Abstract
Leishmania infantum causes Visceral and cutaneous leishmaniasis in northern Morocco. It predominantly affects children under 5 years with incidence of 150 cases/year. Genetic variability and population structure have been investigated for 33 strains isolated from infected dogs and humans in Morocco. A multilocus microsatellite typing (MLMT) approach was used in which a MLMtype based on size variation in 14 independent microsatellite markers was compiled for each strain. MLMT profiles of 10 Tunisian, 10 Algerian and 21 European strains which belonged to zymodeme MON-1 and non-MON-1 according to multilocus enzyme electrophoresis (MLEE) were included for comparison. A Bayesian model-based approach and phylogenetic analysis inferred two L.infantum sub-populations; Sub-population A consists of 13 Moroccan strains grouped with all European strains of MON-1 type; and sub-population B consists of 15 Moroccan strains grouped with the Tunisian and Algerian MON-1 strains. Theses sub-populations were significantly different from each other and from the Tunisian, Algerian and European non MON-1 strains which constructed one separate population. The presence of these two sub-populations co-existing in Moroccan endemics suggests multiple introduction of L. infantum from/to Morocco; (1) Introduction from/to the neighboring North African countries, (2) Introduction from/to the Europe. These scenarios are supported by the presence of sub-population B and sub-population A respectively. Gene flow was noticed between sub-populations A and B. Five strains showed mixed A/B genotypes indicating possible recombination between the two populations. MLMT has proven to be a powerful tool for eco-epidemiological and population genetic investigations of Leishmania.
Introduction
Leishmaniasis constitutes a group of diseases caused by obligatory, intracellular, protozoan parasites of the genus Leishmania, that cause a spectrum of diseases, ranging from self-limiting, self-curing cutaneous leishmaniasis (CL) to visceral leishmaniasis (VL) with fatal spontaneous evolution [1]. In areas bordering the Mediterranean Sea, Leishmania infantum is the etiologic agent of VL, but can also cause CL. VL is endemic in these countries, affecting the most vulnerable young children and adults, normally associated with HIV/AIDS [2]. The recent global VL and CL incidence were estimated by Alvar et al in 2012. Based on these estimates, approximately 0.2 to 0.4 million VL cases and 0.7 to 1.2 million CL cases occur each year. More than 90% of global VL cases occur in just six countries: India, Bangladesh, Sudan, South Sudan, Brazil and Ethiopia [2].Most strains of L. infantum isolated from Mediterranean foci belong to the predominant zymodeme MON-1 [3], despite their very wide geographical distribution. However, other L. infantum zymodemes have been identified in this region: MON-11, MON-24, MON-27, MON-33, MON-34, MON-37, MON-72, MON-77, MON-80, MON-98, MON-105, MON-108 and MON-199. Epidemiological patterns of leishmaniasis in Mediterranean area are changing dramatically, due to several factors, such as widespread migration from rural to urban and peri-urban areas, socio-political situation [4,5], climatic changes increasing exposure to the sandfly and also, in urban areas, increasing HIV infection.
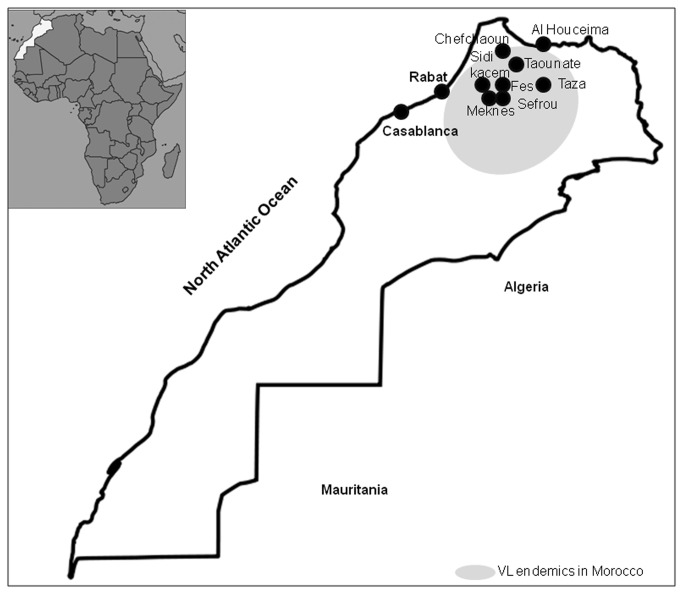
VL is widespread in northern Morocco (Chefchaoun, Taounate, Taza, Fes, My yacoub, Meknes, Sefrou, Al Houceima and sidi kacem) (Figure 1). Few epidemiological data are available concerning its epidemiology and clinical features. Also a new focus of CL due to L. Infantum was recently described [6]. Before 1995, human VL was not an obligatory reportable disease. The incidence of VL was over 150 cases/years in 2006-2008 [2] of which children under 4 years old are most affected. Expansion of arid zone and increase of global temperature increases the incidence of leishmaniasis in this region [7]. Dog is the main reservoir of L.infantum in Morocco as in all the Mediterranean basin [8,9]. The sand fly vectors of L. infantum in this region are: P. perniciosus [10], P.longicuspis [11] and P. ariasi [9]. Zymodeme MON-1 is predominant in Morocco, however, zymodeme MON-24 has been occasionally isolated from a dog [12]. This zymodeme was considered to cause CL sporadically.
Figure 1. Map of Morocco with all areas endemic for VL (shadowed areas).
Different methods have been used for the identification and classification of Leishmania parasites as reviewed [5]. These methods, including multilocus enzyme electrophoresis (MLEE) which is still considered as the gold standard for species and strain typing of Leishmania, are limited in the intrinsic level of polymorphism they can detect and are, only in exceptional cases, able to differentiate strains in the zymodeme MON-1 [5]. Epidemiological studies on VL caused by L. infantum require the use of highly discriminative techniques that are able to differentiate MON-1 strains. Multilocus Microsatellite Typing (MLMT) has proven to be a powerful tool for population genetic and epidemiological studies of Leishmania spp. For review see Schoenian et al.[5]. Ochsenreither et al. [13], Kuhls et al. [14,15] developed panel of 14 microsatellite markers which are able to discriminate L. Infantum strains and have been useful for investigating genetic polymorphism and population structure of L.infantum in various endemic areas [16-18].
In the present study, we used the above mentioned 14 microsatellites markers to investigate the genetic polymorphism and population structure of Moroccan L. infantum strains isolated from human and canine cases (CanL). We have compared the Moroccan MLMT profiles to those of Tunisian, Algerian and European strains belonging to previously characterized MON-1 and non-MON-1 isoenzyme groups. This comparison was done to understand VL transmissions scenarios in North Africa and South Europe.
Material and Methods
1: Source of Leishmania and DNA extraction
Thirty-three strains of Moroccan L. infantum were analysed. Thirty strains were isolated from human VL cases aged between 1-7 years old, and three strains from canine cases. The strains came from all endemic areas in Northern Morocco; 11 from Taounate, eight from Fes, seven from My Yacoub, one from Al Houceima, one from Taza, and for five strains the exact origin was unknown. Table 1 list the strains, their origin and geographical distribution, the clinical form of the disease, and the source of strains used in this study and the reference strains. Isoenzyme profiles were available for only 2 Moroccan strains which belonged to the MON-1 zymodeme.
Table 1. L.infantum strains that have been analysed in this study.
| WHO-Code | Country | Origin | Zymodeme | clinical form | Age | Structure | Source |
|---|---|---|---|---|---|---|---|
| MRO 1 | Morocco | Fes | nd | VL | 2 years | Sub-pop A | This study |
| MRO 7 | Morocco | Fes | nd | VL | 3 years | Sub-pop A | This study |
| MRO 8 | Morocco | Taounate | nd | VL | nd | A/B mix | This study |
| MRO 10 | Morocco | ND | nd | VL | nd | Sub-pop A | This study |
| MRO 13 | Morocco | ND | nd | CanL | nd | Sub-pop A | This study |
| MRO 15 | Morocco | ND | nd | CanL | nd | Sub-pop A | This study |
| MHOM/MA/2004/BOU | Morocco | My Yacoub | nd | VL | nd | Sub-pop B | This study |
| MHOM/MA/2004/CHOUI | Morocco | Taounate | nd | VL | 2 years | Sub-pop A | This study |
| MHOM/MA/2003/NAB | Morocco | Taounate | nd | VL | 1 years | Sub-pop B | This study |
| MHOM/MA/2004/SER | Morocco | My Yacoub | nd | VL | 2 years | A/B mix | This study |
| MHOM/MA/2000/KHAL | Morocco | Fes | MON-1 | VL | nd | Sub-pop B | This study |
| MHOM/MA/2003/LAKH | Morocco | Taounate | nd | VL | 2 years | Sub-pop B | This study |
| MHOM/MA/2003/ZAHI | Morocco | Taounate | nd | VL | 3 years | Sub-pop B | This study |
| MHOM/MA/2003/NAW | Morocco | Taounate | nd | VL | 3 years | Sub-pop B | This study |
| MHOM/MA/2004/NAY | Morocco | My Yacoub | nd | VL | 1 years | Sub-pop A | This study |
| MHOM/MA/2003/CHOU | Morocco | Taounate | nd | VL | 2 years | Sub-pop B | This study |
| MHOM/MA/2003/BER | Morocco | Fes | nd | VL | 2 years | Sub-pop B | This study |
| MHOM/MA/2003/ELO | Morocco | Taounate | nd | VL | 2 years | Sub-pop B | This study |
| MHOM/MA/2003/ANS | Morocco | Fes | nd | VL | 1 years | Sub-pop A | This study |
| MCAN/MA/1996/77 | Morocco | My Yacoub | MON-1 | CanL | 4 years | A/B mix | This study |
| MHOM/MA/2005/HAJ | Morocco | Taounate | nd | VL | 2 years | Sub-pop B | This study |
| MHOM/MA/2005/CHA | Morocco | My Yacoub | nd | VL | 2 years | Sub-pop B | This study |
| MHOM/MA/2005/ZAG | Morocco | My Yacoub | nd | VL | 3 years | Sub-pop A | This study |
| MHOM/MA/2005/ZEM | Morocco | Taounate | nd | VL | 3 years | A/B mix | This study |
| MHOM/MA/2006/RZI | Morocco | Fes | nd | VL | 7 years | Sub-pop B | This study |
| MHOM/MA/2006/NAB | Morocco | Fes | nd | VL | 2 years | Sub-pop B | This study |
| MHOM/MA/2003/BOUZ | Morocco | ND | nd | VL | 4 years | Sub-pop A | This study |
| MHOM/MA/2003/TAB | Morocco | Taza | nd | VL | 3 years | Sub-pop A | This study |
| MHOM/MA/2003/BOUS | Morocco | ND | nd | VL | 2 years | Sub-pop A | This study |
| MHOM/MA/2004/FAT | Morocco | Fes | nd | VL | 3 years | Sub-pop B | This study |
| MHOM/MA/2005/MAR | Morocco | Al Houceima | nd | VL | 1 years | A/B mix | This study |
| MHOM/MA/2003/BEN | Morocco | Taounate | nd | VL | 1 years | Sub-pop B | This study |
| MHOM/MA/2005/HMI | Morocco | My Yacoub | nd | VL | 5 years | Sub-pop A | This study |
| MCAN/FR/1987/RM1 | France | Marseille | 108 | CanL | nd | Sub-pop A | Control Strain |
| MHOM/FR/1978/LEM75 | France | Languedoc | 1 | VL | nd | Sub-pop A | Control Strain |
| MHOM/FR/1995/LPN114 | France | Cote d'Azur | 1 | VL | nd | Sub-pop A | Control Strain |
| MHOM/FR/1997/LSL29 | France | Languedoc | 1 | CL | nd | Sub-pop A | Control Strain |
| MHOM/GR/2001/GH1 | Greece | Athens | 1 | VL | nd | Sub-pop A | Control Strain |
| MHOM/GR/2001/GH2 | Greece | Athens | 1 | VL | nd | Sub-pop A | Control Strain |
| MHOM/GR/2001/GH5 | Greece | Crete | 1 | VL | nd | Sub-pop A | Control Strain |
| MHOM/GR/2001/GH6 | Greece | Athens | 98 | VL | nd | Sub-pop A | Control Strain |
| MHOM/PT/2000/IMT260 | Portugal | Lisbon | 1 | CL | nd | Sub-pop A | Control Strain |
| MCAN/ES/1986/LEM935 | Spain | Poboleda | 77 | CanL | nd | Sub-pop A | Control Strain |
| MHOM/ES/1993/PM1 | Spain | Mallorca | 1 | VL/HIV+ | nd | Sub-pop A | Control Strain |
| MHOM/ES/1986/BCN16 | Spain | Catalonia | 1 | CL | nd | Sub-pop A | Control Strain |
| MHOM/FR/1962/LRC-L47 | France | NA | NA | VL | nd | Population 2 | Control Strain |
| MHOM/FR/1996/LEM3249 | France | Roussillion | 29 | CL | nd | Population 2 | Control Strain |
| MHOM/FR/1980/LEM189 | France | Roussillion | 11 | CL | nd | Population 2 | Control Strain |
| MHOM/IT/1994/ISS1036 | Italy | NA | 228 | VL | nd | Population 2 | Control Strain |
| MHOM/ES/1987/Lombardi | Spain | NA | 24 | CL | nd | Population 2 | Control Strain |
| MHOM/ES/1988/LLM175 | Spain | Madrid | 198 | VL/HIV+ | nd | Population 2 | Control Strain |
| MHOM/ES/1991/LEM2298 | Spain | Valencia | 183 | VL/HIV+ | nd | Population 2 | Control Strain |
| MHOM/ES/1992/LLM373 | Spain | Madrid | 199 | VL/HIV+ | nd | Population 2 | Control Strain |
| MHOM/DZ/1999/LIPA979 | Algeria | Tizi Ouzou | MON-1 | VL | nd | Sub-pop B | Control Strain |
| MHOM/DZ/1999/LIPA1002 | Algeria | Tizi Ouzou | MON-1 | VL | nd | Sub-pop B | Control Strain |
| MCAN /DZ/2000/LIPA1109 | Algeria | Alger | MON-1 | CanL | nd | Sub-pop B | Control Strain |
| MCAN /DZ/2000/LIPA1113 | Algeria | Alger | MON-1 | CanL | nd | Sub-pop B | Control Strain |
| MCAN /DZ/2000/LIPA1117 | Algeria | Alger | MON-1 | CanL | nd | Sub-pop B | Control Strain |
| MHOM/DZ/1996/LIPA477 | Algeria | Ain Defla | MON-24 | CL | nd | Population 2 | Control Strain |
| MHOM/DZ/1999/LIPA1058 | Algeria | Lakhdaria | MON-80 | CL | nd | Population 2 | Control Strain |
| MHOM/DZ/2001/LIPA1140 | Algeria | Boumerdes | MON-24 | CL | nd | Population 2 | Control Strain |
| MHOM/DZ/2001/LIPA1226 | Algeria | Alger | MON-24 | CL | nd | Population 2 | Control Strain |
| MHOM/DZ/1995/LIPA459 | Algeria | Lakhdaria | MON-24 | CL | nd | Population 2 | Control Strain |
| MHOM/TN/2001/Tus167 | Tunisia | Monastir | 1 | VL | 4 years | Sub-pop B | Control Strain |
| MHOM/TN/2002/20S | Tunisia | Beja | 1 | VL | 9 months | Sub-pop B | Control Strain |
| MHOM/TN/2002/Tus221 | Tunisia | Monastir | 1 | VL | 3 years | Sub-pop B | Control Strain |
| MHOM/TN/2002/Tum222 | Tunisia | Monastir | 1 | VL | 3 years | Sub-pop B | Control Strain |
| MCAN/TN/2002/LCnJ20S | Tunisia | Tunis | 1 | CanL | 2 years | Sub-pop B | Control Strain |
| MHOM/TN/2004/LC64 | Tunisia | Tunis | 24 | CL | 6 years | Population 2 | Control Strain |
| MHOM/TN/2002/LC95 | Tunisia | Béja | 24 | CL | 15 years | Population 2 | Control Strain |
| MHOM/TN/2002/SFC89 | Tunisia | Sfax | 24 | CL | 54 years | Population 2 | Control Strain |
| MHOM/TN/2005/SFC51 | Tunisia | Sfax | 24 | CL | 50 years | Population 2 | Control Strain |
| MHOM/TN/2004/TLC3 | Tunisia | Siliana | 24 | CL | 10 years | Population 2 | Control Strain |
VL, visceral leishmaniasis; CL, cutaneous leishmaniasis; CanL, canine leishmaniasis. Sub-pop A, Sub-pop B, mixed A/B and Population 1 represent populations according to STRUCTURE. WHO, World Health Organization
For 27 strains, DNA was extracted from promastigotes grown in Novye MacNealeNicolle biphasic culture medium (NNN) [19]. Furthermore, DNA was isolated from amastigotes in bone marrow aspirates spotted on six glass slides. DNA extraction from cultures was done by using phenol—chloroform extraction method and from slides, with some modifications, as described previously [20,21]. Figure 1 shows geographical distribution of the studies strains and VL endemic areas in Morocco.
The microsatellite profiles obtained previously for ten strains from Tunisia, ten from Algeria and 20 from different European countries were used as controls for comparison [15-17] and to investigate gene flow amongst North African and South European strains. These strains represent different clinical forms (CL and VL) and belonged to zymodeme MON-1 and different non- MON-1 zymodemes, see Table 1.
2: L. infantum identification and microsatellite genotyping
The internal transcribed spacer 1 (ITS-1) was amplified for all samples. The PCR product was digested with the restriction endonuclease HaeIII as described previously [22]. The resulting RFLP profile was identical to those of the WHO reference strain of L. infantum MHOM/TN/1980/IPT1 (data not shown).
Microsatellite genotyping was done using the dinucleotide microsatellite markers: Lm2TG, TubCA, Lm4TA, Li41-56, Li46-67, Li22-35, Li23-41, Li45-24, Li71-33, Li71-5/2, Li71-7, LIST7031, LIST7039 and CS20. PCR conditions were described previously [13,14]. To expose microsatellite profiles, the microsatellite-containing fragments were analyzed by capillary electrophoresis (SMB Services in Molecular Biology Berlin) with an automated ABI PRISM GeneMapper sequencer (Applied Biosystems).
3: Microsatellite data analysis
Two models were used to analyse the microsatellite data: (1) Population structure analysis was investigated using the program STRUCTURE 2.2, which implements a Bayesian model-based clustering method, using genotype data consisting of unlinked markers [23]. The admixture model was used. Markov chain Monte Carlo (MCMC) searches consisted of a burn-in length of 10,000 iterations followed by a run of 100,000 replications for each setting of K (the number of populations) from 1 to 10 with 10 replicate runs of each. The most appropriate number of populations was determined based upon ad hoc statistic ⎢K that evaluates the second order rate of change of the likelihood function with respect to the number of populations (K) [24]. (2) Microsatellite-based genetic distances analysis was based on the proportion of shared alleles distances. Phylogenetic trees were constructed using Neighbour-joining (NJ) method by the help of the softwares MSA 3.0, POPULATIONS 1.2.28 and MEGA version 3.1, as described previously [16].
Descriptive statistics for the observed genetic populations were calculated with the help of GDA software [24]. This includes allelic diversity (number of allelic variants per marker and mean number of alleles (MNA) per population), proportion of polymorphic loci (P), expected (He) and observed (Ho) heterozygosity and inbreeding coefficient (F IS). The degree of genetic differentiation and gene flow among populations were assessed by calculating F ST values with corresponding p-values, FST values higher than 0.25 indicate strong genetic differentiation [25].
4: Ethics statement
According to ethical approval of this study, all samples were anonymized. Written informed consent was obtained from each study participant or the parents/guardians on behalf of the children under 15 years old. Study design and procedures were revised and approved by the Institutional Review Board (IRB), Institut Pasteur du Maroc, Casablanca, Morocco, and the Ethical Committee of Charitè University Medicine, Berlin, Germany.
Results
Every analysed Moroccan strain had an individual microsatellite profile (33 genotypes). Thirteen markers were polymorphic, only marker Li 71-33 was monomorphic. Marker Lm4TA was the most polymorphic one presenting seven alleles, whereas markers Li41-56, Li46-67, Li 71-5/2 and LIST7039 were least polymorphic presenting only two alleles for each. For the 33 Moroccan strains, the mean number of allelic variants per locus (A) was 3.21. The observed heterozygosity (Ho) was between 0 and 0.15 and the expected heterozygosity (He), representing the probability that an individual will be heterozygous over the loci tested, ranged from 0 to 0.81 and was in most cases much higher than Ho. Inbreeding coefficient per locus (F IS) was positive for 11 markers and ranges from 0.93 to .037, for the other three markers (F IS) values was zero. This indicates a large number of homozygotes in the investigated strains (data not shown).
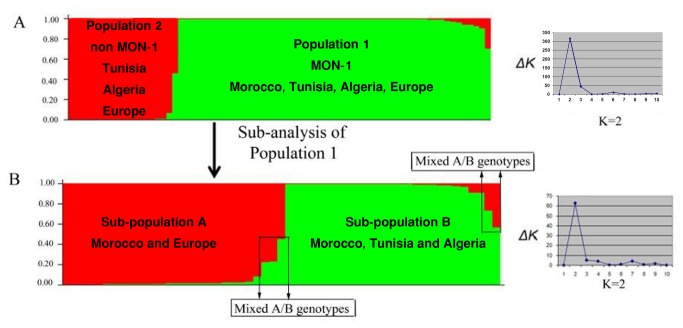
Two main populations were detected by Structure for the data set analysed; Population 1 consisted of all strains from Morocco and the Tunisian, Algerian and European strains of MON-1 type and those of MON-77, 89 and 108, closely related to MON-1 as described previously [15,18]. Population 2 consisted of all non-MON-1 strains from Tunisia, Algeria and Europe which were added as controls; see Figure 2A and Table 1. These two populations were genetically different as shown by their F ST value (0.35) and P-value 0.0001.
Figure 2. Estimated population structure for 33 Moroccan L. infantum strains as inferred by STRUCTURE software on the basis of data for 14 microsatellite markers.
Each of the strains is represented by a single vertical line divided into K colors, where K is the number of populations assumed. Each color represents one population, and the length of the colors segment shows the strain’s estimated proportion of membership in that population.(A) the two main populations derived from the whole dataset which divided strains into MON-1 and non-MON-1 populations. (B) Sub-population analysis of Population 1 (MON-1 group) shows two sub-populations. K represents the true number of populations and sub-populations.
When re-analysed separately by Structure, Population 1 was subdivided into two sub-populations (Figure 2B). Sub-population 1A consisted of 13 Moroccan strains; three from Fes, three from My Yacoub, one from Taounate, one from Taza, five of unknown origin in the endemic area and all European strains of MON-1 type that have been added for comparison. Sub-population 1B comprised 15 Moroccan strains; eight from Taounate, five from Fes, two from My Yacoub, and all Tunisian and Algerian MON-1 control strains (Figure 2B and Table 1). These two sub-populations were as well significantly different as shown by their F ST value (0.34) and P-value 0.0001. Five Moroccan strains have shown mixed genotypes between sub-populations 1A and 1B; two strains from My Yacoub, two from Taounate and one from Al Houceima (see Table 1, Figure 2 and Figure 3). Theses strains shared allele’s characteristic for each sub-population and showed significant shared membership in both sub-populations (Table 2).
Table 2. Membership coefficients of mixed genotypes between Sub-population 1A and 1B as calculated by the STRUCTURE program.
| WHO code | Sub-population 1A | Sub-population 1B |
|---|---|---|
| MHOM/MAR/2004/SER | 0.431 | 0.569 |
| MCAN/MAR/1996/77 | 0.267 | 0.733 |
| MHOM/MAR/2005/ZEM | 0.738 | 0.262 |
| MHOM/MAR/2005/MAR | 0.759 | 0.241 |
| MRO 8 | 0.532 | 0.468 |
Measures for genetic diversity have been calculated for the two Populations 1 and 2 and for Sub-populations 1A and 1B, the five mixed genotypes were excluded from this analysis (Table 3). Population 2 (non-MON-1) was found to be more diverse than Population 1, while Sub-population 1A was more diverse than Sub- population 1B (Table 3). Inbreeding coefficient (F IS) was highest for Population 1 (MON-1 strains), and equally for both sub-populations of this population (Table 3).
Table 3. Genetic diversity and characterization of the L. infantum populations and sub-populations found by STRUCTURE in this study.
| Populations | n | p | MNA | He | Ho | F IS |
|---|---|---|---|---|---|---|
| Population 1 (MON-1) | 55 | 0.85 | 4.64 | 0.39 | 0.06 | 0.84 |
| Population 2 (non MON-1) | 18 | 1 | 7.28 | 0.79 | 0.35 | 0.55 |
| Sub-pop alnalysis with mixed strains being excluded | ||||||
| Sub-pop A | 25 | 0.92 | 3.85 | 0.414 | 0.07 | 0.82 |
| Sub-pop B | 25 | 0.5 | 2.28 | 0.2 | 0.03 | 0.83 |
n - number of strains; P - proportion of polymorphic loci; MNA - mean number of alleles; He - expected heterozygosity; Ho - observed heterozygosity; F IS - inbreeding coefficient.
Three main clusters were detected in the NJ distance tree based on the proportion of shared alleles (D AS) measure (Figure 3). The first cluster comprised all non-MON-1 strains from Tunisia, Algeria and Europe and was identical with Population 1. The second cluster consisted of 13 Moroccan and all European MON-1 strains and was congruent with Sub-population 1A. Finally, the third cluster included the same 15 Moroccan strains and Tunisian and Algerian MON-1 strains as Sub-population 1B did. Interestingly, the mixed genotypes (five Moroccan strains) formed a separate cluster which took an intermediate position between and within the two sub-populations A and B.
Figure 3. Unrooted neighbor-joining tree inferred from genetic distances derived from the proportions of alleles shared among 55 Moroccan L. infantum strains based on 14 microsatellites markers.
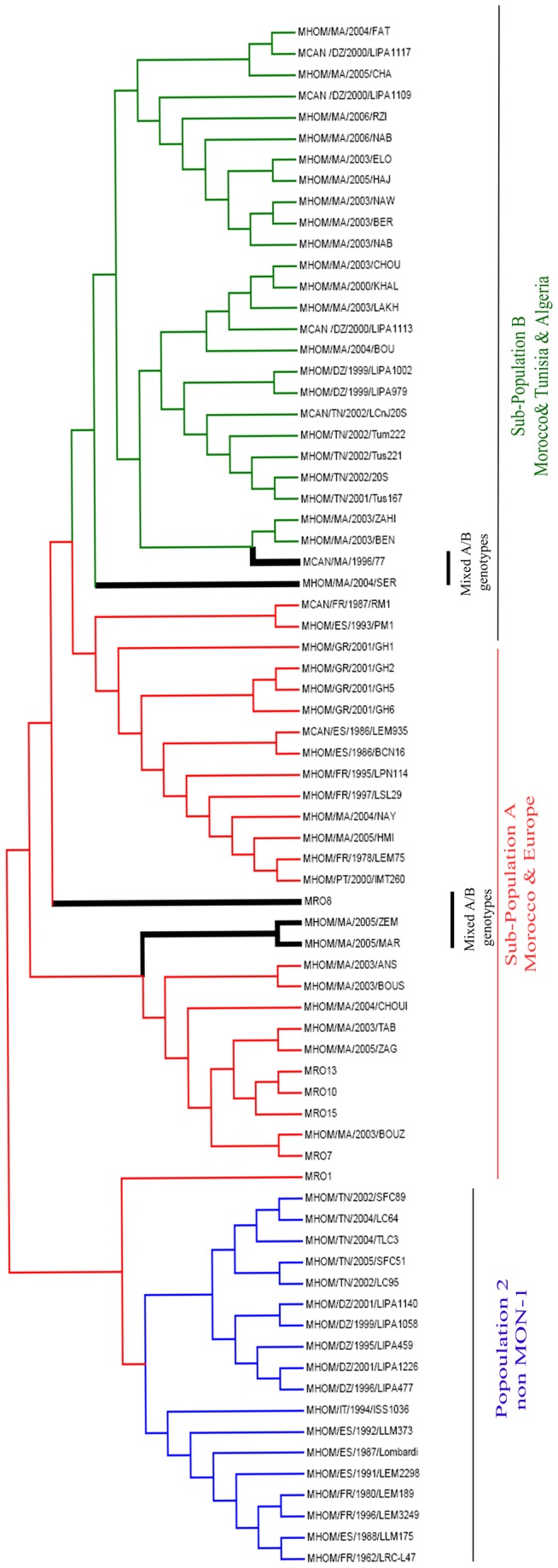
Two main clusters were detected and the “MON-1” cluster was further sub-divided into two sub-clusters. This was in full agreement with the STRUCTURE results (Figure 2). Mixed 1A/1B genotypes were shown in intermediate positions in the tree.
Strains isolated from canine cases grouped together with strains from humans in Sub-population 1A. No correlation was found between a particular MLMT profile and host background.
Discussion
In this study, a panel of 14 microsatellite markers was applied to investigate the genetic polymorphism and population structure of Moroccan L. infantum strains and to compare them with strains from North African and European countries.
Both phylogenetic analysis based on genetic distances as well as Structure analysis using a Bayesian clustering approach grouped the Moroccan strains together with MON-1 strains or strains closely related to MON-1 from Tunisia, Algeria and Europe in Population 1. None of the Moroccan strains were assigned to the non MON-1 population (Population 2). Isoenzyme analysis had been performed for only two of the Moroccan strains studied which were typed as MON-1. MLMT analysis suggests that the remaining Moroccan strains analysed here also represent MON-1 and/or MON-1 closely related zymodyme types.
Surprisingly, two sub-populations were detected in Morocco. Sub-population 1A consisted of 13 Moroccan strains and that were related to the European MON-1 strains added for comparison. On the other hand, Sub-population 1B consisted of 15 Moroccan strains that were more similar to the Tunisian and Algerian MON-1 strains included in this study. The two sub-populations were genetically different from each other as confirmed by their F ST value and the corresponding P-value.
Phylogenetic analysis based on proportion of shared alleles was compatible with structure analysis based on Bayesian clustering model. The detected clusters were identical with identified populations and sub-populations hence confirming the presence of two sub-populations of L. infantum in Morocco and the presence of gene flow and mixed genotypes between them.
The presence of two L. infantum sub-populations co-existing in Moroccan endemic foci suggests multiple introduction of L. infantum from/to Morocco. Two scenarios are possible: (1) introduction from/to the neighboring North African countries, this is supported by the presence of Sub-population 1B (2), introduction from/to the European Mediterranean countries, this is supported by the presence of Sub-population 1A. Since Morocco is the African country closest to the European continent, through the Strait of Gibraltar, and due to the frequent migration of humans and their animals from and to Europe, back and forth introduction of the disease cannot be excluded. Due to their high variability and extensive size homoplasy, microsatellite markers are not suitable for estimating evolutionary history and are not able to confirm the origin and direction of introduction of Moroccan L. infantum from/to Europe. However, genomic sequencing data from North African and European strains is needed to verify these scenarios.
The presence of mixed genotypes between the Moroccan sub-populations indicates the presence of gene flow between them. These strains might represent either mixed infections or 1A/1B hybrid strains. Since all mixed Moroccan genotypes were isolated from amastigotes in bone marrow aspirate spotted on slides, cloning of these strains to test whether they are true hybrids was not possible. Hybrids seem to occur quite frequently. Previous studies in Algeria [16] and Tunisia [17] revealed hybrid genotypes for 12 of the 82 Algerian and Tunisian strains (14.6% ) with one Tunisian strain being cloned and proven as real hybrid. Since multiple heterozygous loci were identified among hybrids and gene flow was detected between different populations, recombination between strains with different alleles seems to be the most parsimonious explanation [26]. The occurrence of gene flow and genetic recombination is increasingly being suggested for different Leishmania species [27] although the mechanism of genetic exchange remains to be established. As L. infantum infects humans and dogs and continues its life cycle in sandfly midgut, it is vital to investigate the compartments where genetic recombination of Leishmania could take place. It was recently reported that L. major is capable of having asexual cycle consistent with a meiotic process in sandfly vector [28].
Three sandfly species transmit L. infantum in Morocco; P. perniciosus [10], P.longicuspis [11] and P. ariasi [9]. One can speculate whether the sub-populations of Moroccan L. infantum might be related to the existence of different sand fly vector species involved in transmitting the respective parasites, as previously described for L .tropica, in North Israel [29]. To prove this hypothesis, it is needed to isolate strains of L. infantum from the respective sandfly species and investigate them with MLMT. Moreover, a new focus of cutaneous leishmaniasis due to L. Infantum was recently described in Morocco. A clear relationship between the clinical presentation (VL vs CL) of leishmaniasis and parasite genotype could be demonstrated in Algeria [16] and Tunisia [17]. All Moroccan VL and CanL cases analyzed in this study have shown typical symptoms of visceral and canine leishmaniasis without significant differences in disease outcome between cases belonging to the two sub-populations, however, L. infantum strains that have caused CL in Morocco have not yet been analyzed by MLMT . Human and canine strains presented identical MLMT profiles. This highlights the key role of dogs as reservoirs of L.infantum in Morocco.
To our knowledge, this is the first study that investigates the population structure and genetic diversity of L. infantum in Morocco by applying MLMT. We were able to detect two different sub-populations of L. infantum in Morocco and correlated them with strains isolated from North African countries and Europe. Putative hybrid strains indicated the presence of gene flow among the two sub-populations. However, it is needed to investigate more strains representing different hosts, clinical forms and zymodyme types to better understand the overall population structure and molecular epidemiology of L. infantum in Morocco and the other North African countries.
Acknowledgments
This study was in partial fulfillment of the requirements of Institut Pasteur Du Maroc toward the PhD degree of Salsabil Hamdi. We are thankful to the Moroccan National Program of Leishmaniasis Control, National Institute of Health for providing samples from different places in Morocco.
Funding Statement
The fieldwork and data collection was granted by Pastur Institute in Morocco for Dr. Meryem Lemrani. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Ashford RW (2000) The leishmaniases as emerging and reemerging zoonoses. Int J Parasitol 30: 1269-1281. doi: 10.1016/S0020-7519(00)00136-3. PubMed: 11113254. [DOI] [PubMed] [Google Scholar]
- 2. Alvar J, Vélez ID, Bern C, Herrero M, Desjeux P et al. (2012) Leishmaniasis worldwide and global estimates of its incidence. PLOS ONE 7: e35671. doi: 10.1371/journal.pone.0035671. PubMed: 22693548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Pratlong F, Rioux JA, Marty P, Faraut-Gambarelli F, Dereure J et al. (2004) Isoenzymatic analysis of 712 strains of Leishmania infantum in the south of France and relationship of enzymatic polymorphism to clinical and epidemiological features. J Clin Microbiol 42: 4077-4082. doi: 10.1128/JCM.42.9.4077-4082.2004. PubMed: 15364993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Azmi K, Schönian G, Nasereddin A, Schnur LF, Sawalha S et al. (2012) Epidemiological and clinical features of cutaneous leishmaniases in Jenin District, Palestine, including characterisation of the causative agents in clinical samples. Trans R Soc Trop Med Hyg 106: 554-562. doi: 10.1016/j.trstmh.2012.06.005. PubMed: 22832019. [DOI] [PubMed] [Google Scholar]
- 5. Schönian G, Mauricio I, Gramiccia M, Cañavate C, Boelaert M et al. (2008) Leishmaniases in the Mediterranean in the era of molecular epidemiology. Trends Parasitol 24: 135-142. doi: 10.1016/j.pt.2007.12.006. PubMed: 18262469. [DOI] [PubMed] [Google Scholar]
- 6. Rhajaoui M, Nasereddin A, Fellah H, Azmi K, Amarir F et al. (2007) New clinico-epidemiologic profile of cutaneous leishmaniasis, Morocco. Emerg Infect Dis 13: 1358-1360. doi: 10.3201/eid1309.060946. PubMed: 18252108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Health Mo (2006) Direction de l'Epidémiologie et de la Lutte contre les maladies. DMT. Service des Maladies Parasitaires. Rapport annuel d'activité. Maroc: Ministère de la santé. [Google Scholar]
- 8. Rami M, Atarhouch T, Sabri M, Cadi Soussi M, Benazzou T et al. (2003) [Canine leishmaniasis in the Rif mountains (Moroccan Mediterranean coast): a seroepidemiological survey]. Parasite 10: 79-85. PubMed: 12669354. [DOI] [PubMed] [Google Scholar]
- 9. Guessous-Idrissi N, Hamdani A, Rhalem A, Riyad M, Sahibi H et al. (1997) Epidemiology of human visceral leishmaniasis in Taounate, a northern province of Morocco. Parasite 4: 181-185. PubMed: 9296060. [DOI] [PubMed] [Google Scholar]
- 10. Benabdennbi I, Pesson B, Cadi-Soussi M, Marquez Morillas F (1999) Morphological and isoenzymatic differentiation of sympatric populations of Phlebotomus perniciosus and Phlebotomus longicuspis (Diptera: Psychodidae) in northern Morocco. J Med Entomol 36: 116-120. PubMed: 10071503. [DOI] [PubMed] [Google Scholar]
- 11. Pesson B, Ready JS, Benabdennbi I, Martín-Sánchez J, Esseghir S et al. (2004) Sandflies of the Phlebotomus perniciosus complex: mitochondrial introgression and a new sibling species of P. longicuspis in the Moroccan Rif. Med Vet Entomol 18: 25-37. doi: 10.1111/j.0269-283x.2004.0471.x. PubMed: 15009443. [DOI] [PubMed] [Google Scholar]
- 12. Haralambous C, Dakkak A, Pratlong F, Dedet JP, Soteriadou K (2007) First detection and genetic typing of Leishmania infantum MON-24 in a dog from the Moroccan Mediterranean coast: genetic diversity of MON-24. Acta Trop 103: 69-79. doi: 10.1016/j.actatropica.2007.05.008. PubMed: 17603990. [DOI] [PubMed] [Google Scholar]
- 13. Ochsenreither S, Kuhls K, Schaar M, Presber W, Schönian G (2006) Multilocus microsatellite typing as a new tool for discrimination of Leishmania infantum MON-1 strains. J Clin Microbiol 44: 495-503. doi: 10.1128/JCM.44.2.495-503.2006. PubMed: 16455904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kuhls K, Keilonat L, Ochsenreither S, Schaar M, Schweynoch C et al. (2007) Multilocus microsatellite typing (MLMT) reveals genetically isolated populations between and within the main endemic regions of visceral leishmaniasis. Microbes Infect 9: 334-343. doi: 10.1016/j.micinf.2006.12.009. PubMed: 17307010. [DOI] [PubMed] [Google Scholar]
- 15. Kuhls K, Chicharro C, Canavate C, Cortes S, Campino L et al. (2008) Differentiation and Gene Flow among European Populations of Leishmania infantum MON-1. PLoS Negl Trop. Drosophila Inf Serv 2: e261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Seridi N, Amro A, Kuhls K, Belkaid M, Zidane C et al. (2008) Genetic polymorphism of Algerian Leishmania infantum strains revealed by multilocus microsatellite analysis. Microbes Infect 10: 1309-1315. doi: 10.1016/j.micinf.2008.07.031. PubMed: 18755285. [DOI] [PubMed] [Google Scholar]
- 17. Chargui N, Amro A, Haouas N, Schönian G, Babba H et al. (2009) Population structure of Tunisian Leishmania infantum and evidence for the existence of hybrids and gene flow between genetically different populations. Int J Parasitol 39: 801-811. doi: 10.1016/j.ijpara.2008.11.016. PubMed: 19211023. [DOI] [PubMed] [Google Scholar]
- 18. Amro A, Schönian G, Al-Sharabati MB, Azmi K, Nasereddin A et al. (2009) Population genetics of Leishmania infantum in Israel and the Palestinian Authority through microsatellite analysis. Microbes Infect 11: 484-492. doi: 10.1016/j.micinf.2009.02.001. PubMed: 19399967. [DOI] [PubMed] [Google Scholar]
- 19. Belkaid PM, Harrat Z, Hamrioui B, Thellier M, Datry A et al. (1996) [A simple media for isolation and culture of Leishmania]. Bull Soc Pathol Exot 89: 276-277. PubMed: 9053049. [PubMed] [Google Scholar]
- 20. Schönian G, Schweynoch C, Zlateva K, Oskam L, Kroon N et al. (1996) Identification and determination of the relationships of species and strains within the genus Leishmania using single primers in the polymerase chain reaction. Mol Biochem Parasitol 77: 19-29. doi: 10.1016/0166-6851(96)02572-8. PubMed: 8784768. [DOI] [PubMed] [Google Scholar]
- 21. Amro A, Azmi K, Schonian G, Nasereddin A, Alsharabati MB et al. (2009) Epidemiology of paediatric visceral leishmaniasis in Hebron district, Palestine. Trans R Soc Trop Med Hyg 103: 731-736 [DOI] [PubMed]
- 22. Schönian G, Nasereddin A, Dinse N, Schweynoch C, Schallig HD et al. (2003) PCR diagnosis and characterization of Leishmania in local and imported clinical samples. Diagn Microbiol Infect Dis 47: 349-358. doi: 10.1016/S0732-8893(03)00093-2. PubMed: 12967749. [DOI] [PubMed] [Google Scholar]
- 23. Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155: 945-959. PubMed: 10835412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14: 2611-2620. doi: 10.1111/j.1365-294X.2005.02553.x. PubMed: 15969739. [DOI] [PubMed] [Google Scholar]
- 25. Wright S (1978) Evolution and the genetics of populations. Variability within and among Natural Populations. Chicago: The University of Chicago Press; p. 4. [Google Scholar]
- 26. Mauricio IL, Yeo M, Baghaei M, Doto D, Pratlong F et al. (2006) Towards multilocus sequence typing of the Leishmania donovani complex: resolving genotypes and haplotypes for five polymorphic metabolic enzymes (ASAT, GPI, NH1, NH2, PGD). Int J Parasitol 36: 757-769. doi: 10.1016/j.ijpara.2006.03.006. PubMed: 16725143. [DOI] [PubMed] [Google Scholar]
- 27. Schönian G, Mauricio I, Cupolillo E (2010) Is it time to revise the nomenclature of Leishmania? Trends Parasitol 26: 466-469. doi: 10.1016/j.pt.2010.06.013. PubMed: 20609626. [DOI] [PubMed] [Google Scholar]
- 28. Akopyants NS, Kimblin N, Secundino N, Patrick R, Peters N et al. (2009) Demonstration of genetic exchange during cyclical development of Leishmania in the sand fly vector. Science 324: 265-268. doi: 10.1126/science.1169464. PubMed: 19359589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Jaffe CL, Baneth G, Abdeen ZA, Schlein Y, Warburg A (2004) Leishmaniasis in Israel and the Palestinian Authority. Trends Parasitol 20: 328-332. doi: 10.1016/j.pt.2004.05.001. PubMed: 15193564. [DOI] [PubMed] [Google Scholar]