Background: Vesiculation of tubular recycling endosomes is essential for the recycling of receptors and lipids to the plasma membrane.
Results: A novel vesiculation assay was used to demonstrate a role for endocytic regulatory proteins in vesiculation.
Conclusion: EHD family proteins play significant roles in both the vesiculation and generation of tubular recycling endosomes.
Significance: This provides the first direct evidence of differential EHD function in vesiculation and tubulation.
Keywords: Cell Biology, Endosomes, Liposomes, Membrane Biogenesis, Membrane Recycling, Membrane Trafficking, Protein Sorting, Receptor Recycling, Subcellular Organelles, Vesicles
Abstract
Endocytic recycling involves the return of membranes and receptors to the plasma membrane following their internalization into the cell. Recycling generally occurs from a series of vesicular and tubular membranes localized to the perinuclear region, collectively known as the endocytic recycling compartment. Within this compartment, receptors are sorted into tubular extensions that later undergo vesiculation, allowing transport vesicles to move along microtubules and return to the cell surface where they ultimately undergo fusion with the plasma membrane. Recent studies have led to the hypothesis that the C-terminal Eps15 homology domain (EHD) ATPase proteins are involved in the vesiculation process. Here, we address the functional roles of the four EHD proteins. We developed a novel semipermeabilized cell system in which addition of purified EHD proteins to reconstitute vesiculation allows us to assess the ability of each protein to vesiculate MICAL-L1-decorated tubular recycling endosomes (TREs). Using this assay, we show that EHD1 vesiculates membranes, consistent with enhanced TRE generation observed upon EHD1 depletion. EHD4 serves a role similar to that of EHD1 in TRE vesiculation, whereas EHD2, despite being capable of vesiculating TREs in the semipermeabilized cells, fails to do so in vivo. Surprisingly, the addition of EHD3 causes tubulation of endocytic membranes in our semipermeabilized cell system, consistent with the lack of tubulation observed upon EHD3 depletion. Our novel vesiculation assay and in vitro electron microscopy analysis, combined with in vivo data, provide evidence that the functions of both EHD1 and EHD4 are primarily in TRE membrane vesiculation, whereas EHD3 is a membrane-tubulating protein.
Introduction
Endocytic recycling is the process of returning membranes and receptors to the plasma membrane following their internalization into the cell. Regulated recycling generally occurs from a series of vesicular and tubular membranes localized to the perinuclear region, collectively known as the endocytic recycling compartment (1). Current models hold that the generation of tubular membranes that extend from this subcellular region facilitate the sorting and trafficking of cargo (2). After sorting, vesiculation takes place, and vesicles containing lipids and receptors are transported along microtubules and returned to the cell surface where they ultimately undergo fusion with the plasma membrane.
Recycling is a tightly regulated event, controlled by a variety of proteins that include small GTPases, their effectors, motor proteins, microtubules, SNARE proteins, and the C-terminal Eps15 homology domain (EHD)3 ATPase protein, EHD1. Indeed, EHD1 and its three mammalian orthologs have all been implicated in regulating various endocytic transport events. EHD1 regulates the exit of proteins from the endocytic recycling compartment to the plasma membrane, whereas EHD3 and EHD4 play roles in transport at the early/sorting endosomes (3). EHD2 is involved in the control of caveolar structures underneath the plasma membrane (4–6).
There have been conflicting reports on the mechanistic roles of EHD proteins. Based on their structure and in vitro studies, EHD proteins can induce membrane bending and tubulation (7, 8). However, a recent study indicates that the EHD1-interaction partners, MICAL-L1 and particularly the F-BAR-domain-containing protein, Syndapin2, are essential proteins responsible for tubule biogenesis in vivo (9). Studies support a major role for EHD1 in membrane vesiculation rather than tubulation (10, 11). However, due to the availability of limited assays to assess vesiculation, the precise functions of these four proteins remain poorly understood.
We now address the physiological roles of all four EHD proteins. Because the existing vesiculation assays are based on in vitro systems and/or complex biophysical measurements (12–14), we have developed a novel semipermeabilized cell system using purified proteins to assess the ability of each of the proteins to vesiculate MICAL-L1-decorated tubular recycling endosomes (TREs). Using this assay along with an in vitro vesiculation assay, we show that EHD1 vesiculates membranes, a role consistent with the enhanced TRE networks observed upon EHD1 depletion. Experiments with EHD4 indicate that this protein plays a role similar to that of EHD1. EHD2, despite being capable of vesiculating TREs in the semipermeabilized cell system, does not induce vesiculation in vivo. Surprisingly, EHD3, the closest paralog to EHD1, plays a role in membrane tubulation, as observed both in our semipermeabilized cell system and in vivo, and fails to vesiculate membranes in vitro. Our data provide evidence that both EHD1 and EHD4 function primarily in TRE membrane vesiculation, whereas EHD3 is a membrane-tubulating protein.
EXPERIMENTAL PROCEDURES
Gene Knockdown by Silencing RNA (siRNA)
Oligonucleotide duplexes targeting EHD1 (15), EHD3 (16), and ON-TARGETplus SMARTpool siRNA targeting EHD2 and EHD4 (Dharmacon) were transfected into HeLa cells for 72 h using Oligofectamine (Invitrogen) as described previously (11).
Antibodies and Reagents
Polyclonal rabbit anti-EHD1, -EHD2, -EHD3, and -EHD4 peptide antibodies were described previously (17). Mouse anti-CD59 antibody was kindly provided by Dr. V. Horejsi and described (11, 18). Commercial antibodies and reagents used in this study: mouse anti-MICAL-L1 and mouse anti-actin (Novus Biologicals), rat anti-HSP70 (Stressgen), rabbit anti-Syndapin2 (Abgent), mouse anti-GAPDH (Protein Tech), goat anti-mouse HRP and goat anti-rat HRP (Jackson ImmunoResearch Laboratories, Inc.), and donkey anti-rabbit HRP (GE Healthcare). Alexa Fluor 568 goat anti-mouse, and Alexa Fluor 568 goat anti-rabbit were purchased from Invitrogen. Digitonin, ATP, creatine phosphate, and creatine phosphokinase were from Sigma.
Immunofluorescence and Uptake Assays
HeLa cells were grown on coverslips, transfected with EHD-siRNAs, and fixed with 4% paraformaldehyde (v/v). Immunostaining was done with anti-MICAL-L1 or anti-Syndapin2 antibodies, followed by appropriate fluorochrome-conjugated secondary antibodies. CD59 and 568-transferrin uptake was performed as described (11). Images were obtained using an LSM5 Pascal confocal microscope (Carl Zeiss) with a 63 × 1.4 numerical aperture objective.
Constructs, Protein Purification, and Circular Dichroism
GST-EH1 was cloned and purified as described previously (9). Pcold-GST was a generous gift from Chojiro Kojima (Osaka, Japan), and all four EHD proteins were cloned into this vector. Pcold-GST-EHDs were purified as described (19), and the purified proteins underwent circular dichroism studies as described previously (9).
Semipermeabilized Cell System Vesiculation Assay
This assay was modified from the methods described by the Balch (20) and Aridor (21) laboratories. HeLa cells were grown on coverslips and at 80–90% confluence, and cytosol was extracted with 20 μg/ml digitonin for 40 s at room temperature. After three washes in KHM buffer containing 110 mm KOAc, 20 mm HEPES (pH 7.2), and 2 mm MgOAc, semipermeabilized cells were incubated with GST proteins in the presence of 25 mm HEPES, 115 mm KCl, 2.5 mm MgCl2, 1 mm ATP, 5 mm creatine phosphate and 0.2 IU of creatine phosphokinase for 30 min at 37 °C. Cells were then washed three times in KHM buffer, fixed, and stained with anti-MICAL-L1 antibody.
Quantification of MICAL-L1 Tubules by ImageJ
Using ImageJ software, the image background was reduced by adjusting the threshold. The particle size was set between 2 μm2 and 150 μm2. All MICAL-L1-containing particles in this range were counted. Ten fields of images from each treatment were analyzed.
Liposome Preparation
Liposomes were prepared by mixing PC (1,2-dioleoyl-sn-glycero-3-phosphocholine), PS (1,2,-dioleoyl-sn-glycero-3-phosphoserine), and PI(4,5)P2 (phosphatidylinositol 4,5-bisphosphate) at an 80/10/10 molar ratio or 100% PC and dried under nitrogen. Lipids were suspended in blocking buffer (176 mm HEPES, 100 mm NaCl, 1 mm EGTA, 1 mm DTT (pH 7)) and subjected to five freeze/thaw cycles on dry ice followed by 1-min sonication at 21 °C. Lipids were then passed through a mini-extruder with 0.4-μm pore filters. Lipids and supplies were from Avanti Polar Lipids.
Liposome Vesiculation Assays and Electron Microscopy
100-μl reactions were mixed with 25 μg/ml liposomes, 2.5 μm total protein, and 1 mm ATPγS and incubated at 37 °C for 15 min. Two 300-mesh copper EM grids coated with formvar and carbon (EMS) were incubated for 15 min with half of the reaction mixture and negatively stained with 2% phosphotungstic acid (aq) three times for 4 s. Samples were visualized using a JEM 1011 transmission electron microscope (JEOL) operated at 100 kV. Images were captured by ORIUS 835.10W CCD camera (Gatan, Inc.).
RESULTS
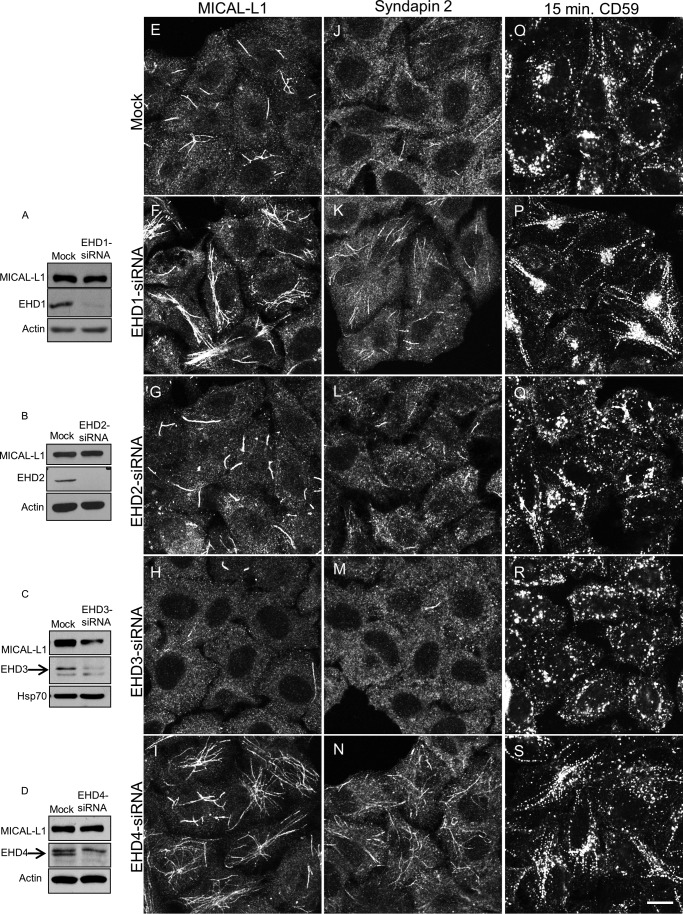
To address the roles of the four human EHD proteins in vivo on TREs, we first used siRNA to knock down their expression. As depicted in Fig. 1, we successfully depleted EHD1 (Fig. 1A), EHD2 (Fig. 1B), EHD3 (Fig. 1C), and EHD4 (Fig. 1D) from cells. MICAL-L1 protein levels remained unaffected upon EHD depletions, except in EHD3-depleted cells where a consistent decrease in expression was observed (Fig. 1C).
FIGURE 1.
Effects of depleting EHD proteins on TREs. HeLa cells were either mock-treated (A–D), treated with EHD1-siRNA (A), EHD2-siRNA (B), EHD3-siRNA (C), or EHD4-siRNA (D) for 72 h. Lysates separated by SDS-PAGE were immunoblotted. Actin (A, B, and D) or HSP70 (C) were used as loading control. Micrographs show representative fields of cells displaying MICAL-L1-containing TREs (E–I), Syndapin2-containing TREs (J–N), or CD59 following 15-min internalization (O–S). Scale bar, 10 μm.
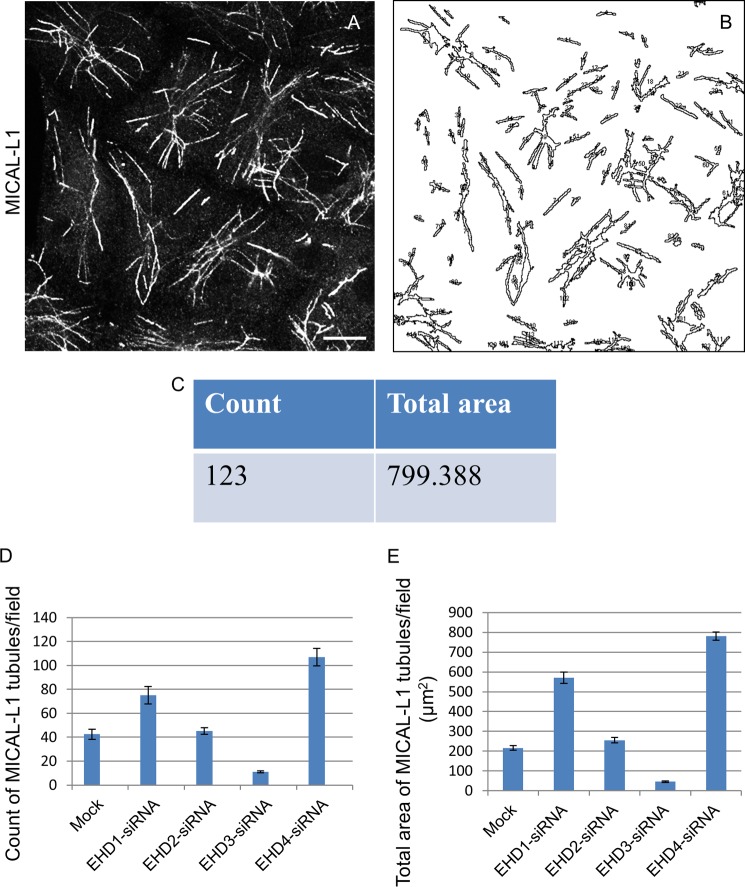
Under these conditions, EHD1 depletion caused an increase in the overall elaborateness, total number, and the total area of MICAL-L1-decorated TREs compared with mock-treated cells (compare Fig. 1, F with E). Quantification of total tubule number and total MICAL-L1-containing tubular area was calculated using ImageJ software (example shown in Fig. 2, A–C) and applied to cells depleted of each EHD protein (quantified in Fig. 2, D and E). EHD2 depletion had little effect on TREs (compare Fig. 1, G and E; see Fig. 2, D and E, for quantification). Surprisingly, EHD3 depletion greatly reduced the number and area of TREs (Fig. 1H; see Fig. 2, D and E, for quantification). EHD4 depletion had an effect similar to that of EHD1 (Fig. 1I; see Fig. 2, D and E, for quantification). Similar results were observed for an additional TRE marker protein, Syndapin2 (Ref. 9 and Fig. 1, J–N). These data support the notion that both EHD1 and EHD4 are involved in the vesiculation of TREs in vivo, whereas EHD3 tubulates recycling endosomes.
FIGURE 2.
Quantification of MICAL-L1-containing TREs upon EHD depletion. ImageJ software was used to analyze fields of cells with MICAL-L1-containing TREs (A–C). The software calculates the number and total mean area of the TRE per micrograph (C). The number of MICAL-L1 tubules (D) or the total area of MICAL-L1 tubules (E) per field upon the depletion of EHD proteins was quantified by ImageJ. Quantification was done from three independent experiments. S.E. (error bars) is shown. Scale bar, 10 μm.
We next tested whether an actual cargo protein that is transported along TREs, CD59 (18), is similarly affected. In mock-treated cells, internalized CD59 partially localized to TREs (Fig. 1O). However, EHD1 depletion induced a dramatic increase in the localization of CD59 to TREs (Fig. 1P). Similar to our results assessing MICAL-L1-decorated TREs, this increase was mirrored by EHD4 depletion (Fig. 1S). Again, EHD2 depletion had little impact on CD59 localization (Fig. 1O), whereas EHD3 depletion, consistent with its effect on MICAL-L1-containing TREs, enhanced vesiculation (Fig. 1R).
We reasoned that if EHD1 depletion indeed led to an enhancement in TREs, then the re-expression of exogenous EHD1 in depleted cells should lead to vesiculation and reduced TREs. As shown in Fig. 3, A and B, upon depletion of EHD1, cells transfected with a siRNA-resistant EHD1 plasmid (Res-GFP-Myc-EHD1; cells within orange boundary) displayed decreased levels of TRE content compared with nontransfected cells. Similar results were observed for EHD4 (Fig. 3, E and F; see cells within orange boundary). On the other hand, when a siRNA-resistant EHD3 plasmid was introduced into EHD3-depleted cells (where no TREs were observed), this induced TRE tubulation (Fig. 3, C and D; see cells within orange boundary).
FIGURE 3.

Rescue experiments for EHD depletions. HeLa cells were treated with either EHD1-siRNA (A and B), EHD3-siRNA (C and D), or EHD4-siRNA (E and F) for 72 h. During the final 24 h, siRNA-resistant EHD1 (Res-GFP-Myc-EHD1) (A), Res-GFP-Myc-EHD3 (C), and Res-HA-EHD4 (E) were transfected into the siRNA-treated cells. Cells were then fixed and stained for MICAL-L1. Transfected cells are bordered by an orange line. Scale bar, 10 μm.
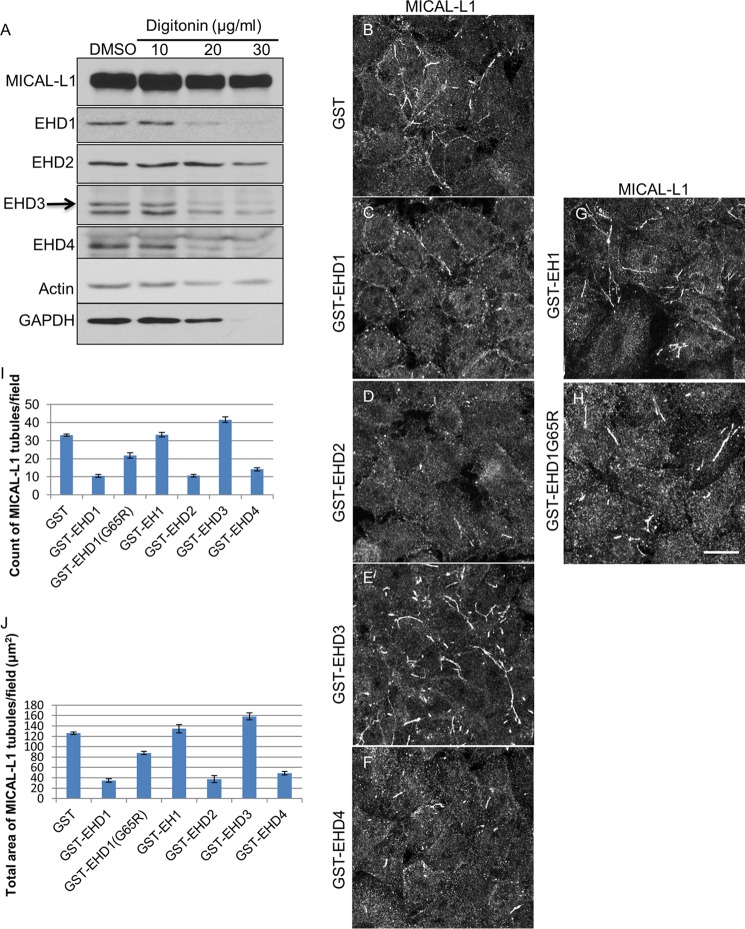
To address definitely the function of the four individual EHD proteins in TRE vesiculation, we first purified GST fusion proteins for all four EHDs and the inactive EHD1-G65R mutant and EH domain only of EHD1 (EH1) (Fig. 4A). Circular dichroism was used to confirm the folding and secondary structure of the purified proteins (Fig. 4B). We then developed a novel semipermeabilized cell system to measure vesiculation. We extracted cytosolic proteins and proteins that were not tightly associated with TRE or the cytoskeleton from live cells. Increasing concentrations of digitonin led to the extraction of EHD1, EHD3, EHD4, and GAPDH (positive control) with a modest impact on actin levels (Fig. 5A). EHD2 was also partially extracted, but remained more resistant than the other EHD proteins, consistent with its association with caveolae (4–6, 22). Only a modest decrease was noted in the level of MICAL-L1, likely due to its tight association with phosphatidic acid on TRE (9).
FIGURE 4.
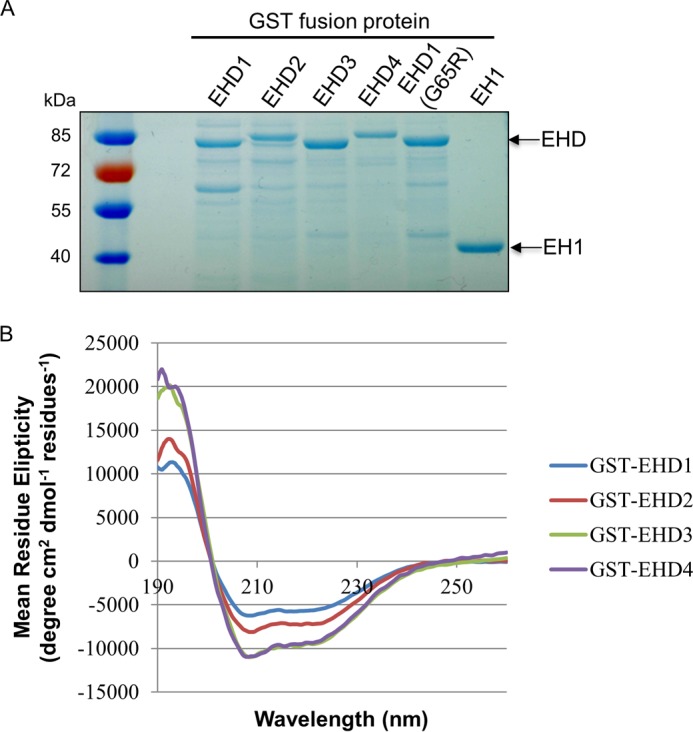
GST fusion protein purification and the secondary structure of purified EHD proteins. A, the purified GST fusion proteins were detected by SDS-PAGE and Coomassie Blue staining. B, the folding of purified EHD proteins was analyzed by circular dichroism.
FIGURE 5.
Semipermeabilized cell system for measuring vesiculation and tubulation of TRE. A, cells were either treated with dimethyl sulfoxide (DMSO) or with 10, 20, or 30 μg/ml digitonin for 40 s. The protein levels of MICAL-L1, EHD1, EHD2, EHD3, EHD4, actin, and GAPDH were detected by immunoblotting. B–H, after permeabilization with 20 μg/ml digitonin, cells were incubated with either GST alone (B), GST-EHD1 (C), GST-EHD2 (D), GST-EHD3 (E), GST-EHD4 (F), GST-EH1 (G), or GST-EHD1G65R (H) for 30 min at 37 °C. The status of TRE (as depicted by endogenous MICAL-L1) was observed under these conditions. I and J, the number of MICAL-L1 tubules (I) and the total area of MICAL-L1 tubules (J) per field were quantified from three independent experiments using ImageJ. Error bars denote S.E. Scale bar, 10 μm.
Semipermeabilized cells on coverslips were incubated with GST (Fig. 5B, negative control), GST bound to the EH1 (Fig. 5G, GST-EH1, negative control), the GST-EHD1 G65R ATP-binding mutant (Fig. 5H, negative control), GST-EHD1 (Fig. 5C), GST-EHD2 (Fig. 5D), GST-EHD3 (Fig. 5E), or GST-EHD4 (Fig. 5F). Little change was observed in TREs with the addition of GST-EH1 (Fig. 5G) compared with GST alone (Fig. 5B). GST-EHD1 G65R showed a modest reduction in TREs, likely due to residual EHD1 activity (Fig. 5H). On the other hand, addition of purified wild-type EHD1 caused a significant reduction in the number and total area of TREs, indicating that EHD1 serves as a TRE vesiculator (Fig. 5C and quantified in I and J). Similarly, EHD2 was capable of vesiculation (Fig. 5D and quantified in I and J). On the other hand, the addition of EHD3 to semipermeabilized cells induced TRE formation, reinforcing the idea that unlike EHD1, EHD3 plays a role in the generation of tubules (Fig. 5E and quantified in I and J). Finally, the addition of EHD4 also induced vesiculation of TREs, similar to EHD1, and similar to the role observed for EHD4 and EHD1 in vivo (Fig. 5F and quantified in I and J).
To further characterize the distinct roles for EHD proteins in membrane modeling using our semipermeabilized cell system, we measured the number of MICAL-L1-containing TREs per field (Fig. 6A) and the total area of MICAL-L1-containing TREs per field (Fig. 6B) with increasing concentrations of EHD proteins. As demonstrated, whereas the addition of GST alone had no impact on MICAL-L1-containing TREs, both EHD1 and EHD4 induced vesiculation in a dose-dependent manner (Fig. 6, A and B). On the other hand, EHD3 increased tubulation in a dose-dependent manner (Fig. 6, A and B), further highlighting the functional differences between these proteins.
FIGURE 6.
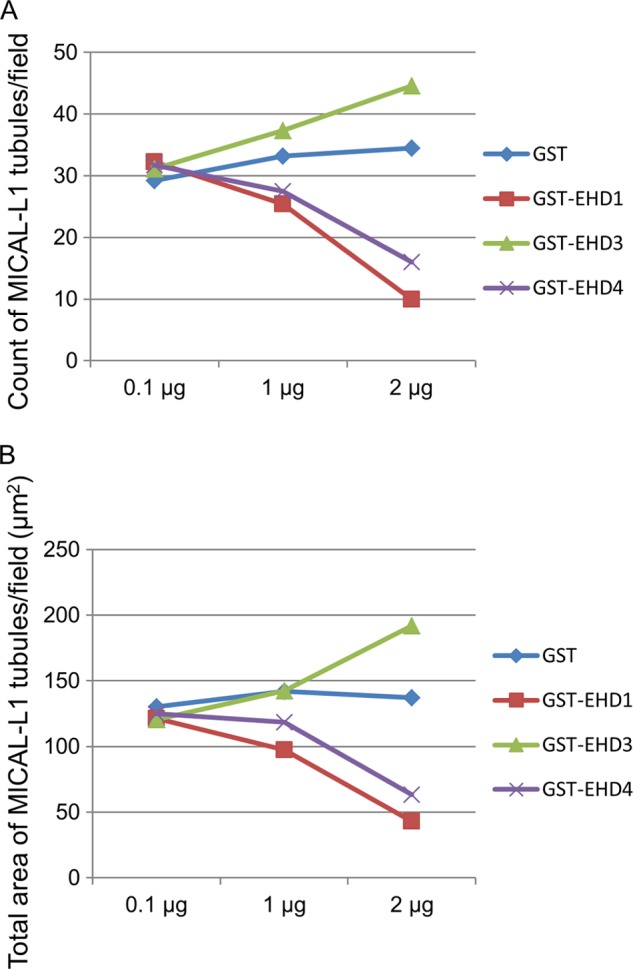
Dose-dependent vesiculation and tubulation of TREs by EHD proteins. 0.1, 1, and 2 μg of either GST alone, GST-EHD1, GST-EHD2, GST-EHD3, or GST-EHD4 was used in the semipermeabilized vesiculation assay. MICAL-L1-containing TREs per field (A) or total area of MICAL-L1-containing TREs were analyzed (B) by ImageJ and plotted.
Given the surprising differences in their effects on membranes both in vivo and in our semipermeabilized cell system, we further addressed the functions of EHD1 and EHD3 in an in vitro vesiculation assay (Fig. 7). As demonstrated in the representative micrographs, addition of EHD1 to PC/PS/PIP2 liposomes in the presence of ATP induced membrane vesiculation (Fig. 7B), whereas liposomes remained unaffected by the addition of EHD1 without ATP (Fig. 7A). Incubation of liposomes in the presence of EHD1 and the nonhydrolyzable ATP analog, ATPγS, induced vesiculation similar to that seen with ATP, suggesting that under these specific in vitro conditions, ATP binding (but not hydrolysis) is sufficient for vesiculation. Indeed, incubation of liposomes in the presence of ATP and EHD1 G65R (a mutant predicted to have defective ATP binding) displayed impaired vesiculation compared with wild-type EHD1 (Fig. 7D). Quantification of these data demonstrated that the mean liposome diameter was ∼280 nm, and this decreased to ∼150 nm in the presence of either EHD1 and ATP or EHD1 and ATPγS (Fig. 8A). On the other hand, incubation of the EHD1 G65R mutant and ATP with liposomes had little effect on the mean liposome diameter (∼250 nm; Fig. 8A).
FIGURE 7.
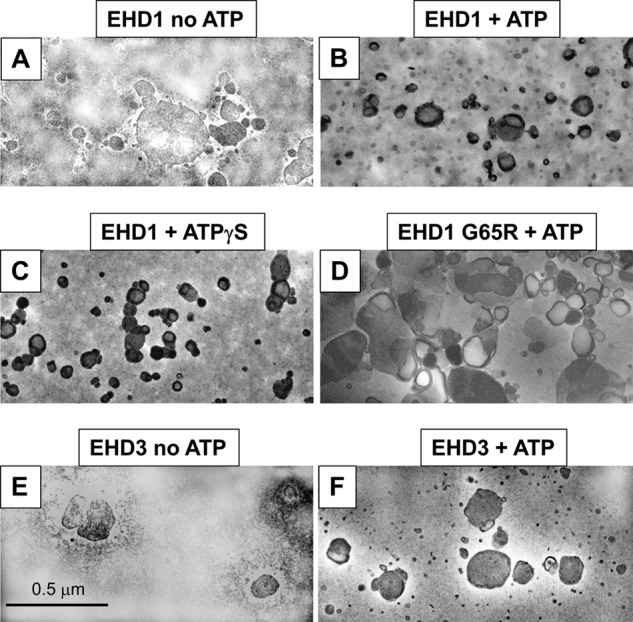
Effect of EHD1 and EHD3 on liposome vesiculation. Liposomes containing 80% PC, 10% PS, and 10% PI(4,5)P2 were incubated for 15 min at 37 °C with the proteins indicated (2.5 μm), and the degree of vesiculation was observed by electron microscopy. A, incubation with EHD1 in the absence of ATP (no vesiculation observed). B, incubation with EHD1 in the presence of ATP (vesiculation observed). C, incubation with EHD1 in the presence of the nonhydrolyzable ATPγS (vesiculation observed). D, incubation with the EHD1 mutant G65R in the presence of ATP (no vesiculation observed). E, incubation with EHD3 in the absence of ATP (no vesiculation observed). F, incubation with EHD3 in the presence of ATP (no vesiculation observed).
FIGURE 8.
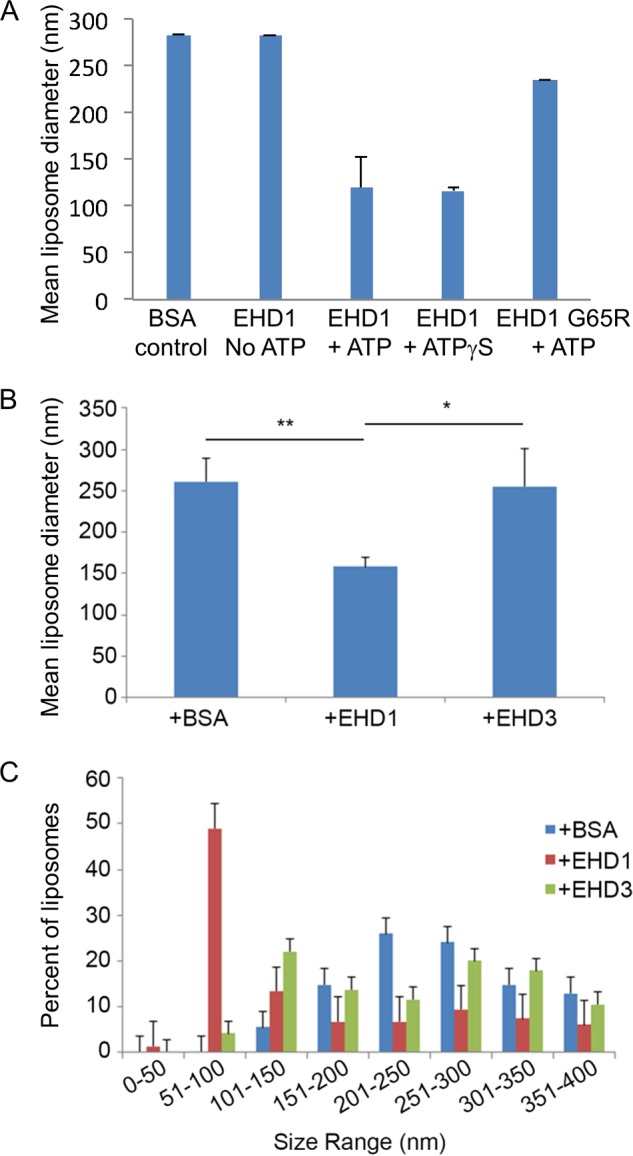
Measurement of the vesiculation of liposomes by EHD1 and EHD3. A, the mean liposome diameter of PC/PS/PI(4,5)P2 liposomes was measured upon incubation with BSA control, EHD1 in the absence of ATP, EHD1 with ATP, EHD1 with the nonhydrolyzable ATPγS analog, and EHD1 G65R in the presence of ATP. B, the mean diameter of PC/PS/PI(4,5)P2 liposomes was measured upon incubation with EHD1, EHD3, or with BSA (control) in the presence of ATP. C, the distribution of vesicle size range upon incubation of PC/PS/PI(4,5)P2 liposomes with EHD1, EHD3, or with BSA (control) is shown. *, p < 0.05; **, p < 0.005 by Student's t test. Error bars, S.D.
In the same experiments, addition of EHD3 (in the absence or presence of ATP; Fig. 7, E and F, quantified in Fig. 8B) did not lead to the vesiculation of PC/PS/PIP2 liposomes. Quantification also demonstrated that incubation of EHD1 with the liposomes caused a dramatic shift in the distribution of vesicle size, with ∼50% of the vesicles found in the 51–100-nm range (compared with <5% for EHD3 or BSA; Fig. 8C). Overall, our results strongly support a role for EHD1 (and EHD4) in TRE vesiculation, whereas EHD3 is likely involved in the tubulation process.
DISCUSSION
C-terminal EHD ATPase proteins play major roles in the regulation of endocytic trafficking. The crystallization of mouse EHD2 (8) and the identification of EHD proteins as ATPases (8, 17, 23) have led to predictions that EHDs act as “pinchases” that constrict and/or vesiculate membranes (Ref. 8; for review see Ref. 3). Experimental evidence also supports a role for EHD proteins in membrane scission (10, 11). However, there is also convincing evidence from in vitro tubulation assays that EHD proteins are capable of bending and tubulating membranes (7, 8).
To clarify the functions of the four EHD proteins, we compared their roles in vivo using a series of knockdowns and developed a novel semipermeabilized cell system to test vesiculation using purified proteins. In vivo, EHD1 depletion caused a more elaborate TRE pattern for the scaffold MICAL-L1, its interaction partner Syndapin2, and the internalized cargo molecule CD59. These data are entirely consistent with the vesiculation of MICAL-L1-containing TREs when purified EHD1 was added to our semipermeabilized cell system, implicating EHD1 as a vesiculator of TREs. In vitro studies with EHD1 further supported its role in vesiculation, and PC/PS/PIP2 liposomes displayed a dramatic decrease in size from ∼250 nm to ∼150 nm when incubated with EHD1, with ∼50% of the vesicles in the 51–100-nm range (as opposed to <5% for BSA control). Whereas a similar role was observed for EHD4 both in vivo and with semipermeabilized cells, the role of EHD2 was more difficult to interpret. In vivo, depletion of EHD2 had little or no effect on TREs, consistent with its association with caveolae (4–6, 22). However, in the semipermeabilized cell system, EHD2 did induce vesiculation. We rationalize that in the latter system, purified EHD2 may be more promiscuous in its localization, and a partial association with TREs (as well as the plasma membrane) may allow it to vesiculate TRE membranes to which it normally does not localize.
Remarkably, despite 86% identity with EHD1, EHD3 functions differently than EHD1. EHD3 depletion led to massive vesiculation of MICAL-L1-, Syndapin2-, and CD59-containing TREs. Consistent with the in vivo data, addition of EHD3 in our semipermeabilized cell system led to greatly enhanced tubulation, pointing to a role for this EHD protein in membrane tubulation. Moreover, in vitro assays showed that EHD3 did not mediate vesiculation.
How do we reconcile the differences in EHD1 and EHD3 function? One possibility is based on recent studies on BAR domain-containing proteins, suggesting that membrane tubulation is effectively an intermediate step in the process of vesiculation. Indeed, modeling by Ayton et al. on N-BAR proteins, and studies by Peter et al. on the N-BAR protein amphiphysin suggest that at intermediate concentrations, N-BARs affect tubule formation, whereas at high concentrations vesiculation of membranes is induced (24, 25). However, because our in vitro studies with controlled concentrations of purified proteins nonetheless show key differences between EHD1 and EHD3, it is more likely that subtle differences in EHD3 versus EHD1 dimers might make the completion of membrane bending to the point of scission more difficult in the former case. For example, several of the helices have amino acid changes that might alter the “width” of the membrane-binding scissors, potentially influencing the degree of membrane bending and perhaps dictating tubulation versus vesiculation. One such example is at the start of α helix 12; in EHD1 and EHD4 residue 376 is a proline, whereas in EHD3 this residue is a serine. However, clarifying the precise mechanism will require a concerted and detailed structural analysis.
Our data clarify the roles of EHD1 and EHD4 as TRE vesiculators. Our findings are also consistent with a function for EHD3 in tubulation rather than vesiculation. Given our recent identification of Syndapin2 as an essential F-BAR protein that tubulates recycling endosomes (9) and its known interaction with all four EHD proteins including EHD3 (26), additional studies will be needed to address the potential synergy of these proteins in TRE generation. Although we speculate that differential interaction partners may also contribute to the functional differences between EHD1 and EHD3, discerning the precise mechanism will await further studies.
This work was supported, in whole or in part, by National Institutes of Health Grants R01GM087455 (to S. C. and P. L. S.), R01GM074876 (to S. C.), and R01GM085146 (to W. G.). This work was also supported by National Research Service Award Postdoctoral Fellowship GM-095094 (to J. A. S.).
- EHD
- Eps15 homology domain
- ATPγS
- adenosine 5′-O-(thiotriphosphate)
- BAR
- Bin-Amphiphysin-Rvs
- F-BAR
- Fes/CIP4 homology-BAR
- N-BAR
- N-terminal amphipathic helix-BAR
- PC
- 1,2-dioleoyl-sn-glycero-3-phosphocholine
- PI(4,5)P2
- phosphatidylinositol 4,5-bisphosphate
- PS
- 1,2,-dioleoyl-sn-glycero-3-phosphoserine
- TRE
- tubular recycling endosome.
REFERENCES
- 1. Grant B. D., Donaldson J. G. (2009) Pathways and mechanisms of endocytic recycling. Nat. Rev. Mol. Cell Biol. 10, 597–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Maxfield F. R., McGraw T. E. (2004) Endocytic recycling. Nat. Rev. Mol. Cell Biol. 5, 121–132 [DOI] [PubMed] [Google Scholar]
- 3. Naslavsky N., Caplan S. (2011) EHD proteins: key conductors of endocytic transport. Trends Cell Biol. 21, 122–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hansen C. G., Howard G., Nichols B. J. (2011) Pacsin 2 is recruited to caveolae and functions in caveolar biogenesis. J. Cell Sci. 124, 2777–2785 [DOI] [PubMed] [Google Scholar]
- 5. Morén B., Shah C., Howes M. T., Schieber N. L., McMahon H. T., Parton R. G., Daumke O., Lundmark R. (2012) EHD2 regulates caveolar dynamics via ATP-driven targeting and oligomerization. Mol. Biol. Cell 23, 1316–1329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Stoeber M., Stoeck I. K., Hänni C., Bleck C. K., Balistreri G., Helenius A. (2012) Oligomers of the ATPase EHD2 confine caveolae to the plasma membrane through association with actin. EMBO J. 31, 2350–2364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Pant S., Sharma M., Patel K., Caplan S., Carr C. M., Grant B. D. (2009) AMPH-1/amphiphysin/Bin1 functions with RME-1/Ehd1 in endocytic recycling. Nat. Cell Biol. 11, 1399–1410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Daumke O., Lundmark R., Vallis Y., Martens S., Butler P. J., McMahon H. T. (2007) Architectural and mechanistic insights into an EHD ATPase involved in membrane remodelling. Nature 449, 923–927 [DOI] [PubMed] [Google Scholar]
- 9. Giridharan S. S., Cai B., Vitale N., Naslavsky N., Caplan S. (2013) Cooperation of MICAL-L1, Syndapin2 and phosphatidic acid in tubular recycling endosome biogenesis. Mol. Biol. Cell 24, 1776–1790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Jakobsson J., Ackermann F., Andersson F., Larhammar D., Löw P., Brodin L. (2011) Regulation of synaptic vesicle budding and dynamin function by an EHD ATPase. J. Neurosci. 31, 13972–13980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cai B., Caplan S., Naslavsky N. (2012) cPLA2α and EHD1 interact and regulate the vesiculation of cholesterol-rich GPI-anchored protein-containing endosomes. Mol. Biol. Cell 23, 1874–1888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Pucadyil T. J., Schmid S. L. (2008) Real-time visualization of dynamin-catalyzed membrane fission and vesicle release. Cell 135, 1263–1275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Pucadyil T. J., Schmid S. L. (2010) Supported bilayers with excess membrane reservoir: a template for reconstituting membrane budding and fission. Biophys. J. 99, 517–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Boucrot E., Pick A., Çamdere G., Liska N., Evergren E., McMahon H. T., Kozlov M. M. (2012) Membrane fission is promoted by insertion of amphipathic helices and is restricted by crescent BAR domains. Cell 149, 124–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Naslavsky N., Boehm M., Backlund P. S., Jr., Caplan S. (2004) Rabenosyn-5 and EHD1 interact and sequentially regulate protein recycling to the plasma membrane. Mol. Biol. Cell 15, 2410–2422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Naslavsky N., McKenzie J., Altan-Bonnet N., Sheff D., Caplan S. (2009) EHD3 regulates early endosome-to-Golgi transport and preserves Golgi morphology. J. Cell Sci. 122, 389–400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Naslavsky N., Rahajeng J., Sharma M., Jovic M., Caplan S. (2006) Interactions between EHD proteins and Rab11-FIP2: a role for EHD3 in early endosomal transport. Mol. Biol. Cell 17, 163–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cai B., Katafiasz D., Horejsi V., Naslavsky N. (2011) Pre-sorting endosomal transport of the GPI-anchored protein, CD59, is regulated by EHD1. Traffic 12, 102–120 [DOI] [PubMed] [Google Scholar]
- 19. Hayashi K., Kojima C. (2008) pCold-GST vector: a novel cold-shock vector containing GST tag for soluble protein production. Protein Expression Purification 62, 120–127 [DOI] [PubMed] [Google Scholar]
- 20. Beckers C. J., Keller D. S., Balch W. E. (1987) Semi-intact cells permeable to macromolecules: use in reconstitution of protein transport from the endoplasmic reticulum to the Golgi complex. Cell 50, 523–534 [DOI] [PubMed] [Google Scholar]
- 21. Long K. R., Yamamoto Y., Baker A. L., Watkins S. C., Coyne C. B., Conway J. F., Aridor M. (2010) Sar1 assembly regulates membrane constriction and ER export. J. Cell Biol. 190, 115–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Simone L. C., Caplan S., Naslavsky N. (2013) Role of phosphatidylinositol 4,5-bisphosphate in regulating EHD2 plasma membrane localization. PLoS ONE 8, e74519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lee D. W., Zhao X., Scarselletta S., Schweinsberg P. J., Eisenberg E., Grant B. D., Greene L. E. (2005) ATP binding regulates oligomerization and endosome association of RME-1 family proteins. J. Biol. Chem. 280, 17213–17220 [DOI] [PubMed] [Google Scholar]
- 24. Ayton G. S., Lyman E., Krishna V., Swenson R. D., Mim C., Unger V. M., Voth G. A. (2009) New insights into BAR domain-induced membrane remodeling. Biophys. J. 97, 1616–1625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Peter B. J., Kent H. M., Mills I. G., Vallis Y., Butler P. J., Evans P. R., McMahon H. T. (2004) BAR domains as sensors of membrane curvature: the amphiphysin BAR structure. Science 303, 495–499 [DOI] [PubMed] [Google Scholar]
- 26. Braun A., Pinyol R., Dahlhaus R., Koch D., Fonarev P., Grant B. D., Kessels M. M., Qualmann B. (2005) EHD proteins associate with syndapin I and II and such interactions play a crucial role in endosomal recycling. Mol. Biol. Cell 16, 3642–3658 [DOI] [PMC free article] [PubMed] [Google Scholar]