Background: Although CTC1-STN1-TEN1 (CST) is considered a specialized replication factor, TEN1 is largely uncharacterized.
Results: Like CTC1 and STN1, TEN1 is needed for telomere replication and genome-wide replication rescue; however, TEN1 depletion causes more severe phenotypes.
Conclusion: TEN1 likely functions with CTC1 and STN1 in most contexts, but TEN1 may have additional roles.
Significance: TEN1/CST help solve a diverse array of replication problems.
Keywords: Chromosomes, DNA-binding Protein, DNA Replication, Genomic Instability, Telomerase, Telomeres, CST, CTC1, OBFC1, STN1
Abstract
TEN1 is a component of the mammalian CTC1-STN1-TEN1 complex. CTC1 and/or STN1 functions in telomere duplex replication, C-strand fill-in, and genome-wide restart of replication following fork stalling. Here we examine the role of human TEN1 and ask whether it also functions as a specialized replication factor. TEN1 depletion causes an increase in multitelomere fluorescent in situ hybridization (FISH) signals similar to that observed after CTC1 or STN1 depletion. However, TEN1 depletion also results in increased telomere loss. This loss is not accompanied by increased telomere deprotection, recombination, or T-circle release. Thus, it appears that both the multiple telomere signals and telomere loss stem from problems in telomere duplex replication. TEN1 depletion can also affect telomere length, but whether telomeres lengthen or shorten is cell line-dependent. Like CTC1 and STN1, TEN1 is needed for G-overhang processing. Depletion of TEN1 does not effect overhang elongation in mid-S phase, but it delays overhang shortening in late S/G2. These results indicate a role for TEN1 in C-strand fill-in but do not support a direct role in telomerase regulation. Finally, TEN1 depletion causes a decrease in genome-wide replication restart following fork stalling similar to that observed after STN1 depletion. However, anaphase bridge formation is more severe than with CTC1 or STN1 depletion. Our findings indicate that TEN1 likely functions in conjunction with CTC1 and STN1 at the telomere and elsewhere in the genome. They also raise the possibility that TEN1 has additional roles and indicate that TEN1/CTC1-STN1-TEN1 helps solve a wide range of challenges to the replication machinery.
Introduction
Telomeres are DNA-protein complexes that protect linear chromosome ends from unwanted repair and recombination. They also provide a means to counteract the loss of DNA sequence that occurs during DNA replication. In mammals, telomeres consist of kilobases of TTAGGG repeats that terminate in a 40–400-nucleotide single-stranded 3′ overhang (1, 2). The overhang is thought to invade the duplex region to produce a lariat-like structure called a T-loop that protects the DNA terminus from degradation or fusion (3, 4). A six-protein complex called shelterin also protects the chromosome end and prevents it from eliciting ATR and ATM-mediated DNA damage responses (3, 5). The repetitive nature and unusual terminal structure of telomeric DNA makes telomeres difficult to replicate. As a result, telomere replication requires additional steps and factors beyond those essential for general genomic DNA replication (6, 7). The initial stage of telomere replication occurs in early to mid-S phase when helicases such as WRN and RTEL, the shelterin component telomeric restriction fragment 1 (TRF1),2 and subunits of the CTC1-STN1-TEN1 (CST) complex act in conjunction with the conventional replication machinery to ensure efficient replication of the duplex region (8–13). Telomerase then extends the 3′ overhang soon after duplex replication is complete (1). Finally, the complementary C-strand is filled in several hours later during late S/G2 phase by polymerase α/primase (pol α) aided by CST (1, 12, 14, 15).
Mammalian CST is homologous to the Cdc13-Stn1-Ten1 complex that protects telomeres in budding yeast (15, 16). In addition to its protective function, Saccharomyces cerevisiae CST plays a key role in telomere replication by recruiting and regulating telomerase activity (17–20). It also interacts with pol α and is thought to facilitate C-strand fill-in (21, 22). Yeast and mammalian CST are clearly related because the STN1 and TEN1 subunits exhibit sequence and/or structural homology to each other and to subunits of RPA (16, 23–27). Also, Cdc13 and CTC1 both appear to harbor oligonucleotide/oligosaccharide binding folds that are used to bind single-stranded DNA (25, 28, 29). One major difference between the yeast and mammalian complexes is that mammalian CST does not appear to be needed for telomere protection. Instead, subunits of CST have evolved a number of roles in telomeric and genome-wide replication. At the telomere, STN1 and CTC1 function both in telomere duplex replication and C-strand fill-in (10–12, 14, 29, 30). Elsewhere in the genome, STN1 and CTC1 participate in the resolution of replication stress (10, 29). Depletion of either subunit leads to formation of anaphase bridges, and STN1 depletion results in reduced replication restart after hydroxyurea (HU)-induced fork stalling (10, 11, 29). Consistent with this genome-wide role in replication, CTC1 and STN1 were originally identified as a pol α accessory factor (AAF) that stimulates template binding and enzyme processivity (31, 32). Recently mutations in CTC1 were found to underlie several diseases including dyskeratosis congenita and Coats plus (33–35). Additionally, single-nucleotide polymorphisms associated with human STN1 (OBFC1) correlate with the presence of short telomeres (36, 37). Thus far, no disease-linked mutations or single-nucleotide polymorphisms have been identified for TEN1.
In vitro studies of mammalian CST indicate that CTC1, STN1, and TEN1 form a trimeric complex, and all three subunits are needed to bind single-stranded DNA (25, 38). However, it is not clear whether the three proteins always function as a complex in vivo. TEN1 was not identified as a component of AAF (31), and the need for TEN1 in telomere replication and resolution of replication stress has not been examined. Observations from yeast and plants suggest that in these organisms, the three subunits of CST may function independently or as subcomplexes. In budding yeast, phosphorylation of Cdc13 by Cdk1 promotes telomerase elongation (19, 39, 40). However, telomeres bound by the entire CST complex cannot be extended by telomerase in vitro (41), and binding of Stn1-Ten1 in vivo is thought to limit telomerase extension and convert the telomere into a closed, protected state (42). Moreover, co-overexpression of Stn1 and Ten1 is sufficient to provide telomere protection in the absence of Cdc13, again suggesting the existence of a functional Stn1-Ten1 subcomplex (43). In Arabidopsis, CTC1 localizes to ∼50% of telomeres, whereas TEN1 is only found at 10–15% (44, 45). This finding suggests the existence of CTC1 subcomplexes that do not contain TEN1. Additionally, Arabidopsis TEN1 inhibits telomerase processivity, whereas CTC1 and STN1 have no effect (29, 44, 45). Here we show that depletion of TEN1 leads to phenotypes similar to those seen after CTC1 or STN1 depletion; however, the severity of a subset of phenotypes is enhanced. Our results indicate that TEN1 likely functions in conjunction with CTC1 and STN1 in most contexts but leave open the possibility of additional roles for this protein in mammalian cells.
EXPERIMENTAL PROCEDURES
Cell Lines and Stable Knockdown of TEN
HeLa1.2.11 cells were grown in RPMI, HT1080 in DMEM, and HCT116 in McCoy's medium supplemented with 10% FBS, antibiotics, and glutamine. Lentivirus encoding shRNAs targeting different regions of TEN1 were produced with the following constructs: shTEN1#1, Sigma TRCN0000337412, PuroR; shTEN1#2, Open Biosystems V3LHS_389003, GFP; and shTEN1#3, Sigma TRCN0000337413, PuroR. The lentivirus encoding nontargeting shRNA (shNT) was as described previously (10). HeLa1.2.11, HCT116, and HT1080 cells were infected with virus and selected with 1 μg/ml puromycin or sorted by FACS to obtain shRNA-expressing cells. The levels of TEN1 or STN1 knockdown were determined by reverse transcription and quantitative real time PCR (RT-qPCR) or by Western blotting. Pools of cells with stable TEN1 knockdown were obtained by infecting HCT116 cells with either shTEN1#1 or shTEN1#3 and HT1080 cells with shTEN1#2. STN1 was depleted in HT1080 cells with Open Biosystems V2LHS_278078. HeLa1.2.11 cells were initially infected with shTEN1#1 virus followed by puromycin selection to give a pool of cells from which single cell clones with ∼50% TEN1 depletion were selected. One clone was then infected with shTEN1#2 virus and sorted for GFP expression. Again both a pool of cells (shTEN1#1+#2) and a clone were isolated (HeLa shTEN1–5). To generate shTEN1 rescue cells (shTEN1–5R), the HeLa shTEN1–5 clone was infected with retrovirus encoding TEN1 with silent mutations (TGTCTGTACGATATGATACAGTCGAGGGTTACTCTCATGGCA, with the mutations underlined) at both the shTEN1#1 and shTEN1#2 target sites. The mutant allele was cloned into the pMSCV-IThy1-1 vector. Viral production, infection, sorting of Thy1-1-expressing cells, and production of clones were as described (10).
RT-qPCR
The efficiency of TEN1 and STN1 depletion was determined by two-step RT-qPCR with SYBR GREEN using the HotStart-It kit (United States Biological) and RNA isolated with the RNeasy kit (Qiagen) (10) and extended DNase digestion to remove genomic DNA. cDNA was made using oligo(dT) primers. Primers for TEN1 amplification, 5′-GGCCAAGTTCCTGATGGG and 5′-CAGTGTTACTCTGGACTGAATCAT, were designed to avoid amplification of a TEN1 pseudogene. GAPDH and STN1 primers were as described (10). Reactions were performed at least in duplicate using GAPDH as the endogenous control.
Antibodies and Western Blots
6HIS-TEN1 was expressed from the pET45 plasmid in bacteria and purified using nickel-nitrilotriacetic acid-agarose (Qiagen). Purified protein was injected into rabbits (Covance), and antibodies from serum were purified using 6HIS-TEN1 covalently linked to NHS Sepharose 4 fast flow (GE Healthcare). For TEN1 and STN1 detection, cells were lysed in low salt buffer (10 mm HEPES, 10 mm KCl, 1.5 mm MgCl2, 0.34 m sucrose, 10% glycerol, 1 mm DTT, 0.1% Triton X-100), and the indicated amounts of protein were separated by SDS-PAGE and transferred to nitrocellulose membrane. The membrane was blocked with 1% milk and incubated with antibody to actinin (Santa Cruz; 1:50,000), TEN1 (1:500), and STN1 (1:1000) (10) overnight followed by incubation with HRP-conjugated secondary antibody (Thermo; 1:1,000) for 30 min and developed using ECL Prime (GE Healthcare). Dilution series were performed to confirm proteins were in the linear range of detection, and bands were quantified using ImageQuant.
Growth Curves
5 × 105 cells were plated in triplicate and allowed to grow for 48 h. Live cells were counted using trypan blue exclusion, and 5 × 105 cells were reseeded for the next time point. Total cell number was extrapolated using a value that was the cell count/fraction of cells used to reseed previous time point. The experiment was performed three times with shNT and shTEN1–5 and twice with shTEN1–5R.
Telomere Length and G-overhang Analysis
Genomic DNA was isolated by proteinase K digestion and phenol chloroform extraction for G-overhang analysis or by high salt precipitation (12) for telomere length analysis, followed by overnight digestion with HinfI and MspI. For telomere length determination, restriction fragments were separated in 1% agarose by pulse-field electrophoresis (HeLa1.2.11) or standard electrophoresis (HCT116 and HT1080). In-gel hybridization was performed using a (TA2C3)4 probe after denaturation with NaOH. Signal was quantified by PhosphorImager, and mean telomere length was determined by dividing each lane into 100 boxes using ImageQuant and applying the formula ΣSig/Σ(SigI/LI), where Sig is the sum of the signal from all 100 boxes, SigI is the signal in an individual box, and LI corresponds to the average length of the DNA in that box as determined using DNA markers and a standard curve (46). For G-overhang analysis, control samples were treated with Exo1 prior to restriction digestion. Samples were separated briefly in 1% agarose gels to keep the TRFs in a tight band. In-gel hybridization was performed under native conditions using (TA2C3)4 probe, the DNA was denatured, and the gel was rehybridized with the same probe. For each lane, the Exo1-resistant signal was subtracted from the untreated nondenatured signal, and the resulting G-overhang signal was normalized for loading using the signal from the denatured sample. For the cell cycle analysis of overhang length, cells were synchronized at the G1/S boundary with a double thymidine block as previously described (12).
Telomere FISH
FISH was performed on metaphase spreads as previously described using FITC-OO-(T2AG3)3 peptide nucleic acid probe (10). To quantify telomere sister chromatid exchange (T-SCE), chromosome orientation FISH was performed as described (47) with the following modifications. Briefly, cells were labeled with a 3:1 mix of BrdU (MP Biomedicals):BrdC (Sigma) (7.5 mm: 2.5 mm) for 16–20 h prior to collection. After preparation of metaphase spreads, the slides were dried overnight, treated with 0.5 mg/ml RNase A for 10 min at 37 °C, and then stained with 0.5 μg/ml Hoechst 33258 (Sigma) in 2× SSC for 15 min at room temperature. The slides were exposed to 365-nm UV light (5.4 × 103 J/m2) using a Stratalinker 1800 for 30 min at room temperature in 2× SSC. BrdU/BrdC-substituted strands were digested with 800 units of Exonuclease III (Promega) for 10 min at room temperature. The slides were then washed with PBS and dehydrated before incubation with probes. Cy3-(C3TA2)3 probe was added for 5 min at 90 °C followed by 2 h at room temperature. The slides were then washed and incubated with FITC-(T2AG3)3 for 2 h at room temperature. The slides were washed, and the FITC signal was amplified as previously described (10).
Combined γH2AX and FISH Staining
Chromosomes were spread and stained as described (48) with the following modifications. Cells were resuspended at 5 × 104 cells/ml in 0.2% KCl, 0.2% sodium citrate and centrifuged onto slides at 1600 rpm for 8 min in a Statspin Cytofuge 2 (Iris Sample Processing). The slides were fixed in 4% formaldehyde for 10 min and then permeabilized with KCM buffer (120 mm KCl, 20 mm NaCl, 10 mm Tris, pH 7.5, 0.1% Triton X-100) for 10 min. The slides were blocked at 37 °C for 15 min in antibody dilution buffer (20 mm Tris, pH 7.5, 2% BSA, 0.2% fish gelatin, 150 mm NaCl, 0.1% Triton X-100, 100 μg/ml RNase A). The slides were then incubated with 1:500 α-γH2AX (Upstate) for 1 h at room temperature, washed three times for 5 min in PBS + 0.1% Tween, and incubated with 1:750 α-mouse Alexa 488 at room temperature for 30 min. After three washes with PBS + 0.1% Tween, antibodies were fixed in 4% formaldehyde for 10 min at room temperature. The slides were dehydrated with ethanol and incubated with Cy3-(C3TA2)3 probe for 3 min at 80 °C, followed by 2 h at room temperature. The slides were washed twice for 5 min in Wash A buffer (70% formamide, 10 mm Tris, pH 7.5) and twice for 5 min in Wash B buffer (50 mm Tris, pH 7.5, 150 mm NaCl, 0.8% Tween). Finally, the slides were mounted in the presence of DAPI.
T-circle Assay
Genomic DNA was isolated and restriction-digested as described for G-overhang assays, denatured, and annealed with (C3TA2)3C3TA*A primer (where the asterisk denotes phosphorothioate) at 95 °C for 5 min followed by 2 h at room temperature. 0.2 mm dNTP was added in the presence or absence of Phi29 polymerase (Thermo) and incubated at 30 °C for 12 h followed by 65 °C for 20 min. Products were separated in 0.6% agarose gels at 100V for 1 h followed by 35 V overnight. The gels were dried, and the products were denatured and detected by in-gel hybridization with G(T2AG3)3 T2AG probe.
Replication Restart
The cells were treated with 2 mm HU for 2 h followed by 50 μm EdU for 30 min and fixed, and EdU was visualized using Click-It (Invitrogen) (10). Mean EdU signal in HU-treated cells was divided by mean EdU signal in untreated cells to normalize for the slower growth of shTEN1–5 cells.
Anaphase Bridges
Cells were treated with 50 ng/ml nocodazole for 3–4 h and then released into fresh medium for 30 min to 1 h. The cells were pre-extracted with 0.05% Triton X-100 for 2 min at 4 °C, washed with PBS, and treated with RNase A for 10 min at 37 °C prior to formaldehyde fixation. Following fixation, the cells were permeabilized in 0.5% Triton X-100 for 10 min at room temperature. The slides were then hybridized with Cy3-(C3TA2)3 for 5 min at 90 °C followed by 2 h at room temperature. Washing, dehydration, and mounting were then performed as previously described (10).
Data Acquisition and Analysis
FISH images were taken at ×1000, and replication restart images were taken at ×200 with a Nikon Eclipse E400 equipped with a Spot 2 digital camera (Diagnostic Instruments Inc.). For each independent experiment, ≥300 metaphase chromosomes were analyzed to quantify multiple telomere signals (MTS) and SFE and ≥250 for T-SCE. Likewise, ≥100 anaphases were counted to quantify anaphase bridges and ≥500 cells for replication restart. p values indicated in the figures were determined using the Student's two-tailed unpaired t test.
RESULTS
TEN1 Depletion Affects Telomere Integrity
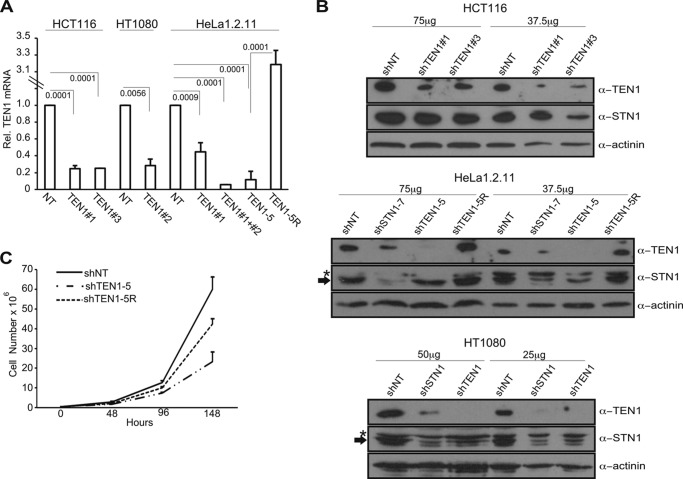
To address the function of TEN1 in human cells, we generated a series of cell lines with stable TEN1 knockdown. We generated two pools of HCT116 cells each expressing a different TEN1 shRNA (shTEN1#1 and shTEN1#3), a single pool of HT1080 cells expressing one shRNA (shTEN1#2) and HeLa1.2.11 pools expressing one or two shRNAs (shTEN1#1 or shTEN1#1+#2), and a clone shTEN1–5 expressing two TEN1 shRNAs (shTEN1#1+#2). The level of knockdown was assessed by RT-qPCR and immunoblotting (Fig. 1). For the HCT116 and HT1080 pools, mRNA levels were depleted by ∼75% (Fig. 1A), and TEN1 protein was depleted by ∼70–80% in HCT116 and ∼95% in HT1080 (Fig. 1B and Table 1). The HeLa shTEN1#1+#2 pool and the clone shTEN1–5 exhibited ∼90% reduction in TEN1 mRNA (Fig. 1A) and ∼95% reduction in protein (Fig. 1B and Table 1). The shTEN1–5 clone was subsequently transfected with a construct expressing an shRNA-resistant TEN1 allele. Individual clones were selected, and one clone, shTEN1–5R, was used to verify that phenotypes were specific to TEN1 depletion and not due to off target effects. The shTEN1–5R clone had nearly endogenous levels of protein expression (Fig. 1, A and B, and Table 1).
FIGURE 1.
TEN1 depletion in human cancer cells. A, RT-qPCR of TEN1 mRNA in various cell lines. Levels are relative to shNT for each cell type with normalization to GAPDH (n = 4 for shTEN1–5; n = 3 for HCT116 and HeLa shTEN1#1; n = 2 for HT1080, HeLa shTEN1#1+#2, and shTEN1–5R; means ± S.D.). Note discontinuous y axis. p values are indicated. B, Western blot showing TEN1 (13 kDa) and STN1 (42 kDa) levels in two HCT116 pools: HeLa1.2.11 shSTN1–7, shTEN1–5, and TEN1–5R clones and HT1080 shSTN1 and shTEN1 pools. Loading control is α-actinin (100 kDa). The total amount of cell lysate is indicated at the top. The asterisk indicates a nonspecific band that cross-reacts with the STN1 antibody, and the arrow indicates STN1. C, representative growth curves for HeLa clones. n = 2 experiments. The cells were counted in triplicate at each time point.
TABLE 1.
Quantification of TEN1 and STN1 protein levels in various cell lines
TEN1 and STN1 were quantified and normalized to actinin in each lane, and this value was normalized to shNT for each cell type. The values represent the means ± S.E. of two different protein concentrations on two separate blots.
| Cell Type | Relative TEN1 levels | Relative STN1 levels |
|---|---|---|
| HCT116 | ||
| shNT | 1 | 1 |
| shTEN1#1 | 0.20 ± 0.05 | 0.82 ± 0.14 |
| shTEN1#3 | 0.34 ± 0.05 | 0.58 ± 0.07 |
| HeLa1.2.11 | ||
| shNT-3 | 1 | 1 |
| shTEN1–5 | 0.04 ± 0.04 | 0.55 ± 0.16 |
| shTEN1–5R | 1.05 ± 0.12 | 0.78 ± 0.19 |
| shSTN1–7 | 0.45 ± 0.11 | 0.14 ± 0.07 |
| HT1080 | ||
| shNT | 1 | 1 |
| shTEN1 | 0.06 ± 0.04 | 0.21 ± 0.16 |
| shSTN1 | 0.16 ± 0.04 | 0.26 ± 0.15 |
In contrast to our previous findings with CTC1- or STN1-depleted HeLa cells, depletion of TEN1 in HeLa caused a significant growth defect that was partially rescued by the TEN1-resistant allele (Fig. 1C, Table 2, and Ref. 10). We also observed a growth arrest when we first isolated the HT1080 shTEN1 pool, but eventually the culture recovered, suggesting a selection for cells with less efficient TEN1 depletion. Neither pool of TEN1-depleted HCT116 showed noticeable growth perturbation (data not shown).
TABLE 2.
Summary of shCST phenotypes in HeLa1.2.11 cells
Comparison of shTEN1 phenotypes identified in this study to previously characterized shCTC1 and shSTN1 cell lines (10, 12). ND, not determined.
| Phenotype | shCTC1 | shSTN1 | shTEN1 |
|---|---|---|---|
| Depletion (%) | 70a | 60–80a | 95 |
| Cell growth | Unchangeda | Unchangeda | Slowc |
| MTS | ↑2–2.5 ×a | ↑2–3 ×a | ↑2× |
| SFE | Unchangeda | Unchangeda | ↑20 ×c |
| T-circles | ND | Unchanged | Unchanged |
| G-overhang | ↑1.25 ×a | ↑1.5–2 ×a,b | ↑2× |
| Replication restart (%) | ND | ↓30a | ↓30 |
| Anaphase bridges | ↑2–2.5 ×a | ↑2–3 ×a | ↑5 ×c |
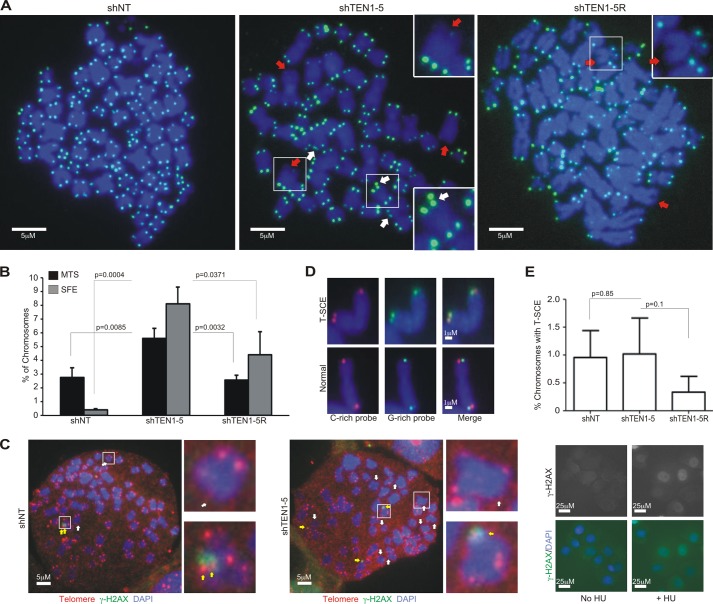
Depletion of mammalian CTC1 or STN1 leads to various telomere abnormalities including the appearance of MTS on individual chromatids during FISH analysis. In STN1-depleted cells, the severity of the MTS phenotype tracks with telomere length and the need for STN1 to facilitate telomere duplex replication (12). When we examined the effect of TEN1 depletion in HeLa cells with long telomeres (HeLa1.2.11, 20–25-kb telomeres), we saw a similar ∼2-fold increase in MTS to that observed after CTC1 or STN1 depletion (Fig. 2, A and B, Table 2, and Ref. 10). This increase was largely rescued by expression of the shRNA-resistant TEN1 allele. We also saw a large (∼20-fold) increase in chromosomes lacking telomere signals (SFE, signal free ends), something that was not observed after stable CTC1 or STN1 depletion. The SFE were rescued by the shRNA-resistant allele but to a lesser extent than the MTS, raising the possibility that SFE and MTS differ in their underlying cause. Interestingly, we did not observe telomere fusions in the shTEN1 cells (Fig. 2A), suggesting that the chromatids with SFE retained sufficient telomeric DNA to maintain telomere protection. To more directly assess whether the SFE remain protected from the DNA damage response, we looked for the presence of γH2AX, a marker for DNA damage signaling. When we performed combined telomere FISH and γH2AX staining on metaphase spreads, we found that most chromatids lacking telomere FISH signals also lacked γH2AX foci (Fig. 2C), indicating that the telomeres remained at least partially protected. The level of telomere dysfunction-induced foci at telomeres with FISH signals was also unchanged. We next explored whether TEN1 depletion leads to an increase in T-SCE by using chromosome orientation FISH (49) to label newly replicated telomeric G- versus C-strands. However, we did not find a significant difference in T-SCE frequency (Fig. 2, D and E), indicating that TEN1 depletion does not lead to increased telomere recombination.
FIGURE 2.
TEN1 is needed for telomere integrity. A, telomere FISH on metaphase spreads of shNT, shTEN1–5, or shTEN1–5R cells showing examples of MTS (white arrows) or SFE (red arrows). Green, FITC-telomere probe; blue, DAPI. Chromosomes in insets are enlarged 2-fold. Scale bars are indicated. B, quantification of MTS and SFE (means ± S.D., n = 3). C, γ-H2AX staining combined with telomere FISH on metaphase spreads from shNT (left) or shTEN1–5 (right) cells. White arrows, SFE; yellow arrows, TIFs; green, γ-H2AX; red, Cy3-(C3TA2)3 probe; blue, DAPI. Chromosomes in insets are enlarged 5-fold. Right panel, positive control showing γ-H2AX staining of nuclei after treatment of shNT cells with 2 mm HU for 24 h. Scale bars are indicated. D, chromosome orientation FISH on metaphase spreads showing examples of chromosomes with and without T-SCE. Red, Cy3-(C3TA2)3 probe; green, FITC-(T2AG3)3 probe; blue, DAPI. Scale bars are indicated. E, quantification of T-SCE (means ± S.D., n = 3).
To ask whether TEN1 depletion leads to similar defects in cells with shorter telomeres, we performed telomere FISH with HCT116 cells (telomere length ∼5 kb). As observed after STN1 depletion in cells with short telomeres (12), we did not observe an increase in MTS (data not shown). However, unlike what was observed after STN1 depletion, TEN1 depletion caused a ∼2-fold increase in SFE (data not shown). The increased MTS in HeLa cells and SFE in HeLa and HCT116 cells after TEN1 depletion suggest that TEN1 facilitates telomere duplex replication and that this role is most important for cells with long telomeres. The appearance of SFE is particularly interesting because this telomere loss phenotype was not previously observed in HeLa1.2.11 cells after stable CTC1 or STN1 depletion (Table 2). A similar level of SFE was observed after STN1 or TEN1 depletion in HEK 293T cells (27), which suggests some cell line-specific variability in the effects of CST loss (see below).
Recently it has been shown that in mouse cells, decreasing the level of one CST subunit can affect the levels of the other subunits, suggesting that CTC1, STN1, and TEN1 mutually stabilize each other (11, 32, 50). We therefore examined whether this is also true for human TEN1 and STN1. Western blot analysis revealed that in both HCT116 and HeLa1.2.11 cells, TEN1 depletion caused ∼45% reduction in STN1. In HeLa cells, STN1 depletion caused a 55% reduction in TEN1 (Fig. 1B and Table 1). Although the reduction in STN1 caused by TEN1 depletion could contribute to the above telomere phenotypes, we have not seen MTS or SFE after this level of STN1 depletion.3 We were not able to examine CTC1 levels because of a lack of antibodies capable of detecting the endogenous protein. However, because CTC1 interacts with STN1 but not TEN1 (25, 38), we would anticipate a ≤45% decline in CTC1 levels in shTEN1 cells because of the decrease in STN1. Again, we have not seen phenotypes with this level of CTC1 depletion.3
Telomere Loss after TEN1 Depletion Is Unlikely to Occur via T-circle Release
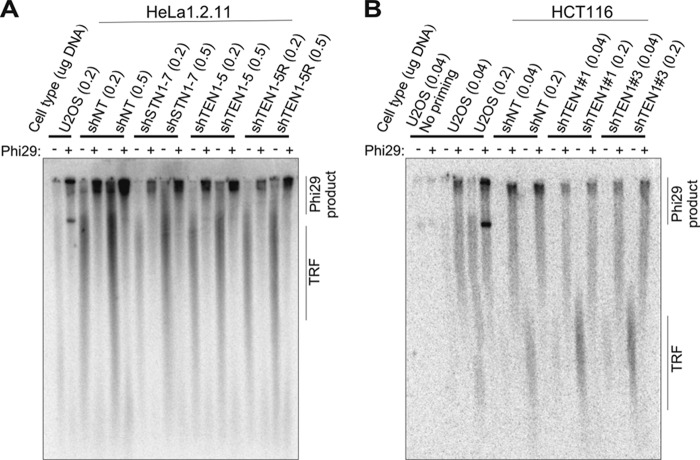
In mouse cells, complete removal of CTC1 by gene disruption results in extensive telomere loss with a concomitant appearance of T-circles (11). T-circles appear to arise from defective processing of T-loops, and the resulting elimination of telomeric DNA can result in telomere loss (51–53). The above findings led us to ask whether the SFE seen after human TEN1 depletion stem from abnormal T-loop processing. We examined this possibility by using a rolling circle amplification assay to test for increased T-circle formation in HeLa shTEN1–5 cells relative to shTEN1–5R or shNT cells. shSTN1 cells were also examined (but not shCTC1 as the knockdown is unstable). U2OS cells were used as a positive control, because they are ALT cells and have a high level of T-circles (54). Genomic DNA was restriction-digested, denatured, and annealed with a telomeric C-strand primer, and any circular molecules were amplified with Phi29 polymerase. Reaction products were separated in agarose gels and hybridized with a probe to the telomeric C-stand (Fig. 3). As previously observed for ALT cells, two types of T-circle amplification products were seen with the U2OS samples incubated with Phi29 polymerase (54): one that sticks in the well and one that migrates into the gel. Only TRFs were visible in the absence of polymerase. Amplification products were also present in the well of Phi29-treated HeLa shTEN1 and shSTN1 samples, but the amount of product did not significantly increase from that obtained with the shTEN1–5R and shNT samples (Fig. 3A). The presence of T-circles in the HeLa cells most likely reflects trimming of the long telomeres by T-loop processing (48). Although the above result suggested that TEN1 and STN1 depletion does not lead to increased T-circle release, the high endogenous background might mask small differences in T-circle levels. Because the level of endogenous telomere trimming can be enhanced in cells with long telomeres (48), we repeated the analysis with DNA samples from shTEN1 HCT116 cells because they have shorter telomeres and hence were expected to have a lower level of endogenous T-circle release (Fig. 3B). However, we again saw amplification products in Phi29-treated samples from the control shNT cells, indicating a background level of T-circles. When we performed the assay with samples from pools of TEN1-depleted cells (shTEN1#1 and shTEN1#3), we also saw Phi29 amplification products, but the levels were generally somewhat lower than that observed with the shNT cells. This is the opposite of what would be expected if TEN1 depletion leads to T-circle release. We therefore conclude that the sudden telomere loss seen after TEN1 depletion is unlikely to result from abnormal T-loop processing.
FIGURE 3.
Effect of TEN1 depletion on T-circle release. A and B, Representative gels showing Phi29 amplification products obtained with DNA from shTEN1 or shSTN1 HeLa 1.2.11 (A) or shTEN1 HCT116 (B) cells relative to shNT, shTEN1 rescue, or U2OS controls. The amounts of DNA used as template are indicated. Products visualized by in-gel hybridization with G(T2AG3)3T2AG probe. The positions of Phi29 amplification products and TRFs are marked. No priming, no primer in Phi29 reaction. Similar results were obtained in three independent experiments.
The Effect of TEN1 Depletion on Telomere Length Is Cell Type-specific
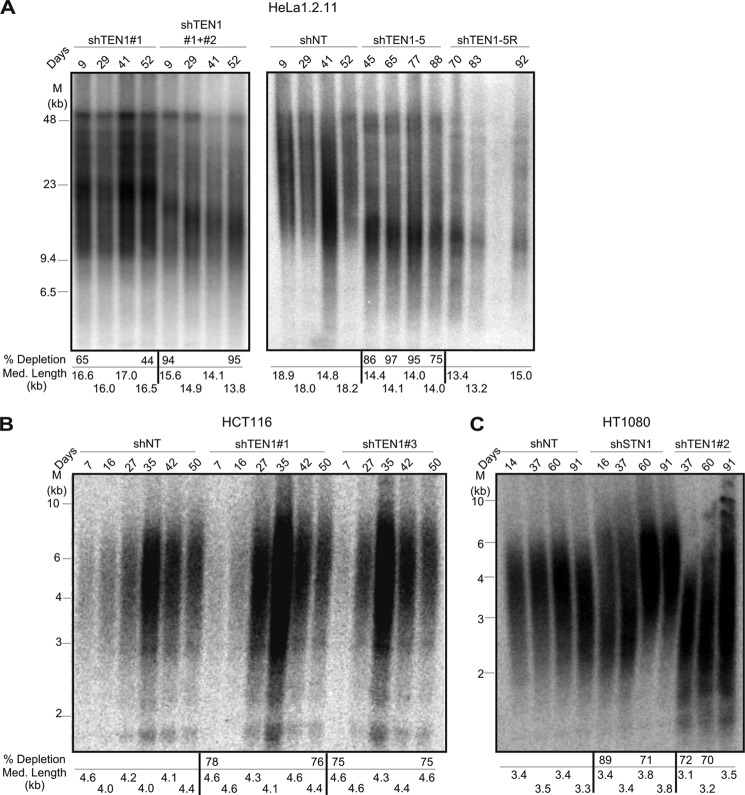
The effect of TEN1 depletion on telomere length was examined by Southern blot analysis of TRFs using DNA samples isolated from shTEN1 and shNT HeLa or HCT116 cells after short and long term culture (Fig. 4). Stable depletion of TEN1 mRNA for the duration of the culture period was verified by RT-qPCR. For the HeLa cells, we analyzed TRFs from shTEN1 pools (shTEN1#1, ∼55% knockdown and shTEN1#1+#2, ∼95% knockdown), the shTEN1–5 clone (∼95% knockdown), and the rescue shTEN1–5R. The shTEN1–5 clone could not be monitored until day 45 after introduction of the second shRNA because of its slow growth (Fig. 1C). TRF analysis revealed that ∼55% TEN1 depletion had a minor effect on telomere length (compare shNT and shTEN1#1), and further reduction in TEN1 levels caused a greater decline in telomere length with eventual stabilization at 14 kb (Fig. 4A). This decrease in telomere length was not rescued by expression of the shRNA-resistant TEN1 allele, suggesting activation of a new telomere length set point following sudden telomere loss. For the HCT116 cells, we examined telomere length in the shTEN1#1 and shTEN1#3 pools (∼75% knockdown) and the shNT control (Fig. 4B). In these cells, TEN1 depletion had negligible effect on telomere length.
FIGURE 4.
The effect of TEN1 depletion on telomere length varies with cell type and level of knockdown. A–C, telomere lengths in HeLa1.2.11 (A), HCT116 (B), or HT1080 (C) cells at various times after introduction of shRNA. Day 0 corresponds to final day of lentivirus transfection. Telomeric restriction fragments were visualized by in-gel hybridization with (TA2C3)4 probe. mRNA depletion and mean telomere length for individual time points are shown below each gel.
Our results showing a decline in telomere length in HeLa cells after deep TEN1 depletion (∼95%) and our previous finding that STN1 depletion (75–80%) has no effect on telomere length (12) are in contrast to a recent publication indicating that depletion of STN1 (70% or 87%) or TEN1 (83% or 92%) in HT1080 cells caused telomere growth (38). To explore this disparity, we generated pools of HT1080 cells depleted for STN1 (∼75% knockdown) or TEN1 (∼95% knockdown) and examined telomere length over the course of 90 days. Interestingly, we also saw a minor increase in telomere length after long term STN1 or TEN1 depletion (Fig. 4C). We then examined the levels of STN1/TEN1 in the knockdown cells and found that they were more tightly co-regulated than in HCT116 or HeLa cells. In HT1080 cells, TEN1 depletion resulted in an ∼80% decline in STN1, whereas STN1 depletion caused an ∼85% decline in TEN1 (Fig. 1B and Table 1). It is unclear whether the telomere growth in HT1080 cells is related to the co-depletion of STN1 and TEN1. However, these findings demonstrate that the effects of STN1 and TEN1 depletion vary with cell type.
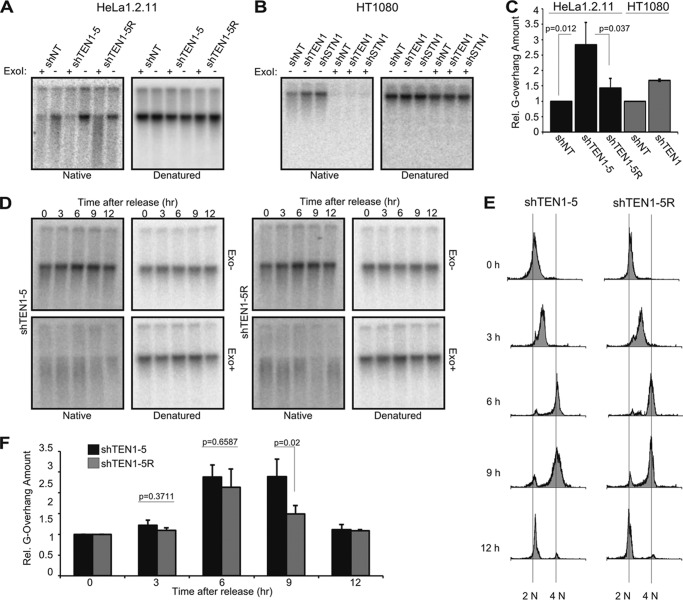
TEN1 Is Needed for C-strand Fill-in
Depletion of CTC1 or STN1 causes a 1.5–2-fold increase in G-overhang length that reflects their role in promoting C-strand fill-in following telomerase action (10, 12). To examine whether TEN1 depletion affects G-overhang dynamics in a similar manner, we examined G-overhang status by using nondenaturing in-gel hybridization to assess relative overhang amount (Fig. 5A). Genomic DNA from HeLa shNT, shTEN1–5, and shTEN1–5R cells was restriction-digested, run briefly in agarose gels, and hybridized with a probe to the G-strand. The DNA was then denatured and rehybridized with the same probe. Quantification of overhang signal relative to total telomere signal for each sample revealed that TEN1 depletion caused a ∼2-fold increase in overhang amount relative to the shTEN1–5R cells and a ∼3-fold increase relative to the shNT control (Fig. 5C). The estimate of a 2-fold increase in overhang signal is probably more accurate because telomere length (and hence the loading control) is similar in the shTEN1–5 and shTEN1–5R cells, whereas the telomeres of the shNT cells are considerably longer (Fig. 4A). Thus, in HeLa cells, CTC1, STN1, and TEN1 knockdown cause a similar increase in overhang abundance (Table 2 and Ref. 10). We repeated the experiment with HT1080 shTEN1 and shNT cells using DNA isolated at 60 days after shRNA transduction (Fig. 4C). We again saw an increase in overhang length in the TEN1-depleted cells (Fig. 5, B and C).
FIGURE 5.
TEN1 is needed for C-strand fill-in. A–F, analysis of overhang abundance by in-gel hybridization with (TA2C3)4 probe in asynchronous (A–C) and synchronous (D–F) cultures. A and B, representative gels showing G-overhang signal from HeLa (A) or HT1080 (B) cells. C, quantification of A (n = 3, means ± S.D.) and B (n = 2, error bars show maximum/minimum values). D, representative experiment showing overhang signal from shTEN1–5 and shTEN1–5R cells at indicated times after release from G1/S block. E, cell cycle analysis for experiment shown in D. F, quantification of data from four independent experiments (means ± S.E.).
Analysis of overhang length and density during progression of STN1-depleted cells through the S phase demonstrated that the longer overhangs observed after STN1 depletion result from a delay in C-strand fill-in rather than increased overhang extension by telomerase (12). To examine whether this is also the case after TEN1 depletion, we synchronized HeLa shTEN1–5 and shTEN1–5R cells at the G1/S boundary by double thymidine block, released them into the S phase, and isolated genomic DNA at various time points for overhang analysis. In-gel hybridization revealed that the overhangs from TEN1-depleted and TEN1 rescue cells exhibited a similar amount and pattern of elongation during early to mid-S phase (Fig. 5, D and F), the period when most telomeres are extended by telomerase (1). However, the shTEN1 cells exhibited a delay in overhang shortening in late S/G2, the time of C-strand fill-in. These results indicate that like STN1, TEN1 also functions in C-strand fill-in.
TEN1 Also Has Nontelomeric Roles in Replication Rescue
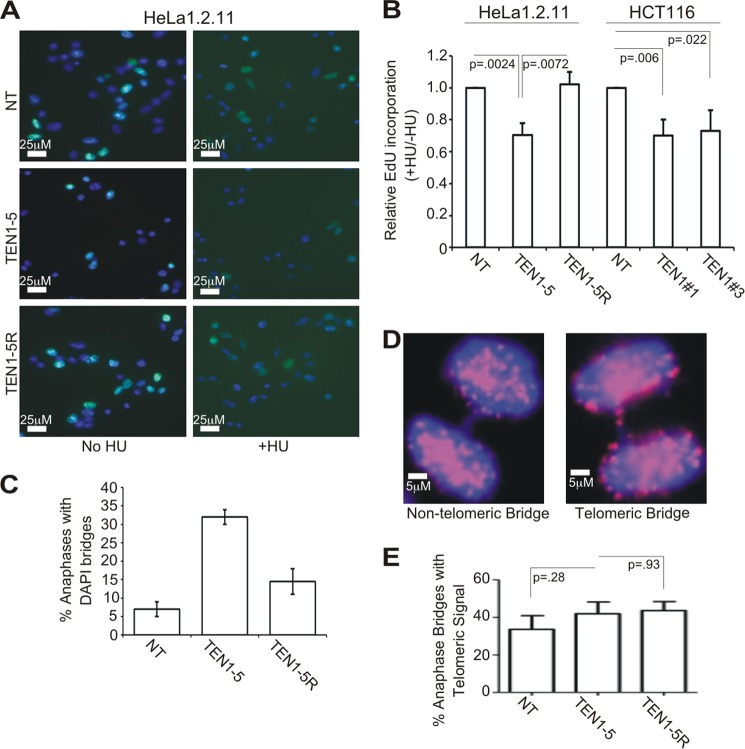
Although the above results indicate that TEN1 functions at telomeres in a similar manner to STN1, TEN1 was not identified as a component of the pol α stimulatory factor AAF (31). We therefore asked whether the role of TEN1 is limited to telomeres or if, like STN1, it has a genome-wide function in replication restart after fork stalling. To test for a role in replication restart, shTEN1 cells (HeLa or HCT116) and control cells were treated with HU for 2 h to stall replication; they were then washed to remove the HU and EdU added to label cells that reinitiated replication. The cells were fixed, the EdU was conjugated with fluorophore, and the EdU signal per nucleus was quantified. To compensate for the slower growth of the shTEN1 HeLa cells relative to control cells, we normalized the EdU uptake by HU-treated cells to that of untreated cells harboring the same shRNA. We could then assess whether TEN1 depletion affected the relative amount of EdU uptake. This experiment revealed that the shTEN1 cells exhibited a 30% decrease in EdU uptake after HU release relative to the corresponding shNT or shRNA-resistant cells (Fig. 6, A and B). This was true both for the HeLa shTEN1–5 clone (95% TEN1 depletion) and for the HCT116 pools (shTEN1#1 or shTEN1#3, ∼75% TEN1 depletion). Interestingly, the magnitude of the replication restart defect in the shTEN1 cells was similar to that seen after STN1 depletion (Table 2 and Ref. 10). Because the restart defect in HU-treated shSTN1 cells was shown by DNA fiber analysis to be due to an inability to activate new origins following fork stalling (10), our results suggest that TEN1 has a similar role in replication rescue. However, definitive proof will require DNA fiber analysis.
FIGURE 6.
TEN1 facilitates replication restart and prevents the accumulation of anaphase bridges. A, EdU incorporation by HeLa clones without HU treatment (left) or after release from HU (right). Blue, DAPI; green, EdU. Scale bars are indicated. B, quantification of EdU incorporation as measured by fluorescence intensity of individual nuclei. The mean level of EdU incorporation was determined for each sample, and the data are expressed as the fractions of EdU incorporated by HU-treated cells relative to untreated cells of the same type (+HU/−HU). The (+HU/−HU) ratio for the shNT of each cell type was set at 1 (n = 3, means ± S.D.). C, quantification of anaphase bridges observed after release of HeLa shNT, shTEN1–5, or shTEN1–5R from nocodazole (mean of n = 2, error bars show maximum/minimum values). D, examples of an anaphase bridge without telomeric signal (left panel) and with telomeric signal (right panel) are shown. Blue, DAPI; red, Cy3-(C3TA2)3 probe. Scale bars are indicated. E, quantification of anaphase bridges with telomere signal by whole cell FISH (means ± S.D., n = 3).
STN1 and CTC1-depleted cells exhibit an increase in anaphase bridges in the absence of telomere fusions, suggesting that both proteins help resolve various genome-wide replication problems (Table 2 and Ref. 10). To examine whether TEN1 depletion also leads to anaphase bridges, we enriched for anaphase cells by treating HeLa shTEN1–5, shTEN1–5R, and shNT cells with nocadozole; the cells were then released into anaphase and fixed. DAPI staining revealed a ∼5-fold increase in anaphase bridges after TEN1 depletion (Fig. 6C). This was largely rescued by the shRNA-resistant allele. Interestingly, the increase in anaphase bridges in the shTEN1 cells was consistently higher than the 2–3-fold increase in STN1-depleted cells (Table 2 and Ref. 10), despite the cells exhibiting a similar decrease in replication restart. It also mirrored the enhanced anaphase bridge phenotype observed in Arabidopsis TEN1 mutants compared with STN1 or CTC1 mutants (44). To assess the extent to which the anaphase bridges might reflect failed sister chromatid separation because of incomplete telomere duplex replication, we performed FISH with a telomere probe to look for the presence of telomeric DNA within the bridges. Although ∼40% of the bridges from the shTEN1 cells contained telomere signals, the frequency was similar in the shNT control cells (Fig. 6, D and E). Moreover, in both cell types, the telomere signal appeared as discrete spots in a well stained DAPI bridge rather than the elongated signals that might be expected if the bridges were caused by replication failure in the telomere duplex and subsequent stretching of the intertwined sisters (55). Overall, our results indicate that TEN1 depletion causes genome-wide instability that leads to defects in chromosome segregation.
DISCUSSION
Here we present the first comprehensive study of human TEN1 and show that depletion of TEN1 leads to a variety of defects commonly associated with telomere duplex replication, telomeric C-strand fill-in, and genome-wide replication rescue. The overall similarity in phenotypes seen in TEN1-, STN1-, and CTC1-depleted cells supports the idea that these three proteins function together as a complex both at the telomere as a key component of the telomere replication machinery and elsewhere in the genome during situations of replication stress. However, there are some intriguing differences in the severity of the phenotypes seen after TEN1 depletion relative to STN1 or CTC1 depletion. Currently, it is not possible to tell whether these differences result from more efficient TEN1 knockdown leading to a lower overall level of CST complex formation or whether they reflect the presence of subcomplexes that are differentially affected by loss of TEN1. Nonetheless, the finding that TEN1 depletion preferentially enhances some phenotypes over others (e.g., SFE versus MTS) points to previously unanticipated differences in the underlying cause and hence in the sensitivity of these phenotypes to TEN1 loss.
The appearance of SFE and MTS after TEN1 depletion indicates that TEN1, like STN1, facilitates replication of the telomere duplex because both phenotypes are indicative of problems with replication fork progression (6, 8). Given that TEN1 depletion does not lead to a significant increase in T-circles, our results indicate that the SFE are unrelated to problems associated with T-loop resolution. Thus, it seems likely that SFE and MTS both reflect difficulties in replicating the repetitive G-rich telomeric sequence. However, our finding that TEN1 depletion results in SFE and MTS, whereas STN1 depletion largely causes MTS (Table 2 and Ref. 10), is striking and points to fundamental differences in the underlying cause of SFE and MTS that are disparate in sensitivity to TEN1/CST levels.
Our finding that TEN1 depletion also delays G-overhang shortening in late S/G2 indicates that TEN1 has an independent role in the final steps of telomere replication at the time of G-overhang maturation. We previously observed a similar delay in overhang shortening in STN1-depleted cells and showed that it is caused by a deficiency in C-strand fill-in. Thus, our current results indicate that TEN1 most likely cooperates with STN1 to facilitate C-strand fill-in by DNA pol α. As we see no change in the rate or extent of overhang elongation in early to mid-S phase, our results do not point to a role for TEN1 in direct regulation of either telomerase or nuclease activities.
Although there is strong evidence that budding yeast CST is responsible for positive and negative regulation of telomerase, our previous studies did not uncover a similar role for the human complex. Specifically, we demonstrated that depletion of STN1 in HeLa cells has negligible effect on overall telomere length or the number of repeats added by telomerase in a single cell cycle (12). However, a subsequent publication found that depletion of CTC1, STN1, or TEN1 causes telomere growth in HT1080 cells, and the authors proposed that human CST acts as a direct inhibitor of telomerase (38). Here we have explored this discrepancy by depleting TEN1 (or STN1) in a selection of cancer cells and then examining the effect on telomere length. Interestingly, we also see an increase in telomere length in HT1080 cells, but in other cell types telomere length is unaffected, or we observe an initial telomere shortening followed by length stabilization. Overall, our results indicate that CST levels may affect telomere length; however, the mechanism appears complex. One way to explain our findings would be if CST plays an indirect role in telomerase regulation, possibly via interactions with TPP1. CST, POT1, and telomerase can each interact with TPP1, suggesting that TPP1 may act as a hub that regulates access of these factors to the overhang. Thus, after CST depletion, cell type-specific variation in the levels of the remaining factors could tip the balance between whether TPP1 recruits telomerase for overhang elongation or POT1 for overhang sequestration.
The discovery that CST subunits have genome-wide roles in DNA replication is consistent with the original identification of CTC1 and STN1 as subunits of the pol α accessory factor AAF (31). Subsequent studies revealed that CTC1 or STN1 depletion/deletion causes hallmarks of general genome instability and revealed a role for STN1 in replication restart (10, 11, 29). Here we show that depletion of TEN1 causes nontelomeric phenotypes that are similar in nature to those seen after STN1 knockdown, suggesting that TEN1 was overlooked in the original AAF preparations because of its small size. Currently, our understanding of the genome-wide role of CST is very limited. The role of STN1 has been examined after HU-induced replication fork stalling, and in this situation, STN1 helps restart replication by promoting the firing of new origins. However, based on the multiple roles of CST at telomeres, it seems likely that CST or CST subunits will be used to solve a variety of replication problems throughout the genome, and it would not be surprising if the mechanism of action varies with genomic location and the type of defect. Our current studies support this concept as TEN1- and STN1-depleted cells show a similar replication restart deficiency, but TEN1 depletion leads to a higher frequency of anaphase bridges. This finding indicates that the process used to enhance restart of replication after HU release is most likely different from the defect that leads to anaphase bridges because the latter is more sensitive to TEN1 depletion. Thus, our results indicate that TEN1/CST is utilized to solve a wider than expected range of challenges to the replication machinery. Dissection of the differential sensitivity of these challenges to CTC1, STN1, or TEN1 loss will provide a firm basis for exploring the role of CST subunits in the resolution of these various replication problems.
Acknowledgments
We thank Jason Stewart and Mary Chaiken for intellectual contributions and Birgit Ehmer for assistance with cell sorting. We also thank Liana Oganesian and Jan Karlseder for advice and T-circle amplification protocol and Susan Waltz for the use of the cytofuge.
This work was supported, in whole or in part, by National Institutes of Health Grants GM041803 (to C. M. P.) and T32CA117846 and F32CA177120 (to C. K.).
C. Kasbek, F. Wang, J. A. Stewart, and C. M. Price, unpublished results.
- TRF
- telomeric restriction fragment
- CST
- CTC1-STN1-TEN1
- BrdC
- bromodeoxycytosine
- EdU
- ethynyl-deoxyuridine
- HU
- hydroxyurea
- MTS
- multiple telomere signal
- SFE
- signal free end
- pol α
- polymerase α
- AAF
- pol α accessory factor
- shNT
- nontargeting shRNA
- RT-qPCR
- quantitative real time PCR
- T-SCE
- telomere sister chromatid exchange
- FISH
- fluorescent in situ hybridization
- RT-qPCR
- reverse transcription and quantitative real time PCR.
REFERENCES
- 1. Zhao Y., Sfeir A. J., Zou Y., Buseman C. M., Chow T. T., Shay J. W., Wright W. E. (2009) Telomere extension occurs at most chromosome ends and is uncoupled from fill-in in human cancer cells. Cell 138, 463–475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Huffman K. E., Levene S. D., Tesmer V. M., Shay J. W., Wright W. E. (2000) Telomere shortening is proportional to the size of the G-rich telomeric 3′-overhang. J. Biol. Chem. 275, 19719–19722 [DOI] [PubMed] [Google Scholar]
- 3. Palm W., de Lange T. (2008) How shelterin protects mammalian telomeres. Annu. Rev. Genet. 42, 301–334 [DOI] [PubMed] [Google Scholar]
- 4. Griffith J. D., Comeau L., Rosenfield S., Stansel R. M., Bianchi A., Moss H., de Lange T. (1999) Mammalian telomeres end in a large duplex loop. Cell 97, 503–514 [DOI] [PubMed] [Google Scholar]
- 5. O'Sullivan R. J., Karlseder J. (2010) Telomeres. Protecting chromosomes against genome instability. Nat. Rev. Mol. Cell Biol. 11, 171–181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Stewart J. A., Chaiken M. F., Wang F., Price C. M. (2012) Maintaining the end. Roles of telomere proteins in end-protection, telomere replication and length regulation. Mutat. Res. 730, 12–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gilson E., Géli V. (2007) How telomeres are replicated. Nat. Rev. Mol. Cell Biol. 8, 825–838 [DOI] [PubMed] [Google Scholar]
- 8. Sfeir A., Kosiyatrakul S. T., Hockemeyer D., MacRae S. L., Karlseder J., Schildkraut C. L., de Lange T. (2009) Mammalian telomeres resemble fragile sites and require TRF1 for efficient replication. Cell 138, 90–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ding H., Schertzer M., Wu X., Gertsenstein M., Selig S., Kammori M., Pourvali R., Poon S., Vulto I., Chavez E., Tam P. P., Nagy A., Lansdorp P. M. (2004) Regulation of murine telomere length by Rtel. A essential gene encoding a helicase-like protein. Cell 117, 873–886 [DOI] [PubMed] [Google Scholar]
- 10. Stewart J. A., Wang F., Chaiken M. F., Kasbek C., Chastain P. D., 2nd, Wright W. E., Price C. M. (2012) Human CST promotes telomere duplex replication and general replication restart after fork stalling. EMBO J. 31, 3537–3549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gu P., Min J. N., Wang Y., Huang C., Peng T., Chai W., Chang S. (2012) CTC1 deletion results in defective telomere replication, leading to catastrophic telomere loss and stem cell exhaustion. EMBO J. 31, 2309–2321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wang F., Stewart J. A., Kasbek C., Zhao Y., Wright W. E., Price C. M. (2012) Human CST has independent functions during telomere duplex replication and C-strand fill-in. Cell Reports 2, 1096–1103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Crabbe L., Verdun R. E., Haggblom C. I., Karlseder J. (2004) Defective telomere lagging strand synthesis in cells lacking WRN helicase activity. Science 306, 1951–1953 [DOI] [PubMed] [Google Scholar]
- 14. Huang C., Dai X., Chai W. (2012) Human Stn1 protects telomere integrity by promoting efficient lagging-strand synthesis at telomeres and mediating C-strand fill-in. Cell Res. 22, 1681–1695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bianchi A., Shore D. (2008) How telomerase reaches its end. Mechanism of telomerase regulation by the telomeric complex. Mol. Cell 31, 153–165 [DOI] [PubMed] [Google Scholar]
- 16. Price C. M., Boltz K. A., Chaiken M. F., Stewart J. A., Beilstein M. A., Shippen D. E. (2010) Evolution of CST function in telomere maintenance. Cell Cycle 9, 3157–3165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chandra A., Hughes T. R., Nugent C. I., Lundblad V. (2001) Cdc13 both positively and negatively regulates telomere replication. Genes Dev. 15, 404–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Evans S. K., Lundblad V. (1999) Est1 and Cdc13 as comediators of telomerase access. Science 286, 117–120 [DOI] [PubMed] [Google Scholar]
- 19. Li S., Makovets S., Matsuguchi T., Blethrow J. D., Shokat K. M., Blackburn E. H. (2009) Cdk1-dependent phosphorylation of Cdc13 coordinates telomere elongation during cell-cycle progression. Cell 136, 50–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Taggart A. K., Teng S. C., Zakian V. A. (2002) Est1p as a cell cycle-regulated activator of telomere-bound telomerase. Science 297, 1023–1026 [DOI] [PubMed] [Google Scholar]
- 21. Puglisi A., Bianchi A., Lemmens L., Damay P., Shore D. (2008) Distinct roles for yeast Stn1 in telomere capping and telomerase inhibition. EMBO J. 27, 2328–2339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Qi H., Zakian V. A. (2000) The Saccharomyces telomere-binding protein Cdc13p interacts with both the catalytic subunit of DNA polymerase α and the telomerase-associated est1 protein. Genes Dev. 14, 1777–1788 [PMC free article] [PubMed] [Google Scholar]
- 23. Gao H., Cervantes R. B., Mandell E. K., Otero J. H., Lundblad V. (2007) RPA-like proteins mediate yeast telomere function. Nat. Struct. Mol. Biol. 14, 208–214 [DOI] [PubMed] [Google Scholar]
- 24. Sun J., Yu E. Y., Yang Y., Confer L. A., Sun S. H., Wan K., Lue N. F., Lei M. (2009) Stn1-Ten1 is an Rpa2-Rpa3-like complex at telomeres. Genes Dev. 23, 2900–2914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Miyake Y., Nakamura M., Nabetani A., Shimamura S., Tamura M., Yonehara S., Saito M., Ishikawa F. (2009) RPA-like mammalian Ctc1-Stn1-Ten1 complex binds to single-stranded DNA and protects telomeres independently of the Pot1 pathway. Mol. Cell 36, 193–206 [DOI] [PubMed] [Google Scholar]
- 26. Gelinas A. D., Paschini M., Reyes F. E., Héroux A., Batey R. T., Lundblad V., Wuttke D. S. (2009) Telomere capping proteins are structurally related to RPA with an additional telomere-specific domain. Proc. Natl. Acad. Sci. U.S.A. 106, 19298–19303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bryan C., Rice C., Harkisheimer M., Schultz D. C., Skordalakes E. (2013) Structure of the human telomeric Stn1-Ten1 capping complex. PLoS One 8, e66756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mitton-Fry R. M., Anderson E. M., Theobald D. L., Glustrom L. W., Wuttke D. S. (2004) Structural basis for telomeric single-stranded DNA recognition by yeast Cdc13. J. Mol. Biol. 338, 241–255 [DOI] [PubMed] [Google Scholar]
- 29. Surovtseva Y. V., Churikov D., Boltz K. A., Song X., Lamb J. C., Warrington R., Leehy K., Heacock M., Price C. M., Shippen D. E. (2009) Conserved telomere maintenance component 1 interacts with STN1 and maintains chromosome ends in higher eukaryotes. Mol. Cell 36, 207–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wu P., Takai H., de Lange T. (2012) Telomeric 3′ overhangs derive from resection by Exo1 and Apollo and fill-In by POT1b-associated CST. Cell 150, 39–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Goulian M., Heard C. J., Grimm S. L. (1990) Purification and properties of an accessory protein for DNA polymerase α/primase. J. Biol. Chem. 265, 13221–13230 [PubMed] [Google Scholar]
- 32. Casteel D. E., Zhuang S., Zeng Y., Perrino F. W., Boss G. R., Goulian M., Pilz R. B. (2009) A DNA polymerase-α·primase cofactor with homology to replication protein A-32 regulates DNA replication in mammalian cells. J. Biol. Chem. 284, 5807–5818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Anderson B. H., Kasher P. R., Mayer J., Szynkiewicz M., Jenkinson E. M., Bhaskar S. S., Urquhart J. E., Daly S. B., Dickerson J. E., O'Sullivan J., Leibundgut E. O., Muter J., Abdel-Salem G. M., Babul-Hirji R., Baxter P., Berger A., Bonafé L., Brunstom-Hernandez J. E., Buckard J. A., Chitayat D., Chong W. K., Cordelli D. M., Ferreira P., Fluss J., Forrest E. H., Franzoni E., Garone C., Hammans S. R., Houge G., Hughes I., Jacquemont S., Jeannet P. Y., Jefferson R. J., Kumar R., Kutschke G., Lundberg S., Lourenço C. M., Mehta R., Naidu S., Nischal K. K., Nunes L., Ounap K., Philippart M., Prabhakar P., Risen S. R., Schiffmann R., Soh C., Stephenson J. B., Stewart H., Stone J., Tolmie J. L., van der Knaap M. S., Vieira J. P., Vilain C. N., Wakeling E. L., Wermenbol V., Whitney A., Lovell S. C., Meyer S., Livingston J. H., Baerlocher G. M., Black G. C., Rice G. I., Crow Y. J. (2012) Mutations in CTC1, encoding conserved telomere maintenance component 1, cause Coats plus. Nat. Genet. 44, 338–342 [DOI] [PubMed] [Google Scholar]
- 34. Keller R. B., Gagne K. E., Usmani G. N., Asdourian G. K., Williams D. A., Hofmann I., Agarwal S. (2012) CTC1 Mutations in a patient with dyskeratosis congenita. Pediatr. Blood Cancer 59, 311–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Polvi A., Linnankivi T., Kivelä T., Herva R., Keating J. P., Mäkitie O., Pareyson D., Vainionpää L., Lahtinen J., Hovatta I., Pihko H., Lehesjoki A. E. (2012) Mutations in CTC1, encoding the CTS telomere maintenance complex component 1, cause cerebroretinal microangiopathy with calcifications and cysts. Am. J. Hum. Genet. 90, 540–549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Levy D., Neuhausen S. L., Hunt S. C., Kimura M., Hwang S. J., Chen W., Bis J. C., Fitzpatrick A. L., Smith E., Johnson A. D., Gardner J. P., Srinivasan S. R., Schork N., Rotter J. I., Herbig U., Psaty B. M., Sastrasinh M., Murray S. S., Vasan R. S., Province M. A., Glazer N. L., Lu X., Cao X., Kronmal R., Mangino M., Soranzo N., Spector T. D., Berenson G. S., Aviv A. (2010) Genome-wide association identifies OBFC1 as a locus involved in human leukocyte telomere biology. Proc. Natl. Acad. Sci. U.S.A. 107, 9293–9298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Codd V., Nelson C. P., Albrecht E., Mangino M., Deelen J., Buxton J. L., Hottenga J. J., Fischer K., Esko T., Surakka I., Broer L., Nyholt D. R., Mateo Leach I., Salo P., Hägg S., Matthews M. K., Palmen J., Norata G. D., O'Reilly P. F., Saleheen D., Amin N., Balmforth A. J., Beekman M., de Boer R. A., Böhringer S., Braund P. S., Burton P. R., de Craen A. J., Denniff M., Dong Y., Douroudis K., Dubinina E., Eriksson J. G., Garlaschelli K., Guo D., Hartikainen A. L., Henders A. K., Houwing-Duistermaat J. J., Kananen L., Karssen L. C., Kettunen J., Klopp N., Lagou V., van Leeuwen E. M., Madden P. A., Mägi R., Magnusson P. K., Männistö S., McCarthy M. I., Medland S. E., Mihailov E., Montgomery G. W., Oostra B. A., Palotie A., Peters A., Pollard H., Pouta A., Prokopenko I., Ripatti S., Salomaa V., Suchiman H. E., Valdes A. M., Verweij N., Vinuela A., Wang X., Wichmann H. E., Widen E., Willemsen G., Wright M. J., Xia K., Xiao X., van Veldhuisen D. J., Catapano A. L., Tobin M. D., Hall A. S., Blakemore A. I., van Gilst W. H., Zhu H., Consortium C., Erdmann J., Reilly M. P., Kathiresan S., Schunkert H., Talmud P. J., Pedersen N. L., Perola M., Ouwehand W., Kaprio J., Martin N. G., van Duijn C. M., Hovatta I., Gieger C., Metspalu A., Boomsma D. I., Jarvelin M. R., Slagboom P. E., Thompson J. R., Spector T. D., van der Harst P., Samani N. J. (2013) Identification of seven loci affecting mean telomere length and their association with disease. Nat. Genet. 45, 422–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Chen L. Y., Redon S., Lingner J. (2012) The human CST complex is a terminator of telomerase activity. Nature 488, 540–544 [DOI] [PubMed] [Google Scholar]
- 39. Smolka M. B., Albuquerque C. P., Chen S. H., Zhou H. (2007) Proteome-wide identification of in vivo targets of DNA damage checkpoint kinases. Proc. Natl. Acad. Sci. U.S.A. 104, 10364–10369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Tseng S. F., Shen Z. J., Tsai H. J., Lin Y. H., Teng S. C. (2009) Rapid Cdc13 turnover and telomere length homeostasis are controlled by Cdk1-mediated phosphorylation of Cdc13. Nucleic Acids Res. 37, 3602–3611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. DeZwaan D. C., Toogun O. A., Echtenkamp F. J., Freeman B. C. (2009) The Hsp82 molecular chaperone promotes a switch between unextendable and extendable telomere states. Nat. Struct. Mol. Biol. 16, 711–716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Teixeira M. T., Arneric M., Sperisen P., Lingner J. (2004) Telomere length homeostasis is achieved via a switch between telomerase-extendible and -nonextendible states. Cell 117, 323–335 [DOI] [PubMed] [Google Scholar]
- 43. Petreaca R. C., Chiu H. C., Eckelhoefer H. A., Chuang C., Xu L., Nugent C. I. (2006) Chromosome end protection plasticity revealed by Stn1p and Ten1p bypass of Cdc13p. Nat. Cell Biol. 8, 748–755 [DOI] [PubMed] [Google Scholar]
- 44. Leehy K. A., Lee J. R., Song X., Renfrew K. B., Shippen D. E. (2013) MERISTEM DISORGANIZATION1 encodes TEN1, an essential telomere protein that modulates telomerase processivity in Arabidopsis. Plant Cell 25, 1343–1354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Song X., Leehy K., Warrington R. T., Lamb J. C., Surovtseva Y. V., Shippen D. E. (2008) STN1 protects chromosome ends in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 105, 19815–19820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Harley C. B., Futcher A. B., Greider C. W. (1990) Telomeres shorten during ageing of human fibroblasts. Nature 345, 458–460 [DOI] [PubMed] [Google Scholar]
- 47. Celli G. B., Denchi E. L., de Lange T. (2006) Ku70 stimulates fusion of dysfunctional telomeres yet protects chromosome ends from homologous recombination. Nat. Cell Biol. 8, 885–890 [DOI] [PubMed] [Google Scholar]
- 48. Pickett H. A., Cesare A. J., Johnston R. L., Neumann A. A., Reddel R. R. (2009) Control of telomere length by a trimming mechanism that involves generation of t-circles. EMBO J. 28, 799–809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Bailey S. M., Williams E. S., Cornforth M. N., Goodwin E. H. (2010) Chromosome Orientation fluorescence in situ hybridization or strand-specific FISH. Methods Mol. Biol. 659, 173–183 [DOI] [PubMed] [Google Scholar]
- 50. Gu P., Chang S. (2013) Functional characterization of human CTC1 mutations reveals novel mechanisms responsible for the pathogenesis of the telomere disease Coats plus. Aging Cell 10.1111/acel.12139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Vannier J. B., Pavicic-Kaltenbrunner V., Petalcorin M. I., Ding H., Boulton S. J. (2012) RTEL1 dismantles T loops and counteracts telomeric G4-DNA to maintain telomere integrity. Cell 149, 795–806 [DOI] [PubMed] [Google Scholar]
- 52. Li B., Jog S. P., Reddy S., Comai L. (2008) WRN controls formation of extrachromosomal telomeric circles and is required for TRF2DeltaB-mediated telomere shortening. Mol. Cell Biol. 28, 1892–1904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Zellinger B., Akimcheva S., Puizina J., Schirato M., Riha K. (2007) Ku suppresses formation of telomeric circles and alternative telomere lengthening in Arabidopsis. Mol. Cell 27, 163–169 [DOI] [PubMed] [Google Scholar]
- 54. Oganesian L., Karlseder J. (2013) 5′ C-rich telomeric overhangs are an outcome of rapid telomere truncation events. DNA Repair 12, 238–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Takai K. K., Kibe T., Donigian J. R., Frescas D., de Lange T. (2011) Telomere protection by TPP1/POT1 requires tethering to TIN2. Mol. Cell 44, 647–659 [DOI] [PMC free article] [PubMed] [Google Scholar]