, JH, MCP, ADF, BO. Wrote the first draft of the manuscript: JB, SB, KR, DP, WH, JH. Contributed to the writing of the second draft manuscript: RK, RN, AI, GS, MCP, MT. Made critical revisions: JB, SB, KR, RK, MT, JH, SA. All authors reviewed and approved of the final manuscript.
Competing Interests
MD, JH, WH, ADF and MT have a patent pending on a reputation system for network verification. GS has a patent pending on evaluation of predictions in the absence of a known ground truth. DP holds shares and has received consulting fees from Selventa, Inc, and holds patents as folows: on a method for quantifying amplitude of a response of a biological network; on a system, method and apparatus for causal implication analysis in biological networks; on a system, method and apparatus for assembling and mining life science data; and on a method, system and apparatus for assembling and using biological knowledge.
Disclosures and Ethics
As a requirement of publication the authors have provided signed confirmation of their compliance with ethical and legal obligations including but not limited to compliance with ICMJE authorship and competing interests guidelines, that the article is neither under consideration for publication nor published elsewhere, of their compliance with legal and ethical guidelines concerning human and animal research participants (if applicable), and that permission has been obtained for reproduction of any copyrighted material. This article was subject to blind, independent, expert peer review. The reviewers reported no competing interests.
Funding
The work presented here was funded by Philip Morris.
References
- 1.Kohavi R, Brodley CE, Frasca B, Mason L, Zheng Z. KDD-Cup 2000 organizers’ report: peeling the onion. ACM SIGKDD Explorations Newsletter. 2000;2(2):86–93. [Google Scholar]
- 2.Shi L, Campbell G, Jones WD, et al. The Microarray Quality Control (MAQC)-II study of common practices for the development and validation of microarray-based predictive models (EI) 2010 doi: 10.1038/nbt.1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Good BM, Su AI. Games with a scientific purpose. Genome Biology. 2011;12(12):135. doi: 10.1186/gb-2011-12-12-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pico AR, Kelder T, van Iersel MP, Hanspers K, Conklin BR, Evelo C. WikiPathways: pathway editing for the people. PLoS biology. 2008 Jul 22;6(7):e184. doi: 10.1371/journal.pbio.0060184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Meyer P, Alexopoulos LG, Bonk T, et al. Verification of systems biology research in the age of collaborative competition. Nat Biotechnol. 2011 Sep;29(9):811–5. doi: 10.1038/nbt.1968. [DOI] [PubMed] [Google Scholar]
- 6.Meyer P, Hoeng J, Rice JJ, et al. Industrial methodology for process verification in research (IMPROVER): toward systems biology verification. Bioinformatics. 2012;28(9):1193–201. doi: 10.1093/bioinformatics/bts116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tarca AL, Lauria M, Unger M, et al. Strengths and limitations of microarray-based phenotype prediction: Lessons learned from the IMPROVER Diagnostic Signature Challenge. Bioinformatics. 2013 Aug 20; doi: 10.1093/bioinformatics/btt492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moult J, Pedersen JT, Judson R, Fidelis K. A large-scale experiment to assess protein structure prediction methods. Proteins. 1995 Nov;23(3):ii–v. doi: 10.1002/prot.340230303. [DOI] [PubMed] [Google Scholar]
- 9.Janin J, Henrick K, Moult J, et al. CAPRI: A Critical Assessment of PRedicted Interactions. Proteins. 2003 Jul 1;52(1):2–9. doi: 10.1002/prot.10381. [DOI] [PubMed] [Google Scholar]
- 10.Hirschman L, Yeh A, Blaschke C, Valencia A. Overview of BioCreAtIvE: critical assessment of information extraction for biology. BMC bioinformatics. 2005;6(Suppl 1):S1. doi: 10.1186/1471-2105-6-S1-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stolovitzky G, Monroe D, Califano A. Dialogue on reverse-engineering assessment and methods: the DREAM of high-throughput pathway inference. Annals of the New York Academy of Sciences. 2007 Dec;1115:1–22. doi: 10.1196/annals.1407.021. [DOI] [PubMed] [Google Scholar]
- 12.Gebel S, Lichtner RB, Frushour B, et al. Construction of a computable network model for DNA damage, autophagy, cell death, and senescence. Bioinformatics and biology insights. 2013;7:97–117. doi: 10.4137/BBI.S11154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Westra JW, Schlage WK, Hengstermann A, et al. A modular cell-type focused inflammatory process network model for non-diseased pulmonary tissue. Bioinformatics and Biology Insights. 2013;7:1–26. doi: 10.4137/BBI.S11509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Park JS, Schlage WK, Frushour BP, et al. Construction of a computable network model of tissue repair and angiogenesis in the lung. Clinical Toxicology. 2013:S12. [Google Scholar]
- 15.Schlage WK, Westra JW, Gebel S, et al. A computable cellular stress network model for non-diseased pulmonary and cardiovascular tissue. BMC Syst Biol. 2011;5:168. doi: 10.1186/1752-0509-5-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mamykina L, Manoim B, Mittal M, Hripcsak G, Hartmann B. Design lessons from the fastest q&a site in the west. Proceedings of the SIGCHI Conference on Human Factors in Computing Systems; 2011; Vancouver, BC, Canada. [Google Scholar]
- 17.Mazumder R, Natale DA, Julio JA, Yeh LS, Wu CH. Community annotation in biology. Biology Direct. 2010;5:12. doi: 10.1186/1745-6150-5-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hoeng J, Deehan R, Pratt D, et al. A network-based approach to quantifying the impact of biologically active substances. Drug Discov Today. 2012 May;17(9–10):413–8. doi: 10.1016/j.drudis.2011.11.008. [DOI] [PubMed] [Google Scholar]
- 19.Clark T, Ciccarese PN, Goble CA. Micropublications: a semantic model for claims, evidence, arguments and annotations in biomedical communications. 2013 doi: 10.1186/2041-1480-5-28. arXiv preprint arXiv:1305.3506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vercruysse S, Kuiper M. Jointly creating digital abstracts: dealing with synonymy and polysemy. BMC Research Notes. 2012;5(1):601. doi: 10.1186/1756-0500-5-601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Demir E, Cary MP, Paley S, et al. The BioPAX community standard for pathway data sharing. Nat Biotechnol. 2010 Sep;28(9):935–42. doi: 10.1038/nbt.1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Westra JW, Schlage WK, Frushour BP, et al. Construction of a computable cell proliferation network focused on non-diseased lung cells. BMC Syst Biol. 2011;5:105. doi: 10.1186/1752-0509-5-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Selventa. Reverse Causal Reasoning Methods Whitepaper. 2011. [Google Scholar]
- 24.Martin F, Thomson TM, Sewer A, et al. Assessment of network perturbation amplitude by applying high-throughput data to causal biological networks. BMC Syst Biol. 2012 May 31;6(1):54. doi: 10.1186/1752-0509-6-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Belcastro V, Poussin C, Gebel S, Mathis C, Schlage WK, Lichtner RB, et al. Systematic Verification of Upstream Regulators of a Computable Cellular Proliferation Network Model on Non-Diseased Lung Cells Using a Dedicated Dataset. Bioinformatics and Biology Insights. 2013;7:217. doi: 10.4137/BBI.S12167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Krewski D, Acosta D, Jr, Andersen M, et al. Toxicity testing in the 21st century: a vision and a strategy. J Toxicol Environ Health B Crit Rev. 2010 Feb;13(2–4):51–138. doi: 10.1080/10937404.2010.483176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ashburner M, Ball CA, Blake JA, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nature Genetics. 2000 May;25(1):25–9. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nédellec C, Bossy R, Kim JD, et al. Overview of BioNLP Shared Task 2013. Paper presented at: BioNLP132013; Sofia, Bulgaria. [Google Scholar]
- 29.Fluck J, Klenner A, Madan S, et al. BEL Networks Derived from Qualitative Translations of BioNLP Shared Task Annotations. Paper presented at: BioNLP132013; Sofia, Bulgaria. [Google Scholar]
- 30.Franceschini A, Szklarczyk D, Frankild S, et al. STRING v9.1: protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Research. 2013 Jan;41(Database issue):D808–15. doi: 10.1093/nar/gks1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Keshava Prasad TS, Goel R, Kandasamy K, et al. Human Protein Reference Database—2009 update. Nucleic Acids Research. 2009 Jan;37(Database issue):D767–72. doi: 10.1093/nar/gkn892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kanehisa M, Goto S, Sato Y, Furumichi M, Tanabe M. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Research. 2012 Jan;40(Database issue):D109–14. doi: 10.1093/nar/gkr988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lopez AD, Murray CC. The global burden of disease 1990–2020. Nat Med. 1998 Nov;4(11):1241–3. doi: 10.1038/3218. [DOI] [PubMed] [Google Scholar]
- 34.Pauwels RA, Buist AS, Calverley PM, Jenkins CR, Hurd SS. Global strategy for the diagnosis, management, and prevention of chronic obstructive pulmonary disease. NHLBI/WHO Global Initiative for Chronic Obstructive Lung Disease (GOLD) Workshop summary. Am J Respir Crit Care Med. 2001 Apr;163(5):1256–76. doi: 10.1164/ajrccm.163.5.2101039. [DOI] [PubMed] [Google Scholar]
- 35.Groza T, Tudorache T, Dumontier M. State of the art and open challenges in community-driven knowledge curation. Journal of Biomedical Informatics. 2013 Feb;46(1):1–4. doi: 10.1016/j.jbi.2012.11.007. [DOI] [PubMed] [Google Scholar]
- 36.Cooper S, Khatib F, Treuille A, et al. Predicting protein structures with a multiplayer online game. Nature. 2010 Aug 5;466(7307):756–60. doi: 10.1038/nature09304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Margolin AA, Bilal E, Huang E, et al. Systematic analysis of challenge-driven improvements in molecular prognostic models for breast cancer. Science Translational Medicine. 2013 Apr 17;5(181):181re181. doi: 10.1126/scitranslmed.3006112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bennet JLS. The Netflix Prize. Proceedings of KDD Cup and Workshop 2007; 8/12/2007; 2007. [Google Scholar]
- 39.Thiele I, Swainston N, Fleming RM, et al. A community-driven global reconstruction of human metabolism Nat Biotechnol March 3, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang Q, Arighi CN, King BL, et al. Community annotation and bioinformatics workforce development in concert—Little Skate Genome Annotation Workshops and Jamborees. Database: The Journal of Biological Databases and Curation. 2012;2012:bar064. doi: 10.1093/database/bar064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang H, Mattes WB, Richter P, Mendrick DL. An omics strategy for discovering pulmonary biomarkers potentially relevant to the evaluation of tobacco products. Biomarkers. 2012;6(6):849–60. doi: 10.2217/bmm.12.78. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1.
| CausalBioNet | KEGG30 (kyoto encyclopedia of genes and genomes) | Reactome | BioCarta | Wikipathways4 | SPIKE31 (signaling pathway integrated knowledge engine) | UCSD Signaling gateway | NCI pathway interaction database* | NetPath32 | |
|---|---|---|---|---|---|---|---|---|---|
| Species human (Hs); Mouse (Mm); Rat (Rn) | Hs, Mm, Rn | >20 species | Hs (curated) +20 species (inferred) |
Hs, Mm | >25 species | Hs | Hs, Mm | Hs | Hs |
| Literature support shown | At edge-level | At pathway-level | At pathway level | At pathway level | At pathway level | At edge level | At molecule/state/transition levels | At edge and pathway level | At edge level |
| Defined biological boundaries | Species Tissue Disease context Biological pathways |
Species Disease context (Hs) Biological pathways |
Biological pathways | Species Biological pathways |
Species Disease context Biological pathways |
Species Biological pathways |
Species Biological pathways |
Species Disease context (cancer) Biological pathways |
|
| Manual curation | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes* | Yes |
| Data-driven enhancement | Yes | No? | No? | No | No | Yes | No | Yes | |
| Crowd curation | Yes | No | No | Partial (comments) | Yes | No | No | No | Through Wikipathways |
| Causal | Yes | No | No | No | No | Yes | No | Yes | Yes |
| Directional edges | Yes | Yes | Partial | Yes | Yes | Yes | Yes | Yes | Yes |
| Multiple types of gene-centric entities | Yes | No | Yes | No | No | No | Yes | Yes | No |
| Interactive visualization | Yes | No | Yes | Yes | Yes | Yes | No | No | No |
| Computable | Yes | Yes/no | Yes (pathway enrichment) | No | No | No | No | No | No but include transcriptionally regulated genes in each pathway |
| Size | >30 networks | >400 signaling modules | 1402 pathways (Hs) | >350 pathways | >430 pathways | 28 curated pathways | ~3500 proteins and their proximal connections | 137 NCI-nature curated pathways (+Reactome + BioCarta) | 32 curated pathways (immune signaling/cancer) |
| Available for download | Yes | Yes | Yes | No | Yes | Yes | No | Yes | Yes |
Notes:
No longer actively curated. The website will be retired in September 2013.