Background: Previous studies show that autophagy deficiency leads to inflammasome activation.
Results: Excessive autophagy activation induces a proinflammatory response.
Conclusion: Our findings provide new insights into the release mechanisms of proinflammatory cytokines regulated by autophagy.
Significance: That autophagy may play a deleterious role in pathogenesis in T2D is a novel finding.
Keywords: Autophagy, Cytokine, Glucose Metabolism, Inflammation, Insulin Resistance, Pancreatic Islets
Abstract
Impairment of glucose-stimulated insulin secretion caused by the lipotoxicity of palmitate was found in β-cells. Recent studies have indicated that defects in autophagy contribute to pathogenesis in type 2 diabetes. Here, we report that autophagy-related 7 (Atg7) induced excessive autophagic activation in INS-1(823/13) cells exposed to saturated fatty acids. Atg7-induced cathepsin B (CTSB) overexpression resulted in an unexpected significant increase in proinflammatory chemokine and cytokine production levels of IL-1β, monocyte chemotactic protein-1, IL-6, and TNF-α. Inhibition of receptor-interacting protein did not affect the inflammatory response, ruling out involvement of necrosis. CTSB siRNA suppressed the inflammatory response but did not affect apoptosis significantly, suggesting that CTSB was a molecular linker between autophagy and the proinflammatory response. Blocking caspase-3 suppressed apoptosis but did not affect the inflammatory response, suggesting that CTSB induced inflammatory effects independently of apoptosis. Silencing of Nod-like receptor 3 (NLRP3) completely abolished both IL-1β secretion and the down-regulation effects of Atg7-induced CTSB overexpression on glucose-stimulated insulin secretion impairment, thus identifying the NLRP3 inflammasome as an autophagy-responsive element in the pancreatic INS-1(823/13) cell line. Combined together, our results indicate that CTSB contributed to the Atg7-induced NLRP3-dependent proinflammatory response, resulting in aggravation of lipotoxicity, independently of apoptosis in the pancreatic INS-1(823/13) cell line.
Introduction
The prevalence of type 2 diabetes (T2D)3 is growing rapidly worldwide, and insulin resistance has been implicated as a major risk factor for the disease. Impaired glucose-stimulated insulin secretion (GSIS) is an essential feature of β-cell dysfunction, which is widely believed to be secondary to prolonged exposure to high glucose and lipid levels termed “glucolipotoxicity.” Palmitate, because of its lipotoxicity, was found to impair the GSIS in β-cells (1). Several lines of evidence appear to prove that diabetes is a low grade inflammatory disease with insulin resistance. The low grade inflammatory state of obesity is characterized by increased infiltration of immune cells and an increase in proinflammatory M1 state macrophages in the metabolic tissue, which in turn has been shown to negatively affect insulin sensitivity (2). It has been shown that elevated levels of IL-1β, IL-6, MCP-1, and C-reactive protein are predictive of T2D (3). The association of cytokines with T2D is also demonstrated by an increased risk of developing the disease for individuals with elevated blood levels of IL-1β, IL-6, and MCP-1 (4). In addition, the IL-1 receptor antagonist effectively counters the effects of IL-1 in diet-induced obesity (5), and T2D patient therapy with IL-1 receptor antagonist significantly improves the proinsulin/insulin ratio and markers of systemic inflammation (6). Thus, understanding the mechanisms involved in the regulation of diabetic inflammation may help to identify novel therapeutic targets with important clinical applications.
Autophagy is an intracellular, lysosome-mediated catabolic mechanism that is responsible for the bulk degradation and recycling of damaged or dysfunctional cytoplasmic components and intracellular organelles (7). Cathepsin B (CTSB) is a lysosomal cysteine protease primarily involved in the degradation or processing of lysosomal proteins. In certain conditions, CTSB has been shown to be involved in additional cellular processes including cell invasion, vesicle trafficking, inflammasome formation, autophagy-based IL-1β secretion (8), and cell death. Induction of autophagy in diabetic and in C57BL/6 mice fed high fat diets revealed active formation of autophagosomes and a higher level of conversion of LC3I to LC3II with low grade systemic inflammation (9). INS-1 cells treated with very high concentrations of palmitic acid (PA) demonstrate increased transcription of CTSB and progressive β-cell apoptosis characterized by cellular hypertrophy followed by depletion of insulin immunoreactivity (10, 11). Recently, studies have clearly indicated that defects in autophagy contribute to the inflammatory response in T2D. For instance, Atg16L1 is an essential component of the autophagic machinery responsible for regulation of endotoxin-induced IL-1β production (12). Defective hypothalamic autophagy directs the central pathogenesis of obesity by induction inflammation and subsequently leads to obesity and insulin resistance in mice fed a high fat diet (13). Depletion of the autophagic proteins LC3B and beclin 1 enhances the activation of caspase-1 and secretion of IL-1β and IL-18 (14). However, several recent studies suggest that autophagy also has a deleterious role at the cellular level. For instance, it has been shown that RNAi knockdown of essential autophagy genes inhibits type II programmed cell death in a variety of cell types under different conditions (15). Excessive levels of autophagy could contribute to cardiomyocyte damage and subsequent death (16). At the organismal level, excessive autophagy activation was found to have deleterious effects in runx1 mutant mice, which exhibited severe wasting of enervated myofibers (17). Although these studies have shown that excessive autophagy activation under different pathological conditions plays different negative roles (18), there is currently no information concerning possible involvement of excessive autophagy activation in the pathogenesis of T2D. In addition, there is increasing evidence that autophagy is activated in failing pancreatic cells, a condition associated with insulin resistance, but the mechanism of the dual role of autophagy in T2D has not been explored. In this study, we show that Atg7 induces excessive autophagy activation in the INS-1(823/13) cell line exposed to PA and that Atg7-induced CTSB overexpression contributes to an NLRP3-dependent proinflammatory response and subsequently impairs GSIS independently of apoptosis. Understanding how excessive autophagy activation increases the risk for diabetic inflammation may provide insight into the development of T2D and have important clinical applications.
EXPERIMENTAL PROCEDURES
Palmitic Acid Solutions
Palmitic (C16:0) acid was purchased from Sigma. Palmitic acid solutions were prepared according to Martino et al. (19). Briefly, PA was dissolved at 70 °C in 0.1 m NaOH to obtain a 100 mm stock solution. A 5% (w/v) solution of PA-free bovine serum albumin (BSA) was prepared in serum-free RPMI 1640 medium. Then a 5 mm PA, BSA mixture was prepared by suitable combination of the two above mentioned solutions. Finally, the PA stock solutions were diluted in RPMI 1640 medium supplemented with 1% fetal bovine serum (FBS) to obtain 0, 0.25, 0.5, and 1 mm final concentrations at a fixed concentration of 0.5% BSA. INS-1(823/13) cells were transfected with construct using Lipofectamine Plus (Invitrogen).
Cell Culture
Rat insulinoma INS-1(823/13) cells were grown in RPMI 1640 medium buffered with 10 mm HEPES containing 10% FBS, 2 mm l-glutamine, 1 mm sodium pyruvate, 50 μm β-mercaptoethanol, and 100 units/ml penicillin/streptomycin. This cell line is capable of insulin release in response to glucose stimulation (20). For the different experiments, cells were cultured in 6-well plates until reaching 80% confluence.
Cell Transfection
INS-1(823/13) cells were transfected with plasmid Atg7-inserted pCMV6-AC-GFP or Jab-1-inserted pCMV6-AC-GFP or with siCTSB or siNLRP3 as appropriate using Lipofectamine 2000 reagent (Invitrogen) according to the manufacturer's suggested protocol.
Glucose-stimulated Insulin Secretion
Expanded INS-1(823/13) cells were preincubated in oxygenated Krebs-Ringer bicarbonate buffer (137 mm NaCl, 4.7 mm KCl, 1.2 mm KH2PO4, 1.2 mm MgSO4-7H2O, 2.5 mm CaCl2-2H2O, 25 mm NaHCO3, 0.25% BSA) containing 3.3 mm glucose at 37 °C for 1 h. Afterward, buffer was replaced with fresh oxygenated Krebs-Ringer bicarbonate buffer containing either 3.3 mm glucose or 16.7 mm glucose, and cells were incubated for 1 h at 37 °C. Supernatant was collected, and cells were lysed by 0.15% HCl. Secreted insulin and cellular insulin content were measured by RIA using a rat insulin RIA kit (Millipore). The insulin secretion index (16.87 mm GSIS over 3 mm GSIS) was calculated. The total intracellular insulin content was extracted by the acid/ethanol method. Briefly, cells were incubated in 1% hydrochloric acid alcohol (ethanol/H2O/HCl, 14:57:3) overnight at 4 °C. The insulin in the supernatant was detected by RIA (Linco Research, St. Charles, MO) and normalized to total protein content.
Quantitative Real Time PCR
Total RNA was extracted using TRIzol reagent (Invitrogen), and cDNA was synthesized using the iScriptTM cDNA synthesis kit (Bio-Rad) according to the manufacturer's instruction. The sequences of probes and primers are summarized in Table 1. TaqMan PCR was performed using the ABI Prism 7700 Sequence Detection System as instructed by the manufacturer (Applied Biosystems). The level of mRNA was normalized to that of 18 S mRNA.
TABLE 1.
Sequences of probes and primers
f, forward; r, reverse.
| Gene | NCBI Reference Sequence | Probe | Primers |
|---|---|---|---|
| IL-1β | NM_012589.2 | CATGGCACATTCTGTTCAAAGAGAGCCTG | f, TCGCTCAGGGTCACAAAGAAA |
| r, CCATCAGAGGCAAGGAGGAA | |||
| TNF-α | NM_012675.3 | CCCGACTACGTGCTCCTCACCCA | f, TCTCTTCAAGGGACAAGGCTG |
| r, ATAGCAAATCGGCTGACGGT | |||
| MCP-1 | NM_031530.1 | CCCCACTCACCTGCTGCTACTCATTCA | f, TTGGCTCAGCCAGATGCA |
| r, CCAGCCTACTCATTGGGATCA | |||
| IL-6 | NM_012589.2 | TGTTACTCTTGTTACATGTCTCCTTTCTCAGGGCT | f, GGTACATCCTCGACGGCATCT |
| r, GTGCCTCTTTGCTGCTTTCAC |
Confocal Microscopy
INS-1(823/13) cells grown in a chamber slide were fixed with paraformaldehyde and permeabilized with 0.5% Triton X-100 for 4 min at room temperature. After washing out primary antibodies, the plates were washed in PBS and incubated with anti-LC3 antibodies diluted in PBA (PBS + 1% (w/v) BSA). Plates were then further washed in PBS and incubated with anti-rabbit Alexa Fluor 633 (Invitrogen). The samples were then further washed in PBS and mounted using Vectashield mounting medium (Abacus ALS, Australia). Images were captured using a Zeiss Meta-510 LSM microscope (excitation, 633 nm; emission, 647 nm). LC3-labeled puncta were counted in individual cells (four to five per field, 20 fields per experiment, three experiments total), and values were averaged. Each value represents a total of 200 cells counted. For the Annexin V/propidium iodide (PI) double staining assay, INS-1(823/13) cells (4 × 105) were cultured in a 6-well plate. After cells were treated with test agents and harvested, the cells were dyed using a FITC-Annexin V/PI kit. The stained cells were centrifuged at 1,000 rpm for 5 min, and the cell pellets were dropped on sterile coverslips and then examined by fluorescence microscopy.
Flow Cytometric Assay of Apoptosis
5 × 105 INS-1(823/13) cells were cultured with or without PA and later incubated for a minimum of 5 h in culture medium at 37 °C with or without additional drug or plasmid treatment. Cells were then harvested and fixed with cold 70% ethanol, stored overnight at −20 °C, and stained with PI staining buffer (1 mg/ml RNase A, 0.1% Triton X-100, 50 g/ml PI in PBS) to determine the apoptotic/necrotic cell population (the sub-G1/G0 cell fraction). To evaluate the early stages of apoptosis, treated cells were stained with Annexin V-FITC (Caltag) in Annexin V staining buffer for 15 min at room temperature and counterstained with 50 g/ml PI in Hanks' balanced salt solution (Stem Cell Technologies) with no phenol red and analyzed with a FACSCalibur flow cytometer (BD Biosciences).
Western Blot Analysis
Cell lysates were separated by electrophoresis prior to transfer to PVDF membranes. The membranes were then probed with the antibody of interest, and immunoreactive bands were detected by chemiluminescence. Images were captured, and data were analyzed using the Bio-Rad ChemiDocTM XRS+ imaging system. Data were normalized relative to actin. The following primary antibodies were used for Western blots and immunostaining: rabbit anti-LC3 (Sigma, L8918), rat monoclonal to lysosome-associated membrane protein 2 (Abcam, ab13524), anti-β-actin antibody (Abcam, ab3280), rabbit polyclonal anti-Atg7 (Cell Signaling Technology, 2631), mouse monoclonal to cathepsin B (Abcam, ab58802), anti-IL-1β antibody (Abcam, ab2105), anti-IL-6 antibody (Abcam, ab6672), anti-MCP-1 antibody (Abcam, ab9669), anti-TNF-α antibody (Abcam, ab9635), and guinea pig polyclonal anti-p62 (ProGen, GP62-N). Alexa Fluor-conjugated antibodies (Molecular Probes, A21057, A21076, and A21096) were used as secondary antibodies. Data for LC3 Western blot analysis were obtained in the presence of the autophagic flux inhibitor bafilomycin (10 nm) to exclude the possibility that defective autophagy/inhibited flux increased LC3II.
Statistics
All values are expressed as mean ± S.D. Differences between means were analyzed using either one-way or two-way analysis of variance followed by Newman-Keuls post hoc testing for pairwise comparison using SPSS. The null hypothesis was rejected when the p value was <0.05.
RESULTS
PA Induced Autophagy Activation in INS-1(823/13) Cells
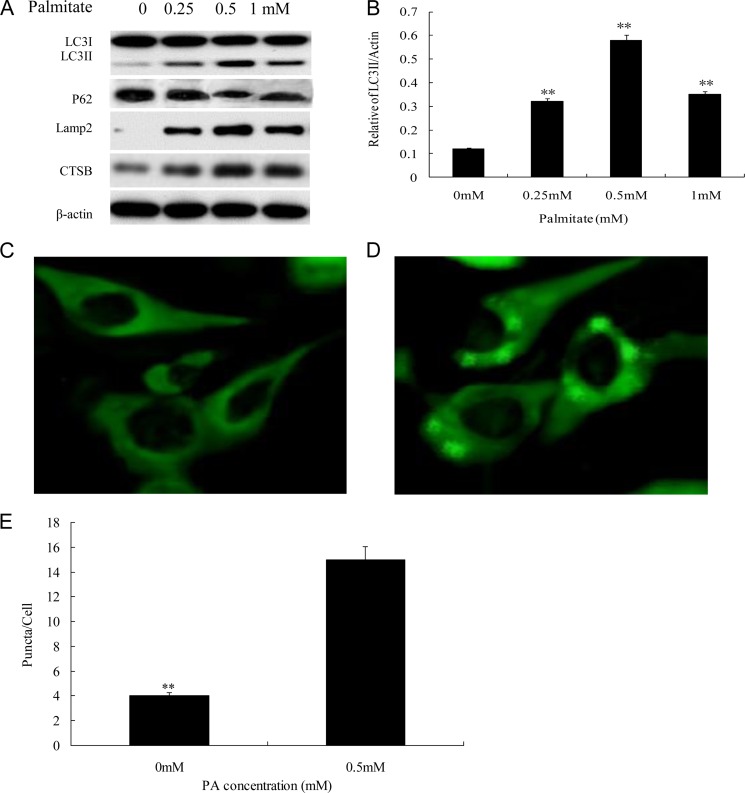
The saturated fatty acid PA was reported to be autophagic to β-cells (19). We first sought to determine which PA concentration caused proper levels of both autophagy (Fig. 1) and apoptosis (Fig. 2) in INS-1(823/13) cells because this was very important for the subsequent experiments. For this purpose, we first investigated the effects of four PA concentrations (0, 0.25, 0.5, and 1.0 mm) on INS-1(823/13) cells at an incubation time of 12 h after which autophagy activation was evaluated. The activation of autophagy by 0.25, 0.5, and 1.0 mm PA in INS-1(823/13) was demonstrated by elevation of LC3II/LC3I, CTSB, and lysosome-associated membrane protein 2 and reduction of p62 after 12 h of incubation in contrast to the control (0 mm PA) (Fig. 1A). We found the maximum effect with 0.5 mm PA, which significantly enhanced INS-1(823/13) cell autophagy activity after 12-h incubation (p < 0.01; Fig. 1, A and B). These results were further confirmed by quantification of LC3II/actin in the 0.5 mm PA group compared with the 0 mm PA group (p < 0.01; Fig. 1B). Notably, the level of LC3II/actin was decreased in the 1 mm PA group in contrast to the 0.5 mm PA group in agreement with previous reports (Fig. 1, A and B) (21, 22). Thus, we chose 0.5 mm PA as the proper PA concentration for autophagic activation. We observed numerous and large puncta in the cytoplasm of INS-1(823/13) cells by LC3II staining in the 0.5 mm PA group (Fig. 1D) compared with the control group, which had no detectable dots (Fig. 1C), and the proportion of INS-1(823/13) cells containing LC3II-stained dots/cell was counted. The number of puncta/cell significantly increased at 12 h after 0.5 mm PA treatment as compared with cells that were not exposed to PA (p < 0.01; Fig. 1E).
FIGURE 1.
0.5 mm PA stimulated proper autophagy activation in INS-1(823/13) cells. A, PA stimulated expression of autophagy activation-associated proteins LC3II/LC3I, p62, lysosome-associated membrane protein (Lamp2), and CTSB. INS-1(823/13) cells treated with 0, 0.25, 0.5, and 1 mm PA were analyzed by immunoblotting. β-Actin was used as a control. B, ratio of LC3II/actin was analyzed in INS-1(823/13) cells treated with 0, 0.25, 0.5, and 1 mm PA for 12 h (**, p < 0.01). C and D, cells stained with LC3 antibody followed by secondary antibody (green) were visualized by confocal microscopy. INS-1(823/13) cells treated with 0.5 mm PA (D) were compared with control (0 mm PA) (C). E, the number of cytoplasmic puncta/cell is shown. Data are expressed as mean ± S.D. (error bars) from three independent experiments (analyzed by t test; **, p < 0.01 compared with control group).
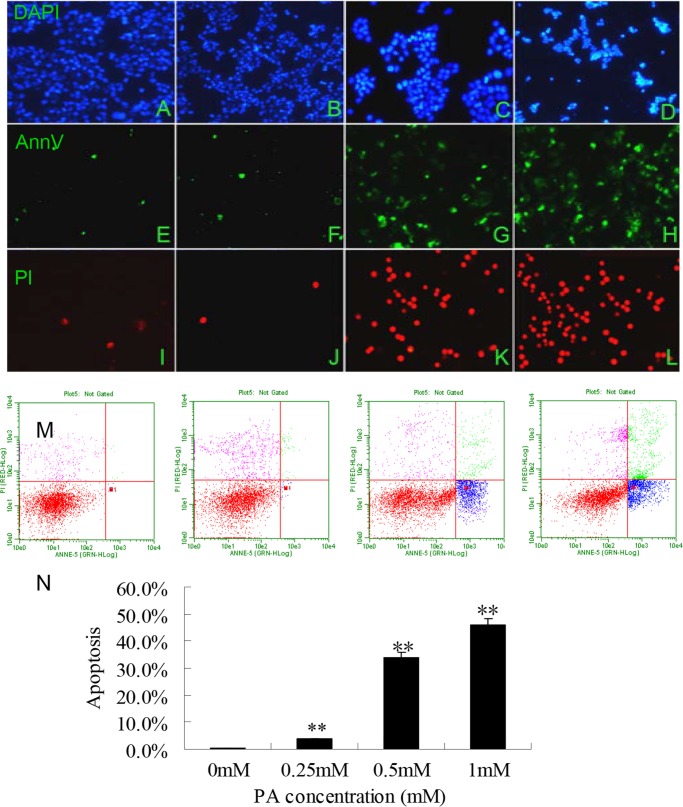
FIGURE 2.
0.5 mm PA induced appropriate INS cell apoptosis as assessed by morphological and FACS analyses. A–L, images of representative fields were captured at a magnification of 40× under fluorescence microscope (Nikon microscope, Japan). INS-1(823/13) cells treated with 0, 0.25, 0.5, and 1 mm PA (from left to right) and stained with DAPI (A–D), Annexin V (AnnV; E–H), and PI (I–L) were observed under a fluorescence microscope. M, the induction of apoptosis was determined by flow cytometric analysis of Annexin V-FITC and PI staining after treatment with 0, 0.25, 0.5, and 1 mm PA (starting from left to right). Cells in the lower right quadrant are Annexin-positive, early apoptotic cells. The cells in the upper right quadrant are Annexin-positive/PI-positive, late apoptotic cells. N, the percentages of INS-1(823/13) cell apoptosis showing a greater but appropriate degree of apoptosis in the 0.5 mm PA group compared with the control and the group treated with 1 mm PA. The data are expressed as the mean ± S.D. (error bars) of three to five independent experiments with each condition performed in duplicate (**, p < 0.01).
Exposure of INS-1(823/13) Pancreatic Cells to 0.5 mm PA Induced Proper Cell Apoptosis
PA has been shown to induce apoptosis in pancreatic β-cell lines as well as in primary cultured β-cells (23). To investigate which of 0, 0.25, 0.5, and 1.0 mm PA induced appropriate apoptosis in INS-1(823/13) cells, we examined the effect of PA treatment on apoptotic cell death in INS-1(823/13) cells by morphological and FACS analyses (Fig. 2). Annexin V/PI staining indicated that 0.5 mm PA could induce proper apoptosis consistent with a previous report (Fig. 2, C, G, and K) (24). Little if any apoptosis was observed in control cells grown in the presence of a concentration lower than 0.5 mm PA (Fig. 2, A, E, and I, 0 mm PA; Fig. 2, B, F, and J, 0.25 mm PA). Excessive loss of cells through apoptosis was induced by 1 mm PA concentration (Fig. 2, D, H, and I). Treatment of the cells with 0.5 mm PA resulted in a great increase in the amount of early apoptotic cells as analyzed by FACS (Fig. 2M) (0, 0.25, 0.5, and 1 mm PA from left to right, respectively). Although early apoptotic cells showed no differences between 0.5 mm PA and 1 mm PA concentrations, the proportion of late apoptotic cells increased significantly with the latter concentration of PA (p < 0.01; Fig. 2N). Thus, we chose 0.5 mm PA for the subsequent experiments.
Atg7-induced Excessive Autophagy Activation Resulted in Enhanced Proinflammatory Cytokine Response
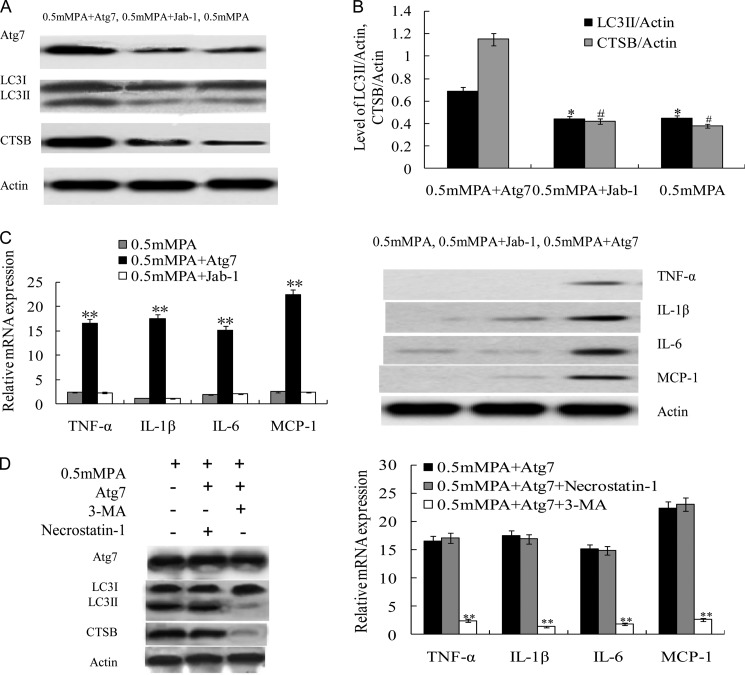
To study the effect of Atg7-induced autophagy activation in INS-1(823/13) exposed to 0.5 mm PA, we transfected cells with Atg7-inserted pCMV6-AC-GFP plasmid and a Jab-1-inserted pCMV6-AC-GFP unrelated plasmid as a control. Cells were incubated for 5 h before experiments. The ability of 0.5 mm PA to induce autophagy activation in INS-1(823/13) cells was revealed by increased expression of Atg7 and CTSB and proteolytic conversion of LC3I into the LC3II isoform (Fig. 3A), consistent with Fig. 1A. Significantly higher Atg7-induced CTSB expression and ratio of conversion of LC3I to LC3II were observed in the 0.5 mm PA + Atg7 group compared with the 0.5 mm PA group, and the relative quantification of LC3II/actin and CTSB/actin is shown in Fig. 3B (p < 0.05) (25). To determine whether the Atg7-induced excessive autophagy activation was an important contributor to the increased expression of proinflammatory genes and that this process was involved in necrosis, we detected the mRNA and protein expression levels of these genes in INS-1(823/13) cells. Surprisingly, as shown in Fig. 3C, Atg7-induced autophagy activation significantly increased mRNA expression of TNF-α, IL-1β, IL-6, and MCP-1 in the 0.5 mm PA + Atg7 group in contrast to the 0.5 mm PA group or the 0.5 mm MPA + Jab-1 group, respectively (Fig. 3C) (p < 0.01). The protein level was higher in the 0.5 mm PA + Atg7 group with respect to the other two groups (Fig. 3C). PA-induced autophagy results in necroptosis in endothelial cells, and it may initiate inflammation (26). To test whether necrosis was involved in Atg7 overexpression in INS-1(823/13) cells, leading to the initiation of inflammation, cells were pretreated with 10 mm 3-methyladenine, a class III phosphatidylinositol 3-kinase autophagy inhibitor, or 30 μm necrostatin-1, a receptor-interacting protein inhibitor of necroptosis. Atg7-induced autophagy overexpression was characterized by increased expression of Atg7 and CTSB and an increased conversion ratio of LC3II/LC3I. 3-Methyladenine reduced the LC3II/LC3I ratio and CTSB expression and blocked the mRNA expression of TNF-α, IL-1β, IL-6, and MCP-1, whereas necrostatin-1 failed to exhibit any effect on Atg7 overexpression-induced autophagy activity or inflammatory response (Fig. 3D), thus further confirming that the mode of Atg7-induced inflammatory response did not involve necrosis.
FIGURE 3.
Atg7-induced excessive autophagy activation potentiated inflammatory cytokine production in INS-1(823/13) cells exposed to PA. A, Atg7 transfection induced excessive autophagy activation. The Atg7-transfected 0.5 mm PA-treated (Atg7 + 0.5 mm) group showed increased Atg7, LC3II, and CTSB expression compared with control treated with 0.5 mm PA or mock control (Jab-1 + 0.5 mm PA). B, relative levels of LC3II/actin and CTSB/actin were significantly higher in the 0.5 mm PA + Atg7 group compared with both the 0.5 mm PA group and the 0.5 mm PA + Jab-1 group (* and #, p < 0.05) (n = 6). C, relative mRNA expression levels of the proinflammatory cytokines TNF-α, IL-1β, IL-6, and MCP-1 were determined by real time PCR. Data are expressed as the mean ± S.D. (error bars) (**, p < 0.01) (n = 6). The protein levels of proinflammatory cytokines were examined by Western blot of the INS-1(823/13) cells in the 0.5 mm PA group, the 0.5 mm PA + Atg7 group, and the 0.5 mm PA + Jab-1 group. D, representative Western blot showing protein expression levels of Atg7 and LC3II/LC3I of cells that were incubated for 8 h with 0.5 mm PA + Atg7 in the presence of 10 mm 3-methyladenine (3-MA) or 30 μm necrostatin-1 (left panel). Relative mRNA expression levels of the proinflammatory cytokines TNF-α, IL-1β, IL-6, and MCP-1 were determined by real time PCR (right panel). Data are expressed as the mean ± S.D. (error bars) (**, p < 0.01) (n = 6).
Blocking CTSB Expression Suppressed Inflammatory Response but Did Not Affect Apoptosis Markedly
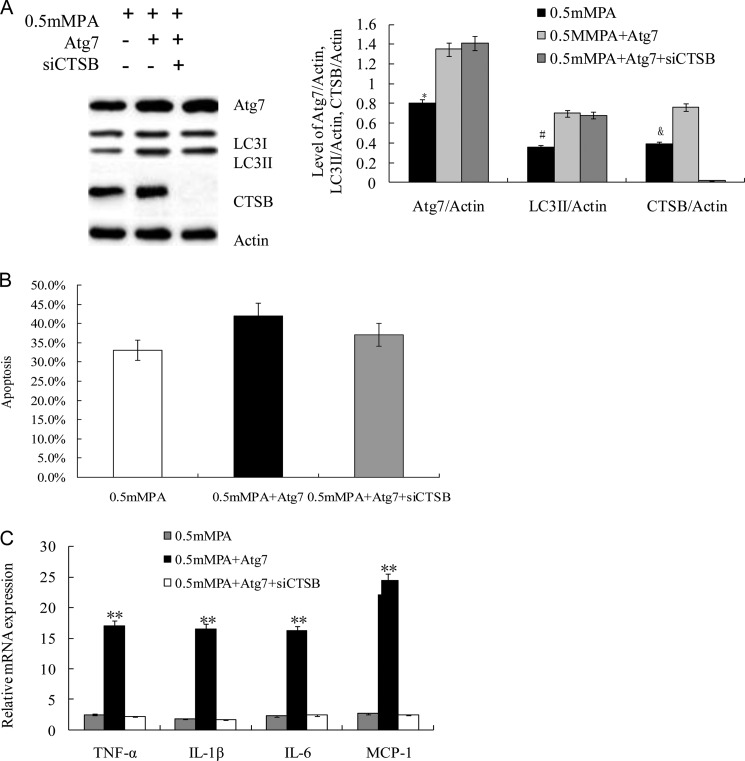
To study whether the induction of proinflammatory cytokine expression in INS-1(823/13) cells was caused by CTSB overexpression, cells were transfected with 1 μm siRNA against CTSB for 5 h prior to additional treatment. As expected, we found significantly higher expression of CTSB and a higher conversion ratio of LC3II/LC3I in Atg7-inserted pCMV6-AC-GFP-transfected INS-1(823/13) cells in the 0.5 mm PA + Atg7 group and the 0.5 mm PA + Atg7 + siCTSB group compared with control (0.5 mm PA) (p < 0.05; Fig. 4A). siCTSB completely inhibited CTSB protein expression in the 0.5 mm PA + Atg7 + siCTSB group (Fig. 4A) (27). These results were further confirmed by quantification of Atg7/actin, LC3II/actin, and CTSB/actin (p < 0.05; Fig. 4A, right panel). To validate whether the role of CTSB in proinflammatory induction is related to cell apoptosis, we analyzed the above four groups of cells by FACS. Fig. 4B demonstrates that CTSB was not involved in PA-induced apoptosis as determined by FACS analysis (p = 0.31; >0.05). However, inhibition of CTSB in the 0.5 mm PA + Atg7 + siCTSB group significantly reduced mRNA expression of TNF-α, IL-1β, IL-6, and MCP-1 compared with the 0.5 mm PA + Atg7 group as determined by real time PCR (p < 0.01; Fig. 4C), suggesting that expression of these genes strongly depends on CTSB protein synthesis in INS-1(823/13) cells. These findings confirmed the pivotal role of Atg7-induced overexpression of CTSB, which contributed to the proinflammatory response in INS-1(823/13) cells.
FIGURE 4.
Blockage of CTSB suppressed Atg7-induced inflammatory response. A, CTSB expression was completely inhibited by siRNA. The protein expression levels of Atg7 and CTSB and the ratio of LC3II/LC3I were assayed by Western blot analysis using cell-free lysates prepared from the culture. Actin was used as a loading control. Levels of Atg7/actin, LC3II/actin, and CTSB/actin were analyzed in INS-1(823/13) cells treated with 0.5 mm PA, Atg7 + 0.5 mm PA, and (Atg7 + 0.5 mm PA + siCTSB, respectively (*, #, and &, p < 0.05) (n = 6). B, cell apoptosis was determined by FACS in the 0.5 mm PA group, Atg7 + 0.5 mm PA group, and Atg7 + 0.5 mm PA + siCTSB group. There were no significant differences among them, although siRNA caused a lower trend of apoptosis. C, relative levels of mRNA of the proinflammatory cytokines were determined by real time PCR. Data are expressed as the mean ± S.D. (error bars) (**, p < 0.01) (n = 6).
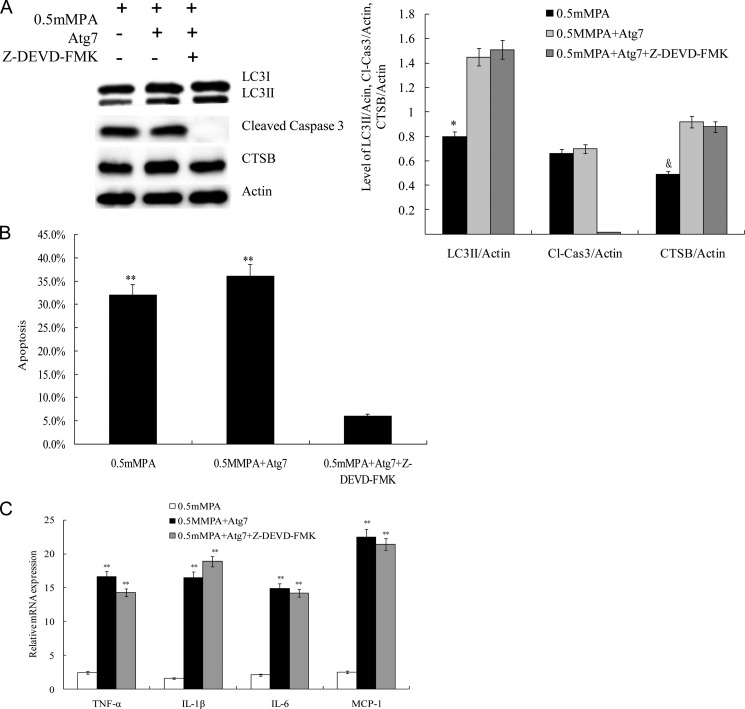
Blocking Caspase-3 Suppressed Apoptosis but Did Not Affect Inflammatory Response
To examine whether the effect of Atg7-induced CTSB overexpression on the proinflammatory cytokine response required caspase-3, INS-1(823/13) cells cultured with 0.5 mm PA were pretreated with a 50 μm concentration of the caspase-3 inhibitor Z-DEVD-fmk (Santa Cruz Biotechnology) before additional treatments. As expected, Z-DEVD-fmk completely inhibited caspase-3 expression in INS-1(823/13) cells, and the protein expression level of CTSB and ratio of LC3II/LC3I was markedly higher in the Atg7 + 0.5 mm PA group and the Atg7 + 0.5 mm PA + Z-DEVD-fmk group compared with the 0.5 mm PA group (Fig. 5A). No differences in the CTSB expression and ratio of LC3II/LC3I were observed between the Atg7 + 0.5 mm PA group and the Atg7 + 0.5 mm PA + Z-DEVD-fmk group (Fig. 5A). These results were further confirmed by quantification of LC3II/actin, cleaved caspase-3/actin, and CTSB/actin (p < 0.05; Fig. 5A, right panel). Apoptosis was markedly reduced in cells treated with Z-DEVD-fmk (p < 0.01; Fig. 5B). To determine whether the induction of the inflammatory response was dependent upon caspase-3 activation, we detected mRNA expression of TNF-α, IL-1β, IL-6, and MCP-1 in INS-1(823/13) cells exposed to PA after pretreatment with Z-DEVD-fmk. The mRNA expression of TNF-α, IL-1β, IL-6, and MCP-1 was not affected in the group treated with Z-DEVD-fmk compared with the control (Fig. 5C) (28). Taken together, these results indicate that blocking caspase-3 suppressed apoptosis but did not affect the proinflammatory response.
FIGURE 5.
The caspase-3 inhibitor Z-DEVD-fmk suppressed apoptosis but did not affect inflammatory response. A, caspase-3 was completely inhibited by Z-DEVD-fmk (50 μm). The protein expression levels of caspase-3 and CTSB and the ratio of LC3II/LC3I were assayed by Western blot analysis using cell-free lysates prepared from the culture. Actin was used as a loading control. Levels of LC3II/actin, cleaved caspase-3 (Cl-Cas3)/actin, and CTSB/actin were analyzed in INS-1(823/13) cells treated with 0.5 mm PA, Atg7 + 0.5 mm PA, and Atg7 + 0.5 mm PA + Z-DEVD-fmk, respectively (* and &, p < 0.05) (n = 6). B, INS-1(823/13) cell apoptosis under three different treatments was determined by FACS (**, p < 0.01) (n = 6). C, relative levels of mRNA of the proinflammatory cytokines of the three groups were determined by real time PCR. Data are expressed as the mean ± S.D. (error bars) (**, p < 0.01) (n = 6).
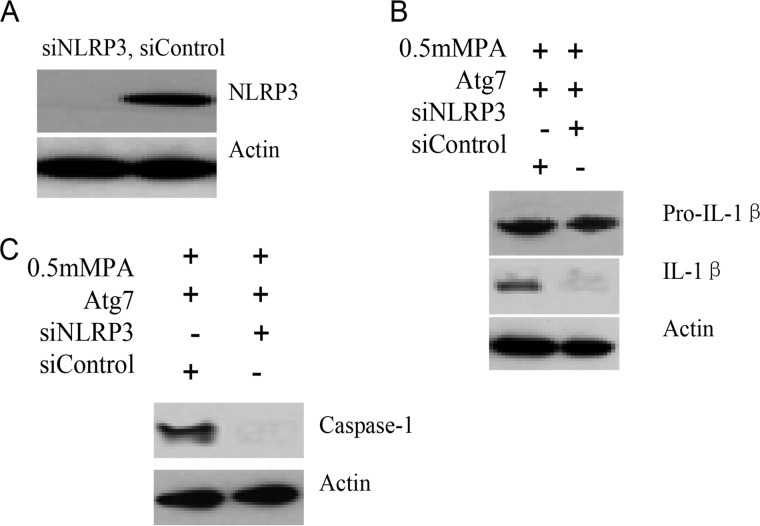
NLRP3 Knockdown Blocked Atg7-induced Inflammatory Response
Caspase-1 itself is synthesized as a zymogen, procaspase-1, that undergoes autocatalytic processing, resulting in two subunits that form the active caspase-1. Activation of caspase-1 occurs within the NLRP3 inflammasome following its assembly, which is essential for the cleavage of pro-IL-1β into its mature, biologically active form. To define the role of NLRP3 inflammasome components in the activation of caspase-1 by excessive autophagic activation, NLRP3 in INS-1(823/13) cells was depleted by siRNA. Compared with non-target siRNA control, protein levels of NLRP3 knockdown cells were totally inhibited (Fig. 6A). NLRP3 and non-target control knockdown INS-1(823/13) cells were then challenged with additional treatments, and caspase-1 activation was revealed by the appearance of caspase-1 and IL-1β in Western blots. When compared with non-target control cells of the 0.5 mm PA + Atg7 + siControl group, depletion of NLRP3 caused a distinct reduction of IL-1β in the 0.5 mm PA + Atg7 + siNLRP3 group (Fig. 6B). Concomitantly, a distinct reduction of caspase-1 levels was also observed (Fig. 6C). Taken together, the results show that NLRP3 was required for Atg7-induced caspase-1 activation and IL-1β processing in INS-1(823/13) cells.
FIGURE 6.
NLRP3 knockdown blocked Atg7-induced proinflammatory response. A, INS-1(823/13) cells were stably transfected with siRNA that targets NLRP3, and protein levels of NLRP3 knockdown cells were analyzed by Western blot analysis. NLRP3 or non-target control (siControl) knockdown cells were treated with 0.5 mm PA and transfected with Atg7. Secreted IL-1β (B) and activated caspase-1 (C) were examined by Western blot analysis. Taken together, these data reveal that excessive autophagy activity induced caspase-1 activation and cytokine secretion through an NLRP3-dependent pathway.
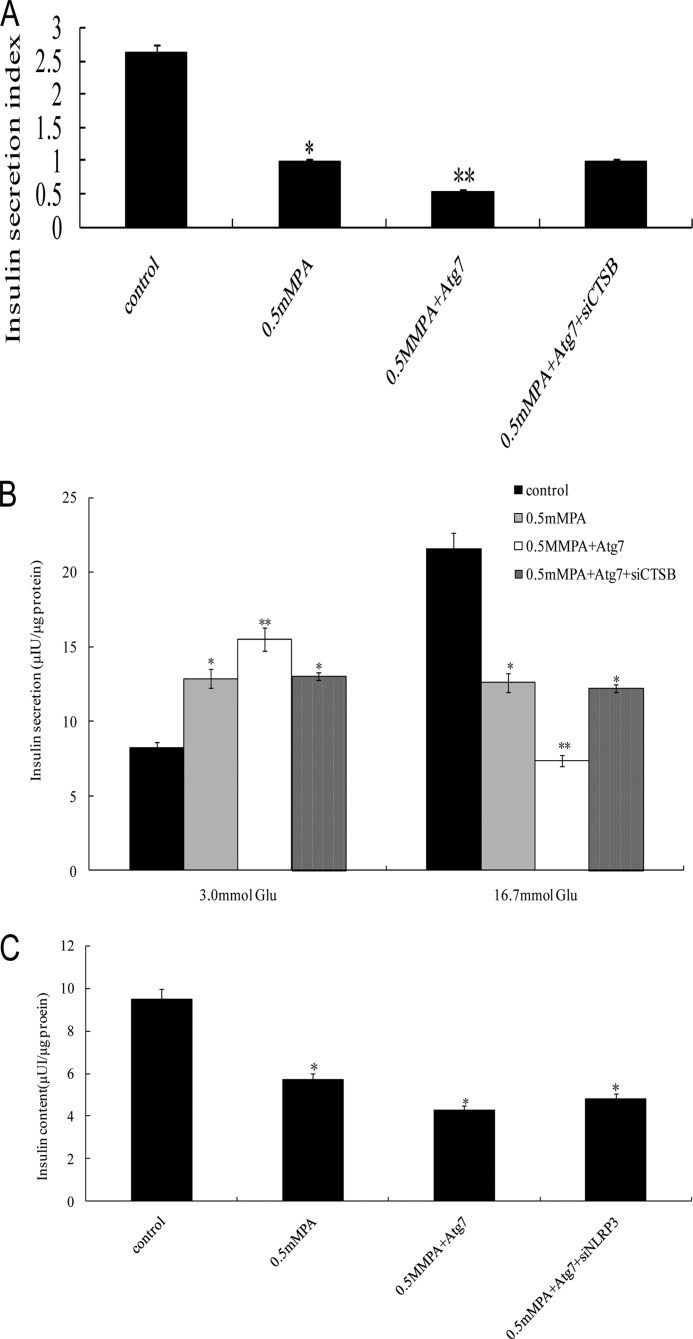
Atg7-induced Excessive Autophagy Activation Induced Severe Impairment of Glucose-stimulated Insulin Secretion and Reduced Insulin Content in INS-1(823/13) Cells
To determine the effect of Atg7-induced excessive autophagy activation on the insulin secretion function in INS-1(823/13) cells, the insulin secretion of cells under different treatments was measured by an RIA kit, and then insulin secretion indexes were calculated. In the presence of 16.7 mm glucose, the insulin release in INS-1(823/13) cells exposed to Atg7 + 0.5 mm PA was markedly reduced compared with the control group (0 mm PA) (p < 0.01; Fig. 7A and B). There was similar insulin release between the 0.5 mm PA group and the 0.5 mm PA + Atg7 + siNLRP3 group (Fig. 7, A and B), but these two groups showed a significantly lower insulin release compared with the control group. 0.5 mm PA elevated insulin secretion at low glucose (basal insulin release; 3 mm glucose) (p < 0.05; Fig. 7B), whereas GSIS at high glucose (16.7 mm glucose) was markedly decreased (p < 0.05; Fig. 7B) in both the 0.5 mm PA group and the 0.5 mm PA + Atg7 + siNLRP3 group. In the Atg7- and 0.5 mm PA-treated group (0.5 mm PA + Atg7), the GSIS in INS-1(823/13) cells was severely decreased compared with the control (p < 0.01; Fig. 7B). These results suggest that Atg7-induced excessive autophagy activity was deleterious, impairing GSIS in INS-1(823/13) β-cells exposed to PA, and blockade of the proinflammatory response attenuated this effect. To further determine the toxic effects of Atg7-induced excessive autophagy activation, we next investigated the total insulin content in INS-1(823/13) cells (Fig. 7C). INS-1(823/13) cells were treated for 24 h with 0.5 mm PA, 0.5 mm PA + Atg7, and 0.5 mm PA + Atg7 + siNLRP3, which reduced the total insulin content by 38.3, 34.5, and 36.2%, respectively, compared with the control group (0 mm PA) (p < 0.05), suggesting that INS-1(823/13) cells may be damaged by the toxic effects of Atg7-induced excessive autophagy activation.
FIGURE 7.
Atg7-induced excessive autophagy activation induced severe impairment of GSIS function. After equilibration at 3.3 mm glucose, INS-1(823/13) cells were stimulated with high glucose (16.7 mm) for 1 h. A, the glucose-stimulated insulin secretion in INS-1(823/13) cells was measured by RIA, and then the insulin secretion index was calculated (insulin release at high glucose/insulin release at basal glucose) (*, p < 0.05; **, p < 0.01) (n = 6). B, the insulin secretion in INS-1(823/13) cells in response to 3.0 and 16.7 mm glucose stimulation was examined (*, p < 0.05; **, p < 0.01) (n = 6). C, total insulin content in INS-1(823/13) cells was extracted by the acid/ethanol method and detected using an RIA kit. Data are expressed as the mean ± S.D. (error bars) (*, p < 0.05) (n = 6).
DISCUSSION
The present study was designed to examine the pathophysiological role of Atg7-induced excessive autophagy activation in INS-1(823/13) cells. Several observations emerged. 1) Treatment of INS-1(823/13) cells with 0.5 mm PA induced autophagy activation (29). 2) Atg7-induced CTSB overexpression induced MCP-1, TNF-α, IL-1β, and IL-6 expression in INS-1(823/13) cells exposed to PA. 3) CTSB-induced MCP-1, TNF-α, IL-1β, and IL-6 expression was independent of apoptosis; expression of inflammatory markers upon inhibition of necrosis was not changed, ruling out the involvement of necrosis. 4) siCTSB suppressed the inflammatory response but did not affect apoptosis. 5) Caspase-3 inhibitor Z-DEVD-fmk suppressed apoptosis but did not affect the inflammatory response (30). 6) siNLRP3 blocked the Atg7-induced inflammatory response. 7) Atg7-induced excessive autophagy activation aggravated the resultant GSIS impairment, and siNLRP3 attenuated this effect. Thus, this study has identified for the first time a molecular target of CTSB that contributed to the Atg7-induced NLRP3-dependent proinflammatory response and subsequently led to GSIS aggravation independent of apoptosis.
The results in the present study may have an important implication in the cell biology of CTSB. CTSB, a lysosomal cysteine protease, has been suggested to be involved in autophagy-induced inflammasome formation (31, 32). We report here that the increased level of CTSB induced by Atg7 transfection acted as a stimulus, which was demonstrated to trigger NLRP3 activation and promote caspase-1-dependent proinflammatory cytokine secretion and may enhance pathogenesis in type 2 diabetes (33). Further evidence from other researchers has indicated that release of CTSB after lysosomal damage can activate NLRP3, consistent with our results (27). In macrophages, excessive autophagy activation enhances lysosomal destabilization and is also associated with NLRP3 activation induced by cholesterol crystals, which are probably involved in the inflammation that promotes atherosclerosis. Inhibition of CTSB suppresses its release from destabilized lysosomes, attenuating the inflammatory response in this report (35) in agreement with our study.
Recent reports reveal a complex interplay between inflammation and autophagic pathways. Blockade of autophagy by genetic ablation of the autophagy regulator Atg16L1 or Atg7 enables LPS-dependent inflammasome activation, suggesting that autophagy normally counters inflammasome activation by LPS (12). It has been suggested that autophagosomes may target inflammasomes for degradation, and possible target reactive oxygen species may indirectly inhibit inflammasome activity (36). However, our results suggest a cross-talk between the toxic effect of excessive autophagy activation and the expression of the proinflammatory cytokines. Our observations are partly congruent with a recent study demonstrating in macrophages that PA could activate NLRP3, whereas there was no response to the unsaturated oleate (37). The two seemingly contradictory functions of autophagy suggest that the complex roles of autophagy can act as either a friend or foe of T2D, depending on its level of activation or the different stages of disease. At the beginning, autophagy provides a protective role of survival advantage to β-cells where they suffer from metabolic stress. After induction, excessive autophagy activation instead causes an elevation in the production of CTSB, which can stimulate intracellular inflammasomes, and then activates NLRP3 and subsequently inflammatory caspases (38), which stimulate procytokine secretion or possibly cause autophagic cell death resulting in GSIS impairment. The results reported herein are also consistent with recent studies demonstrating that autophagy might play a deleterious role in acute pancreatitis via the maturation of autophagosomes, which result in CTSB-mediated trypsinogen activation (39).
Previous reports have shown that autophagy is regulated by immunologically relevant cytokines with activation by TNF-α and MCP-1, major Th1 cytokines, and inhibition by IL-4 and IL-13, major Th2 cytokines (40). Thus, autophagy is an effector of Th1/Th2 polarization where either Th1 cytokines are protective or Th2 cytokines are permissive when it comes to controlling intracellular pathogens (34). Proinflammatory cytokines usually observed in T2D ultimately disrupt the balance of autophagy in the affected organ. Our findings provide new insights into the release mechanisms of proinflammatory cytokines regulated by autophagy, showing that autophagy also has negative effects on secretion of the proinflammatory Th1 cytokines integrated with aggravation of lipotoxicity. With respect to this possibility, it would be interesting to examine the complex interplay between autophagy and the NLRP3-dependent proinflammatory Th1 response during T2D progression in islets of diabetic patients and animal models.
Footnotes
- T2D
- type 2 diabetes
- Atg7
- autophagy-related 7
- CTSB
- cathepsin B
- LC3
- light chain 3
- MCP-1
- monocyte chemotactic protein-1
- NLRP3
- Nod-like receptor 3
- PA
- palmitic acid
- INS-1
- rat insulinoma cell line
- GSIS
- glucose-stimulated insulin secretion
- PI
- propidium iodide
- Z
- benzyloxycarbonyl
- fmk
- fluoromethyl ketone.
REFERENCES
- 1. Shao S., Liu Z., Yang Y., Zhang M., Yu X. (2010) SREBP-1c, Pdx-1, and GLP-1R involved in palmitate-EPA regulated glucose-stimulated insulin secretion in INS-1 cells. J. Cell. Biochem. 111, 634–642 [DOI] [PubMed] [Google Scholar]
- 2. Gregor M. F., Hotamisligil G. S. (2011) Inflammatory mechanisms in obesity. Annu. Rev. Immunol. 29, 415–445 [DOI] [PubMed] [Google Scholar]
- 3. Donath M. Y., Shoelson S. E. (2011) Type 2 diabetes as an inflammatory disease. Nat. Rev. Immunol. 11, 98–107 [DOI] [PubMed] [Google Scholar]
- 4. Festa A., D'Agostino R., Jr., Tracy R. P., Haffner S. M. (2002) Elevated levels of acute-phase proteins and plasminogen activator inhibitor-1 predict the development of type 2 diabetes: the insulin resistance atherosclerosis study. Diabetes 51, 1131–1137 [DOI] [PubMed] [Google Scholar]
- 5. Osborn O., Brownell S. E., Sanchez-Alavez M., Salomon D., Gram H., Bartfai T. (2008) Treatment with an Interleukin 1β antibody improves glycemic control in diet-induced obesity. Cytokine 44, 141–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Larsen C. M., Faulenbach M., Vaag A., Ehses J. A., Donath M. Y., Mandrup-Poulsen T. (2009) Sustained effects of interleukin-1 receptor antagonist treatment in type 2 diabetes. Diabetes Care 32, 1663–1668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Komatsu M. (2012) Liver autophagy: physiology and pathology. J. Biochem. 152, 5–15 [DOI] [PubMed] [Google Scholar]
- 8. Dupont N., Jiang S., Pilli M., Ornatowski W., Bhattacharya D., Deretic V. (2011) Autophagy-based unconventional secretory pathway for extracellular delivery of IL-1β. EMBO J. 30, 4701–4711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ebato C., Uchida T., Arakawa M., Komatsu M., Ueno T., Komiya K., Azuma K., Hirose T., Tanaka K., Kominami E., Kawamori R., Fujitani Y., Watada H. (2008) Autophagy is important in islet homeostasis and compensatory increase of β cell mass in response to high-fat diet. Cell Metab. 8, 325–332 [DOI] [PubMed] [Google Scholar]
- 10. Lai E., Bikopoulos G., Wheeler M. B., Rozakis-Adcock M., Volchuk A. (2008) Differential activation of ER stress and apoptosis in response to chronically elevated free fatty acids in pancreatic β-cells. Am. J. Physiol. Endocrinol. Metab. 294, E540–E550 [DOI] [PubMed] [Google Scholar]
- 11. Liang H., Zhong Y., Zhou S., Li Q. Q. (2011) Palmitic acid-induced apoptosis in pancreatic β-cells is increased by liver X receptor agonist and attenuated by eicosapentaenoate. In Vivo 25, 711–718 [PubMed] [Google Scholar]
- 12. Saitoh T., Fujita N., Jang M. H., Uematsu S., Yang B. G., Satoh T., Omori H., Noda T., Yamamoto N., Komatsu M., Tanaka K., Kawai T., Tsujimura T., Takeuchi O., Yoshimori T., Akira S. (2008) Loss of the autophagy protein Atg16L1 enhances endotoxin-induced IL-1β production. Nature 456, 264–268 [DOI] [PubMed] [Google Scholar]
- 13. Meng Q., Cai D. (2011) Defective hypothalamic autophagy directs the central pathogenesis of obesity via the IκB kinase β (IKKβ)/NF-κB pathway. J. Biol. Chem. 286, 32324–32332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Nakahira K., Haspel J. A., Rathinam V. A., Lee S. J., Dolinay T., Lam H. C., Englert J. A., Rabinovitch M., Cernadas M., Kim H. P., Fitzgerald K. A., Ryter S. W., Choi A. M. (2011) Autophagy proteins regulate innate immune responses by inhibiting the release of mitochondrial DNA mediated by the NALP3 inflammasome. Nat. Immunol. 12, 222–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Levine B., Yuan J. (2005) Autophagy in cell death: an innocent convict? J. Clin. Investig. 115, 2679–2688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Pattingre S., Tassa A., Qu X., Garuti R., Liang X. H., Mizushima N., Packer M., Schneider M. D., Levine B. (2005) Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell 122, 927–939 [DOI] [PubMed] [Google Scholar]
- 17. Wang X., Blagden C., Fan J., Nowak S. J., Taniuchi I., Littman D. R., Burden S. J. (2005) Runx1 prevents wasting, myofibrillar disorganization, and autophagy of skeletal muscle. Genes Dev. 19, 1715–1722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Matsui Y., Takagi H., Qu X., Abdellatif M., Sakoda H., Asano T., Levine B., Sadoshima J. (2007) Distinct roles of autophagy in the heart during ischemia and reperfusion: roles of AMP-activated protein kinase and Beclin 1 in mediating autophagy. Circ. Res. 100, 914–922 [DOI] [PubMed] [Google Scholar]
- 19. Martino L., Masini M., Novelli M., Beffy P., Bugliani M., Marselli L., Masiello P., Marchetti P., De Tata V. (2012) Palmitate activates autophagy in INS-1E β-cells and in isolated rat and human pancreatic islets. PLoS One 7, e36188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Cao L. Z., Tang D. Q., Horb M. E., Li S. W., Yang L. J. (2004) High glucose is necessary for complete maturation of Pdx1-VP16-expressing hepatic cells into functional insulin-producing cells. Diabetes 53, 3168–3178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Choi S. E., Lee S. M., Lee Y. J., Li L. J., Lee S. J., Lee J. H., Kim Y., Jun H. S., Lee K. W., Kang Y. (2009) Protective role of autophagy in palmitate-induced INS-1 β-cell death. Endocrinology 150, 126–134 [DOI] [PubMed] [Google Scholar]
- 22. Tan S. H., Shui G., Zhou J., Li J. J., Bay B. H., Wenk M. R., Shen H. M. (2012) Induction of autophagy by palmitic acid via protein kinase C-mediated signaling pathway independent of mTOR (mammalian target of rapamycin). J. Biol. Chem. 287, 14364–14376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Shimabukuro M., Zhou Y. T., Levi M., Unger R. H. (1998) Fatty acid-induced β cell apoptosis: a link between obesity and diabetes. Proc. Natl. Acad. Sci. U.S.A. 95, 2498–2502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Karaskov E., Scott C., Zhang L., Teodoro T., Ravazzola M., Volchuk A. (2006) Chronic palmitate but not oleate exposure induces endoplasmic reticulum stress, which may contribute to INS-1 pancreatic β-cell apoptosis. Endocrinology 147, 3398–3407 [DOI] [PubMed] [Google Scholar]
- 25. Kabeya Y., Mizushima N., Ueno T., Yamamoto A., Kirisako T., Noda T., Kominami E., Ohsumi Y., Yoshimori T. (2000) LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 19, 5720–5728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Khan M. J., Rizwan Alam M., Waldeck-Weiermair M., Karsten F., Groschner L., Riederer M., Hallström S., Rockenfeller P., Konya V., Heinemann A., Madeo F., Graier W. F., Malli R. (2012) Inhibition of autophagy rescues palmitic acid-induced necroptosis of endothelial cells. J. Biol. Chem. 287, 21110–21120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Masters S. L., Dunne A., Subramanian S. L., Hull R. L., Tannahill G. M., Sharp F. A., Becker C., Franchi L., Yoshihara E., Chen Z., Mullooly N., Mielke L. A., Harris J., Coll R. C., Mills K. H., Mok K. H., Newsholme P., Nuñez G., Yodoi J., Kahn S. E., Lavelle E. C., O'Neill L. A. (2010) Activation of the NLRP3 inflammasome by islet amyloid polypeptide provides a mechanism for enhanced IL-1β in type 2 diabetes. Nat. Immunol. 11, 897–904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Song M. K., Roufogalis B. D., Huang T. H. (2012) Reversal of the caspase-dependent apoptotic cytotoxicity pathway by taurine from Lycium barbarum (goji berry) in human retinal pigment epithelial cells: potential benefit in diabetic retinopathy. Evid. Based Complement. Alternat. Med. 2012, 323784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Martino L., Masini M., Novelli M., Beffy P., Bugliani M., Marselli L., Masiello P., Marchetti P., De Tata V. (2012) Palmitate activates autophagy in INS-1E β-cells and in isolated rat and human pancreatic islets. PLoS One 7, e36188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lightfoot Y. L., Chen J., Mathews C. E. (2011) Role of the mitochondria in immune-mediated apoptotic death of the human pancreatic β cell line βLox5. PLoS One 6, e20617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Caglič D., Globisch A., Kindermann M., Lim N. H., Jeske V., Juretschke H. P., Bartnik E., Weithmann K. U., Nagase H., Turk B., Wendt K. U. (2011) Functional in vivo imaging of cysteine cathepsin activity in murine model of inflammation. Bioorg. Med. Chem. 19, 1055–1061 [DOI] [PubMed] [Google Scholar]
- 32. Sun L., Wu Z., Hayashi Y., Peters C., Tsuda M., Inoue K., Nakanishi H. (2012) Microglial cathepsin B contributes to the initiation of peripheral inflammation-induced chronic pain. J. Neurosci. 32, 11330–11342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zhou R., Yazdi A. S., Menu P., Tschopp J. (2011) A role for mitochondria in NLRP3 inflammasome activation. Nature 469, 221–225 [DOI] [PubMed] [Google Scholar]
- 34. Deretic V. (2009) Multiple regulatory and effector roles of autophagy in immunity. Curr. Opin. Immunol. 21, 53–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Rajamäki K., Lappalainen J., Oörni K., Välimäki E., Matikainen S., Kovanen P. T., Eklund K. K. (2010) Cholesterol crystals activate the NLRP3 inflammasome in human macrophages: a novel link between cholesterol metabolism and inflammation. PLoS One 5, e11765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bauernfeind F. G., Horvath G., Stutz A., Alnemri E. S., MacDonald K., Speert D., Fernandes-Alnemri T., Wu J., Monks B. G., Fitzgerald K. A., Hornung V., Latz E. (2009) Cutting edge: NF-κB activating pattern recognition and cytokine receptors license NLRP3 inflammasome activation by regulating NLRP3 expression. J. Immunol. 183, 787–791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wen H., Gris D., Lei Y., Jha S., Zhang L., Huang M. T., Brickey W. J., Ting J. P. (2011) Fatty acid-induced NLRP3-ASC inflammasome activation interferes with insulin signaling. Nat. Immunol. 12, 408–415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Niemi K., Teirilä L., Lappalainen J., Rajamäki K., Baumann M. H., Öörni K., Wolff H., Kovanen P. T., Matikainen S., Eklund K. K. (2011) Serum amyloid A activates the NLRP3 inflammasome via P2X7 receptor and a cathepsin B-sensitive pathway. J. Immunol. 186, 6119–6128 [DOI] [PubMed] [Google Scholar]
- 39. Orlichenko L., Stolz D. B., Noel P., Behari J., Liu S., Singh V. P. (2012) ADP-ribosylation factor 1 protein regulates trypsinogen activation via organellar trafficking of procathepsin B protein and autophagic maturation in acute pancreatitis. J. Biol. Chem. 287, 24284–24293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kolattukudy P. E., Niu J. (2012) Inflammation, endoplasmic reticulum stress, autophagy, and the monocyte chemoattractant protein-1/CCR2 pathway. Circ. Res. 110, 174–189 [DOI] [PMC free article] [PubMed] [Google Scholar]